Figure S3.
RPB1 Degradation Is a Major Determinant of UV-Induced Shutdown of Transcription Initiation, Related to Figure 3
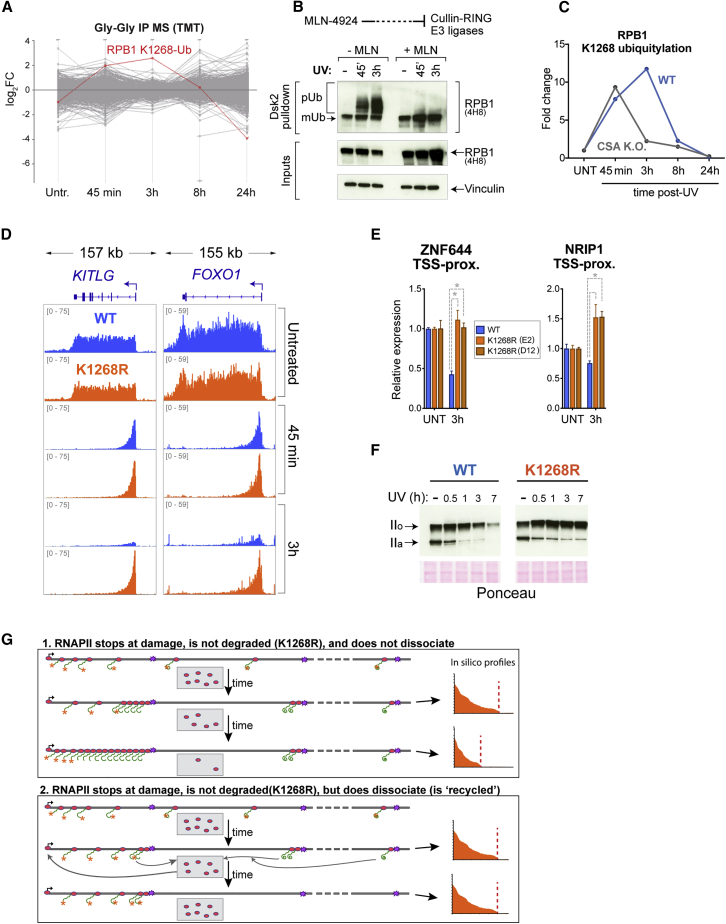
(A) Abundance of all detected ubiquitylation sites in the proteome of WT cells, at different times after UV irradiation (20 J/m2). Each ubiquitylation site is represented as one gray line connecting different time points. K1268 ubiquitylation is marked as a red line. Also see Table S2.
(B) RPB1 poly-ubiquitylation is inhibited by the NEDDylation inhibitor MLN4924, showing that it requires a cullin E3 ligase
(C) K1268 ubiquitylation in WT and CSA knock-out cells, before, and at different times after UV irradiation (20 J/m2), quantified by TMT Gly-Gly IP mass-spectrometry, normalized to untreated condition. WT only is also shown in Figure 3A. Note that CSA KO cells have normal K1268R ubiquitylation at the earliest time-point, strongly indicating that CUL4CSA plays no direct role in it. However, CSA KO cells have defective transcription after UV, potentially explaining why the CSA KO cells have decreased K1268 ubiquitylation at this time-point: only transcribing RNAPII is ubiquitylated (Anindya et al., 2007).
(D) Browser tracks from the TTchem-seq experiment, KITLG and FOXO1 genes. The data are normalized to yeast spike-in.
(E) RT-qPCR measuring nascent RNA production at TSS-proximal regions of ZNF644 and NRIP1 genes. WT and K1268R CRISPR knock-in cells were either untreated, or collected 3 h post UV irradiation (20 J/m2). Primer locations are indicated in Figure 3C. Data are normalized to the mature GAPDH expression, and to untreated condition for each cell line. Representative experiments of three biological replicates are shown, with data represented as mean ± SD. Statistically significant differences (p < 0.05, multiple t tests, Holm-Sidak correction) in all three biological replicates are indicated with asterisks.
(F) Western blot analysis of chromatin fractions assessing UV-induced RPB1 degradation after 20 J/m2 UV irradiation. RNAPII half-life was estimated to be ∼1.5 h in WT cells.
(G) Outline of two different scenarios for RNAPII fate at DNA damage when it cannot be degraded (K1268R mutant cells), with the predicted transcription activity profiles on the right – in the case where RNAPII dissociation at DNA damage does not take place (upper) or where it does (lower).