Abstract
Aims
We conducted a systematic review assessing the reporting quality of studies validating models based on machine learning (ML) for clinical diagnosis, with a specific focus on the reporting of information concerning the participants on which the diagnostic task was evaluated on.
Method
Medline Core Clinical Journals were searched for studies published between July 2015 and July 2018. Two reviewers independently screened the retrieved articles, a third reviewer resolved any discrepancies. An extraction list was developed from the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis guideline. Two reviewers independently extracted the data from the eligible articles. Third and fourth reviewers checked, verified the extracted data as well as resolved any discrepancies between the reviewers.
Results
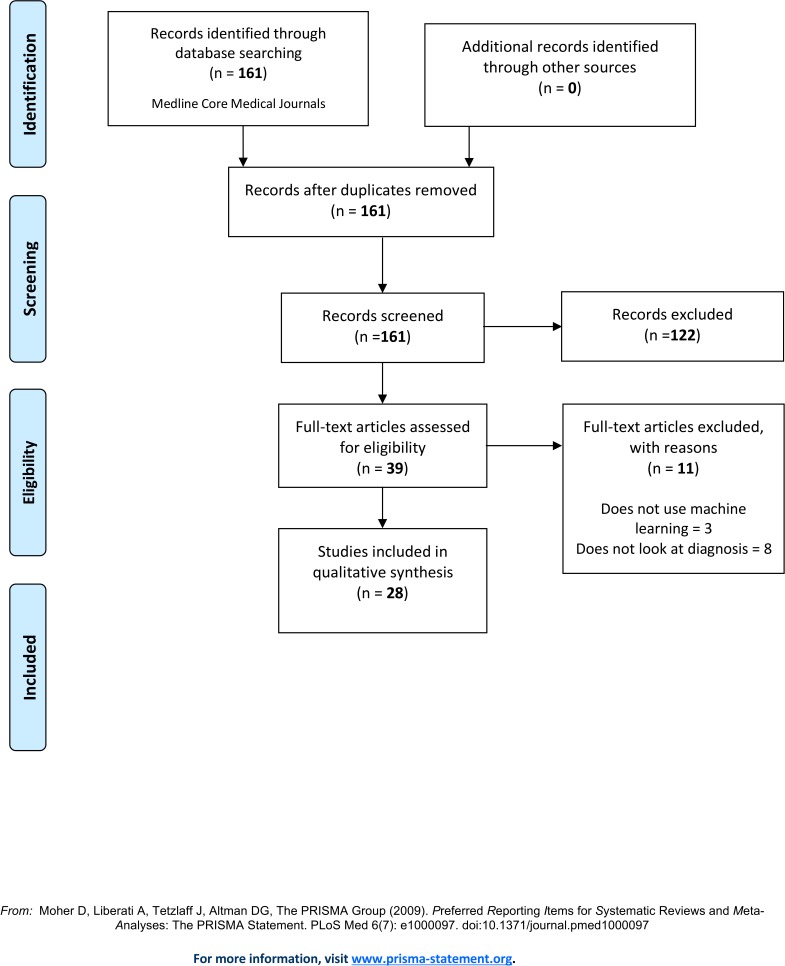
The search results yielded 161 papers, of which 28 conformed to the eligibility criteria. Detail of data source was reported in 24 of the 28 papers. For all of the papers, the set of patients on which the ML-based diagnostic system was evaluated was partitioned from a larger dataset, and the method for deriving such set was always reported. Information on the diagnostic/non-diagnostic classification was reported well (23/28). The least reported items were the use of reporting guideline (0/28), distribution of disease severity (8/28 patient flow diagram (10/28) and distribution of alternative diagnosis (10/28). A large proportion of studies (23/28) had a delay between the conduct of the reference standard and ML tests, while one study did not and four studies were unclear. For 15 studies, it was unclear whether the evaluation group corresponded to the setting in which the ML test will be applied to.
Conclusion
All studies in this review failed to use reporting guidelines, and a large proportion of them lacked adequate detail on participants, making it difficult to replicate, assess and interpret study findings.
PROSPERO registration number
Keywords: machine learning, medical diagnosis, clinical prediction, reporting quality
Strengths and limitations of this study.
This is the first systematic review evaluating the reporting quality of studies developing and/or validating machine learning (ML) methods for medical diagnosis within the medical literature.
Using a systematic approach, this review included studies published within the Medline Core Clinical Journals, these journals cover all areas of clinical and public health.
The review used Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis to help extract information concerning the participants within the reviewed studies.
This review only focused on the reporting quality and, therefore, did not evaluate the statistical methodology and conduct of studies using ML diagnostic systems.
Although a risk of bias assessment is not essential for research on research, the following review did not use the risk of bias assessment tool.
Introduction
Machine learning (ML) is a rapidly developing area, characterised as the science of training computers to conduct specific tasks, such as classification or prediction, without explicit programming, but where decisions are taken based on patterns and relationships within large and complex datasets.1 Over the past decade, access to large amounts of clinical data and the development of new ML techniques have led to a rise in the application of ML methods to medicine.2 3 Due to their propensity to facilitate and promote timely and objective clinical decision-making, ML methods have been applied to gain valuable insights into clinical diagnoses. For example, ML methods have been used to diagnose skin cancer using skin lesion images,4 diagnose cerebral aneurysms using clinical notes5 and diagnose stroke using neuroimaging data6—see box 1 for an example of ML-based diagnostic system.
Box 1.
Machine learning (ML) is the ability to create algorithms to accomplish specific tasks without explicitly programming them, but rather take decisions based on previously seen data. Here is a summary of the steps when creating ML algorithms:
Model development
Step 1: Defining the research problem. This could be broken down to either a classification, regression or a clustering problem.
Step 2: Identification of data sources and formats. Data could be in various formats (eg, images, text, speech or numerical), and data could come from various sources, such as hospital, insurance databases or previous research projects.
Step 3: Training and test set derivation. Here, the data could be broken down into two independent components: the training set and test set. The training set is used to create the ML algorithm and the test set is used to evaluate the ML algorithm.
Step 4: Model development. The model is developed using the training dataset. The model could be either supervised or unsupervised (supervised models require labelled data, whereas unsupervised models do not). The loss function and the methods for handling outliers and missing data are also described. A portion of the training set, the model selection set, is often withheld from model training to allow for model selection and to avoid overfitting.
Step 5: Evaluation of the model. The test set is used to evaluate the ML algorithm using a variety of metrics to compare the prediction with the gold standard outcome label (often referred to as ground truth).
Model validation
To obtain an accurate assessment of model’s performance in a clinical setting, the model must be validated against data, which is drawn from a clinical cohort. Internal validation refers to a model being evaluated on a cohort taken from the same setting as the data used to develop the model. External validation is where the cohort data are taken from a separate setting, which overcomes any systematic biases present in the data source used for model validation.
It is worth noting that one potential area for confusion is the differing meanings of the terms test set and validation set between the ML and medical research community. A medical researchers validation set is an ML test set.
While there is no consensus on the definition, a key principle of ML models is that they are developed based on the automatic extraction of patterns from data.7 In contrast to traditional statistics, whereby models are explicitly programmed based on statistical theory and assumptions, ML models learn from examples without the need for explicit rules to make decisions.8 Generally, a researcher developing an ML model has access to a large dataset that is divided into a training set and a test set (see box 1). The training set is used to develop an ML model that will learn the relationships between available clinical data and an outcome of interest (eg, a diagnosis). The performance of the ML model is then evaluated by applying it to the test set. As ML models are only as good as the data used to train them, it is vital to emphasise the importance of data quality.9
Despite their popularity, the promising applications of ML-based diagnostic systems come with its own set of pitfalls. Studies using ML for medical diagnosis may contain systematic errors in both the design and execution.10–12 For instance, selection bias can occur if the sample used to produce the ML-based diagnostic system is not entirely representative of the population on which the model may be used in the future.11 Repeated evaluation of model performances against the same test set may result in the selected model overfitting the test set, resulting in an over-optimistic assessment of model performance.13 These methodological biases can make it difficult to generalise conclusions from the results yielded. This could lead to erroneous yet devastating clinical decisions, that is, recommending a medical treatment to an individual that is different from those in the population the that treatment was developed and validated on.14
There is a parallel between what ML researchers refer to as ‘test set’ and the ‘population on which a diagnostic test is evaluated’ within diagnostic accuracy studies. The diagnostic accuracy of an ML-based systems is reliant on demographic and clinical characteristics of the population in which it was applied on, therefore, if the cohorts are not a representative sample of the targeted population, then the generalisability of the study results may be limited. A further hindrance to the application of ML methods for medical diagnosis (and more generally in biomedical research) is that ML researchers may not be familiar with the requirements and guidelines that biomedical research have collectively established to ensure transparent and unbiased evidence-based knowledge accumulation.15 16
Clinical prediction models undergo a scientifically rigorous process to establish their diagnostic accuracy, which encompasses their safety, validity, reproducibility, usability and reliability. Highlighting the importance of transparent and rigorous reporting of clinical predictions models accuracy studies, particularly as the diagnostic prediction models of an instrument can vary greatly due to factors such as population characteristics, clinical setting, disease prevalence and severity as well as aspects of test execution and interpretation.16 To aide with and standardise this process, Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) guideline was set in place. The TRIPOD is an internationally accepted reporting guideline that was developed to improve the reliability and value of clinical prediction models through the promotion of transparent and accurate reporting.16
In medical research, reporting guidelines are implemented to aid in the transparent evaluation, usability and reproducibility of a diagnostic instrument.17 Luo et al18 have constructed a reporting guideline for the development and usage of ML predictive models in biomedical research. This is an important step towards a rigorous and robust approach to the usage of ML methods in medical research. Since publication in December 2016 (up to May 2019), the guideline of Simera et al, which is currently available on The Enhancing the QUAlity and Transparency Of Health Research network website,19 has garnered only ~50 citations. Additionally, in 2015, a more specific and robust guideline was developed to aid the reporting of prediction models used in prognostic and diagnostic studies (TRIPOD).16 TRIPOD has ~1000 citations demonstrating that is has been accepted by the community as a useful set of guidelines for diagnostic/prognostic prediction. In this work, we evaluate whether ML studies make use of these guidelines.
To date, there have been no studies evaluating the reporting quality of studies using ML methods, particularly diagnostic studies. Knowing this may aid in the evaluation of reporting standards employed by ML researchers. In this review, we focus on medical research studies that used ML methods to aid clinical diagnosis. Further, we have narrowed our review to applied ML methods, which are envisaged to be clinically useful, in which the end users are practitioners and research consumers.
We aimed to produce a systematic review assessing the reporting quality of studies developing or validating ML models for clinical diagnosis, with a specific focus on the reporting of information concerning the participants on which the diagnostic task was evaluated.
Methods
This review was registered with International prospective register of systematic reviews on 30 July 2018. The framework used for this methodological systematic review is Preferred Reporting Items for Systematic reviews and Meta-Analyses guideline for systematic reviews.20
Literature search
On July 2018, two authors (MY and JL) independently searched through the Medline Core Clinical Journals for articles developing or validating ML models for clinical diagnosis. Core Clinical Journals, also known as Abridged Index Medicus, is a filter option within Medline that limits to clinically useful journals. This is a selection of 119 English-language journals that focus on clinical studies and that are considered to be of immediate interest to practising physicians.21 Using this filter excludes journals in bioinformatics or computational biology, which are highly likely to include articles explaining the development of ML-based diagnosis systems. However, these journals might not target clinicians. In addition to this, due to the ever-expanding ML literature, we have narrowed our review to studies published between July 2015 and 1 July 2018. See online supplementary file 1 for the search strategy.
bmjopen-2019-034568supp001.pdf (98.5KB, pdf)
Subsequent to the literature search, the two reviewers (MY and JL) screened the title and abstracts of the search results. Once the eligible papers were identified and retrieved, both the first reviewer (MY) and the second reviewer (JL) independently screened the full articles for eligibility. Discrepancies between the two reviewers were discussed and resolved by a third reviewer (IA).
Inclusion and exclusion
Studies were included if they used ML for clinical diagnosis, for example if they used statistical techniques to conduct classification, regression or clustering based on clinical data for disease diagnosis without being explicitly programmed. Other inclusions were primary study designs that evaluated the accuracy of such ML-based systems for diagnostic tasks, and articles in the English language. Studies were excluded if they did not report original research, if they were systematic reviews, had no abstract or did not specify the type of ML model adopted.
Extraction list
For studies developing, evaluating or updating clinical prediction models (this includes diagnosis), the TRIPOD provides guidance on reporting the key items. As it stands, TRIPOD is the most rigorous and relevant guideline for evaluating the use of ML methods for medical diagnosis. As such, an extraction list based on the TRIPOD checklist was developed. The focus of the extraction list was to extract information about the participants on which the diagnostic task was evaluated on, namely selection method and population characteristics. We additionally extracted general information concerning the diagnostic tasks, namely the target condition and the target population. The extraction list was tested and validated by two reviewers (MY and JL), by applying it on a random sample of the eligible papers.
Data extraction
Two reviewers (JL and PS) independently extracted the data from the eligible articles based on the items listed in table 1. For each of the items, reviewers declared if the item was clearly reported (yes/no/unclear) and justify the declaration by citing the manuscript verbatim, as well as providing a written explanation if the reporting was considered unclear. The third and fourth reviewers (MY and IA) checked and verified the extracted data and resolved any disagreements between the reviewers through discussion.
Table 1.
Item list used to extract eligible papers
| Item groups | Item list | Detailed items |
| General characteristics | Diagnostic task | What is the target condition? |
| Study objective | Is the study aiming at the development of a diagnostic method, evaluation of a diagnostic method or both? | |
| Target population | What is the population targeted by the diagnostic test? | |
| Methods | Data sources | Where and when potentially eligible participants were identified (setting, location and dates) |
| Data split | Method for partitioning the evaluation set from the training data. To assess whether participants formed a consecutive, random or convenience series. | |
| Test dataset eligibility criteria | On what basis potentially eligible participants were identified within the test dataset (such as symptoms, results from previous tests, inclusion in registry). | |
| Results | Baseline characteristics | Baseline demographic and clinical characteristics of participants |
| Diagnosis/non-diagnosis classification | Classification of the diagnosed and non-diagnosed patients within the test set. | |
| Flow diagram | Flow of participants, using a diagram. | |
| Severity | Distribution of severity of disease in those with the target condition. | |
| Alternative diagnosis | Distribution of alternative diagnoses in those without the target condition. | |
| Difference between reference test and ML test | Is there a time interval between index test and reference standard? | |
| Applicability | Does the evaluation population correspond to the setting in which the diagnosis test will be applied? |
ML, machine learning.
Data analysis
Findings from the included studies demonstrating study characteristics, reporting quality and presence of bias were presented in descriptive statistics and figures.
Patient and public involvement
There was no patient or public involvement in any phase of this study, this included the development of the research question, the analysis and the conclusions.
Results
The search yielded 161 papers, of which 28 conformed to the eligibility criteria, see figure 1. During the screening of the title and abstract, most papers were excluded due to the search term ‘CAD’ being analogous to both ‘computer-aided detection’ and coronary artery disease’. During the full-text review, eleven papers were excluded because they did not use ML methods for medical diagnosis, and three papers were excluded because they did not use ML method but were captured in the search because they studied coronary artery disease (CAD).
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses flow diagram. From: Moher D et al.34
Study characteristics
The study characteristics of the all eligible studies are presented in table 2 (see online supplementary file 1 for list of studies). From the papers extracted, majority of the studies were published in 2017 (43%) and mostly in the Radiology journal (29%). Oncology was the most researched domain (47%), followed by Neurology (18%). The majority of studies focused on model development (97%), with only one study looking at model validation.
Table 2.
Study characteristics
| Items | Total n (%) |
| Year | |
| 2015 | 4 (14) |
| 2016 | 9 (32) |
| 2017 | 12 (43) |
| 2018 | 3 (11) |
| Journals | |
| Radiology | 8 (29) |
| JAMA | 2 (7) |
| Brain | 2 (7) |
| American Journal of Roentgenology | 2 (7) |
| Neurology | 1 (3) |
| Medicine | 1 (3) |
| Surgery | 1 (3) |
| Chest | 1 (3) |
| Gastroenterology | 1 (3) |
| Journal of the American College of Cardiology | 1 (3) |
| Journal of Allergy and Clinical Immunology | 1 (3) |
| American Journal of Clinical Pathology | 1 (3) |
| American Journal of Ophthalmology | 1 (3) |
| The Journal of Infectious Diseases | 1 (3) |
| Digestive Diseases and Sciences | 1 (3) |
| The British Journal of Radiology | 1 (3) |
| The Journal of Pediatrics | 1 (3) |
| Clinical Specialty | |
| Oncology | 13 (47) |
| Neurology | 5 (18) |
| Immunology | 2 (7) |
| Ophthalmology | 2 (7) |
| Others specialties* | 6 (21) |
| Task | |
| Development and evaluation | 27 (97) |
| Evaluation | 1 (3) |
*Other clinical specialities include cardiology, gastroenterology, infectious disease, psychiatry, endocrinology and various.
Reporting quality
Detail of the data source was reported in 86% of the papers, with all studies providing information on the separation method for deriving the evaluation set from the larger dataset. Eighty-two per cent of studies reported eligibility criteria for both evaluation set. Information on the diagnostic/non-diagnostic classification evaluation metric used was included in 82% of all papers. The least-reported items were use of reporting guideline (0%), distribution of disease severity (29%), patient flow diagram (36%), distribution of alternative diagnosis (36%) and baseline characteristic (61%). See table 3 for a full breakdown of reporting quality.
Table 3.
Reporting quality
| Items | Reported, n (%) | Not reported, n (%) | Unclear, n (%) |
| Methods | |||
| Data source | 24 (86) | 0 (0) | 4 (14) |
| Data split methods | 28 (100) | 0 (0) | 0 (0) |
| Test set eligibility criteria (evaluation set) | 23 (82) | 5 (18) | 0 (0) |
| Results | |||
| Baseline characteristic | 17 (61) | 11 (39) | 0 (0) |
| Diagnosis/non-diagnosis classification | 23 (82) | 4 (14) | 1 (4) |
| Flow diagram | 10 (36) | 18 (64) | 0 (0) |
| Disease severity | 8 (29) | 18 (64) | 2 (7) |
| Alternative diagnosis | 10 (36) | 18 (64) | 0 (0) |
| Use of reporting guideline | 0 (0) | 28 (100) | 0 (0) |
Presence of bias
Within the eligible studies, 82% had a a time interval between the conduct of the reference standard and ML test (table 4). Within 54% of studies, it was unclear whether the study populations corresponded to the setting in which the diagnostic test will be applied to. However, in 29% of studies, the clinical setting of the gathered evaluation dataset did not correspond to the clinical setting in which study authors hoped it would be applied.
Table 4.
Presence of bias
| Items | Yes, n (%) | No, n (%) | Unclear, n (%) |
| Is there a time interval between reference standard ML test? | 23 (82) | 1 (4) | 4 (14) |
| Does the test population correspond to the population/setting in which the diagnosis test will be applied? | 5 (18) | 8 (29) | 15 (54) |
Discussion
This review found that studies developing or validating ML-based systems for clinical diagnosis failed to use reporting guidelines and lacked adequate detail for assessment, interpretation and reproducibility. With nearly all studies providing detail on data sources, eligibility criteria and diagnosis classification, only a few studies reported study participant flow diagram, distribution of disease severity and distribution of alternative diagnosis. Our findings are in line with those of Faes et al recent systematic reviews22 in which they found poor reporting and potential biases arising from study design in studies using ML methods for classifying diseases from medical imaging. Similarly, in another systematic review, Christodoulou et al23 found studies comparing the performance of logistic regression models with ML models for clinical prediction to have poor methodology and reporting quality.
A high number of studies reviewed had a time difference between the conduct of the reference test and that of ML-based diagnostic systems, suggesting the potential for incorporation bias.24 25 This is largely an issue in ML-based diagnostic systems where labelling is the gold standard, but patient data are labelled retrospectively. This may happen several years after initial data collection and in a different setting.
In more than half of the studies, it was unclear whether the study population corresponded to the setting in which the ML diagnostic system will be used in. However, in a third of the reviewed studies, the test populations did not correspond to the populations in which tests were hoped to be applied to, further limiting their generalisability. In addition to this, studies utilising ML-based diagnostics systems fail to report baseline characteristics. This could be problematic; within diagnostic accuracy studies it is imperative to report sample characteristics as this aid researchers, research consumers and practitioners in determining the relevancy and applicability of study findings to a wider setting.
Information on data source was unclear in four studies; this is vital in evaluating the source and methods used to derive study samples. In diagnostic studies the use of different methods to derive the evaluation sample from the wider population could lead to more or less accurate estimation of the diagnostic performance. The ideal method for sampling should be based on probability and not convenience, as this allows for a representative sample to be selected from a sampling frame whereby all eligible individuals have an equal chance of being selected. In addition to this, ML-based diagnostic systems that are evaluated using internal validation, where the evaluation set is partitioned from the same cohort as the training set, risk learning the systematic biases in the data of the particular centre from which the cohort was drawn. Such methods only address the systems internal validity, and model performance may deteriorate when deployed on an cohort drawn from a different centre.8 External validation, where the ML-based diagnostic systems are evaluated on a cohort that has played no role in model development, is an important step to verify and asses the whether the system is reliable and deployable on potential populations for clinical use.26–28 This is further highlighted in a recent systematic review evaluating the performance of ML algorithms for the diagnostic analysis of medical images; within this review, Kim et al29 found only 6% (31 out of 516 studies) had externally validated their algorithms.
Low-quality clinical research that is reported inadequately or that offers invalid data and distorted outcomes are deemed wasteful; such research is non-replicable and unusable.30 31 One way to increase the value and reusability of these novel and promising ML-based systems is through complete, accurate and transparent reporting.19 31. Some of the methodological and reporting issues facing the studies reviewed in this systematic review can be mitigated through the use of reporting guidelines such as TRIPOD guideline. More specifically, there has been a recent initiative to develop an extension of the TRIPOD statement which is specific to ML studies (TRIPOD-ML).32 Such guidelines aid researchers developing ML-based diagnostic systems in addressing the important aspects of design, execution and complete and reliable reporting of studies. However, no reporting guideline can salvage a poorly designed and executed study. To prevent flawed design and execution of ML methods, informatic and biomedical researchers looking to develop ML-based diagnostic systems should consult with a methodologist, epidemiologist or a statistician. Having input from such experts will aid in research that is methodological robust in design and execution—resulting in research that it is reliable, reproducible and that adds scientific value.33
Strengths and limitations of study
To our knowledge, this is the first systematic review evaluating the reporting quality of studies developing and/or validating ML methods for medical diagnosis within the medical literature. A possible limitation within this review is that we have not included all medical journals and, therefore, our findings may not be applicable to all journals. Despite this, we have included studies published within the Medline Core Clinical Journals, these journals cover all areas of clinical and public health.
This review did not evaluate the statistical methodology and conduct of studies using ML diagnostic systems. This could be considered a limitation as a transparent reporting that does not guarantee a quality study. Nevertheless, this review shows that these studies do not comply with TRIPOD guideline on the reporting this considerably affects the trust we have in the estimates they are giving concerning the efficacy of their diagnostic methods. Another potential limitation is that the following review did not use risk of assessment tool, however, this is not an essential component for this type of review, as the main objective is to determine the reporting quality of studies and not synthesis research evidence .
Conclusion
We found that all eligible studies in this review failed to use reporting guidelines and the studies lacked adequate detail on the participants on which the diagnostic task was evaluated on, thus making it difficult to replicate, assess and interpret study findings.
Diagnostic studies using ML methods have great potential to improve clinical decision-making and take the load off health systems. However, studies with poor reporting can be more problematic than of help. Within biomedical research, there is an already established framework and guidelines in which ML researchers can use to aid the execution and reporting of ML methods for clinical diagnosis, with the TRIPOD guideline being the most robust and widely used.
Supplementary Material
Footnotes
Contributors: JS is a guarantor of this review. All authors have made substantive intellectual contributions to the development of this review. MY and IA were involved in conceptualising the review. MY, IA and PR developed the protocol. MY, JL and IA did the literature search, MY, JL, PS and IA carried out the study selection and data extraction. MY, IA, JS, PS, MF and MC were involved in the writing and editing of the manuscript.
Funding: This review was conducted independently by the research team.
Disclaimer: There is no funding attached to this systematic review.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: No data are available. All data are freely available within the appendices. No additional available.
References
- 1.Paliwal M, Kumar UA. Neural networks and statistical techniques: a review of applications. Expert Syst Appl 2009;36:2–17. 10.1016/j.eswa.2007.10.005 [DOI] [Google Scholar]
- 2.Cleophas TJ, Zwinderman AH. Machine Learning in Medicine - a Complete Overview. Springer International Publishing, 2015. [Google Scholar]
- 3.Topol EJ. High-Performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25:44–56. 10.1038/s41591-018-0300-7 [DOI] [PubMed] [Google Scholar]
- 4.Esteva A, Kuprel B, Novoa RA, et al. . Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–8. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin H, Kim KH, Song C, et al. . Electrodiagnosis support system for localizing neural injury in an upper limb. J Am Med Inform Assoc 2010;17:345–7. 10.1136/jamia.2009.001594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehme AK, Volz LJ, Feis D-L, et al. . Identifying neuroimaging markers of motor disability in acute stroke by machine learning techniques. Cereb Cortex 2015;25:3046–56. 10.1093/cercor/bhu100 [DOI] [PubMed] [Google Scholar]
- 7.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference and prediction. 27 The Mathematical Intelligencer, 2005: 83–5. [Google Scholar]
- 8.Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. Springer Science & Business Media, 2008. [Google Scholar]
- 9.Obermeyer Z, Emanuel EJ. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. N Engl J Med 2016;375:1216–9. 10.1056/NEJMp1606181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 2018;286:800–9. 10.1148/radiol.2017171920 [DOI] [PubMed] [Google Scholar]
- 11.Bone D, Goodwin MS, Black MP, et al. . Applying machine learning to facilitate autism diagnostics: pitfalls and promises. J Autism Dev Disord 2015;45:1121–36. 10.1007/s10803-014-2268-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster KR, Koprowski R, Skufca JD. Machine learning, medical diagnosis, and biomedical engineering research - commentary. Biomed Eng Online 2014;13:94 10.1186/1475-925X-13-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian J, Simon R. Overfitting in prediction models – is it a problem only in high dimensions? Contemp Clin Trials 2013;36:636–41. 10.1016/j.cct.2013.06.011 [DOI] [PubMed] [Google Scholar]
- 14.Greenhouse JB, Kaizar EE, Kelleher K, et al. . Generalizing from clinical trial data: a case study. the risk of suicidality among pediatric antidepressant users. Stat Med 2008;27:1801–13. 10.1002/sim.3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen JF, Korevaar DA, Altman DG, et al. . Stard 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016;6:e012799 10.1136/bmjopen-2016-012799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moons KGM, Altman DG, Reitsma JB, et al. . Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1–73. 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
- 17.Simera I, Moher D, Hirst A, et al. . Transparent and accurate reporting increases reliability, utility, and impact of your research: reporting guidelines and the EQUATOR network. BMC Med 2010;8:24 10.1186/1741-7015-8-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo W, Phung D, Tran T, et al. . Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res 2016;18:e323 10.2196/jmir.5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simera I, Moher D, Hoey J, et al. . The EQUATOR network and reporting guidelines: helping to achieve high standards in reporting health research studies. Maturitas 2009;63:4–6. 10.1016/j.maturitas.2009.03.011 [DOI] [PubMed] [Google Scholar]
- 20.Liberati A, Altman DG, Tetzlaff J, et al. . The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 2009;339:b2700 10.1136/bmj.b2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Committed selection: abridged index Medicus. N Engl J Med 1970;282:220–1. 10.1056/NEJM197001222820410 [DOI] [PubMed] [Google Scholar]
- 22.Faes L, Liu X, Kale A. Deep Learning Under Scrutiny: Performance Against Health Care Professionals in Detecting Diseases from Medical Imaging - Systematic Review and Meta-Analysis, 2019. [DOI] [PubMed] [Google Scholar]
- 23.Christodoulou E, Ma J, Collins GS, Jie M, Steyerberg EW, et al. . A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol 2019;110:12-22. 10.1016/j.jclinepi.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 24.Kramer MS, Roberts-Bräuer R, Williams RL. Bias and 'overcall' in interpreting chest radiographs in young febrile children. Pediatrics 1992;90:11–13. [PubMed] [Google Scholar]
- 25.Whiting P, Rutjes AWS, Reitsma JB, et al. . Sources of variation and bias in studies of diagnostic accuracy: a systematic review. Ann Intern Med 2004;140:189–202. 10.7326/0003-4819-140-3-200402030-00010 [DOI] [PubMed] [Google Scholar]
- 26.Steyerberg E. Overfitting and optimism in prediction models. Clinical Prediction Models: Springer, 2009: 83–100. [Google Scholar]
- 27.Riley RD, Ensor J, Snell KIE, et al. . External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ 2016;353:i3140. 10.1136/bmj.i3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steyerberg EW, Harrell FE, Borsboom GJ, et al. . Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol 2001;54:774–81. 10.1016/s0895-4356(01)00341-9 [DOI] [PubMed] [Google Scholar]
- 29.Kim DW, Jang HY, Kim KW, et al. . Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol 2019;20:405–10. 10.3348/kjr.2019.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan A-W, Song F, Vickers A, et al. . Increasing value and reducing waste: addressing inaccessible research. The Lancet 2014;383:257–66. 10.1016/S0140-6736(13)62296-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glasziou P, Altman DG, Bossuyt P, et al. . Reducing waste from incomplete or unusable reports of biomedical research. The Lancet 2014;383:267–76. 10.1016/S0140-6736(13)62228-X [DOI] [PubMed] [Google Scholar]
- 32.Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet 2019;393:1577–9. 10.1016/S0140-6736(19)30037-6 [DOI] [PubMed] [Google Scholar]
- 33.Ioannidis JPA, Greenland S, Hlatky MA, et al. . Increasing value and reducing waste in research design, conduct, and analysis. Lancet 2014;383:166–75. 10.1016/S0140-6736(13)62227-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moher D, Liberati A, Tetzlaff J, et al. . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2019-034568supp001.pdf (98.5KB, pdf)