Abstract
Inositol polyphosphate multikinase (IPMK) is required for the biosynthesis of inositol phosphates (IPs) through the phosphorylation of multiple IP metabolites such as IP3 and IP4. The biological significance of IPMK’s catalytic actions to regulate cellular signaling events such as growth and metabolism has been studied extensively. However, pharmacological reagents that inhibit IPMK have not yet been identified. We employed a structure-based virtual screening of publicly available U.S. Food and Drug Administration-approved drugs and chemicals that identified the antidepressant, vilazodone, as an IPMK inhibitor. Docking simulations and pharmacophore analyses showed that vilazodone has a higher affinity for the ATP-binding catalytic region of IPMK than ATP and we validated that vilazodone inhibits IPMK’s IP kinase activities in vitro. The incubation of vilazodone with NIH3T3-L1 fibroblasts reduced cellular levels of IP5 and other highly phosphorylated IPs without influencing IP4 levels. We further found decreased Akt phosphorylation in vilazodone-treated HCT116 cancer cells. These data clearly indicate selective cellular actions of vilazodone against IPMK-dependent catalytic steps in IP metabolism and Akt activation. Collectively, our data demonstrate vilazodone as a method to inhibit cellular IPMK, providing a valuable pharmacological agent to study and target the biological and pathological processes governed by IPMK.
Keywords: drug repositioning, inositol phosphate, IPMK, vilazodone, virtual screening
INTRODUCTION
Inositol phosphates (IP) are a group of inositol metabolites that contain multiple phosphates. IPs act as second messengers for external signals and are highly conserved across all eukaryotes from budding yeasts to humans (Berridge et al., 2000; Streb et al., 1983). Biosynthesis of IP metabolites begins with the synthesis of inositol 1,4,5-trisphosphate (InsP3, IP3), which is produced by phospholipid-specific phospholipase C (PLC). PLCs are activated by external stimuli such as growth factors to cleave phosphatidylinositol 4,5-bisphosphate and produce IP3, which is then released from the cell membrane into the cytosol. In higher eukaryotes, IP3 stimulates Ca2+ release from the endoplasmic reticulum by binding to IP3 receptors, which are IP3-gated Ca2+ channels. Higher inositol polyphosphates, including IP4, IP5, and IP6, are generated by IP kinase phosphorylation of the inositol ring at each of six hydroxyl groups (Fig. 1A). IP6 can be further metabolized by the inositol-hexakisphosphate kinases (IP6Ks), which add a phosphate at the 5-position to produce 5-diphosphoinositol pentakisphosphate (5-PP-IP5,5-IP7). The identification of diphosphoinositol-pentakisphosphate kinases (PP-IP5Ks), which phosphorylate the 1/3-position of IP6 to produce 1/3-PP-IP5 (1/3-IP7), demonstrates the complex control of IP metabolism in eukaryotes (Chakraborty et al., 2011; Park et al., 2018).
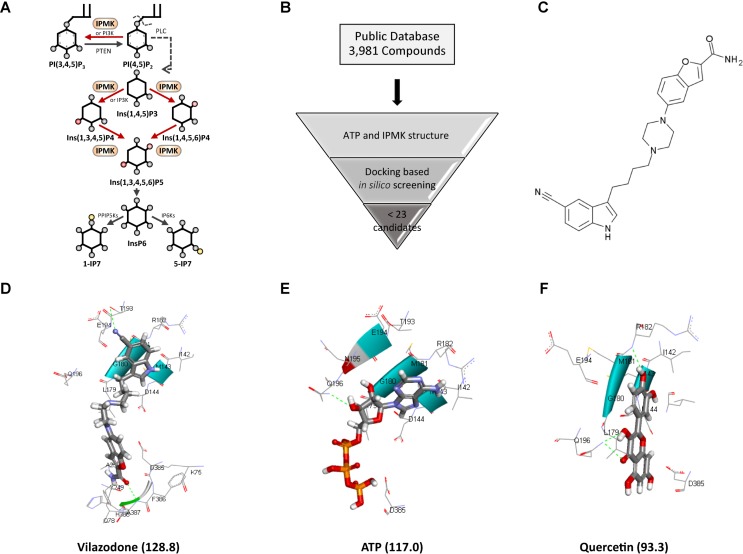
Fig. 1. Discovery of vilazodone as an IPMK inhibitor.
(A) IP metabolic pathway. (B) In silico virtual screening workflow. (C) Structure of vilazodone. (D-F) Predicted binding modes: docking scores are indicated in parentheses, respectively. The docking poses of vilazodone (D), ATP (E), and quercetin (F) within the ATP-binding core of human IPMK.
Insights into the biological functions of IPs have come from genetic studies in yeast and by manipulating inositol polyphosphate multikinase (IPMK) expression in mammalian cells. IPMK is essential for the production of IP4 and IP5, making it crucial for the synthesis of other highly phosphorylated IP metabolites (Odom et al., 2000; Saiardi et al., 1999). As IPMK is the only enzyme in the mammalian genome that mediates IP5 synthesis (Lee et al., 2012), its deletion in mammalian cells leads to marked depletion of IP5 and downstream IPs, including IP6 and IP7. In yeast, the kinase activity of IPMK regulates the activities of the chromatin remodeling SWI/SNF and Ino80 complexes in response to phosphate availability (Shen et al., 2003; Steger et al., 2003). IPMK products like Ins(1,4,5,6)P4 suppress human cancer cell growth by inhibiting the activation of Akt/PKB (Jackson et al., 2011; Piccolo et al., 2004; Razzini et al., 2000). Conversely, IPMK can also function as (Lee et al., 2012) a phosphatidylinositol 3-kinase (PI3K) to phosphorylate phosphatidylinositol 4,5-bisphosphate and produce phosphatidylinositol 3,4,5-trisphosphate, a major growth signal for activation of Akt kinase. Ins(1,4,5,6)P4 was also found to bind tightly to the highly basic interface between HDAC3 and the SMRT-DAD domain when they form a complex (Watson et al., 2012), suggesting that IPMK functions as a primary regulator of class I HDACs in mammalian cells. Ins(1,3,4,5,6)P5, also produced by IPMK (Watson et al., 2012), is important in the Wnt signaling cascade for initiating translocation of β-catenin to the nucleus (Gao and Wang, 2007; Wang and Wang, 2012). Recent studies on IPMK knock-down in mammalian cells have found that decreased levels of IP6 mediated the activation of necroptosis and B cell receptor signaling via IP6-binding targets like MLKL (Dovey et al., 2018) and Btk kinase (Kim et al., 2019). Together, these studies reveal the critical roles of IPMK in the regulation of IP signaling.
Due to the lack of a reliable IPMK inhibitor, however, we cannot fully understand the effects of IPMK and related IP metabolic actions. Computational drug repositioning or repurposing is a promising and efficient strategy for discovering new uses for existing drugs. To develop pharmacological tools for IPMK research, we investigated the possibility that existing drugs inhibit IPMK via structure-based virtual screening, which led to our identification of vilazodone, an U.S. Food and Drug Administration (FDA)-approved antidepressant (Cruz, 2012), as an IPMK inhibitor.
MATERIALS AND METHODS
Structure modeling
We performed in silico structural modeling on > 3,000 compounds from public database Drugcentral using Discovery Studio 3.1 (DS) from Accelrys (USA). Quercetin was used as a reference structure model as its efficacy has been reported recently (Gu et al., 2019). We analyzed two-dimensional (2D) structural similarities between vilazodone and ATP using pipeline pilots. We converted the 2D structures to extended connectivity fingerprints (ECFP) and functional class fingerprints (FCFP) molecular descriptors (Rogers and Hahn, 2010), standard circular fingerprints based on the Morgan algorithm (Morgan, 1965). The similarity between compound pairs was calculated by the Tanimoto similarity. The Tanimoto similarity is calculated with the bits of the binary fingerprint vectors
where A and B are the number of bits present in compounds A and B, respectively, and C is the number of bits shared by A and B.
Docking simulations using the Libdock algorithm (Rao et al., 2007) in DS were performed with three compounds, vilazodone, quercetin, and ATP. The X-ray crystal structure complex of IPMK with ATP was obtained from the protein data bank (PDB ID: 6M88) (Gu et al., 2019). The proposed binding site was centered on the ligand and a site sphere was created at coordinates 4.717, 27.452, 23.075 with a 14.01Å diameter for IPMK. The protocol included 100 hotspots with a docking tolerance of 0.25. The FAST conformation method was also used with CHARMM. For the similarity analysis of the binding modes, we analyzed the hydrogen bonds and van der Waals interactions, key factors for binding, from the docking simulation results, and used the results to construct a binary matrix. Hierarchical clustering was performed with the constructed matrix.
The receptor-ligand pharmacophore generation module in DS was used to generate a set of selective pharmacophore models from the target-compound interactions in the complex structures of ATP and IPMK. This protocol considered the hydrogen bond acceptor/donor, hydrophobic feature, negative/positive ionizable feature, aromatic rings, and exclusion volumes for feature types to generate a selective pharmacophore model from the interactions. A set of candidate pharmacophore models were enumerated from the interactions.
IPMK protein purification
Human IPMK was expressed in Sf9 cells using a baculovirus expression vector system. Cells were harvested in 300 mM NaCl, 50 mM Tris pH 8.0, 5% glycerol and 1 mM phenylmethylsulfonylfluoride (PMSF) and lysed by 3 freeze-thaw cycles in liquid nitrogen. Lysates were cleared by centrifugation at 18,000 rpm for 90 min. Supernatants were applied to Ni-NTA resins (Qiagen, Germany) with 20 mM imidazole and incubated for 2 h. The N-terminal his-tagged human IPMK was eluted with 100 mM imidazole and treated with TEV protease to remove the tag, followed by HiTrap SP HP (GE Healthcare, USA) and Superdex 200 60 600 (GE Healthcare) in a buffer containing 300 mM NaCl, 50 mM Tris-HCl (pH 8.0).
IPMK enzymatic activity assays
Inositol kinase activity of IPMK was measured using the ADP-Glo Kinase kit (V9101; Promega, USA) followed by the manufacturer’s protocol. Briefly, assays were carried out in 96 well immuno plates with 50 ng of recombinant IPMK, 10 μM ATP, and 100 μM of either vilazodone or quercetin in kinase assay buffer (50 mM HEPES [pH 7.4], 10 mM MgCl2, 50 mM KCl) at 30°C with shaking for 30 min. Reactions were quenched with ADP-Glo reagent at room temperature for 40 min. The kinase detection substrate was added, and the reaction incubated for another 30 min. The luminescence was detected with a plate reader.
Immunoblotting
HCT116 cells (4 × 105 cells) were seeded into 6 well at the day before treatment. After 4 h of treatment with 10 μM vilazodone, cells were lysed with 1% NP-40 buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1 mM EDTA [pH 8.0], 50 mM NaF, 10 mM Na-pyrophosphate, and protease inhibitor cocktail [Roche, Switzerland]). Whole cell lysates (20 μg) were loaded into lands of 4% to 12% Bis-Tris SDS-PAGE gels in 1× glycine buffer, and transferred to nitrocellulose membrane. Anti-phospho-S473 Akt, total Akt (Cell Signaling Technology, USA), and α-tubulin (Sigma Aldrich, USA) antibodies were diluted 1:1,000 in 1× TBST with 5% BSA. Secondary antibodies were diluted in 1:2,000 in 1× TBST with 5% nonfat dry milk. Proteins were detected with chemiluminescence immunoblot detection system (Bio-Rad, USA).
Cell culture and high-performance liquid chromatography analysis of intracellular inositol phosphates
NIH3T3-L1 fibroblasts (2 × 105 cells) were seeded into 60 mm dishes in DMEM supplemented with 10% bovine calf serum. The cells were labeled with 60 μCi [3H]-myo-inositol for 3 days. Vilazodone (Sellekchem, USA) and quercetin (Sigma Aldrich) were dissolved in dimethyl sulfoxide (DMSO) so that the final concentration of DMSO in the cell cultures would be 0.1% and were treated for 4 h at 10 μM concentration. Soluble inositol metabolites were extracted with perchloric acid buffer and the remaining insoluble inositol metabolites were extracted with 0.1 M NaOH and 0.1% Triton X-100 for normalization as previously described (Lee et al., 2016). 3H-labeled IPs were resolved by high-performance liquid chromatography (HPLC), and each fraction was counted in a liquid scintillation counter.
RESULTS
To screen potential IPMK inhibitors, we performed an in silico structure-based modeling approach using > 3,000 compounds available from public database (Fig. 1B). Twenty-two candidate compounds with a 2.5-fold higher than quercetin which was used as a positive control compound in analysis scores and included in the same cluster, were derived which determined to be potent from in silico assay. From these candidates, we selected vilazodone, an FDA-approved serotonin transporter inhibitor for the treatment of major depressive disorder for the further study (Fig. 1C). To investigate whether vilazodone is structurally similar to ATP, we performed a structural similarity analysis based on the ECFP and FCFP. Tanimoto scores were calculated to measure the similarities of compound pairs. In the similarity analysis, the Tanimoto scores of ECFP and FCFP were only 0.10 and 0.11, respectively, which indicates that vilazodone is structurally dissimilar from ATP.
Next, we performed docking simulations with IPMK that predicted the docking scores of vilazodone and ATP to be 128.8 and 117.0, respectively. These results suggest that vilazodone binds to the kinase pocket of IPMK more stable than ATP. For further analyses, we included quercetin as a reference compound, as it has shown potent inhibition for IPMK in vitro (Gu et al., 2019). The docking score of quercetin with IPMK was 93.3, suggesting that the interaction between vilazodone and IPMK is stronger than that of quercetin and IPMK (Figs. 1D-1F). In our docking models for IPMK with vilazodone, ATP, and quercetin, we found that vilazodone has hydrogen bonds with T193 and A387; quercetin with R182 and Q196; and ATP with Q196. These results suggest that vilazodone binds to the IPMK’s ATP-binding catalytic core including the residues I142, M143, D144, G180, R182, T193, E194, and A387.
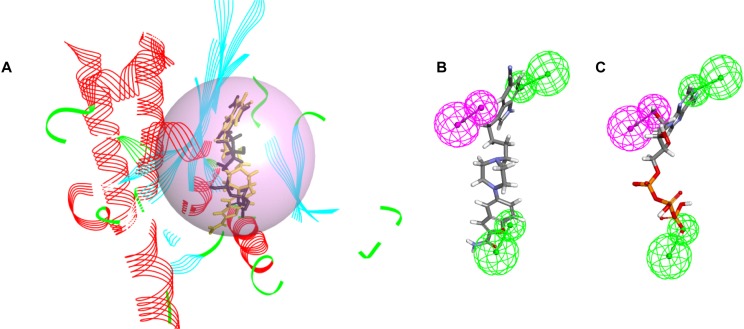
We analyzed the binding modes of vilazodone from the docking simulation results and found that the binding modes of vilazodone showed similar patterns to that of ATP, and belonged to the same cluster as ATP (Fig. 2A). In addition, considering the complementary pharmacophores of IPMK and its ligand, we performed a receptor-ligand pharmacophore model analysis. Pharmacophore models were derived for ATP-bound IPMK with two dominant hydrogen bonding acceptors, one hydrogen bonding donor pharmacophores, and their excluded volumes. The spatial distances between the features in the generated model are 10.43, 13.03, and 18.53 (Supplementary Fig. S1). Vilazodone fitted the models well, since all three dominant pharmacophores were mapped to IPMK. Since ATP is well fitted to the same pharmacological model (Figs. 2B and 2C), we predict that vilazodone targets the ATP-binding pocket of IPMK.
Fig. 2. Clustered models and the best pharmacophore model of IPMK aligned to either vilazodone or ATP.
(A) Docking simulation results indicate that villazodone and ATP have similar binding modes in the same cluster. The black compound is ATP and yellow is villazodone. (B and C) Generated pharmacophore models. Fitted pharmacophore model of vilazodone (B) and ATP (C), respectively. The hydrogen bond acceptor is green and the hydrogen bond donor variables are magenta.
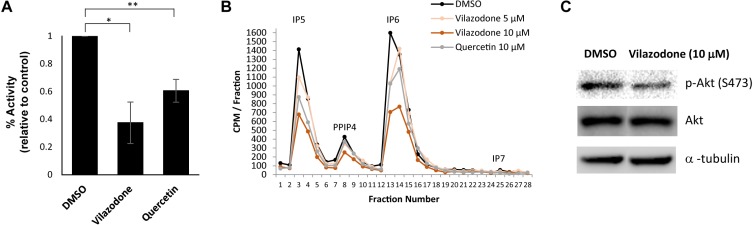
To confirm the inhibitory effect of vilazodone on the enzymatic activity of IPMK, we set up a kinase assay system using 10 μM (1,4,5,6) IP4 as a substrate and 10 μM ATP. By measuring the amount of ATP exhausted during the kinase reaction, we found that vilazodone lowers the catalytic activity of IPMK by 60%, compared to the DMSO control (Fig. 3A). To validate the assay, we used quercetin as a positive control inhibitor of IPMK (Gu et al., 2019). Next, we sought to characterize the cellular effect of vilazodone on inositol phosphate metabolism. Similar to the in vitro assays, the levels of highly phosphorylated IP metabolites synthesized by IPMK, including IP5, IP6, and IP7, were significantly diminished in the presence of 10 μM vilazodone for 4 h (Fig. 3B, Supplementary Fig. S2). However, IP4 level was not altered by vilazodone which suggests a selective inhibition of IPMK at IP5 generation. We confirmed vilazodone treatment did not influence the expression of IPMK protein (Supplementary Fig. S3). From further analyses on the effect of vilazodone in cellular signaling events, we observed that in HCT116 cells, Akt phosphorylation level was decreased following 10 μM vilazodone treatment for 4 h (Fig. 3C), which implies an inhibition of IPMK in Akt activation. Taken together, vilazodone exposure results in the typical cellular phenotypes of IPMK-depleted cells in a catalytically dependent fashion (Frederick et al., 2005; Kim et al., 2011; Maag et al., 2011; Morgan-Lappe et al., 2006), suggesting its potential as a novel IPMK inhibitor.
Fig. 3. The inhibitory effects of vilazodone on IPMK in vitro and in cellulo.
(A) In vitro IP kinase assays of IPMK. Enzymatic activity was measured in the absence or presence of 100 μM vilazodone or quercetin. Data are represented as mean ± SE (n = 3); *P < 0.05; **P < 0.01 (Student’s t-test). (B) HPLC profile of extracts from NIH3T3 L1 cells treated 4 h with DMSO (black), 5 μM vilazodone (light orange), 10 μM vilazodone (orange), or 10 μM quercetin (grey). (C) Immunoblotting of Akt S473 phosphorylation and total Akt in HCT116 cells treated with DMSO or 10 μM vilazodone for 4 h. Shown are representative HPLC profiles (B) and Western blots (C) from multiple independent experiments.
DISCUSSION
Despite long-standing efforts, the discovery of pharmacological interventions to inhibit IP kinases proceeds slowly. We performed a computer-aided virtual screening to find small molecular inhibitors of IPMK. We identified vilazodone, an FDA-approved drug, as an inhibitor of IPMK both in vitro and in cell culture. Vilazodone-treated NIH3T3-L1 fibroblasts resulted in the significant reduction of IP5 and its downstream IP metabolites, such as IP6 and IP7. The level of cellular IP4 was not influenced by vilazodone. Given that IPMK and other IP3 kinases are involved in the production of IP4, no changes in IP4 by vilazodone treatment suggest a selective action against IPMK and not other IP3Ks. Importantly, the cellular effects of vilazodone on IP metabolism (e.g., specific depletion of IP5, IP6, IP7 but not IP4) are identical to the phenotypes reported from IPMK-depleted mouse fibroblasts and IPMK-deleted stem cells, further supporting the selective inhibitory effect of vilazodone on IPMK.
Vilazodone was originally approved by the FDA for use as a therapy for the treatment of major depressive disorders. Mechanistically, vilazodone is known to inhibit serotonin transporters, increasing the bioavailability of serotonin and extending its duration of action at the synapse in the brain. Based on our findings, vilazodone’s unexpected activity in the suppression of IPMK and IP homeostasis might be critical to interpreting vilazodone’s pharmacological actions.
Our data suggest that the pharmaceutical drug vilazodone could serve as a useful compound to study the functions of IPMK and related IP metabolism. Further studies will need to characterize the pharmacological actions of vilazodone on the PI3 kinase activities of IPMK in vitro and in cells. It will also be advantageous to develop vilazodone analogs with improved pharmacokinetic properties for the selective and potent inhibition of IPMK. Future research could use the binding characteristics of our proposed IPMK-bound vilazodone model structure as a foundation to design a new generation of IPMK or other IP kinase inhibitors.
In conclusion, our discovery of vilazodone as an IPMK inhibitor greatly augments the available strategies for studying the significance of IPMK metabolism and complements genetic approaches. Vilazodone rapidly depletes IP5 and other downstream IP levels in mammalian cells, allowing us to quantitatively and dynamically manipulate the IP pathway. This will ultimately contribute to identifying the molecular mechanisms it controls.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by TJ Park Science Fellowship of the POSCO TJ Park Foundation (to S.E.P.), KAIST Advanced Institute for Science-X Fellowship (to S.J.P.) and the National Research Foundation of Korea (NRF-2019R1A6A1A03031807 to Y.B.; NRF-2018R1A5A1024261 and NRF-2018R1A2B2005913 to S.K.). We thank Yumi Shin in the Song lab for technical support for protein purification.
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Chakraborty A., Kim S., Snyder S.H. Inositol pyrophosphates as mammalian cell signals. Sci. Signal. 2011;4:1–12. doi: 10.1126/scisignal.2001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz M.P. Vilazodone HCL (Viibryd): a serotonin partial agonist and reuptake inhibitor for the treatment of major depressive disorder. P. T. 2012;37:28–31. [PMC free article] [PubMed] [Google Scholar]
- Dovey C.M., Diep J., Clarke B.P., Hale A.T., McNamara D.E., Guo H., Brown N.W., Cao J.Y., Grace C.R., Gough P.J., et al. MLKL requires the inositol phosphate code to execute necroptosis. Mol. Cell. 2018;70:936–948.:e7. doi: 10.1016/j.molcel.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick J.P., Mattiske D., Wofford J.A., Megosh L.C., Drake L.Y., Chiou S.T., Hogan B.L.M., York J.D. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang H.Y. Inositol pentakisphosphate mediates Wnt/β-catenin signaling. J. Biol. Chem. 2007;282:26490–26502. doi: 10.1074/jbc.M702106200. [DOI] [PubMed] [Google Scholar]
- Gu C., Stashko M.A., Puhl-Rubio A.C., Chakraborty M., Chakraborty A., Frye S.V., Pearce K.H., Wang X., Shears S.B., Wang H. Inhibition of inositol polyphosphate kinases by quercetin and related flavonoids: a structure-activity analysis. J. Med. Chem. 2019;62:1443–1454. doi: 10.1021/acs.jmedchem.8b01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.G., Al-Saigh S., Schultz C., Junop M.S. Inositol pentakisphosphate isomers bind PH domains with varying specificity and inhibit phosphoinositide interactions. BMC Struct. Biol. 2011;11:1–9. doi: 10.1186/1472-6807-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim S.F., Maag D., Maxwell M.J., Resnick A.C., Juluri K.R., Chakraborty A., Koldobskiy M.A., Cha S.H., Barrow R., et al. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13:215–221. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Kim E., Min H., Kim M.G., Eisenbeis V.B., Dutta A.K., Pavlovic I., Jessen H.J., Kim S., Seong R.H. Inositol polyphosphates promote T cell-independent humoral immunity via the regulation of Bruton's tyrosine kinase. Proc. Natl. Acad. Sci. U. S. A. 2019;116:12952–12957. doi: 10.1073/pnas.1821552116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Kim Y.R., Park J., Kim S. Inositol polyphosphate multikinase signaling in the regulation of metabolism. Ann. N. Y. Acad. Sci. 2012;1271:68–74. doi: 10.1111/j.1749-6632.2012.06725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.S., Lee J.Y., Kyung J.W., Yang Y., Park S.J., Lee S., Pavlovic I., Kong B., Jho Y.S., Jessen H.J., et al. Inositol pyrophosphates inhibit synaptotagmin-dependent exocytosis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:8314–8319. doi: 10.1073/pnas.1521600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D., Maxwell M.J., Hardesty D.A., Boucher K.L., Choudhari N., Hanno A.G., Ma J.F., Snowman A.S., Pietropaoli J.W., Xu R., et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H.L. The generation of a unique machine description for chemical structures-a technique developed at chemical abstracts service. J. Chem. Doc. 1965;5:107–113. doi: 10.1021/c160017a018. [DOI] [Google Scholar]
- Morgan-Lappe S., Woods K.W., Li Q., Anderson M.G., Schurdak M.E., Luo Y., Giranda V.L., Fesik S.W., Leverson J.D. RNAi-based screening of the human kinome identifies Akt-cooperating kinases: a new approach to designing efficacious multitargeted kinase inhibitors. Oncogene. 2006;25:1340–1348. doi: 10.1038/sj.onc.1209169. [DOI] [PubMed] [Google Scholar]
- Odom A.R., Stahlberg A., Wente S.R., York J.D. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Park S.J., Lee S., Park S.E., Kim S. Inositol pyrophosphates as multifaceted metabolites in the regulation of mammalian signaling networks. Anim. Cells Syst. 2018;22:1–6. doi: 10.1080/19768354.2017.1408684. [DOI] [Google Scholar]
- Piccolo E., Vignati S., Maffucci T., Innominato P.F., Riley A.M., Potter B.V.L., Pandolfi P.P., Broggini M., Iacobelli S., Innocenti P., et al. Inositol pentakisphosphate promotes apoptosis through the PI 3-K/Akt pathway. Oncogene. 2004;23:1754–1765. doi: 10.1038/sj.onc.1207296. [DOI] [PubMed] [Google Scholar]
- Rao S.N., Head M.S., Kulkarni A., LaLonde J.M. Validation studies of the site-directed docking program LibDock. J. Chem. Inf. Model. 2007;47:2159–2171. doi: 10.1021/ci6004299. [DOI] [PubMed] [Google Scholar]
- Razzini G., Berrie C.P., Vignati S., Broggini M., Mascetta G., Brancaccio A., Falasca M. Novel functional PI 3-kinase antagonists inhibit cell growth and tumorigenicity in human cancer cell lines. FASEB J. 2000;14:1179–1187. doi: 10.1096/fasebj.14.9.1179. [DOI] [PubMed] [Google Scholar]
- Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Saiardi A., Erdjument-Bromage H., Snowman A.M., Tempst P., Snyder S.H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999;9:1323–1326. doi: 10.1016/S0960-9822(00)80055-X. [DOI] [PubMed] [Google Scholar]
- Shen X., Xiao H., Ranallo R., Wu W.H., Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Steger D.J., Haswell E.S., Miller A.L., Wente S.R., O'Shea E.K. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb H., Irvine R.F., Berridge M.J., Schulz I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–69. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang H.Y. Dvl3 translocates IPMK to the cell membrane in response to Wnt. Cell. Signal. 2012;24:2389–2395. doi: 10.1016/j.cellsig.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P.J., Fairall L., Santos G.M., Schwabe J.W.R. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.