Abstract
Post-translational modifications play major roles in the stability, function, and localization of target proteins involved in the nervous system. The ubiquitin-proteasome pathway uses small ubiquitin molecules to degrade neuronal proteins. Deubiquitinating enzymes (DUBs) reverse this degradation and thereby control neuronal cell fate, synaptic plasticity, axonal growth, and proper function of the nervous system. Moreover, mutations or downregulation of certain DUBs have been found in several neurodegenerative diseases, as well as gliomas and neuroblastomas. Based on emerging findings, DUBs represent an important target for therapeutic intervention in various neurological disorders. Here, we summarize advances in our understanding of the roles of DUBs related to neurobiology.
Keywords: Alzheimer’s disease, deubiquitinating enzyme inhibitors, epilepsy, neural stem cells, Parkinson’s disease
INTRODUCTION
Post-translational modifications (PTMs) play key regulatory roles in development and function of the nervous system. Ubiquitination is one of the most important PTMs mediated by specific ligases for proteasomal degradation of target proteins. Expression of intercellular proteins is tightly regulated by a process of synthesis and degradation mediated directly or indirectly by the proteasomal pathway. The small regulatory protein ubiquitin (Ub) targets specific proteins for degradation by the ubiquitin proteasome system (UPS) or the lysosomal system. Conjugation of Ub to proteins, or ubiquitination, occurs through a series of events mediated by three enzymes: E1 (Ub activating enzyme), E2 (Ub conjugating enzyme), and E3 (Ub ligase) (Fig. 1A). Covalent attachment of one ubiquitin molecule to the lysine site of the target protein residue (monoubiquitination) can regulate its subcellular localization, activity, and interacting affinity. Similarly, ubiquitin molecules can bind to each other, forming an Ub-chain to a substrate protein and leading to its degradation by polyubiquitination. Ubiquitin has seven lysine residues (K6, K11, K27, K29, K33, K48, and K63), which together form numerous branched or linear chains that are involved in determination of the fate of target proteins (Kulathu and Komander, 2012; Ye and Rape, 2009). Erroneous ubiquitination of a protein could be detrimental for cells, as proteins may degrade prematurely, leading to autophagy and unintended cell death. Ubiquitin molecules are abundantly expressed in neurodegenerative disorders: as neurofibrillary tangles in Alzheimer’s disease (AD), Lewy bodies in Parkinson’s disease (PD), and intranuclear inclusions in hereditary polyglutamine diseases (Lennox et al., 1988; Mori et al., 1987; Paulson et al., 1997). Moreover, ubiquitin controls diverse neuronal processes including cell survival, cell fate determination, neurite outgrowth, morphogenesis, synapse development, and synaptic functions (DiAntonio et al., 2001; Ding and Shen, 2008; Jason and Ehlers, 2007; Tai and Schuman, 2008). Ubiquitination of synaptic proteins can be controlled by acute or chronic changes in synaptic activity (Chen et al., 2003; Ehlers, 2003).
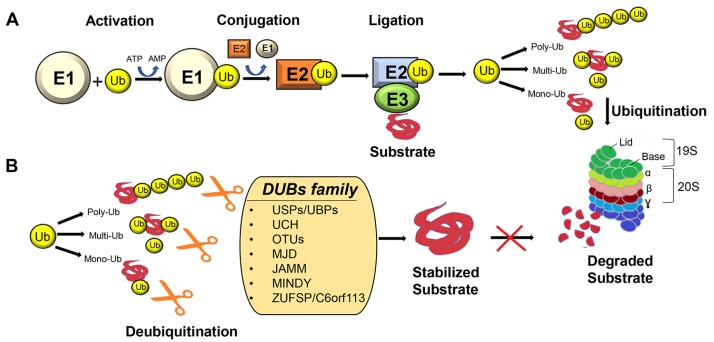
Fig. 1. The ubiquitin proteasome system.
(A) Ubiquitination is catalyzed by organized events mediated by E1, E2, and E3 ligases which promote the ligation of ubiquitin molecule to the lysine residues in the target protein substrates. Ubiquitin(s) attached proteins undergo the 26s proteasome degradation. (B) Deubiquitinating enzyme can remove the poly-, multi-, mono-ubiquitin molecules attached to the target substrate and inhibits its degradation and thereby stabilizes it.
Ubiquitination is antagonized by ubiquitin proteases, referred to as deubiquitinating enzymes (DUBs), which counteract the action of ligases by removing ubiquitin chains or maintaining the cellular pool of free ubiquitin monomer (Fig. 1B). DUBs are categorized into seven subfamilies: ubiquitin-specific proteases/ubiquitin-specific processing proteases (USPs/UBPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases, Josephin or Machado–Joseph disease protein domain proteases, Jab1/MPN domain-associated metalloisopeptidase (JAMM) domain proteins, motif interacting with Ub-containing novel DUB family (MINDY), and ZUFSP/C6orf113 (Abdul Rehman et al., 2016; Kwasna et al., 2018; Nishi et al., 2014) (Fig. 1B). The role of DUBs in the nervous system can be estimated by growing evidence on mouse (Saigoh et al., 1999; Wilson et al., 2002) and human (Kawaguchi et al., 1994) neurological disorders associated with mutations in certain DUBs. Several DUBs are involved in regulation of proteins involved in nervous system functions, neurodegenerative diseases, and brain cancers. In this review, we focus on DUBs that regulate intra-cellular processes for proper function of the nervous system and are implicated in several neurological disorders and brain tumors.
UBIQUITIN SPECIFIC PROTEASES
USP4
Ubiquitin-specific protease 4 (USP4) directly interacts with and deubiquitinates a specific G-protein coupled receptor, adenosine A2 (A2A), regulating the level of A2A receptors through the endoplasmic reticulum-associated protein degradation (ERAD) pathway. Overexpression of USP4 depletes ubiquitinated A2A and increases the number of functional receptors in hippocampal neurons (Milojević et al., 2006). Adenosine receptors mediate neuroprotection in the brain and are important targets for treatment of chronic neurodegenerative diseases (Abbracchio and Cattabeni, 1999). USP4, a deubiquitinase of A2AR, plays an important role in regulating its subcellular localization for ligand binding and signal generation (Toews, 2006). Therefore, pharmacological manipulation of USPs (like USP4) to regulate the expression of GPCRs could be a novel therapeutic strategy for various ailments including neurological disorders (Toews, 2006) (Fig. 2F).
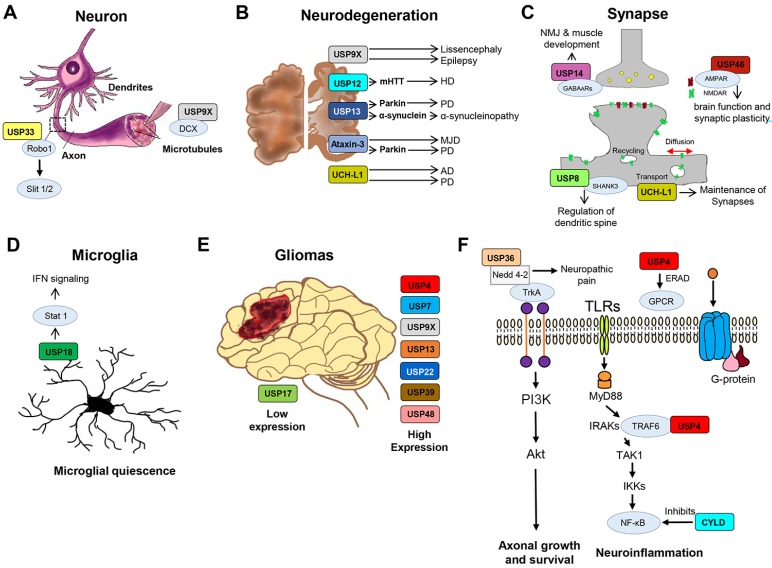
Fig. 2. Deubiquitinating enzymes involved in the regulation of nervous system and neurological disorders.
(A) USP9X is associated with microtubule-associated protein DCX and USP33 regulates axonal guidance receptor Robo1 and Slit 1/2 pathway. (B) USP9X, USP12, USP13, Ataxin-3, and UCH-L1 are directly or indirectly involved in the regulation of various neurodegenerative disorders. (C) USP8 interacts with synaptic protein SHANK3, USP14 stabilizes GABAARs to regulate NMJ & muscle development, USP46 regulates AMPARs to modulate the synaptic plasticity and UCH-L1 is involved in regulation of synaptic development, synaptic function, synaptic plasticity and synaptic transmission. (D) USP18 causes microglial quiescence and regulates IFN signaling via Stat 1. (E) USP4, USP7, USP9X, USP13, USP22, USP39, and USP48 are highly expressed in human gliomas and regulates cell survival. USP17 expression is low in gliomas and reduces tumorigenesis. (F) USP36 regulates Nedd 4-2, an E3 ligase of TrKA, involved in axonal growth and survival, whereas USP4 regulates GPCR and stabilizes TRAF6 in NF-κB pathway. CYLD is involved in the regulation of neuro-inflammation via NF-κB pathway.
Neurological inflammation can be induced by the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway upon activation of microglia, an active inducer of secondary spinal cord injury. The level of microglia is tightly maintained in the central nervous system (CNS); however, during traumatic conditions, microglia are excessively activated, inducing inflammatory cytokines and thereby aggravating secondary neuronal injury and inflammation. Tumor necrosis factor receptor associated factor 6 (TRAF6) is an essential adaptor protein for NF-κB signaling and has a major role in inflammation and immune response. In the microglial cells of rats, the expression of USP4 decreases after spinal cord injury (SCI). USP4 deubiquitinates TRAF6 and inhibits the TRAF6-stimulated NF-κB reporter gene and regulates the activation of NF-κB (Xiao et al., 2012) (Fig. 2F). USP4 might participate in promoting the activation of microglial-mediated neuronal inflammation by deubiquitinating TRAF6 via regulating the NF-κB signaling pathway (Jiang et al., 2017). Additionally, USP4 is frequently expressed in glioblastoma tissues and cell lines (Fig. 2E). Upon treatment with the anti-glioblastoma drug Temozolomide, USP4 knockdown cells undergo apoptosis in a p53 dependent manner (Qin et al., 2019). Taken together, these reports highlight the dynamic role of USP4 and likely other DUBs in regulation of diverse neurological pathways and disease pathologies.
USP7
USP7, or herpesvirus-associated ubiquitin-specific protease (HAUSP), regulates various brain functions and neurodegenerative disorders. USP7 interacts with Ataxin-1, which is associated with the spinocerebellar ataxia type 1 (SCA1), an autosomal-dominant neurodegenerative disorder characterized by several neurological defects and symptoms (Hong et al., 2002). Another important role of USP7 is maintenance of the level of repressor element 1-silencing transcription factor (REST), which is involved in inhibition of neural cell differentiation (Huang et al., 2011). During proteasomal degradation of REST, multiple E3 ligases are actively involved in maintaining REST expression. β-TrCP, an E3 ligase of REST, is abundantly expressed during neuronal differentiation, while the levels of USP7 and REST are low, suggesting a critical role for USP7 in neuronal differentiation and cell proliferation (Huang et al., 2011) . Deletion of USP7 in neural cells causes neonatal lethality, hypoplasia, and deficiencies in development, primarily due to USP7-p53-mediated apoptosis (Kon et al., 2011). USP7-mediated p53 stability is highly crucial for brain development, while p53 dependent or independent functions of USP7 contribute largely to mice lethality (Kon et al., 2011).
USP7 also deubiquitinates N-MYC proto-oncogene, which is amplified in numerous advanced stage tumors such as neuroblastomas. Knockdown of USP7 in neuroblastoma cancer cells and genetic manipulation of USP7 expression in mouse brain inhibit the stability and activity of N-MYC, suggesting USP7 as a potential therapeutic target for N-MYC-amplified tumors (Tavana et al., 2016). Lysinespecific demethylase 1 (LSD1) and USP7 are frequently overexpressed in aggressive brain tumors such as gliomas (Fig. 2E). USP7 inhibits the ubiquitination of LSD1 and regulates its protein turnover in A172 and T98G cells leading to rapid proliferation and invasion of glioblastoma cells (Yi et al., 2016). Thus, it could be estimated that USP7 demonstrates a crucial prognostic marker and potential therapeutic target for neuroblastomas and gliomas (Tavana et al., 2016; Yi et al., 2016).
USP8
PD is pathologically characterized by neuronal death and formation of inclusions known as Lewy bodies (LBs). Misfolding of α-synuclein is a common feature of PD (Spillantini et al., 1997) which leads to cognitive dysfunction (Schneider et al., 2012) as well as risk of dementia at an early age (Ross et al., 2008). Recently, α-synuclein inclusion was found to contain K63-linked ubiquitin chains, and USP8 present in Lewy bodies controls the K63-linked ubiquitination of α-synuclein in dopaminergic neurons to regulate PD state (Alexopoulou et al., 2016). USP8 also deubiquitinates LepRb, a receptor of leptin that has been implicated in regulation of synapses, neuronal plasticity, cognition, cortical volume, memory function, and depression-related phenomena. Additionally, the abundance of USP8 increases glutamatergic synapse formation in hippocampal structures (Bland et al., 2019).
Regulation of SHANK3 protein level is important to maintain synaptic density and levels of multiple synaptic proteins. SHANK3 mutations or any alterations in its expression often lead to neurodevelopmental disorders such as Phelan–McDermid syndrome, autism spectrum disorders, and schizophrenia. USP8/UBPY enhances the protein levels of SHANK3 and SHANK1 and subsequently enhances dendritic spine density in primary rat neurons (Kerrisk Campbell and Sheng, 2018) (Fig. 2C). USP8 also plays a key role in trafficking and stabilization of β-site amyloid precursor protein-cleaving (BACE1) enzyme, which is involved in the production of amyloid-β that accumulates in the brains of Alzheimer’s patients (Yeates and Tesco, 2016).
The tropomyosin-related kinase (Trk) family of receptor tyrosine kinases control synaptic plasticity, morphology, functions, and neuronal cell survival. USP8 interacts with TrkA receptor in a nerve growth factor (NGF)-dependent manner and inhibits neuronal differentiation in PC12 cells. Additionally, overexpression of USP8 blocks neurite outgrowth, which highlights the importance of USP8 in brain development (Ceriani et al., 2015). Considering the diverse role of USP8 in synaptic development and neurological disorders, USP8 could be a key target for future research on neurobiology.
USP9X
Faf (fat facets) play a major role in maintaining synaptic span, synaptic branching, and boutons in Drosophila (DiAntonio et al., 2001) and deubiquitinates liquid facets (Lqf) that are implicated in endocytosis (Cadavid et al., 2000; Chen et al., 2002). USP9X is the mammalian ortholog of Faf and binds to the Lqf ortholog epsin-1 and regulates its function and protein stability (Chen et al., 2003). USP9X is often overexpressed in glioblastomas, the most common primary brain tumors (Fig. 2E). A recent study showed that inhibition of USP9X by small-molecule inhibitor WP1130 decreases stem-cell-like glioblastoma cells, patient-derived xenografts, and cell viability of glioblastomas (Karpel-Massler et al., 2016), suggesting that USP9X could be a potential therapeutic target for glioblastomas.
USP9X is also correlated to lissencephaly, epilepsy (Friocourt et al., 2005) (Fig. 2B), and an X-linked intellectual disability candidate gene (Tarpey et al., 2009). USP9X regulates the stability of substrates involved in neurodevelopment signaling pathways such as Notch (Chastagner et al., 2008; Overstreet et al., 2004; Qiu et al., 2000), Wnt (Taya et al., 1999), and transforming growth factor beta (TGF-β) (Dupont et al., 2009). USP9X regulates the stability of ubiquitin ligases Mind Bomb1 (Choe et al., 2007; Yoon and Gaiano, 2005) and intracellular domain E3 ligase, Itch in the Notch pathway (Mouchantaf et al., 2006), which plays a major role in early neurodevelopment, learning, memory, and certain neurological diseases in adults (Lasky and Wu, 2005). USP9X also interacts with acute lymphoblastic leukemia-1 fusion partner chromosome 6 (AF-6), which is involved in establishment of adherens junctions and polarity in neural progenitor cells (Ikeda et al., 1999; Zhadanov et al., 1999).
USP9X is very important for development of the human CNS due to its association with the microtubule-associated protein doublecortin (DCX) (Friocourt et al., 2005) (Fig. 2A), which is implicated in neuronal migration, protein sorting, and trafficking of vesicles (Francis et al., 1999). Mutations in PRICKLE genes often cause epilepsy-related seizures. USP9X deubiquitinates PRICKLE and regulate PRICKLE-mediated seizures (Paemka et al., 2015), delineating the significance of USP9X in epilepsy. During the development of PD, USP9X regulates the level of α-synuclein and activates SMAD4 by stabilizing it at K519 and subsequently promoting the TGF-β pathway (often correlated with several neurodegenerative diseases) (Valderrama-Carvajal et al., 2002). Additionally, the Huntington's disease protein has also been associated with USP9X in mouse brain (Kaltenbach et al., 2007). Recently, the expression of USP9X or Mcl-1 (an anti-apoptotic member of the Bcl-2 family and a substrate of USP9X) has been shown to cause rapid death in malignant peripheral nerve sheath tumors (MPNSTs) (Bianchetti et al., 2018). Considering the diverse role of USP9X in nervous system, it could be a promising therapeutic target in neurogenerative disorders and malignancies (Li et al., 2017).
USP13
Glioblastoma harbors glioma stem cells (GSCs) are key players in tumor propagation and maintained by core transcriptional factors such as SOX2 and C-MYC. USP13 stabilizes C-Myc by inhibiting the E3 ligase and FBXL14-mediated ubiquitination and thereby maintains GSC self-renewal and tumorigenic potential (Fang et al., 2017) (Fig. 2E). Moreover, MYC proteins, which include L-MYC, C-MYC, and N-MYC, are also involved in development of the mid-, fore-, and hind-brain (Wey and Knoepfler, 2010). USP13 is abundantly expressed in the brain of PD patients (Fig. 2B), and deubiquitinates Parkin (an E3 ligase that targets certain neurological protein for degradation) and α-synuclein to regulate their metabolism in α-synucleinopathies (Liu et al., 2018). Missense mutations in the α-synuclein gene are common in PD, Lewy body dementia, and multiple system atrophy (Spillantini and Goedert, 2000). Thus, being a regulator of parkin and α-synuclein, USP13 could be a novel therapeutic target in α-synucleinopathies (Liu et al., 2018) (Fig. 2B).
USP14
USP14 is a key DUB involved in maintaining monoubiquitin level at developing synapses and is indispensable for development of synapses and proper function of neuromuscular junctions (NMJs). Loss of USP14 causes developmental defects at motor neurons. In ataxia (axJ) mice, the Purkinje cells in the cerebellum highly express GABAA receptors (GABAARs), which undergo proteasomal degradation (Saliba et al., 2007). GABAARs are one of the most studied neurotransmitter receptors and contribute to regulation of various brain functions and brain-related disorders (Everington et al., 2018). USP14 interacts with GABAARs and regulates their stability and cell surface distribution (Lappe-Siefke et al., 2009) (Fig. 2C). Moreover, depletion of USP14 in (axJ) mice not only results in perinatal lethality, reduced muscle development, structural and functional defects at the NMJ; but also leads to depletion of free ubiquitin in the brain and spinal cord (Anderson et al., 2005; Chen et al., 2009). The dominantly-negative, catalytically mutant USP14 in the murine nervous system mimics many defective phenotypes such as NMJ structure defects, reduced muscle development, and reduced motor performance (Vaden et al., 2015). However, restoring free ubiquitin level in the USP14 catalytically-inactive mice causes improvements in NMJ structure and reduction in pJNK accumulation in the motor neurons as well as negatively affects muscle development and motor functions (Vaden et al., 2015), indicating a crucial role for USP14 in synaptic development and functions.
USP46
USP46 was identified as the first DUB regulating glutamate receptors (GLuRs) by an RNAi-based screen in Caenorhabditis elegans. USP46 inhibits proteasomal degradation of glutamate receptor 1 (GLR-1) at synapses by deubiquitinating the receptor and stabilizing it in the lysosome (Kowalski et al., 2011). GLR-1 encodes a receptor subunit of a non-NMDA excitatory ionotropic glutamate receptor subtype and has 40% homology with mammalian (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) AMPA, which controls the majority of excitatory transmissions in the brain, as well as synaptic development and functions (Anggono and Huganir, 2012). Mammalian USP46 is expressed throughout the brain, including the hippocampus, amygdala, cerebellum, and prefrontal cortex, and stabilizes AMPARs (GluA1 and GLuA2) (Huo et al., 2015) (Fig. 2C). Thus, it could be predicted that USP46 might be involved in regulation of synaptic plasticity, brain functions, and synaptic transmission by stabilizing glutamatergic AMPARs (Huo et al., 2015). USP46 has also been associated with regulation of the GABAergic system in mice, which maintains fast-inhibitory transmission in the brain (Tomida et al., 2009). Loss of USP46 results in depression-like behaviors in mice (Imai et al., 2012) as well as reduction in expression of GABA synthesis enzyme glutamic acid decarboxylase (GAD67) (Tomida et al., 2009). Taken together, the involvement of USP46 in regulation of diverse synaptic receptors, underscores the importance of USP46 in synaptic formation and neuronal morphogenesis (Huo et al., 2015).
CYLD
CYLD negatively regulates NF-κB (Fig. 2F), which is involved in neuroinflammation, by reducing NF-κB activity in ischemic stroke (Kovalenko et al., 2003). NF-κB transcription factors are abundantly expressed in glial cells and cerebral blood vessels and protect neurons against different injuries or neuronal inflammatory reactions (Shih et al., 2015).
Other USPs
USP12 is a potent inducer of neuronal autophagy and regulates neuronal proteostasis and mutant huntingtin (mHTT), one of the main causes of the neurodegenerative disorder Huntington’s disease (Aron et al., 2018) (Fig. 2B).
USP17 is downregulated in glioma tissues (Fig. 2E), but overexpression of USP17 reduces tumorigenesis and cell proliferation in gliomas by reducing Ras and MYC protein levels (Hu et al., 2016).
Tissue homeostasis in the CNS is usually controlled by microglia, and dysregulation of microglia often leads to neuropsychiatric, neurodegenerative, and neuroinflammatory diseases known as microgliopathies. USP18 in the white matter of microglia contributes to microglial quiescence (Fig. 2D), and activates Stat 1 or other interferon genes, controlling IFN signaling (Goldmann et al., 2015). Moreover, USP18-deleted mouse brains exhibit microglial clusters in white matter, similar to the state in several human microgliopathies (Schwabenland et al., 2019).
USP22 gene is highly expressed in human brain glioma cells (Fig. 2E) and is associated with several neurological disorders (Li et al., 2013; Melo-Cardenas et al., 2016). Knockdown of USP22 effectively inhibits the cell viability of brain glioma cells, resulting in apoptosis and cell cycle arrest (Li et al., 2013).
USP33 is associated with the axonal guidance receptor Roundabout (Robo) 1 (Fig. 2A), which plays a major role in controlling axon crossing across the midline between brain hemispheres and neuronal dendrites (Yuasa-Kawada et al., 2009). USP33 protects the signal-competent Robo1 receptor complex from degradation and promotes the Slit signaling pathway (Yuasa-Kawada et al., 2009), which is essential for axon pathfinding in the CNS (Guan and Rao, 2003; Tessier-Lavigne and Goodman, 1996).
USP36 binds to one of the E3 ligases of TrkA neurotrophin receptor Nedd 4-2 and thereby regulates the association of Nedd 4-2 and TrkA (Anta et al., 2016). Nedd 4-2 has been directly implicated in the growth of peripheral neuropathic pain (Laedermann et al., 2013).
In addition to its role on neuronal differentiation (Tang, 2009), REST also controls proliferation of medulloblastoma cells (Das et al., 2013). REST expression downregulates CDKNIB/p27 (a cyclin-dependent kinase inhibitor) and promotes the proliferation of medulloblastomas. REST transcriptionally represses USP37 expression, whereas USP37 stabilizes p27 protein and block cell proliferation. Thus, REST and USP37 play an important role in regulating the stability of p27 and thereby controlling the proliferation of medulloblastomas (Das et al., 2013). Recently, USP37 has been reported to have tumor-suppressive properties in neural cancers; the level of USP37 was found to be downregulated in human medulloblastoma specimens (Dobson et al., 2017). Moreover, G9a (a histone methyltransferase) promotes USP37 depletion in a REST-dependent manner and causes growth and proliferation of medulloblastoma cells (Dobson et al., 2017).
USP39 has been implicated in human glioma by regulating the TAZ proteins in orthotopic xenografts (Fig. 2E). Knockdown of USP39 causes downregulation of TAZ pre-mRNA splicing efficiency in glioma cells, leading to depletion of TAZ protein (Ding et al., 2019).
Gli1 is an important downstream target of the Hedgehog (Hh) signaling pathway implicated in cell proliferation and tumorigenesis. USP48 has been shown to stabilize Gli1, whereas the Hh pathway has been shown to induce USP48 expression by transactivating Gli1, forming a reciprocal feedback loop. Depletion of USP48 inhibits glioma cell viability and tumor generation by partially stabilizing Gli1, which incidates a critical role of the USP48‐Gli1 axis in glioblastoma tumorigenesis (Zhou et al., 2017) (Fig. 2E).
UCH family
The carboxyl-terminal hydrolase UCH-L1 is highly expressed in the brain and is often implicated in neurodegenerative disorders in both mice and humans. UCH-L1 is abundantly found in the protein aggregates and inclusion bodies associated with PD and AD (Lowe et al., 1990; Setsuie and Wada, 2007; Wilkinson et al., 1992) (Fig. 2B). UCH-L1 plays a significant role in synaptic remodeling by maintaining synaptic structure in hippocampal neurons and modulating level of free monoubiquitin pools in an activity-dependent manner (Cartier et al., 2009). Downregulation of UCH-L1 causes synaptic defects such as decreased spine density, accumulation of pre- and post-synaptic proteins, and increased spine size (Cartier et al., 2009). Interestingly, pharmacological inhibition of UCH-L1 increases spine size and pre-synaptic and post-synaptic protein clusters and decreases spine density, implying an important role of UCH-L1 in regulation of brain functions (Setsuie and Wada, 2007). Moreover, UCH-L1 is correlated with decrease in synaptic vesicle number, increases in tubulovesicular structures in axons, and denervation of muscles (Chen et al., 2010) (Fig. 2C). In a mouse model of AD, reductions of monomeric ubiquitin and long-term potentiation (LTP) were observed due to loss of UCH-L1 in the brain (Gong et al., 2006).
Ap-UCH removes ubiquitin from polyubiquitinated substrates during proteasomal degradation and thereby maintains synaptic activity in Aplysia (Hegde et al., 1997). Similarly, in mammals, UCH-L3, an orthologue of AP-UCH, plays a major role in maintaining synaptic plasticity. Deficiency of UCH-L3 causes significant deficits in learning and memory in homozygous mice without any developmental, histological, or fertile abnormalities (Wood et al., 2005). Considering the importance of UCH sub-family of DUBs in synaptic development and regulation, further investigation is required to understand their role in synaptic defects related neurodegenerative disorders.
Ataxin-3
Ataxin-3, a member of the MJD sub-family of DUBs, was first implicated in the neurodegenerative disorder spinocerebellar ataxia type 3 (SCA3), also known as Machado-Joseph Disease (Fig. 2B). SCA3 is the most aggressive inherited ataxic age-related disorder and often leads to difficulties in speech and swallowing, impaired eye movements, neuropathy, and sometimes dystonia or parkinsonism (Todi et al., 2007; Williams and Paulson, 2008). Several studies have associated Ataxin-3 with several E3 ligases such as the carboxyl-terminus of HSC70-interacting protein (CHIP) (Jana et al., 2005), ubiquitination factor E4B (E4B/Ufd2) (Matsumoto et al., 2004), and parkin (Durcan et al., 2010). Parkin is a multifunctional ubiquitin ligase associated with maintenance of neuronal survival. Loss of Parkin increases risks of certain neurodegenerative diseases such as PD, AD, and amyotrophic lateral sclerosis (ALS) (Zhang et al., 2016).
The development and integrity of the nervous system are dependent on the balance between various components of Ub-dependent pathways, particularly DUBs. The tight regulation of ubiquitination and deubiquitination and the availability of mono-ubiquitins are critical for synapse structure and function. Several studies have explored the importance of DUBs in neurodevelopmental disorders and brain tumors. However, the search for DUB-based therapies is still in its infancy, and structural studies, enzymatic assays, and extensive research into the DUBs involved in diverse neurological ailments are needed to support drug development.
DUBs AS THERAPEUTIC TARGETS FOR NEURODEGENERATIVE DISEASES
Over the past decade, therapies focusing on the proteasomal pathway have shown huge promise due to their critical roles in protein regulation and several signaling pathways. Among post-translational regulators, DUBs offer several advantages as therapeutic targets due to their cell-type or substrate specificity. Although DUBs exhibit strong similarities between the active-enzyme site cysteine and histidine boxes, several DUBs demonstrate critical differences in accessibility to the catalytic pocket (Colland, 2010). Thus, developing DUB-specific inhibitors may be an attractive alternative for design of novel therapeutics to treat malignancies and neurodegenerative disorders. Several specific-DUB inhibitors, including USP7 and UCH-L1, have been developed to date (Colland, 2010; Todi and Das, 2012). Moreover, inhibition of USP14 by 1-[1-(4-fluorophenyl)-2,5-dimethylpyrrol-3-yl]-2-pyrrolidin-1-ylethanone (IU1) has been found to enhance the degradation of several proteins related to neurodegenerative diseases (Lee et al., 2010). A small-molecule inhibitor of USP9X, WP1130, demonstrated anti-proliferative properties by attenuating the growth of glioblastoma cells (Karpel-Massler et al., 2016). Although DUBs are a promising target in neurobiology, pharmacological inhibition of DUBs provides several challenges for the scientific community.
CURRENT CHALLENGES IN DUB-BASED THERAPEUTICS FOR NEUROBIOLOGY
DUB activity in cells is specific to the target substrates undergoing mono-ubiquitination or poly-ubiquitination, which comprise ubiquitin-chains bearing linkages or mixed chains containing ubiquitin and UBLs (Hospenthal et al., 2015; Komander and Rape, 2012). Although proteasomal inhibition is an important therapeutic strategy for various disorders related to neurobiology, indiscriminate inhibition of DUB activity might affect other cellular processes that rely on the 26S proteasomal system. Several DUBs are involved in the regulation of normal brain function as well as neurodegenerative disorders. For instance, USP14 inhibitor has a significant effect in regulating the level of proteins involved in neurodegeneration (Lee et al., 2010), but also functions in synaptic development and plasticity (Vaden et al., 2015; Wilson et al., 2002). Similarly, USP8 regulates the synapses by deubiquitinating LepRb and SHANK3 (Kerrisk Campbell and Sheng, 2018), but also stabilizes the BACE1 enzyme involved in AD (Yeates and Tesco, 2016). Thus, full or near-complete pharmacological inhibition of DUBs to control a neurological condition may have adverse effects.
Despite having druggable catalytic pockets, there are several challenges to developing potent compounds for inhibiting DUB expression. First, several DUBs share similar structure and properties and there are several limitations to developing classical small-molecule chemical inhibitors specifically targeting a particular DUB without affecting the expression of other DUBs. Second, most of the standard assays used to identify DUB inhibitors are prone to non-selective redox or alkylating false positives as DUB activity on ubiquitin molecules is dependent on a reactive thiol group (Wrigley et al., 2011). Additionally, the mode of action of several DUBs comprises complex enzymatic activity via allosteric sites, substrate catalysis, and involvement of both the active and non-active forms of DUBs (Mevissen and Komander, 2017; Sahtoe and Sixma, 2015). Therefore, before targeting DUBs as a therapeutic strategy, we need to address the limitations associated with the development of DUB-based therapies.
CONCLUSION AND PERSPECTIVE
In nervous system, PTMs play critical roles in proteasomal degradation and vesicular trafficking of intracellular proteins. Degradation of proteins is critical for the structure, function, and plasticity of synaptic connections (DiAntonio and Hicke, 2004). Ubiquitin protein-aggregates are found in a wide spectrum of neurodegenerative diseases, including AD, Huntington’s disease, and PD. Ubiquitin protease system (UPS) components like E3 ligases, DUBs, chaperons, shuttling factors, and various subtypes of proteasomes form complex networks in neurons (Tai and Schuman, 2008). Moreover, several neurodegenerative disorders are characterized by the accumulation of misfolded proteins, which affects nerve cell function and survival. DUBs have been reported to maintain healthy nerve cells by regulating the degradation of toxic proteins (Todi and Paulson, 2011). DUBs have also been implicated in the regulation of synapses, synaptic plasticity, and several neurodegenerative diseases such as AD, PD, and epilepsy, as well as neuroblastoma and glioblastomas (Kowalski and Juo, 2012; Ristic et al., 2014; Todi and Paulson, 2011). Recently, several studies have identified the role of DUBs in the regulation of various functions of the nervous system and in neurological disorders (Anta et al., 2016; Aron et al., 2018; Ding et al., 2019; Fang et al., 2017; Goldmann et al., 2015; Hu et al., 2016; Melo-Cardenas et al., 2016; Schwabenland et al., 2019; Zhou et al., 2017) (Fig. 2).
To date, several DUBs have been shown to regulate the nervous system, which makes them an attractive therapeutic target for disease intervention. However, there are several questions that must be addressed before targeting the DUBs involved in neurological disorders. For instance, neurons are highly polarized cells and the DUB activity in neurons depends on localization or re-localization of DUBs to different cellular or sub-cellular compartments. Thus, before pharmacologically targeting DUBs, it is important to understand which DUBs are shared among excitatory and inhibitory synapses as well as between sensory and motor neurons (Todi and Paulson, 2011). Moreover, we lack information about the expression pattern of DUBs during development and in adults, during activity and resting, in different areas of the brain and spinal cord, and in different types of neuronal and glial cells (Ristic et al., 2014). Given the importance of DUBs in the nervous system and in neurological disorders including cancers, further investigation is needed to improve our understanding of the role of DUBs and UPP in neuronal physiology and pathophysiology (Ristic et al., 2014).
In this review, we summarized the roles of DUBs, which regulate the stability and function of several neuronal proteins (Table 1). Major advances have been made over the past decade in identifying the critical roles of DUBs in nervous system development, function, and disease. Given the importance of PTMs in the nervous system, DUBs could be an excellent target for treatments aimed at maintaining appropriate function of the nervous system and controlling brain-related disorders including cancers.
Table 1.
List of DUBs and their functions in neurobiology
| DUBs | Functions | Reference | |
|---|---|---|---|
| USPs | |||
| Biological functions | Clinical significance | ||
| USP4 | Regulates the stability of G-protein coupled receptor, adenosine A2 (A2A) | Abundantly expressed in glioblastoma | (Jiang et al., 2017; Milojević et al., 2006; Qin et al., 2019; Toews, 2006) |
| Regulates functional receptors in neurons | Regulates the cell viability in glioblastomas | ||
| Involved in neuro-inflammation by deubiquitinatingTRAF6 | |||
| USP7 | Involved in transcription of Ataxin-1 | Deubiquitinates N-MYC and regulates neuroblastomas | (Hong et al., 2002; Huang et al., 2011; Kon et al., 2011; Tavana et al., 2016; Yi et al., 2016) |
| Regulates neuronal differentiation by stabilizing REST | Abundantly expressed in gliomas | ||
| Regulates neonatal lethality, hypoplasia, and developmental defects | Inhibits LSD1 to regulate glioblastomas | ||
| USP8 | Controls ubiquitination of α-synuclein | Regulates the stability of SHANK3, involved in several neurodegenerative disorders | (Alexopoulou et al., 2016; Bland et al., 2019; Ceriani et al., 2015; Kerrisk Campbell and Sheng, 2018; Yeates and Tesco, 2016) |
| Deubiquitinates LepRb receptor | |||
| Regulates glutamatergic synapse formation in the hippocampus | Stabilizes the BACE1 enzyme involved in production of amyloid-β in the AD-affected brain | ||
| Inhibits neuronal differentiation by interacting with TrkA | |||
| Blocks neurite outgrowth | |||
| USP9X | Regulates the stability of substrates involved in neurodevelopment signaling pathways (Notch, Wnt, TGF-β, and Itch) | Highly expressed in glioblastomas | (Abbracchio and Cattabeni, 1999; Chastagner et al., 2008; Dupont et al., 2009; Friocourt et al., 2005; Karpel-Massler et al., 2016; Mouchantaf et al., 2006; Paemka et al., 2015; Tarpey et al., 2009) |
| Involved in lissencephaly, epilepsy and X-linked intellectual disability | |||
| Interacts with AF-6, involved in development of neural progenitor cells | Regulates cell death and apoptosis in glioblastomas | ||
| Associated with neuronal protein DCX | Regulates seizures by deubiquitinating PRICKLE | ||
| Regulates α-synuclein, SMAD4 and TGF-β pathway (involved in neurodegenerative disorders) | |||
| USP13 | Stabilizes C-MYC and maintains glioma stem cells | Overexpressed in the brain of PD patients | (Fang et al., 2017; Liu et al., 2018) |
| Stabilizes Parkin and α-synuclein (involved in PD, dementia and neurological disorders) | |||
| USP14 | Maintains the synaptic structure and function | Inhibition of USP14 leads to degradation of several proteins involved in neurodegenerative disorders | (Lee et al., 2010; Vaden et al., 2015; Wilson et al., 2002) |
| Interacts with neurotransmitter receptor, GABAARs | |||
| Mutations in USP14 lead to defects in NMJ structure and reduction in motor performance | |||
| USP46 | Deubiquitinates GLR-1 at synapses | Highly expressed throughout the brain | (Huo et al., 2015; Imai et al., 2012; Kowalski et al., 2011; Tomida et al., 2009) |
| Stabilizes AMPARs (GLuA1 and GLuA2) | Depletion of USP46 leads to depression-like behaviors | ||
| Involved in mice GABAergic system | |||
| CYLD | Negatively regulates NF-κB (involved in neuroinflammation) | (Kovalenko et al., 2003) | |
| Other USPs | |||
| USP12 | Regulates neuronal autophagy and neuronal proteostasis | Regulates the expression of mHTT (involved in Huntington’s disease) | (Aron et al., 2018) |
| USP17 | Low expression in glioma cells | (Hu et al., 2016) | |
| Reduces tumorigenesis and proliferation of gliomas | |||
| USP18 | Causes microglial quiescence | Associated with the formation of microglial clusters (like state in human microgliopathies) | (Goldmann et al., 2015; Schwabenland et al., 2019) |
| Activates Stat 1 or other interferons and controls IFN signaling | |||
| USP22 | Overexpressed in brain gliomas and regulates proliferation of gliomas | (Li et al., 2013; Melo-Cardenas et al., 2016) | |
| Involved in various neurodegenerative disorders | |||
| USP33 | Associated with axonal guidance receptor Robo1 | (Yuasa-Kawada et al., 2009) | |
| USP36 | Interacts with E3 ligases of TrkA neurotrophin receptor, Nedd 4-2 | (Anta et al., 2016) | |
| USP37 | Associated with the stabilization of p27 | Associated with the suppression of growth and proliferation of medulloblastomas | (Das et al., 2013; Dobson et al., 2017) |
| USP39 | Regulates TAZ protein in human gliomas | (Ding et al., 2019) | |
| USP48 | Stabilizes Gli1 (a downstream target of Hedgehog signaling pathway) | Regulates the viability of gliomas | (Zhou et al., 2017) |
| UCH family | |||
| UCH-L1 | Regulates synaptic mono-ubiquitination | Highly expressed in protein aggregates and inclusion bodies associated with PD and AD | (Cartier et al., 2009; Gong et al., 2006; Lowe et al., 1988) |
| Maintains synaptic structure | |||
| Maintains spine density and size | |||
| Regulates pre- and postsynaptic protein levels | |||
| Regulates monomeric Ub an LTP | |||
| UCL-L3 | Regulates memory and conditions related to memory defects in mice | (Wood et al., 2005) | |
| Ataxin-3 | Implicated in Machado–Joseph Disease | (Durcan et al., 2010; Jana et al., 2005; Matsumoto et al., 2004) | |
| Associated with E3 ligases (CHIP, E4B, Parkin) of proteins involved in the regulation of several neurological conditions | |||
ACKNOWLEDGMENTS
This research was supported by a grant from National Research Foundation of Korea (2018M3A9H3022412 and 20 17R1A2B2008727) and Medical Research Center (2017 R1A5A2015395), funded by the National Research Foundation of Korea (NRF) of the Ministry of Science, ICT and Future Planning, Republic of Korea.
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Abbracchio M.P., Cattabeni F. Brain adenosine receptors as targets for therapeutic intervention in neurodegenerative diseases. Ann. N. Y. Acad. Sci. 1999;890:79–92. doi: 10.1111/j.1749-6632.1999.tb07983.x. [DOI] [PubMed] [Google Scholar]
- Abdul Rehman S.A., Kristariyanto Y.A., Choi S.Y., Nkosi P.J., Weidlich S., Labib K., Hofmann K., Kulathu Y. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou Z., Lang J., Perrett R.M., Elschami M., Hurry M.E.D., Kim H.T., Mazaraki D., Szabo A., Kessler B.M., Goldberg A.L., et al. Deubiquitinase Usp8 regulates α-synuclein clearance and modifies its toxicity in Lewy body disease. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E4688–E4697. doi: 10.1073/pnas.1523597113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C., Crimmins S., Wilson J.A., Korbel G.A., Ploegh H.L., Wilson S.M. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J. Neurochem. 2005;95:724–731. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- Anggono V., Huganir R.L. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr. Opin. Neurobiol. 2012;22:461–469. doi: 10.1016/j.conb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anta B., Martín-Rodríguez C., Gomis-Perez C., Calvo L., López-Benito S., Calderón-García A.A., Vicente-García C., Villarroel Á., Arévalo J.C. Ubiquitin-specific protease 36 (USP36) controls neuronal precursor cell-expressed developmentally down-regulated 4-2 (Nedd4-2) actions over the neurotrophin receptor TrkA and potassium voltage-gated channels 7.2/3 (Kv7.2/3) J. Biol. Chem. 2016;291:19132–19145. doi: 10.1074/jbc.M116.722637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron R., Pellegrini P., Green E.W., Maddison D.C., Opoku-Nsiah K., Wong J.S., Daub A.C., Giorgini F., Finkbeiner S. Deubiquitinase Usp12 functions noncatalytically to induce autophagy and confer neuroprotection in models of Huntington's disease. Nat. Commun. 2018;9:3191. doi: 10.1038/s41467-018-05653-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchetti E., Bates S.J., Carroll S.L., Siegelin M.D., Roth K.A. Usp9X regulates cell death in malignant peripheral nerve sheath tumors. Sci. Rep. 2018;8:17390. doi: 10.1038/s41598-018-35806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland T., Sahin G.S., Zhu M., Dillon C., Impey S., Appleyard S.M., Wayman G.A. USP8 deubiquitinates the leptin receptor and is necessary for leptin-mediated synapse formation. Endocrinology. 2019;160:1982–1998. doi: 10.1210/en.2019-00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadavid A., Ginzel A., Fischer J.A. The function of the Drosophila fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development. 2000;127:1727–1736. doi: 10.1242/dev.127.8.1727. [DOI] [PubMed] [Google Scholar]
- Cartier A.E., Djakovic S.N., Salehi A., Wilson S.M., Masliah E., Patrick G.N. Regulation of synaptic structure by ubiquitin C-terminal hydrolase L1. J. Neurosci. 2009;29:7857–7868. doi: 10.1523/JNEUROSCI.1817-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani M., Amigoni L., D'Aloia A., Berruti G., Martegani E. The deubiquitinating enzyme UBPy/USP8 interacts with TrkA and inhibits neuronal differentiation in PC12 cells. Exp. Cell Res. 2015;333:49–59. doi: 10.1016/j.yexcr.2015.01.019. [DOI] [PubMed] [Google Scholar]
- Chastagner P., Israël A., Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Sugiura Y., Myers K.G., Liu Y., Lin W. Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1636–1641. doi: 10.1073/pnas.0911516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Polo S., Di Fiore P.P., De Camilli P.V. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14908–14913. doi: 10.1073/pnas.2136625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.C., Qin L.N., Li X.M., Walters B.J., Wilson J.A., Mei L., Wilson S.M. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J. Neurosci. 2009;29:10909–10919. doi: 10.1523/JNEUROSCI.2635-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang B., Fischer J.A. A specific protein substrate for a deubiquitinating enzyme: liquid facets is the substrate of fat facets. Genes Dev. 2002;16:289–294. doi: 10.1101/gad.961502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe E.A., Liao L., Zhou J.Y., Cheng D., Duong D.M., Jin P., Tsai L.H., Peng J. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J. Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colland F. The therapeutic potential of deubiquitinating enzyme inhibitors. Biochem. Soc. Trans. 2010;38:137–143. doi: 10.1042/BST0380137. [DOI] [PubMed] [Google Scholar]
- Das C.M., Taylor P., Gireud M., Singh A., Lee D., Fuller G., Ji L., Fangusaro J., Rajaram V., Goldman S., et al. The deubiquitylase USP37 links REST to the control of p27 stability and cell proliferation. Oncogene. 2013;32:1691–1701. doi: 10.1038/onc.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A., Haghighi A.P., Portman S.L., Lee J.D., Amaranto A.M., Goodman C.S. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412:449. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- DiAntonio A., Hicke L. Ubiquitin-dependent regulation of the synapse. Annu. Rev. Neurosci. 2004;27:223–246. doi: 10.1146/annurev.neuro.27.070203.144317. [DOI] [PubMed] [Google Scholar]
- Ding K., Ji J., Zhang X., Huang B., Chen A., Zhang D., Li X., Wang X., Wang J. RNA splicing factor USP39 promotes glioma progression by inducing TAZ mRNA maturation. Oncogene. 2019;38:6414–6428. doi: 10.1038/s41388-019-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M., Shen K. The role of the ubiquitin proteasome system in synapse remodeling and neurodegenerative diseases. Bioessays. 2008;30:1075–1083. doi: 10.1002/bies.20843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson T.H., Hatcher R.J., Swaminathan J., Das C.M., Shaik S., Tao R.H., Milite C., Castellano S., Taylor P.H., Sbardella G. Regulation of USP37 expression by REST-associated G9a-dependent histone methylation. Mol. Cancer Res. 2017;15:1073–1084. doi: 10.1158/1541-7786.MCR-16-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M.J., Soligo S., Morsut L. FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- Durcan T.M., Kontogiannea M., Thorarinsdottir T., Fallon L., Williams A.J., Djarmati A., Fantaneanu T., Paulson H.L., Fon E.A. The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum. Mol. Genet. 2010;20:141–154. doi: 10.1093/hmg/ddq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M.D. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 2003;6:231. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Everington E.A., Gibbard A.G., Swinny J.D., Seifi M. Molecular characterization of GABA-A receptor subunit diversity within major peripheral organs and their plasticity in response to early life psychosocial stress. Front. Mol. Neurosci. 2018;11:18. doi: 10.3389/fnmol.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Zhou W., Wu Q., Huang Z., Shi Y., Yang K., Chen C., Xie Q., Mack S.C., Wang X., et al. Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. J. Exp. Med. 2017;214:245–267. doi: 10.1084/jem.20151673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M.C., Friocourt G., McDonnell N., Reiner O., Kahn A. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/S0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- Friocourt G., Kappeler C., Saillour Y., Fauchereau F., Rodriguez M.S., Bahi N., Vinet M.C., Chafey P., Poirier K., Taya S. Doublecortin interacts with the ubiquitin protease DFFRX, which associates with microtubules in neuronal processes. Mol. Cell. Neurosci. 2005;28:153–164. doi: 10.1016/j.mcn.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Goldmann T., Zeller N., Raasch J., Kierdorf K., Frenzel K., Ketscher L., Basters A., Staszewski O., Brendecke S.M., Spiess A., et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34:1612–1629. doi: 10.15252/embj.201490791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B., Cao Z., Zheng P., Vitolo O.V., Liu S., Staniszewski A., Moolman D., Zhang H., Shelanski M., Arancio O. Ubiquitin hydrolase Uch-L1 rescues β-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126:775–788. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- Guan K.L., Rao Y. Signalling mechanisms mediating neuronal responses to guidance cues. Nat. Rev. Neurosci. 2003;4:941. doi: 10.1038/nrn1254. [DOI] [PubMed] [Google Scholar]
- Hegde A.N., Inokuchi K., Pei W., Casadio A., Ghirardi M., Chain D.G., Martin K.C., Kandel E.R., Schwartz J.H. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in aplysia. Cell. 1997;89:115–126. doi: 10.1016/S0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- Hong S., Kim S.J., Ka S., Choi I., Kang S. USP7, a ubiquitin-specific protease, interacts with ataxin-1, the SCA1 gene product. Mol. Cell. Neurosci. 2002;20:298–306. doi: 10.1006/mcne.2002.1103. [DOI] [PubMed] [Google Scholar]
- Hospenthal M.K., Mevissen T.E., Komander D. Deubiquitinase-based analysis of ubiquitin chain architecture using ubiquitin chain restriction (UbiCRest) Nat. Protoc. 2015;10:349–361. doi: 10.1038/nprot.2015.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Chen H., Han C., Lan J., Xu Y., Li C., Xue Y., Lou M. Expression and functional implications of USP17 in glioma. Neurosci. Lett. 2016;616:125–131. doi: 10.1016/j.neulet.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Huang Z., Wu Q., Guryanova O.A., Cheng L., Shou W., Rich J.N., Bao S. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat. Cell Biol. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y., Khatri N., Hou Q., Gilbert J., Wang G., Man H.Y. The deubiquitinating enzyme USP46 regulates AMPA receptor ubiquitination and trafficking. J. Neurochem. 2015;134:1067–1080. doi: 10.1111/jnc.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda W., Nakanishi H., Miyoshi J., Mandai K., Ishizaki H., Tanaka M., Togawa A., Takahashi K., Nishioka H., Yoshida H. Afadin: a key molecule essential for structural organization of cell-cell junctions of polarized epithelia during embryogenesis. J. Cell Biol. 1999;146:1117–1132. doi: 10.1083/jcb.146.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Mamiya T., Tsukada A., Sakai Y., Mouri A., Nabeshima T., Ebihara S. Ubiquitin-specific peptidase 46 (Usp46) regulates mouse immobile behavior in the tail suspension test through the GABAergic system. PLoS One. 2012;7:e39084. doi: 10.1371/journal.pone.0039084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana N.R., Dikshit P., Goswami A., Kotliarova S., Murata S., Tanaka K., Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- Jason J.Y., Ehlers M.D. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacological Rev. 2007;59:14–39. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
- Jiang X., Yu M., Ou Y., Cao Y., Yao Y., Cai P., Zhang F. Downregulation of USP4 promotes activation of microglia and subsequent neuronal inflammation in rat spinal cord after injury. Neurochem. Res. 2017;42:3245–3253. doi: 10.1007/s11064-017-2361-2. [DOI] [PubMed] [Google Scholar]
- Kaltenbach L.S., Romero E., Becklin R.R., Chettier R., Bell R., Phansalkar A., Strand A., Torcassi C., Savage J., Hurlburt A., et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpel-Massler G., Banu M.A., Shu C., Halatsch M.E., Westhoff M.A., Bruce J.N., Canoll P., Siegelin M.D. Inhibition of deubiquitinases primes glioblastoma cells to apoptosis in vitro and in vivo. Oncotarget. 2016;7:12791–12805. doi: 10.18632/oncotarget.7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32. 1. Nat. Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- Kerrisk Campbell M., Sheng M. USP8 deubiquitinates SHANK3 to control synapse density and SHANK3 activity-dependent protein levels. J. Neurosci. 2018;38:5289–5301. doi: 10.1523/JNEUROSCI.3305-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kon N., Zhong J., Kobayashi Y., Li M., Szabolcs M., Ludwig T., Canoll P.D., Gu W. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ. 2011;18:1366–1375. doi: 10.1038/cdd.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Kowalski J.R., Dahlberg C.L., Juo P. The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans. J. Neurosci. 2011;31:1341–13. doi: 10.1523/JNEUROSCI.4765-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski J.R., Juo P. The role of deubiquitinating enzymes in synaptic function and nervous system diseases. Neural. Plast. 2012;2012:892749. doi: 10.1155/2012/892749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y., Komander D. Atypical ubiquitylation-the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012;13:508. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- Kwasna D., Abdul Rehman S.A., Natarajan J., Matthews S., Madden R., De Cesare V., Weidlich S., Virdee S., Ahel I., Gibbs-Seymour I., et al. Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol. Cell. 2018;70:150–164.:e6. doi: 10.1016/j.molcel.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laedermann C.J., Cachemaille M., Kirschmann G., Pertin M., Gosselin R.D., Chang I., Albesa M., Towne C., Schneider B.L., Kellenberger S. Dysregulation of voltage-gated sodium channels by ubiquitin ligase NEDD4-2 in neuropathic pain. J. Clin. Invest. 2013;123:3002–3013. doi: 10.1172/JCI68996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C., Loebrich S., Hevers W., Waidmann O.B., Schweizer M., Fehr S., Fritschy J.M., Dikic I., Eilers J., Wilson S.M. The ataxia (axJ) mutation causes abnormal GABAA receptor turnover in mice. PLoS Genet. 2009;5:e1000631. doi: 10.1371/journal.pgen.1000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky J.L., Wu H. Notch signaling, brain development, and human disease. Pediatr. Res. 2005;57:104–109. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., Gartner C., Dimova N., Hanna J., Gygi S.P. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox G., Lowe J., Morrell K., Landon M., Mayer R.J. Ubiquitin is a component of neurofibrillary tangles in a variety of neurodegenerative diseases. Neurosci. Lett. 1988;94:211–217. doi: 10.1016/0304-3940(88)90297-2. [DOI] [PubMed] [Google Scholar]
- Li Z., Cheng Z., Raghothama C., Cui Z., Liu K., Li X., Jiang C., Jiang W., Tan M., Ni X., et al. USP9X controls translation efficiency via deubiquitination of eukaryotic translation initiation factor 4A1. Nucleic Acids Res. 2017;46:823–839. doi: 10.1093/nar/gkx1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.H., Yu Y., Du C., Fu H., Wang J., Tian Y. RNA interference-mediated USP22 gene silencing promotes human brain glioma apoptosis and induces cell cycle arrest. Oncol. Lett. 2013;5:1290–1294. doi: 10.3892/ol.2013.1188. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu X., Hebron M., Shi W., Lonskaya I., Moussa C.E.H. Ubiquitin specific protease-13 independently regulates parkin ubiquitination and alpha-synuclein clearance in alpha-synucleinopathies. Hum. Mol. Genet. 2018;28:548–560. doi: 10.1093/hmg/ddy365. [DOI] [PubMed] [Google Scholar]
- Lowe J., Blanchard A., Morrell K., Lennox G., Reynolds L., Billett M., Landon M., Mayer R.J. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson's disease, Pick's disease, and Alzheimer's disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J. Pathol. 1988;155:9–15. doi: 10.1002/path.1711550105. [DOI] [PubMed] [Google Scholar]
- Lowe J., McDermott H., Landon M., Mayer R.J., Wilkinson K.D. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J. Pathol. 1990;161:153–160. doi: 10.1002/path.1711610210. [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Yada M., Hatakeyama S., Ishimoto H., Tanimura T., Tsuji S., Kakizuka A., Kitagawa M., Nakayama K.I. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23:659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Cardenas J., Zhang Y., Zhang D.D., Fang D. Ubiquitin-specific peptidase 22 functions and its involvement in disease. Oncotarget. 2016;7:44848–44856. doi: 10.18632/oncotarget.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen T.E., Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- Milojević T., Reiterer V., Stefan E., Korkhov V.M., Dorostkar M.M., Ducza E., Ogris E., Boehm S., Freissmuth M., Nanoff C. The ubiquitin-specific protease Usp4 regulates the cell surface level of the A2A receptor. Mol. Pharmacol. 2006;69:1083–1094. doi: 10.1124/mol.105.015818. [DOI] [PubMed] [Google Scholar]
- Mori H., Kondo J., Ihara Y. Ubiquitin is a component of paired helical filaments in Alzheimer's disease. Science. 1987;235:1641–1644. doi: 10.1126/science.3029875. [DOI] [PubMed] [Google Scholar]
- Mouchantaf R., Azakir B.A., McPherson P.S., Millard S.M., Wood S.A., Angers A. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem. 2006;281:38738–38747. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- Nishi R., Wijnhoven P., le Sage C., Tjeertes J., Galanty Y., Forment J.V., Clague M.J., Urbé S., Jackson S.P. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol. 2014;16:1016–1018. doi: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet E., Fitch E., Fischer J.A. Fat facets and liquid facets promote delta endocytosis and delta signaling in the signaling cells. Development. 2004;131:5355–5366. doi: 10.1242/dev.01434. [DOI] [PubMed] [Google Scholar]
- Paemka L., Mahajan V.B., Ehaideb S.N., Skeie J.M., Tan M.C., Wu S., Cox A.J., Sowers L.P., Gecz J., Jolly L., et al. Seizures are regulated by ubiquitin-specific peptidase 9 X-linked (USP9X), a de-ubiquitinase. PLoS Genet. 2015;11:e1005022. doi: 10.1371/journal.pgen.1005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson H.L., Das S.S., Crino P.B., Perez M.K., Patel S.C., Gotsdiner D., Fischbeck K.H., Pittman R.N. Machado-Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann. Neurol. 1997;41:453–462. doi: 10.1002/ana.410410408. [DOI] [PubMed] [Google Scholar]
- Qin N., Han F., Li L., Ge Y., Lin W., Wang J., Wu L., Zhao G., Deng Y., Zhang J. Deubiquitinating enzyme 4 facilitates chemoresistance in glioblastoma by inhibiting P53 activity. Oncol. Lett. 2019;17:958–964. doi: 10.3892/ol.2018.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L., Joazeiro C., Fang N., Wang H.Y., Elly C., Altman Y., Fang D., Hunter T., Liu Y.C. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- Ristic G., Tsou W.L., Todi S.V. An optimal ubiquitin-proteasome pathway in the nervous system: the role of deubiquitinating enzymes. Front. Mol. Neurosci. 2014;7:72. doi: 10.3389/fnmol.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross O.A., Braithwaite A.T., Skipper L.M., Kachergus J., Hulihan M.M., Middleton F.A., Nishioka K., Fuchs J., Gasser T., Maraganore D.M. Genomic investigation of α-synuclein multiplication and Parkinsonism. Ann. Neurol. 2008;63:743–750. doi: 10.1002/ana.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahtoe D.D., Sixma T.K. Layers of DUB regulation. Trends Biochem. Sci. 2015;40:456–467. doi: 10.1016/j.tibs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Saigoh K., Wang Y.L., Suh J.G., Yamanishi T., Sakai Y., Kiyosawa H., Harada T., Ichihara N., Wakana S., Kikuchi T. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat. Genet. 1999;23:47. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- Saliba R.S., Michels G., Jacob T.C., Pangalos M.N., Moss S.J. Activity-dependent ubiquitination of GABAA receptors regulates their accumulation at synaptic sites. J. Neurosci. 2007;27:13341–13351. doi: 10.1523/JNEUROSCI.3277-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J., Arvanitakis Z., Yu L., Boyle P., Leurgans S., Bennett D. Cognitive impairment, decline and fluctuations in older community-dwelling subjects with Lewy bodies. Brain. 2012;135:3005–3014. doi: 10.1093/brain/aws234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabenland M., Mossad O., Peres A.G., Kessler F., Maron F.J.M., Harsan L.A., Bienert T., von Elverfeldt D., Knobeloch K.P., Staszewski O., et al. Loss of USP18 in microglia induces white matter pathology. Acta Neuropathol. Commun. 2019;7:106. doi: 10.1186/s40478-019-0757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsuie R., Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem. Int. 2007;51:105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shih R.H., Wang C.Y., Yang C.M. NF-kappaB signaling pathways in neurological inflammation: a mini review. Front. Mol. Neurosci. 2015;8:77. doi: 10.3389/fnmol.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini M.G., Goedert M. The α-synucleinopathies: Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy. Ann. N. Y. Acad. Sci. 2000;920:16–27. doi: 10.1111/j.1749-6632.2000.tb06900.x. [DOI] [PubMed] [Google Scholar]
- Spillantini M.G., Schmidt M.L., Lee V.M.Y., Trojanowski J.Q., Jakes R., Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Tai H.C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008;9:826. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- Tang B.L. REST regulation of neural development: from outside-in? Cell Adh. Migr. 2009;3:1–2. doi: 10.4161/cam.3.2.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey P.S., Smith R., Pleasance E., Whibley A., Edkins S., Hardy C., O'meara S., Latimer C., Dicks E., Menzies A. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat. Genet. 2009;41:535. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavana O., Li D., Dai C., Lopez G., Banerjee D., Kon N., Chen C., Califano A., Yamashiro D.J., Sun H., et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat. Med. 2016;22:1180–1186. doi: 10.1038/nm.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taya S., Yamamoto T., Kanai-Azuma M., Wood S.A., Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes β-catenin. Genes Cells. 1999;4:757–767. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M., Goodman C.S. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Todi S., Das C. Should deubiquitinating enzymes be targeted for therapy. Clin. Pharmacol. Biopharm. 2012;1:1000e108. doi: 10.4172/2167-065X.1000e108. [DOI] [Google Scholar]
- Todi S.V., Paulson H.L. Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci. 2011;34:370–382. doi: 10.1016/j.tins.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi S.V., Williams A.J., Paulson H.L. Polyglutamine disorders including Huntington's disease. In: Waxman S.G., editor. Molecular Neurology. Academic Press; Cambridge: 2007. pp. 257–275. [DOI] [Google Scholar]
- Toews M.L. Adenosine receptors find a new partner and move out. Mol. Pharmacol. 2006;69:1075–1078. doi: 10.1124/mol.106.022699. [DOI] [PubMed] [Google Scholar]
- Tomida S., Mamiya T., Sakamaki H., Miura M., Aosaki T., Masuda M., Niwa M., Kameyama T., Kobayashi J., Iwaki Y. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat. Genet. 2009;41:688. doi: 10.1038/ng.344. [DOI] [PubMed] [Google Scholar]
- Vaden J.H., Bhattacharyya B.J., Chen P.C., Watson J.A., Marshall A.G., Phillips S.E., Wilson J.A., King G.D., Miller R.J., Wilson S.M. Ubiquitin-specific protease 14 regulates c-Jun N-terminal kinase signaling at the neuromuscular junction. Mol. Neurodegener. 2015;10:3. doi: 10.1186/1750-1326-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama-Carvajal H., Cocolakis E., Lacerte A., Lee E.H., Krystal G., Ali S., Lebrun J.J. Activin/TGF-β induce apoptosis through Smad-dependent expression of the lipid phosphatase SHIP. Nat. Cell Biol. 2002;4:963. doi: 10.1038/ncb885. [DOI] [PubMed] [Google Scholar]
- Wey A., Knoepfler P.S. C-myc and N-myc in the developing brain. Aging (Albany NY) 2010;2:261–262. doi: 10.18632/aging.100151. [DOI] [Google Scholar]
- Wilkinson K.D., Deshpande S., Larsen C.N. Comparisons of neuronal (PGP 9.5) and non-neuronal ubiquitin C-terminal hydrolases. Biochem. Soc. Trans. 1992;20:631–637. doi: 10.1042/bst0200631. [DOI] [PubMed] [Google Scholar]
- Williams A.J., Paulson H.L. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.M., Bhattacharyya B., Rachel R.A., Coppola V., Tessarollo L., Householder D.B., Fletcher C.F., Miller R.J., Copeland N.G., Jenkins N.A. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat. Genet. 2002;32:420. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- Wood M.A., Kaplan M.P., Brensinger C.M., Guo W., Abel T. Ubiquitin C-terminal hydrolase L3 (Uchl3) is involved in working memory. Hippocampus. 2005;15:610–621. doi: 10.1002/hipo.20082. [DOI] [PubMed] [Google Scholar]
- Wrigley J.D., Eckersley K., Hardern I.M., Millard L., Walters M., Peters S.W., Mott R., Nowak T., Ward R.A., Simpson P.B., et al. Enzymatic characterisation of USP7 deubiquitinating activity and inhibition. Cell Biochem. Biophys. 2011;60:99. doi: 10.1007/s12013-011-9186-4. [DOI] [PubMed] [Google Scholar]
- Xiao N., Li H., Luo J., Wang R., Chen H., Chen J., Wang P. Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFα-induced cancer cell migration. Biochem. J. 2012;441:979–987. doi: 10.1042/BJ20111358. [DOI] [PubMed] [Google Scholar]
- Ye Y., Rape M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009;10:755. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates E.F.A., Tesco G. The endosome-associated deubiquitinating enzyme USP8 regulates BACE1 enzyme ubiquitination and degradation. J. Biol. Chem. 2016;291:15753–15766. doi: 10.1074/jbc.M116.718023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., Cui Y., Xu Q., Jiang Y. Stabilization of LSD1 by deubiquitinating enzyme USP7 promotes glioblastoma cell tumorigenesis and metastasis through suppression of the p53 signaling pathway. Oncol. Rep. 2016;36:2935–2945. doi: 10.3892/or.2016.5099. [DOI] [PubMed] [Google Scholar]
- Yoon K., Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 2005;8:709. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Yuasa-Kawada J., Kinoshita-Kawada M., Wu G., Rao Y., Wu J.Y. Midline crossing and Slit responsiveness of commissural axons require USP33. Nat. Neurosci. 2009;12:1087–1089. doi: 10.1038/nn.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhadanov A.B., Provance D.W., Jr., Speer C., Coffin J.D., Goss D., Blixt J., Reichert C.M., Mercer J.A. Absence of the tight junctional protein AF-6 disrupts epithelial cell-cell junctions and cell polarity during mouse development. Curr. Biol. 1999;9:880–882. doi: 10.1016/S0960-9822(99)80392-3. [DOI] [PubMed] [Google Scholar]
- Zhang C.W., Hang L., Yao T.P., Lim K.L. Parkin regulation and neurodegenerative disorders. Front. Aging Neurosci. 2016;7:248. doi: 10.3389/fnagi.2015.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Lin K., Zhang S., Ma L., Xue J., Morris S.A., Aldape K.D., Huang S. Gli1-induced deubiquitinase USP48 aids glioblastoma tumorigenesis by stabilizing Gli1. EMBO Rep. 2017;18:1318–1330. doi: 10.15252/embr.201643124. [DOI] [PMC free article] [PubMed] [Google Scholar]