Abstract
Objective
This study aims to evaluate the correlation between viral clearance and blood biochemical index of 94 discharged patients with COVID-19 infection in Shenzhen Third People’s Hospital, enrolled from Jan 5 to Feb 13, 2020.
Methods
The clinical and laboratory findings were extracted from the electronic medical records of the patients. The data were analysed and reviewed by a trained team of physicians. Information on clinical signs and symptoms, medical treatment, virus clearance, and laboratory parameters including interleukin 6 (IL-6) and C-reactive protein were collected.
Results
COVID-19 mRNA clearance ratio was identified significantly correlated with the decline of serum creatine kinase (CK) and lactate dehydrogenase (LDH) levels. Furthermore, COVID-19 mRNA clearance time was positively correlated with the length of hospital stay in patients treated with either IFN-α + lopinavir/ritonavir or IFN-α + lopinavir/ritonavir + ribavirin.
Conclusions
Therapeutic regimens of IFN-α + lopinavir/ritonavir and IFN-α + lopinavir/ritonavir + ribavirin might be beneficial for treatment of COVID-19. Serum LDH or CK decline may predict a favorable response to treatment of COVID-19 infection.
Keywords: COVID-19, Clearance ratio, LDH, CK
Introduction
Since Dec 8, 2019, the sudden outbreak of a mysterious pneumonia cases, infected with a novel coronavirus known as COVID-19, have rapidly spread from Wuhan [1] to other areas, causing staggering number of infections and deaths. As of this writing, a total of 77,272 cases including 2597 deaths have been reported in China. Based on the phylogenetic analysis, COVID-19 belongs to a distinct clade of beta-coronavirus similar to human severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), two species outbreaking in the latest decade and causing severe human deaths [2]. Despite the estimated fatality rate of COVID-19 is lower than that of SARS or MERS, the scale of the COVID-19 contagion has caused more casualties than either of them as of this writing [3]. In addition, human to human transmission of COVID-19 has been confirmed [4]. A number of cases of COVID-19 infection has been reported by investigators who described the epidemiological and clinical characteristics of hospitalized patients [5–8]. However, there is a limited number of detailed analysis on cytokines, blood chemicals or viral mRNA clearance rate on discharged patients.
Here, we aim to describe the changes and correlation of clinical features and candidate molecular markers, as well as potent therapeutic regimen in view of 94 discharged patients. We hope that our findings might be useful to inspire future clinical research associated with COVID-19 infection.
Methods
Data sources
In this retrospective, single-centre study, all data were collected from a total of 94 patients enrolled from Jan 11th 2020 to Feb 4th 2020 in Shenzhen Third People’s Hospital. All of them were confirmed with COVID-19 infection and discharged up to Feb 13th, 2020. The epidemiological, demographic, clinical, and laboratory findings were extracted from the electronic medical records of the patients. The data were analysed and reviewed by a trained team of physicians. Information including demographic data, exposure history, clinical signs and symptoms, underling comorbidities, medical treatment, laboratory parameters were collected. Nasal and pharyngeal swab specimens, sputum and bronchoalveolar lavage fluid (BALF) were obtained from patients on admission. The study was approved by Shenzhen Third People’s Hospital Ethics Committee.
COVID-19 was confirmed by real-time RT-PCR as described previously [6]. Other respiratory viruses including influenza, avian influenza, respiratory syncytial virus, mycoplasma and chlamydia were also examined with real-time RT-PCR kits approved by the China Food and Drug Administration. Antiviral drugs of interferon-α, lopinavir/ritonavir, ribavirin, arbidol, favipiravir, human γ-globulin and glucocorticoid were used for treatment of these patients.
Statistical analyses
Mean [standard deviation (SD)] and range were reported for normally distributed, continuous variables. Frequencies and percentages were reported for categorical variables. The one way ANOVA was used to compare continuous variable among three group, while independent samples t test was used between two groups. The Mann–Whitney U test was utilized to compare significant differences among continuous data. All statistical tests were two-tailed, and the p value less than 0.05 was considered statistically significant. All analyses were done with SPSS software, version 22.0.
Results
Presenting characteristics
Table 1 lists the characteristics of all discharged patients. The median age for moderate symptom patients was 40 years (range 1–78 years), while 19 years for mild-symptom patients (range 7–39 years), as well as 63 years (range 32–69) for critical patients (one-way ANOVA, p < 0.0001). Approximately 48 patients (51%) were the patients aged ≤ 40 years, including 2 children under 3 years. Overall, 15 patients had one or two types of chronic cardiovascular diseases, including coronary heart disease (CHD), hypertension (HBP) or diabetes (T2DM). 79 of them had resided in Hubei province, while another 13 had the history of contact with Hubei residents. But none of them were exposed to the Huanan seafood market. According to our research, 36 of the 94 patients (38%) were associated with familial clusters. There was no significant difference among mild, moderate or severe group of patients by their epidemiology history. However, three groups of patients indeed presented significantly different illness onset on the most common symptoms like fever (one-way ANOVA, p = 0.0089), fatigue (one-way ANOVA, p = 0.0001) and diarrhea (one-way ANOVA, p = 0.0486), but not pharyngalgia or dry cough (Table 1). Besides, the average length of hospital stay and interferon treatment duration of moderate group were 14.12 (13.34–14.90) days and 14.24 (13.45–15.03) days, respectively, while those of the severe group took average 2.08 days and 1.44 days longer than total average levels.
Table 1.
Baseline characteristics of 94 patients infected with COVID-19
| Total, N = 94 | Mild group, N = 8 | Moderate group, N = 75 | Severe group, N = 11 | 1-way ANOVA | |||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Percentage | Number | Percentage | Number | Percentage | Number | Percentage | P value | |
| Age, years | |||||||||
| Median (range) | 40 (1–78) | 19 (7–39) | 40 (1–78) | 63 (32–69) | 0.0001 | ||||
| ≤ 20 | 12 | 12.77 | 4 | 50.00 | 7 | 9.33 | 0 | ||
| 21–40 | 36 | 38.30 | 4 | 50.00 | 32 | 42.67 | 1 | 9.09 | |
| 41–6 0 | 28 | 29.79 | 0 | 25 | 33.33 | 3 | 27.27 | ||
| ≥ 61 | 18 | 19.15 | 0 | 11 | 14.67 | 7 | 63.64 | ||
| Sex | |||||||||
| Male | 42 | 45.00 | 3 | 38.00 | 34 | 45.00 | 5 | 46.00 | |
| Female | 52 | 55.00 | 5 | 62.00 | 41 | 55.00 | 6 | 54.00 | |
| Comorbidities | |||||||||
| HBP | 9 | 9.60 | 0 | 6 | 8.00 | 3 | 27.27 | ||
| CHD | 6 | 6.40 | 0 | 2 | 2.67 | 4 | 36.36 | ||
| Diabetes | 5 | 5.30 | 0 | 3 | 4.00 | 2 | 18.18 | ||
| Cardiovascular diseases events (CHD/T2DM/HBP) | |||||||||
| None | 79 | 84.04 | 8 | 100.00 | 65 | 87.00 | 6 | 55.00 | 0.0001 |
| One type | 10 | 10.64 | 0 | 9 | 12.00 | 1 | 9.00 | ||
| Combined | 5 | 5.32 | 0 | 1 | 1.00 | 4 | 36.00 | ||
| Epidemiology history | |||||||||
| Resided in Wuhan | 79 | 84.04 | 7 | 87.50 | 64 | 85.33 | 8 | 72.73 | |
| Contacted with Wuhan resident | 13 | 13.83 | 1 | 12.50 | 10 | 13.33 | 2 | 18.18 | |
| Not clear | 2 | 2.13 | 0 | 1 | 1.33 | 1 | 9.09 | ||
| Onset of signs and symptoms | |||||||||
| Fever | 69 | 73.40 | 3 | 37.50 | 55 | 73.33 | 11 | 100.00 | 0.0089 |
| Fatigue | 28 | 29.79 | 1 | 12.50 | 18 | 24.00 | 9 | 81.82 | 0.0001 |
| Dry cough | 46 | 48.94 | 2 | 25.00 | 36 | 48.00 | 8 | 72.73 | 0.1157 |
| Diarrhea | 8 | 8.51 | 0 | 5 | 6.67 | 3 | 27.27 | 0.0486 | |
| Pharyngalgia | 18 | 19.15 | 2 | 25.00 | 14 | 18.67 | 2 | 18.18 | 0.9099 |
| Hospital admission | |||||||||
| Length of hospital stay [mean (day); (95% CI)] | 14.28 | (13.61–14.95) | 12.88 | (11.36–14.39) | 14.12 | (13.34–14.90) | 16.36 | (14.79–17.93) | 0.045 |
| Interferon treatment duration [mean (day); (95% CI)] | 14.11 | (13.39–14.82) | 10.88 | (8.00–13.75) | 14.24 | (13.45–15.03) | 15.55 | (13.84–17.25) | 0.01 |
Data are given as counts with percentage, mean with 95% CI, or median with range. Determined using one-way analysis of variance
CHD coronary heart disease, T2DM type 2 diabetes, HBP high blood pressure
Differences on laboratory parameters
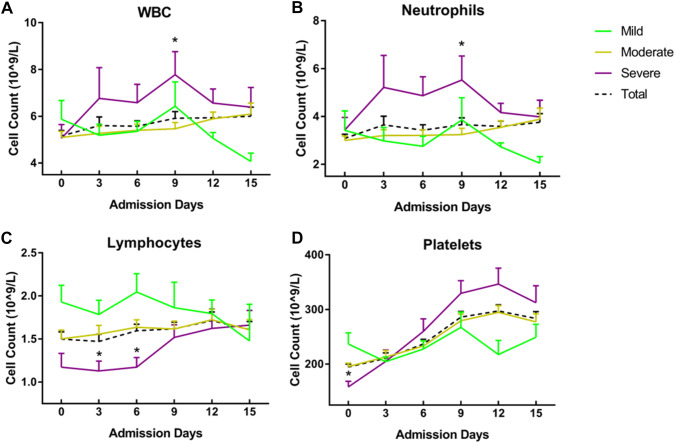
Figure 1 showed the outcomes of blood cell counting. On admission, only the amount of platelets (Fig. 1d) exhibited obvious differences among three groups (p = 0.006, one way ANOVA; p < 0.05, severe group vs moderate group). For the following 6 days, there was no significant difference on numbers of immune cell types until Day 9, when the increased white blood cell (WBC) and neutrophil cell numbers were significantly higher in severe group comparing with moderate group (Fig. 1a, b). Shown in Fig. 1c with quite distinguishable difference on cell accounts among three group, the lymphocyte number in severe group remained lower within the first week of hospitalization (p < 0.05, vs moderate group on day 3 and 6, respectively). From then on, the lymphocyte number increased in severe group and turned to as similar level as other patients.
Fig. 1.
Changes in the levels of the immune cell types and platelets. Timeline charts illustrate the accounts of immune cell numbers in different admission day. Error bars are SEM. The solid lines in green, yellow and purple represent the mild, moderate and severe group of patients respectively. And the dotted line in black shows the average levels of all patients in each day. One asterisk stands for p < 0.05 for severe group vs moderate group; two stand for p < 0.01, while three stand for p < 0.001 (Color figure online)
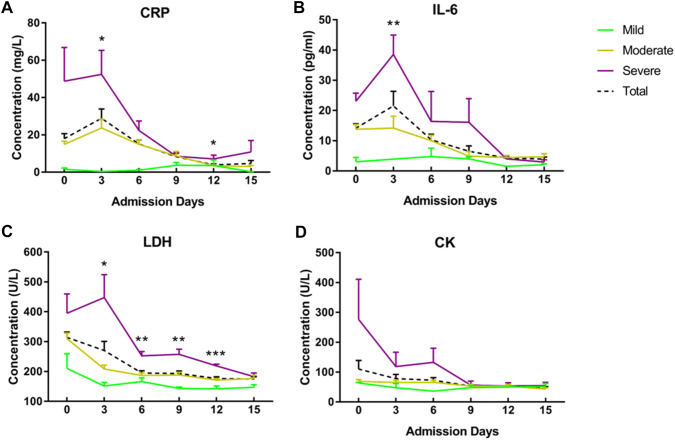
Figure 2 showed the blood biochemical test at different time points. Among the first 6–9 days of hospitalization, patients in severe group showed significantly higher levels of serum interleukin 6 (IL-6) and LDH than those in moderate group (Fig. 2b, c). Except for mild group, there was a fluctuation around day 3 on hospital in serum C-reactive protein (CRP) and IL-6 levels, which then dramatically dropped down around Day 6 to 9 in both moderate and severe group of patients (Fig. 2a, b). Besides, the CK concentration declined rapidly in severe group (Fig. 2d). In contrast, the changes of serum LDH concentration slipped smoother during hospital stay and the differences remained significance among three groups until day 12, approaching similar level after that (Fig. 2c).
Fig. 2.
Dynamic Profile of blood biochemical factors. Timeline charts illustrate the blood biochemical factors in different admission day. Error bars are SEM. The solid lines in green, yellow and purple represent the mild, moderate and severe group of patients respectively. And the dotted line in black shows the average levels of all patients in each day. One asterisk stands for p < 0.05 for severe group vs moderate group; two stand for p < 0.01, while three stand for p < 0.001 (Color figure online)
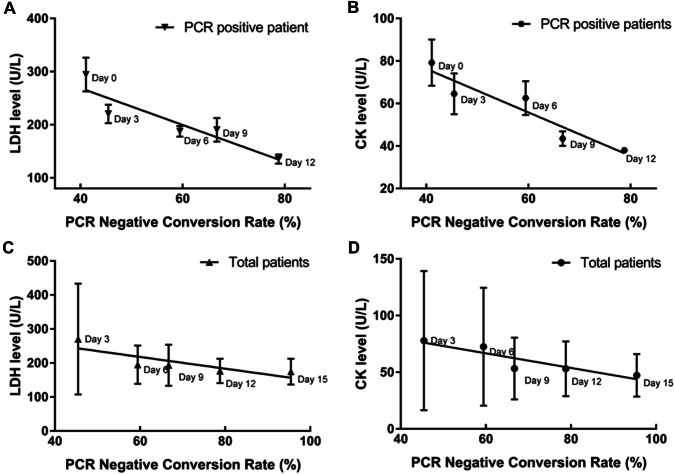
Correlations between PCR negative conversion rate and serum LDH or CK level
PCR negative conversion rate was calculated by the rate of PCR negative to all undischarged patients by PCR detection of viral mRNA. The results indicated that the PCR negative conversion rate was over 50% since Day 6 (59.46%) and kept increasing up to 95.45% on Day 15 (Table 2). Linear regression analysis was used to study the association between serum LDH or CK concentrations and PCR negative conversion rate. Figure 3a, b showed that there existed a significant inverse correlation between PCR negative conversion rate and average CK level (r2 = 0.822, p = 0.0337) instead of LDH level (r2 = 0.696, p = 0.079). Further revised average LDH and CK concentrations in PCR positive patients were all inversely correlated with the negative conversion rate (r2 = 0.843, p = 0.0279 and r2 = 0.890, p = 0.0161) (Fig. 3c, d).
Table 2.
PCR negative conversion rate of inpatients
| Admission day | Day 0 | Day 3 | Day 6 | Day 9 | Day 12 | Day 15 |
|---|---|---|---|---|---|---|
| PCR negative conversion rate | 41.07% | 45.45% | 59.46% | 66.70% | 78.79% | 95.45% |
PCR negative conversion rate is calculated by the ratios of PCR negative patients to total detected patients in each day by virus mRNA RT-PCR
Fig. 3.
Regression analysis for serum LDH and CK levels with PCR negative conversion rate. Regression analysis for serum LDH or CK level and virus mRNA elimination rate. a, b The correlation of PCR negative conversion rate and average CK level (r2 = 0.822, p = 0.0337) and LDH level (r2 = 0.696, p = 0.079) in total patients in each day, respectively. c, d Both significantly inversed correlation between the negative conversion rate and average CK level (r2 = 0.843, p = 0.0279) and LDH level (r2 = 0.890, p = 0.0161) of PCR positive patients in each day respectively
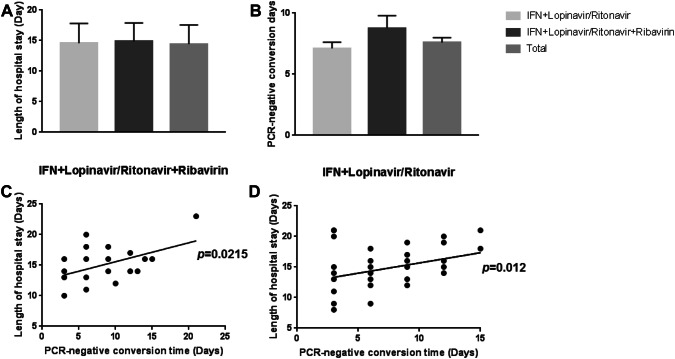
Correlation between lengths of hospital stay and PCR negative conversion times in different antiviral treatments
There were 46 patients treated with IFN-α + lopinavir/ritonavir. Another 21 patients were treated with IFN-α + lopinavir/ritonavir combined with ribavirin. As shown in Fig. 4a, b, there was no significant difference on average lengths of hospital stay or PCR negative conversion times among different antivirus treatment groups. On the other hand, correlation analysis (Fig. 4c, d) indicated that the duration of hospital stay was significantly correlated with PCR negative conversion times in IFN-α + lopinavir/ritonavir + ribavirin group (p = 0.0215), as well as IFN-α + lopinavir/ritonavir group (p = 0.012).
Fig. 4.
Correlation between length of hospital stay and PCR negative conversion time in different antivirus treatment groups. a, b The average lengths of hospital stay and PCR negative conversion times in IFN + lopinavir/ritonavir combined with ribavirin treatment group and IFN + lopinavir/ritonavir treatment group. Error bars are SEM. c, d The correlation of PCR negative conversion time and length of hospital stay (days) in IFN + lopinavir/ritonavir combined with ribavirin treatment group (p = 0.0215) and IFN + lopinavir/ritonavir treatment group (p = 0.012)
Discussion
In this study, we characterized the changes of several immune cell types and serum biochemical factors during hospitalization in 94 discharged patients with COVID-19 infection. We found that the decline of LDH and CK was correlated with viral mRNA elimination, especially the serum LDH and CK level in virus mRNA positive patients, suggesting that constitutive decrease of LDH or CK levels probably predict a favorable response to course of COVID-19 infected patients. This phenomenon promoted us to analyze the correlation between COVID-19 clearance ratio and each of these parameters.
On admission, patients were generally showing symptoms of fever, dry cough, fatigue and sore throat. Most of the patients had mild to moderate symptoms. Patients with one or two underlying chronic diseases tended to develop severe symptoms. Notably, life-threatening complications such as acute respiratory distress syndrome, sepsis or multiple organ failure were not observed in this cohort. Besides, consistent with previous reports [9, 10], our results showed that 36 patients from 14 different families were clustered, suggesting that person-to-person transmission was efficient in case of COVID-19.
So far, no antiviral therapy has been proved to be effective for the treatment of patients with COVID-19 infection. A couple of therapeutic interventions for coronavirus were investigated during the outbreak of SARS-CoV and MERS-CoV [11, 12], and combination of ribavirin and interferon has been proved beneficial in patients with SARS-CoV or MERS-CoV infection [13, 14]. In addition, the potent efficacy of remdesivir [15], lopinavir and ritonavir [16], as well as intravenous immunoglobulin [17, 18] have been documented for MERS-CoV treatment. As previously reported, early administration of interferon protected mice from lethal MERS-CoV infection, while late administration of exogenous interferon promoted the proinflammatory cytokine response and inhibited the optimal virus-specific T cell response [19, 20]. In this cohort, 89% of the patients were hospitalized in 7 days after onset and 71% of them were treated with interferon and lopinavir/ritonavir, or combined with ribavirin. As showed in our study, COVID-19 mRNA conversion time was correlated with the hospital stay length in both IFN-α + lopinavir/ritonavir and IFN-α + lopinavir/ritonavir + ribavirin treatment group, indicating that these two therapeutic regimens might be beneficial for treatment of COVID-19 infected patients.
This study has several limitations. First, only 94 discharged patients were included. Collection of data from a lager cohort would be better to get a more comprehensive understanding on discharged patients. Second, we only performed qualitative analysis on viral mRNA. Further quantitative analysis on COVID-19 could be more precise on evaluations of the efficiency of different therapeutic regimen. However, we will keep moving on this project in future as well.
Acknowledgements
We acknowledge the following staff members of Shenzhen Third People's Hospital, Shenzhen, China, Zhiyi Ke, Dongli Lian, Wenting Zhang for their contribution to the diagnosis.
Author contributions
JY: conception and design, data collection. RZ and LZ: data collection and analysis. SK: data analysis and review. JL, XL, YL, XD, GT and ST: data collection. LL and YL: intellectual input and critical review. YP: data analysis and drafting. ZW: data collection, analysis and critical review.
Funding
This work was supported by Sanming Project of Medicine in Shenzhen (SZSM201512005).
Compliance with ethical standards
Conflict of interest
The findings and conclusions are those of the authors and do not necessarily represent the views of the Shenzhen Third People's Hospital. All authors declare that they have no conflict of interest exists.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jing Yua, Rougrong Zou, Lijiao Zeng made equal contribution to this work.
Contributor Information
Yanchao Pan, Email: panychao@126.com.
Zhaoqin Wang, Email: zhaoqinw108@126.com.
References
- 1.Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang S, Xia S, Ying T, Lu L. A novel coronavirus (2019-nCoV) causing pneumonia-associated respiratory syndrome. Cell Mol Immunol. 2020. [DOI] [PMC free article] [PubMed]
- 4.Phan LT, Nguyen TV, Luong QC, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed]
- 5.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020. [DOI] [PMC free article] [PubMed]
- 6.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu XW, Wu XX, Jiang XG, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020. [DOI] [PMC free article] [PubMed]
- 11.Omrani AS, Saad MM, Baig K, et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis. 2014;14:1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khalid M, Al Rabiah F, Khan B, Al Mobeireek A, Butt TS, Al ME. ribavirin and interferon-alpha2b as primary and preventive treatment for Middle East respiratory syndrome coronavirus: a preliminary report of two cases. Antivir Ther. 2015;20:87–91. doi: 10.3851/IMP2792. [DOI] [PubMed] [Google Scholar]
- 14.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9. [DOI] [PMC free article] [PubMed]
- 16.Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arabi YM, Arifi AA, Balkhy HH, et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med. 2014;160:389–397. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 18.Kapoor M, Pringle K, Kumar A, et al. Clinical and laboratory findings of the first imported case of Middle East respiratory syndrome coronavirus to the United States. Clin Infect Dis. 2014;59:1511–1518. doi: 10.1093/cid/ciu635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Channappanavar R, Fehr AR, Zheng J, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Investig. 2019;130:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyall J, Gross R, Kindrachuk J, et al. Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs. 2017;77:1935–1966. doi: 10.1007/s40265-017-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]