Abstract
The attenuated avian infectious bronchitis virus (IBV), derived from a wild strain (TW2575/98w) in chicken embryos after 75 passages, is designed as a commercial vaccine strain (TW2575/98vac) to control the disease in Taiwan. The differences in viral infectivity, replication efficiency, and genome sequences between TW2575/98w and TW2575/98vac were determined and compared. TW2575/98vac caused earlier death of chicken embryos and had higher viral replication efficiency. Thirty amino acid substitutions resulting from 44 mutated nucleotides in the viral genome were found in TW2575/98vac. All of the molecular variations lead to attenuation, found in TW2575/98, were not observed consistently in the other IBVs (TW2296/95, Ark/Ark-DPI/81, the Massachusetts strain, GA98/CWL0470/98, and CK/CH/LDL/97I) and vice versa. After further comparisons and evaluations from three aspects: (1) longitudinal analysis on the timing of variations appeared in specific homologous strain passages, (2) horizontal evaluations with the amino acid changes between wild and vaccine strains among the other 5 IBVs, and (3) inspection on alterations in the chemical characteristics of substituted amino acid residues in viral proteins, four amino acid substitutions [V342D in p87, S1493P and P2025S in HD1, as well as F2308Y in HD1(P41)] were selected as highly possible candidates for successful TW2575/98w attenuation. Our findings imply that molecular variations, which contribute to the successful attenuation of different IBVs, are diverse and not restricted to a fixed pattern or specific amino acid substitutions in viral proteins. In addition, four amino acid changes within the replicase gene-encoded proteins might be associated with TW2575/98 virus virulence.
Electronic supplementary material
The online version of this article (10.1007/s11262-020-01753-5) contains supplementary material, which is available to authorized users.
Keywords: Virulence, Infectivity, Vaccine, Genome, Substitutions
Introduction
Avian infectious bronchitis virus (IBV), which belongs to gammacoronavirus, is worldwide in distribution and causes a highly contagious disease in chickens. IBV has a single-stranded, positive-sense RNA genome of 27.6 kb in size. Two polyproteins 1a and 1ab account for approximately two-thirds of the viral genome-coding region and make up the replication transcription complex (RTC). The polyprotein 1ab is translated through the -1 frame-shift translation mechanism [1]. The 1ab protein is further cleaved into 15 nonstructural proteins (NSPs), which may account for virus pathogenicity [2]. The IBV genome encodes four major structural proteins: the spike glycoprotein (S), the small envelope protein (E), the membrane glycoprotein (M), and the nucleocapsid protein (N) [3].
Several IBV strains have been attenuated as vaccines for controlling the disease. The sequence changes in viral genome associated with attenuation were studied in some wild strains, such as the pathogenic Arkansas-Delmarva Poultry Industry Ark/Ark-DPI/81 (ArkDPI) [4, 5], the Massachusetts strain (Mass), and the pathogenic Georgia 98 virus GA98/CWL0470/98 (GA98) in the US [5] as well as the CK/CH/LDL/97I strain [6], the YX10 strain [7], and SDZB0808 strain [8] in China. Comparison of the genome sequences of virulent and attenuated avian IBV strain Ark DPI viruses [4] revealed that most of these substitutions were located in the replicase 1a and spike genes. Analyses on the consensus full-length genome sequences of three different IBV serotypes (Ark, GA98, and Mass41) suggesting the potential role of NSP3 in the attenuation procedure [5]. Sequence variations in different genes, especially the S gene and NSP3, in the genomes of CK/CH/LDL/97I viruses might contribute to differences in viral replication, pathogenicity, antigenicity, immunogenicity, and tissue tropism [6]. In the live-attenuated vaccine candidate strain (YX10p90), 26 amino acid substitutions in 6 proteins and a 110-bp deletion in the 3′ untranslated region were identified [7]. The study on virulent QX genotype IBV strain (SDZB0808) attenuation mechanism found that the replicase 1a sequence was truncated by 30 bp in the same gene region in the passage 60–110 viruses [8].
Several studies used the avirulent IBV Beaudette strain as a backbone to generate the recombinant IBVs with genes from a virulent M41 strain [2, 9–11] or a QX-like nephropathogenic isolate [12] for investigating the sequences related to the attenuation. Gene 1 (replicase gene) and the accessory protein genes (genes 3 and 5) could be targets for rational attenuation for vaccine development. One or more of the gene 1 (replicase gene)-encoded proteins (NSP2-NSP16) are determinants of pathogenicity [10]. Very few amino acid substitutions within the replicase gene can result in attenuation following serial passage in embryonated eggs [2]. Four accessory proteins (3a, 3b, 5a, and 5b), whose functions may include antagonism of innate immune responses, encoded by genes 3 and 5, are not essential for replication [10]. Spike (S) protein is a determinant of cell tropism and could induce protective immunity but not alter the viral pathogenicity [2, 9, 12]. None of the structural or accessory genes derived from an IBV virulent isolate were able to restore virulence and the loss of virulence associated with the Beaudette strain resides in the replicase gene [2]. However, another study using the reverse genetics system based on virulent QX-like YN IBV strain in China [13] indicated that the S gene and 5a accessory gene are responsible for the attenuation. The S gene mutations and the 5a accessory gene of the attenuated IBV strain (aYN) were both accountable for the decreased pathogenicity of virulent IBV, which was not due to a replication deficiency in the recombinant viruses.
In addition, sequence changes in several other regions of viral genome might be related to the attenuation of IBVs. Some studies [7, 14–16] found that a fragment of different lengths (49, 58, 109, or 110 nucleotides) in 3′-UTR downstream of the N protein gene was deleted in the attenuated strain of several IBVs. Its correlation to the attenuation has been proposed. A study [17] indicated that the avirulent strain H120 developed a shorter poly(A) tail, especially in the early infection, than the virulent strain TW1. It may also be possible to employ viral poly(A) tail length as an indicator of virulence.
The mutations associated with pathogenicity attenuation in chickens could therefore be summarized in several genes (replicase gene seems to be the main target) and are variable leading to the differing efficacies associated with different vaccines. However, the results from current studies were still unable to confirm which amino acid changes were responsible for loss of pathogenicity in different IBVs.
Taiwan IBVs can be divided into two groups, Taiwan Group I (TW-I) and Taiwan Group II (TW-II) [18, 19]. One of the IBVs in each group, TW2575/98 strain for the TW-I group and TW2296/95 for the TW-II group, had been attenuated as vaccines in Taiwan. The TW2575/98 vaccine strain is more pathogenic in chicken embryos than the wild strain [20]. Sequence changes in the 3′ 7.3 kb of TW2575/98 and TW2296/95 viruses after attenuating passage in embryonated eggs were identified [15].
We compared the sequence differences between the wild TW2575/98 virus (TW2575/98w) and the attenuated vaccine strain (TW2575/98vac). Molecular variations in both strains were identified. The differences between them in terms of viral infectivity and replication efficiency in chicken embryos were also determined. Additionally, in comparison with other attenuated IBV strains, the mutations related to successful attenuation on TW2575/98 are proposed.
Material and methods
Viruses
Two strains, TW2575/98 (TW-I) and TW2296/95 (TW-II), were passaged in specific-pathogen-free (SPF) chicken embryos after 74 and 76 times to obtain attenuated vaccine strains, respectively [19]. The fourth passage of TW2575/98 in SPF chicken embryos (Animal Health Research Institute, Chidin, Taiwan) was used as the wild strain (TW2575/98w) and the 77th passage as the vaccine strain (TW2575/98vac) in this study. The third TW2296/95 passage was used as the wild strain (TW2296/95w) and the 79th passage as the vaccine strain (TW2296/95vac) for genome sequencing in this study.
Comparison of the differences in the ability to infect chicken embryos between wild and vaccine strains
One 50% embryo infectious dose (EID50)/0.1 ml of each virus (TW2575/98w and TW2575/98vac) was inoculated into 26 chicken embryos, respectively, in experiment 1. In experiment 2, three concentrations (102.7 EID50/0.1 ml, 103.7 EID50/0.1 ml, and 104.7 EID50/0.1 ml) of two viruses (TW2575/98w and TW2575/98vac) were inoculated into the allantoic cavities of three 9- to 11-day-old SPF chicken embryos, respectively. The inoculated chicken embryo eggs were candled twice a day for 7 days in two experiments. All inoculated embryos were sacrificed on 7th day post-inoculation (DPI) if not dead. The collected embryo eggs were stored at 4 °C for overnight and their allantoic fluids were harvested for IBV gene quantification using real-time reverse transcription (RT)-polymerase chain reaction (PCR). Student's t test was used to determine the differences between wild and vaccine strains at the α error of 0.05. In Fig. 1, the regression lines revealed from wild and vaccine strain data were drawn using excel and the slopes (regression coefficients) for virus replication vs DPI from wild and attenuated strains were compared using Student’s t test [21].
Fig. 1.
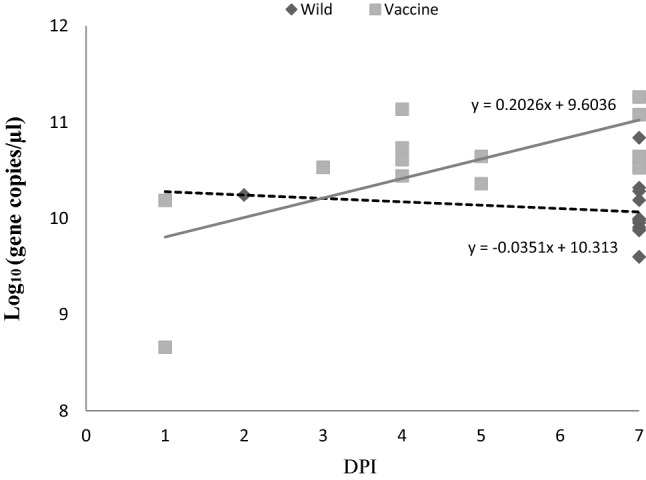
Viral genome copies detected in allantoic fluid and number of days to embryonic death with a low concentration (1 EID50) of wild and vaccine TW2575/98 strains. DPI means day post-inoculation. The regression lines were drawn using excel and the virus replication slopes (regression coefficients) vs DPI from wild and attenuated strains were significantly different (p < 0.05) using Student’s t test [21]. Solid trend line stands for vaccine strain and dashed trend line for wild strain
Quantitation of viral genomes using real-time RT-PCR
Viral RNA in allantoic fluid harvested from inoculated embryonated eggs was extracted using TRIzol Reagent (Life Technologies, Frederick, Maryland, USA) according to earlier studies [22, 23]. The virus quantities harvested from allantoic fluid in embryos were determined basing on the hypervariable regions 1 (HVR1) of S1 gene of viral RNA amplified by using real-time RT-PCR with specific primers. The primer sequences were rC2U: 5′TGGTT GGCA(T/C) TTACA (A/C/T)GG(A/G/T)3′, rC3L: 5′(A/G)CAAT GTGTA ACAAA (T/C)ACT3′ [18, 23]. At first, viral RNA samples were reverse-transcribed, according to the supplier’s standard protocol, for 120 min at 37 °C with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, California, USA). Quantitative PCR was performed under the following conditions: 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 1 min at 60 °C using 2X Power SYBR Green PCR Master Mix (Applied Biosystems), and 200 nM of forward and reverse primers. Each assay was run on an Applied Biosystems 7300 Real-Time PCR system in triplicate. The exact copy numbers of the target gene were determined by absolute quantification relating the cutoff threshold (CT) value to a standard curve. The standard curve was constructed using measurement in triplicate on tenfold serial dilutions of the cloned plasmid prepared from a yT&A vector (Yeastern Biotech, Taipei, Taiwan) harbored RT-PCR product amplified using the primers rC2U and rC3L, ranging from 1 × 105 to 1 × 109 copies/µl.
Gene sequencing and comparison
The full-length sequence of genomes from TW2757/98vac, TW2296/95w, and TW2296/95vac was determined in this study. The primers for direct sequencing are shown in the supplementary information (sTable 1). A total volume of 50 µl of reaction mixture was prepared by adding 5 µl of 10 × DNA polymerase buffer, 0.5 µl of 5U/µl Taq DNA polymerase (Promega, Madison, Wisconsin, USA), 16 µl of 1.25 mM dNTPs (Promega), 0.5 µl of 50 pmol/µl upstream primer, 0.5 µl of 50 pmol/µl downstream primer, 0.4 µl of 40 U/µl Recombinant RNasin ribonuclease inhibitor (Promega), 0.1 µl of 10 U/µl of Moloney Murine Leukemia Virus reverse transcriptase (Invitrogen, Carlsbad, California, USA), 20 µl of viral RNA solution, and 7 µl of diethyl pyrocarbonate-treated water. RT-PCR was performed in one step and conducted in the GeneAmp PCR System 2700 (Applied Biosystems). Reverse transcription was performed at 40 °C for 30 min. PCR was then performed for 35 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 1 min and polymerization at 72 °C for 100 s [15]. The initial denaturation step was conducted at 94 °C for 3 min, and the final polymerization step was at 72 °C for 10 min. The amplified RT-PCR products were analyzed on a 1.5% agarose gel (electrophoresis grade; Gibco BRL, Life Technologies, Grand Island, New York, USA) and stained with ethidium bromide. Nucleotide sequences of the resultant RT-PCR products were determined by commercial service with an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (V3.1) in an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA).
To determine the terminal genome sequences from TW2757/98w, TW2757/98vac, TW2296/95w, and TW2296/95vac, two primers, IB5′F (5′-ACT GAA AAT AGA TAT TAA TAT AT) and IB5′R (5′-CCG ACC ACG GCG CC), were used to amplify a 268 bp fragment containing the 5′ end of IBV. The amplified PCR products were purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany) and cloned into the pCR4-Topo vector. The clone was sequenced from both directions with primers T3 (5′-GCA ATT AAC CCT CAC TAA AGG) and T7 (5′-TAA TAC GAC TCA CTA TAG GG). All sequencing works were performed using the Big Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) in an automatic sequencer. Rapid amplification of cDNA ends (RACE) was used as recommended by the RACE kit manufacturer (Invitrogen). For this, the first-strand cDNA was made with oligonucleotide IBV101 (+) (2 µM), and the product was C-tailed and then PCR amplified with oligonucleotides RACE-C (−) (10 µM) and IBV88 (+) (10 µM) for determining 5′ terminal genome sequences, the bridge anchor primer supplied with the kit. The products were cloned into the TOPO XL vector (Invitrogen) and the resulting plasmids were sequenced by automated sequencing. To determine the 3′-terminal genome sequences, cDNA was made using oligonucleotide DI3′( +), which binds to the 3′-poly(A) tail and provides BamHI, SalI, AccI, HincI, and MluI restriction endonuclease cloning sites, amplified by PCR with oligonucleotides DI3′ (+) and IBV3UTR-2 (−), and the PCR products were cloned into TOPO XL cloning vector (Invitrogen). The resulting plasmids were sequenced by automated DNA sequencer.
The RACE primer sequences used for 5′ terminal genome sequences were the following. RACE-C (−): 5′-GGCCACGCGTCGACTAGTACCCCCCCCCCC; IBV88 (+): 5′-gCAATGGACAACCTAGGGGACC; IBV101 (+): 5′-GCACTAAAGTGCACTGCAATGGAAC. There was a poly(A) tail at the end of the IBV genome. The RACE primers used for 3′ terminal genome sequences were DI3′ (+): CGGGGATCCGTCGACACGCGTTTTTTTTTTTTTTTTTTTT and IBV3UTR-2 (−): GCCAGTGCCGGGGCCACGCGG.
Gene sequences were compiled and edited using programs in the LaserGene biocomputing software package (DNASTAR, Madison, Wisconsin, USA). Amino acid sequences were deduced and further analyzed by an open bioinformatics database and analysis resource for virology research, The Virus Pathogen Database and Analysis Resource (ViPR, www.ViPRbrc.org) [24]. The sequence differences between the wild TW2575/98 virus (TW2575/98w) and the attenuated vaccine strain (TW2575/98vac) were compared. The switch of chemical characteristics (polarity, charge, and hydrophobicity) of 30 amino acid residues due to substitution found in both strains was also checked.
For further analyses on the timing of genetic changes during attenuation of IBVs, several variations of amino acid sequence deduced from viral genomes of 4 previous passages (19th, 35th, 53rd, and 69th) of TW2575/98 were determined and compared. The 3′-UTR downstream sequence of the N protein gene in 4 other specific passages of TW2296/95 (22nd, 34th, 46th, and 56th) was determined and compared.
Sequence accession number
GenBank accession numbers for genome sequences of the reference IBVs were TW2575/98w (DQ646405), TW2575/98vac (MN128087), TW2296/95w (MN128088), TW2296/95vac (MN128086), ArkDPI (GQ504720), ArkDPI-attenuated (GQ504721), GA98 (GQ504722), GA98-attenuated (GQ504723), Mass (GQ504724), Mass-attenuated (GQ504725), CK/CH/LDL/971 (JX195177), and CK/CH/LDL/971-attenuated (JX195178).
Results
Replication efficiency and embryonic death revealed from TW2575/98w and TW2575/98vac
Although both wild and vaccine viruses caused the same 50% infection of chicken embryos, the viral gene copy number in the allantoic fluid infected with TW2575/98vac was higher than that infected with TW2575/98w under a low infection concentration (1 EID50) (p < 0.05) in Table 1, suggesting the TW2575/98vac replicated more efficiently than TW2575/98w. In addition, chicken embryos infected with TW2575/98vac died earlier (4.54 ± 2.11 DPI) than that infected with TW2575/98w (6.62 ± 1.39 DPI) (p < 0.05). For TW2575/98w, only one infected chicken embryo died on the second-day after inoculation and the other 12 were sacrificed or dead on day 7 post-inoculation (DPI 7). The viral gene copies in infected chicken embryos on DPI 7 are not increased significantly than those on DPI 2. For TW2575/98vac, 2, 1, 4, and 2 infected chicken embryos died on DPI 1, 3, 4 and 5, respectively, and 4 infected chicken embryos were sacrificed or dead on DPI 7.
Table 1.
Mean viral gene copies detected in allantoic fluid and mean days of embryonic death by infection with a low concentration (1 EID50) of wild and vaccine strains of TW2575/98
| Viruses | Viral gene-detected chicken embryos (n/26)a | Mean of the viral gene copy number (n × 109/µl) | Mean days of embryonic death after DPI |
|---|---|---|---|
| TW2575/98w | 13 | 15.90 ± 16.67b* | 6.62 ± 1.39* |
| TW2575/98vac | 13 | 57.99 ± 53.64* | 4.54 ± 2.11* |
DPI means day post-inoculation
*Significant difference between wild and vaccine strains (p < 0.05)
a26 chicken embryos were inoculated with each virus (TW2575/98w and TW2575/98vac) and incubated for 7 days
bMean ± SD
In Fig. 1, the virus replication lines formulae from wild and attenuated strains were y = 10.31–0.035x and y = 9.60 + 0.203x with slopes of − 0.035 and 0.203, respectively. The slopes were significantly different using Student’s t test (p < 0.05). The results indicated the viral gene copies increased with the increase in incubation time for virus replication in chicken embryos infected with TW2575/98vac but not TW2575/98w.
As shown in Table 2, the mean viral genome copies detected from allantoic fluid of chicken embryos inoculated with the three concentrations (102.7 EID50/0.1 ml, 103.7 EID50/0.1 ml, and 104.7 EID50/0.1 ml) of TW2575/98w were lower than those of TW2575/98 vac. Additionally, the time leading to the death of embryo is less for TW2575/98vac than for TW2575/98w (p < 0.05) at concentrations of 102.7 and 103.7 EID50/0.1 ml.
Table 2.
Viral gene copies detected in allantoic fluid and days of embryonic death by infection with different concentrations of wild and vaccine strains of TW2575/98
| Viruses | Virus titer (10n EID50/0.1 ml) | Mean of viral gene copies (n × 109)/µl among 7 days | Mean days of embryonic death after DPI |
|---|---|---|---|
| TW2575/98w | 4.7 | 14.0 ± 7.67a | 5.0 ± 1.73 |
| 3.7 | 12.6 ± 7.02* | 5.7 ± 1.15* | |
| 2.7 | 3.3 ± 4.18* | 6.0 ± 1.73* | |
| TW2575/98vac | 4.7 | 46.2 ± 46.53 | 3.3 ± 1.15 |
| 3.7 | 81.9 ± 51.14* | 4.0 ± 1.00* | |
| 2.7 | 30.3 ± 15.18* | 3.0 ± 1.00* |
DPI day post-inoculation
*Significant difference between wild and vaccine strains (p < 0.05)
aMean ± SD
Sequence analyses and comparisons
The TW2575/98w and TW2575/98vac genomes consist of 27,739 nucleotides (nts) and a poly(A) tail and contain 10 ORFs flanked by 5′ and 3′ untranslated regions (UTR). The genome organization is 5′-1a-(1ab)-S-3a-3b-E-M-5a-5b-N-3′. Six leader transcriptional regulatory sequences (TRS) in the genome of TW2575/98 are listed in Table 3. Five of them are the same as the consensus CT(T/G)AACAA(A/T) [25]. However, the TRS of the S gene in TW2575/98w and TW2575/98vac was AAGAACAAA, which was different from the consensus. The TW2296/95 has the same TRS (AAGAACAAA) in the S gene of TW2575/98.
Table 3.
The transcriptional regulatory sequences (TRS) of TW2575/98
| Gene | Sequence | Position (nt) |
|---|---|---|
| Leader | CCAACTTAACAAAACGGAC | 53–71 |
| S | AAGAAAGAACAAAAGACCG | 20,375–20,393 |
| Gene 3 | GTAACTGAACAATACAGAC | 23,901–23,919 |
| M | AAAACTTAACAATCCGGAG | 24,496–24,514 |
| Gene 5 | AACGCTTAACAAATACAGA | 25,601–25,619 |
| N | CTTTCTTAACAAAGCAGGA | 25,902–25,920 |
The nucleotides in TRS are in bold. The nucleotides of TRS in the S gene are different to the consensus CT(T/G)AACAA(A/T) and underlined
The detailed amino acid positions and cleavage sites of 1ab replicase of TW2575/98 compiled from wild and vaccine strains are the same as shown in Table 4. The replicase was cleaved at 14 sites by 2 enzymes into 15 nonstructural proteins, predicted by comparison with IBV strain BJ and Beaudette genome data (GenBank Sequence Accession: AY319651 and NC_001451) referred from the Virus Pathogen Resource (ViPR) [24]. The first two sites in the N-terminal of replicase are cleaved by papain-like protease (PLpro). The other 12 sites in the remainder are cleaved by coronavirus main proteinase (3CLpro). The length of poly(A) tails in the genomes of TW2575/98w and TW2575/98vac was the same (data not shown).
Table 4.
Different proteins cleaved from the 1ab protein of TW2575/98
| Proteinsa | Amino acid Positions (residue) | Cleavage siteb | Cleavage enzyme |
|---|---|---|---|
| Leader protein P87 | 1–674 | P87-G/G-HD1 | PLpro |
| HD1 (Nsp1) | 675–2802 | ||
| Ac domain | 675–833 | ||
| PL1pro | 834–1040 | ||
| X domain | 1041–1194 | ||
| PL2 pro | 1201–1491 | ||
| Y domain | 1823–2288 | Y domain -G/G- P41 | PLpro |
| P41 | 2289–2802 | ||
| 3CL(Nsp2) | 2803–3109 | P41-Q/A-3CL | 3CLpro |
| HD2(Nsp3) | 3110–3403 | 3CL-Q/S-HD2 | 3CLpro |
| Nsp4 | 3404–3486 | HD2-Q/S-Nsp4 | 3CLpro |
| Nsp5 | 3487–3696 | Nsp4-Q/S-Nsp5 | 3CLpro |
| Nsp6 | 3697–3807 | Nsp5-Q/N-Nsp6 | 3CLpro |
| GLF(Nsp7) | 3808–3952 | Nsp6-Q/S-GLF | 3CLpro |
| Nsp8 | 3953–3975 | GLF-Q/S-23aac | 3CLpro |
| RdRp(Nsp9) | 3953–4892 | GLF-Q/S-RdRp | 3CLpro |
| Helicase(Nsp10) | 4893–5492 | RdRp-Q/S-Helicase | 3CLpro |
| ExoN(Nsp11) | 5493–6013 | Helicase-Q/G-Nsp11 | 3CLpro |
| XendoU(Nsp12) | 6014–6351 | Nsp11-Q/S-Nsp12 | 3CLpro |
| 2′-O-MT(Nsp13) | 6352–6653 | Nsp12-Q/S-Nsp13 | 3CLpro |
In comparison with the genome sequence between TW2575/98w and TW2575/98vac, 44 nucleotides mutated was found and led to 30 changes of amino acid residues. Among the amino acid changes, 21 substitutions occurred in nonstructural proteins and the other 9 substitutions were in structural proteins. Two of 44 mutated nucleotides were located in untranslated regions (UTR): one was in 5′ UTR and the other was in 3′ UTR. The other 12 nucleotides resulted in silence mutation. The switch of chemical characteristics (polarity, charge, and hydrophobicity) of 30 amino acid residues due to substitution was also checked. Eleven amino acid substitutions in TW2575/98vac did not alter the chemical characteristics of residue (Table 5).
Table 5.
Differences between wild and vaccine strains of TW2575/98 at the nucleotide and deduced amino acid
| No. | Nucleotide Substitution from TW2575/98w to TW2575/98vac | Deduced amino acid substitution | Switch of chemical characteristics of amino acid residue due to substitution | Specific passage which amino acid substitution was found | Gene involved |
|---|---|---|---|---|---|
| 1 | T147C | None | 5′UTR | ||
| 2 | T684C | F53La | Np–Np | 77b | p87 |
| 3 | T737A | Silent | |||
| 4 | C976T | A150V | Np–Np | 35 | p87 |
| 5 | A1380G | I285V | Np–Np | 77 | p87 |
| 6 | C1388T | Silent | |||
| 7 | T1552A | V342D | Np → Ch | 77 | p87 |
| 8 | T1588C | L354P | Np–Np | 77 | p87 |
| 9 | T1616C | Silent | |||
| 10 | G3005A | Silent | |||
| 11 | T3013A | I829N | Np → Pu | 19 | HD1 (Ac domain) |
| 12 | G3021C | Silent | |||
| 13 | G3022C | G832P | Pu → Np | 19 | HD1 (Ac domain) |
| 14 | G3379A | G951E | Pu → Ch | 35 | HD1 (PL1pro) |
| 15 | G4365A | V1280I | Np–Np | 35 | HD1 (PL2pro) |
| 16 | T5004C | S1493P | Pu → Np | 77 | HD1 |
| 17 | A5210G | Silent | |||
| 18 | T5445C | Silent | |||
| 19 | C5605T | A1693V | Np–Np | 53 | HD1 |
| 20 | C6600T | P2025S | Np → Pu | 69 | HD1 |
| 21 | T7450A | F2308Y | Np → Pu | 69 | HD1 (P41) |
| 22 | T9018G | W2831G | Np → Pu | 53 | 3CLPro |
| 23 | A10137G | I3204V | Np–Np | 53 | HD2 |
| 24 | G10565T | K3346N | Ch → Pu | 53 | HD2 |
| 25 | G12025A | G3833D | Pu → Ch | 35 | GLF |
| 26 | C13645G | Silent | |||
| 27 | T13886G | F453V | Np–Np | 19 | RdRp |
| 28 | T13950C | L474P | Np–Np | 53 | RdRp |
| 29 | A14737T | Silent | |||
| 30 | T14971C | Silent | |||
| 31 | C16596T | P1356L | Np–Np | 19 | Helicase |
| 32 | C17332T | Silent | |||
| 33 | T18104C | Y1859H | Pu → Ch | 19 | ExoN |
| 34 | C20604A | P56T | Np → Pu | 35 | S1 |
| 35 | G20718T | A94S | Np → Pu | 35 | S1 |
| 36 | A22192G | E585G | Ch → Pu | 35 | S2 |
| 37 | G22223T | L595F | Np–Np | 69 | S2 |
| 38 | A22273T | N612I | Pu → Np | 35 | S2 |
| 39 | C24535T | Silent | |||
| 40 | A24595T | E4V | Ch → Np | 77 | M |
| 41 | C24616T | S11F | Pu → Np | 19 | M |
| 42 | C25128T | R182C | Ch → Pu | 35 | M |
| 43 | T25641G | F7C | Np → Pu | 35 | 5a |
| 44 | C27250T | None | 3′UTR |
Np nonpolar group, Ch charged group, Pu polar uncharged group
aAmino acid changes in the specific residue
bThe 77th passage of virus is vaccine strain
The timings of 30 amino acid substitutions during IBV attenuation are listed in Table 5. Six substitutions (I829N and G832P in HD1 (Ac domain), F453V in RdRp, P1356L in Helicase, Y1859H in ExoN, and S11F in M) were found in the 19th and the 4 later specific passages, which also means that the substitution occurred during the 5th to 19th passages. Ten substitutions (A150V in p87, G951E and V1280I in HD1 (PL1pro), G3833D in GLF, P56T and A94S in S1, E585G and N612I, R182C in M and F7C in 5a) were found in the 35th and the 3 later specific passages. Five substitutions (A1693V in HD1, W2831G in 3CLPro, I3204V and K3346N in HD2, and L474P in RdRp) were found in the 53rd and the 2 later specific passages. Three substitutions (P2025S in HD1, F2308Y in HD1 (P41), and L595F in S2) were found in the 69th and 77th specific passages. Six substitutions (F53L, I285V, V342D, and L354P in p87, S1493P in HD1, and E4V in M) were found in the 77th passage.
Amino acid changes at the 30 corresponding sites in proteins between wild and vaccine strains of TW2575/98 and the other 5 IB viruses (TW2296/95, ArkDPI, Mass, GA98, and CK/CH/LDL/971) were checked comparatively and listed in Table 6. Amino acid substitutions in the 30 corresponding sites between wild and vaccine TW2575/98 strains were not the same as those from the other 5 IBVs. Thirteen out of 30 TW2575/98vac substituted amino acid residues located in the AC domain, PL1pro, PL2 pro, HD1, 3CLpro, GLF, RdRp, nsp11, S1, and M, respectively, are the same as those at the corresponding sites in wild strains from at least one of the other 5 IBVs.
Table 6.
Comparison of amino acid changes in viral proteins between wild and vaccine strains from the 30 corresponding sites in TW2575/98 and the other 5 IB viruses
| Proteins | TW2575/98 | TW2296/95 | ArkDPI | Mass | GA98 | CK/CH/LDL/971 |
|---|---|---|---|---|---|---|
| P87 | F53La | (F)b | (F) | (F) | (F) | (F) |
| A150V | (A) | (A) | (A) | (A) | (A) | |
| I285V | (I) | (I) | (I) | (I) | (I) | |
| V342D | (V) | (V) | (V) | (V) | (V) | |
| L354P | (L) | (L) | (L) | (L) | (L) | |
| AC domain | I829Ne | (N) | (N) | (N) | (N) | (N) |
| G832Pe | (P) | (P) | (P) | (P) | (P) | |
| PL1pro | G951Ee | (E) | (A) | –c → Ed | (E) | (D) |
| PL2 pro | V1280Ie | (V) | (V) | (V) | (V) | (I) |
| HD1 | S1493P | (S) | (S) | (S) | (S) | (S) |
| A1693Ve | (F) | (V) | (V) | (V) | (V) | |
| P2025S | Y → Hd | (H) | (H) | (H) | (H) | |
| F2308Y | (F) | (F) | (F) | (F) | (F) | |
| 3CLpro | W2831Ge | (G) | (G) | (G) | (G) | (G) |
| HD2 | I3204V | (I) | (I) | (I) | (I) | (I) |
| K3346N | (K) | (K) | (K) | (K) | (K) | |
| GLF | G3833De | (D) | (D) | (D) | (D) | (D) |
| RdRp | F453Ve | (V) | (V) | (V) | (V) | (V) |
| L474P | (L) | (L) | (L) | (L) | (L) | |
| nsp10 | P1356L | (P) | (P) | (P) | (P) | (P) |
| nsp11 | Y1859He | (H) | (H) | (H) | (H) | (H) |
| S1 | P56Te | (L) | (N) | (S) | (S) | (T) |
| A94Se | (S) | (S) | (S) | (V) | (S) | |
| S2 | E585G | (E) | (E) | (E) | (D) | (E) |
| L595F | (L) | (L) | (L) | (L) | (L) | |
| N612I | (N) | (N) | (N) | (N) | (N) | |
| M | E4Ve | (V) | (M) | (E) | (E) | (S) |
| S11Fe | (T) | (L) | (F) | (F) | (T) | |
| R182C | (R) | (R) | (R) | (R) | (R) | |
| 5a | F7C | (F) | (F) | (F) | (F) | (L) |
aAmino acid changes in the specific site of residue
bAmino acid in parentheses stands for none of the alterations between wild and vaccine strains
cDash means none of the amino acids existed in the site
dMeans the substitution of amino acid from wild to vaccine strains
eIndicates the 13 sites were found that at least one of the other 5 IBVs’ wild strains has the same amino acid residue in TW2575/98vac
The 3′-UTR (nt 27,217–27,283, 67 bp) sequence comparison in 6 specific passages (3rd, 22nd, 34th, 46th, 56th, and 79th) of TW2296/95 are shown in Fig. 2. It was found that a fragment deletion (nt 27,233–27,271, 49 bp) was observed in 4 passages (34th, 46th, 56th, and 79th). However, such deletion was not found during the TW2575/98 passages.
Fig. 2.
Sequences of 3′-UTR downstream (the compared region is from nt 27,217 to nt 27,283, 67 bp according to TW2296/95w) of the N protein gene from different passages of strain TW2296/95 were compared. − stands for deletion. Changed nucleotides are underlined. Specific passages of the virus are noted with numbers at the end of the virus name (TW2296/95). w the 3rd passage, vac the 79th passage
Discussion
Contrary to avian influenza viruses and Newcastle disease viruses, the IBV vaccine strain is more pathogenic than the wild strain in chicken embryos although its pathogenicity is lower than the wild strain in chickens [26]. This phenomenon is explained as the vaccine strain replication efficiency in chicken embryos is better than that of the wild strain [20].
This study determined that TW2575/98vac has higher replication efficiency than TW2575/98w in chicken embryo inoculated with lower titer (1 EID50) of virus inoculation (Table 1 and Fig. 1). However, in the previous study for two other IBVs (Ark/Ark-DPI/81 and Mass/Mass41/41) [5], no statistical difference was found between the wild and attenuated viruses at any of the time points tested when a higher virus titer (105 EID50/egg) was inoculated into chicken embryos. To clarify the difference from our observations, we used 3 different virus concentrations to inoculate chicken embryos in another experiment (Table 2). At the higher virus inoculation concentration (104.7 EID50/0.1 ml), we had similar findings like ArkDPI and Mass with no statistical difference between TW2575/98vac and TW2575/98w. At two lower virus inoculation concentrations (103.7 and 102.7 EID50/0.1 ml), the results revealed a similar observation in Table 1 that TW2575/98vac is significantly higher than TW2575/98w (p < 0.05). Therefore, the difference in replication efficiency between wild and vaccine IBV strains in chicken embryos could be observed when the virus inoculation concentration was decreased to a lower level. In addition, earlier embryonic death revealed from TW2575/98vac (Tables 1 and 2) may be the result of the higher replication efficiency and therefore account for the higher pathogenicity in chicken embryos.
The question raised from our results on viral infectivity is why replication efficiency and pathogenicity of TW2575/98vac is higher than that for TW2575/98w in chicken embryos. Furthermore, what molecular variations on specific gene locus during attenuation might be responsible for the virulence alterations between them? It is speculated that the amino acid substitutions within the replicase of TW2575/98vac, when compared with TW2575/98w, may be the possible reason. We discuss this question in 3 aspects: (1) the timing for the appearance of individual amino acid substitutions in specific TW2575/98 passages, (2) the comparisons of genetic changes acquired from the other vaccines attenuated from wild IBVs at the 30 corresponding sites in the TW2575/98, and (3) the switch on the substituted amino acid residue chemical characteristics from possible candidates for contributing virulence.
In the first aspect, amino acid substitutions in viral proteins would appear and cumulate gradually during continuous IBV passages in chicken embryos. Those substitutions appeared and cumulated in later passages may strongly contribute to the loss of virulence and lead to successful attenuation. Two of the other attenuated IBVs, the ArkDPI vaccine [25] and QX-like nephropathogenic vaccine [7] have been developed after continuous passages in chicken embryos. The decline in virulence of the wild ArkDPI and QX-like viruses for chickens could be observed gradually and became significantly after the 51st and 58th passages, respectively. In our laboratory, 1-day-old birds given 1 × 107 EID50 of passage 65 of TW2575/98 via the intranasal route had 20% (2/10) mortality [23]. Although the passage 65 of the IBV stock is not fully attenuated, it indicates the later substitutions appear after the 65th passage may have direct associations with attenuation. The data from 30 amino acid substitutions found in specific TW2575/98w passages are listed in Table 5. Nine amino acid substitutions that appear in the 69th and 77th specific passages may be potential candidates contributing to successful TW2575/98w to TW2575/98vac attenuation. Of the 9 candidates (amino acid substitutions) are divided into 4 in p87 (F53L, I285V, V342D, and L354P), 2 in HD1 (S1493P and P2025S), 1 in HD1 (P41) (F2308Y), 1 in S2 (L595F), and 1 in M (E4V), respectively.
Furthermore, the significance of these 9 candidates was considered and evaluated under the second aspect concerning comparisons with available genetic information related to the attenuation of other wild IBVs. The amino acid changes in proteins between wild and vaccine strains from the 30 corresponding sites in TW2575/98 and the other 5 IB viruses (TW2296/95, ArkDPI, Mass, GA98, and CK/CH/LDL/971) were checked by the horizontal evaluations (Table 6). Those 13 amino acid substitutions in TW2575/98vac are the same as one of the wild strains at least from the other 5 IB viruses, which can explain that these amino acids have existed early in wild strains of the other 5 IB viruses. This kind of amino acid change may not be necessary for successful attenuation. Accordingly, one candidate (E4V) in M (one of 13 corresponding sites) could be excluded from the list of 9 candidates for contributing successful attenuation.
The remaining 8 candidates were evaluated further under the third aspect that the chemical characteristics (polarity, charge, and hydrophobicity) of amino acid residue resulting from substitution may change the function of viral proteins and then have impacts on the viral virulence. Among the 8 candidates, the alternations of chemical characteristics occur in the 4 candidates [V342D in p87, S1493P and P2025S in HD1, as well as F2308Y in HD1 (P41)] (Table 5). Based on the study of Cavanagh et al. and Armesto et al., gene 1 is the target of rational attenuation [2, 10]. Phillips et al. proposed further in 2012 that changes in nonstructural protein 3 (NSP3) are associated with attenuation [5]. Considering that the four amino acid substitutions (V342D, S1493P, P2025S, and F2308Y) are located in gene 1 and the latter 3 ones belong to the NSP3 in our study, these four candidates therefore may have great highly direct contributions for successful attenuation of the wild IB viruses (TW2575/98). However, the involvement of the rest four amino acids in the process of attenuation still cannot be excluded definitely.
In addition, although the replicase cleavage sites of 1ab and the TRSs in viral genome may play some of key roles in virus replication, the affection in the difference between wild and vaccine TW2575/98 strains cannot be considered because their sequences are the same as each other. Meanwhile, the TRS (AAGAACAAA) of the S gene in TW2575/98 was the same as the other Taiwanese. Therefore, this unique TRS may be used as a maker to differentiate Taiwanese IBVs from the IBVs in other countries including some of Taiwan-like IBVs in China [27].
Furthermore, a 30-bp deletion was observed in the replicase 1a sequence when the virulent IBV strain SDZB0808 was passaged to the 60th generation and this deletion was inherited in subsequent generations [8]. This kind of deletion was not found among the TW2575/98 passages during attenuation. The deletion (a fragment of different lengths) in 3′-UTR downstream of the N protein gene has been found in the attenuated strain of several IBVs, such as TW2296/95 (TW-II group) in Taiwan (a 49-bp deletion at the 76th passage) [15], QX-like strain CK/CH/LHLJ/04 V (a 109-bp deletion at the 110th passage) [28] and YX10p90 (a 110-bp deletion at the 90th passage) in China [7], as well as Vic S, I, and S strains (a 58-bp deletion) in Australia [14], respectively. Therefore, the deletion is proposed to be correlated to the attenuation. In this study, the deletion of a fragment (nt 27,233–27,271, 49 bp) has occurred early before the 34th passage of TW2296/95 during the attenuation processes (Fig. 2). However, this deletion is not observed in the viral genome of TW2575/98 during the passages. Therefore, it may not be required for the successful attenuation of the TW2575/98w (TW-I group). Besides, no difference in poly(A) tail lengths was observed between the wild and vaccine TW2575/98 strains. Hence, poly(A) tail length is not related to a change in TW2575/98 virulence.
Finally, although the methodology about the determinants related to successful attenuation was confirmed by reverse genetics and supported by direct evidence [10], further study for specific amino acid substitutions may be still required. Therefore, the methodology used in our study, which analyzes the appearance of amino acid substitutions in different homologous strain passages attenuated gradually from TW2575/98w to TW2575/98vac (longitudinal selection) and comparisons between amino acid substitutions observed after attenuation of TW2575/98 (TW-I group) and TW2296/95 (TW-II group) in Taiwan as well as the heterologous IBVs in several countries (horizontal comparison), may contribute to supplement the reverse genetics methodology for targeting virulence determinants. Therefore, further study to clarify the roles of the aforementioned four candidates (amino acid substitutions) involved in attenuation is potentially valuable in the future.
In summary, the change in viral virulence is associated with the virus proliferation effectiveness, because in the virus selection process, the viruses that have the early rapid growth ability are easily selected. Because of the ability to grow rapidly, vaccine strains often cause early embryonic death. The virus strain attenuation process selected by domesticating seems to be similar to that of unlocking the number-lock. Each number-lock can be set with a different code combination. That is why our findings in this study indicated that any one of all molecular variations found in TW2575/98 were not observed at the corresponding site of other IBVs and vice versa. Therefore, molecular variations contributed to the successful attenuation of different wild IBVs. IBVs are diverse and not restricted to a fixed pattern or specific amino acid alterations in viral proteins. Additionally, four amino acid changes within the replicase gene-encoded proteins might be associated with TW2575/98 virus virulence.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to greatly thank Yeou-Liang Lin and Ching-Ping Tsai for their helps in manuscript preparation. Financial support from the Ministry of Science and Technology and The Bureau of Animal and Plant Health Inspection and Quarantine, Taiwan is highly appreciated.
Author contributions
All authors (C-TT, H-YW, and C-HW) contributed to the study conception and design. Material preparation, data collection and analysis were performed by C-TT. The first draft of the manuscript was written by C-TT and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal participants
This article does not contain any studies with human participants performed by any of the authors. The use of chicken embryos in this study was approved by the Institutional Animal Care and Use Committee, National Taiwan University (Approval Number: 104-L-00053).
Informed consent
This article does not contain any information of human participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References:
- 1.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armesto M, Cavanagh D, Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS ONE. 2009;4(10):e7384. doi: 10.1371/journal.pone.0007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spaan W, Cavanagh D, Horzinek MC. Coronaviruses: structure and genome expression. J Gen Virol. 1988;69(Pt 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 4.Ammayappan A, Upadhyay C, Gelb J, Jr, Vakharia VN. Identification of sequence changes responsible for the attenuation of avian infectious bronchitis virus strain Arkansas DPI. Arch Virol. 2009;154(3):495–499. doi: 10.1007/s00705-009-0325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips JE, Jackwood MW, McKinley ET, Thor SW, Hilt DA, Acevedol ND, Williams SM, Kissinger JC, Paterson AH, Robertson JS, Lemke C. Changes in nonstructural protein 3 are associated with attenuation in avian coronavirus infectious bronchitis virus. Virus Genes. 2012;44(1):63–74. doi: 10.1007/s11262-011-0668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao F, Han Z, Zhang T, Shao Y, Kong X, Ma H, Liu S. Genomic characteristics and changes of avian infectious bronchitis virus strain CK/CH/LDL/97I after serial passages in chicken embryos. Intervirology. 2014;57(6):319–330. doi: 10.1159/000365193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng K, Xue Y, Wang J, Chen W, Chen F, Bi Y, Xie Q. Development and efficacy of a novel live-attenuated QX-like nephropathogenic infectious bronchitis virus vaccine in China. Vaccine. 2015;33(9):1113–1120. doi: 10.1016/j.vaccine.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huo YF, Huang QH, Lu M, Wu JQ, Lin SQ, Zhu F, Zhang XM, Huang YY, Yang SH, Xu CT. Attenuation mechanism of virulent infectious bronchitis virus strain with QX genotype by continuous passage in chicken embryos. Vaccine. 2016;34(1):83–89. doi: 10.1016/j.vaccine.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Hodgson T, Casais R, Dove B, Britton P, Cavanagh D. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J Virol. 2004;78(24):13804–13811. doi: 10.1128/jvi.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavanagh D, Casais R, Armesto M, Hodgson T, Izadkhasti S, Davies M, Lin F, Tarpey I, Britton P. Manipulation of the infectious bronchitis coronavirus genome for vaccine development and analysis of the accessory proteins. Vaccine. 2007;25(30):5558–5562. doi: 10.1016/j.vaccine.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keep S, Bickerton E, Armesto M, Britton P. The ADRP domain from a virulent strain of infectious bronchitis virus is not sufficient to confer a pathogenic phenotype to the attenuated Beaudette strain. J Gen Virol. 2018;99(8):1097–1102. doi: 10.1099/jgv.0.001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shan D, Fang S, Han Z, Ai H, Zhao W, Chen Y, Jiang L, Liu S. Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res. 2018;250:104–113. doi: 10.1016/j.virusres.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Cheng J, Yan S, Jia W, Zhang K, Zhang G. S gene and 5a accessory gene are responsible for the attenuation of virulent infectious bronchitis coronavirus. Virology. 2019;533:12–20. doi: 10.1016/j.virol.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mardani K, Browning GF, Ignjatovic J, Noormohammadi AH. Rapid differentiation of current infectious bronchitis virus vaccine strains and field isolates in Australia. Austral Vet J. 2006;84(1–2):59–62. doi: 10.1111/j.1751-0813.2006.tb13130.x. [DOI] [PubMed] [Google Scholar]
- 15.Huang YP, Wang CH. Sequence changes of infectious bronchitis virus isolates in the 3' 73 kb of the genome after attenuating passage in embryonated eggs. Avian Pathol. 2007;36(1):59–67. doi: 10.1080/03079450601110015. [DOI] [PubMed] [Google Scholar]
- 16.Casais R, Davies M, Cavanagh D, Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J Virol. 2005;79(13):8065–8078. doi: 10.1128/jvi.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shien JH, Su YD, Wu HY. Regulation of coronaviral poly(A) tail length during infection is not coronavirus species- or host cell-specific. Virus Genes. 2014;49(3):383–392. doi: 10.1007/s11262-014-1103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang CH, Tsai CT. Genetic grouping for the isolates of avian infectious bronchitis virus in Taiwan. Arch Virol. 1996;141(9):1677–1688. doi: 10.1007/bf01718291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang CH, Huang YC. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch Virol. 2000;145(2):291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai CT, Chiou HY, Wang CH. Infectious bronchitis vaccine is more pathogenic in chicken embryos than its wild strain. Vet Arhiv. 2016;86(5):699–709. [Google Scholar]
- 21.Andrade JM, Estevez-Perez MG. Statistical comparison of the slopes of two regression lines: a tutorial. Anal Chim Acta. 2014;838:1–12. doi: 10.1016/j.aca.2014.04.057. [DOI] [PubMed] [Google Scholar]
- 22.Huang YP, Chang WC, Wang CH. Evaluation of an attenuated TW I infectious bronchitis vaccine. Taiwan Vet J. 2005;31:148–154. [Google Scholar]
- 23.Huang YP, Wang CH. Development of attenuated vaccines from Taiwanese infectious bronchitis virus strains. Vaccine. 2006;24(6):785–791. doi: 10.1016/j.vaccine.2005.08.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, Zhou L, Larson CN, Dietrich J, Klem EB, Scheuermann RH. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ammayappan A, Upadhyay C, Gelb J, Jr, Vakharia VN. Complete genomic sequence analysis of infectious bronchitis virus Ark DPI strain and its evolution by recombination. Virol J. 2008;5:157. doi: 10.1186/1743-422X-5-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bijlenga G, Cook JK, Gelb J, Jr, de Wit JJ. Development and use of the H strain of avian infectious bronchitis virus from the Netherlands as a vaccine: a review. Avian Pathol. 2004;33(6):550–557. doi: 10.1080/03079450400013154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia J, He X, Yao KC, Du LJ, Liu P, Yan QG, Wen YP, Cao SJ, Han XF, Huang Y. Phylogenetic and antigenic analysis of avian infectious bronchitis virus in southwestern China, 2012–2016. Infect Genet Evol. 2016;45:11–19. doi: 10.1016/j.meegid.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Zhang X, Gong L, Yan B, Li C, Han Z, Shao Y, Li H, Kong X. Altered pathogenicity, immunogenicity, tissue tropism and 3'-7kb region sequence of an avian infectious bronchitis coronavirus strain after serial passage in embryos. Vaccine. 2009;27(34):4630–4640. doi: 10.1016/j.vaccine.2009.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



