Summary
In the 50 years since acute respiratory distress syndrome (ARDS) was first described, substantial progress has been made in identifying the risk factors for and the pathogenic contributors to the syndrome and in characterising the protein expression patterns in plasma and bronchoalveolar lavage fluid from patients with ARDS. Despite this effort, however, pharmacological options for ARDS remain scarce. Frequently cited reasons for this absence of specific drug therapies include the heterogeneity of patients with ARDS, the potential for a differential response to drugs, and the possibility that the wrong targets have been studied. Advances in applied biomolecular technology and bioinformatics have enabled breakthroughs for other complex traits, such as cardiovascular disease or asthma, particularly when a precision medicine paradigm, wherein a biomarker or gene expression pattern indicates a patient's likelihood of responding to a treatment, has been pursued. In this Review, we consider the biological and analytical techniques that could facilitate a precision medicine approach for ARDS.
Introduction
In the 50 years since Ashbaugh and colleagues1 first described acute respiratory distress syndrome (ARDS), substantial progress has been made in identifying the pathogenic contributors to the syndrome and in improving the ventilatory support of patients.2 Concurrent improvements in the management of sepsis,3, 4 the most common precipitant of ARDS, and ventilator liberation, sedation practices, and preventive therapies,5, 6 have also contributed to the improved survival of patients with ARDS. Nonetheless, in observational studies7, 8 from across the world, the mortality of patients with ARDS remains high; about one in three patients diagnosed with ARDS will not survive 60 days. Pharmacological treatments for ARDS remain scarce, with only one drug (cisatracurium besilate) showing a potential, albeit probably non-specific, benefit in a randomised trial of 340 patients. 9 The need for new therapies for ARDS is indisputable, yet numerous well conceived studies have been unable to identify an effective treatment.10, 11, 12, 13, 14 Drugs might have been ineffective because responses to, or the toxic effects of, therapy are non-uniform; because of problems with drug timing, duration, or delivery; or because of incomplete understanding of the pathogenesis or heterogeneity of ARDS.15, 16, 17 A precision approach, whereby therapies are specifically targeted to patients most likely to benefit, might overcome such limitations and is broadly advocated for ARDS.18, 19 In this Review, we consider tools and approaches that might facilitate the identification of novel and potential precision therapies for ARDS by identifying the mechanisms and therapeutic relevance of specific biological pathways, by identifying subgroups of patients likely to respond to a particular therapy based on their biology, or (perhaps ideally) both.
Discovery and bias-free approaches to identify new biological mechanisms and drug candidates in ARDS
Substantial inflammation in the alveolar and plasma compartments has been a recognised characteristic of patients with ARDS since the 1980s. High concentrations of inflammatory cytokines have been associated with both injurious ventilator strategies and worse outcomes.20, 21, 22, 23 Beyond inflammation, mechanisms including endothelial activation, respiratory epithelial dysfunction, and surfactant depletion have been established as major contributors to the pathogenesis of ARDS,24, 25 and candidate studies26, 27, 28, 29, 30 of individual biomarkers and genes in these pathways have yielded and continue to yield some important associations (table 1 ). Novel analytical techniques to be discussed in this Review might highlight new uses for some of these accepted markers to refine ARDS endotypes or to serve as enrichment markers in future trials. To date, however, improved molecular understanding of the pathogenic contributors to ARDS has yet to translate into mortality reductions for patients with the syndrome.
Table 1.
Selected key biological features, and their related pathways, associated with ARDS risk or mortality by candidate and discovery approaches
| Genes (DNA) | Transcripts (RNA) | Metabolites | Proteins (biomarkers) | |
|---|---|---|---|---|
| Inflammation | CXCL8 (IL8), IL6, IL1B, TNF, CSF2 (GM-CSF), IL10, IL1RN, MBL2, NFKBIA, TIRAP, TLR1, PI3, IRAK3, DARC, ADIPOQ | IL1R2, FTL, PI3, S100A2 | .. | IL8, IL6, IL1B, IL18, CSF2 (GM-CSF), CSF3 (G-CSF), TNFRSR1A (sTNFR1), IL10, TNF (TNF-α) |
| Endothelial injury or function | ANGPT2, TEK (TIE2), ACE, NAMPT (PBEF) VEGFA, DIO2, PLAU, SERPINE1 (PAI1), MYLK, SIP3, FVL | .. | Sphingomyelin | ANGPT1 (ANG1), ANGPT2 (ANG2), VEGF-A, VWF, apolipoproteins, cell-free haemoglobin, PAI1, endothelial glycocalyx, THBD (sTM), ICAM1 (sICAM), PROC |
| Epithelial injury or function | SFTPB, EGF | .. | .. | AGER (sRAGE), SFTPD, FGF7 (KGF), HGF, SCGB1A (CC16), MUC1 (KL6) |
| Extracellular matrix injury | .. | .. | Desmosine (urine) | COL1A2 (PCPII), COL3A1 (PCP III) |
| Platelet count or function | LRRC16A | .. | .. | .. |
| Oxidant stress pathway | HMOX1, HMOX2, NFE2L2 (NRF2), NQO1 | .. | Glutathione, nitric oxide (urine) | .. |
| Iron homoeostasis | HMOX1, HMOX2 | FTL | .. | FTL, FTH1 |
| Apoptosis | FAS | .. | Phosphatidylserine | FASLG (FASL) |
| Purine metabolism | .. | .. | Hypoxanthine, xanthine, guanosine | .. |
| Urea cycle | .. | .. | Glutamate, creatinine, creatine | .. |
| Glycolysis or carbohydrate metabolism | .. | .. | Lactate, citrate, adenosine | .. |
| Lipid metabolism | .. | .. | Phosphatidylcholine, sphingomyelin | Apolipoproteins |
| Unclassified role in ARDS | PPFIA, POPDC3, FAAH, XKR3, ASRD | .. | .. | .. |
Genes and transcripts are shown by their HUGO gene nomenclature committee symbol, with a common alternative name in parentheses if appropriate.26, 27, 30, 31, 32, 33 Metabolites34, 35 and proteins17, 29, 36, 37, 38 are shown using Reference Sequence abbreviations, with common alternative names in parentheses. ARDS=acute respiratory distress syndrome.
How could a paradigm-shifting breakthrough be facilitated? Perhaps the field of ARDS has persisted in looking under the light, restricting inquiry to genes and proteins already hypothesised to affect lung injury.39 Although continued investigation of selected candidate pathways, identified from experimental and observational investigations of ARDS, does and should continue, a complementary approach is to use discovery methods independent of any understanding of the pathogenesis of ARDS. Such “bias-free” approaches include assessing large-scale variation in genomic DNA, transcriptome (mRNA) expression, non-coding RNA, proteins, lipids, or metabolites and comparing patients with ARDS with at-risk controls. Generally, the goal of such approaches is to identify novel candidates that can then be investigated in more traditional hypothesis-testing experiments (figure 1 ). We stress that discovery approaches are not inherently superior to hypothesis-driven, candidate investigations, but rather that discovery approaches can bring new candidates to the pipeline of investigation. Discovery approaches are not predicated on our current understanding of ARDS biology but rather on the investigator's ability to select representative biospecimens and populations of patients and unaffected controls. Thus, such approaches are free from the biases introduced by our current molecular understanding of ARDS, such as which pathways are activated and which molecules incite or perpetuate lung injury. We refer to discovery approaches as bias-free when they are also independent of known genetic or proteomic sequences; for example, sequencing is bias-free, whereas a whole-genome gene expression array requires knowledge about transcripts to design specific probes (table 2 ). Although a sceptic might view discovery approaches as akin to a fishing expedition, the broad goal of these studies is not to merely identify the most discriminatory marker between samples, but also to expand our consideration of pathways and mechanisms not presently thought to contribute to disease and to explain why an unexpected marker might differ significantly between cases and non-cases or between target tissues.
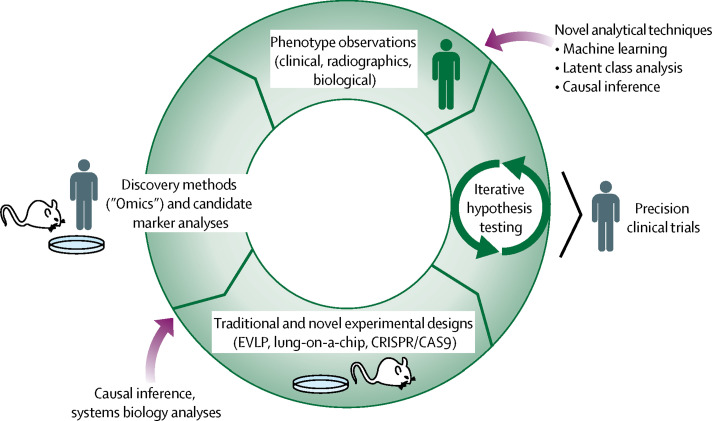
Figure 1.
Integrative approaches to identify and test new precision therapies for acute respiratory distress syndrome
Observations made from candidate and discovery methods fuel traditional hypothesis testing and inform novel designs such as EVLP or lung-on-a-chip. Drug candidates with a consistent signal for benefit across multiple study designs are the most promising for biomarker-enriched clinical trials. EVLP=ex-vivo lung perfusion. CRISPR=clustered regularly interspaced short palindromic repeats. CAS9=CRISPR-associated protein 9.
Table 2.
Candidate and discovery methods to interrogate different biological features
| Hypothesis testing (candidate) | Medium-to-high-throughput discovery | Bias-free discovery | ||
|---|---|---|---|---|
| DNA (genomics) | ||||
| Genomic DNA (all cells the same); somatic DNA (eg, tumour, individual); epigenomics (methylation, acetylation) | Candidate genotyping; methylation-specific PCR | DNA array; genome-wide association studies; bisulfite-array (methylation status) | Next-generation sequencing; chromatin-immunoprecipitation sequencing (protein–DNA binding sites)*; bisulfite sequencing (methylation sites)† | |
| RNA‡ (transcriptomics) | ||||
| Transcriptome; mRNA or cDNA; miRNA; ncRNA | Candidate PCR | cDNA microarray (also known as gene expression microarray) | RNA sequencing | |
| Proteins‡ (proteomics) | Candidate (antibody-based) ELISA; western blot | Multiplex ELISA; bead-based assays; DNA-aptamer array | MALDI-TOF mass spectrometry | |
| Metabolites§ (metabolomics) | Aminoacids; carbohydrates; lipids; glycoproteins; peptidoglycans; small molecules; drugs or toxins | Target gas chromatography-mass spectrometry | Gas, liquid, or solid chromatography (for separation); mass spectrometry or nuclear magnetic resonance spectroscopy (for detection) | Gas, liquid, or solid chromatography (for separation); mass spectrometry or nuclear magnetic resonance spectroscopy (for detection) |
| Microbiota | Candidate PCR; culture detection | Targeted amplicon sequencing | Next-generation sequencing (also known as metagenomics) | |
We refer to candidate testing when an individual known entity (SNP, specific transcript, protein, etc) is quantified, whereas medium-throughput or high-throughput discovery approaches rely on known features but are capable of multiplex assays. For example, genome-wide association studies or cDNA microarrays rely on the knowledge of a genetic sequence to generate probes that will assay each SNP or transcript. By contrast, bias-free methods imply that the features being detected might be unknown—for example, next-generation sequencing sequences all nucleic acids detected, regardless of whether a sequence is recognised. MALDI-TOF=matrix-assisted laser desorption/ionisation time-of-flight. cDNA=complementary DNA. miRNA=microRNA. ncRNA=non-coding RNA. SNP=single-nucleotide polymorphism.
Chromatin-immunoprecipitation sequencing is a method to detect and sequence areas of the genome where proteins interact with DNA to, for example, understand transcription factor binding.
Bisulfite sequencing is a method that uses bisulfite treatment of DNA to uncover DNA methylation patterns that might give clues to DNA regulation.
Vary by cell type and time.
Vary by location and time.
Key messages.
-
•
Effective pharmacotherapies for acute respiratory distress syndrome (ARDS) are still elusive, and the clinical and biological heterogeneity of ARDS suggests that precision approaches might be helpful.
-
•
Novel approaches to understanding ARDS biology, identifying discrete subgroups of ARDS, or both are needed to facilitate precision therapies for ARDS.
-
•
Discovery-based approaches, combined with candidate marker analyses, might help to identify new pathways relevant to ARDS for subsequent testing in hypothesis-driven experiments, potentially including novel preclinical models of ARDS.
-
•
New approaches to analyses of complex clinical and biological data might help to identify distinct ARDS subgroups that might be suitable for targeted therapies.
Genome-wide association studies
Genome-wide association studies attempt to find regions of the genome in which genetic variation among unrelated cases is consistently skewed compared with the background population.40 An extension of classic family-based genetic linkage studies, genome-wide association studies rely on very large populations to detect common genetic variants with small-to-moderate effect sizes and stipulate an extreme statistical imbalance (p values of less than 5 × 10−18) to declare significance. One genome-wide association study of trauma-associated ARDS has been reported;41 no single-nucleotide polymorphism (SNP) achieved genome-wide significance, although a replicating variant in the liprin-α gene that warrants further mechanistic investigation was identified. This first attempt at a genome-wide association study of ARDS was instructive for highlighting its insufficient power despite including 600 patients with ARDS and more than 2000 healthy controls in a discovery population, followed by an additional 500 critically ill patients in a replication population.41 Although a genome-wide association study is a powerful tool with reasonable consensus about analytical strategy, additional considerations might restrict its application to a trait such as ARDS. For example, although ARDS is the cause of many thousands of deaths each year, clinical recognition of ARDS is poor,7 and no validated method exists to diagnose ARDS with electronic medical record coding.42 Additionally, because ARDS is a complication of serious insults, such as sepsis, pneumonia, aspiration, trauma, and ventilator exposure,8 the use of healthy controls can be problematic. Controls carrying a high-risk genetic variant might not develop ARDS if they do not develop sepsis, and thus would be classified as non-cases, attenuating any potential association signal.43 By selecting controls with a risk factor for ARDS, studies might be less susceptible to potential misclassification. Similarly, when evaluating variants for association with ARDS mortality, it would be reasonable to require that both cases and controls have ARDS. Few populations exist that are adequately powered to test or replicate genome-wide ARDS variants. Furthermore, heterogeneous patients are currently labelled as ARDS cases despite different precipitants, sites of injury, and biological profiles. Combining such patients in an effort to maximise sample size might have the unfortunate effect of drowning out statistical signals that might only be active in one subset of the population.15, 26, 36, 44
Next-generation sequencing
Whereas genome-wide association studies search for common variants that are involved in common diseases, each variant conferring just a small fraction of altered risk, rare-variant analyses use DNA sequencing to detect polymorphisms that could have a large effect but are highly uncommon.45 Sequencing of genomic coding regions (so-called exome sequencing) was done in a collaborative effort sponsored by the National Heart, Lung, and Blood Institute46 and included 96 cases of ARDS. When ARDS cases were compared with presumably healthy controls in the 1000 Genomes Project, two SNPs in the genes XK related 3 and arylsulfatase D showed differential expression in ARDS cases,31 although both findings still require replication. Perhaps surprisingly, exome sequencing, which aims to detect major deleterious genetic disruptions, has shown that, for a fraction of the population, the clinical phenotype is explained by multiple major genetic variants rather than just one.47 The potential for multiple variants that affect ARDS coexisting in one patient is even more relevant for common (non-coding) genetic variation and, if unrecognised, this issue might contribute to imprecise risk estimates or restricted reproducibility.
Gene expression profiling
The search for new candidates involved in the pathogenesis of ARDS is not restricted to genomic DNA. Discovery approaches exist to interrogate transcript expression (so-called transcriptomics), DNA modifications (so-called epigenomics), proteins, metabolites, and microbiota. The key technological shift that facilitated the explosion of these ‘omic fields was the ability to perform high-throughput feature capture and identification without a-priori designation of which features would be sampled. In the case of transcriptomics, in which mRNA is the feature under investigation, the field has progressed from whole-genome arrays—ie, microchips coated with thousands of oligonucleotide probes specific to the complementary DNA (cDNA) sequence of roughly 22 000 known genes—to RNA sequencing, whereby cDNA or occasionally RNA is sequenced using next-generation sequencing technology.48 The main advantage of RNA sequencing compared with microarrays is that RNA sequencing does not rely on probes designed to capture cDNA variation based on previous knowledge of the genome, but instead applies sequencing to all cDNA captured from the sample.49 As a result of RNA sequencing investigations, we are gaining insight into the regulatory roles of non-coding RNA and small RNA species, the potential for alleles that do not change protein sequence to affect expression, and the complexity involved in isoform determination.50 The challenges involved in applying RNA sequencing to a phenotype such as ARDS are not trivial and can be summarised as obstacles facing the broad field of transcriptomics—including developing bioinformatic solutions to high-dimensional data analysis, quality assurance, and achieving a consensus on significance testing—and burdens that might be unique to ARDS. Burdens unique to ARDS are fundamental questions about study design: which tissue(s) should be profiled by RNA sequencing to discover new markers and inform about ARDS pathogenesis? Lung tissue is so rarely available while the patient is alive with early ARDS,51 so are alveolar macrophages an adequate substitute? Kovach and colleagues52 reported the first microarray analysis of human alveolar macrophages collected from bronchoalveolar lavage fluid samples from patients with early ARDS (days 0–4) and identified a pattern of relative immune tolerance distinguishing ARDS-derived alveolar macrophages from those of healthy controls. Although the contribution of alveolar macrophages in lung host defence, injury, and resolution is established,53 it would seem unwise to ignore the potential contributions of lung-resident neutrophils and lymphocytes, as well as the endothelial, epithelial, and stromal cells of the lungs. Attempts to characterise gene expression in whole-blood or circulating mononuclear cells have also been done for ARDS54, 55 and have implicated dysregulation in neutrophil-expressed genes and the ferritin heavy chain, although these findings await replication.
Discovery proteomics and metabolomics
High-throughput approaches are also feasible for analysing the expression patterns of proteins or small-molecule metabolites in body fluid or tissue. Rather than attempting to quantify a protein or molecule of known identity, these techniques first separate the population of unknown proteins or small molecules, then quantify and identify them. Advances in the granular separation of proteins or small molecules with differential light absorption and activation (ie, matrix-assisted laser desorption/ionisation time-of-flight [MALDI-TOF] mass spectrometry), and the resolution of compounds with gas and liquid chromatography, have led to substantial improvements compared with two-dimensional gel electrophoresis. Additionally, mass spectrometry and nuclear magnetic resonance spectroscopy have improved the accuracy and sensitivity of the identification of unknown proteins and metabolites.56 A high-dimensional proteomic analysis57 of bronchoalveolar lavage fluid from patients with ARDS identified differential protein expression between ARDS survivors and non-survivors over the first 4 days of ARDS, with non-survivors manifesting decreased expression of proteins related to coagulation, iron homoeostasis, and immune activation and increased expression of proteins related to glycolysis, collagen metabolism, and the actin cytoskeleton. High-throughput proteomic screening of plasma from patients with ARDS identified increased expression of glycoproteins and serum amyloid A and decreased expression of complement factor H and apolipoprotein A, B, and C compared with plasma from healthy controls.58 However, discovery proteomics in lung injury has been hampered by several factors, including the complexity of the plasma proteome and overwhelming signals from high-abundance plasma proteins (eg, albumin), which have limited the insights gained to date;59 thus, most of the recent advances in this area have come from candidate marker analyses. Indeed, the expectation is that each promising feature identified through a discovery approach should then be replicated as a candidate marker, as is highlighted by the iterative example in figure 1; the most compelling associations will show converging support from multiple lines of evidence.
Large-scale metabolomics has also been done with both bronchoalveolar lavage fluid and plasma from patients with ARDS compared with healthy controls.34, 35 Whereas both bronchoalveolar lavage fluid and plasma showed increased expression of lactate, citrate, and creatine, other metabolites were specific to each fluid. For example, bronchoalveolar lavage fluid from patients with ARDS was characterised by increased expression of guanosine, xanthine, and hypoxanthine—metabolites of guanosine and uric acid metabolism pathways—and decreased expression of phosphatidylcholines, the phospholipids that constitute pulmonary surfactant.34 Plasma from patients with ARDS was characterised by metabolites that might indicate disrupted oxidant stress signalling (glutathione), energy homoeostasis (adenosine), endothelial barrier function (sphingomyelin), and apoptosis (phosphatidylserine).35 Although exciting, each of these proteomic and metabolomic investigations were done with very small sample sizes (n<20 individuals), and the direct translation from protein or metabolite identification to ARDS diagnosis or therapy is not obvious.34, 35, 57, 58 By contrast, another group examined the metabolome of exhaled breath condensate collected from the ventilators of more than 100 critically ill patients and identified three volatile compounds—octane, acetaldehyde, and 3-methylheptane—in the exhaled breath condensate of patients with ARDS compared with ventilated controls.60 By use of training and validation sets, the authors of that study showed that these three metabolites had a reasonable diagnostic performance on their own and that discrimination was improved when they were added to the Lung Injury Prediction score compared with the score used alone.60, 61, 62
Microbiome analysis
The lung microbiome represents another opportunity to apply cutting-edge sequencing techniques in a bias-free manner. Dickson and colleagues63 showed that, in a mouse model of sepsis induced by caecal ligation and puncture, the lung microbiota was rapidly modified by sepsis and gut-associated bacteria were the dominant community for 5 days. The gastrointestinal tract, and not the upper respiratory tract, was shown to be the source of sepsis-induced lung microbiome alterations for mice with sepsis from systemic insults, whereas this shift from typical lung-resident microbes to gut-associated bacteria was not observed in mice exposed to intratracheal lipopolysaccharide to model direct lung injury.63 In support of these findings,63 sequencing of bacterial ribosomal RNA from the bronchoalveolar lavage fluid of 68 patients with ARDS identified gut-associated Bacteroides species in 28 (41%) patients, compared with only one (3%) of 26 healthy controls. Enrichment of gut bacteria in bronchoalveolar lavage fluid was associated with the plasma concentration of inflammatory markers. Thus, for non-pulmonary sepsis-associated ARDS, translocation of bacteria from the gut to the lungs might have a more prominent role than previously recognised, because traditional culture methods are relatively insensitive to anaerobic bacteria. Whether this mechanism can be targeted to prevent or treat ARDS, however, remains untested.
New analytical approaches
As the potential for each ‘omic method to be applied to ARDS is considered, the sheer volume of data generated is potentially staggering. How should this wealth of biological data, often coupled with extensive clinical data from the critical care setting, be analysed to maximise our potential insight into novel treatment approaches for ARDS? Even for candidate marker analyses, novel analytical approaches might be needed to maximise the insight gained and integrate with complex molecular and clinical data. Traditionally, biological data from human beings with ARDS have been analysed in one of two ways: by identifying the biological phenotypes most strongly associated with a clinical outcome of interest (often death), with the idea that targeting these pathways might thereby reduce mortality, or by dividing patients into categories based on clinically evident or pre-supposed characteristics and then comparing their clinical and biological phenotypes to identify distinct subgroups that might respond differently to treatment.
A common approach has been to measure biomarkers thought to reflect specific biological pathways and test their association with poor outcomes, typically with regression-based methods. This approach is appealingly straightforward; it is intuitive and, at least in its simplest form, requires minimal advanced training in statistical analysis for either the investigator or the reader of the final product. Moreover, this approach has led to substantial advances in our understanding of the biology of human ARDS. Over the past three decades, studies ranging from small, single-centre investigations to secondary analyses of large multicentre clinical trials have reported associations between poor clinical outcomes and biomarkers of key pathways of lung injury—for example, injury to the lung epithelium and endothelium—as well as numerous inflammatory pathways. Although a comprehensive review of these studies is beyond the scope of this Review, several of the key studies are cited in table 1. This approach is particularly well suited to confirming the human relevance of biological pathways identified in laboratory models of ARDS. This approach has most commonly been used to analyse candidate plasma proteins and genetic polymorphisms, although more recent investigations have focused on unbiased discovery methods such as DNA and RNA sequencing, metabolomics, and proteomics. This approach also has notable limitations. First and most important is the old adage that correlation does not imply causation; put differently, biological markers associated with mortality in ARDS might not be causal drivers of ARDS mortality and, therefore, might not be good therapeutic targets. Studying these associations in the setting of a randomised trial, and incorporating analysis of the response of the biomarker in question to the randomly assigned therapy, might partially mitigate but not eliminate this issue.21 Second, the regression-based models typically used for these types of analyses do not mandate or necessarily facilitate analyses of heterogeneity within ARDS, so as to identify treatment-responsive subgroups. Alternative analytical approaches are better suited than regression methods for this particular question.
A second approach often applied to patient data to search for novel treatment strategies for ARDS is to divide patients into subgroups based on pre-existing hypotheses or clinically evident features and then to compare the biology of these subgroups. For example, for decades, investigators have focused on whether direct lung injury (ie, ARDS resulting from direct damage to the lung parenchyma, such as in pneumonia or aspiration of gastric contents) differs from indirect lung injury (ie, ARDS resulting from an insult distant to the lung parenchyma, such as non-pulmonary sepsis or massive blood transfusion).64 Clinical studies36, 65 have supported this distinction, identifying differences in endothelial injury and inflammation between patients with direct and indirect lung injury. Other similar types of analyses have subdivided ARDS into diffuse versus focal radiographic changes, or into subgroups based on clinical risk factors (eg, trauma vs sepsis vs other).66, 67, 68 Similar to outcome-focused methods, these approaches have the advantage of being intuitive and logical, particularly for clinicians who can recognise in their own practice the obvious clinical differences between subgroups of patients with different clinical features or natural histories. However, these approaches rely on our pre-existing biases about disease classification and, to date, have not led to successful clinical trials targeting specific ARDS subgroups.
As our ability to quantify biological complexity has advanced over the past several decades, so too have novel statistical methods designed to disentangle heterogeneity within complex datasets. The use of these novel statistical methods in other disease phenotypes, such as asthma, has led to substantial progress towards the discovery of disease endotypes—that is, specific subgroups of a syndrome with distinct pathobiologies and differential responses to treatment. Some of these methods are now being used in translational studies of critical illness, with a similar goal.
Cluster-based methods encompass various analytical techniques that share the broad goal of identifying groups (clusters) of observations with similar characteristics. These types of methods, such as hierarchical clustering and κ-means clustering, have commonly been applied to genomic data, with the aim to identify clusters of patients with similar gene expression patterns. Once biological data has been used to identify patient clusters, the clusters can subsequently be analysed for differences in clinical outcomes, clinical phenotypes, or other variables of interest. Multiple examples exist of the use of these techniques in asthma, in which they have helped to identify the T-helper-2-high endotype in several studies.69, 70 In critical care, cluster-based methods have been used to identify subclasses of paediatric septic shock.71, 72 These methods have several advantages, including an explicit focus on reducing heterogeneity by identifying subgroups that are internally similar, an unbiased approach free from presupposition, and facile visual representation (ie, the classic heat map). Another advantage for use of clustering in ARDS is that it can be done on baseline characteristics without consideration of outcome. However, one notable disadvantage of cluster-based methods is that they will always identify clusters73 and do not test a specific hypothesis regarding the number of clusters in a particular class. This parameter must be set externally and, as such, might be prone to bias regarding the number of clusters present, although methods exist to estimate the optimal number of clusters from the data.74
Another advanced analytical approach, which, similar to cluster analysis, often falls under the rubric of machine learning, is classification trees, often referred to as classification and regression tree analysis. This approach is designed to identify unexpected cut-points in a set of data that permit classification into subgroups that might not be readily apparent. This approach generates a branching tree-like structure that branches at specific cut-points of a given variable and ends in multiple terminal nodes, which are often distinguished by differences in outcomes. This method has the advantage of being able to identify patterns in the data that would otherwise be unapparent. However, classification trees are also notorious for overfitting the model to the data and, as such, mandate external validation in independent datasets. Tree-based models have been used in hospital inpatients to identify predictors of clinical deterioration,75 in adult septic shock to refine prognostic stratification based on plasma biomarkers,76 and in ARDS to identify clinical features associated with poor outcomes.77 These trees are constructed based on the association between the measured variables and a specific clinical outcome, so the caveats regarding outcome-based models also apply to these trees—ie, that the variables which identify the branches in the tree might not be causally related to the observed differences in outcomes. Similar to cluster-based models, tree-based models also require external and potentially arbitrary decisions regarding the number of branches and terminal nodes, although methods with re-sampling and cross-validation have been developed to inform these decisions.75
Latent class analysis is an approach derived from mixture modelling that is explicitly designed to identify hidden (so-called latent) subgroups within a larger group. Latent class analysis has been widely applied in psychiatric research and has been an important contributor to the study of asthma endotypes.78, 79 One major strength of latent class modelling is that it can test the hypothesis that a certain number of classes (k) fits the data better than one fewer class (k – 1), providing more objective confirmation of the optimal number of classes for a given dataset. In ARDS, use of latent class analysis has identified two distinct subphenotypes in independent analyses of three randomised trials.15, 17 The two subphenotypes were clearly distinguished in part on the basis of biomarker profiles, with one subphenotype characterised as hyper-inflammatory compared with the other subphenotype, and by their responses to randomly assigned positive end-expiratory pressure and a fluid-conservative management strategy. When applied to trauma cohorts, latent class analysis also identified distinct subgroups of ARDS that were distinguished largely by time to ARDS development, but also by clinical characteristics and plasma biomarker expression.80 One disadvantage of latent class analysis is that relatively large datasets (n>300 individuals) are typically needed to confidently fit latent class models.
Quantitative trait analysis
Recognising that heterogeneity is a cardinal feature of ARDS that might confound traditional genomic approaches, an alternative approach is to apply genomic tools (eg, genome-wide association studies) to an intermediate trait that has meaning in the context of ARDS (eg, a plasma biomarker or expression of an mRNA transcript) to apply causal inference methods that might help to evaluate one aspect of the syndrome. For example candidate studies81, 82, 83 have shown plasma concentrations of angiopoietin-2 (ANG2)—a protein secreted by activated endothelium that potentiates vascular permeability—to be a strong marker for ARDS diagnosis, prediction, and prognosis. Discovery genetic approaches also implicate this pathway in ARDS risk,84 making ANG2 an attractive candidate ARDS intermediate protein. If we consider that every biological process is, to some degree, controlled by our DNA, the genetic regulation of a plasma protein such as ANG2 is far less complex than that of ARDS, because the phenotype of plasma ANG2 concentration is regulated by fewer genes and has a stronger correlation between genetic variation and measured output (ANG2) than does a disease phenotype such as ARDS. Thus, the likelihood of finding genetic variants that explain a high proportion of variance in plasma ANG2 concentration is greater than finding variants that explain a large proportion of ARDS risk.50 Furthermore, quantitative traits are statistically more powerful than dichotomous traits, such that statistically significant results might be detected with only 100 or 200 patients, rather than with the 1000s necessitated by traditional genome-wide association studies,85 and such sample sizes are far more achievable for populations of critically ill patients. Genomics can then be used to apply causal inference methods—techniques borrowed from the econometric and social science fields—to infer causality from observational data. The advantage of identifying causal biomarkers in ARDS, rather than markers that merely correlate with ARDS, is that modifying the causal intermediate is more likely to influence disease outcome. Experimental evidence from animal or in-vitro models allows us to directly test whether intermediates have direct causal effects; however, it would be unethical to randomly assign people to receive doses of a potentially injurious marker. Thus, methods to infer the effect of a potential mediator—for example, a plasma marker such as ANG2, a bronchoalveolar lavage fluid metabolite, or a whole blood transcript—on ARDS risk or mortality are desirable. Once a marker is suggested to have a causal role, rigorous experiments in model systems should be done to more definitively show causality.
Mediation analysis is a formal method to explain the mechanism by which a potential explanatory variable influences an outcome via an intermediate, or mediator variable. Many quantitative traits have a strong genetic component; thus, mediation analysis can be applied to associations between SNPs and disease outcomes to test whether a significant portion of the association is mediated by change in a third variable, the quantitative trait.86 If the intermediate trait (biomarker, transcript, or metabolite) mediates a significant portion of the observed risk, this provides good evidence that the marker might be mechanistically linked to the disease outcome. Wei and colleagues87 used mediation analysis to address whether changes in platelet count might explain ARDS risk and mortality. They first identified genetic variants that strongly influenced baseline platelet count during critical illness and then assessed whether a SNP strongly associated with platelet count was also associated with ARDS risk; they found that a small but significant portion of the association between a SNP and ARDS was mediated indirectly through changes in baseline platelet count. In a related analysis,88 the same group used this method to show that a decrease in platelet count mediates a portion of ARDS mortality. The associations between genetic variants and a quantitative trait such as a plasma marker can also be harnessed for instrumental variable analysis, whereby SNPs that are strongly associated with a biomarker are used to genetically predict the biomarker, and then the association between the genetically predicted biomarker and the outcome is assessed. This approach is sometimes called a mendelian randomisation analysis, because each individual is considered randomised by random assortment of parental alleles to a genotype that might express high or low levels of a biomarker. Mendelian randomisation studies have provided strong support for the causal contribution of LDL-cholesterol to risk of coronary artery disease; genetic variants associated with the plasma concentration of LDL-cholesterol were also associated with coronary artery disease, and when the concentration of LDL-cholesterol was predicted by genotype, genetically predicted LDL-cholesterol was associated with this disease.89 Furthermore, dissecting the genetic determinants of LDL-C concentration has uncovered novel treatment platforms for coronary artery disease,89, 90 offering an attractive template for ARDS genetic research if appropriate intermediates can be identified.
Novel preclinical models of ARDS
Another approach to determining the therapeutic relevance of pathways identified with ‘omics methods is to test them in novel preclinical models of ARDS. Although a thorough review of human, animal, and in-vitro models of lung injury is beyond the scope of this Review, several developments that are facilitating the translation from bedside to bench and back again are worth highlighting and are shown in figures 2 and 3. These techniques can interrogate candidates identified in bias-free methods with a rigorous experimental design and, in some cases, can model treatment, while also monitoring the variation in quantitative markers, physiology, or gene expression.
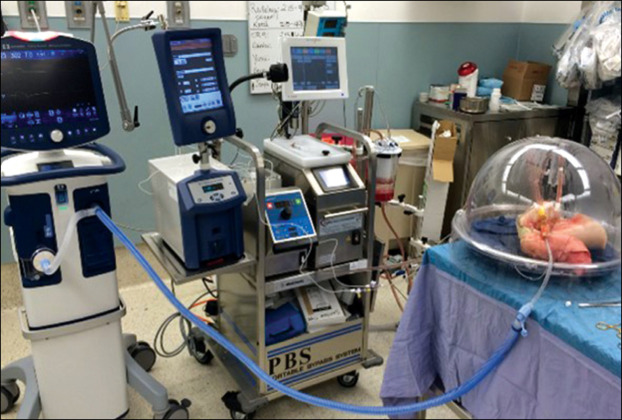
A recognised shortcoming in human ARDS research is the rarity of lung tissue from acute ARDS, which then restricts applications that are cell-specific, such as transcriptomic, epigenomic, and proteomic methods. One solution to this scarcity of tissue is to use human lungs that were declined for transplantation. Up to 80% of evaluated lungs are deemed unsuitable for transplant because of poor oxygenation, visible injury, or poor compliance, features that are common to ARDS.92, 93 Furthermore, to better simulate conditions in vivo, these human lungs can be ventilated and perfused in an ex-vivo lung perfusion (EVLP) system for several hours (figure 2 ), allowing the observation of physiological measures such as compliance, alveolar fluid clearance, and oxygenation and the sampling of lung tissue, perfusate, and alveolar fluid.94, 95 Clinical trials (eg, NCT01365429)92 are being done to test whether EVLP can increase the number of suitable lungs for transplantation while maintaining optimal transplant outcomes. However, these sophisticated systems can also be optimal ARDS preclinical models in which to screen potential ARDS therapies,94, 95 because they have the advantages of safety (no human exposure) and access to human lung tissue. By taking biopsy samples of the tissue and using pharmacological inhibitors or agonists, the mechanism of a drug's action can be investigated. The EVLP system can also be adapted to model a uniform injury by applying bacteria or endotoxin to the lung, making this an adaptable model for hypothesis-testing for ARDS prevention and therapy.95
Figure 2.
Ex-vivo lung perfusion system
Human lungs under the dome are ventilated and perfused.
© 2017 Ed Cantu III
Another potential therapeutic screening method uses microfluidic bioengineering and three-dimensional cell culture to produce a microengineered lung-on-a-chip, complete with an alveolar–capillary interface, cyclic stretch to mimic breathing, and perfusion to model circulation (figure 3 ).91 Although this system relies on human cell lines that are capable of persisting in long-term culture, and thus, to date, alveolar epithelial cells derived from a lung cancer cell line rather than primary human alveolar epithelial cells have been used, drug-induced permeability pulmonary oedema was modelled with this system and was shown to share many features with clinical toxicity from interleukin 2, including reduced oxygen tension and endothelial and epithelial paracellular gap formation.91, 96 Most impressively, that model could be used to test whether co-administration of preventive agents—for example, angiopoietin-1 or a transient receptor potential cation channel subfamily V member 4 channel inhibitor—could block the interleukin 2-mediated permeability and, via immunohistochemistry, the mechanism of action could be evaluated.91 The microfluidic lung-on-a-chip could also be adapted to study therapeutic applications and might be considered a more high-throughput method for drug screening than EVLP because it does not rely on the scarce resource of human lungs. Additionally, if a quantitative trait such as a plasma protein concentration was identified as having causal significance through causal inference modalities or endotype identification, then drug screening could proceed with the secreted protein as an outcome, improving efficiency. Although promising compounds would still need to be tested in both small and large animal models of ARDS before proceeding to human trials, both EVLP and lung-on-a-chip microdevices might help to select agents most likely to benefit patients.
Figure 3.
Illustration of a microfabricated lung-on-a-chip
The chip consists of a three-dimensional microchannel system with culture chambers for both epithelium and endothelium at a porous interface. The epithelium and endothelium are capable of being stretched and perfused.91
© 2017 Donguen (Dan) Huh
Finally, the field of genome editing has been revolutionised in the past 5 years by the elucidation of a family of endonucleases, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) systems, that are capable of site-specific DNA cleavage.97 The simplicity by which the CRISPR/Cas9 system can be adapted to recognise and cleave a specific genomic sequence has been harnessed for gene silencing, to identify transcriptional repressors or enhancers, and even to insert specific point mutations into the genomes of human tissue. To date, CRISPR/Cas9 editing has been used to make mice susceptible to the coronavirus responsible for Middle East respiratory syndrome, overcoming the relative species restriction that Middle East respiratory syndrome coronavirus exhibits and allowing the testing of potential vaccine and antibody treatments.98 Future uses of this technology in ARDS could be to interrogate the functional significance of polymorphisms or genes identified through genome-wide association studies or transcriptomic screens, to provide or refute evidence for pathogenic causality for ARDS biomarkers, and to potentially allow testing of a precision paradigm whereby a targeted treatment could be tested in different genetic backgrounds.
The next step: biologically informed trials
In this age of rapid technological and analytical advances, the promise of matching patients to therapies most likely to help, and least likely to harm, seems more attainable than at any previous point in history. Numerous challenges remain, however, particularly in identifying new drug targets and selecting patients most likely to benefit. The approaches reviewed here have already identified plasma biomarkers that could serve as prognostic enrichment factors; markers that identify patients more likely to experience ARDS or who are at an increased risk of dying if ARDS is present.15, 19 Enrichment strategies that select such high-risk patients should improve power for clinical trials by ensuring a population with sufficient outcomes to analyse a potential treatment effect. If causal inference methods or preclinical models of ARDS suggest that a marker has a causal role in the development or progression of ARDS, then the marker might actually identify a biological endotype of ARDS. Endotype-defining markers might be useful for predictive enrichment, whereby a plasma marker could inform about a patient's likely response to a specific therapy.15, 17 As a direct extension, one could then imagine clinical trials for which eligibility might be predicated on the potential patient expressing a specific plasma marker. This approach has been shown to be highly successful in cancer therapy, especially when using tumour gene expression profiles to select targeted therapy.99, 100 In asthma, endotype recognition has similarly stimulated new therapies that might be more successful when restricted to patients predicted to benefit.101
To realise the potential of biomarker-driven clinical trials for ARDS, however, a few things must happen.102, 103 First, markers must be rapidly available for hospital inpatients to inform potential trial eligibility. By contrast with cancer, a biopsy is unlikely to be available for ARDS and patients and providers cannot afford to wait long for potential genetic results. The technology to measure protein biomarkers by ELISA in real time is not a technical barrier, but such assays are generally not commercially available. Microfluidic cell separation coupled with multiplexed, colour-coded probes might eventually allow rapid gene expression tests at the bedside;104 however, this technology has not been widely applied to critical illness.105 Second, biomarker-driven trials would require more knowledge about the performance of potential plasma markers over time during the natural course of ARDS, because the concentration of many markers is highly variable by time.82, 106 Third, in many cases, biomarker-directed trials would ideally be preceded by testing of potential therapies in preclinical models of ARDS such as EVLP or lung-on-a-chip, in addition to animal models of ARDS, to better predict whether an endotype-defining marker denotes a pathway that is modified by the potential therapy. We acknowledge that, even when a precision paradigm for ARDS is well supported by consistent findings across endotype identification, genomic or proteomic profiling, and even experimental designs such as EVLP, a potential therapy might not improve mortality if it has a deleterious effect on a comorbid condition during critical illness or if it has utility for ARDS prevention but not progression, particularly if ARDS develops rapidly. Well designed studies will be needed to selectively target ARDS prevention and therapy. Clinical trial considerations are addressed more comprehensively in a related review about this issue.107
In summary, precision medicine is a realistic paradigm for ARDS that could begin to be tested in the near future. As ‘omic methods, novel models of lung injury, and new analytical approaches improve the resolution of ARDS endotypes and suggest new mechanisms of treatment, pharmacological breakthroughs might be closer than we realise.
Search strategy and selection criteria
We searched PubMed from inception to Jan 15, 2017, for the search terms “genomic”, “polymorphism”, “gene expression”, “proteomic”, “metabolomic”, “microbiome”, “unsupervised learning”, “cluster analysis”, and “latent class analysis” combined with “acute respiratory distress syndrome” and “acute lung injury”. The search was limited to studies of human beings. Returned lists of articles were then screened manually by reading abstracts to exclude neonatal lung injury and neonatal respiratory distress syndrome. The remaining manuscripts were read in full and their reference lists were reviewed when appropriate. When possible, we have cited comprehensive recent reviews.
Acknowledgments
Acknowledgments
We would like to thank Edward Cantu III (Department of Surgery, University of Pennsylvania) and Dongeun Huh (Department of Bioengineering, University of Pennsylvania) for sharing their images, and Kevin Delucchi (Department of Psychiatry, University of California) for his critical review of the manuscript. This Review was funded by grants from the National Institutes of Health to CSC (HL131621) and NJM (HL122474).
Contributors
NJM and CSC both contributed to the literature search, figure design, and manuscript drafting.
Declaration of interests
CSC reports grant funding and consulting fees from GlaxoSmithKline and consulting fees from Boehringer Ingelheim and Bayer. NJM received grant funding from GlaxoSmithKline and consulting fees for serving on an advisory board from Sobi.
References
- 1.Ashbaugh DG, Bigelow DB, Petty TL, Levine BE. Acute respiratory distress in adults. Lancet. 1967;290:319–323. [Google Scholar]
- 2.Network ARDS. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. The Acute Respiratory Distress Syndrome Network. N Engl J Med. 2000;342:1301–1308. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 3.Dellinger RP, Levy MM, Rhodes A. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhodes A, Evans LE, Alhazzani W. Surviving Sepsis Campaign: international guidelines for management of sepsis and septic shock: 2016. Crit Care Med. 2017;45:486–552. doi: 10.1097/CCM.0000000000002255. [DOI] [PubMed] [Google Scholar]
- 5.Ouellette DR, Patel S, Girard TD. Liberation from mechanical ventilation in critically ill adults: an official American College of Chest Physicians/American Thoracic Society clinical practice guideline: inspiratory pressure augmentation during spontaneous breathing trials, protocols minimizing sedation, and noninvasive ventilation immediately after extubation. Chest. 2017;151:166–180. doi: 10.1016/j.chest.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Girard TD, Alhazzani W, Kress JP. An official American Thoracic Society/American College of Chest Physicians clinical practice guideline: liberation from mechanical ventilation in critically ill adults. Rehabilitation protocols, ventilator liberation protocols, and cuff leak tests. Am J Respir Crit Care Med. 2017;195:120–133. doi: 10.1164/rccm.201610-2075ST. [DOI] [PubMed] [Google Scholar]
- 7.Bellani G, Laffey JG, Pham T. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315:788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 8.Ranieri VM, Rubenfeld GD, Thompson BT, for the ARDS Definition Task Force Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 9.Papazian L, Forel JM, Gacouin A. Neuromuscular blockers in early acute respiratory distress syndrome. N Engl J Med. 2010;363:1107–1116. doi: 10.1056/NEJMoa1005372. [DOI] [PubMed] [Google Scholar]
- 10.Truwit JD, Bernard GR, Steingrub J, for the National Heart, Lung, and Blood Institute ARDS Clinical Trials Network Rosuvastatin for sepsis-associated acute respiratory distress syndrome. N Engl J Med. 2014;370:2191–2200. doi: 10.1056/NEJMoa1401520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAuley DF, Laffey JG, O'Kane CM. Simvastatin in the acute respiratory distress syndrome. N Engl J Med. 2014;371:1695–1703. doi: 10.1056/NEJMoa1403285. [DOI] [PubMed] [Google Scholar]
- 12.Smith FG, Perkins GD, Gates S, for the BALTI-2 study investigators Effect of intravenous β-2 agonist treatment on clinical outcomes in acute respiratory distress syndrome (BALTI-2): a multicentre, randomised controlled trial. Lancet. 2012;379:229–235. doi: 10.1016/S0140-6736(11)61623-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthay MA, Brower RG, Carson S, for the National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome (ARDS) Clinical Trials Network Randomized, placebo-controlled clinical trial of an aerosolized β2-agonist for treatment of acute lung injury. Am J Respir Crit Care Med. 2011;184:561–568. doi: 10.1164/rccm.201012-2090OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeiher BG, Artigas A, Vincent JL. Neutrophil elastase inhibition in acute lung injury: results of the STRIVE study. Crit Care Med. 2004;32:1695–1702. doi: 10.1097/01.ccm.0000133332.48386.85. [DOI] [PubMed] [Google Scholar]
- 15.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA, for the NHLBI ARDS Network Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenner JS, Greineder C, Shuvaev V, Muzykantov V. Endothelial nanomedicine for the treatment of pulmonary disease. Expert Opin Drug Deliv. 2015;12:239–261. doi: 10.1517/17425247.2015.961418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Famous KR, Delucchi K, Ware LB. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med. 2017;195:331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beitler JR, Goligher EC, Schmidt M. Personalized medicine for ARDS: the 2035 research agenda. Intensive Care Med. 2016;42:756–767. doi: 10.1007/s00134-016-4331-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prescott HC, Calfee CS, Thompson BT, Angus DC, Liu VX. Toward smarter lumping and smarter splitting: rethinking strategies for sepsis and acute respiratory distress syndrome clinical trial design. Am J Respir Crit Care Med. 2016;194:147–155. doi: 10.1164/rccm.201512-2544CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranieri VM, Suter PM, Tortorella C. Effect of mechanical ventilation on inflammatory mediators in patients with acute respiratory distress syndrome: a randomized controlled trial. JAMA. 1999;282:54–61. doi: 10.1001/jama.282.1.54. [DOI] [PubMed] [Google Scholar]
- 21.Parsons PE, Eisner MD, Thompson BT. Lower tidal volume ventilation and plasma cytokine markers of inflammation in patients with acute lung injury. Crit Care Med. 2005;33:1–6. doi: 10.1097/01.ccm.0000149854.61192.dc. [DOI] [PubMed] [Google Scholar]
- 22.Meduri GU, Headley S, Kohler G. Persistent elevation of inflammatory cytokines predicts a poor outcome in ARDS. Plasma IL-1 beta and IL-6 levels are consistent and efficient predictors of outcome over time. Chest. 1995;107:1062–1073. doi: 10.1378/chest.107.4.1062. [DOI] [PubMed] [Google Scholar]
- 23.Meduri GU, Kohler G, Headley S, Tolley E, Stentz F, Postlethwaite A. Inflammatory cytokines in the BAL of patients with ARDS. Persistent elevation over time predicts poor outcome. Chest. 1995;108:1303–1314. doi: 10.1378/chest.108.5.1303. [DOI] [PubMed] [Google Scholar]
- 24.Katzenstein AL, Bloor CM, Leibow AA. Diffuse alveolar damage—the role of oxygen, shock, and related factors. A review. Am J Pathol. 1976;85:209–228. [PMC free article] [PubMed] [Google Scholar]
- 25.Wiener-Kronish JP, Albertine KH, Matthay MA. Differential responses of the endothelial and epithelial barriers of the lung in sheep to Escherichia coli endotoxin. J Clin Invest. 1991;88:864–875. doi: 10.1172/JCI115388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer NJ, Christie JD. Genetic heterogeneity and risk for ARDS. Semin Respir Crit Care Med. 2013;34:459–474. doi: 10.1055/s-0033-1351121. [DOI] [PubMed] [Google Scholar]
- 27.Meyer NJ. Beyond single-nucleotide polymorphisms: genetics, genomics, and other ‘omic approaches to acute respiratory distress syndrome’. Clin Chest Med. 2014;35:673–684. doi: 10.1016/j.ccm.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blondonnet R, Constantin JM, Sapin V, Jabaudon M. A pathophysiologic approach to biomarkers in acute respiratory distress syndrome. Dis Markers. 2016;2016:3501373. doi: 10.1155/2016/3501373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ware LB, Calfee CS. Biomarkers of ARDS: what's new? Intensive Care Med. 2016;42:797–799. doi: 10.1007/s00134-015-3973-0. [DOI] [PubMed] [Google Scholar]
- 30.Brown SM, Grissom CK, Rondina MT. Polymorphisms in key pulmonary inflammatory pathways and the development of acute respiratory distress syndrome. Exp Lung Res. 2015;41:155–162. doi: 10.3109/01902148.2014.983281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shortt K, Chaudhary S, Grigoryev D. Identification of novel single nucleotide polymorphisms associated with acute respiratory distress syndrome by Exome-Seq. PLoS One. 2014;9:e111953. doi: 10.1371/journal.pone.0111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sapru A, Liu KD, Wiemels J. Association of common genetic variation in the protein C pathway genes with clinical outcomes in acute respiratory distress syndrome. Crit Care. 2016;20:151. doi: 10.1186/s13054-016-1330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh CC, David S, Zhang R. Gene control of tyrosine kinase TIE2 and vascular manifestations of infections. Proc Natl Acad Sci USA. 2016;113:2472–2477. doi: 10.1073/pnas.1519467113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans CR, Karnovsky A, Kovach MA, Standiford TJ, Burant CF, Stringer KA. Untargeted LC–MS metabolomics of bronchoalveolar lavage fluid differentiates acute respiratory distress syndrome from health. J Proteome Res. 2014;13:640–649. doi: 10.1021/pr4007624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stringer KA, Serkova NJ, Karnovsky A, Guire K, Paine R, Standiford TJ. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma 1H-nuclear magnetic resonance quantitative metabolomics and computational analysis. Am J Physiol Lung Cell Mol Physiol. 2011;300:L4–11. doi: 10.1152/ajplung.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calfee CS, Janz DR, Bernard GR. Distinct molecular phenotypes of direct versus indirect ARDS in single-center and multicenter studies. Chest. 2015;147:1539–1548. doi: 10.1378/chest.14-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blondonnet R, Constantin JM, Sapin V, Jabaudon M. A pathophysiologic approach to biomarkers in acute respiratory distress syndrome. Dis Markers. 2016;2016:20. doi: 10.1155/2016/3501373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forel JM, Guervilly C, Hraiech S. Type III procollagen is a reliable marker of ARDS-associated lung fibroproliferation. Intensive Care Med. 2015;41:1–11. doi: 10.1007/s00134-014-3524-0. [DOI] [PubMed] [Google Scholar]
- 39.Battaglia M, Atkinson MA. The streetlight effect in type 1 diabetes. Diabetes. 2015;64:1081–1090. doi: 10.2337/db14-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manolio TA, Collins FS, Cox NJ. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christie JD, Wurfel MM, Feng R. Genome wide association identifies PPFIA1 as a candidate gene for acute lung injury risk following major trauma. PLoS One. 2012;7:e28268. doi: 10.1371/journal.pone.0028268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 43.Copeland KT, Checkoway H, McMichael AJ, Holbrook RH. Bias due to misclassification in the estimation of relative risk. Am J Epidemiol. 1977;105:488–495. doi: 10.1093/oxfordjournals.aje.a112408. [DOI] [PubMed] [Google Scholar]
- 44.Tejera P, Meyer NJ, Chen F. Distinct and replicable genetic risk factors for acute respiratory distress syndrome of pulmonary or extrapulmonary origin. J Med Genet. 2012;49:671–680. doi: 10.1136/jmedgenet-2012-100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tennessen JA, Bigham AW, O'Connor TD. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Posey JE, Harel T, Liu P. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Djebali S, Davis CA, Merkel A. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson G, Powell JE, Marigorta UM. Expression quantitative trait locus analysis for translational medicine. Genome Med. 2015;7:60. doi: 10.1186/s13073-015-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palakshappa JA, Meyer NJ. Which patients with ARDS benefit from lung biopsy? Chest. 2015;148:1073–1082. doi: 10.1378/chest.15-0076. [DOI] [PubMed] [Google Scholar]
- 52.Kovach MA, Stringer KA, Bunting R. Microarray analysis identifies IL-1 receptor type 2 as a novel candidate biomarker in patients with acute respiratory distress syndrome. Respir Res. 2015;16:29. doi: 10.1186/s12931-015-0190-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aggarwal NR, King LS, D'Alessio FR. Diverse macrophage populations mediate acute lung inflammation and resolution. Am J Physiol Lung Cell Mol Physiol. 2014;306:L709–L725. doi: 10.1152/ajplung.00341.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kangelaris KN, Prakash A, Liu KD. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. Am J Physiol Lung Cell Mol Physiol. 2015;308:L1102–L1113. doi: 10.1152/ajplung.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howrylak JA, Dolinay T, Lucht L. Discovery of the gene signature for acute lung injury in patients with sepsis. Physiol Genomics. 2009;37:133–139. doi: 10.1152/physiolgenomics.90275.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers AJ, Matthay MA. Applying metabolomics to uncover novel biology in ARDS. Am J Physiol Lung Cell Mol Physiol. 2014;306:L957–L961. doi: 10.1152/ajplung.00376.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhargava M, Becker TL, Viken KJ. Proteomic profiles in acute respiratory distress syndrome differentiates survivors from non-survivors. PLoS One. 2014;9:e109713. doi: 10.1371/journal.pone.0109713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X, Shan Q, Jiang L, Zhu B, Xi X. Quantitative proteomic analysis by iTRAQ for identification of candidate biomarkers in plasma from acute respiratory distress syndrome patients. Biochem Biophys Res Commun. 2013;441:1–6. doi: 10.1016/j.bbrc.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 59.Gerszten RE, Accurso F, Bernard GR. Challenges in translating plasma proteomics from bench to bedside: update from the NHLBI Clinical Proteomics Programs. Am J Physiol Lung Cell Mol Physiol. 2008;295:16–22. doi: 10.1152/ajplung.00044.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bos LDJ, Weda H, Wang Y. Exhaled breath metabolomics as a noninvasive diagnostic tool for acute respiratory distress syndrome. Eur Respir J. 2014;44:188–197. doi: 10.1183/09031936.00005614. [DOI] [PubMed] [Google Scholar]
- 61.Gajic O, Dabbagh O, Park PK. Early identification of patients at risk of acute lung injury: evaluation of lung injury prediction score in a multicenter cohort study. Am J Respir Crit Care Med Medicine. 2011;183:462–470. doi: 10.1164/rccm.201004-0549OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trillo-Alvarez C, Cartin-Ceba R, Kor DJ. Acute lung injury prediction score: derivation and validation in a population-based sample. Eur Respir J. 2011;37:604–609. doi: 10.1183/09031936.00036810. [DOI] [PubMed] [Google Scholar]
- 63.Dickson RP, Singer BH, Newstead MW. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernard GR, Artigas A, Brigham KL. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149:818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt EP, Li G, Li L. The circulating glycosaminoglycan signature of respiratory failure in critically ill adults. J Biol Chem. 2014;289:8194–8202. doi: 10.1074/jbc.M113.539452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mrozek S, Jabaudon M, Jaber S. Elevated plasma levels of sRAGE are associated with nonfocal CT-based lung imaging in patients with ARDS: a prospective multicenter study. Chest. 2016;150:998–1007. doi: 10.1016/j.chest.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 67.Calfee CS, Eisner MD, Ware LB. Trauma-associated lung injury differs clinically and biologically from acute lung injury due to other clinical disorders. Crit Care Med. 2007;35:2243–2250. doi: 10.1097/01.ccm.0000280434.33451.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Constantin JM, Grasso S, Chanques G. Lung morphology predicts response to recruitment maneuver in patients with acute respiratory distress syndrome. Crit Care Med. 2010;38:1108–1117. doi: 10.1097/CCM.0b013e3181d451ec. [DOI] [PubMed] [Google Scholar]
- 69.Haldar P, Pavord ID, Shaw DE. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–224. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore WC, Meyers DA, Wenzel SE. Identification of asthma phenotypes using cluster analysis in the severe asthma research program. Am J Respir Crit Care Med. 2010;181:315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong HR, Cvijanovich N, Lin R. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong HR, Cvijanovich NZ, Anas N. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med. 2015;191:309–315. doi: 10.1164/rccm.201410-1864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 74.Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Softw. 2014;61:36. [Google Scholar]
- 75.Churpek MM, Yuen TC, Winslow C, Meltzer DO, Kattan MW, Edelson DP. Multicenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med. 2016;44:368–374. doi: 10.1097/CCM.0000000000001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong HR, Lindsell CJ, Pettila V. A multi-biomarker-based outcome risk stratification model for adult septic shock. Crit Care Med. 2014;42:781–789. doi: 10.1097/CCM.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown LM, Calfee CS, Matthay MA, Brower RG, Thompson BT, Checkley W. A simple classification model for hospital mortality in patients with acute lung injury managed with lung protective ventilation. Crit Care Med. 2011;39:2645–2651. doi: 10.1097/CCM.0b013e3182266779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 79.Howard R, Rattray M, Prosperi M, Custovic A. Distinguishing asthma phenotypes using machine learning approaches. Curr Allergy Asthma Rep. 2015;15:38. doi: 10.1007/s11882-015-0542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reilly JP, Bellamy S, Shashaty MGS. Heterogeneous phenotypes of the acute respiratory distress syndrome after major trauma. Ann Am Thorac Soc. 2014;11:728–736. doi: 10.1513/AnnalsATS.201308-280OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Agrawal A, Matthay MA, Kangelaris KN. Plasma angiopoietin-2 predicts the onset of acute lung injury in critically ill patients. Am J Respir Crit Care Med. 2013;187:736–742. doi: 10.1164/rccm.201208-1460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calfee CS, Gallagher D, Abbott J, Thompson BT, Matthay MA. Plasma angiopoietin-2 in clinical acute lung injury: prognostic and pathogenetic significance. Crit Care Med. 2012;40:1731–1737. doi: 10.1097/CCM.0b013e3182451c87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fremont RD, Koyama T, Calfee CS. Acute lung injury in patients with traumatic injuries: utility of a panel of biomarkers for diagnosis and pathogenesis. J Trauma. 2010;68:1121–1127. doi: 10.1097/TA.0b013e3181c40728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meyer NJ, Li M, Feng R. ANGPT2 genetic variant is associated with trauma-associated acute lung injury and altered plasma angiopoietin-2 isoform ratio. Am J Respir Crit Care Med. 2011;183:1344–1353. doi: 10.1164/rccm.201005-0701OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Westra HJ, Peters MJ, Esko T. Systematic identification of trans-eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Imai K, Keele L, Tingley D. A general approach to causal mediation analysis. Psychol Methods. 2010;15:309–334. doi: 10.1037/a0020761. [DOI] [PubMed] [Google Scholar]
- 87.Wei Y, Wang Z, Su L. Platelet count mediates the contribution of a genetic variant in LRRC 16A to ARDS risk. Chest. 2015;147:607–617. doi: 10.1378/chest.14-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wei Y, Tejera P, Wang Z. A missense genetic variant in LRRC16A/CAMIL1 improves ARDS survival by attenuating platelet count decline. Am J Respir Crit Care Med. 2016 doi: 10.1164/rccm.201605-0946OC. published online Oct 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 90.Cohen JC. Emerging LDL therapies: using human genetics to discover new therapeutic targets for plasma lipids. J Clin Lipidol. 2013;7(suppl 3):S1–S5. doi: 10.1016/j.jacl.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huh D, Leslie DC, Matthews BD. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cypel M, Yeung JC, Liu M. Normothermic ex vivo lung perfusion in clinical lung transplantation. N Engl J Med. 2011;364:1431–1440. doi: 10.1056/NEJMoa1014597. [DOI] [PubMed] [Google Scholar]
- 93.Chaney J, Suzuki Y, Cantu E, van Berkel V. Lung donor selection criteria. J Thorac Dis. 2014;6:1032–1038. doi: 10.3978/j.issn.2072-1439.2014.03.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McAuley DF, Curley GF, Hamid UI. Clinical grade allogeneic human mesenchymal stem cells restore alveolar fluid clearance in human lungs rejected for transplantation. Am J Physiol Lung Cell Mol Physiol. 2014;306:L809–L815. doi: 10.1152/ajplung.00358.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee JW, Krasnodembskaya A, McKenna DH, Song Y, Abbott J, Matthay MA. Therapeutic effects of human mesenchymal stem cells in ex vivo human lungs injured with live bacteria. Am J Respir Crit Care Med. 2013;187:751–760. doi: 10.1164/rccm.201206-0990OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gores KM, Delsing AS, Kraus SJ. Plasma angiopoietin-2 concentrations are related to impaired lung function, and organ failure in a clinical cohort receiving high dose interleukin-2 therapy. Shock. 2014;42:115–120. doi: 10.1097/SHK.0000000000000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cockrell AS, Yount BL, Scobey T. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol. 2016;2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flaherty KT, Robert C, Hersey P. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 100.Kris MG, Johnson BE, Berry LD. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ortega HG, Liu MC, Pavord ID. Mepolizumab treatment in patients with severe eosinophilic asthma. N Engl J Med. 2014;371:1198–1207. doi: 10.1056/NEJMoa1403290. [DOI] [PubMed] [Google Scholar]
- 102.Freidlin B, Korn EL. Biomarker enrichment strategies: matching trial design to biomarker credentials. Nat Rev Clin Oncol. 2014;11:81–90. doi: 10.1038/nrclinonc.2013.218. [DOI] [PubMed] [Google Scholar]
- 103.Mehta C, Schafer H, Daniel H, Irle S. Biomarker driven population enrichment for adaptive oncology trials with time to event endpoints. Stat Med. 2014;33:4515–4531. doi: 10.1002/sim.6272. [DOI] [PubMed] [Google Scholar]
- 104.Geiss GK, Bumgarner RE, Birditt B. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 105.Cuenca AG, Gentile LF, Lopez MC. Development of a genomic metric that can be rapidly used to predict clinical outcome in severely injured trauma patients. Crit Care Med. 2013;41:1175–1185. doi: 10.1097/CCM.0b013e318277131c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Calfee CS, Ware LB, Eisner MD. Plasma receptor for advanced glycation end products and clinical outcomes in acute lung injury. Thorax. 2008;63:1083–1089. doi: 10.1136/thx.2008.095588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matthay MA, McAuley DF, Ware LB. Clinical trials in ARDS: challenges and opportunities. Lancet Respir Med. 2017;5:524–534. doi: 10.1016/S2213-2600(17)30188-1. [DOI] [PubMed] [Google Scholar]