Highlights
-
•
Long-term multicentre surveillance of antimicrobial susceptibility patterns in Shigella sonnei.
-
•
Epidemic clones and integron types and resistance gene cassettes were characterised.
-
•
PFGE indicated large-scale clonal transmission among different cities occurred several times during 10 years.
-
•
Class 1, 2 and atypical class 1 integrons were detected in S. sonnei.
-
•
High prevalence of integrons and gene cassettes was related to the increasing antimicrobial resistance.
Keywords: Shigella sonnei, Resistance, Multidrug-resistant, PFGE, Integron
Abstract
Objectives
The rapid emergence of drug-resistant Shigella sonnei is a serious public health problem. This study aimed to characterise the antimicrobial resistance patterns, molecular subtypes, and integron types and resistance gene cassettes in S. sonnei from Jiangsu Province, China.
Methods
In total, 340 S. sonnei were collected in 2002–2011 throughout Jiangsu Province. Antimicrobial susceptibility testing, pulsed-field gel electrophoresis (PFGE), PCR amplification of integrons, restriction fragment length polymorphism (RFLP) and DNA sequencing of cassette regions were performed.
Results
Resistance rates to ampicillin (67.7%), nalidixic acid (75.2%), tetracycline (73.7%) and trimethoprim/sulfamethoxazole (68.7%) remained high. Strains from Centre and South Jiangsu showed higher resistance and multiresistance rates compared with the North. PFGE analysis indicated that large-scale clonal transmission among different cities occurred several times during 10 years. Among all strains, 55.9% (190/340) harboured class 1 integrons, 80.3% (273/340) harboured class 2 integrons and 49.4% (168/340) harboured an atypical class 1 integron. Resistance rates to nine antimicrobials in the class 1 integron-positive group were significantly higher than in the negative group (P < 0.05). Seven different gene cassettes were detected in class 1 integrons. The most prevalent type was aacA4–cmlA1 (114/286). Class 2 integrons carried the gene cassette array dfrA1–sat1–aadA1, and the atypical class 1 integron carried blaOXA-30–aadA1.
Conclusions
The increasing antimicrobial resistance and significant clonal transmission of S. sonnei circulating in Jiangsu were closely related to the high prevalence of integrons and gene cassettes. Long-term cross-regional monitoring of antimicrobial resistance is urgently required for S. sonnei.
1. Introduction
Shigella is a major gastrointestinal pathogen accounting for 5–10% of diarrhoeal diseases or bacillary dysentery throughout the world [1]. In China, up to 1.7 million episodes of shigellosis occurred each year, with up to 0.20 million patients hospitalised [2]. Among the four species of Shigella (Shigella dysenteriae, Shigella flexneri, Shigella boydii and Shigella sonnei), S. sonnei has been the predominant species in developed countries and is often the second most prevalent species in developing countries for many years [3], [4], [5]. Recently, S. sonnei has become increasingly important due to the growing rate of this species in many developing countries [6], [7].
Antimicrobial treatment is usually recommended for shigellosis as it can reduce the duration of symptoms and the severity of illness by shortening the period of pathogen excretion [8]. However, owing to long-term overuse of antimicrobials, drug-resistant and multidrug-resistant (MDR) Shigella isolates prevail worldwide [9], [10]. Evidence from China [11], Bangladesh [5], Belgium [12], Gabon [13] and the USA [14] indicated that China has a much higher prevalence of resistance to most commonly used antimicrobials.
The close correlation between antimicrobial resistance and the presence of integrons containing gene cassettes has been frequently reported in studies from various countries and regions [10], [15], [16]. However, limited information is available on the following three aspects: the regional level of the antimicrobial resistance profile; the genotype level of the dominating endemic strains; and long-term surveillance and comparison of integrons and gene cassettes.
Therefore, this study was designed to investigate the antimicrobial resistance patterns, epidemic clones and integrons characteristics of strains collected during a 10-year period (2002–2011) in Jiangsu Province, China.
2. Materials and methods
2.1. Sample collection
A total of 340 S. sonnei were collected between 2002 and 2011 from outpatients and inpatients in hospitals throughout Jiangsu Province. Jiangsu Province, a populous and prosperous province in China, was divided into three regions comprised of 13 provincial cities, the South (Nanjing, Suzhou, Wuxi, Changzhou and Zhenjiang), the Centre (Yangzhou, Taizhou and Nantong) and the North (Xuzhou, Suqian, Huai’an, Yancheng and Lianyungang). As shown in Table 1 , South Jiangsu exhibited the highest isolation rate of S. sonnei (186/340; 54.7%), followed by the Centre (84/340; 24.7%) and the North (70/340; 20.6%).
Table 1.
Source of Shigella sonnei strains (n = 340), by year and by city, 2002–2011.
| Characteristic | No. of strains | % |
|---|---|---|
| Year | ||
| 2002 | 6 | 1.8 |
| 2003 | 2 | 0.6 |
| 2004 | 6 | 1.8 |
| 2005 | 7 | 2.1 |
| 2006 | 19 | 5.6 |
| 2007 | 48 | 14.1 |
| 2008 | 89 | 26.2 |
| 2009 | 98 | 28.8 |
| 2010 | 33 | 9.7 |
| 2011 | 32 | 9.4 |
| City | ||
| Changzhou | 75 | 22.1 |
| Huai’an | 15 | 4.4 |
| Lianyungang | 21 | 6.2 |
| Nanjing | 36 | 10.6 |
| Nantong | 63 | 18.5 |
| Suqian | 0 | 0.0 |
| Suzhou | 22 | 6.5 |
| Taizhou | 4 | 1.2 |
| Wuxi | 28 | 8.2 |
| Xuzhou | 27 | 7.9 |
| Yancheng | 7 | 2.1 |
| Yangzhou | 17 | 5.0 |
| Zhenjiang | 25 | 7.4 |
All strains were identified by API system (bioMérieux, Marcy-l’Étoile, France) and were confirmed as S. sonnei by slide agglutination with Shigella antisera (Lanzhou Institute of Biological Products, Lanzhou, China).
2.2. Antimicrobial susceptibility testing
Susceptibility to nine commonly used antimicrobials was determined by the disk diffusion method. The antimicrobial agents were as follows: ampicillin (10 μg); amoxicillin/clavulanic acid (AMC) (30 μg); cefalotin (30 μg); cefotaxime (30 μg); gentamicin (120 μg); nalidixic acid (30 μg); norfloxacin (10 μg); tetracycline (30 μg); and trimethoprim/sulfamethoxazole (SXT) (25 μg). Results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [17]. Escherichia coli ATCC 25922 was used as a quality control strain.
In view of the small sample size (n = 21 strains) and different antimicrobials used in antimicrobial susceptibility testing in 2002–2005, this study only compared the resistance rates of S. sonnei collected in 2006–2011.
2.3. Pulsed-field gel electrophoresis (PFGE)
To determine DNA fingerprinting profiles, the 340 S. sonnei strains were analysed by PFGE according to the US Centers for Disease Control and Prevention (CDC) PulseNet protocol [18] and were digested with the restriction enzyme Xbal (Takara, Dalian, China). Electrophoresis was performed with a CHEF-DR II system (Bio-Rad, Hercules, CA) under the following conditions: switching time from 2.2 s to 54.2 s at 6 V for 17 h with an angle of 120°. Salmonella enterica serovar Braenderup H9812 was used as a molecular weight standard. PFGE banding patterns were compared using BioNumerics software v.4.0 (Applied Maths, Sint-Martens-Latem, Belgium) based on the Dice coefficient. Dendrograms were constructed by the unweighted-pair group method with arithmetic mean (UPGMA), with a position tolerance of 1% and optimisation of 0.5%. Strains sharing ≥90% similarity in DNA fingerprinting profiles were defined as a cluster [19].
2.4. Identification of integrons and gene cassettes
Class 1, 2 and 3 integrons were detected by PCR using degenerate primers hep35 and hep36 targeted to conserved regions of integrase genes intI1, intI2 and intI3 [20]. PCR products were analysed by restriction fragment length polymorphism (RFLP) to classify integrons [21]. Atypical class 1 integrons were also screened in S. sonnei using primers int1f and is16 [20].
The variable region of class 1 and 2 integrons was amplified using the primers 5′CS1/3′CS1 and 5′CS2/3′CS2, respectively, as described previously [22]. PCR products of variable regions were digested by the restriction enzyme HinfI (Takara). Same-sized amplicons with the same RFLP pattern were considered as one type. At least one representative of each type of amplicon was sequenced to identify the gene cassettes. Sequence analysis and alignment was performed using BLAST programs available at the National Center for Biotechnology Information (NCBI) website.
2.5. Statistical analysis
Statistical analysis was performed with Stata v.7.0 software (StataCorp LP, College Station, TX). Pearson’s χ2 test was used to analyse the antimicrobial resistance of isolates from different regions. Fisher’s exact test was applied to compare antimicrobial resistance between integron-positive and integron-negative groups. A P-value of <0.05 was considered statistically significant.
3. Results
3.1. Resistance trends from 2006 to 2011
A wide of spectrum of antimicrobial resistance was observed in 319 S. sonnei strains isolated from 2006 to 2011 (Table 2 ). The resistance rates to ampicillin (67.7%), nalidixic acid (75.2%), tetracycline (73.7%) and SXT (68.7%) remained at a high level during 6 years, whereas resistance to cefotaxime and norfloxacin was relatively lower (17.9% and 21.0%, respectively). Of the nine antimicrobials tested, a sharp resistance change was observed for gentamicin, with a three-fold increase from 26.3% in 2006 to 78.1% in 2011. A decrease in resistance to AMC (from 52.6% to 18.8%) was observed.
Table 2.
Antimicrobial resistance of Shigella sonnei, 2006–2011.
| Antimicrobial agent | Resistant [% (n)] |
||||||
|---|---|---|---|---|---|---|---|
| 2006 (N = 19) | 2007 (N = 48) | 2008 (N = 89) | 2009 (N = 98) | 2010 (N = 33) | 2011 (N = 32) | Overall (N = 319) | |
| Ampicillin | 52.6 (10) | 81.3 (39) | 48.3 (43) | 77.6 (76) | 60.6 (20) | 87.5 (28) | 67.7 (216) |
| AMC | 52.6 (10) | 97.9 (47) | 39.3 (35) | 30.6 (30) | 0.0 (0) | 18.8 (6) | 40.1 (128) |
| Cefalotin | 52.6 (10) | 33.3 (16) | 15.7 (14) | 45.9 (45) | 27.3 (9) | 25.0 (8) | 32.0 (102) |
| Cefotaxime | 5.3 (1) | 14.6 (7) | 12.4 (11) | 30.6 (30) | 21.2 (7) | 3.1 (1) | 17.9 (57) |
| Gentamicin | 26.3 (5) | 8.3 (4) | 39.3 (35) | 34.7 (34) | 33.3 (11) | 78.1 (25) | 35.7 (114) |
| Nalidixic acid | 78.9 (15) | 95.8 (46) | 61.8 (55) | 76.5 (75) | 63.6 (21) | 87.5 (28) | 75.2 (240) |
| Norfloxacin | 26.3 (5) | 25.0 (12) | 4.5 (4) | 46.9 (46) | 0.0 (0) | 0.0 (0) | 21.0 (67) |
| Tetracycline | 73.7 (14) | 93.8 (45) | 58.4 (52) | 73.5 (72) | 72.7 (24) | 87.5 (28) | 73.7 (235) |
| SXT | 68.4 (13) | 72.9 (35) | 64.0 (57) | 65.3 (64) | 81.8 (27) | 71.9 (23) | 68.7 (219) |
AMC, amoxicillin/clavulanic acid; SXT, trimethoprim/sulfamethoxazole.
3.2. Regional distribution of resistance rates
Of the 319 S. sonnei strains isolated from 2006 to 2011, 176 were collected from South Jiangsu, 78 from the Centre and 65 from the North (Table 3 ). The proportions of resistant strains from these three regions showed a significant difference for eight of the nine antimicrobials tested, the only exception being tetracycline. Resistance to ampicillin, cefalotin, cefotaxime and norfloxacin was higher in Centre Jiangsu, whilst resistance to gentamicin, nalidixic acid and SXT was higher in the South. The North isolates showed low rates of antimicrobial resistance.
Table 3.
Antimicrobial resistance of Shigella sonnei from South, Centre and North Jiangsu, 2006–2011.
| Antimicrobial agent | Resistant [% (n)] |
P-value | ||
|---|---|---|---|---|
| South (N = 176) | Centre (N = 78) | North (N = 65) | ||
| Ampicillin | 69.9 (123) | 76.9 (60) | 50.8 (33) | 0.003 |
| AMC | 38.1 (67) | 33.3 (26) | 53.8 (35) | 0.032 |
| Cefalotin | 24.4 (43) | 51.3 (40) | 29.2 (19) | <0.001 |
| Cefotaxime | 13.6 (24) | 30.8 (24) | 13.8 (9) | 0.003 |
| Gentamicin | 40.9 (72) | 38.5 (30) | 18.5 (12) | 0.005 |
| Nalidixic acid | 79.0 (139) | 78.2 (61) | 61.5 (40) | 0.016 |
| Norfloxacin | 9.1 (16) | 48.7 (38) | 20.0 (13) | <0.001 |
| Tetracycline | 76.7 (135) | 75.6 (59) | 63.1 (41) | 0.093 |
| STX | 76.1 (134) | 65.4 (51) | 52.3 (34) | 0.001 |
AMC, amoxicillin/clavulanic acid; SXT, trimethoprim/sulfamethoxazole.
3.3. Time and regional distribution of multidrug-resistant (MDR) Shigella sonnei
Multidrug resistance (resistance to three or more classes of antimicrobial agents) was detected in 74.3% (237/319) of the strains, and the MDR rates remained at a high level from 2006 to 2011 (78.9%, 93.8%, 61.8%, 75.5%, 60.6% and 87.5%, respectively).
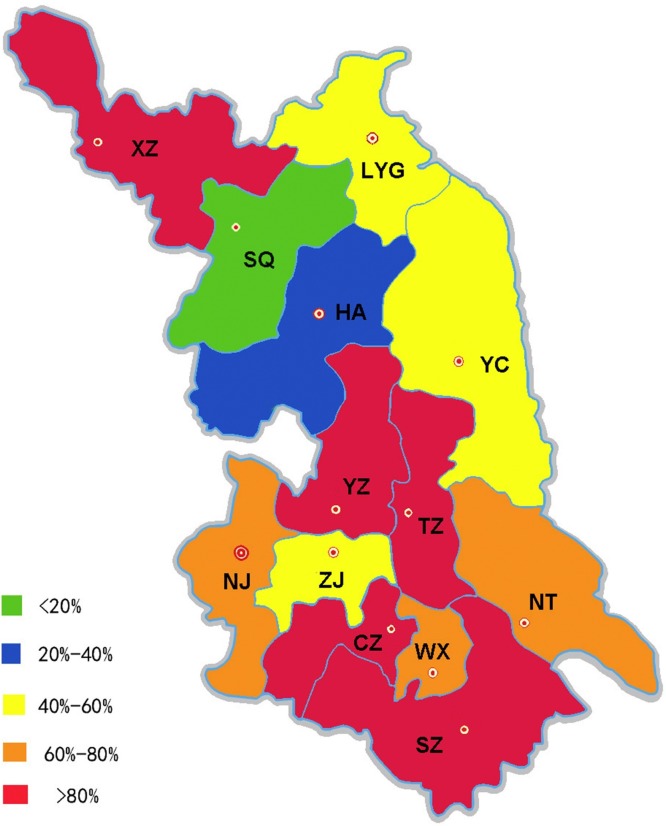
This study analysed the MDR rates of different regions in Jiangsu Province (Fig. 1 ). MDR strains were most frequent among South Jiangsu (77.3%), followed by the Centre (76.9%) and the North (63.1%). MDR rates in 13 cities are displayed in Fig. 1: Taizhou (100.0%), Xuzhou (91.7%) and Suzhou (90.5%) showing the highest MDR rates.
Fig. 1.
Multidrug resistance rates in different cities of Jiangsu Province. CZ, Changzhou; HA, Huai’an; LYG, Lianyungang; NJ, Nanjing; NT, Nantong; SQ, Suqian; SZ, Suzhou; TZ, Taizhou; WX, Wuxi; XZ, Xuzhou; YC, Yancheng; YZ, Yangzhou; ZJ, Zhenjiang.
3.4. Pulsed-field gel electrophoresis profile
Among 32 S. sonnei isolated in 2011, three PFGE clusters were identified. A total of 31 isolates were grouped into cluster SA1 (24 isolates), SA2 (2 isolates) and SA4 (5 isolates). These clusters spread in five, one and two cities, respectively (Supplementary Fig. S1A).
Among 33 S. sonnei isolated in 2010, five PFGE clusters were observed. All 33 isolates were grouped into cluster SB1 (15 isolates), SB2 (3 isolates), SB3 (2 isolates), SB4 (6 isolates) and SB5 (7 isolates). Two (40.0%) of the five clusters spread in two cities, one (20.0%) in four cities, one (20.0%) in five cities and, notably, there was one PFGE cluster spread in eight different cities (Supplementary Fig. S1B).
During 10 years, similar clonal transmission among different cities was revealed several times in Jiangsu Province. The dendrograms of PFGE patterns of all strains are displayed in Supplementary material.
3.5. Integrons detected in Shigella sonnei
The detection rate of integrons in S. sonnei was generally high (Table 4 ). Among all strains, 55.9% (190/340) harboured class 1 integrons, 80.3% (273/340) harboured class 2 integrons and 49.4% (168/340) harboured an atypical class 1 integron. None of the strains harboured a class 3 integron.
Table 4.
Rates of integron-positive strains from 2002–2011.
| Year | Class 1 integron | Class 2 integron | Atypical class 1 integron |
|---|---|---|---|
| 2002 | 0.0 (0/6) | 100.0 (6/6) | 100.0 (6/6) |
| 2003 | 50.0 (1/2) | 50.0 (1/2) | 100.0 (2/2) |
| 2004 | 50.0 (3/6) | 100.0 (6/6) | 100.0 (6/6) |
| 2005 | 100.0 (7/7) | 100.0 (7/7) | 100.0 (7/7) |
| 2006 | 57.9 (11/19) | 94.7 (18/19) | 84.2 (16/19) |
| 2007 | 77.1 (37/48) | 93.8 (45/48) | 87.5 (42/48) |
| 2008 | 33.7 (30/89) | 75.3 (67/89) | 23.6 (21/89) |
| 2009 | 66.3 (65/98) | 74.5 (73/98) | 56.1 (55/98) |
| 2010 | 48.5 (16/33) | 72.7 (24/33) | 18.2 (6/33) |
| 2011 | 62.5 (20/32) | 81.3 (26/32) | 21.9 (7/32) |
| Total | 55.9 (190/340) | 80.3 (273/340) | 49.4 (168/340) |
To determine the role of class 1 integron in drug resistance, strains were divided into two groups (class 1 integron-positive and class 1 integron-negative). Resistance rates to the nine antimicrobials in the class 1 integron-positive group were significantly higher than the class 1 integron-negative group (P < 0.05) (Table 5 ).
Table 5.
Correlation between antibiogram profile and class 1 integrons in Shigella sonnei, 2006–2011.
| Antimicrobial agent | Class 1 integron-positive (N = 179) |
Class 1 integron-negative group (N = 140) |
P-value | ||||
|---|---|---|---|---|---|---|---|
| %R (n) | %I (n) | %S (n) | %R (n) | %I (n) | %S (n) | ||
| Ampicillin | 86.6 (155) | 0.6 (1) | 12.8 (23) | 43.6 (61) | 2.9 (4) | 53.6 (75) | <0.001 |
| AMC | 50.3 (90) | 15.1 (27) | 34.6 (62) | 27.1 (38) | 6.4 (9) | 66.4 (93) | <0.001 |
| Cefalotin | 39.7 (71) | 22.9 (41) | 37.4 (67) | 22.1 (31) | 25.7 (36) | 52.1 (73) | 0.003 |
| Cefotaxime | 23.5 (42) | 14.0 (25) | 62.6 (112) | 10.7 (15) | 9.3 (13) | 80.0 (112) | 0.002 |
| Gentamicin | 40.8 (73) | 6.7 (12) | 52.5 (94) | 29.3 (41) | 2.9 (4) | 67.9 (95) | 0.015 |
| Nalidixic acid | 88.8 (159) | 2.2 (4) | 8.9 (16) | 57.9 (81) | 9.3 (13) | 32.9 (46) | <0.001 |
| Norfloxacin | 31.3 (56) | 5.0 (9) | 63.7 (114) | 7.9 (11) | 5.0 (7) | 87.1 (122) | <0.001 |
| Tetracycline | 87.7 (157) | 0.6 (1) | 11.7 (21) | 55.7 (78) | 2.9 (4) | 41.4 (58) | <0.001 |
| SXT | 78.8 (141) | 2.8 (5) | 18.4 (33) | 55.7 (78) | 10.0 (14) | 34.3 (48) | <0.001 |
%R/I/S, percent resistant/intermediate/susceptible; AMC, amoxicillin/clavulanic acid; SXT, trimethoprim/sulfamethoxazole.
3.6. Resistance gene cassettes of integrons
The variable regions of class 1, class 2 and atypical class 1 integrons were amplified and analysed in all integrase-positive strains. Five amplicons (0.15, 0.75, 1.5, 1.6 and 2.2 kb) were identified in variable regions of class 1 integron. Sequencing of the variable region revealed that the 0.75-kb amplicon carried one gene cassette (dfrA5); the 1.5-, 1.6- and 2.2-kb amplicons carried two gene cassettes (dfrA1–aadA1, dfrA17–aadA5 and aacA4–cmlA1, respectively); the 0.15-kb amplicon carried 5′CS and 3′CS (Table 6 ). In total, seven different gene cassettes were detected in class 1 integrons. The most prevalent type was aacA4–cmlA1 (114/286).
Table 6.
Gene cassette arrays in class 1 integrons from Shigella sonnei, 2002–2011.
| Approximate length (kb) | No. of isolates | Gene cassette array |
|---|---|---|
| 0.15 | 22 | 5′CS and 3′CS |
| 1.5 | 5 | dfrA1–aadA1 |
| 1.6 | 4 | dfrA17–aadA5 |
| 2.2 | 42 | aacA4–cmlA1 |
| 0.15 + 1.5 | 1 | 5′CS and 3′CS, dfrA1–aadA1 |
| 0.15 + 2.2 | 52 | 5′CS and 3′CS, aacA4–cmlA1 |
| 0.75 + 2.2 | 1 | dfrA5, aacA4–cmlA1 |
| 1.5 + 2.2 | 13 | dfrA1–aadA1, aacA4–cmlA1 |
| 1.6 + 2.2 | 1 | dfrA17–aadA5, aacA4–cmlA1 |
| 0.15 + 0.75 + 2.2 | 1 | 5′CS and 3′CS, dfrA5, aacA4–cmlA1 |
| 0.15 + 1.5 + 2.2 | 4 | 5′CS and 3′CS, dfrA1–aadA1, aacA4–cmlA1 |
Only one amplicon of 2.0 kb in size was identified in class 2 integrons, which carried dfrA1–sat1–aadA1. The gene cassette array bla OXA-30–aadA1 was detected in a 2.4-kb amplicon of an atypical class 1 integron.
4. Discussion
In this study, only 40 (11.8%) of 340 S. sonnei isolates were obtained from 2002 to 2006, whereas 300 isolates (88.2%) were obtained in the following 5 years in Jiangsu Province, which was a rapidly developing area in China. The noticeable increase of S. sonnei has also been observed in other fast-developing countries [23], and in some areas such as Beijing and Vietnam [6], [24]. S. sonnei has even replaced S. flexneri to become the predominant subgroup. The pattern shift might due to improvements in the economic situation, environmental conditions and hygiene habits in these regions. Another finding was that only two S. sonnei isolates were collected in 2003 during the severe acute respiratory syndrome (SARS) epidemic. Since the main transmission mode of Shigella is person-to-person contact or consumption of contaminated food [25], fewer people getting out and eating out in order to avoid SARS infections may have resulted in the obvious decrease of S. sonnei isolates.
Antimicrobial resistance of Shigella has become one of the most serious global public health concerns [14]. Over the past few decades, Shigella have become resistant to most of the widely used antimicrobials [26], [27]. In this study, the resistance patterns of S. sonnei were analysed from three different angles: changing resistance categorised by year; changing resistance categorised by geographical region; and MDR analysis.
In this study during the period 2006–2011, more than one-half of S. sonnei isolates were resistant to ampicillin (67.7%), nalidixic acid (75.2%), tetracycline (73.7%) and SXT (68.7%). Resistance to these antimicrobials is commonly reported and the frequency of resistance differs by country. In Belgium, an 18-year research showed that resistance rates to these four antimicrobials were 19.2%, 3.8%, 59.3% and 85.9%, respectively [12]. In the USA during 2000–2010 [28], resistance rates to the common drugs nalidixic acid (1%), tetracycline (18%) and SXT (33%) were dramatically lower than those observed in the current study, whilst ampicillin had a similar resistance rate of 74%. In addition, in Eastern China high levels of resistance to ampicillin (70.8%), nalidixic acid (69.3%), tetracycline (74.8%) and SXT (73.8%) were reported [9]. The widespread nature of nalidixic acid and fluoroquinolone resistance has been documented in many countries [29], [30]. In the current study, resistance to nalidixic acid was found to be very high, with a peak at 95.8% in 2007, whilst the third-generation fluoroquinolone norfloxacin showed a lower resistance rate of 21.0% on average. Cefotaxime also had a lower frequency of resistance compared with cefalotin. Thus, the third-generation fluoroquinolones and cephalosporins are recommended as first-line drugs to treat infections caused by S. sonnei.
This study further analysed the resistance of S. sonnei in different regions of Jiangsu Province. Resistance rates of isolates from the South, Centre and North differed significantly for eight of nine antimicrobials tested. North Jiangsu exhibited low resistance rates compared with the Centre and the South. The great geographical variation found in resistance patterns may be ascribed to the various prescribing practises of doctors in different regions. This calls for strong monitoring of the local resistance patterns in order to provide effective empirical treatment regimens.
Together with the increased single-drug resistance, the frequency of MDR isolates during 6 years remained at a high level, slowly rising from 78.9% in 2006 to 87.5% in 2011. The same trend had already been revealed a few years ago in a long-term surveillance in Belgium, where the MDR rate of S. sonnei increased from 55.9% in 1990 to 80.2% in 2007, reaching a peak of 89.0% during 2004–2005 [12]. Geographical distribution of multidrug resistance indicated that South Jiangsu represented the predominant MDR region (77.3%), followed by the Centre (76.9%) and the North (63.1%).
The declining susceptibility to commonly used antimicrobials and the emergence of MDR S. sonnei could be highly related to inappropriate prescription and easy access to antimicrobials among outpatients. It is therefore important to strictly control antimicrobial application and to monitor local resistance in order to facilitate the rational use of antimicrobial agents.
This high frequency of MDR S. sonnei strains has led to tremendous interest in molecular typing and determination of resistance mechanisms. Bacterial molecular typing is frequently applied for outbreak surveillance and phylogenetic investigation. Strain-specific fingerprints generated are used to facilitate the identification of disease transmission routes and sources [7], [8], [9], [10], [11]. To date, various genotyping methods including biotyping, multilocus variable-number tandem-repeat analysis (MLVA) and PFGE have been used for the epidemiological investigation of S. sonnei [31], [32]. Biotyping is a relatively earlier phenotypic method with insufficient discrimination.
Although MLVA has a higher discriminatory power for distinguishing epidemiological relationships, it is not a universal method and is not 100% reproducible, and it has no well-established protocol for surveillance networks [5]. On the other hand, PFGE, as the acceptable gold-standard technique for molecular typing, has the standardised PulseNet PFGE protocol for interlaboratory comparison of results [18] and has been successfully used for subtyping sporadic or epidemic isolates of Shigella [11], [24]. PFGE is an electrophoresis-based method that can directly or indirectly reflect pathogen variation, in other words, the changing in DNA sequence.
In this study, PFGE was used to characterise the homology of all 340 S. sonnei isolates. The results revealed that many PFGE clusters contained strains from multiple geographical regions; some clusters even contained strains from six to seven cities throughout the province. Large-scale transmission of different clones was found in different years. In 2009, cluster SC6 containing 33 isolates spread between Changzhou and Nantong; in 2008, cluster SD5 containing 35 isolates was prevalent in Wuxi, Zhenjiang, Suzhou, Lianyungang, Nanjing and Xuzhou; and in 2007, cluster SE1 containing 35 isolates was present in Xuzhou, Lianyungang, Changzhou, Taizhou, Suzhou, Yancheng and Huai'an, a total of 7 cities. Similarly, the clonal spread of S. sonnei has also been described by other studies in Malaysia during 1997–2009 [32] and on a global scale [33]. These observations emphasise the necessity of monitoring the molecular subtypes of S. sonnei from endemic regions and controlling outbreaks of S. sonnei.
The most commonly applied method of integron detection is to use the specific primer of various integrons for single PCR and to examine them separately, which is time consuming and complicated. This study used degenerate primers combined with restriction enzyme digestion to detect class 1, 2 and 3 integrons concurrently. A total of 55.9% of S. sonnei isolates carried class 1 integrons in Jiangsu Province, perceptibly higher than that in Bangladesh (2.5%) [5], South Korea (14.9%) [16] and Zhejiang Province of China (12.9%) [21]. This difference may due to geographical variation or the increased prevalence of class 1 integrons in recent years. Class 2 integrons were detected in 80.3% of S. sonnei isolates, which was in concordance with previous reports (100.0% in Gabon [13], 100.0% in South Korea [16], 81.0% in Uzbekistan [34] and 80.6% in Zhejiang Province [21]). Class 3 integrons were not detected in any isolates. Furthermore, an atypical class 1 integron, ever present in Africa, Europe and America [35], was also detected with a positive rate of 49.4%. Overall, the current study searched for four classes of integron (classes 1, 2 and 3 and atypical class 1 integrons) in all isolates obtained. The results showed that class 2 integrons remained the preponderant integron among S. sonnei strains but it was not as dominant as reported previously. It is well known that classes 1 and 2 and atypical class 1 integrons play an important role in increased antimicrobial resistance. This was supported by the fact that the resistance rates of the class 1 integron-positive group were all statistically higher than the class 1 integron-negative group for all nine antimicrobials, thus integrons were associated with resistance of S. sonnei to multiple antimicrobials at the phenotype level.
Further analysis of the variable region sequencing showed the close correlation between drug resistance and the presence of integrons at the genotype level. Four of five amplicons carried one or two gene cassettes; only one amplicon (0.15 kb) carried the skeletal structure of 5′CS and 3′CS. The class 1 integron containing 0.15-kb and 2.2-kb amplicons was the most common type in this study, and aacA4–cmlA1 was the most prevalent gene cassette, conferring resistance to aminoglycoside and chloramphenicol, respectively. The gene cassette array dfrA1–sat1–aadA in class 2 integrons conferred resistance to trimethoprim, streptothricin and streptomycin [16], [36]. The gene cassette bla OXA-30–aadA1, conferring resistance to β-lactams and aminoglycosides, was detected in an atypical class 1 integron. The decrease of AMC resistance is probably due to the decrease of the number of strains harbouring an atypical class 1 integron, which contains bla OXA-30 encoding an enzyme conferring resistance to AMC. So far, a similar atypical class 1 integron has been reported in China, India and France [21], [37], [38], [39].
In conclusion, this study systematically analysed the subtype shift, antimicrobial resistance patterns, genetic homology and molecular characteristics of integrons in 340 S. sonnei collected from 2002 to 2011 in Jiangsu Province, China. The multicentre, large-sample research, globally unusual, is of great significance for the clinical therapy, prevention and control of S. sonnei infection in Jiangsu Province, China.
Funding
This research was supported by the National Natural Science Foundation of China [81471994], Jiangsu Provincial Medical Talent, Six Talent Peaks Project of Jiangsu Province [WSN-135] and Advanced Health Talent of Six-One Project of Jiangsu Province [LGY2016042].
Competing interests
None declared.
Ethical approval
Not required.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jgar.2017.03.009.
Contributor Information
Huimin Qian, Email: jsqhm@126.com.
Ping Ma, Email: pingm62@aliyun.com.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Ahmed A.M., Furuta K., Shimomura K., Kasama Y., Shimamoto T. Genetic characterization of multidrug resistance in Shigella spp. from Japan. J Med Microbiol. 2006;55:1685–1691. doi: 10.1099/jmm.0.46725-0. [DOI] [PubMed] [Google Scholar]
- 2.Wang X.Y., Tao F., Xiao D., Lee H., Deen J., Gong J. Trend and disease burden of bacillary dysentery in China (1991–2000) Bull World Health Organ. 2006;84:561–568. doi: 10.2471/blt.05.023853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sivapalasingam S., Nelson J.M., Joyce K., Hoekstra M., Angulo F.J., Mintz E.D. High prevalence of antimicrobial resistance among Shigella isolates in the United States tested by the National Antimicrobial Resistance Monitoring System from 1999 to 2002. Antimicrob Agents Chemother. 2006;50:49–54. doi: 10.1128/AAC.50.1.49-54.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan E., Jabeen K., Ejaz M., Siddiqui J., Shezad M.F., Zafar A. Trends in antimicrobial resistance in Shigella species in Karachi, Pakistan. J Infect Dev Ctries. 2009;3:798–802. doi: 10.3855/jidc.500. [DOI] [PubMed] [Google Scholar]
- 5.Ud-Din A.I., Wahid S.U., Latif H.A., Shahnaij M., Akter M., Azmi I.J. Changing trends in the prevalence of Shigella species: emergence of multi-drug resistant Shigella sonnei biotype g in Bangladesh. PLoS One. 2013;8:e82601. doi: 10.1371/journal.pone.0082601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinh H., Nhu N.T., Nga T.V., Duy P.T., Campbell J.I., Hoang N.V. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 2009;9:204. doi: 10.1186/1471-2334-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei H.L., Wang Y.W., Li C.C., Tung S.K., Chiou C.S. Epidemiology and evolution of genotype and antimicrobial resistance of an imported Shigella sonnei clone circulating in central Taiwan. Diagn Microbiol Infect Dis. 2007;58:469–475. doi: 10.1016/j.diagmicrobio.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Salam M.A., Bennish M.L. Antimicrobial therapy for shigellosis. Rev Infect Dis. 1991;13(Suppl. 4):S332–S341. doi: 10.1093/clinids/13.supplement_4.s332. [DOI] [PubMed] [Google Scholar]
- 9.Shen Y., Qian H., Gong J., Deng F., Dong C., Zhou L. High prevalence of antibiotic resistance and molecular characterization of integrons among Shigella isolates in Eastern China. Antimicrob Agents Chemother. 2013;57:1549–1551. doi: 10.1128/AAC.02102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke X., Gu B., Pan S., Tong M. Epidemiology and molecular mechanism of integron-mediated antibiotic resistance in Shigella. Arch Microbiol. 2011;193:767–774. doi: 10.1007/s00203-011-0744-3. [DOI] [PubMed] [Google Scholar]
- 11.Xia S., Xu B., Huang L., Zhao J.Y., Ran L., Zhang J. Prevalence and characterization of human Shigella infections in Henan Province, China, in 2006. J Clin Microbiol. 2011;49:232–242. doi: 10.1128/JCM.01508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vrints M., Mairiaux E., Van Meervenne E., Collard J.M., Bertrand S. Surveillance of antibiotic susceptibility patterns among Shigella sonnei strains isolated in Belgium during the 18-year period 1990 to 2007. J Clin Microbiol. 2009;47:1379–1385. doi: 10.1128/JCM.02460-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaumburg F., Alabi A.S., Kaba H., Lell B., Becker K., Grobusch M.P. Molecular characterization of Shigella spp. from patients in Gabon 2011–2013. Trans R Soc Trop Med Hyg. 2015;109:275–279. doi: 10.1093/trstmh/tru175. [DOI] [PubMed] [Google Scholar]
- 14.Folster J.P., Pecic G., Bowen A., Rickert R., Carattoli A., Whichard J.M. Decreased susceptibility to ciprofloxacin among Shigella isolates in the United States, 2006 to 2009. Antimicrob Agents Chemother. 2011;55:1758–1760. doi: 10.1128/AAC.01463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tariq A., Haque A., Ali A., Bashir S., Habeeb M.A., Salman M. Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can J Microbiol. 2012;58:1047–1054. doi: 10.1139/w2012-085. [DOI] [PubMed] [Google Scholar]
- 16.Oh J.Y., Yu H.S., Kim S.K., Seol S.Y., Cho D.T., Lee J.C. Changes in patterns of antimicrobial susceptibility and integron carriage among Shigella sonnei isolates from southwestern Korea during epidemic periods. J Clin Microbiol. 2003;41:421–423. doi: 10.1128/JCM.41.1.421-423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute . CLSI; Wayne, PA: 2012. Performance standards for antimicrobial susceptibility testing; twenty-second information supplement. Document M100-S22. [Google Scholar]
- 18.Ribot E.M., Fair M.A., Gautom R., Cameron D.N., Hunter S.B., Swaminathan B. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W., Luo Y., Li J., Lin L., Ma Y., Hu C. Wide dissemination of multidrug-resistant Shigella isolates in China. J Antimicrob Chemother. 2011;66:2527–2535. doi: 10.1093/jac/dkr341. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y., Zhuang L., Kang H., Ma P., Xu T., Pan S. Prevalence, resistance patterns, and characterization of integrons of Shigella flexneri isolated from Jiangsu Province in China, 2001–2011. Eur J Clin Microbiol Infect Dis. 2016;35:1347–1353. doi: 10.1007/s10096-016-2671-3. [DOI] [PubMed] [Google Scholar]
- 21.White P.A., McIver C.J., Rawlinson W.D. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother. 2001;45:2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu B., Pan S., Wang T., Zhao W., Mei Y., Huang P. Novel cassette arrays of integrons in clinical strains of Enterobacteriaceae in China. Int J Antimicrob Agents. 2008;32:529–533. doi: 10.1016/j.ijantimicag.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 23.Chompook P., Samosornsuk S., von Seidlein L., Jitsanguansuk S., Sirima N., Sudjai S. Estimating the burden of shigellosis in Thailand: 36-month population-based surveillance study. Bull World Health Organ. 2005;83:739–746. [PMC free article] [PubMed] [Google Scholar]
- 24.Qu F., Bao C., Chen S., Cui E., Guo T., Wang H. Genotypes and antimicrobial profiles of Shigella sonnei isolates from diarrheal patients circulating in Beijing between 2002 and 2007. Diagn Microbiol Infect Dis. 2012;74:166–170. doi: 10.1016/j.diagmicrobio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niyogi S.K. Increasing antimicrobial resistance: an emerging problem in the treatment of shigellosis. Clin Microbiol Infect. 2007;13:1141–1143. doi: 10.1111/j.1469-0691.2007.01829.x. [DOI] [PubMed] [Google Scholar]
- 27.Haukka K., Siitonen A. Emerging resistance to newer antimicrobial agents among Shigella isolated from Finnish foreign travellers. Epidemiol Infect. 2008;136:476–482. doi: 10.1017/S0950268807008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiferaw B., Solghan S., Palmer A., Joyce K., Barzilay E.J., Krueger A. Antimicrobial susceptibility patterns of Shigella isolates in Foodborne Diseases Active Surveillance Network (FoodNet) sites, 2000–2010. Clin Infect Dis. 2012;54(Suppl. 5):S458–S463. doi: 10.1093/cid/cis230. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharya D., Bhattacharya H., Sayi D.S., Bharadwaj A.P., Singhania M., Sugunan A.P. Changing patterns and widening of antibiotic resistance in Shigella spp. over a decade (2000–2011), Andaman Islands, India. Epidemiol Infect. 2015;143:470–477. doi: 10.1017/S0950268814000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirose K. Antimicrobial susceptibility of Shigella sonnei isolates in Japan and molecular analysis of S. sonnei isolates with reduced susceptibility to fluoroquinolones. Antimicrob Agents Chemother. 2005;49:1203–1205. doi: 10.1128/AAC.49.3.1203-1205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Belkum A., Tassios P.T., Dijkshoorn L., Haeggman S., Cookson B., Fry N.K. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13(Suppl. 3):1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 32.Koh X.P., Chiou C.S., Ajam N., Watanabe H., Ahmad N., Thong K.L. Characterization of Shigella sonnei in Malaysia, an increasingly prevalent etiologic agent of local shigellosis cases. BMC Infect Dis. 2012;12:122. doi: 10.1186/1471-2334-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filliol-Toutain I., Chiou C.S., Mammina C., Gerner-Smidt P., Thong K.L., Phung D.C. Global distribution of Shigella sonnei clones. Emerg Infect Dis. 2011;17:1910–1912. doi: 10.3201/eid1710.101486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madiyarov R.S., Bektemirov A.M., Ibadova G.A., Abdukhalilova G.K., Khodiev A.V., Bodhidatta L. Antimicrobial resistance patterns and prevalence of class 1 and 2 integrons in Shigella flexneri and Shigella sonnei isolated in Uzbekistan. Gut Pathog. 2010;2:18. doi: 10.1186/1757-4749-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubois V., Parizano M.P., Arpin C., Coulange L., Bezian M.C., Quentin C. High genetic stability of integrons in clinical isolates of Shigella spp. of worldwide origin. Antimicrob Agents Chemother. 2007;51:1333–1340. doi: 10.1128/AAC.01109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLappe N., O’Halloran F., Fanning S., Corbett-Feeney G., Cheasty T., Cormican M. Antimicrobial resistance and genetic diversity of Shigella sonnei isolates from western Ireland, an area of low incidence of infection. J Clin Microbiol. 2003;41:1919–1924. doi: 10.1128/JCM.41.5.1919-1924.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang C.Y., Lu P.L., Lin C.C., Lee T.M., Tsai M.Y., Chang L.L. Integron types, gene cassettes, antimicrobial resistance genes and plasmids of Shigella sonnei isolates from outbreaks and sporadic cases in Taiwan. J Med Microbiol. 2011;60:197–204. doi: 10.1099/jmm.0.022517-0. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh S., Pazhani G.P., Niyogi S.K., Nataro J.P., Ramamurthy T. Genetic characterization of Shigella spp. isolated from diarrhoeal and asymptomatic children. J Med Microbiol. 2014;63:903–910. doi: 10.1099/jmm.0.070912-0. [DOI] [PubMed] [Google Scholar]
- 39.Dubois V., Parizano M.P., Arpin C., Coulange L., Bezian M.C., Quentin C. High genetic stability of integrons in clinical isolates of Shigella spp. of worldwide origin. Antimicrob Agents Chemother. 2007;51:1333–1340. doi: 10.1128/AAC.01109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.