Abstract
A number of PVs have been described in bats but to the best of our knowledge not from feces. Using a previously described NetoVIR protocol, Eidolon helvum pooled fecal samples (Eh) were treated and sequenced by Illumina next generation sequencing technology. Two complete genomes of novel PVs (EhPV2 and EhPV3) and 3 partial sequences (BATPV61, BATPV890a and BATPV890b) were obtained and analysis showed that the EhPV2 and EhPV3 major capsid proteins cluster with and share 60–64% nucleotide identity with that of Rousettus aegyptiacus PV1, thus representing new species of PVs within the genus Psipapillomavirus. The other PVs clustered in different branches of our phylogenetic tree and may potentially represent novel species and/or genera. This points to the vast diversity of PVs in bats and in Eidolon helvum bats in particular, therefore adding support to the current concept that PV evolution is more complex than merely strict PV-host co-evolution.
Keywords: Bat, Papillomavirus, Feces
Highlights
-
•
First characterization of papillomavirus from stools of bats.
-
•
Novel bat papillomaviruses occupy diverse polyphyletic phylogenetic positions.
-
•
Observed diversity of bat papillomaviruses, supports existence of a complex papillomavirus–host co-evolution.
1. Introduction
The family Papillomaviridae comprises a large family of non-enveloped icosahedral viruses with a circular double stranded DNA genome (Bernard et al., 2010) ranging from 6953 bp (Chelonia mydas papillomavirus type 1) to 8607 bp (Canine papillomavirus type 1 (CPV1)) in length (Van Doorslaer, 2013). Typically, the genome contains six canonical genes that are located in 2 regions: genes coding for early proteins (E1, E2, E6 and E7, functional proteins) and genes encoding late proteins (L1 and L2, capsid proteins). The non-coding region is required for control of replication and transcription. All these genes are expressed in a temporal and highly regulated manner (Bernard et al., 2010, Howley and R., 2006). Generally, papillomaviruses (PVs) infect the stratified squamous epithelium of the skin and of the mucosa in various vertebrate species persisting asymptomatically or causing neoplasia (Van Ranst et al., 1992b). The host range of PVs expands across a wide array of vertebrates and PVs have been characterized from humans, domestic and wild mammals and also in birds and reptiles (turtle and snake) (Rector and Van Ranst, 2013). Recently bats have come to the limelight as an important reservoir of mammalian viruses including very pathogenic ones like Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS)-related coronaviruses, as well as Filoviridae, such as Marburgvirus, and Henipaviruses. Despite the identification and characterization of several novel viruses infecting bats, only a few PVs have been completely sequenced. Our group sequenced the first bat PV (RaPV1) from an Egyptian fruit bat Rousettus aegyptiacus (Rector et al., 2006) and thereafter PVs were isolated from a Rickett's bigfooted bat Myotis ricketti (MrPV1) and a Schreiber's long-fingered bat Miniopterus schreibersii, (MscPV1, MscPV2) (Tse et al., 2012, Wu et al., 2012). The most recent ones (EhPV1, from Eidolon helvum [Eh]; EsPV1, EsPV2, and EsPV3, from Eptesicus serotinus; and RfPV1, from Rhinolophus ferrumequinum) were sequenced and analyzed by García-Pérez (Garcia-Perez et al., 2013, Garcia-Perez et al., 2014), whereas Wang and colleagues (2015) identified two novel divergent incomplete PV sequences from New Zealand in lesser short-tailed bats Mystacina tuberculate. Also, Baker and colleagues identified two partial sequences of papillomaviruses isolated from Eidolon helvum from Ghana (Baker et al., 2013). Most of these bat PVs are distantly related to each other and therefore belong to different phylogenetic clades (genera). De Villiers and colleagues (2004) established the criteria for the classification of PVs: different genera share less than 60% nucleotide sequence identity and species within a genus share between 60% and 70% nucleotide (nt) identity of the ORF L1 (Garcia-Perez et al., 2014, Mengual-Chulia et al., 2012).
Here, we describe the complete sequences of two novel divergent PVs (Eidolon helvum papillomavirus 2 [EhPV2] and Eidolon helvum papillomavirus 3 [EhPV3]) and three partially sequenced PVs (BATPV61, BATPV890a and BATPV890b) obtained from the fecal samples of straw-colored fruit bats Eidolon helvum in Cameroon. The partial genome analyses also showed that they could be new species of PVs.
2. Methods
2.1. Sample collection
Bat fecal samples were collected between December 2013 and May 2014 by a previously described method developed by Donaldson and colleagues (2010), after obtaining an authorization from the Delegation of Public Health for South West Region. Briefly, bats were captured in 3 different regions (Lysoka, Muyuka and Limbe) of the South West Region of Cameroon using mist nets around fruit trees and around human dwellings. Captured bats were retrieved from the net traps and held in paper sacks for 10 to 15 min, allowing enough time for the excretion of fresh fecal boluses. Sterile disposable spatulas were used to retrieve feces from the paper sacks, and placed into tubes containing 1 ml of universal transport medium (UTM). Labeled samples were put on ice and then transferred to the Cell and Molecular biology Laboratory, Biotechnology Unit, University of Buea, Cameroon and stored at − 20 °C, until they were shipped to the Laboratory of Clinical and Epidemiological Virology, Leuven, Belgium where they were stored at − 80 °C.
2.2. Sample preparation and sequencing
To reduce the sequencing cost, three to five fecal samples were pooled (87 samples and 25 pools in total), and treated to enrich viral particles using the previously described NetoVIR protocol (Conceicao-Neto et al., 2015): fecal suspensions (10% w/v in universal transport medium) were homogenized for 1 min at 3000 rpm with a MINILYS homogenizer (Bertin Technologies) and filtered using a 0.8 μm PES filters (Sartorius). The filtrate was then treated with a cocktail of Benzonase (Novagen) and Micrococcal Nuclease (New England Biolabs) at 37 °C for 2 h to digest free-floating nucleic acids. Samples were extracted using the QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer's instructions but without addition of carrier RNA to the lysis buffer. First and second strand synthesis and random PCR amplification for 17 cycles were performed using a slightly modified Whole Transcriptome Amplification (WTA2) Kit procedure (Sigma-Aldrich), with a denaturation temperature of 95 °C instead of 72 °C to allow for the denaturation of dsDNA and dsRNA. WTA2 products were purified with MSB Spin PCRapace spin columns (Stratec) and the libraries were prepared for Illumina sequencing using a modified version of the NexteraXT Library Preparation Kit (Illumina). Sequencing of the samples was performed on a HiSeq 2500 platform (Illumina) for 300 cycles (2 × 150 bp paired ends).
PV reads were identified in 10 pools out of 25. PCR primers (Supplementary data S1) were designed to identify the exact sample(s) in which the PVs were present. After identification of the samples of interest, the PV genomes were then amplified using the multiply-primed rolling circle amplification (RCA) method with the TempliPhi 100 Amplification kit (Amersham Biosciences) as previously described by Rector et al., 2004a, Rector et al., 2004b. Briefly, 1 μl of extracted DNA or water (negative control) was transferred into a 0.5 ml tube with 5 μl of TempliPhi sample buffer, containing exonuclease-protected random hexamers. The samples were denatured at 95 °C for 3 min, and afterwards placed on ice. A premix was prepared on ice by mixing for each sample 5 μl of TempliPhi reaction buffer, 0.2 μl of TempliPhi enzyme mix containing the ϕ29 DNA polymerase and exonuclease-protected random hexamers in 50% glycerol, and 0.2 μl of extra dNTPs (at a stock concentration of 25 mM of each dNTP). After mixing by vortexing, 5 μl of this premix was added to the cooled samples. The reactions were incubated overnight (approximately 16 h) at 30 °C. Afterwards, the reactions were placed on ice, subsequently heated to 65 °C for 10 min to inactivate the ϕ29 DNA polymerase, and stored at − 20 °C awaiting further analysis. To investigate whether PV DNA was amplified, 2 μl of the RCA products were digested with 10 U of BamHI, EcoRI, HindIII, SalI, and XbaI. After digestion, the products were run on a 0.8% agarose gel to check for the presence of a DNA band consistent with full length PV DNA (~ 8 kb), or multiple bands with sizes adding up to this length. Furthermore, primers (Supplementary data S1) were designed and used to amplify the complete genomes with the Expand Long Template PCR System (Roche Diagnostics) using the RCA product, according to the manufacturer's instructions. PCR products were separated on a 0.8% agarose gel. The sequence of the genomes was then obtained by primer walking (list of primers in supplementary data S1) on the expand long-template PCR product. Sequencing was performed on an ABI Prism 3130xl Genetic Analyzer (Applied Biosystems). The chromatogram sequencing files were inspected with Chromas 2.4.3 (Technelysium) and the contigs prepared using MEGA6.06 (Tamura et al., 2013).
Open reading frame (ORF) analysis was performed with the ORF Finder tool on the NCBI server of the National Institutes of Health (http://www.ncbi.nlm.nih.gov/orffinder). The L1 sequences of PVs described here and 106 other animal and human PV types representing the different genera of the family Papillomaviridae and all bat PVs were imported from GenBank after they were compared to the reference on the Papillomavirus Episteme website (PaVE, https://pave.niaid.nih.gov/#explore/reference:genomes). The amino acid alignment was done using MUSCLE and then reverse translated (Edgar, 2004). After testing for the best DNA/protein model (supplementary data S2), a phylogenetic tree was constructed using the Maximum Likelihood method based on the General Time Reversible (GTR) model gamma distributed with Invariant sites (G + I) (Nei and Kumar, 2000) in MEGA6.06 (Tamura et al., 2013). Bootstrap support values were obtained for 10 00 replicates. P-distances were also constructed in MEGA6.06 and were used to calculate percentage identities. All sequences were deposited in GenBank under the accession numbers KX276953–KX276957.
3. Results
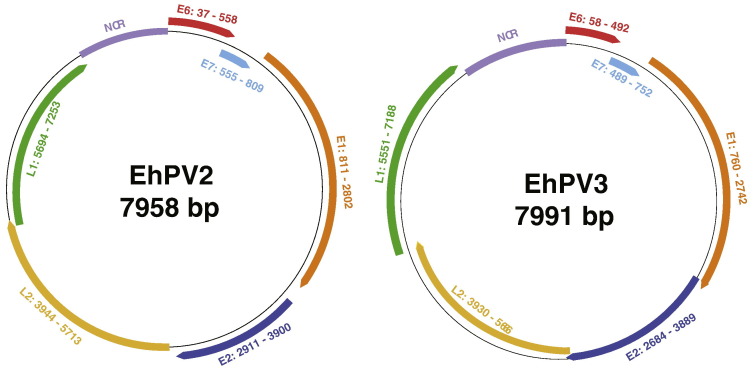
Fecal samples from healthy straw-colored fruit bats (E. helvum) were pooled and sequenced by Illumina next generation sequencing (NGS) technology. Ten out of the twenty-four pools contained papillomavirus reads, ranging from 0.01–13.3% of viral reads (Supplementary data S3). NGS alone was insufficient to produce contigs representing a complete PV genome. Therefore, from each pool, individual sample(s) containing the PV were identified on which subsequent experiments were carried out. For pool three (P3) rolling-circle amplification (RCA) and the Expand Long Template PCR system allowed us to amplify two complete PV genomes (EhPV2 and EhPV3). Subsequently, primer walking (and resequencing with designed primers at different regions for validation) was used to get the complete sequence of the genomes. Several attempts to amplify the other three PVs either by RCA and/or Expand Long Template PCR system failed. Therefore, only partial genomes (covered by 50–100 reads) from pools P5 (BATPV61, KX276955) and P25 (BATPV890a, KX276954 and BATPV890b, KX276953) are presented here. Only very few reads in pools 2, 4, 10, 14, and 17 were classified as PVs, indicating low viral load and so no further attempts were made to amplify their full genomes. EhPV1 and EhPV2 have a length of 7958 and 7991 base pair (bp) with a GC content of 51% and 50%, respectively. Like all PVs that have been characterized so far these PVs have all their ORFs on the same coding strand and in the same orientation of their dsDNA genome. They both contain the typical early proteins E6, E7, E1, and E2, the late proteins L2 and L1 and the non-coding regulatory region (NCR) between E6 and L1 of 741 and 860 bp, respectively (Fig. 1 ).
Fig. 1.
Circular representation of the open reading frames of the genomes of Eidolon helvum papillomaviruses 2 and 3 (EhPV2 and EhPV3). Numbers indicate the nucleotide positions of the start and end of the ORFs.
The nt and amino acids (aa) locations of various predicted features (motifs) of EhPV2 and EhPV3 are shown in Table 1 . The E6 ORFs of EhPV2 and EhPV3 are composed of 179 and 174 amino acids (aa), respectively, with both containing two zinc-binding domains with the sequence C-X-X-C-X29-C-X-X-C and C-X-X-C-X30-C-X-X-C separated by 36 aa. The E7 protein of both viruses also contain a zinc binding domain (CX2CX29CX2C) and a pRb binding motif (L-X-C-X-E), with sizes of 179 and 174 aa, respectively. The E1 ORFs encode the largest protein, 644 and 680 aa in EhPV2 and EhPV3, respectively and the ATP-binding site of the ATP-dependent helicase (GPPNTGKS) is conserved in the C-terminal part of both E1 proteins. A leucine zipper domain (LX6LX6LX6L), a major characteristic of E2, was not present in the carboxyl-terminal part of both E2 proteins. A distinct E4 could not be identified, but there is however a proline rich portion within the E2 ORFs (proline content of 11% and 13.5% respectively).
Table 1.
Nucleotide and amino acid features of EhPV2 and EhPV3 genomes. Values in bracket indicate nucleotide or amino acid positions of genomic feature. -: feature absent.
| EhPV2 | EhPV3 | |
|---|---|---|
| Predicted nucleotide features | ORF (nt positions) | ORF (nt positions) |
| E2 binding site (ACCG-N4-CGGT) | E6 (73–84); L2 (5108–5119) | NCR (2–13 and 7904–7915); E2 (1323–1334) |
| Tata box (TATAAA or TATA[A/T]A[A/T]) | – | NCR (25–30 and 7733–7738); E1 (2379–2384); E2 (3248–3253) |
| SP1 binding sites (GGCGGG) | E2 (1259–1264 and 1746–1751) | – |
| NF1 binding sites (CGGAA) | E2 (3421–3425); L2 (4052–4056); L1 (6687–6691 and 7188–7192) | E1 (1184–1188 and 1325–1329); E2 (3210–3214); L1 (6029–6033); NCR (7755–7759) |
| AP1 binding site (TGANTCA) | E1 (2790–2796); E2 (3807–3813) | – |
| Predicted amino acid feature | Protein (aa positions) | Protein (aa positions) |
| Zinc-binding motifs (CX2CX29CX2C) | E6 (56–92; 129–166); E7 (126–162) | E6 (27–63;100–137); E7 (47–83) |
| Retinoblastoma binding motif (DLXCXE) | E7 (21–26) | E7 (21–26) |
| ATP-dependent helicase motifs (GPPNTGKS) | E1 (493–500) | E1 (486–493) |
The late regions of the genomes contained the major (L1) and minor (L2) capsid protein genes and both contained a series of arginine and lysine residues (RKRRKRKR and KRKRR; RKKRKRK and KRRRR, respectively) at their 3′ end, which functions as a nuclear localization signal.
On the nucleotide level, EhPV3 contains the polyadenylation signals (AATAAA) for the early and late transcripts at the beginning of the L2 gene and in the NCR (nt position 4026–4031 and 7612–7617), respectively, whereas in the EhPV2 genome it is located in the E1 and NCR (nt position 1459–1464 and 7683–7688), respectively. In the EhPV3 sequences, two TATA boxes (TATAAA) of the E6 promoter were located close to the 3′ end of the NCR (nt positions 25–30 and 7733–7738, respectively) and two others were located on E1 and E2 (nt position 2379–2384 and 3248–3253, respectively). No TATA box was present in the genome of EhPV2.
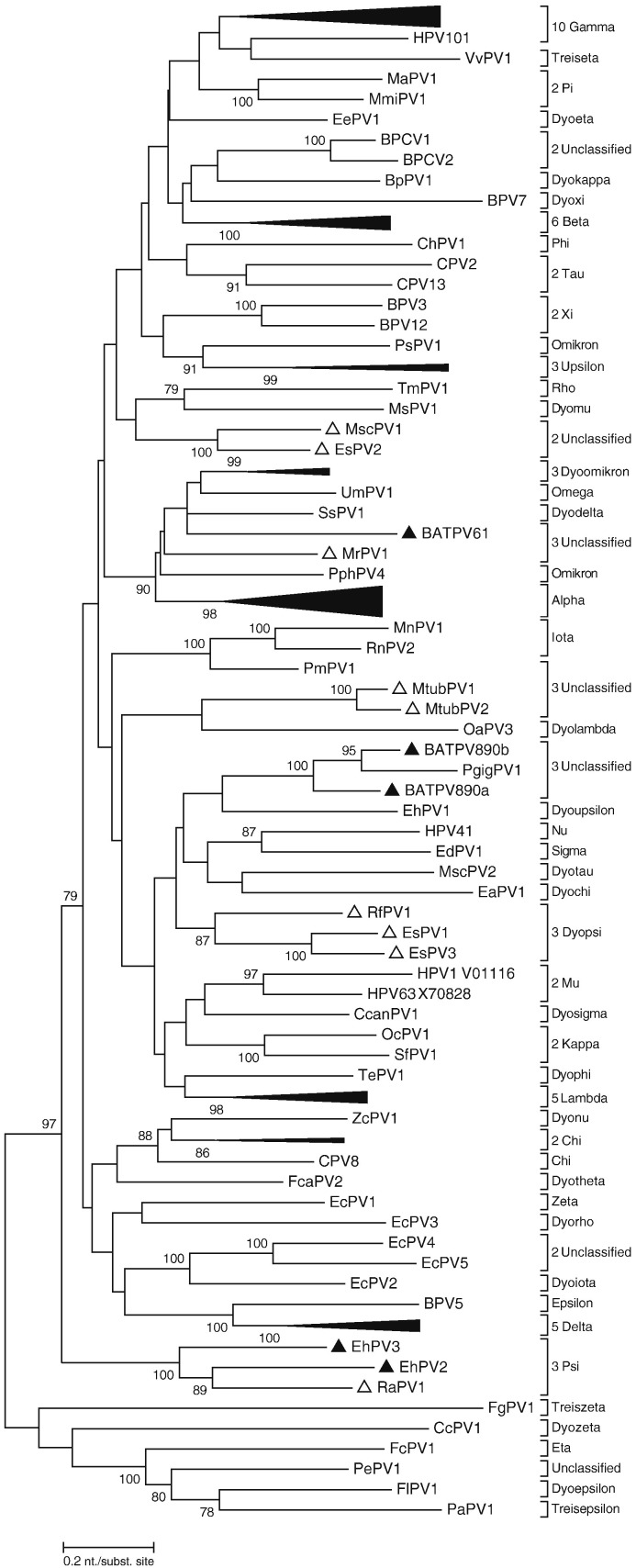
Phylogenetic analysis was based on the complete L1 sequences of EhPV2 and EhPV3, nearly complete L1 of the 3 partial bat PVs (1053, 1306 and 1260 nt in length for BAT61, BAT890a and BAT890b, respectively), and 97 other animal and human PVs (Fig. 2 ). Both EhPV2 and EhPV3 L1 clustered distantly together with the Rousettus aegyptiacus papillomavirus type 1 (RaPV1), classified in the genus Psipapillomavirus. The other bat PVs in this study clustered at different positions of the tree. BATPV61 clustered distantly with the PVs of the genera Dyodeltapapillomavirus (represented by SsPV1, isolated from domestic pig), Omegapapillomavirus (containing UmPV1, isolated from polar bear), an unclassified genus (containing MrPV1 from rickett's big-footed bat) and Dyopipapillomavirus (containing PphPV4, isolated from harbour porpoise). Furthermore, BATPV890a and BATPV890b clustered distantly with the unclassified Eidolon helvum papillomavirus type 1 (EhPV1) isolated from the hair follicle of straw-colored fruit bat from Germany.
Fig. 2.
Maximum Likelihood phylogenetic tree, based on a L1 nucleotide sequence alignment of EhPV2, EhPV3, BATPV61, BATPV890a and BATPV890b and 99 other PV type species of the different PV genera and species. The recognized PV genera are indicated with their full name. Previously known bat PVs are indicated with open triangles, whereas those described in this paper are indicated with filled triangle. The numbers at the internal nodes represent the bootstrap probabilities (in percent), as determined for 1000 iterations. Only bootstrap values greater than 70% are shown. The scale bar indicates the genetic distance (nucleotide substitutions per site) and * indicate PVs with only a partial L1 gene. NCBI GenBank accession numbers for all the PVs used in the phylogenetic tree are in Supplementary data S4.
The L1 sequences of EhPV2 and EhPV3 share nucleotide similarities ranging from 60 to 64% with RaPV1. According to the recent classification criteria, PV types belong to different PV genera when they share less than 60% nucleotide sequence identity across the entire L1 ORF and species within a genus, 60–70% nucleotide identity (10), therefore these 3 isolates may represent distinct species in the genus Psipapillomavirus. Other PVs identified in this study had a similarity between 55 and 64% with the closest mammalian PVs. However, since we were unable to retrieve their complete L1 sequences, the classification of these partially sequenced PVs remains preliminary.
4. Discussion
Here we present the genetic characterization of two complete PV genomes (EhPV2 and EhPV3) from the faces of an adult straw-colored fruit bat (Eidolon helvum). Both EhPV2 and EhPV3 have the typical genetic characteristics of a PV genome, containing four early genes, two late genes, and a non-coding region. Several nucleotide and amino acid consensus motifs known to play a role in the PV life cycle are also present in their genomes (Table 1). We also report partial sequences from 3 other PVs (BATPV61, BATPV890a and BATPV890b), for which we were not able to obtain the complete genome. The reason for the failure of the RCA procedure is not clear, however, we suppose that it might have been as a result of degraded viral particles and genomes, since the success of the method is based on the presence of intact episomal PV genomes.
Different PVs have been described from different bat species. The first one, RaPV1, was characterized from basosquamous carcinoma in an Egyptian fruit bat (R. aegyptiacus) (Rector et al., 2006), followed by MrPV1 isolated from a rectal swabs of a Rickett's bigfooted bat (Myotis ricketti) (Wu et al., 2012), and MscPV1 and MscPV2 detected from rectal swabs of Schreiber's long-fingered bats (Miniopterus schreibersii) (Tse et al., 2012, Wu et al., 2012). Also, EhPV1 and PgigPV1 (just about 35% of the L1 gene was sequenced) were isolated from the hair bulbs of E. helvum and Indian flying fox (Pteropus giganteus) (Garcia-Perez et al., 2013) while EsPV1, EsPV2 and EsPV3 were detected from the mucosa of serotine bat (Eptesicus serotinus), (Garcia-Perez et al., 2014). Additionally, RfPV1 was also described from the mucosa of greater horseshoe bat (R. ferrumequinum) (Garcia-Perez et al., 2014); while partial genomes (about 37% of the L1 gene sequence) of Mystacina tuberculate PV1 (MtubPV1) and PV2 (MtubPV2) were identified in a New Zealand lesser short-tailed bat (Wang et al., 2015). These bat PVs have been retrieved from different tissues and geographic locations and are polyphyletic (Garcia-Perez et al., 2013) as seen in Fig. 2. This indicates that different ancient papillomavirus lineages are present in chiroptera, as has previously also been reported for papillomaviruses in for instance humans (and other primates), artiodactyls, carnivores and rodents (Burk et al., 2013, Rector and Van Ranst, 2013).
Moreover, our partial BATPV890a and BATPV890b sequences clusters together with unclassified bat PVs EhPV1, PgigPV1, MscPV2, EsPV1, EsPV3 and RfPV1 and are most closely related to the established genera nu and sigma, while BATPV61 clustered together with bat MrPV1 and other PVs (SsPV1, UmPV1, and PphPV4), representing distinct genera. Therefore, different bat PVs occupy diverse phylogenetic positions thereby supporting the existence of several bat PV lineages. This distribution supports the hypothesis of the diversification of PVs in different ancestral lineages, after which lineages could have co-diversified with their amniote hosts (Gottschling et al., 2007, Gottschling et al., 2011). Previously, it was generally accepted that the different ancient PV lineages have co-evolved and co-speciated with their vertebrate hosts, a hypothesis which is said to be supported by the fact that PVs of closely related host species are generally cluster together in the PV phylogenetic tree (Tachezy et al., 2002, Van Ranst et al., 1992a). However, the fact that bat PVs from even the same species are scattered across the phylogenic tree supports the currently held concept that PV evolution is more complex than merely strict PV-host co-evolution.
EhPV2 and EhPV3 cluster together with the Rousettus aegyptiacus papillomavirus 1 (RaPV1) and based on the nt level, EhPV2 and EhPV3 share at least 60% similarity with RaPV1. According to the PV classification criteria (de Villiers et al., 2004) these two PVs constitute new species and together with RaPV1 make up the genus Psipapillomavirus. The other partial PVs identified clustered elsewhere. BATPV890a and BATPV890b clusters with EhPV1 and PgigPV1, while BATPV61 clustered with a non-bat PV Sus scrofa papillomavirus 1 (SsPV1). Given that we are missing only about 8%, 10% and 26% of L1 of BATPV890a, BATPV890b and BATPV61 (i.e., 1306, 1260 and 1053 nt in length), respectively, it implies that they could constitute new species in their respective genera. This therefore suggests that bats can be the hosts to a plethora of different PVs occupying different phylogenetic positions (Garcia-Perez et al., 2014).
Generally PVs infect stratified squamous epithelium of the skin and of the mucosa in various vertebrate species (Van Ranst et al., 1992b) and the detection of PVs in stools or their shedding through feces is still unusual. However, given that this has also been observed in humans, coupled with the fact that oncogenic, mucosal and cutaneous PVs have been detected in raw sewage samples (Di Bonito et al., 2015, Fratini et al., 2014, La Rosa et al., 2013), seem to suggest that like enteric viruses, epitheliotropic viruses can find their way into sewage from skin washes, mucus membranes, urine and feces. Those found in feces are likely infecting tissues of the alimentary canal which therefore lead to a sheading in feces. It is possible that the virus may be infecting the endothelial cells of anal region of bats (Varsani et al., 2014) without causing clinical syndromes especially as the bats from which these viruses were isolated appeared healthy. This is not surprising as most bat viruses known today were discovered in apparently healthy bats and therefore it has been suggested that they may have a specific immune system or antiviral activity against virus infections (Shi, 2010). Generally, most PV infections do not elicit host immune responses, primarily because the virus is confined to epithelial cells and the PV proteins that have the highest antigenic properties (L1 and L2) are only expressed in the uppermost layers of an infected mucosa or epithelium; and secondarily because PV infections are mostly non-lytic (Feller et al., 2010).
5. Conclusion
Here we report the characterization of two complete and 3 near complete genomes of Cameroonian Eidolon helvum PVs. It indicates that Eidolon helvum can host several papillomaviruses. Further, these viruses occupy diverse phylogenetic positions, pointing to the diversity of bat PVs, and backs existence of a complex PV–host co-evolution. However, some of the sequences were incomplete and therefore systematic sampling of more viruses from more host ranges would eventually help in reconciling this complexity.
Authors' contributions
CKY, SMG, MVR and JM conceived and designed the study; CKY and SMG collected the samples; CKY, AR, MZ, NCN and EH performed the experiments; CKY, AR, MZ, NCN, EH, PM and JM analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Conflict of interests
The authors declare that they have no competing interests.
Funding
This work was supported by KU Leuven (grant no EJX-C9928-StG/15/020BF, 2015).
Acknowledgments
CKY was supported by the Interfaculty Council for Development Cooperation (IRO) from the KU Leuven. NCN was supported by the Flanders Innovation & Entrepreneurship (VLAIO) (141330).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.virep.2016.08.001.
Contributor Information
Claude Kwe Yinda, Email: kweclaude.yinda@kuleuven.be.
Annabel Rector, Email: annabel.rector@kuleuven.be.
Mark Zeller, Email: mark.zeller@kuleuven.be.
Nádia Conceição-Neto, Email: nadia.neto@kuleuven.be.
Elisabeth Heylen, Email: elisabeth.heylen@kuleuven.be.
Piet Maes, Email: piet.maes@kuleuven.be.
Stephen Mbigha Ghogomu, Email: stephen.ghogomu@ubuea.cm.
Marc Van Ranst, Email: marc.vanranst@kuleuven.be.
Jelle Matthijnssens, Email: jelle.matthijnssens@kuleuven.be.
Appendix A. Supplementary data
Supplementary data S1: Primers sequence used in the study.
Supplementary data S2: Output of test for best DNA/protein model.
Supplementary data S3: Number or papillomavirus reads and percentages to total number of non-phage viral reads. * Indicates pool from which papillomavirus reads were described in this paper and + indicates pool with complete genomes of papillomavirus.
Supplementary data S4: GenBank accession numbers of papillomavirus type used for phylogenetic analysis.
References
- Baker K.S., Leggett R.M., Bexfield N.H., Alston M., Daly G., Todd S., Tachedjian M., Holmes C.E., Crameri S., Wang L.F., Heeney J.L., Suu-Ire R., Kellam P., Cunningham A.A., Wood J.L., Caccamo M., Murcia P.R. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441:95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H.U., Burk R.D., Chen Z., van Doorslaer K., zur Hausen H., de Villiers E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk R.D., Harari A., Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao-Neto N., Zeller M., Lefrere H., De Bruyn P., Beller L., Deboutte W., Yinda C.K., Lavigne R., Maes P., Van Ranst M., Heylen E., Matthijnssens J. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci. Rep. 2015;5:16532. doi: 10.1038/srep16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers E.M., Fauquet C., Broker T.R., Bernard H.U., zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Di Bonito P., Della Libera S., Petricca S., Iaconelli M., Sanguinetti M., Graffeo R., Accardi L., La Rosa G. A large spectrum of alpha and beta papillomaviruses are detected in human stool samples. J. Gen. Virol. 2015;96:607–613. doi: 10.1099/vir.0.071787-0. [DOI] [PubMed] [Google Scholar]
- Donaldson E.F., Haskew A.N., Gates J.E., Huynh J., Moore C.J., Frieman M.B. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 2010;84:13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller L., Wood N.H., Khammissa R.A., Chikte U.M., Meyerov R., Lemmer J. HPV modulation of host immune responses. SADJ. 2010;65:266–268. [PubMed] [Google Scholar]
- Fratini M., Di Bonito P., La Rosa G. Oncogenic papillomavirus and polyomavirus in water environments: is there a potential for waterborne transmission? Food Environ. Virol. 2014;6:1–12. doi: 10.1007/s12560-013-9134-0. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez R., Gottschling M., Wibbelt G., Bravo I.G. Multiple evolutionary origins of bat papillomaviruses. Vet. Microbiol. 2013;165:51–60. doi: 10.1016/j.vetmic.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez R., Ibanez C., Godinez J.M., Arechiga N., Garin I., Perez-Suarez G., de Paz O., Juste J., Echevarria J.E., Bravo I.G. Novel papillomaviruses in free-ranging Iberian bats: no virus-host co-evolution, no strict host specificity, and hints for recombination. Genome Biol. Evol. 2014;6:94–104. doi: 10.1093/gbe/evt211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling M., Stamatakis A., Nindl I., Stockfleth E., Alonso A., Bravo I.G. Multiple evolutionary mechanisms drive papillomavirus diversification. Mol. Biol. Evol. 2007;24:1242–1258. doi: 10.1093/molbev/msm039. [DOI] [PubMed] [Google Scholar]
- Gottschling M., Goker M., Stamatakis A., Bininda-Emonds O.R., Nindl I., Bravo I.G. Quantifying the phylodynamic forces driving papillomavirus evolution. Mol. Biol. Evol. 2011;28:2101–2113. doi: 10.1093/molbev/msr030. [DOI] [PubMed] [Google Scholar]
- Howley P.M., R. L.D. Papillomaviridae. In: Knipe D.M., Howley P.M., editors. Virology. 5th ed. Lippincott/The Williams & Wilkins Co.; Philadelphia: 2006. pp. 2299–2354. [Google Scholar]
- La Rosa G., Fratini M., Accardi L., D'Oro G., Della Libera S., Muscillo M., Di Bonito P. Mucosal and cutaneous human papillomaviruses detected in raw sewages. PLoS One. 2013;8 doi: 10.1371/journal.pone.0052391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengual-Chulia B., Garcia-Perez R., Gottschling M., Nindl I., Bravo I.G. Novel animal papillomavirus sequences and accurate phylogenetic placement. Mol. Phylogenet. Evol. 2012;65:883–891. doi: 10.1016/j.ympev.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S. Cambridge University Press; New York: 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- Rector A., Van Ranst M. Animal papillomaviruses. Virology. 2013;445:213–223. doi: 10.1016/j.virol.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Rector A., Bossart G.D., Ghim S.J., Sundberg J.P., Jenson A.B., Van Ranst M. Characterization of a novel close-to-root papillomavirus from a Florida manatee by using multiply primed rolling-circle amplification: Trichechus manatus latirostris papillomavirus type 1. J. Virol. 2004;78:12698–12702. doi: 10.1128/JVI.78.22.12698-12702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A., Tachezy R., Van Ranst M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 2004;78:4993–4998. doi: 10.1128/JVI.78.10.4993-4998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A., Mostmans S., Van Doorslaer K., McKnight C.A., Maes R.K., Wise A.G., Kiupel M., Van Ranst M. Genetic characterization of the first chiropteran papillomavirus, isolated from a basosquamous carcinoma in an Egyptian fruit bat: the Rousettus aegyptiacus papillomavirus type 1. Vet. Microbiol. 2006;117:267–275. doi: 10.1016/j.vetmic.2006.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z. Bat and virus. Protein Cell. 2010;1:109–114. doi: 10.1007/s13238-010-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy R., Rector A., Havelkova M., Wollants E., Fiten P., Opdenakker G., Jenson B., Sundberg J., Van Ranst M. Avian papillomaviruses: the parrot Psittacus erithacus papillomavirus (PePV) genome has a unique organization of the early protein region and is phylogenetically related to the chaffinch papillomavirus. BMC Microbiol. 2002;2:19. doi: 10.1186/1471-2180-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse H., Tsang A.K., Tsoi H.W., Leung A.S., Ho C.C., Lau S.K., Woo P.C., Yuen K.Y. Identification of a novel bat papillomavirus by metagenomics. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K. Evolution of the papillomaviridae. Virology. 2013;445:11–20. doi: 10.1016/j.virol.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Van Ranst M., Fuse A., Fiten P., Beuken E., Pfister H., Burk R.D., Opdenakker G. Human papillomavirus type 13 and pygmy chimpanzee papillomavirus type 1: comparison of the genome organizations. Virology. 1992;190:587–596. doi: 10.1016/0042-6822(92)90896-w. [DOI] [PubMed] [Google Scholar]
- Van Ranst M., Kaplan J.B., Burk R.D. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J. Gen. Virol. 1992;73(Pt 10):2653–2660. doi: 10.1099/0022-1317-73-10-2653. [DOI] [PubMed] [Google Scholar]
- Varsani A., Kraberger S., Jennings S., Porzig E.L., Julian L., Massaro M., Pollard A., Ballard G., Ainley D.G. A novel papillomavirus in Adelie penguin (Pygoscelis adeliae) faeces sampled at the Cape Crozier colony, Antarctica. J. Gen. Virol. 2014;95:1352–1365. doi: 10.1099/vir.0.064436-0. [DOI] [PubMed] [Google Scholar]
- Wang J., Moore N.E., Murray Z.L., McInnes K., White D.J., Tompkins D.M., Hall R.J. Discovery of novel virus sequences in an isolated and threatened bat species, the New Zealand lesser short-tailed bat (Mystacina tuberculata) J. Gen. Virol. 2015;96:2442–2452. doi: 10.1099/vir.0.000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ren X., Yang L., Hu Y., Yang J., He G., Zhang J., Dong J., Sun L., Du J., Liu L., Xue Y., Wang J., Yang F., Zhang S., Jin Q. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012;86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data S1: Primers sequence used in the study.
Supplementary data S2: Output of test for best DNA/protein model.
Supplementary data S3: Number or papillomavirus reads and percentages to total number of non-phage viral reads. * Indicates pool from which papillomavirus reads were described in this paper and + indicates pool with complete genomes of papillomavirus.
Supplementary data S4: GenBank accession numbers of papillomavirus type used for phylogenetic analysis.