Abstract
Strains of human coronavirus (HCoV), namely HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1, primarily infect the upper respiratory and gastrointestinal tracts and are the most common cause of non-rhinovirus-induced common cold in humans. Although the manifestations of coronavirus infection (i.e., rhinorrhea, sneezing, cough, nasal obstruction, and bronchitis) are generally self-limiting in healthy adults, certain strains such as HCoV-NL63 and HCoV-HKU1 can cause severe lower respiratory tract infection and febrile seizure, especially in infants, people of advanced age, and immunocompromised hosts. In 2003, a novel HCoV strain was identified as the causative agent of the severe acute respiratory syndrome (SARS) epidemic that began in Asia in 2002. The strain has hence been referred to as SARS-CoV. In addition, as recently as September 2012, another novel HCoV, human betacoronavirus 2c EMC2012, was identified as being the cause of fever, renal failure, pneumonia, and severe respiratory distress in two patients in the Middle East. Phylogenetic analysis has revealed highly conserved sequences of ORF1ab, spike, nucleocapsid, and envelope protein genes, but not membrane protein genes, between human betacoronavirus 2c EMC2012 and SARS-CoV. This review focuses on the differences in the genomes of certain HCoV strains, the pathogenesis of said strains, and recent developments in the establishment of therapeutic agents that might aid in the treatment of patients with such infections.
Keywords: human betacoronavirus 2c EMC2012, human coronavirus, phylogenetic tree, severe acute respiratory syndrome coronavirus (SARS-CoV)
1. Coronaviruses
Coronaviruses, which are enveloped, single-stranded, positive-sense RNA viruses belonging to the subfamily Coronavirinae in the family Coronaviridae, primarily cause respiratory and enteric diseases in mammals and birds [1], [2], [3], [4], [5], [6], [7], [8], [9]. The genus is subdivided into three main groups (alpha, beta, and gamma) based on the genetic and serological properties of each virus [10], [11], [12], [13]. Most coronaviruses in the three groups infect animals or birds only. Of the viruses that infect human hosts, human coronavirus (HCoV)-229E and HCoV-OC43, both alphacoronaviruses, are the best characterized and are responsible for most non-rhinovirus-induced cases of the common cold [14]. Other respiratory pathogens in the coronavirus genera that have recently been characterized include the alpha virus HCoV-NL63 and the beta virus HCoV-HKU1. Those two viruses tend to be isolated from young patients, patients of advanced age, and immunocommpromised hosts [13], [15], [16]. Recently, however, more virulent strains have emerged, including severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV), a betacoronovirus that was responsible for the outbreak of SARS in Asia and other countries in 2002–2003, and a novel HCoV, 2c EMC2012, which was identified as the causative agent of rapidly progressive acute respiratory infection in two men from the Middle East in 2012 [17], [18], [19].
Coronavirus virions typically range in size from 80 nm to 100 nm in diameter, contain an RNA genome of approximately 26–32 kb in size, and are characterized microscopically by their crown- or halo-shaped appearance [20], [21], [22], [23], [24], [25], [26]. The genome of most coronaviruses encodes two replicases, expressed in the form of two polyproteins, open reading frame (ORF)1a, measuring approximately 450 kDa, and ORF1ab, measuring approximately 750 kDa. These polyproteins are processed into a number of non-structural (NS) proteins and four structural proteins, namely spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein (Fig. 1 ) [3], [27]. ORF1a and ORF1ab are predecessors of NS proteins, being cleaved by papain-like protease (PLpro, nsp3) and 3C-like protease (3CLpro, nsp5) to generate the final NS proteins [28], [29]. Some members of betacoronavirus subgroup A also code for a shorter spike-like protein called hemagglutinin esterase [30], [31], [32]. Studies have shown that, at least in SARS-CoV, a receptor-binding domain on the S protein mediates the attachment of said virus to its cellular receptor, angiotensin-converting enzyme 2 (ACE2) [33], [34]. Lin et al found that HCoV-NL63 also makes use of the S protein-ACE2 mechanism for cell entry [35].
Fig. 1.

Coronavirus genome. ORF1a and 1b are located at the 5'-terminal 2/3 gene of the coronavirus and encode two polyproteins, namely pp1a (∼450 kDa) and pp1ab (∼750 kDa). The four structural proteins in coronavirus include the spike (S) protein, envelope (E) protein, membrane (M) protein, and nuclepcapsid (N) protein.
Very recently, van Boheemen et al. and Xingyi Ge et al. constructed a phylogenetic tree of conronaviruses using the polymerase gene nsp12 and found that coronaviruses could be grouped according to the presence of ORFs, namely Groups 1, 2a, 2b, 2c, 2d, and 3 [36], [37]. According to their data, HCoV-229E, HCoV-NL63, porcine epidemic diarrhea virus, and transmissible gastroenteritis virus belong to Group 1; HCoV-OC43 and mouse hepatitis virus belong to Group 2a; SARS-CoV belongs to Group 2b; human betacoronavirus 2c EMC2012 belongs to Group 2c; and avian infectious bronchitis virus belongs to Group 3 [36], [37]. Other phylogenetic trees that have been constructed in our laboratory have revealed that 53% of the ORF1ab sequences, 33% of the envelope protein sequences, 45% of the nucleocapsid protein sequences, 34% of the spike protein sequences, and 41% of the membrane protein sequences in SARS-CoV are conserved in human betacoronavirus 2c EMC2012, indicating that SARS-CoV and betacoronavirus 2c EMC2012 are very similar (Fig. 2 ) [36], [37], [38], [39].
Fig. 2.
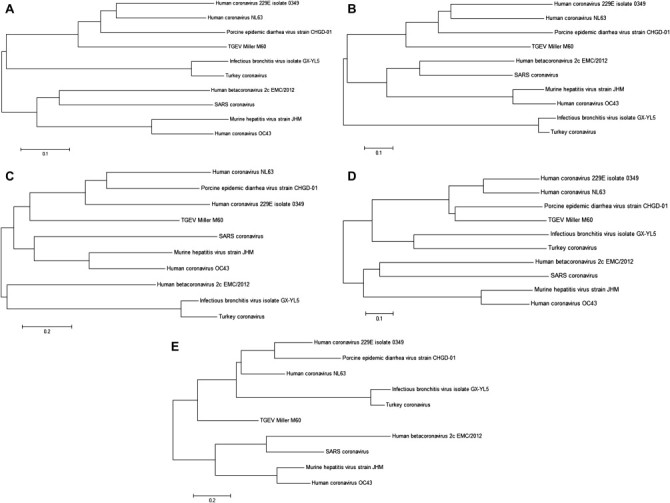
Phylogenetic trees. Phylogenetic trees were constructed with (A) ORF1ab, (B) nucleocapsid, (C) membrane, (D) spike, and (E) envelope protein. The phylogenetic trees were constructed using MEGA5 software [36], [37], [38], [39].
2. Epidemiology and pathogenesis of SARS coronavirus
SARS-CoV is the causative agent of SARS and causes an atypical pneumonia that spread rapidly throughout parts of Asia, North America, and Europe during 2002–2003 [37], [38]. Shortly after the first case of SARS was reported in the Chinese province of Guangdong in November 2002, the disease quickly spread worldwide, with cases having been reported in some 30 countries in 2003. By the time the outbreak had been contained, over 8000 cases had been confirmed and 916 people had died due to the disease [40], [41], [42], [43], [44], [45]. According to the World Health Organization, the mortality rate associated with SARS was >10% [46], [47]. People of advanced age were at the greatest risk of death, with a case-fatality rate approaching 50% in people >65 years who were infected by the virus [48].
The major route of transmission of SARS-CoV is close person-to-person contact, primarily via contact with aerosolized droplets or other bodily fluids. Many studies have shown that this virus can survive for more than 3 days outside the human body in sputum and feces, resulting in the potential for continuous infection during an epidemic. In addition to causing respiratory symptoms, chills, headache, muscle ache, and fever, infection with SARS-CoV has been shown to result in severe diffuse pneumonia, pulmonary fibrosis, and in the most severe cases, death. Pathological examinations of specimens taken from patients with SARS-related acute lung injury, acute respiratory distress syndrome, and pulmonary fibrosis show bronchial epithelial cell exfoliation, cilia loss, multinucleated syncytia cells, squamous epithelial tissue deformation, and increased levels of immune response cells in lung tissue [49], [50]. In addition, clinical laboratory tests have shown that patients with SARS often present with thrombocytopenia and leukopenia [51], [52] as well as overexpression of serum inflammatory cytokines, including interferon (IFN)-γ, interleukin (IL)-18, transforming growth factor (TGF)-β1, tumor necrosis factor (TNF)-α, and IL-6 [50], [53].
SARS-CoV infection can cause bronchial epithelial cell peeling, cilia damage, the formation of multinucleated giant cells, squamous cell aplasia, alveolar interstitial fiber cell hyperplasia, and fibrotic lung disease [54], [55]. Although blood tests in some patients with SARS-CoV infection show a normal white blood cell count, most show evidence of leukopenia, lymphopenia, and thrombocytopenia. Other biochemical signs typically exhibited in blood samples from patients with SARS include abnormal levels of proteins indicative of abnormal liver function, increased C-reactive protein levels, increased levels of creatine phophokinase and lactate dehydrogenase, overexpression of inflammatory cytokines IL-1, IL-6, IL-8, IL-12, IL-18, TGF-β1, TNF-α, IFN-γ-inducible Protein 10 (IP-10), IFN-γ, monokine induced by IFN-γ (MIG) as well as overexpression of chemokines chemokine [C-C motif] ligand 2 (CCL2)/monocyte chemoattractant protein-1 (MCP-1), CCL3/MIP-1α, and chemokine [C-X-C motif] (CXCL10)/IP-10 [50], [53], [56], [57], [58], [59], [60], [61], [62]. In the early stage of infection, TGF-β1 has been shown to be markedly elevated in serum and in lung tissue [50], [63], [64]. In addition, patients with late-phase SARS-related acute respiratory distress syndrome and pulmonary fibrosis often present with activated T-helper type-1 cells [53], [65], neutrophil aggregation, the accumulation of immune response cells, such as monocytes, macrophages, T cells, and B cells in the lungs [50], and rapid depletion of T cells [57], [58], [59], [60], [61], [62].
The most effective treatment for patients with SARS-CoV infection is the combination of ribavirin and steroids; however, this treatment is associated with a number of side effects. Ribavirin can cause red blood cell rupture, anemia, and death, while steroid treatment can cause stomach ulcers, edema, cardiac stress, and osteoporosis [66]. Controlled trials of drugs for the treatment of patients with SARS are urgently needed.
The innate immune response of type I IFN has a vital role in anti-viral replication, but SARS-CoV was demonstrated to antagonize type I IFN in mouse models [67], [68]. Mice lacking innate immunity suffer from signal transducer and activator of transcription-1 (STAT1) or myeloid primary response gene 88 (MyD88) deficiency and higher SARS-CoV infection rates [67], [68]. In addition, it has been reported that SARS-CoV-infected macrophages release CXCL10/IFN-γ-inducible protein 10 and CCL2/monocyte chemotactic protein 1, but do not induce the production of IFN-β [69].
Several SARS-CoV structural and NS proteins and genomes have been shown to participate in immune regulation and to induce cytokine expression. For example, the nucleocapsid of the SARS-CoV virion has been shown to induce the expression of plasminogen activator-1 via a Smad3-dependent induction of TGF-β1 expression [70], [71], [72], where Smad3 is an intracellular protein that transduces extracellular signals from TGF-β ligands to the nucleus, where they activate downstream TGF-β gene transcription. Nuclepcapsid protein was also identified as antagonizing IFN-β [73]. Furthermore, baculovirus-synthesized SARS-CoV spike protein has been shown to induce activator protein-1 (AP-1) and IL-8 upregulation by activating mitogen-activated protein kinases (MAPKs) in lung cells [74]. Moreover, it was found that spike protein induces innate immune response through the activation of the nuclear factor-kappaB (NF-κB) pathway [75]. In addition, Law et al found that SARS-CoV NSP1 induces CCL5, CXVL10, and CCL3 mRNA expression via the activation of NF-κB [76]. The 3a DNA vaccine is reported to stimulate IFN-γ production mainly by stimulating the production of CD8+ T cells, and IL-2 mainly by stimulating the production of CD4+ T cells [77]. Overall, structural and NS proteins and also SARS genomes are demonstrated to interfere with cell immunity.
Unlike other coronaviruses, the SARS-CoV genome only codes for one papain-like protease (PLpro). PLpro recognizes the amino acid fragments of LNGG and cleaves polyprotein replicase 1a at NS1/2, NS2/3, and NS3/4 boundaries, resulting in a mature viral protein [29], [78]. In a cell model of SARS-CoV infection, Speigal et al showed that PLpro de-ubiquitinating/de-ISGylating activity was associated with the inhibition of type I IFN expression [79]. Other studies have demonstrated that PLpro inhibits polyI:C-induced activation of IFN-β, NF-κB, and IFN-stimulated response element by blocking the export of phosphorylated regulatory factor 3 into the nucleus, thereby preventing the biosynthesis of type I IFN [80], [81]. In addition, Li et al showed that elevated levels of PLpro result in an attenuation of type I IFN-induced activation of the IFN-stimulated response element and AP-1, and that PLpro downregulates ERK1 (MAPK3) by upregulating the ubiquitin–proteasome system and suppressing the interaction between ERK1 and STAT1, resulting in type I IFN antagonism of SARS-CoV PLpro [82]. Furthermore, PLpro has been shown to induce TGF-β1, IL-1α, and CCL5 expression as well as triggering TGF-β1 production via ubiquitin proteasome-, p38 MAPK-, and ERK1/2-mediated signaling [46]. Therefore, it appears that PLpro plays an important role in the maturation process of SARS-CoV and in avoiding detection by the innate immune response system.
3. HCoV-NL63
HCoV-NL63, a Group 1 coronavirus, was first identified in a child with bronchiolitis in the Netherlands in 2004. Since then, cases of HcoV-NL63 have been documented worldwide. Infection is most prevalent during the winter months, tends to affect children under the age of 6 years, normally manifests as cough, fever, sore throat, rhinitis, expectoration, and upper and lower respiratory tract infection, such as bronchitis, bronchiolitis or pneumonia and has been shown to be associated with croup [83], [84], [85].
4. HCoV-HKU1
HCoV-HKU1, a Group 2 coronavirus that was first identified in an adult with chronic pulmonary disease in Hong Kong in 2005 [86], [87], causes rhinorrhea, fever, coughing, and wheezing, and can result in bronchiolitis and pneumonia if left untreated [15], [88].
5. Human betacoronavirus 2c EMC/2012
Human betacoronavirus 2c EMC2012 first appeared in the Middle East in April 2012 and was laboratory proven in nine patients by December 30, 2012 [17], [18], [19], [89], [90], [91], [92]. All of the laboratory-confirmed cases have been reported in Qatar (two cases), Saudi Arabia (five cases) and Jordan (two cases) [92]. All patients were severely ill, and five have died. The first confirmed case of betacoronavirus 2c EMC2012 infection occurred in Saudi Arabia. The patient presented with symptoms and signs of acute respiratory illness (high fever, cough, shortness of breath, and difficulty in breathing) and died in hospital. The second confirmed case was diagnosed in a man of Qatari origin who had recently travelled to Saudi Arabia. The patient presented with respiratory disease and renal failure. He was sent to London for treatment and recovered from the disease in hospital [89], [90]. The recent three confirmed cases in Saudi Arabia are epidemiologically linked and occurred in one family living within the same household in Saudi Arabia, and two of these have died [92]. In these three cases, at least two family members with direct personal contact increases the suspicion that person-to-person transmission may have occurred [92]. Two confirmed cases in Jordan were discovered through the testing of stored samples from a cluster of pneumonia cases in healthcare workers that occurred in April 2012. Both of these patients died [92].
This novel coronavirus was identified by real-time polymerase chain reaction based on pan-coronavirus primers [93], [94], [95] and there is evidence that the virus might have originated from bats [94]. It is not clear whether the symptoms and signs in these patients are typical of infection with this virus and no specific treatment recommendations have been made thus far. Acute respiratory support is advised for hospitalized patients with severe symptoms.
Like other coronaviruses, 2c EMC2012 is a very fragile virus, with a survival time not exceeding 24 hours outside the body. The pathogenic function of human betacoronavirus 2c EMC2012 PLpro(s) is not clear. Preliminary studies suggest that maturation of viral replication involves the cleavage of polyprotein replicase 1a at NS1/2, NS2/3, and NS3/4 boundaries, a mechanism shared by other coronaviruses [29], [78]. Moreover, phylogenetic tree analyses have shown that human betacoronavirus 2c EMC2012 and SARS-CoV have a similar ORF1ab subgenome (Fig. 2). Further studies are needed to differentiate between 2c EMC2012 and SARS-CoV.
6. Conclusion
HCoVs cause upper respiratory and gastrointestinal tract problems and the common cold in humans in worldwide, 2012 HCoVs cause upper respiratory and gastrointestinal tract problems and the common cold in humans in worldwide. The major method of transmission of HCoVs is from person-to-person and by droplet infection. Since the first two HCoVs, HCoV-229E and HCoV-OC43, were identified and studied extensively from the 1960s to 1980s, SARS-CoV has caused a mortality rate >10% globally in 2003 [14], [45]. HCoV-NL63 and HCoV-HKU1 were identified in 2004 and 2005 [13], [15], [16]. Since then, a novel coronavirus, human betacoronavirus 2c EMC2012, appeared in Saudi Arabia in April 2012, that has so far infected nine people and resulted in five deaths [92]. In the phylogenetic trees constructed with ORF1ab, E, N, S, and M, human betacoronavirus 2c EMC2012, has shown high similarities with SARS-CoV. All of these facts prove that novel coronaviruses have appeared in recent years and are becoming more severe, causing high mortality in humans.
References
- 1.Lai M.M. Coronavirus: organization, replication and expression of genome. Ann Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 2.Lai M.M. Background paper. Transcription and replication of coronavirus RNA: a 1989 update. Adv Exp Med Biol. 1990;276:327–333. doi: 10.1007/978-1-4684-5823-7_44. [DOI] [PubMed] [Google Scholar]
- 3.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prentice E., Denison M.R. The cell biology of coronavirus infection. Adv Exp Med Biol. 2001;494:609–614. doi: 10.1007/978-1-4615-1325-4_90. [DOI] [PubMed] [Google Scholar]
- 5.Loa C.C., Lin T.L., Wu C.C., Bryan T.A., Thacker H.L., Hooper T. Purification of turkey coronavirus by Sephacryl size-exclusion chromatography. J Virol Methods. 2002;104:187–194. doi: 10.1016/S0166-0934(02)00069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wunschmann A., Frank R., Pomeroy K., Kapil S. Enteric coronavirus infection in a juvenile dromedary (Camelus dromedarius) J Vet Diagn Invest. 2002;14:441–444. doi: 10.1177/104063870201400518. [DOI] [PubMed] [Google Scholar]
- 7.Theamboonlers A., Samransamruajkit R., Thongme C., Amonsin A., Chongsrisawat V., Poovorawan Y. Human coronavirus infection among children with acute lower respiratory tract infection in Thailand. Intervirology. 2007;50:71–77. doi: 10.1159/000097392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedersen N.C., Allen C.E., Lyons L.A. Pathogenesis of feline enteric coronavirus infection. J Feline Med Surg. 2008;10:529–541. doi: 10.1016/j.jfms.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Hoek L., Ihorst G., Sure K., Vabret A., Dijkman R., de Vries M. Burden of disease due to human coronavirus NL63 infections and periodicity of infection. J Clin Virol. 2010;48:104–108. doi: 10.1016/j.jcv.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- 11.Esper F., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Coronavirus HKU1 infection in the United States. Emerg Infect Dis. 2006;12:775–779. doi: 10.3201/eid1205.051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijgen L., Keyaerts E., Zlateva K., Van Ranst M. Identification of six new polymorphisms in the human coronavirus 229E receptor gene (aminopeptidase N/CD13) Int J Infect Dis. 2004;8:217–222. doi: 10.1016/j.ijid.2004.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 15.Sloots T.P., McErlean P., Speicher D.J., Arden K.E., Nissen M.D., Mackay I.M. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo P.C., Lau S.K., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia. Arch Virol. 2005;150:2299–2311. doi: 10.1007/s00705-005-0573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Health Protection Agency, HPA Press release. Acute respiratory illness associated with a new virus identified in the UK. HPA; London: 2012. http://www.hpa.org.uk/NewsCentre/NationalPressReleases/2012PressReleases/120923acuterespiratoryillnessidentified/ Available at: [accessed 06.01.13] [Google Scholar]
- 18.European Centre for Disease Prevention and Control, Rapid Risk Assessment: Severe respiratory disease associated with a novel coronavirus, ECDPC, September 24, 2012. Available at: http://ecdc.europa.eu/en/publications/Publications/RRA-Novel-coronavirus-final20120924.pdf [accessed 06.01.13].
- 19.Centers for Disease Control and Prevention (CDC). Severe respiratory illness associated with a novel coronavirus - Saudi Arabia and Qatar, 2012. MMWR Morb Mortal Wkly Rep 2012;61:820. [PubMed]
- 20.Herold J., Raabe T., Siddell S. Molecular analysis of the human coronavirus (strain 229E) genome. Arch Virol. 1993;7:63–74. doi: 10.1007/978-3-7091-9300-6_6. [DOI] [PubMed] [Google Scholar]
- 21.Herold J., Raabe T., Siddell S.G. Characterization of the human coronavirus 229E (HCV 229E) gene 1. Adv Exp Med Biol. 1993;342:75–79. doi: 10.1007/978-1-4615-2996-5_12. [DOI] [PubMed] [Google Scholar]
- 22.Lai M.M., Liao C.L., Lin Y.J., Zhang X. Coronavirus: how a large RNA viral genome is replicated and transcribed. Infect Agents Dis. 1994;3:98–105. [PubMed] [Google Scholar]
- 23.Haijema B.J., Volders H., Rottier P.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J Virol. 2003;77:4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 25.Havlickova M. [Newly identified representatives of the Coronaviridae family.] Klin Mikrobiol Infekc Lek. 2005;11:70. [PubMed] [Google Scholar]
- 26.Gonzalez J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 29.Ziebuh J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr Opin Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X.M., Kousoulas K.G., Storz J. The hemagglutinin/esterase gene of human coronavirus strain OC43: phylogenetic relationships to bovine and murine coronaviruses and influenza C virus. Virology. 1992;186:318–323. doi: 10.1016/0042-6822(92)90089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klausegger A., Strobl B., Regl G., Kaser A., Luytjes W., Vlasak R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol. 1999;73:3737–3743. doi: 10.1128/jvi.73.5.3737-3743.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popova R., Zhang X. The spike but not the hemagglutinin/esterase protein of bovine coronavirus is necessary and sufficient for viral infection. Virology. 2002;294:222–236. doi: 10.1006/viro.2001.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W., Choe H., Farzan M. Insights from the association of SARS-CoV S-protein with its receptor, ACE2. Adv Exp Med Biol. 2006;581:209–218. doi: 10.1007/978-0-387-33012-9_36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin H.X., Feng Y., Tu X., Zhao X., Hsieh C.H., Griffin L. Characterization of the spike protein of human coronavirus NL63 in receptor binding and pseudotype virus entry. Virus Res. 2011;160:283–293. doi: 10.1016/j.virusres.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge X., Li Y., Yang X., Zhang H., Zhou P., Zhang Y. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol. 2012;86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D.M.E. Genomic characterization of newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3 doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gholizadeh S., Djadid N.D., Nouroozi B., Bekmohammadi M. Molecular phylogenetic analysis of Anopheles and Cellia subgenus Anophelines (Diptera: Culicidae) in temperate and tropical regions of Iran. Acta Tropica. 2012 doi: 10.1016/j.actatropica.2012.11.003. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 39.Guzmán C., Caballero L., Martín L.M., Alvarez J.B. Waxy genes from spelt wheat: new alleles for modern wheat breeding and new phylogenetic inferences about the origin of this species. Ann Bot. 2012;110:1161–1171. doi: 10.1093/aob/mcs201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Booth C.M., Matukas L.M., Tomlinson G.A., Rachlis A.R., Rose D.B., Dwosh H.A. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 2003;289:2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 41.Peiris J.S. Severe acute respiratory syndrome (SARS) J Clin Virol. 2003;28:245–247. doi: 10.1016/j.jcv.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rickerts V., Wolf T., Rottmann C., Preiser W., Drosten C., Jakobi V. [Clinical presentation and management of the severe acute respiratory syndrome (SARS)] Dtsch Med Wochenschr. 2003;128:1109–1114. doi: 10.1055/s-2003-39253. [DOI] [PubMed] [Google Scholar]
- 45.World Health Organization . WHO; Geneva: 2003. Summary table of SARS cases by country, 1 November 2002 - 7 August 2003.http://www.who.int/csr/sars/country/country2003_08_15.pdf Available at: [accessed 06.01.13] [Google Scholar]
- 46.Li S.W., Yang T.C., Wan L., Lin Y.J., Tsai F.J., Lai C.C. Correlation between TGF-beta1 expression and proteomic profiling induced by severe acute respiratory syndrome coronavirus papain-like protease. Proteomics. 2012;12:3193–3205. doi: 10.1002/pmic.201200225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai C.C., Jou M.J., Huang S.Y., Li S.W., Wan L., Tsai F.J. Proteomic analysis of up-regulated proteins in human promonocyte cells expressing severe acute respiratory syndrome coronavirus 3C-like protease. Proteomics. 2007;7:1446–1460. doi: 10.1002/pmic.200600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan-Yeung M., Xu R.H. SARS: epidemiology. Respirology. 2003;8(Suppl):5. doi: 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L., Ding Y., Zhang Q., Che X., He Y., Shen H. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J Pathol. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K. SARS coronavirus induces apoptosis in Vero E6 cells. J Med Virol. 2004;73:323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W.K., Chen S.Y., Liu I.J., Chen Y.C., Chen H.L., Yang C.F. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg Infect Dis. 2004;10:1213–1219. doi: 10.3201/eid1007.031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang K.J., Su I.J., Theron M., Wu Y.C., Lai S.K., Liu C.C. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicholls J., Dong X.P., Jiang G., Peiris M. SARS: clinical virology and pathogenesis. Respirology. 2003;8(Suppl):S6–S8. doi: 10.1046/j.1440-1843.2003.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lang Z., Zhang L., Zhang S., Meng X., Li J., Song C. Pathological study on severe acute respiratory syndrome. Chin Med J (Engl) 2003;116:976–980. [PubMed] [Google Scholar]
- 56.Wang M., Xu H.F., Mo Z.Y., Zheng B.J., Gu J., Qin P.Z. Anti-SARS virus antibody responses against human SARS-associated coronavirus and animal SARS-associated coronavirus-like virus. Chin Med J (Engl) 2004;117:1723–1725. [PubMed] [Google Scholar]
- 57.Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 60.Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li T., Qiu Z., Zhang L., Han Y., He W., Liu Z. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189:648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pang C., Gu D.L. [Some problems about detecting the suspected cases of SARS according to the local skin temperatures on face] Space Med Med Eng (Beijing) 2003;16:231–234. [PubMed] [Google Scholar]
- 64.Baas T., Taubenberger J.K., Chong P.Y., Chui P., Katze M.G. SARS-CoV virus-host interactions and comparative etiologies of acute respiratory distress syndrome as determined by transcriptional and cytokine profiling of formalin-fixed paraffin-embedded tissues. J Interferon Cytokine Res. 2006;26:309–317. doi: 10.1089/jir.2006.26.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang W.K., Chen S.Y., Liu I.J., Kao C.L., Chen H.L., Chiang B.L. Temporal relationship of viral load, ribavirin, interleukin (IL)-6, IL-8, and clinical progression in patients with severe acute respiratory syndrome. Clin Infect Dis. 2004;39:1071–1075. doi: 10.1086/423808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morgenstern B., Michaelis M., Baer P.C., Doerr H.W., Cinatl J., Jr. Ribavirin and interferon-beta synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun. 2005;326:905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J Virol. 2004;78:11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4:e1000240. doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheung C.Y., Poon L.L., Ng I.H., Luk W., Sia S.F., Wu M.H. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin X., Chen Y., Meng A., Feng X. Termination of TGF-beta superfamily signaling through SMAD dephosphorylation–a functional genomic view. JJ Genet Genomics. 2007;34:1–9. doi: 10.1016/S1673-8527(07)60001-0. [DOI] [PubMed] [Google Scholar]
- 71.Ray S., Broor S.L., Vaishnav Y., Sarkar C., Girish R., Dar L. Transforming growth factor beta in hepatitis C virus infection: in vivo and in vitro findings. J Gastroenterol Hepatol. 2003;18:393–403. doi: 10.1046/j.1440-1746.2003.02985.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhao X., Nicholls J.M., Chen Y.G. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-beta signaling. J Biol Chem. 2008;283:3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu X., Pan J., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang Y.J., Liu C.Y., Chiang B.L., Chao Y.C., Chen C.C. Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J Immunol. 2004;173:7602–7614. doi: 10.4049/jimmunol.173.12.7602. [DOI] [PubMed] [Google Scholar]
- 75.Dosch S.F., Mahajan S.D., Collins A.R. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142:19–27. doi: 10.1016/j.virusres.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Law A.H., Lee D.C., Cheung B.K., Yim H.C., Lau A.S. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation. J Virol. 2007;81:416–422. doi: 10.1128/JVI.02336-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu B., Tao L., Wang T., Zheng Z., Li B., Chen Z. Humoral and cellular immune responses induced by 3a DNA vaccines against severe acute respiratory syndrome (SARS) or SARS-like coronavirus in mice. Clin Vaccine Immunol. 2009;16:73–77. doi: 10.1128/CVI.00261-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J Virol. 2005;79:4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A., Haller O. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li S.W., Lai C.C., Ping J.F., Tsai F.J., Wan L., Lin Y.J. Severe acute respiratory syndrome coronavirus papain-like protease suppressed alpha interferon-induced responses through downregulation of extracellular signal-regulated kinase 1-mediated signalling pathways. J Gen Virol. 2011;92:1127–1140. doi: 10.1099/vir.0.028936-0. [DOI] [PubMed] [Google Scholar]
- 83.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han T.H., Chung J.Y., Kim S.W., Hwang E.S. Human Coronavirus-NL63 infections in Korean children, 2004-2006. J Clin Virol. 2007;38:27–31. doi: 10.1016/j.jcv.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vabret A., Mourez T., Dina J., van der Hoek L., Gouarin S., Petitjean J. Human coronavirus NL63, France. Emerg Infect Dis. 2005;11:1225–1229. doi: 10.3201/eid1108.050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Woo P.C., Huang Y., Lau S.K., Tsoi H.W., Yuen K.Y. In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiol Immunol. 2005;49:899–908. doi: 10.1111/j.1348-0421.2005.tb03681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pyrc K., Berkhout B., van der Hoek L. The novel human coronaviruses NL63 and HKU1. J Virol. 2007;81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lau S.K., Woo P.C., Yip C.C., Tse H., Tsoi H.W., Cheng V.C. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Health Protection Agency, Novel Coronavirus 2012, HPA, 2012. Available at: http://www.hpa.org.uk/Topics/InfectiousDiseases/InfectionsAZ/NovelCoronavirus2012/ [accessed 06.01.13].
- 90.World Health Organization . WHO; 2012. World Health Organization: global alert and response (GAR) - Novel coronavirus infection 2012.http://www.who.int/csr/don/2012_09_25/en/index.html Available at: [accessed 06.01.13] [Google Scholar]
- 91.Danielsson N., ECDC Internal Response Team. Catchpole M. Novel coronavirus associated with severe respiratory disease: case definition and public health measures. Euro Surveill. 2012;17(39) doi: 10.2807/ese.17.39.20282-en. [DOI] [PubMed] [Google Scholar]
- 92.World Health Organization . WHO; 2012. Background and summary of novel coronavirus infection – as of 21 December 2012.http://www.who.int/csr/disease/coronavirus_infections/update_20121221/en/index.html Available at: [accessed 06.01.13] [Google Scholar]
- 93.Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17(39) doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 94.Balboni A., Gallina L., Palladini A., Prosperi S., Battilani M. A real-time PCR assay for bat SARS-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. ScientificWorldJournal. 2012;2012:989514. doi: 10.1100/2012/989514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moës E., Vijgen L., Keyaerts E., Zlateva K., Li S., Maes P. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


