Abstract
Coronavirus disease 2019 (COVID-19) is a kind of viral pneumonia which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The emergence of SARS-CoV-2 has been marked as the third introduction of a highly pathogenic coronavirus into the human population after the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) in the twenty-first century. In this minireview, we provide a brief introduction of the general features of SARS-CoV-2 and discuss current knowledge of molecular immune pathogenesis, diagnosis and treatment of COVID-19 on the base of the present understanding of SARS-CoV and MERS-CoV infections, which may be helpful in offering novel insights and potential therapeutic targets for combating the SARS-CoV-2 infection.
Keywords: Coronavirus, SARS-CoV-2, SARS-CoV, MERS-CoV, Pathogenesis
Graphical abstract
Highlights
-
•.
The highly pathogenic SARS-CoV-2 appearing in December 2019 can cause COVID-19 and even death in infected persons.
-
•.
Coronavirus infections led to the damage of lung, while imbalanced and excessive immune responses may cause pneumonia.
-
•.
RT-PCR and CT scans are significant for the diagnosis of SARS-CoV-2 infection, and drugs and vaccines against SARS-CoV-2 are being developed.
1. Introduction
Novel coronavirus-induced pneumonia, which was named as coronavirus disease 2019 (COVID-19) by the WHO on the February 11, 2020, has rapidly increased in epidemic scale [1]. On the same day, the international virus classification commission announced that the novel coronavirus was named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 is not the first severe respiratory disease outbreak caused by the coronavirus. Just in the past two decades, coronaviruses have caused three epidemic diseases, namely, COVID-19, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) [2]. At present, the cases of COVID-19 have been found in many countries around the world [3]. According to the latest data, up to the March 1, 2020, the number of confirmed cases in China reached 79,968, of which 2,873 were dead, and 41,681 were cured. In addition to China, the number of confirmed cases in other countries also reached 7,041, of which 105 were dead, and 459 were cured. On the 31st of January 2020, the World Health Organization (WHO) announced that COVID-19 was listed as the Public Health Emergency of International Concern (PHEIC), meaning that it may pose risks to multiple countries and requires a coordinated international response. The review tries to explain the molecular immune pathogenesis and diagnosis of COVID-19 and provide a reference for the prevention and drug development of SARS-CoV-2 infection, based on the recent research progress of SARS-CoV-2 and the knowledge from researches on SARS-CoV and MERS-CoV.
2. Virology of SARS-CoV-2
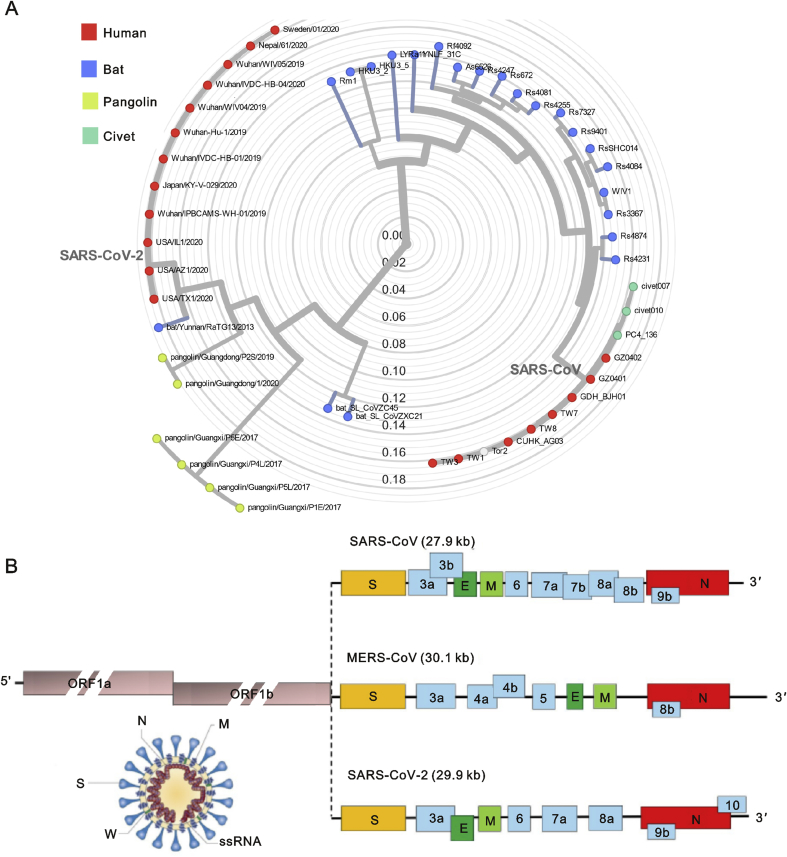
Coronaviruses are enveloped viruses with a positive sense single-stranded RNA genome (26–32 kb) [4]. Four coronavirus genera (α, β, γ, δ) have been identified so far, with human coronaviruses (HCoVs) detected in the α coronavirus (HCoV-229E and NL63) and β coronavirus (MERS-CoV, SARS-CoV, HCoV-OC43 and HCoV-HKU1) genera [5]. In late December 2019, patients presenting with cough, fever, and dyspnea with acute respiratory distress syndrome (ARDS) due to an unidentified microbial infection were reported in Wuhan, China. Virus genome sequencing of five patients with pneumonia hospitalized from December 18 to December 29, 2019, revealed the presence of a previously unknown β-CoV strain in all of them [6]. This isolated novel β-CoV shows 88% identity to the sequence of two bat-derived severe acute respiratory syndromes (SARS)-like coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, and about 50% identity to the sequence of MERS-CoV [6]. The novel β-CoV was then named “SARS-CoV-2” by the International Virus Classification Commission. The phylogenetic tree of SARS-like coronaviruses complete genome sequences is clearly depicted in Fig. 1A.
Fig. 1.
The phylogenetic tree of SARS-like coronaviruses complete genome sequences and genome of SARS-CoV, MERS-CoV and SARSCoV- 2.
(A) This phylogeny shows evolution of SARS-like β-coronaviruses including samples from human (n = 20), bat (n = 22), civet (n = 3) and pangolin (n = 6). The phylogenetic tree of complete genome sequences of coronaviruses was obtained and analyzed with Nextstrain (https://github.com/blab/sars-like-cov). (B) Coronaviruses form enveloped and spherical particles of 100–160 nm in diameter. They contain a positivesense single stranded RNA (ssRNA) genome of 26–32 kb in size. In SARS-CoV, MERS-CoV and SARS-CoV-2, the 5′-terminal two-thirds of the genome ORF1a/b encodes polyproteins, which form the viral replicase transcriptase complex. The other ORFs on the one-third of the genome encode four main structural proteins: spike (S), envelope (E), nucleocapsid (N) and membrane (M) proteins, as well as several accessory proteins.
The genome of SARS-CoV-2 is similar to that of typical CoVs and contains at least ten open reading frames (ORFs). The first ORFs (ORF1a/b), about two-thirds of viral RNA, are translated into two large polyproteins. In SARS-CoV and MERS-CoV, two polyproteins, pp1a and pp1ab, are processed into 16 non-structural proteins (nsp1-nsp16), which form the viral replicase transcriptase complex [7]. Those nsps rearrange membranes originating from the rough endoplasmic reticulum (RER) into double-membrane vesicles where viral replication and transcription occur [8,9]. The other ORFs of SARS-CoV-2 on the one-third of the genome encode four main structural proteins: spike (S), envelope (E), nucleocapsid (N) and membrane (M) proteins, as well as several accessory proteins with unknown functions which do not participate in viral replication (Fig. 1B).
Several groups of scientists in China have all discovered that SARS-CoV-2, just like SARS-CoV, requires the angiotensin-converting enzyme 2 (ACE2) [1] as a receptor to enter cells [10]. The binding of the virus with host cell receptors is a significant determinant for the pathogenesis of infection. SARS-CoV most likely originated in bats [11]and adapted to non-bat ACE2 variants as it crossed species to infect humans [12]. Dipeptidyl peptidase 4 (DPP4, also known as CD26) was identified as a functional receptor for MERS-CoV, because the receptor-binding S1 domain of the MERS-CoV spike protein was copurified with DPP4 specifically from lysates of susceptible Huh-7 cells [13]. MERS-CoV can bind DPP4 from multiple species, which promotes the transmission to humans and other species, and infection of cells from a large number of species [14]. A better understanding of the relative effects of receptor binding and protease action will help predict whether specific zoonotic coronaviruses infect humans and the possibility of adaptation.
3. Pathogenesis of COVID-19
Patients with COVID-19 show clinical manifestations including fever, nonproductive cough, dyspnea, myalgia, fatigue, normal or decreased leukocyte counts, and radiographic evidence of pneumonia [15], which are similar to the symptoms of SARS-CoV and MERS-CoV infections [16]. Hence, although the pathogenesis of COVID-19 is poorly understood, the similar mechanisms of SARS-CoV and MERS-CoV still can give us a lot of information on the pathogenesis of SARS-CoV-2 infection to facilitate our recognition of COVID-19.
3.1. Coronavirus entry and replication
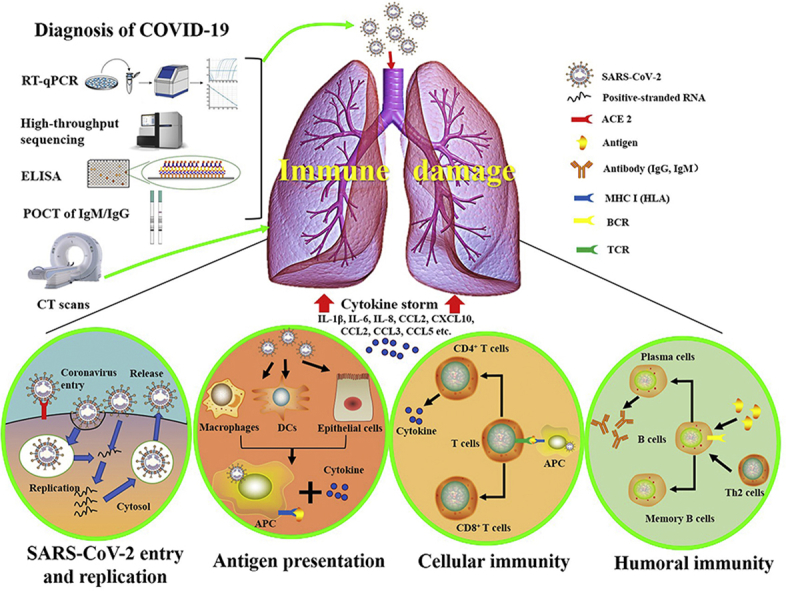
Coronavirus S protein has been reported as a significant determinant of virus entry into host cells [2]. The envelope spike glycoprotein binds to its cellular receptor, ACE2 for SARS-CoV [10] and SARS-CoV-2 [17], CD209L(a C-type lectin, also called L-SIGN) for SARS-CoV [18], DPP4 for MERS-CoV [13]. The entry of SARS-CoV into cells was initially identified to be accomplished by direct membrane fusion between the virus and plasma membrane [19]. Belouzard et al. [20] found that a critical proteolytic cleavage event occurred at SARS-CoV S protein at position (S2′) mediated the membrane fusion and viral infectivity. MERS-CoV also has evolved an abnormal two-step furin activation for membrane fusion [21]. Besides membrane fusion, the clathrin-dependent and -independent endocytosis mediated SARS-CoV entry too [22,23]. After the virus enters the cells, the viral RNA genome is released into the cytoplasm and is translated into two polyproteins and structural proteins, after which the viral genome begins to replicate [5]. The newly formed envelope glycoproteins are inserted into the membrane of the endoplasmic reticulum or Golgi, and the nucleocapsid is formed by the combination of genomic RNA and nucleocapsid protein. Then, viral particles germinate into the endoplasmic reticulum-Golgi intermediate compartment (ERGIC). At last, the vesicles containing the virus particles then fuse with the plasma membrane to release the virus [2].
3.2. Antigen presentation in coronavirus infection
While the virus enters the cells, its antigen will be presented to the antigen presentation cells(APC), which is a central part of the body’s anti-viral immunity. Antigenic peptides are presented by major histocompatibility complex (MHC; or human leukocyte antigen (HLA) in humans) and then recognized by virus-specific cytotoxic T lymphocytes (CTLs). Hence, the understanding of antigen presentation of SARS-CoV-2 will help our comprehension of COVID-19 pathogenesis. Unfortunately, there is still lack of any report about it, and we can only get some information from previous researches on SARS-CoV and MERS-CoV. The antigen presentation of SARS-CoV mainly depends on MHC I molecules [24], but MHC II also contributes to its presentation. Previous research shows numerous HLA polymorphisms correlate to the susceptibility of SARS-CoV, such as HLA-B∗4601, HLA-B∗0703, HLA-DR B1∗1202 [25] and HLA-Cw∗0801 [26], whereas the HLA-DR0301, HLA-Cw1502 and HLA-A∗0201 alleles are related to the protection from SARS infection [27]. In MERS-CoV infection, MHC II molecules, such as HLA-DRB1∗11:01 and HLA-DQB1∗02:0, are associated with the susceptibility to MERS-CoV infection [28]. Besides, gene polymorphisms of MBL (mannose-binding lectin) associated with antigen presentation are related to the risk of SARS-CoV infection [29]. These researches will provide valuable clues for the prevention, treatment, and mechanism of COVID-19.
3.3. Humoral and cellular immunity
Antigen presentation subsequently stimulates the body’s humoral and cellular immunity, which are mediated by virus-specific B and T cells. Similar to common acute viral infections, the antibody profile against SARS-CoV virus has a typical pattern of IgM and IgG production. The SARS-specific IgM antibodies disappear at the end of week 12, while the IgG antibody can last for a long time, which indicates IgG antibody may mainly play a protective role [30], and the SARS-specific IgG antibodies primarily are S-specific and N-specific antibodies [2]. Comparing to humoral responses, there are more researches on the cellular immunity of coronavirus. The latest report shows the number of CD4+ and CD8+ T cells in the peripheral blood of SARS-CoV-2-infected patients significantly is reduced, whereas its status is excessive activation, as evidenced by high proportions of HLA-DR (CD4 3.47%) and CD38 (CD8 39.4%) double-positive fractions [31]. Similarly, the acute phase response in patients with SARS-CoV is associated with severe decrease of CD4+ T and CD8+ T cells. Even if there is no antigen, CD4+ and CD8+ memory T cells can persist for four years in a part of SARS-CoV recovered individuals and can perform T cell proliferation, DTH response and production of IFN-γ [32]. Six years after SARS-CoV infection, specific T-cell memory responses to the SARS-CoV S peptide library could still be identified in 14 of 23 recovered SARS patients [33]. The specific CD8+ T cells also show a similar effect on MERS-CoV clearance in mice [34]. These findings may provide valuable information for the rational design of vaccines against SARS-CoV-2.
3.4. Cytokine storm in COVID-19
The report in Lancet shows ARDS is the main death cause of COVID-19. Of the 41 SARS-CoV-2-infected patients admitted in the early stages of the outbreak, six died from ARDS [15]. ARDS is the common immunopathological event for SARS-CoV-2, SARS-CoV and MERS-CoV infections [31]. One of the main mechanisms for ARDS is the cytokine storm, the deadly uncontrolled systemic inflammatory response resulting from the release of large amounts of pro-inflammatory cytokines (IFN-α, IFN-γ, IL-1β, IL-6, IL-12, IL-18, IL-33, TNF-α, TGFβ, etc.) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, etc.) by immune effector cells in SARS-CoV infection [15,[35], [36], [37]]. Similar to those with SARS-CoV, individuals with severe MERS-CoV infection show elevated levels of IL-6, IFN-α, and CCL5, CXCL8, CXCL-10 in serum compared to those with the mild-moderate disease [38]. The cytokine storm will trigger a violent attack by the immune system to the body, cause ARDS and multiple organ failure, and finally lead to death in severe cases of SARS-CoV-2 infection, just like what occurs in SARS-CoV and MERS-CoV infection [31].
3.5. Coronavirus immune evasion
To better survive in host cells, SARS-CoV and MERS-CoV use multiple strategies to avoid immune responses. The evolutionarily conserved microbial structures called pathogen-associated molecular patterns (PAMPs) can be recognized by pattern recognition receptors (PRRs). However, SARS-CoV and MERS-CoV can induce the production of double-membrane vesicles that lack PRRs and then replicate in these vesicles, thereby avoiding the host detection of their dsRNA [39]. IFN-I(IFN-α and IFN-β) has a protective effect on SARS-CoV and MERS-CoV infection, but the IFN-I pathway is inhibited in infected mice [40,41]. Accessory protein 4a of MERS-CoV may block the induction of IFN at the level of MDA5 activation through direct interaction with double-stranded RNA [42]. Besides, ORF4a, ORF4b, ORF5, and membrane proteins of MERS-CoV inhibit nuclear transport of IFN regulatory factor 3 (IRF3) and activation of IFN β promoter [43]. The antigen presentation can also be affected by the coronavirus. For example, gene expression related to antigen presentation is down-regulated after MERS-CoV infection [44]. Therefore, destroying the immune evasion of SARS-CoV-2 is imperative in its treatment and specific drug development.
4. Diagnosis of COVID-19
Clinical diagnosis of COVID-19 is mainly based on epidemiological history, clinical manifestations and some auxiliary examinations, such as nucleic acid detection, CT scan, immune identification technology (Point-of-care Testing (POCT) of IgM/IgG, enzyme-linked immunosorbent assay (ELISA)) and blood culture. However, the clinical symptoms and signs of patients infected with SARS-CoV-2 are highly atypical, including respiratory symptoms, cough, fever, dyspnea, and viral pneumonia. Therefore, auxiliary examinations are necessary for the diagnosis of COVID-19, just as the epidemiological history.
4.1. Nucleic acid detection technology
The two commonly used nucleic acid detection technologies for SARS-CoV-2 are real-time quantitative polymerase chain reaction (RT-qPCR) and high-throughput sequencing. The authoritative identification method for SARS-CoV-2 is virus blood culture and high-throughput sequencing of the whole genome [1]. However, the application of high-throughput sequencing technology in clinical diagnosis is limited because of its equipment dependency and high cost. So RT-qPCR is the most common, effective and straightforward method for detecting pathogenic viruses in respiratory secretions and blood [45].
After the outbreak of SARS-CoV-2 in China, many companies soon launched RT-qPCR test kits for clinical diagnosis. The Chinese Center for Disease Control and Prevention (China CDC) recommends the use of specific primers and probes in the ORF1ab and N gene regions for SARS-CoV-2 detection by RT-qPCR. The patient is defined as having a laboratory-confirmed infection when both targets are positive (http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html). Chu et al. [46] described two 1-step RT-qPCR assays (TaqMan-based fluorescence signal) to detect two different regions (ORF1b and N) of the viral genome separately. The negative control samples were all confirmed as negative ones, while samples from two SARS-CoV-2 infected patients were confirmed as positive ones in respiratory specimens by this method. Another study showed that the positive rate of SARS-CoV-2 was 91.7% (11/12) in the patients’ self-collected saliva by using RT-qPCR (non-probes SYBR based fluorescence signal), which suggests that saliva is a promising non-invasive specimen for the diagnosis, monitoring, and infection control of patients with SARS-CoV-2 infection [47]. RT-qPCR detection also showed high sensitivity and specificity for SARS-CoV and MERS-CoV infection [48]. However, five patients with negative results of RT-qPCR for SARS-CoV-2 may present with positive chest CT findings, and repeated swab tests (RT-qPCR) eventually confirmed that all patients were infected by SARS-CoV-2 [49]. The detection of SARS-CoV using RT-qPCR can only achieve a sensitivity of 50%–79%, depending on the protocol used the sample type and number of clinical specimens collected [50]. Thus, it is essential to improve the detection rate of RT-qPCR for SARS-CoV-2 infection. Besides, RT-qPCR has some other shortcomings, including certain biological safety hazards brought by the retention and operation of patient samples, cumbersome nucleic acid detection operations, and long waiting time for results.
4.2. CT scans and other diagnostic methods
For the diagnosis of COVID-19, although RT-qPCR is specific, its false-negative rate cannot be ignored because of the severe consequences of missed diagnosis. So many clinicians proposed CT scans should be one necessary auxiliary diagnostic method because it is more sensitive. For individuals with a high clinical suspicion of SARS-CoV-2 infection with negative RT-qPCR screening, a combination of repeated RT-qPCR tests and chest CT scan may be helpful. Especially the high-resolution CT (HRCT) for the chest is essential for early diagnosis and evaluation of disease severity of patients with SARS-CoV-2 [51]. Several studies have analyzed chest CT images of patients infected with SARS-CoV-2 [52,53]. The typical CT images show bilateral pulmonary parenchymal ground-glass and consolidative pulmonary opacities, sometimes with a rounded morphology and a peripheral lung distribution. Lung involvement with a peripheral predominance was also seen in patients with SARS-CoV and MERS-CoV infections, and the chest CT showed that disease progressed with ground-glass opacities and consolidation, which is similar to that of SARS-CoV-2 infection [54,55]. According to those findings, CT scans have a great clinical diagnostic value for COVID-19, especially in the high prevalence area of SARS-CoV-2 infection. However, CT scans also have some shortcomings, such as indistinguishability from other viral pneumonia and the hysteresis of abnormal CT imaging.
Given the shortcomings of the currently used nucleic acid detection and CT scans for the diagnosis of COVID-19, clinical laboratories should apply some immunological detection kits that target viral antigens or antibodies as soon as possible. Currently, POCT of IgM/IgG and ELISA kits for SARS-CoV-2 have been developed and pre-tested by some companies and have shown higher detection rates than nucleic acid detection, but there is still no product or published article. The sensitivity of SARS-CoV N-based IgG ELISA (94.7%) is significantly higher than that of SARS-CoV S-based IgG ELISA (58.9%) [48], but the sensitivity of SARS-CoV-2 IgG/IgM remains to be studied. Hence, developing other sensitive and specific auxiliary methods is necessary and urgent for the diagnosis of COVID-19.
5. Current treatment strategies for COVID-19
Just like SARS-CoV and MERS-CoV [2,56], there is currently no clinically proven specific antiviral agent available for SARS-CoV-2 infection. The supportive treatment, including oxygen therapy, conservation fluid management, and the use of broad-spectrum antibiotics to cover secondary bacterial infection, remains to be the most important management strategy [15]. According to the research on molecular mechanisms of coronavirus infection [57] and the genomic organization of SARS-CoV-2 [6], there are several potential therapeutic targets to repurpose the existing antiviral agents or develop effective interventions against this novel coronavirus.
5.1. Virally targeted inhibitors
Remdesivir, an adenosine analogue that can target the RNA-dependent RNA polymerase and block viral RNA synthesis, has been a promising antiviral drug against a wide array of RNA viruses (including SARS/MERS-CoV) infections in cultured cells [58], mice [59] and nonhuman primate models [60,61]. The Washington Department of Health administrated remdesivir intravenously first and found that remdesivir might have potential protection from SARS-CoV-2 infection [62]. Then remdesivir and chloroquine have been demonstrated to inhibit SARS-CoV-2 effectively in vitro [63]. Hence, other nucleoside analogues, such as favipiravir, ribavirin and galidesivir [56,64], may be potentially clinically applicable against SARS-CoV-2. Chymotrypsin-like (3C-like protease, 3CLpro) and papain-like protease (PLP) are non-structural proteins, which have an essential function for coronaviral replication and can inhibit the host innate immune responses [65]. So 3CLpro inhibitors, such as cinanserin [66] and flavonoids [67], and PLP inhibitors, such as diarylheptanoids [68], are other attractive choices to fight against SARS-CoV-2. ACE2 mediates SARS-CoV-2 entry into the cell as a functional receptor of coronaviruses. So blocking the binding of S protein with ACE2 is also a meaningful strategy against SARS-CoV-2 infection [10].
5.2. Antibody and plasma therapy
It has also been reported that there are many convalescent patients donating plasma against SARS-CoV-2, just as SARS-CoV [69] and MERS-CoV [70] trials. It has preliminary acquired favorable results in acute, severe SARS-CoV-2 patients. Moreover, the generation of recombinant human monoclonal antibody (mAb) is a fairly straightforward path to neutralize SARS-CoV. CR3022, a SARS coronavirus-specific human monoclonal antibody, can bind potently with the receptor-binding domain(RBD) of SARS-CoV-2 and has the potential to be developed as candidate therapeutics of SARS-CoV-2 infections [71]. Other monoclonal antibodies neutralizing SARS-CoV, such as m396, CR3014, could be an alternative for the treatment of SARS-CoV-2 [72].
5.3. Vaccines
Effective SARS-CoV-2 vaccines are essential for reducing disease severity, viral shedding and transmission, thus helping to control the coronavirus outbreaks. There are several vaccination strategies against SARS-CoV, MERS-CoV tested in animals, including a live-attenuated virus, viral vectors, inactivated virus, subunit vaccines, recombinant DNA, and proteins vaccines [73]. These studies are in progress, but it requires months to years to develop the vaccines for SARS-CoV-2.
Currently, there may be many promising targets for SARS-CoV-2, but more laboratory and clinical evidence still should be explored. The WHO is working with Chinese scientists to launch more than 80 clinical trials on potential treatments for SARS-CoV-2. Traditional Chinese medicine seems to have some effects in the supportive treatments. Some new pharmaceutical drugs, including HIV drugs and stem cells, were testified in those clinical trials.
6. Conclusion
In conclusion, the occurrence and development of SARS-CoV-2 depend on the interaction between the virus and the individual’s immune system. Viral factors include virus type, mutation, viral load, viral titer, and viability of the virus in vitro. The individual’s immune system factors include genetics (such as HLA genes), age, gender, nutritional status, neuroendocrine-immune regulation, and physical status. These factors all contribute to whether an individual is infected with the virus, the duration and severity of the disease, and the reinfection. In the early stages of the epidemic, accurate diagnosis helps control the spread of the disease. It is imperative to develop new, safe, accurate, fast and simple new technologies for detecting SARS-CoV-2. Of course, physicians will intentionally intervene in the two factors to make them develop into a direction beneficial to human health, which can help patients recover as soon as possible. However, it must not be considered that medical intervention can achieve a 100% curative effect.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (project No.81970029) and Fundamental Research Funds for the Central Universities of China (The Emergency Projects on COVID-19. project No.xzy032020042). We sincerely thank the individuals who contributed to this work including Xudong Yang, Wenhua Zhu, Jing Xu, Fumeng Huang, Muhammad Saadiq Khan, Jiajun Ren, and Xipeng Wang.
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.03.001.
Contributor Information
Liesu Meng, Email: mengliesu@xjtu.edu.cn.
Shemin Lu, Email: lushemin@xjtu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Wit E., van Doremalen N., Falzarano D. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S., Wong G., Shi W. Epidemiology, Genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlman S. J. Netland Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knoops K., Kikkert M., Worm S.H. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060226. e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., Moore M.J., Vasilieva N. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge X.Y., Li J.L., Yang X.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinese SARS Molecular Epidemiology Consortium Chinese Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 13.Raj V.S., Mou H., Smits S.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barlan A., Zhao J., Sarkar M.K. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J. Virol. 2014;88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffers S.A., Tusell S.M., Gillim-Ross L. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmons G., Reeves J.D., Rennekamp A.J. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., Yang P., Liu K. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuba K., Imai Y., Ohto-Nakanishi T. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol. Ther. 2010;128:119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Wu P., Gao F. Novel immunodominant peptide presentation strategy: a featured HLA-A∗2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Virol. 2010;84:11849–11857. doi: 10.1128/JVI.01464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keicho N., Itoyama S., Kashiwase K. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum. Immunol. 2009;70:527–531. doi: 10.1016/j.humimm.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y.M., Liang S.Y., Shih Y.P. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J. Clin. Microbiol. 2006;44:359–365. doi: 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S.F., Chen K.H., Chen M. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol. 2011;24:421–426. doi: 10.1089/vim.2011.0024. [DOI] [PubMed] [Google Scholar]
- 28.Hajeer A.H., Balkhy H., Johani S. Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Ann. Thorac. Med. 2016;11:211–213. doi: 10.4103/1817-1737.185756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu X., Chong W.P., Zhai Y. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J. Infect. 2015;71:101–109. doi: 10.1016/j.jinf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 31.Xu Z., Shi L., Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Resp. Med. 2020 doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan Y.Y., Huang Z.T., Li L. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch. Virol. 2009;154:1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang F., Quan Y., Xin Z.T. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 34.Zhao J., Li K., Wohlford-Lenane C. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U.S.A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams A.E., Chambers R.C. The mercurial nature of neutrophils: still an enigma in ARDS? Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306:L217–L230. doi: 10.1152/ajplung.00311.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameron M.J., Bermejo-Martin J.F., Danesh A. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min C.K., Cheon S., Ha N.Y. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci. Rep. 2016;6:25359. doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Channappanavar R., Fehr A.R., Vijay R. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Channappanavar R., Fehr A.R., Zheng J. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;130:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niemeyer D., Zillinger T., Muth D. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y., Zhang L., Geng H. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menachery V.D., Schafer A., Burnum-Johnson K.E. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E1012–E1021. doi: 10.1073/pnas.1706928115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corman V.M., Landt O., Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu D.K.W., Pan Y., Cheng S.M.S. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.To K.K., Tsang O.T., Chik-Yan Yip C. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo P.C., Lau S.K., Wong B.H. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 2005;43:3054–3058. doi: 10.1128/JCM.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie X., Zhong Z., Zhao W. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yam W.C., Chan K.H., Poon L.L. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 2003;41:4521–4524. doi: 10.1128/jcm.41.10.4521-4524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan Y., Guan H., Zhou S. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur. Radiol. 2020 doi: 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi H., Han X., Zheng C. Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology. 2020 doi: 10.1148/radiol.2020200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung M., Bernheim A., Mei X. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020 doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ooi G.C., Khong P.L., Muller N.L. Severe acute respiratory syndrome: temporal lung changes at thin-section CT in 30 patients. Radiology. 2004;230:836–844. doi: 10.1148/radiol.2303030853. [DOI] [PubMed] [Google Scholar]
- 55.Ajlan A.M., Ahyad R.A., Jamjoom L.G. Middle East respiratory syndrome coronavirus (MERS-CoV) infection: chest CT findings. AJR Am. J. Roentgenol. 2014;203:782–787. doi: 10.2214/AJR.14.13021. [DOI] [PubMed] [Google Scholar]
- 56.Zumla A., Chan J.F., Azhar E.I. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groneberg D.A., Hilgenfeld R., Zabel P. Molecular mechanisms of severe acute respiratory syndrome (SARS) Respir. Res. 2005;6:8. doi: 10.1186/1465-9921-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lo M.K., Jordan R., Arvey A. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheahan T.P., Sims A.C., Leist S.R. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Wit E., Feldmann F., Cronin J. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 2020 doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo M.K., Feldmann F., Gary J.M. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aau9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holshue M.L., DeBolt C., Lindquist S. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Clercq E. New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chem. Asian J. 2019;14:3962–3968. doi: 10.1002/asia.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L., Gui C., Luo X. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jo S., Kim S., Shin D.H. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park J.Y., Jeong H.J., Kim J.H. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012;35:2036–2042. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- 69.Mair-Jenkins J., Saavedra-Campos M., Baillie J.K. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koenig K.L. Identify-Isolate-Inform: a modified tool for initial detection and management of Middle East Respiratory Syndrome patients in the emergency department. West. J. Emerg. Med. 2015;16:619–624. doi: 10.5811/westjem.2015.7.27915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian X., Li C., Huang A. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microb. Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020 doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.