Significance
Nematode-trapping fungi (NTF) are carnivorous microbes that hold potential to be used as biological control agents due to their ability to consume nematodes. In this work, we show that NTF are ubiquitous generalist predators found in sympatry with their prey in soil samples. Wild isolates of NTF displayed a naturally diverse ability to execute their predatory lifestyle. We generated a large whole-genome sequencing dataset for many of the fungal isolates that will serve as the basis of future projects. In particular, we establish TWF154, a highly responsive strain of Arthrobotrys oligospora, as a model strain to study the genetics of NTF. Lastly, we provide evidence that the G-protein β-subunit Gpb1 plays an important role in trap induction in NTF.
Keywords: nematode-trapping fungi, predator–prey interaction, natural population, G-protein signaling, trap morphogenesis
Abstract
Nematode-trapping fungi (NTF) are a group of specialized microbial predators that consume nematodes when food sources are limited. Predation is initiated when conserved nematode ascaroside pheromones are sensed, followed by the development of complex trapping devices. To gain insights into the coevolution of this interkingdom predator–prey relationship, we investigated natural populations of nematodes and NTF that we found to be ubiquitous in soils. Arthrobotrys species were sympatric with various nematode species and behaved as generalist predators. The ability to sense prey among wild isolates of Arthrobotrys oligospora varied greatly, as determined by the number of traps after exposure to Caenorhabditis elegans. While some strains were highly sensitive to C. elegans and the nematode pheromone ascarosides, others responded only weakly. Furthermore, strains that were highly sensitive to the nematode prey also developed traps faster. The polymorphic nature of trap formation correlated with competency in prey killing, as well as with the phylogeny of A. oligospora natural strains, calculated after assembly and annotation of the genomes of 20 isolates. A chromosome-level genome assembly and annotation were established for one of the most sensitive wild isolates, and deletion of the only G-protein β-subunit–encoding gene of A. oligospora nearly abolished trap formation. In summary, our study establishes a highly responsive A. oligospora wild isolate as a model strain for the study of fungus–nematode interactions and demonstrates that trap formation is a fitness character in generalist predators of the nematode-trapping fungus family.
The living components of soils are in permanent interaction and play central roles in various aspects of biogeochemistry, including nutrient cycling and transport across distances (1). Specifically, nematodes are the most abundant animals in nature, amounting to a staggering 0.3 gigatonnes or 4.5 × 1020 individuals, many of which are devastating parasites of plants, animals, and humans (2). Although available, nematicide chemicals pose severe environmental and health risks and therefore biological control agents such as carnivorous predatory fungi hold potential as alternative pest control tools. Predatory fungi that prey and consume nematodes have been described from several major fungal phyla, suggesting that fungal carnivorism has arisen independently multiple times during evolution (3, 4). In particular, nematode-trapping fungi (NTF) constitute a group of nematophagous organisms that includes a large number of species that rely on the formation of traps to ambush their prey (5, 6). We have previously described that NTF eavesdrop on an evolutionarily conserved family of nematode pheromones, the ascarosides, that are used as a signal to switch from saprophytism to predation via the induction of trap morphogenesis (7), and that NTF use olfactory mimicry to lure their prey into traps (8). Upon being trapped, the nematodes are paralyzed, pierced, invaded, and digested by the fungus (5, 6). Both nematodes and NTF can be found in a wide range of habitats (9, 10); however, sympatrism and adaptation in the context of their relationship are yet to be investigated and little is known about the natural history of this example of a predator–prey interaction.
In this study, we investigated the ecology of natural populations of nematodes and NTF and found that Arthrobotrys species were sympatric with diverse nematodes and behaved as generalist predators. The degree of predation varied greatly among strains and several wild isolates were substantially more efficient predators than the currently used laboratory strain. We sequenced and assembled the genomes of a number of wild Arthrobotrys oligospora strains and demonstrated a correlation between the levels of trap formation, prey killing efficiency, and phylogeny. Lastly, targeted gene deletion of a vG-protein β-subunit–encoding gene revealed that the predatory lifestyle of A. oligospora is dependent upon G-protein signaling.
Results and Discussion
Soil Samples Reveal a Rich Diversity of Nematodes and NTF.
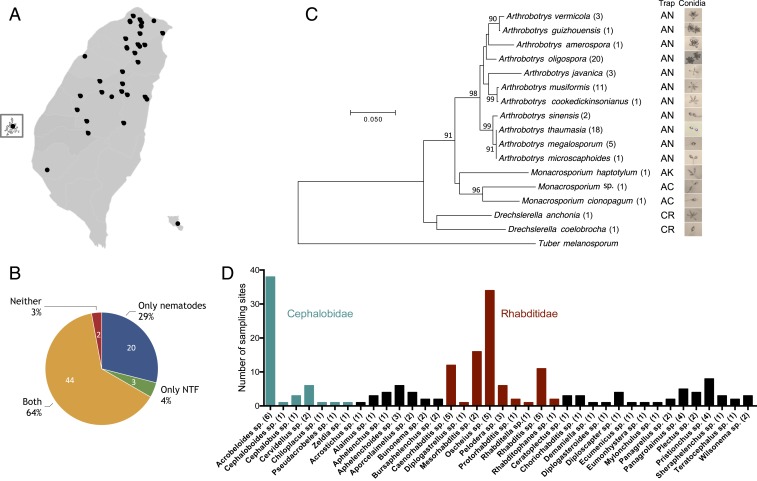
With the goal of isolating sympatric nematodes and NTF from natural habitats, we collected 178 soil samples from 69 ecologically diverse sites in Taiwan (Fig. 1A). Sampling sites were mainly located in northern and central Taiwan, and included forest, lake, agricultural, and coastal environments as well as educational campuses. More than 66% of the sampling sites were mountainous areas, with altitudes ranging from ∼300 to 3,400 m (SI Appendix, Table S3). We established pure cultures of both organisms and their identities were confirmed by sequencing the internal transcribed spacer (ITS) region of the fungal strains and the small-subunit (SSU) ribosomal DNA (rDNA) regions of the nematodes. By not applying a metagenomic sequencing approach, we have likely underestimated the fungal and nematode biodiversity in our samples. Nevertheless, our approach enabled us to establish cultivable isolates of NTF and nematodes and examine their interactions in the laboratory. We identified nematodes and NTF in the vast majority of the 178 soil samples and ∼92 and ∼68% of the sampling sites contained nematodes and/or NTF, respectively. They were sympatric in more than 63% of the sites, indicating that these organisms share the same niches in soil environments and interact closely in nature (Fig. 1B). Only 2 sampling sites (the Xiaoyoukeng fumarole at Yangmingshan National Park and the coastline of Penghu Island) were found to be free of nematodes and NTF, presumably due to extreme environments (high temperature and high salt concentration, respectively) that likely impede survival. In total, we identified 16 previously described species of NTF, as well as a few strains that could represent new species. Among the 16 known species, 11 were identified as belonging to the genus Arthrobotrys, which develops 3D adhesive nets as a trapping device to capture prey (5, 6) (Fig. 1C). A. oligospora was the most commonly found species, followed by Arthrobotrys thaumasia and Arthrobotrys musiformis; combined, these 3 species were present in ∼70% of the sampling sites that had been identified as harboring NTF (Fig. 1C). The remaining 5 species were of the Monacrosporium and Drechslerella genera, which develop adhesive columns and constricting rings as trapping devices (4), and were each isolated from 1 sampling site only (Fig. 1C). Approximately 35% of the sampling sites contained 2 or more species of NTF, indicating that different species of NTF can occupy the same ecological niches (SI Appendix, Table S3). Conidial morphology was diverse among the isolated NTF (Fig. 1C).
Fig. 1.
High diversity of nematodes and nematode-trapping fungus species are sympatric in soil samples. (A) Geographic distribution of the sampling sites in Taiwan. (B) Nematodes and NTF are sympatric in more than 60% of our sampling sites. (C) Phylogeny of the different species of NTF isolated in this study based on their ITS sequences. The numbers represent the prevalence of the difference species of NTF in soil samples. Trap types and images of conidia are shown (Right). AC, adhesive columns; AN, adhesive networks; AK, adhesive knobs; CR, constricting rings. (D) Prevalence and diversity of nematode species in Taiwanese soil samples.
Nematodes were present in ∼93% of the soil samples (Fig. 1B). We cultured the wild nematodes on standard Caenorhabditis elegans nematode growth media (NGM) with Escherichia coli (OP50) as food source, and directly amplified and sequenced the SSU rDNA regions from those able to grow on OP50. We identified more than 74 Nematoda species (Fig. 1D), covering clades II, IV, and V (SI Appendix, Fig. S1), demonstrating a rich nematode biodiversity in Taiwanese soil environments. Oscheius tipulae, Acrobeloides apiculatus, and Acrobeloides nanus were found in ∼40, ∼30, and ∼9% of the sampling sites, respectively (Fig. 1D), in line with previous surveys that showed that these nematodes, especially O. tipulae, are widespread cosmopolitan soil nematodes (11–15).
Arthrobotrys Fungi Behave as Generalist Predators.
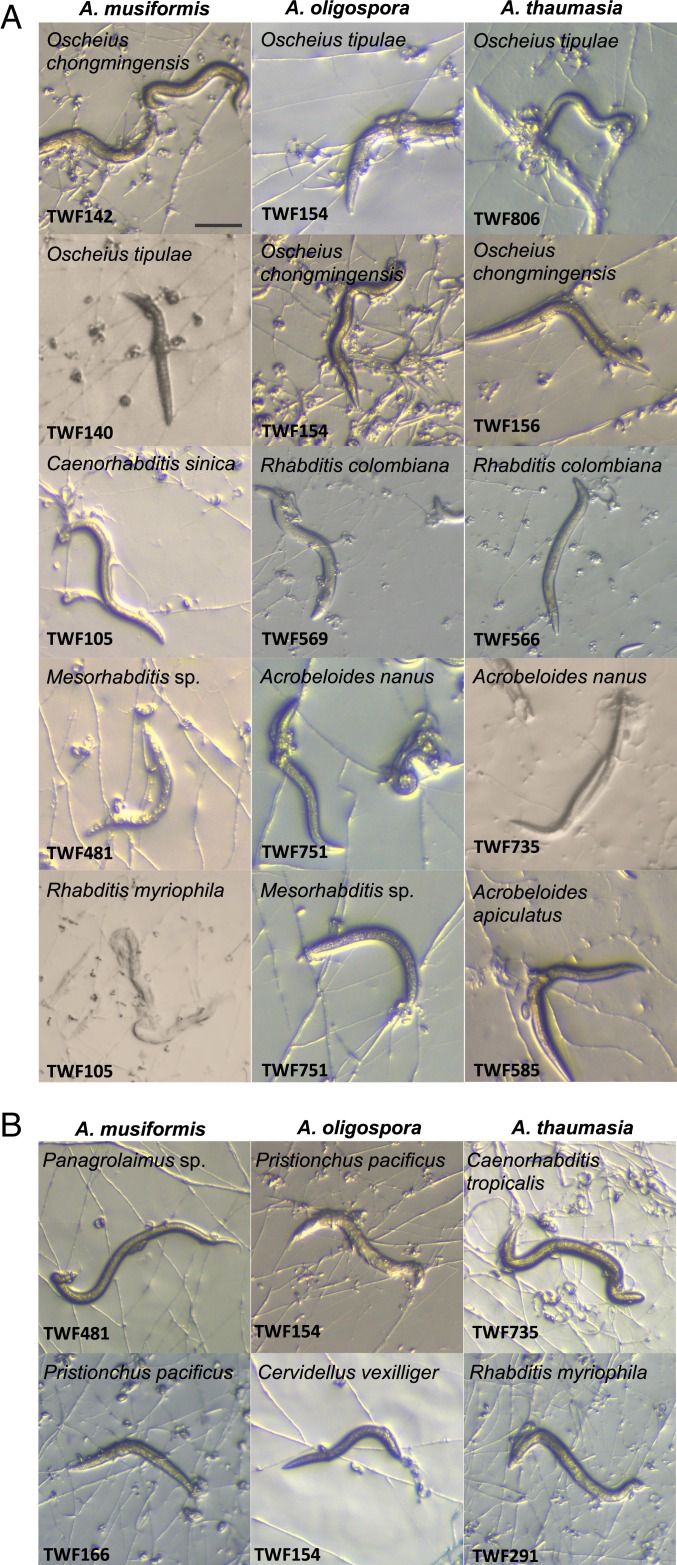
A. oligospora, A. thaumasia, and A. musiformis, the 3 most commonly identified NTF in our soil samples, were sympatric with at least 50, 29, and 31 species of nematodes, respectively, indicating that NTF naturally encounter a broad range of nematodes, including several Caenorhabditis species (SI Appendix, Table S4). C. elegans is known to be associated with rotten fruits (16), and in 1 soil sample collected from an apple orchard, we identified both C. elegans and A. oligospora, providing direct evidence that C. elegans encounters A. oligospora in its natural habitat. To assess nematode predation, we tested the trapping ability of A. oligospora, A. thaumasia, and A. musiformis on sympatric nematode species and observed that all 3 Arthrobotrys preyed on all of the nematode species for which we were able to establish cultures (Fig. 2A). Importantly, when nematodes and NTF from distant sampling sites were cocultured, trap formation and carnivorism were still observed. For example, the nematodes Pristionchus pacificus and Cervidellus vexilliger were not sympatric with A. oligospora in any of the samples, yet A. oligospora still developed traps and consumed isolates of these 2 nematode species (Fig. 2B). Thus, we conclude that A. oligospora does not specifically recognize and prey on particular species of nematodes but instead behaves as a generalist predator. These data support previous findings that highly conserved nematode signals such as ascarosides play a critical role in this predator–prey interaction (7). Additionally, our observation that multiple nematode-trapping fungus species coexist in the same sample suggests that they might compete for prey in their natural environments.
Fig. 2.
A. oligospora, A. musiformis, and A. thaumasia are generalist predators. (A) NTF preying on different sympatric nematode species. (Scale bar, 200 µm.) (B) NTF preying on allopatric nematode species.
Predation Is a Highly Polymorphic Trait in Arthrobotrys Species.
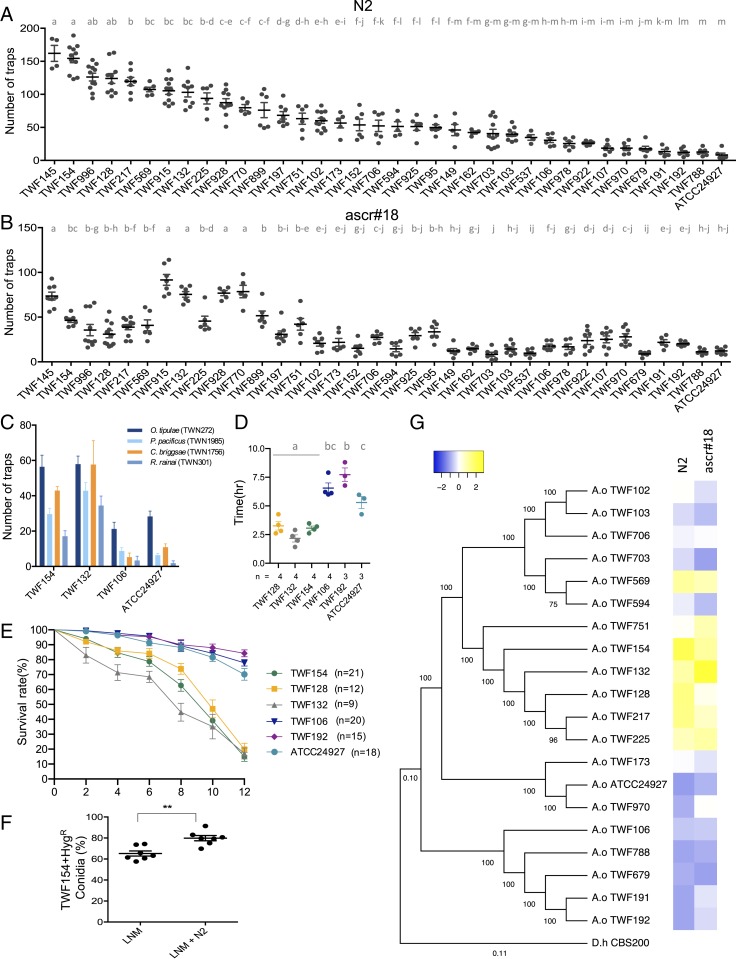
Phenotypic diversity is commonly observed in natural populations of fungi. For example, variations in stress sensitivity have been observed in Saccharomyces cerevisiae (17) and Neurospora crassa (18). Two previous studies have surveyed A. oligospora in the wild (19, 20) but no phenotypic characterization has been performed. Thus, we designed an assay to quantitatively measure the ability of individual isolates to sense prey and develop traps, the most distinctive feature of NTF, and examined the natural diversity of nematode-trapping performance. Wild NTF were exposed to 30 C. elegans L4 larvae for 6 h and the number of traps developed in cultures at 24 h postexposure to nematodes was determined. The average prey-sensing ability of 35 of the A. oligospora wild isolates was highly polymorphic for this particular trait, ranging from ∼8 to 162 traps (Fig. 3A; raw data are available in SI Appendix, Table S5). It is noteworthy that even TWF788, the lowest-performing isolate from our samples, formed, on average, a higher number of traps than the most commonly studied laboratory strain of A. oligospora, ATCC24927 (21), while the highest-performing strains, TWF132 and TWF154 (SI Appendix, Fig. S2), exhibited almost 20-fold more traps than ATCC24927. In fact, our quantitative approach revealed several statistically different groups and at least 21 wild isolates outperformed ATCC24927. A polymorphic response that ranged from ∼8 to 92 traps was also observed when fungal hyphae were exposed to synthesized ascaroside #18 (Fig. 3B; raw data are available in SI Appendix, Table S6).
Fig. 3.
Prey-sensing ability varies considerably among wild isolates of A. oligospora. (A) Quantification of trap number induced by C. elegans WT strain N2 among wild isolates of A. oligospora (mean ± SEM, n = 6; different letters represent significant differences from Tukey test). (B) Quantification of trap number induced by ascarosides among wild isolates of A. oligospora (mean ± SEM, n = 6; different letters represent significant differences from Tukey test). (C) Quantification of trap number induced by C. elegans in wild isolates of A. musiformis and A. thaumasia (mean ± SEM, n = 6). (D) Time of trap emergence after exposition to C. elegans among different A. oligospora wild isolates. (Mean ± SEM; n = 4; different letters represent significant differences from Tukey test.) (E) Percentage of live C. elegans after exposition to different strains of A. oligospora over 12 h. (Mean ± SEM.) (F) Competition fitness assay of A. oligospora isolates TWF154 and TWF106, computed as the percentage of conidia produced by TWF154 against the total number of conidia in the presence and absence of C. elegans (N2). (Mean ± SEM; n = 7; asterisks represent significance levels of unpaired t test.) (G) Phylogeny of 19 A. oligospora wild isolates sequenced and assembled for this study. The phylogenetic tree was constructed using 500 random single orthologs and the heatmap summarizes the trapping response of the isolates toward N2 and ascarosides.
While some wild isolates were highly responsive, others were almost insensitive to this cue, and ATCC24927 was again among the lowest-performing statistical group (Fig. 3B). This finding demonstrates that ascaroside-triggered trap morphogenesis is highly polymorphic among our A. oligospora wild isolates. Responsiveness to C. elegans was positively correlated with responsiveness to ascarosides (SI Appendix, Fig. S3). This correlation, although robust (Spearman, r = 0.7556, P < 0.0001), was not observed throughout all of the strains, suggesting that prey sensing is a multifactorial process. In addition, we studied the amount of phenotypic variation due to genetic variation by means of the SNP heritability in the response of A. oligospora to nematode cues using the genomes of a subpopulation of 18 wild isolates (SI Appendix). We obtained preliminary values of 0.473 and 0.463 for the response to nematodes and ascarosides, respectively, suggesting that genetic differences influenced the response of A. oligospora to prey signals. We did not observe a clear pattern when the association between geographic location and prey-sensing phenotypes was examined (SI Appendix, Fig. S4). We further determined trap formation in 10 A. thaumasia and 10 A. musiformis wild isolates in response to C. elegans (SI Appendix, Fig. S5) and found that this trait was also highly polymorphic in these 2 species, thus providing evidence that prey sensing is likely highly polymorphic in natural populations of the Arthrobotrys genus and that A. thaumasia strains formed substantially fewer traps than A. oligospora and A. musiformis (SI Appendix, Fig. S5). We next investigated if the polymorphic prey-sensing phenotype displayed by A. oligospora wild isolates was nematode species-dependent. We selected 3 A. oligospora strains that were strongly responsive (TWF132, TWF154, and TWF145) and 4 strains that were weakly responsive (TWF106, TWF788, TWF191, and ATCC24927) to C. elegans and measured their ability to form traps in response to 4 other nematode species (Caenorhabditis briggsae, O. tipulae, Rhabditis rainai, and P. pacificus). Overall, the levels of trap formation were diverse among these strains and if responsiveness to C. elegans was higher, the sensitivity to other nematode species was similarly higher and vice versa (Fig. 3C). Thus, natural diversity in the quantities of trap formation is not restricted to just a single species of NTF nor is it restricted to the response to a single species of nematode.
We next assessed whether the onset of trap morphogenesis and the ability to kill nematode prey varied among the natural population of A. oligospora in addition to the variation in trap numbers developed in response to nematode prey. We found that strains TWF154, TWF128, and TWF132, which formed more traps in response to nematode prey (Fig. 3A), developed the initial traps as soon as ∼2.5 h after exposure to C. elegans whereas strains forming fewer traps (TWF106, TWF192, and ATCC24927) exhibited slower onset of trap morphogenesis (∼5 to ∼7.5 h) (Fig. 3D). In line with these results, strains that developed traps faster and greater in number captured the nematode prey significantly faster (Fig. 3E). In our killing assays, only ∼20% of nematodes were still alive 12 h after being added to the fungal cultures in the presence of highly responsive wild isolates in contrast to ∼75 to 85% survival in the case of the weakly responsive wild isolates (Fig. 3E). Altogether, these results indicate that predation on nematodes is a naturally diverse trait in populations of generalist NTF and that some strains have evolved to become intrinsically more robust and efficient in sensing nematodes, executing the trapping developmental program, and consuming prey. Hence, trap morphogenesis seems to function as a fitness characteristic in NTF.
Competition Assays Link Predation Ability to Fitness in A. oligospora.
After consuming the nematode prey, A. oligospora develop chains of conidia (asexual spores) (SI Appendix, Fig. S6). To test the hypothesis that predation ability contributes to the fitness of A. oligospora, measured by the number of conidia produced, we genetically marked the strong predator TWF154 with a hygromycin-resistant marker (TWF154-HYGR) and competed this strain with the weak predator TWF106 in the presence or absence of nematode prey. We found that in the absence of nematode prey, the strong predatory strain exhibited a moderate fitness advantage compared with the weak predatory strain; ∼62% of the conidia harvested from the competition assay were hygromycin-resistant (Fig. 3F). In contrast, in the presence of nematode prey, the fitness advantage was further increased by ∼15%, since 78% of the conidia harvested from the competition assay were produced by the strong predatory strain (Fig. 3F). These results suggest that higher predation ability results in better fitness in the nematode-trapping fungus A. oligospora.
The Nuclear and Mitochondrial Genome of a Highly Responsive A. oligospora Isolate.
In order to generate a genomic resource for future studies on the molecular basis of trap morphogenesis in NTF, we assembled the genomes of 19 A. oligospora wild isolates (Fig. 3G). Interestingly, a phylogenetic analysis using 500 random genes from these 19 genomes (and ATCC24927) showed a clustering pattern that was correlated with the levels of trap formation (Fig. 3G). For example, TWF751, TWF154, TWF132, TWF128, TWF217, and TWF225 formed a separate clade and belonged to the highest-performing group, which were most sensitive to the nematode prey (Fig. 3A). We anticipate that future genome-wide association studies and comparative genomics analyses with additional genomes from the natural population will disclose multiple candidate genetic loci that govern the predation trait.
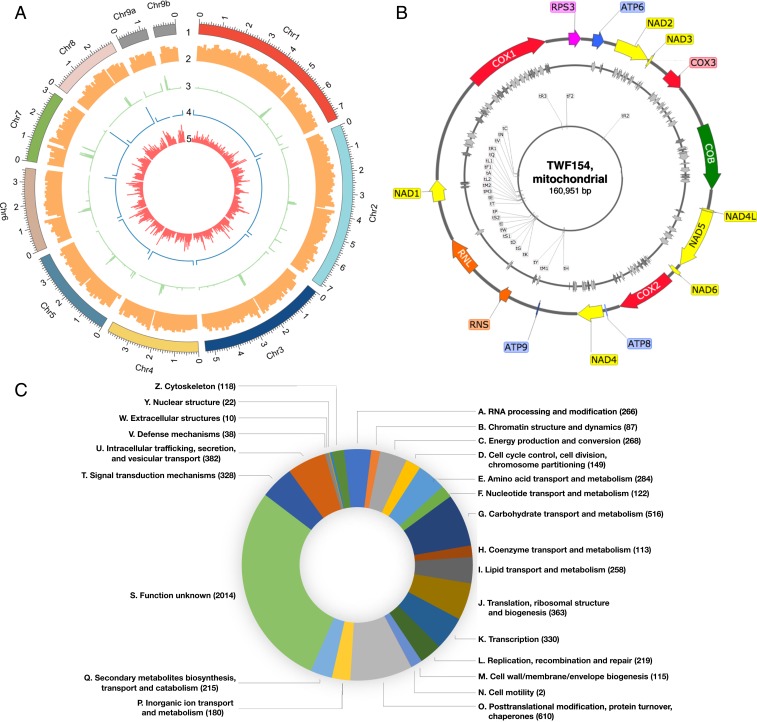
We decided to focus on TWF154, a high-performing wild isolate, in response to C. elegans, other nematode species, and ascarosides (Fig. 3 A–C), that we intend to establish as the preferred A. oligospora model wild-type (WT) strain for further molecular mechanistic studies. We used the PacBio RSII platform to obtain high-quality long reads from strain TWF154 with 131× coverage and assembled a complete and contiguous genome of 39.6 Mbp distributed over 10 scaffolds (Fig. 4A). Eight out of 10 contigs represented a telomere-to-telomere complete chromosome (Chr) assembly (Chr1 to Chr8; Fig. 4A). The remaining 2 contigs (Chr9a and Chr9b) reflect 2 telomere-to-centromere assemblies based on the telomeric repeat sequences identified at one end and the transposable element clusters at the other consisting predominantly of long terminal repeat retrotransposons and DNA transposons, which are typical signatures of centromeres in fungi (22), suggesting that Chr9a and Chr9b represent 2 arms of the same chromosome (Fig. 4A). Genes were distributed evenly along the chromosomes with an average of 247 genes per Mbp, except for regions of low gene distribution at the centromeres (Fig. 4A). Predicted gene function cataloging using the cluster of orthologous groups database showed the presence of all major categories, although the largest group of genes was classified as “function unknown” (Fig. 4B). The quality of the A. oligospora TWF154 genome sequence is among the most contiguous and complete of the sequenced ascomycetes. General features of the genome assemblies for the A. oligospora wild isolates TWF154 and ATCC24927 (23) are presented in Table 1. For example, the number of scaffolds and N50 in our annotation of TWF154 (10 scaffolds and N50 = 5,390,931) were improved in comparison with ATCC24927 (215 and 2,037,373, respectively). Using RNA-seq data from 5 different culture conditions as guidance and the proteomes of representative fungi (SI Appendix), the program Funannotate predicted 12,107 gene models in our improved A. oligospora genome assembly; in comparison, 11,479 genes had previously been found for ATCC24927 (Table 1). The proteome is 97.7% complete based on the BUSCO (24) assessment using the Ascomycota dataset, which is comparable to other published reference fungal genomes. Furthermore, we also assembled and annotated a complete mitochondrial genome (Fig. 4C). Notably, the size of the mitochondrial genome of TWF154 (∼161 kb) is much larger than that of the human (∼17 kb), N. crassa (∼65 kb), or S. cerevisiae (∼79 kb) mitochondrial genomes. The expansion of the mitochondrial genome of TWF154 and other fungal mitochondrial genomes, namely in nematode-trapping fungus species (25–32), can be attributed to the presence of a substantial number of genes encoding homing endonucleases (HEGs) of the LAGLI-DADG and GIY-YIG families in intronic and intergenic regions (Fig. 4C). It will be interesting to evaluate whether the distribution of introns and HEGs varies in diverse Arthrobotrys strains and, more generally, the role of these genes during fungus–nematode interactions.
Fig. 4.
G-protein signaling is required for prey sensing in A. oligospora. (A) Genome architecture of A. oligospora strain TWF154. Tracks (outer to inner) represent the distribution of genomic features: 1) positions (in Mb) of the 10 TWF154 contigs, with numbers indicating the order of scaffold size; 2) gene density (along a 100-kb sliding window); 3) distribution of transposable elements (along a 10-kb sliding window); 4) telomere repeat frequency (along a 1-kb sliding window), showing that 8 contigs have 2 telomeric ends; and 5), A. oligospora species-specific gene content (along a 100-kb sliding window). (B) Predicted function of genes in the TWF154 genome cataloged using the cluster of orthologous groups database. (C) Circular map of the mitochondrial genome of A. oligospora TWF154. Tracks (outer to inner) show: 1) annotation of mitochondrial DNA-encoded genes: subunits of NADH dehydrogenase/complex I (yellow), cytochrome c oxidase/complex IV (red), and ATP synthase/complex V (blue); apocytochrome b COB (green), ribosomal protein S3 RPS3 (pink), and ribosomal RNA genes RNS and RNL (orange); 2) approximate location of LAGLI-DADG (light gray) and GIY-YIG endonucleases (dark gray); 3) annotation of transfer RNA, where “t” stands for “tRNA” followed by the respective amino acid 1-letter code and number of copies.
Table 1.
Genomic features of the 2 assemblies of A. oligospora genomes
| General features | ATCC24927 | TWF154 |
| Size of assembled genome, bp | 40,072,829 | 39,619,008 |
| Number of scaffolds | 215 | 10 |
| N50 scaffold size, bp | 2,037,373 | 5,390,931 |
| N90 scaffold size, bp | 691,077 | 2,932,219 |
| GC content, % | 44.45 | 43.96 |
| Number of genes | 11,479 | 12,107 |
| Number of tRNA genes | 145 | 102 |
| Busco completeness, % | 97.4 | 97.7 |
G-Protein Signaling Is Required for Prey Sensing in A. oligospora.
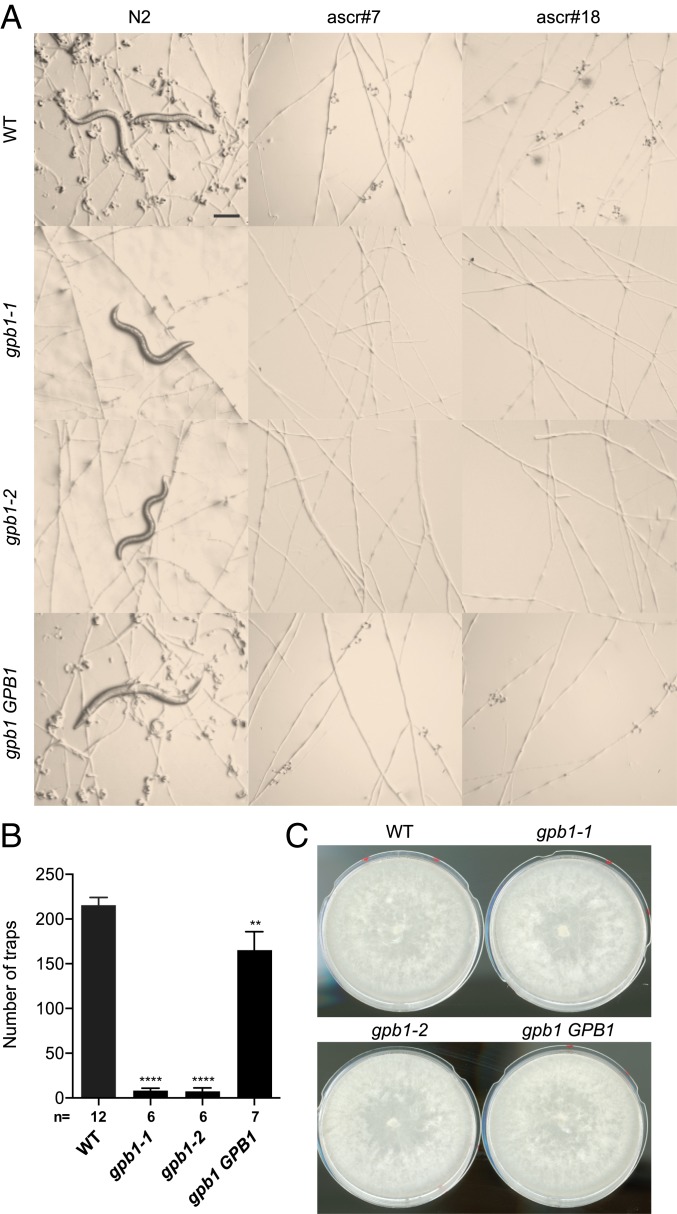
We next performed functional studies of candidate genes that might be involved in sensing the nematodes. In C. elegans, ascarosides are sensed by G protein-coupled receptors (GPCRs) (33, 34) and in fungi, G-protein signaling and GPCRs are critical for sensing environmental stimuli, including sex pheromones, and are required for virulence and morphogenesis in various fungal pathogens (35, 36). It had been suggested via chemical inhibitors that G proteins might be involved in the closure of the constriction ring of another nematode-trapping fungus, Drechslerella dactyloides (37). Furthermore, a recent study showed that the disruption of a Rab GTPase, AoRab-7A, abolished trap formation in A. oligospora (38). For these reasons, we investigated the role of G-protein signaling in prey sensing in A. oligospora. We identified a single G-protein β-subunit gene (GPB1, EYR41_009480) and 3 G-protein α-subunit genes (EYR41_010456, EYR41_011543, and EYR41_011720) in the genome of A. oligospora. Since the Gαs are known to have overlapped functions in many fungi (39), we decided to test the function of the G-protein β-subunit first. We obtained 2 independent gpb1 deletion mutants generated via homologous recombination and determined that the efficiency of homologous recombination was extremely low (∼3%). The 2 gpb1 mutants were strongly defective in trap morphogenesis in response to C. elegans and ascarosides, and exhibited no obvious growth defects (Fig. 5). This phenotype was complemented by reintroducing the WT copy of the GPB1 gene (Fig. 5 A and B). These results demonstrate that G-protein signaling is required for ascaroside sensing and/or trap formation in A. oligospora and that the ascaroside receptors in A. oligospora might be GPCRs.
Fig. 5.
G-protein β-subunit Gpb1 plays an important role in trap induction in A. oligospora. (A) Images of the traps induced by C. elegans laboratory strain N2 or ascarosides (ascr#7 and ascr#18) in the WT (TWF154), gpb1 mutants, and gpb1 GPB1 rescued strain. (Scale bar, 200 µm.) (B) Quantification of the trap numbers induced by C. elegans WT strain N2 or ascarosides (ascr#7 and ascr#18) in the WT (TWF154), gpb1 mutants, or gpb1 GPB1 reconstituted strain (n shown along the x axis). (Mean + SEM; n shown along the x axis; asterisks represent significance levels of unpaired t test compared to the WT.) (C) Fungal colonies developed by the WT (TWF154), gpb1 mutants, and gpb1 GPB1 rescued strain after 5 d of growth on PDA plates (5-cm diameter).
Conclusion
Our study revealed that NTF are widespread in soils of distinct ecological provenances, behave as generalist predators, are sympatric with diverse species of nematodes, and vary considerably in their spatial and temporal ability to capture the nematode prey. NTF hold great potential to be utilized as biological control agents in agricultural settings. However, only recently have the molecular mechanisms underlying the singular biology of these specialized predators begun to be uncovered. For example, a recent work has generated a strain lacking the signaling scaffold protein Soft and shown that cell–cell fusion is required for ring closure in Duddingtonia flagrans (40). In A. oligospora, ATCC24927-background trapping-deficient mutations have been isolated: adhesin protein Mad1 (41), autophagy protein Atg8 (42), mitogen-activated protein kinase Slt2 (43), pH sensor PalH (44), NADPH oxidase NoxA (45), low-affinity calcium uptake system proteins (46), Rab GTPase Rab7A (38), actin-associated protein Crn1 (47), Woronin body component Hex1 (48), malate synthase Mls (49), glycogen phosphorylase Gph1 (50), transcription factors VelB (51) and StuA (52), and microRNA-processing protein Qde2 (53), to which we add the G-protein β-subunit Gpb1. However, a mechanistic view of how these and other molecular players are interconnected during the interaction of A. oligospora with their prey is totally lacking. The establishment of a high-efficiency A. oligospora model strain (TWF154) and the generation of publicly available whole-genome sequencing resources for an ample number of wild isolates will pave the way to the identification of novel loci underlying phenotypic traits and, more generally, to the fast advancement of research with NTF.
Materials and Methods
Isolation of Nematodes and NTF from Soil Samples.
Nematodes and NTF were isolated using the soil sprinkle method (54) with some modifications. Briefly, soil samples were sprinkled on low-nutrient medium (LNM) and, after 2 to 7 d, nematodes were transferred to NGM containing E. coli OP50. SSU rDNA sequences were PCR-amplified directly from the nematodes using the single-worm PCR protocol with primers G18S4a (5′-GCTCAAGTAAAAGATTAAGCCATGC-3′) and DF18S-8 (5′-GTTTACGGTCAGAACTASGGCGG-3′) (55, 56). The National Center for Biotechnology Information (NCBI) basic local alignment search tool (BLAST) (57) was used to determine the identity of the nematode isolates to species level (if the identity was higher than or equal to 97% when compared with a known species) or to the genus level (if the identity was lower than 97%). Once nematodes had been isolated from the LNM plates, C. elegans were added to the same plates to induce trap formation in the NTF. After 3 to 7 d, the plates were examined under a stereo microscope and single conidia of the NTF were isolated and transferred individually to potato dextrose agar (PDA) to establish pure cultures. ITS regions were directly amplified from fungal hyphae by PCR with the universal primers ITS1 5′-TCCGTAGGTGAACCTGCGG-3′ and ITS4 5′-TCCTCCGCTTATTGATATGC-3′ following the single-worm PCR protocol (56, 58). Species identity was assigned if the ITS sequence identity was 97% or higher from NCBI BLAST (57) analyses. Strains of NTF and nematodes used in this study are listed in SI Appendix, Table S7. The SSU rDNA and ITS sequences are available in the NCBI with the accession numbers provided in SI Appendix, Table S8. C. elegans animals employed in this study were maintained following standard procedures (55).
Phenotypical Characterization.
To quantify trap formation, fungal isolates were grown on LNM for 5 d, transferred to a fresh LNM plate, and, after ∼48 h (25 °C, dark), exposed to 30 N2 C. elegans L4-stage larvae for 6 h (after which the animals were removed with a pick) or 1 µL 1 mM ascaroside #18 (ascr#18) (59) that was added ∼2 mm away from the border of hyphal tips. We focused on ascr#18 because it is widespread among both free-living and parasitic nematodes. It is the most abundantly excreted ascaroside of all plant-parasitic nematodes analyzed so far (60, 61) and it is the most abundant in several entomopathogenic nematodes (62). Ascaroside #18 was synthesized as reported (63) and was of greater than 98% purity. Micrographs were taken after 24 h after the addition of nematodes or ascaroside using a Nikon SMZ 745T stereo microscope. For each plate, 3 images were randomly taken within 0.5 cm from the edge of the plate using a 40× magnification, and the sum of traps in the 3 images was recorded. To estimate the survival rate of C. elegans after exposure to A. oligospora, the number of moving nematodes was computed every 2 h for a total of 12 h using a stereo microscope. Nematodes crawling on the wall of the plate, and therefore not in contact with the fungal hyphae, were excluded. Survival rates are presented as the percentage of moving worms over the total number of worms. The time for trap emergence after contact with nematodes was measured in the same conditions as described for the previous assays but 5-cm LNM plates, 3 d of growth (25 °C, dark), and 300 adult N2 C. elegans were used. A Zeiss Stemi 305 microscope with a Zeiss Axiocam ERc 5s camera was used to automatically capture images of the colony edge every 60 s for a total of 12 h. Either 3 or 4 time-lapse videos were taken for each strain consisting of 2,560- × 1,920-pixel images representing an area of about 5.7 × 4.3 mm of the edge of the fungal colony. The time-lapse videos were visually inspected to record the time point at which the first trap arose in the imaged areas. For the competition assay, a hygromycin B (HygB)-resistant TWF154 strain was constructed by transforming a HygB resistance cassette [amplified from vector pAN7-1 (64)] into TWF154 protoplasts (SI Appendix). The HygB-resistant TWF154 strain showed no defect in growth, conidiation, and trap morphogenesis compared with WT. Single spores of both HygB-resistant TWF154 and TWF106 (HygB-sensitive) were collected and transferred together to the center of replicated 5-cm LNM plates. After 4 d of growth, 300 adult N2 C. elegans were added to half of the plates and all fungal colonies were further grown for 3 more days to allow for trapping and digestion of the nematodes by the fungi. A spore suspension was then collected from the plates by adding 2 mL ddH2O and scratching the surface of the fungal colony, followed by centrifugation (1 min, 13,200 rpm) and removal of the supernatant, yielding ∼0.2 mL highly concentrated spore solutions. Between 8 and 12 µL of each spore suspension were added to 9-cm plates containing PDA and PDA with 50 μL/mL HygB, respectively; all plates were treated with 90 µL 11 mg/mL ampicillin before the spore solution was added to prevent bacterial contamination. After 24 h, the germinated spores in each plate were counted using a Zeiss Stemi 305 microscope.
TWF154 Genome Assembly and Annotation and Phylogenetic Analyses of Nematode-Trapping Fungi.
The TWF154 reference genome was sequenced and assembled from ∼0.5 million long reads with an average length of 11,258 bp sequenced from the Pacbio RSII platform and polished with 18 million 250-bp Illumina reads. Annotation was done using Funannotate (65) and the pipeline described at https://funannotate.readthedocs.io/en/latest/tutorials.html. Annotation of the mitochondrial genome was performed using a combination of MFannot (66) and MITOS2 (67). Details can be found in SI Appendix. The neighbor-joining algorithm of MEGA7 or MEGAX was used to construct maximum-likelihood phylogenetic trees. The tree of 16 different nematode-trapping fungus species isolated from soil samples was constructed based on ITS nucleotide sequences. Tuber melanosporum was the outgroup. The tree of the 20 A. oligospora isolates was constructed using 500 random single-copy orthologs from ATCC24927, the TWF154 reference genome, and 18 additional wild isolate genomes. Dactylellina haptotyla (strain CBS200) was used as the outgroup. The genomes of 18 wild isolates of A. oligospora were assembled from 18 million 250-bp Illumina reads using AAFTF (68) and annotated using Funannotate (65). Further details about the assembly and annotation of the fungal isolates, as well as tree construction, can be found in SI Appendix.
Identification and Deletion of the GPB1 Homolog in A. oligospora.
The GPB1 homolog of A. oligospora was identified with Blast2GO 5 Pro (69) by performing a local BLAST using the amino acid sequence of Gpb1 orthologs of N. crassa (UniProt ID Q7RWT0), Aspergillus nidulans (Q5BH99), Fusarium oxysporum (Q96VA6), and S. cerevisiae (A6ZP55) as the query and the proteome of the T727 strain of A. oligospora as the database. GPB1 was deleted by a homologous recombination-based strategy (70). Detailed methods including primers used for generating the gpb1 deletion mutant are described in SI Appendix.
Data Availability.
The genomic data presented in this paper have been deposited to National Center for Biotechnology Information GenBank. The accession numbers are SOZJ00000000 (TWF154 reference genome), MN972613 (TWF154 mitochondria), WIQW00000000 (TWF102), WIQX00000000 (TWF103), WIWS00000000 (TWF106), JAABOH000000000 (TWF128), JAABLO000000000 (TWF132), JAAGKP000000000 (TWF173), WIPF00000000 (TWF191), WIPG00000000 (TWF192), JAABOI000000000 (TWF217), WIWR00000000 (TWF225), WIRA00000000 (TWF569), WIRB00000000 (TWF594), WIWT00000000 (TWF679), WIQZ00000000 (TWF703), WIQY00000000 (TWF706), WIWQ00000000 (TWF751), JAABOE000000000 (TWF788), and JAABOJ000000000 (TWF970). References to these accession numbers can also be found throughout this paper and in SI Appendix.
Supplementary Material
Acknowledgments
We thank John Wang, Jun-Yi Leu, and Sheng-Feng Shen for their helpful comments and suggestions about this work. We also thank Ling-Mei Hsu and A-Mei Yang for their technical assistance. This work was supported by the start-up fund of Academia Sinica and Taiwan Ministry of Science and Technology 106-2311-B-001-039-MY3 (to Y.-P.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The genomic data presented in this paper have been deposited to National Center for Biotechnology Information GenBank. The accession numbers are SOZJ00000000 (TWF154 reference genome), MN972613 (TWF154 mitochondria), WIQW00000000 (TWF102), WIQX00000000 (TWF103), WIWS00000000 (TWF106), JAABOH000000000 (TWF128), JAABLO000000000 (TWF132), JAAGKP000000000 (TWF173), WIPF00000000 (TWF191), WIPG00000000 (TWF192), JAABOI000000000 (TWF217), WIWR00000000 (TWF225), WIRA00000000 (TWF569), WIRB00000000 (TWF594), WIWT00000000 (TWF679), WIQZ00000000 (TWF703), WIQY00000000 (TWF706), WIWQ00000000 (TWF751), JAABOE000000000 (TWF788), and JAABOJ000000000 (TWF970). References to these accession numbers can also be found throughout this paper and in SI Appendix.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919726117/-/DCSupplemental.
References
- 1.Bardgett R. D., van der Putten W. H., Belowground biodiversity and ecosystem functioning. Nature 515, 505–511 (2014). [DOI] [PubMed] [Google Scholar]
- 2.van den Hoogen J., et al. , Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Yang E., et al. , Origin and evolution of carnivorism in the Ascomycota (fungi). Proc. Natl. Acad. Sci. U.S.A. 109, 10960–10965 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y., Yang E., An Z., Liu X., Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc. Natl. Acad. Sci. U.S.A. 104, 8379–8384 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vidal-Diez de Ulzurrun G., Hsueh Y. P., Predator-prey interactions of nematode-trapping fungi and nematodes: Both sides of the coin. Appl. Microbiol. Biotechnol. 102, 3939–3949 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Jiang X., Xiang M., Liu X., Nematode-trapping fungi. Microbiol. Spectr. 5, FUNK-0022-2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsueh Y. P., Mahanti P., Schroeder F. C., Sternberg P. W., Nematode-trapping fungi eavesdrop on nematode pheromones. Curr. Biol. 23, 83–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsueh Y. P., et al. , Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. eLife 6, e20023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray N. F., Ecology of nematophagous fungi: Distribution and habitat. Ann. Appl. Biol. 102, 501–509 (1983). [Google Scholar]
- 10.Bongers T., Ferris H., Nematode community structure as a bioindicator in environmental monitoring. Trends Ecol. Evol. 14, 224–228 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Azizoglu U., Karabörklü S., Ayvaz A., Yilmaz S., Phylogenetic relationships of insect-associated free-living Rhabditid nematodes from eastern Mediterranean region of Turkey. Appl. Ecol. Environ. Res. 14, 93–103 (2016). [Google Scholar]
- 12.Baïlle D., Barrière A., Félix M. A., Oscheius tipulae, a widespread hermaphroditic soil nematode, displays a higher genetic diversity and geographical structure than Caenorhabditis elegans. Mol. Ecol. 17, 1523–1534 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Félix M. A., Oscheius tipulae. WormBook, 1–8 (2006). [DOI] [PMC free article] [PubMed]
- 14.Bird A. F., Ryder M. H., Feeding of the nematode Acrobeloides nanus on bacteria. J. Nematol. 25, 493–499 (1993). [PMC free article] [PubMed] [Google Scholar]
- 15.Renco M., Murin J., Soil nematode assemblages in natural European peatlands of the Horna Orava protected landscape area, Slovakia. Wetlands 33, 459–470 (2013). [Google Scholar]
- 16.Félix M. A., Braendle C., The natural history of Caenorhabditis elegans. Curr. Biol. 20, R965–R969 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Kvitek D. J., Will J. L., Gasch A. P., Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 4, e1000223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonçalves A. P., Hall C., Kowbel D. J., Glass N. L., Videira A., CZT-1 is a novel transcription factor controlling cell death and natural drug resistance in Neurospora crassa. G3 (Bethesda) 4, 1091–1102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., et al. , Genetic diversity and recombination in natural populations of the nematode-trapping fungus Arthrobotrys oligospora from China. Ecol. Evol. 3, 312–325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Yu Z. F., Xu J., Zhang K. Q., Divergence and dispersal of the nematode-trapping fungus Arthrobotrys oligospora from China. Environ. Microbiol. Rep. 3, 763–773 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Nordbring-Hertz B., Influence of medium composition and additions of animal origin on formation of capture organs in Arthrobotrys oligospora. Physiol. Plant. 21, 52–65 (1968). [Google Scholar]
- 22.Stajich J. E., et al. , Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. U.S.A. 107, 11889–11894 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., et al. , Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 7, e1002179 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Deng Y., et al. , Comparison of the mitochondrial genome sequences of six Annulohypoxylon stygium isolates suggests short fragment insertions as a potential factor leading to larger genomic size. Front. Microbiol. 9, 2079 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang M. L., et al. , Characterization of the complete mitochondrial genome of Drechslerella brochopaga, a fungal species trapping nematodes with constricting rings. Mitochondrial DNA B Resour. 4, 858–859 (2019). [Google Scholar]
- 27.Kolesnikova A. I., et al. , Mobile genetic elements explain size variation in the mitochondrial genomes of four closely-related Armillaria species. BMC Genomics 20, 351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S. J., et al. , Complete mitochondrial genome and phylogenetic analysis of Orbilia dorsalia, a species producing mature sexual structures on culture. Mitochondrial DNA B Resour. 4, 573–574 (2019). [Google Scholar]
- 29.Zhang Y. J., Zhang S., Liu X. Z., The complete mitochondrial genome of the nematode endoparasitic fungus Hirsutella minnesotensis. Mitochondrial DNA A. DNA Mapp. Seq. Anal. 27, 2693–2694 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y. Q., Yu Z. F., The complete mitochondrial genomes of the nematode-trapping fungus Arthrobotrys musiformis. Mitochondrial DNA B Resour. 4, 979–980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou D. Y., et al. , The complete mitochondrial genome of the nematode-trapping fungus Dactylellina haptotyla. Mitochondrial DNA B Resour. 3, 964–965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zubaer A., Wai A., Hausner G., The mitochondrial genome of Endoconidiophora resinifera is intron rich. Sci. Rep. 8, 17591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGrath P. T., et al. , Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477, 321–325 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim K., et al. , Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science 326, 994–998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lengeler K. B., et al. , Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvaro C. G., Thorner J., Heterotrimeric G protein-coupled receptor signaling in yeast mating pheromone response. J. Biol. Chem. 291, 7788–7795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen T. H., Hsu C. S., Tsai P. J., Ho Y. F., Lin N. S., Heterotrimeric G-protein and signal transduction in the nematode-trapping fungus Arthrobotrys dactyloides. Planta 212, 858–863 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Yang X., et al. , Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 102, 4601–4613 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Hsueh Y. P., Xue C., Heitman J., G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans. Mol. Biol. Cell 18, 3237–3249 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youssar L., et al. , Intercellular communication is required for trap formation in the nematode-trapping fungus Duddingtonia flagrans. PLoS Genet. 15, e1008029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang L., et al. , A proposed adhesin AoMad1 helps nematode-trapping fungus Arthrobotrys oligospora recognizing host signals for life-style switching. Fungal Genet. Biol. 81, 172–181 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Chen Y. L., Gao Y., Zhang K. Q., Zou C. G., Autophagy is required for trap formation in the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. Rep. 5, 511–517 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Zhen Z., et al. , MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 116, 42–50 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Li J., et al. , The pH sensing receptor AopalH plays important roles in the nematophagous fungus Arthrobotrys oligospora. Fungal Biol. 123, 547–554 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Li X., et al. , The NADPH oxidase AoNoxA in Arthrobotrys oligospora functions as an initial factor in the infection of Caenorhabditis elegans. J. Microbiol. 55, 885–891 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Zhang W., et al. , Role of low-affinity calcium system member Fig1 homologous proteins in conidiation and trap-formation of nematode-trapping fungus Arthrobotrys oligospora. Sci. Rep. 9, 4440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang D., et al. , The roles of actin cytoskeleton and actin-associated protein Crn1p in trap formation of Arthrobotrys oligospora. Res. Microbiol. 168, 655–663 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Liang L., et al. , The Woronin body in the nematophagous fungus Arthrobotrys oligospora is essential for trap formation and efficient pathogenesis. Fungal Biol. 121, 11–20 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Zhao X., et al. , Malate synthase gene AoMls in the nematode-trapping fungus Arthrobotrys oligospora contributes to conidiation, trap formation, and pathogenicity. Appl. Microbiol. Biotechnol. 98, 2555–2563 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Wu Q. Y., Zhu Y. Y., Zou C. G., Kang Y. Q., Liang L. M., GPH1 is involved in glycerol accumulation in the three-dimensional networks of the nematode-trapping fungus Arthrobotrys oligospora. J. Microbiol. 54, 768–773 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Zhang G., et al. , The velvet proteins VosA and VelB play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 10, 1917 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie M., et al. , AoStuA, an APSES transcription factor, regulates the conidiation, trap formation, stress resistance and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. 21, 4648–4661 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Ji X., et al. , The lifestyle transition of Arthrobotrys oligospora is mediated by microRNA-like RNAs. Sci. China Life Sci., 10.1007/s11427-018-9437-7 (23 April 2019). [DOI] [PubMed]
- 54.Gray N. F., Ecology of nematophagous fungi—Methods of collection, isolation and maintenance of predatory and endo-parasitic fungi. Mycopathologia 86, 143–153 (1984). [Google Scholar]
- 55.Barrière A., Félix M. A., Isolation of C. elegans and related nematodes. WormBook, 1–19 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Fay D., Bender A., Genetic mapping and manipulation: Chapter 4—SNPs: Introduction and two-point mapping. WormBook, 1–7 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Martin K. J., Rygiewicz P. T., Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5, 28 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choe A., et al. , Ascaroside signaling is widely conserved among nematodes. Curr. Biol. 22, 772–780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manosalva P., et al. , Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat. Commun. 6, 7795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klessig D. F., et al. , Nematode ascaroside enhances resistance in a broad spectrum of plant–pathogen systems. J. Phytopathol. 167, 265–272 (2019). [Google Scholar]
- 62.Manohar M., et al. , Plant metabolism of nematode pheromones mediates plant-nematode interactions. Nat. Commun. 11, 208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y. K., Sanchez-Ayala M. A., Sternberg P. W., Srinivasan J., Schroeder F. C., Improved synthesis for modular ascarosides uncovers biological activity. Org. Lett. 19, 2837–2840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Punt P. J., Oliver R. P., Dingemanse M. A., Pouwels P. H., van den Hondel C. A., Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56, 117–124 (1987). [DOI] [PubMed] [Google Scholar]
- 65.Palmer J., Funannotate. http://funannotate.readthedocs.io. Accessed 2 June 2019.
- 66.Beck N., Lang B., MFannot, Organelle genome annotation webserver. https://megasun.bch.umontreal.ca/cgi-bin/dev_mfa/mfannotInterface.pl. Accessed 15 June 2019.
- 67.Bernt M., et al. , Mitos: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Stajich J. E., Palmer J., AAFTF: Automated assembly for the fungi. GitHub. https://github.com/stajichlab/AAFTF. Accessed 1 March 2019.
- 69.Conesa A., Götz S., Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szewczyk E., et al. , Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic data presented in this paper have been deposited to National Center for Biotechnology Information GenBank. The accession numbers are SOZJ00000000 (TWF154 reference genome), MN972613 (TWF154 mitochondria), WIQW00000000 (TWF102), WIQX00000000 (TWF103), WIWS00000000 (TWF106), JAABOH000000000 (TWF128), JAABLO000000000 (TWF132), JAAGKP000000000 (TWF173), WIPF00000000 (TWF191), WIPG00000000 (TWF192), JAABOI000000000 (TWF217), WIWR00000000 (TWF225), WIRA00000000 (TWF569), WIRB00000000 (TWF594), WIWT00000000 (TWF679), WIQZ00000000 (TWF703), WIQY00000000 (TWF706), WIWQ00000000 (TWF751), JAABOE000000000 (TWF788), and JAABOJ000000000 (TWF970). References to these accession numbers can also be found throughout this paper and in SI Appendix.