Summary
The major mechanism of antibody-mediated neutralization of the Middle East respiratory syndrome coronavirus (MERS-CoV) involves competition with the cellular receptor dipeptidyl peptidase 4 (DPP4) for binding to the receptor-binding domain (RBD) of the spike (S) glycoprotein. Here, we report a unique epitope and unusual neutralizing mechanism of the isolated human antibody MERS-4. Structurally, MERS-4 approached the RBD from the outside of the RBD-DPP4 binding interface. Such binding resulted in the folding of the β5-β6 loop toward a shallow groove on the RBD interface critical for accommodating DPP4. The key residues for binding are identified through site-directed mutagenesis. Structural modeling revealed that MERS-4 binds to RBD only in the “up” position in the S trimer. Furthermore, MERS-4 demonstrated synergy with several reported antibodies. These results indicate that MERS-4 neutralizes MERS-CoV by indirect rather than direct competition with DPP4. This mechanism provides a valuable addition for the combined use of antibodies against MERS-CoV infection.
Keywords: Middle East respiratory syndrome, coronavirus, crystal structure, neutralizing antibody, antibody epitope
Graphical Abstract

Highlights
-
•
MERS-4 binds RBD from the outside of the RBD–DPP4 binding interface
-
•
MERS-4 favors binding to the RBD in the “up” position in the S trimer
-
•
MERS-4 neutralizes MERS-CoV by indirect rather than direct competition with DPP4
-
•
MERS-4 is a valuable addition to the combined use of MERS-CoV antibodies
Zhang et al. report the structural and functional analysis of the potent MERS-CoV neutralizing antibody MERS-4. MERS-4 recognizes a unique epitope and indirectly disrupts interaction between the receptor binding domain and the receptor DPP4. This mechanism provides a valuable addition for the combined use of antibodies against MERS-CoV infection.
Introduction
The 2012 emergence of the Middle East respiratory syndrome coronavirus (MERS-CoV) in Saudi Arabia was the second major introduction of a highly pathogenic coronavirus into the human population since the outbreak of SARS-CoV in China in 2003 (Assiri et al., 2013, Zaki et al., 2012). The global spread of SARS-CoV resulted in more than 8,000 infections and nearly 800 deaths worldwide (Peiris et al., 2004). Fortunately, the SARS-CoV epidemic rapidly died off because of conventional public health measures, even if the exact mechanism of its introduction into, and disappearance from, the human population still remains a mystery (Graham et al., 2013). By contrast, the MERS-CoV epidemic has persisted for more than 5 years with no signs of abating (http://www.who.int/csr/don/10-november-2017-mers-oman/en/). Apart from the sudden initial outbreak in Saudi Arabia, MERS-CoV has spread to other countries outside the Arabian Peninsula carried by infected travelers (Bermingham et al., 2012), leading to an outbreak in South Korea in 2015 (Choi, 2015). These are continuous reports of human MERS-CoV infection in affected regions, largely due to contact with dromedary camels, which are believed to be a major reservoir host for MERS-CoV and the immediate source of human infection (Haagmans et al., 2014, Hemida et al., 2014). Compared with SARS-CoV with a fatality rate of approximately 10% (Peiris et al., 2004), MERS-CoV appears to be more deadly, with its fatality rate reaching as high as 35% (de Wit et al., 2016). All of these facts indicate that MERS-CoV will remain as a severe and long-time threat to global health and highlight the urgent need for effective prophylactic and therapeutic measures.
Scientific progress achieved since the SARS-CoV epidemic has greatly increased our capacity to respond to MERS-CoV. Like that of SARS-CoV, the spike (S) glycoprotein of MERS-CoV plays a critical role in mediating viral entry and in inducing a protective antibody response in infected individuals (Raj et al., 2013, Zumla et al., 2016). The isolation of potent neutralizing monoclonal antibodies has been reported shortly after the identification of MERS-CoV. This was achieved by using various technological platforms such as phage or yeast display of antibody library (Jiang et al., 2014, Tang et al., 2014, Ying et al., 2014), immunization of experimental animals (Chen et al., 2017a, Du et al., 2014, Li et al., 2015, Pascal et al., 2015, Wang et al., 2015a), and direct isolation from human survivors of MERS-CoV infection (Chen et al., 2017b, Corti et al., 2015, Wang et al., 2018). Currently, close to 20 different neutralizing monoclonal antibodies have been characterized in cell culture and experimental animal models. A large majority target the receptor-binding domain (RBD) of the MERS-CoV S glycoprotein and interfere with the binding of the cellular receptor dipeptidyl peptidase 4 (DPP4). We previously reported the isolation of the two neutralizing antibodies MERS-4 and MERS-27 via screening of a yeast-displayed library of human scFv (single-chain variable domain fragment) using the MERS-CoV RBD as bait (Jiang et al., 2014). Both antibodies demonstrated potent neutralizing activity against live and pseudotyped MERS-CoV, and a high level of synergy when used together.
Structural determination of antibodies and their epitope specificities provides a critical foundation for a better understanding of their mechanisms of neutralization. We and others have demonstrated that MERS-CoV RBD possesses core and receptor-binding subdomains (Chen et al., 2013, Lu et al., 2013, Wang et al., 2013, Wang et al., 2015a). A large majority of neutralizing antibodies against MERS-CoV, including MERS-27, 4C2, D12, JC57-14, m336, MCA1, and CDC-C2, have been found to target the receptor-binding subdomain and overlap with the DPP4 binding surface (Chen et al., 2017b, Li et al., 2015, Wang et al., 2015a, Wang et al., 2018, Ying et al., 2015, Yu et al., 2015). Therefore, these antibodies share at least one similar aspect of neutralization by directly competing with the cellular receptor DPP4 for binding to RBD.
Here, we report the structural and functional analysis of our previously isolated potent neutralizing antibody MERS-4 and its variant MERS-4V2, which reveals their unique epitope specificity and unusual mechanism of action. In contrast to all the reported RBD-targeting neutralizing antibodies that compete with DPP4 for binding to the RBD, MERS-4 and MERS-4V2 approached the RBD from the outside of the RBD-DPP4 binding interface. Site-directed mutagenesis identified several key residues critical for binding and the neutralizing activity of MERS-4 and MERS-4V2. Structural comparisons of the RBD in unbound, DPP4-bound, and antibody-bound states revealed significant conformational changes in the RBD when bound to MERS-4 or MERS-4V2. To the best of our knowledge, this is the first report demonstrating that a MERS-CoV neutralizing antibody can indirectly interfere with DPP4 binding through conformational changes. Its unique epitope specificity and unusual mechanism of action enable MERS-4 to synergize with other antibodies, providing a valuable addition for the combined use of antibodies against MERS-CoV infection.
Results
Overall Structure of MERS-4 and Its MERS-4V2 Variant Bound to the RBD
We previously reported that the human monoclonal antibody MERS-4 targets the RBD of the spike glycoprotein and exhibits strong neutralization activity against live and pseudotyped MERS-CoV infection (Jiang et al., 2014). However, the yield of MERS-4 from the transfected HEK293F cells was rather low (<1 mg/L), hampering our efforts toward detailed structural analysis. To identify a variant with improved productivity, we generated a library of mutant MERS-4 comprising random replacements in the 5-residue-long CDR3 region, displayed on the surface of yeast, and selected for binding to the RBD through fluorescence-activated cell sorting (FACS)-based sorting as described previously (Jiang et al., 2014). Among a total of 17 selected mutants, only the variant MERS-4V2 demonstrated a substantial improvement of productivity (>10-fold) while maintaining potency as strong as that of the original MERS-4 in neutralizing pseudotyped virus and binding the S trimer (Figure S1A). Sequence analysis showed that the original CDR3 residues Ala-Gly-Asn-Asp (AGND) of MERS-4 were replaced by Thr-Asn-Thr-Tyr (TNTY) in MERS-4V2 (Figure S1B).
To study the overall structure of antibodies bound to the RBD, we expressed MERS-4 and MERS-4V2 in HEK293F cells and obtained the corresponding Fab fragments. The antibody and RBD complexes were formed by mixing the MERS-4 or MERS-4V2 Fab with the RBD. However, despite our repeated efforts in optimizing and screening more than 200 crystals, we were only able to obtain X-ray diffraction data of RBD in complex with MERS-4 Fab at 4.5 Å and with MERS-4V2 Fab at 7 Å. Nevertheless, we solved the structure of the MERS-4 Fab/RBD complex (PDB: 5ZXV) and refined it to R work and R free factors of 30.2% and 34.4%, respectively (Figure S2A; Table S1). We went further to construct an scFv version of MERS-4V2 and replaced the MERS-4V2 Fab during complex formation with RBD. The structure of the MERS-4V2 scFv/RBD complex (PDB: 5YY5) was successfully solved to a resolution of 2.8 Å with R work and R free factors of 24.7% and 27.7%, respectively (Figure S2B; Table S1).
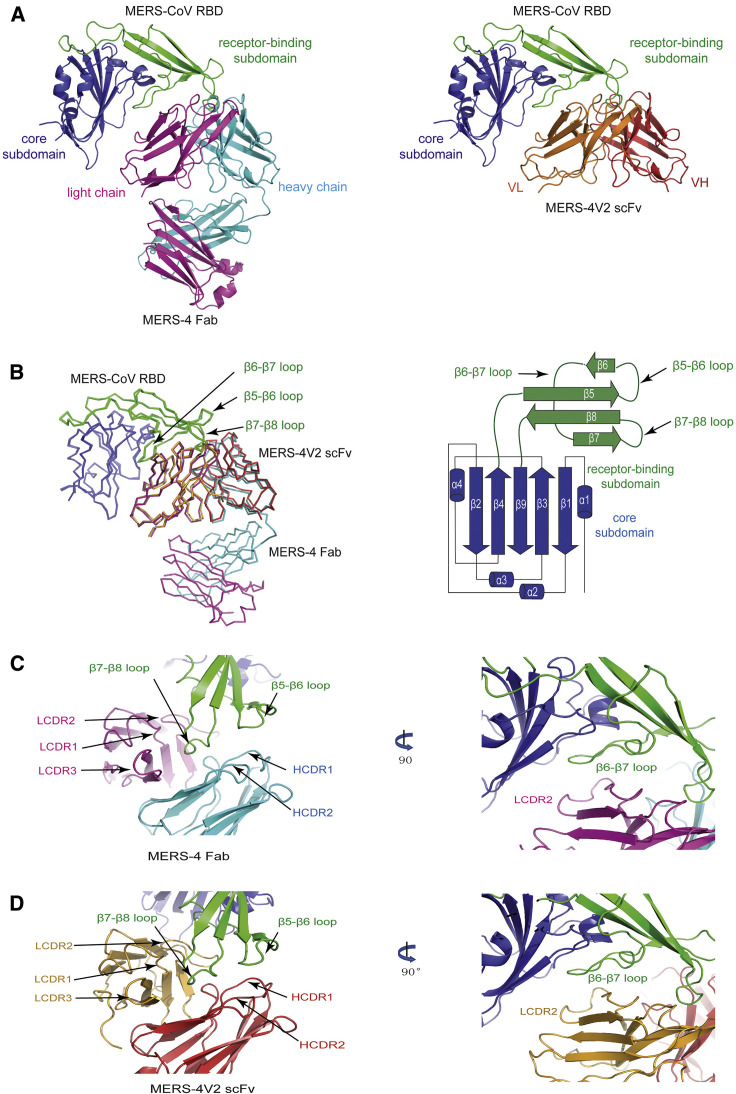
As shown in Figure 1 A, MERS-4 Fab and MERS-4V2 scFv shared the same mode of binding to the RBD. A superimposition of MERS-4 Fab with MERS-4V2 scFv revealed that their respective recognition was largely mediated through interactions with the β5-β6, β6-β7, and β7-β8 loops in the receptor-binding subdomain of the RBD (Figure 1B). Negligible structural differences were found between the two, with an overall root-mean-square deviation (RMSD) of 0.6 Å for 204 aligned Cα atoms. At the binding interface, the β7-β8 loop of RBD inserted into the cavity between the antibody heavy and light chains, forming interactions with all CDR loops of the antibody except for HCDR3 (Figures 1C and 1D). In particular, the short β5-β6 loop interacted with the HCDR1 and HCDR2 loops of the antibody heavy chain, and the long β6-β7 loop predominately interacted with the LCDR2 loop of the antibody light chain (Figures 1C and 1D).
Figure 1.
Crystal Structures of MERS-CoV RBD in Complex with MERS-4 and Its Variant MERS-4V2
(A) Overall structures of the RBD/MERS-4 Fab and the RBD/MERS-4V2 scFv (right) complexes. The RBD core subdomain was in blue, while the receptor-binding subdomain was in green, the MERS-4 light chain in magenta, the MERS-4 heavy chain in cyan, the MERS-4V2 VL in orange, and the MERS-4V2 VH in red.
(B) Structural superimposition of the RBD/MERS-4 and the RBD/MERS-4V2 complexes and schematic illustration of the MERS-CoV RBD (right).
(C and D) MERS-4 (C) and MERS-4V2 (D) interact with the β5-β6, β6-β7, and β7-β8 loops of the RBD.
Structural Features at the Binding Interface between MERS-4V2 and the RBD
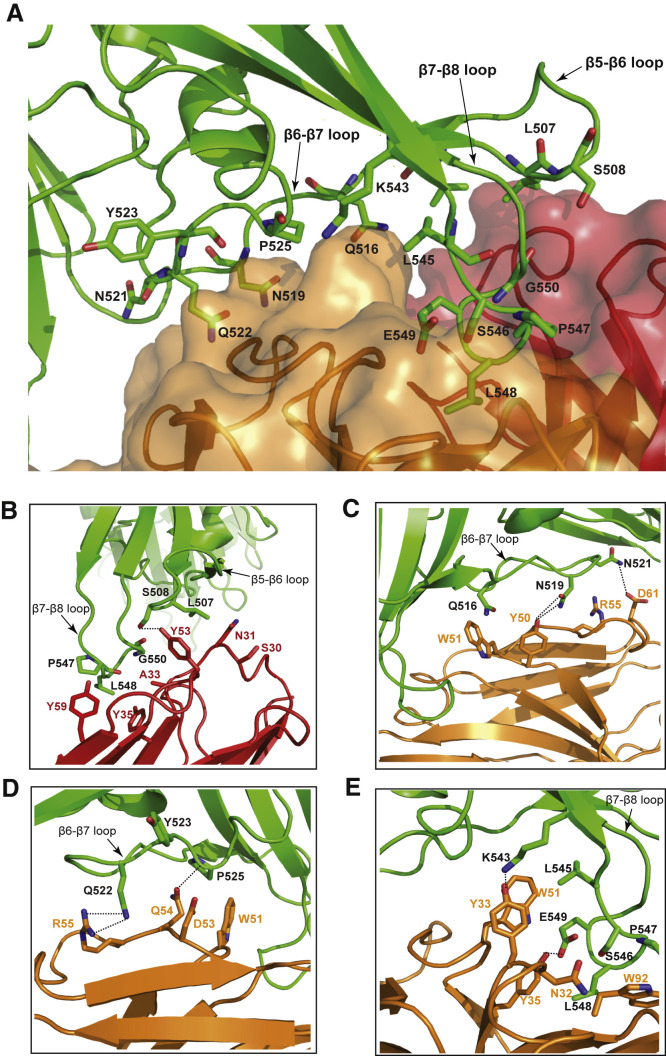
We conducted a detailed analysis of structural features at the binding interface derived from the MERS-4V2 scFv/RBD complex (Figures 2 and S2C). At the binding interface, 16 MERS-4V2 scFV residues from all 6 CDR except for HCRD3 formed contacts with 15 residues from the β5-β6, β6-β7, and β7-β8 loops of the receptor-binding subdomain of the RBD (Figure 2A; Table S2). Specifically, the RBD residues Leu507 and Ser508 from the β5-β6 loop interacted with Ser30, Asn31, and Tyr53 from the heavy chain (Figure 2B; Table S2). The RBD residues Gln516, Asn519, Asn521, Gln522, Tyr523, and Pro525 from the β6-β7 loop interacted with Tyr50, Trp51, Asp53, Gln54, Arg55, and Asp61 from the antibody light chain (Figures 2C and 2D; Table S2). Furthermore, the RBD residues Lys543, and Leu545 to Gly550 from the β7-β8 loop interacted with Ala33, Tyr35, Tyr53, and Tyr59 from the heavy chain, as well as Asn32, Tyr33, Tyr35, Trp51, and Trp92 from the light chain (Figures 2B and 2E; Table S2). Among all interactions at the interface, one example of hydrophobic interactions involved Leu548 (RBD β7-β8 loop), Tyr35 (heavy chain), Tyr35 (light chain), and Trp92 (light chain) (Figures 2B and 2E; Table S2). Examples of hydrogen bonds included Ser508 (RBD β5-β6 loop) to Tyr53 (heavy chain), Asn519 (RBD β6-β7 loop) to Tyr50 (light chain), Asn521 (RBD β6-β7 loop) to Asp61 (light chain), and Gln522 (RBD β6-β7 loop) to Arg55 (light chain) (Figures 2B–2D; Table S2). As indicated above, the HCDR3 appeared not to be involved in the RBD interaction within a distance cutoff of 4.0 Å. This may explain the unchanged neutralizing and binding activities of MERS-4V2 compared with the parental MERS-4 despite the four-residue replacement in the HCDR3 (Figures S1A and S1B).
Figure 2.
The Binding Interface between MERS-CoV RBD and MERS-4V2
(A) Overall view of the interface showing the MERS-4V2 epitope consisting of residues from the β5-β6, β6-β7, and β7-β8 loops of the RBD.
(B) Interactions between the RBD residues from the β5-β6 loop, the β7-β8 loop, and MERS-4V2 heavy chain.
(C–E) Interactions between the RBD residues from the β6-β7 loop (C and D), β7-β8 loop (E), and the corresponding residues of MERS-4V2 light chain.
Structural Alterations in the RBD Bound to MERS-4 and MERS-4V2
We have previously shown that MERS-4 interfered with the interaction between soluble RBD and the cellular receptor DPP4 expressed on the surface of Huh7 cells (Jiang et al., 2014). Surface plasmon resonance (SPR) analysis showed that the binding of soluble RBD to chip-coupled DPP4 was reduced in the presence of increasing concentrations of MERS-4 in a dose-dependent manner (Figure S3). However, structural superimposition demonstrated that the epitope targeted by MERS-4 and MERS-4V2 is located outside the DPP4 binding surface on the RBD (Figure 3 ). This apparent disconnect prompted us to compare all of the available RBD structures in the unbound (PDB: 4ZPW, 4KQZ, and 4L3N) (Chen et al., 2013, Lu et al., 2013, Wang et al., 2015a), DPP4-bound (PDB: 4L72 and 4KR0) (Lu et al., 2013, Wang et al., 2013), MERS-4-bound, and MERS-4V2-bound states (Figure 3A). While the overall structure of RBD remained relatively stable in all the different states, the MERS-4- or MERS-4V2-bound RBD was found to have a conformational change of the β5-β6 loop (Leu506 to Glu513) (Figure 3A). This particular change involved the folding of the β5-β6 loop toward a shallow groove on the RBD interface critical for accommodating a short helix of DPP4 (Figure 3A). The maximum distance change occurred at Asp510, whose Cα atom moved more than 3 Å into the groove (Figure 3B). Our previous study revealed that the β5-β6 loop participates in the formation of a shallow groove to accommodate the docking of a short helix of DPP4 in patch 2 of the binding interface (Figure 3C) (Wang et al., 2013). Residues Leu506, Asp510, and Glu513 from this loop were found to be involved in forming the core hydrophobic and peripheral hydrophilic interactions in patch 2 (Figure 3C), and mutations of these residues decreased the binding affinity between RBD and DPP4 significantly. The conformational change identified here is therefore expected to bring steric clashes between Asp510 from RBD and Ser292 and Arg317 from DPP4 (Figure 3D), thereby preventing the docking of the short helix of DPP4 into the shallow groove of RBD.
Figure 3.
Comparisons of the DPP4 Binding Site with the Epitopes of MERS-4 and MERS-4V2, and Conformational Change of the RBD β5-β6 Loop in the Antibody-Bound State
(A) Structural superimposition showing that the epitopes of MERS-4 and MERS-4V2 (right) are distinct from the DPP4 binding site. A significant conformational difference was found in the RBD β5-β6 loop between antibody-bound and DPP4-bound states.
(B) Zoom-in view of the aligned RBD β5-β6 loops in unbound (4KQZ: blue; 4L3N: magenta; 4ZPW: wheat) and DPP4-bound (4L72: cyan; 4KR0: yellow) with either the MERS-4-bound (green) or MERS-4V2-bound (green) (right) states.
(C) Patch 2 of the RBD/DPP4 binding interface in which residues Leu506, Asp510, and Glu513 from the RBD β5-β6 loop are critical for DPP4 binding.
(D) The steric clashes in the red circle between the β5-β6 loop of the RBD and the DPP4 receptor upon antibody binding.
See also Figure S3.
Binding of MERS-4 to the Different Conformational States of RBD in the Context of the S Trimer
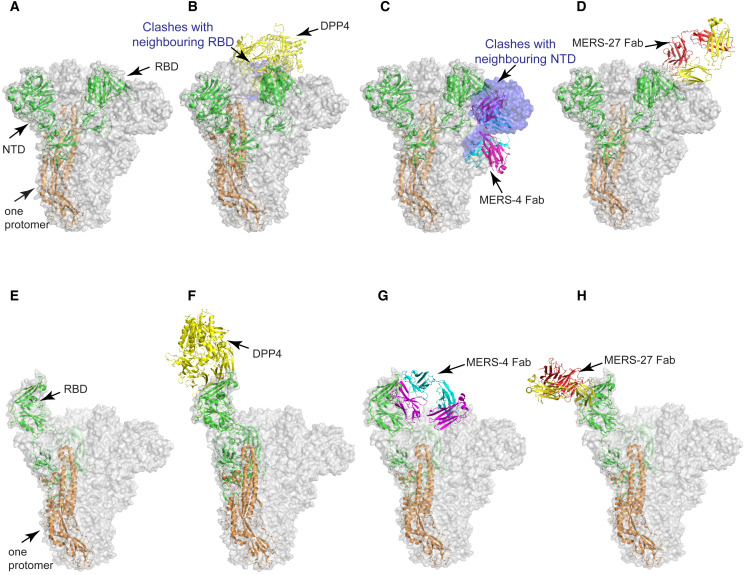
We and others have recently demonstrated that MERS-CoV and SARS-CoV S trimers existed in various conformational states involving the “up” or “down” positions of RBD (Gui et al., 2017, Pallesen et al., 2017, Yuan et al., 2017). The working hypothesis is that all three RBDs in the “down” position keep the S trimer in an inactive, non-receptor-binding state, whereas at least one RBD in the “up” position is required for the S trimer to be in an activated, receptor-binding state (Pallesen et al., 2017, Yuan et al., 2017). To study which state MERS-4 could bind and access its epitope, we superimposed MERS-4 Fab/RBD crystal structures onto the MERS-CoV S trimer structure in the two major conformational states. At the same time, we also analyzed the binding of receptor DPP4 and MERS-27 to the S trimer glycoprotein. As shown in Figure 4 , severe steric clashes would be expected between MERS-4 Fab and the N-terminal domain (NTD) of the neighboring S monomer when all three RBD are in the “down” position, suggesting that the epitope of MERS-4 is covered and inaccessible in the inactivated state. By contrast, when there was one RBD in the “up” position, the epitope of MERS-4 became readily exposed and accessible. Similarly, the receptor DPP4 also favored RBD in the “up” over the one in the “down” position (Pallesen et al., 2017, Yuan et al., 2017). However, the MERS-27 epitope was readily accessible regardless of the state of the RBD. Taken together, these results indicate that MERS-4 binds to the RBD in the “up” position when the virus becomes partially activated. MERS-27, however, binds to the RBD irrespective of the conformational states within the S trimer.
Figure 4.
Structural Superimpositions of the RBD/DPP4, RBD/MERS-4, and RBD/MERS-27 Crystal Structures onto the MERS-CoV S Trimer Glycoprotein in Receptor-Binding Inactivated and Activated States
(A) MERS-CoV S trimer in receptor-binding inactivated state with all three RBDs in the “down” positions (PDB: 5w9j). The S trimer is shown with semi-transparent surface, in which one S protomer (S1 subunit in green and S2 subunit in orange) is shown as a cartoon.
(B–D) Structural superimpositions of the RBD/DPP4 (B), RBD/MERS-4 (C), and RBD/MERS-27 (D) crystal structures onto the S trimer in receptor-binding inactivated state. DPP4 and MERS-4 Fab have steric clashes with the RBD and NTD of the neighboring S protomer, respectively.
(E) MERS-CoV S trimer in receptor-binding activated state with one RBD in the “up” positions (PDB: 5w9h).
(F–H) Structural superimpositions of the RBD/DPP4 (F), RBD/MERS-4 (G), and RBD/MERS-27 (H) crystal structures onto the S trimer in receptor-binding activated state. The epitope is exposed and readily accessible for binding.
Confirmation of the Unique Neutralizing Epitope
To further confirm the unique epitope in the context of binding and neutralization, we mutated a panel of the RBD residues to alanine, including Leu507, Ser508 in the β5-β6 loop, as well as Leu545, Ser546, Pro547, Leu548, and Glu549 in the β7-β8 loop. All of the mutants except for Leu548 could be expressed and purified in insect cells as the same way as the wild-type RBD. The SPR analysis showed that mutations at Leu507, Leu545, Ser546, Pro547, and Glu549 in the RBD significantly decreased the binding affinity to MERS-4 (Figures 5 A and S4A) and MERS-4V2 (Figures 5A and S4B) by more than 10-fold. The mutation at Ser508 decreased the binding affinity to both antibodies by less than 3-fold (Figures 5A and S4). We next measured the neutralization activity of MERS-4 against pseudotyped viruses bearing wild-type or mutant S glycoprotein (Leu507Ala, Leu545Ala, Ser546Ala, and Pro547Ala) in the cell entry assay. Consistent with the binding changes, pseudotyped viruses bearing these mutations became less sensitive to MERS-4 neutralization (Figure 5B). Those bearing Ser508Ala, Leu548Ala, or Glu549Ala mutations failed to produce detectable amounts of infectious viral particles and therefore were excluded from our experiments. Lastly, we went further to study whether the binding of MERS-4 to the mutant S glycoprotein was also reduced when the protein was expressed on the surface of cells. Specifically, transfected HEK293T cells expressing either the wild-type or mutant S glycoprotein were harvested, stained with MERS-4 or antibody 5F9 specific for the NTD of S glycoprotein as a control (Chen et al., 2017a), and analyzed by FACS. As shown in Figure 5C, MERS-4 demonstrated variable levels of reduction in its binding to the mutant S glycoprotein, while the control antibody 5F9 remained virtually unchanged. Furthermore, the L545A mutation completely abolished the binding activity of MERS-4, which was consistent with the results obtained using the pseudotyped viruses. These results suggest that reduced binding affinity of MERS-4 to the mutant S glycoprotein is one of the major contributors to viral resistance. Collectively, we confirmed the unique epitope targeted by MERS-4 and MERS-4V2 observed in the crystal structures and highlighted the unusual mechanism of neutralization by these antibodies.
Figure 5.
Impact of Mutations in the RBD on Binding and Neutralization Sensitivity to MERS-4 and MERS-4V2
(A) Binding affinities of the wild-type RBD and its mutants (L507A, S508A, L545A, S546A, P547A, and E549A) to MERS-4 and MERS-4V2.
(B) Neutralizing activity of MERS-4 against MERS-CoV pseudotyped with wild-type or mutant S glycoprotein (L507A, L545A, S546A, and P547A). The fold changes in IC50 of mutant viruses relative to the wild-type (WT) (>1: increase resistance and <1: increase sensitivity) (right).
(C) MERS-4 staining of HEK293T cells expressing wild-type or mutant S glycoprotein. The fold changes in MFI of mutant viruses relative to the wild-type were listed in the table.
MFI, median fluorescence intensity. See also Figure S4.
Binding of MERS-4 and MERS-27 to the RBD
The epitope of MERS-4 is different from those of other reported antibodies including MERS-27, which we isolated and defined previously (Figure S5). It prompted us to study the combined binding of MERS-4 and MERS-27 to the RBD. Gel filtration analysis showed that the peak of the mixture comprising RBD, MERS-4 Fab, and MERS-27 Fab had a forward shift compared with that of the RBD/MERS-4 Fab complex (Figure S6A), indicating that MERS-4 and MERS-27 could bind to the RBD at the same time. To confirm this, we used small-angle X-ray scattering (SAXS) study on the purified ternary complex in solution. We initially built a monomeric model of the RBD/MERS-4/MERS-27 ternary complex, which was generated by superimposing the RBD/MERS-4 and RBD/MERS-27 crystal structures. However, the monomeric model fits poorly to the SAXS data with a χ value of 7.0 (Figure 6 ). To investigate the assembly of the complex, we built a dimeric model of the ternary complex using the interface observed in the RBD/MERS-27 crystal structure. To understand the dynamic assembly, MultiFoXS was applied to define the population of the models in solution (Pelikan et al., 2009). SAXS fitting with multi-state models (31% monomer model, 18% dimer model of the RBD/MERS-4/MERS-27, and 51% RBD/MERS-27) significantly improved the goodness of the fit with a χ value of 2.0 (Figure 6), revealing the transient complexation of Fabs. The transient behavior was also observed from the SEC-SAXS (size exclusion chromatography in line with SAXS) (Figure S6). The broad distribution of radius of gyration (Rg) values across the SEC-SAXS peak, ranging from 47 to 37 Å (Figure S6B), suggests a multi-state mixture of the complex, which was further confirmed by the good fits obtained for various sections of the SEC-SAXS peak using ensemble models (Figures S6C and S6D). These results collectively showed that MERS-4 and MERS-27 are capable of binding to RBD at different epitopes to form a ternary complex.
Figure 6.
Combination of MERS-4 and MERS-27 in Binding to the RBD
Comparison of the experimental SAXS data (black dots) with the theoretical scattering curve calculated from the full-atomic RBD/MERS-4/MERS-27 monomer model (red line) and the theoretical scattering curve calculated from an ensemble consisting of 31% monomer model, 18% dimer model of the RBD/MERS-4/MERS-27, and 51% RBD/MERS-27 (green line). Residuals calculated as I (q) experimental/I (q) model are shown below the scattering curves.
See also Figure S6.
Synergistic Neutralization Effects of MERS-4 with Other Antibodies against RBD and NTD
We have previously shown that MERS-4 and MERS-27 exhibited a synergistic effect by titrating the neutralizing potency of an equimolar mixture of the two antibodies and comparing the dose response with that of neutralization assays performed with the individual antibody alone (Chou, 2010, Chou and Talalay, 1984, Keck et al., 2013). The synergy between them was consistent with the unique epitopes of MERS-4 and MERS-27 on the RBD (Figure 7 C). Here, we further tested whether MERS-4 could synergize with additional antibodies targeting either the RBD (m336) or NTD (5F9) of the S glycoprotein. As shown in Figure 7A, the percent neutralization obtained using combined MERS-4 and m336 demonstrated a 2.60-fold reduction of half maximal inhibitory concentration (IC50) and 2.73-fold reduction of IC80 compared with that of either MERS-4 or m336 alone. Furthermore, the combination index (CI) values of combined MERS-4 and m336 at fractional effect values of effective dose 50%, 75%, 90%, and 95% (ED50, ED75, ED90, and ED95) were 0.48, 0.38, 0.30, and 0.26, respectively. As a CI value of 1 indicates an additive effect, <1 indicates synergism, and >1 indicates antagonism, the combination of MERS-4 and m336 works in a clearly synergistic manner. Furthermore, the combination of MERS-4 and 5F9 demonstrated better synergy in particular at relatively lower concentrations (Figure 7B). The percent neutralization of combined MERS-4 and 5F9 demonstrated a 15.21-fold reduction in IC50 and 52.7-fold reduction in IC80 compared with that of either antibody alone. Furthermore, the CI values of combined MERS-4 and 5F9 at fractional effect values of ED 50%, 75%, 90%, and 95% (ED50, ED75, ED90, and ED95) were 0.06, 0.05, 0.06, and 0.06, respectively. These results showed that MERS-4 can act in synergy with RBD-specific m336 as well as NTD-specific 5F9 antibodies.
Figure 7.
Effects of MERS-4 Combined with m336, 5F9, and MERS-27, Respectively, in Neutralizing Pseudotyped MERS-CoV
(A) Effects of MERS-4 combined with m336 in neutralizing pseudotyped MERS-CoV. Percent neutralization was calculated for serial 3-fold dilutions of each antibody alone and in combination at constant ratios in a range of concentrations from 27 times to 1/81 of IC50s. The constant ratios of the combined antibodies were their IC50s. On the x axis, a dose of 1 was at the IC50 concentration. Fractional effect (FA) plots generated by the CompuSyn program for MERS-4, m336, and their combination showing dosage versus effect. Median effect plot of calculated CI values (logarithmic) versus FA values, in which a log CI of <0 is synergism and a log CI of >0 is antagonism. Data shown are average values from four independent experiments.
(B and C) The percent neutralization, fractional effect, and CI values for MERS-4 combined with 5F9 (B) and MERS-4 combined with MERS-27 (C) were calculated and generated using the same method.
See also Figures S5 and S7.
Discussion
We report the structural and functional analysis of the potent neutralizing antibody MERS-4 and its variant MERS-4V2, which revealed a unique epitope specificity and novel mechanism of neutralization. The structure of MERS-4 Fab bound to RBD was determined at a resolution of 4.5 Å, and that of MERS-4V2 scFv bound to RBD was solved at 2.8 Å. MERS-4 and MERS-4V2 demonstrated the same binding mode and epitope specificity. In contrast to all other RBD-targeting neutralizing antibodies, which directly compete with DPP4 for binding to the RBD, MERS-4 and MERS-4V2 approached the RBD outside the RBD-DPP4 binding interface by targeting the β5-β6, β6-β7, and β7-β8 loops. MERS-4- and MERS-4V2-bound RBD demonstrated significant conformational changes largely involving the folding of the β5-β6 loop toward a shallow groove on the RBD interface critical for accommodating a short helix of DPP4. In the context of the S trimer, MERS-4 prefers binding to the RBD in the “up” rather than the “down” position when virus becomes partially activated. Site-directed mutagenesis confirmed the key residues critical for binding and neutralizing activities of MERS-4 and MERS-4V2. Reduced affinity for the RBD appeared to be the major contributor to the compromised neutralizing activities against the mutant viruses. A synergistic effect was observed between MERS-4 and RBD-specific (m336) as well as NTD-specific (5F9) antibodies, although the exact mechanism remains unknown. Taken together, our study reveals that MERS-4 and MERS-4V2 recognize a unique neutralizing epitope with an unusual mechanism of action. Such special features will enable MERS-4 to synergize with other antibodies and provide a valuable addition for the combined use of antibodies against MERS-CoV infection.
Since the identification of the highly pathogenic MERS-CoV in 2012 (Bermingham et al., 2012, Zaki et al., 2012), great efforts have been made to develop prophylactic and therapeutic interventions against this virus. In particular, monoclonal antibodies and vaccines targeting the S glycoprotein are a major research focus due to its critical role in mediating viral entry and its great potency in inducing protective antibody response in infected and naive individuals (Chen et al., 2009, Zumla et al., 2016). Among close to 20 reported neutralizing antibodies, the large majority (MERS-27, m336, D12, 4C2, MCA1, CDC-C2, and JC57-14) were shown to directly compete with the cellular receptor DPP4 for binding to the RBD (Chen et al., 2017b, Li et al., 2015, Wang et al., 2015a, Wang et al., 2018, Ying et al., 2015, Yu et al., 2015). This dominant mechanism of action is further supported by structural studies in which their epitopes were found to clearly, although variably, overlap with the DPP4 binding surface (Figure S7). Because MERS-4 shares many biochemical and biological features with the abovementioned antibodies, it was expected that its mechanism of neutralization would follow the same suit. However, a novel epitope specificity and unusual mechanism of action came as a pleasant surprise. This not only reveals a new vulnerable site on the RBD, but also provides a novel target for future vaccine design and development.
To the best of our knowledge, this is the first report demonstrating that antibody binding can indirectly disrupt the interaction between RBD with DPP4. While the exact process requires further study, there are at least two possibilities. One is that the antibody captures and fixes one of the RBD conformational states when the β5-β6 loop spontaneously bends toward the receptor binding interface. Given the highly dynamic nature of RBD in the context of the S trimer, it is plausible that some of the loop structures in the RBD could also be quite flexible, providing opportunities for antibody binding during the structural transformation. The other is that the antibody itself triggers such a structural alteration. If this were the case, the heavy chain of MERS-4 or MERS-4V2 would likely push and fold the β5-β6 loop toward the receptor binding interface in order to achieve optimal binding. This hypothesis is supported by atomic analysis of the interaction between MERS-4V2 and the RBD. Among a total of 16 interactive binding residues, 14 from the β6-β7 and β7-β8 loops interacted with the light chain, whereas 2 in the β5-β6 loop participated in binding with the heavy chain. The interactions between the β6-β7 and β7-β8 loops of the RBD and the light chain of MERS-4V2 are therefore the most likely the driving force of antigen-antibody binding. However, optimal binding of MERS-4 would require the heavy chain to overcome the steric obstruction by pushing and folding the β5-β6 loop toward the binding interface, resulting in a distorted conformation as shown in the crystal structure. Regardless of the exact process, the observed folding of the β5-β6 loop toward the binding interface would be expected to disrupt the docking of the short helix of DPP4 onto the binding surface of RBD, thereby blocking virus entry. Of note, such unique mechanism of action has not been reported for other viruses. Antibodies against the receptor binding site (RBS) of influenza virus and the CD4 binding site (CD4bs) of HIV type I (HIV-1) exert their neutralizing activities largely through direct competition with their respective receptors (Wu and Kong, 2016, Ren and Zhou, 2016). Perhaps the closest scenario to MERS-4-mediated inhibition is found in antibodies targeting the V3 region of the HIV-1 envelope where binding may capture or induce conformational changes that block the subsequent interaction between the V3 region and the co-receptor CCR5 or CXCR4. Certainly, such a hypothesis would have to be verified in the future (Barnes et al., 2018, Mouquet et al., 2012, Pejchal et al., 2011).
The MERS-CoV spike glycoprotein showed limited sequence variation among different strains, especially in the RBD that is responsible for receptor binding. However, this does not mean the spike glycoprotein will remain unchanged as the virus continues to spread among multiple animal species and to probe and adapt in human population. For stronger and broader protective effect against MERS-CoV, a combined use of two or more antibodies will provide a superior alternative (Wang et al., 2018). However, any effective combination would require the candidate antibodies to recognize distinct epitopes for additive or synergistic effect. The unique epitope of MERS-4 and MERS-4V2 therefore makes them good candidates for combination use with those reported elsewhere (Chen et al., 2017b, Li et al., 2015, Wang et al., 2015a, Wang et al., 2018, Ying et al., 2015, Yu et al., 2015). Indeed, combinations of MERS-4 and MERS-27, MERS-4 and m336, and MERS-4 and 5F9 demonstrated impressive synergy in the pseudotyped MERS-CoV assay. The exact mechanism in achieving the synergy, however, is uncertain, particularly for those sharing the same mechanism in disrupting interaction between RBD and DPP4 (MERS-4 and MERS-27, and MERS-4 and m336). Presumably, the two antibodies may preferentially act on RBD at the different spatial and temporal points during interaction with the receptor DPP4, allowing better exposure of otherwise less accessible epitopes. If this is the case, the observed synergy would be most likely derived from the recognitions of distinct epitopes rather than the same neutralization mechanism. Obviously, antibodies with distinct mechanisms and binding at disparate epitopes would be more likely to have synergy than those shared mechanism and overlapped epitopes. Synergy showed here between MERS-4 and 5F9 is a good example. Nevertheless, the exact mechanisms underlying synergy must be complex and should be treated differently from case to case. The preliminary results presented here do offer some rationales for MERS-4 as a valuable addition for the combined use of antibodies against MERS-CoV infection.
Experimental Procedures
Protein Expression and Purification
The MERS-CoV RBD (residues 367–588) and the ectodomain of human DPP4 (residues 39–766) were expressed in Sf9 insect cells. The purified RBD was digested with endoglycosidase F1 and F3 at room temperature overnight and was then further purified through gel-filtration chromatography. The MERS-4 and MERS-4V2 IgG were expressed in HEK293F cells. The purified MERS-4 and MERS-4V2 were digested with endoproteinase Lys-C at 37°C, and the Fab and Fc fragments were separated by loading samples onto a diethylaminoethyl (DEAE) ion-exchange column. The genes encoding the scFv of MERS-4V2 were cloned with a C-terminal His-tag in the heavy chain variable region and light chain variable region (VH-VL) orientation, linked together by a (G4S)3. The MERS-4V2 scFv was expressed in HEK293F cells through transient transfection. The scFv was captured by nickel beads and purified through gel-filtration chromatography.
Crystallization and Data Collection
Crystals of the RBD/MERS-4 Fab complex were successfully grown at 18°C using the sitting drop vapor diffusion method, which involved mixing equal volume of protein and reservoir solution containing 2% v/v tacsimate (pH 5.0), 0.1 M sodium citrate tribasic dihydrate (pH 5.6), 16% w/v polyethylene glycol 3350, and 2 M sodium thiocyanate. Crystals of the RBD/MERS-4V2 scFv complex were successfully grown at 18°C using the sitting drop vapor diffusion method, which involved mixing equal volumes of protein and reservoir solution containing 0.1 M Tris (pH 7.5), 10% w/v polyethylene glycol 8000, and 8% (v/v) ethylene glycol. Diffraction data were collected on the BL17U beamline at Shanghai Synchrotron Research Facility (Wang et al., 2015b) and processed with HKL2000 (Otwinowski and Minor, 1997). All data collection and processing statistics are listed in Table S1.
Structural Determination and Refinement
The structure was determined by molecular replacement with the crystallographic software PHASER (McCoy et al., 2007). The search models are the MERS-CoV RBD structure (PDB: 4L72) and the structures of the variable and constant domain of the heavy and light chains available in the PDB with the highest sequence identities. Iterative refinement with the program PHENIX and model building with the program COOT were performed to complete the structure refinement (Adams et al., 2002, Emsley and Cowtan, 2004). Structure validation was performed with the program PROCHECK (Laskowski et al., 1993). All structure refinement statistics are listed in Table S1.
SPR Analysis
Real-time binding and analysis by SPR were conducted on a BIAcore T200 instrument. MERS-4 IgG (20 μg/mL) and MERS-4V2 (20 μg/mL) in 10 mM sodium acetate (pH 5.0) was immobilized to 600 response units on the flow cell. For the collection of data, RBD and its mutants were injected in a buffer of 10 mM HEPES (pH 7.2), 150 mM NaCl, and 0.005% (v/v) Tween 20 over the flow cells at various concentrations. Data were analyzed with the BIAcore T200 evaluation software by fitting to a 1:1 Langmuir binding model. For the DPP4 binding inhibition assay, RBD (100 nM) and MERS-4 Fab were mixed in advance at a molar ratio 1:1, 1:2, 1:5, and 1:10. DPP4 (10 μg/mL) in 10 mM sodium acetate (pH 4.5) was immobilized to 600 response units on the flow cell. For the collection of data, MERS-CoV RBD alone and its various complex with MERS-4 Fab were injected in a buffer of 10 mM HEPES (pH 7.2), 150 mM NaCl, and 0.005% (v/v) Tween 20 over the flow cells.
MERS-CoV Pseudotyped Virus Production and Neutralization Assay
The MERS-CoV pseudotyped virus was generated by cotransfecting pcDNA 3.1 expression vectors encoding the wild-type or mutant MERS-CoV S glycoprotein, with a pNL4-3R-Eluciferase viral backbone plasmid into 293T cells as described previously (Shang et al., 2011). The viral titers of the pseudotyped virus were measured as luciferase activity in relative light units 48 hr after transfection. The mutant S glycoprotein expression vector was generated by the site-directed mutagenesis kit and confirmed by sequencing. Neutralization assays were performed by incubating 100 TCID50 (median tissue culture infectious dose) of pseudotyped virus with 16 serial 1:3 dilutions of purified antibody at 37°C for 1 hr; then Huh7 cells (about 1.5 × 104 per well) were added. Infectivity was quantified by the luciferase activity 48 hr after infection. IC50s were calculated with the dose-response inhibition model in GraphPad Prism (GraphPad Software).
Cell Surface Staining
HEK293T cells were transfected by wild-type or mutant MERS-CoV spike expression plasmids. After 48 hr, cells were harvested and washed using PBS and incubated with monoclonal antibodies (mAbs) for 30 min at room temperature. Cells were then washed and stained with fluorescently labeled anti-human IgG-PE or anti-mouse IgG-FITC secondary antibody. Cells were then washed by PBS and analyzed with FACSCalibur and FACSComp software (BD Biosciences).
Cooperativity of the Two Neutralizing mAbs for Virus Neutralization
Synergistic, additive, and antagonistic interaction between MERS-4 and 5F9, MERS-4 and m336, and MERS-4 and MERS-27 for virus neutralization were evaluated by the median effect analysis method by the CompuSyn software as previously reported. The measured neutralization values were input to the program as fractional effects (FA) ranging between 0.01 and 0.99 for each of the two antibodies and for both in combination. CI values were calculated in relation to FA values. A logarithmic CI value of 0 indicates an additive effect, <0 indicates synergism, and >0 indicates antagonism.
SAXS Data Collection and Analysis
SAXS data were collected at the SIBLYS beamline 12.3.1 of the Advanced Light Source at the Lawrence Berkeley National Laboratory using 1.0 Å wavelength and Pilatus 2M detector at a 1.5-m sample-to-detector distance (Classen et al., 2013), resulting in scattering vectors ranging from 0.01 to 0.5 Å−1. The scattering vector is defined as q = 4π sinθ/λ, where 2θ is the scattering angle. SEC in line with SEC-SAXS was performed to ensure the aggregation-free state of the sample. The SEC column was equilibrated with running buffer (50 mM Tris-HCl [pH 7.3], 100 mM NaCl, 3% glycerol, and 0.01% sodium azide) with a flow rate of 0.5 mL/min. The 50-μL sample was run through the SEC and 2-s X-ray exposures were collected continuously during an ∼25-min elution. The SAXS frames recorded prior to the protein elution peak were used to subtract all other frames. The subtracted frames were investigated by R g and scattering intensity at q = 0 Å−1 (I(0)) derived by the Guinier approximation I(q) = I(0) exp(−q 2 R g 2/3) with the limits qR g < 1.5 (Guinier and Fournet, 1955). I (0) and R g values were compared for each collected SAXS curve across the entire elution peak. The elution peak was mapped by plotting the scattering intensity at q = 0 Å−1 (I (0)) relative to the recorded frame. Graduate decreasing of R g values across an elution peak indicates transient sample behavior. SAXS was also acquired in the high-throughput modality at sample concentrations between 1 and 5 mg/mL to compare with the SEC-SAXS profile (Hura et al., 2009). The full atomic model was built with the program MODELER to construct the missing loops and linkers (Sali and Blundell, 1993). The theoretical SAXS profile and the corresponding fit to the experimental data were calculated using the program FoXS (Schneidman-Duhovny et al., 2013). A multistate model of complexes coexisting in solution was selected by MultiFoXS (Schneidman-Duhovny et al., 2016). The size of the multistate model was selected based on the level of improvement in the SAXS fit.
Acknowledgments
We thank Dr. Jianhua He and staff scientists at the SSRF BL17U beam line and Dr. Shilong Fan at the X-ray crystallography platform of the Tsinghua University Technology Center for assistance in diffraction data collection. We thank Prof. Tianlei Ying and Prof. Shibo Jiang (Fudan University) for providing the m336. We thank Prof. Wenjie Tan (Chinese Center for Disease Control and Prevention) for providing the 5F9. This work was supported by the National Key Plan for Scientific Research and Development of China (grants 2016YFD0500307 and 2016YFC1200902), the National Natural Science Foundation of China (grants 31470751, U1405228, 81530065, and 81471929), and the Grand Challenges China (grant 81661128042).
Author Contributions
S.Z., P.W., P.Z., and L.J. expressed, purified, and crystallized the protein, and carried out the SPR analysis with help from D.W.; P.Z., Y.L., W.J., H.W., and X.S. carried out the pseudotyped virus entry and antibody neutralization analyses; S.Z. and P.W. collected and processed the diffraction data; X.W. carried out structural determination and refinement; M.H. collected and analyzed the SAXS data with input from S.W. and X.F.; X.W. and L.Z. supervised the project and wrote the manuscript with help from S.Z., P.Z., P.W., and A.F.
Declaration of Interests
The authors declare no competing interests.
Published July 10, 2018
Footnotes
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.06.041.
Contributor Information
Xinquan Wang, Email: xinquanwang@mail.tsinghua.edu.cn.
Linqi Zhang, Email: zhanglinqi@mail.tsinghua.edu.cn.
Data and Software Availability
The accession numbers for the atomic coordinates reported in this paper are PDB: 5ZXV and 5YY5.
Supplemental Information
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., KSA MERS-CoV Investigation Team Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Gristick H.B., Freund N.T., Escolano A., Lyubimov A.Y., Hartweger H., West A.P., Jr., Cohen A.E., Nussenzweig M.C., Bjorkman P.J. Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope. Nat. Commun. 2018;9:1251. doi: 10.1038/s41467-018-03632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- Chen P.R., Groff D., Guo J., Ou W., Cellitti S., Geierstanger B.H., Schultz P.G. A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chem. Int. Engl. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Rajashankar K.R., Yang Y., Agnihothram S.S., Liu C., Lin Y.-L., Baric R.S., Li F. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lu S., Jia H., Deng Y., Zhou J., Huang B., Yu Y., Lan J., Wang W., Lou Y. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg. Microbes Infect. 2017;6:e37. doi: 10.1038/emi.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Bao L., Chen C., Zou T., Xue Y., Li F., Lv Q., Gu S., Gao X., Cui S. Human neutralizing monoclonal antibody inhibition of Middle East respiratory syndrome coronavirus replication in the common marmoset. J. Infect. Dis. 2017;215:1807–1815. doi: 10.1093/infdis/jix209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.Y. An outbreak of Middle East respiratory syndrome coronavirus infection in South Korea, 2015. Yonsei Med. J. 2015;56:1174–1176. doi: 10.3349/ymj.2015.56.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- Chou T.C., Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Classen S., Hura G.L., Holton J.M., Rambo R.P., Rodic I., McGuire P.J., Dyer K., Hammel M., Meigs G., Frankel K.A., Tainer J.A. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Cryst. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Zhao J., Pedotti M., Simonelli L., Agnihothram S., Fett C., Fernandez-Rodriguez B., Foglierini M., Agatic G., Vanzetta F. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Yang Y., Qiu H., Wang L., Kou Z., Tao X., Yu H., Sun S., Tseng C.T. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinier A., Fournet F. Wiley Interscience; 1955. Small Angle Scattering of X-rays. [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Chu D.K., Poon L.L., Perera R.A., Alhammadi M.A., Ng H.Y., Siu L.Y., Guan Y., Alnaeem A., Peiris M. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014;20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura G.L., Menon A.L., Hammel M., Rambo R.P., Poole F.L., 2nd, Tsutakawa S.E., Jenney F.E., Jr., Classen S., Frankel K.A., Hopkins R.C. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang N., Zuo T., Shi X., Poon K.M., Wu Y., Gao F., Li D., Wang R., Guo J. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6:234ra59. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- Keck Z., Wang W., Wang Y., Lau P., Carlsen T.H., Prentoe J., Xia J., Patel A.H., Bukh J., Foung S.K. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J. Virol. 2013;87:37–51. doi: 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Li Y., Wan Y., Liu P., Zhao J., Lu G., Qi J., Wang Q., Lu X., Wu Y., Liu W. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015;25:1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Scharf L., Euler Z., Liu Y., Eden C., Scheid J.F., Halper-Stromberg A., Gnanapragasam P.N., Spencer D.I., Seaman M.S. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal K.E., Coleman C.M., Mujica A.O., Kamat V., Badithe A., Fairhurst J., Hunt C., Strein J., Berrebi A., Sisk J.M. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA. 2015;112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(Suppl 12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R., Doores K.J., Walker L.M., Khayat R., Huang P.S., Wang S.K., Stanfield R.L., Julien J.P., Ramos A., Crispin M. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelikan M., Hura G.L., Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Zhou P. Epitope-focused vaccine design against influenza A and B viruses. Curr. Opin. Immunol. 2016;42:83–90. doi: 10.1016/j.coi.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A. FoXS, FoXSDock and MultiFoXS: single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016;44(W1):W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang H., Han X., Shi X., Zuo T., Goldin M., Chen D., Han B., Sun W., Wu H., Wang X., Zhang L. Genetic and neutralization sensitivity of diverse HIV-1 env clones from chronically infected patients in China. J. Biol. Chem. 2011;286:14531–14541. doi: 10.1074/jbc.M111.224527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S., Sheehan J., Zhu Q. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. USA. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., Lees C.R., Zhou T., Yassine H.M., Kanekiyo M. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.S., Yu F., Huang S., Sun B., Zhang K.H., Liu K., Wang Z.J., Xu C.Y., Wang S.S., Yang L.F. The macromolecular crystallography beamline of SSRF. Nucl. Sci. Tech. 2015;26:12–17. [Google Scholar]
- Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on MERS-CoV Spike to avoid neutralization escape. J. Virol. 2018;92 doi: 10.1128/JVI.02002-17. e02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Kong X.P. Antigenic landscape of the HIV-1 envelope and new immunological concepts defined by HIV-1 broadly neutralizing antibodies. Curr. Opin. Immunol. 2016;42:56–64. doi: 10.1016/j.coi.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C., Lu L., Liu Q., Wang L., Feng Y., Wang Y. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Prabakaran P., Du L., Shi W., Feng Y., Wang Y., Wang L., Li W., Jiang S., Dimitrov D.S., Zhou T. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zhang S., Jiang L., Cui Y., Li D., Wang D., Wang N., Fu L., Shi X., Li Z. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci. Rep. 2015;5:13133. doi: 10.1038/srep13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.