Significance
Many mobile genetic elements contain anti-CRISPRs (Acrs) to evade host CRISPR defenses. Acrs have been discovered that inhibit therapeutically relevant CRISPR-Cas gene editors such as Cas12 and Cas9, including many inhibitors for Streptococcus pyogenes Cas9 (SpyCas9). However, there are few inhibitors known for the Cas9 from Staphylococcus aureus (SauCas9), which is both highly active in human cells and easier to deliver than SpyCas9 due to its smaller size. In this work, we search within Staphylococcus genomes for Acrs able to inhibit SauCas9. We discover three Acrs (AcrIIA13, AcrIIA14, and AcrIIA15) and demonstrate all three are able to inhibit CRISPR-Cas9 gene editing. Our findings highlight the utility of our Acr search method for finding Acrs against specific Cas nucleases.
Keywords: CRISPR, anti-CRISPR, self-targeting, Cas9, genome editing
Abstract
Anti-CRISPRs (Acrs) are small proteins that inhibit the RNA-guided DNA targeting activity of CRISPR-Cas enzymes. Encoded by bacteriophage and phage-derived bacterial genes, Acrs prevent CRISPR-mediated inhibition of phage infection and can also block CRISPR-Cas-mediated genome editing in eukaryotic cells. To identify Acrs capable of inhibiting Staphylococcus aureus Cas9 (SauCas9), an alternative to the most commonly used genome editing protein Streptococcus pyogenes Cas9 (SpyCas9), we used both self-targeting CRISPR screening and guilt-by-association genomic search strategies. Here we describe three potent inhibitors of SauCas9 that we name AcrIIA13, AcrIIA14, and AcrIIA15. These inhibitors share a conserved N-terminal sequence that is dispensable for DNA cleavage inhibition and have divergent C termini that are required in each case for inhibition of SauCas9-catalyzed DNA cleavage. In human cells, we observe robust inhibition of SauCas9-induced genome editing by AcrIIA13 and moderate inhibition by AcrIIA14 and AcrIIA15. We also find that the conserved N-terminal domain of AcrIIA13–AcrIIA15 binds to an inverted repeat sequence in the promoter of these Acr genes, consistent with its predicted helix-turn-helix DNA binding structure. These data demonstrate an effective strategy for Acr discovery and establish AcrIIA13–AcrIIA15 as unique bifunctional inhibitors of SauCas9.
CRISPR systems are RNA-guided, adaptive immune systems that defend prokaryotes against invading mobile genetic elements (MGEs) (1). However, some MGEs, particularly phages, have evolved anti-CRISPRs (Acrs), peptide inhibitors of Cas proteins that block CRISPR defense systems (2, 3). Acrs have been discovered to inhibit distinct CRISPR systems, including type I (4–8), type II (9–16), type III (17, 18), and type V (7, 19). Strategies for identifying new Acrs include testing genes of unknown function that are proximal to anti-CRISPR–associated (aca) genes (5–7, 9, 15) and screening genes in organisms with self-targeting CRISPR systems (7, 19).
The frequent clustering of Acr and aca genes together allows for a “guilt-by-association” approach that quickly identifies potential Acr candidates for experimental testing, but requires a known Acr or aca gene to seed the search (5–7, 9, 15). Conversely, self-targeting CRISPR systems are present in diverse genomes that could encode corresponding CRISPR-Cas inhibitors to block autoimmunity (20) (Fig. 1A), but finding relevant Acr-encoding genes within these genomes can be daunting. Nonetheless, analysis of known Acrs suggests that self-targeting CRISPR-containing genomes are attractive to study because they often encode at least one corresponding Acr (19).
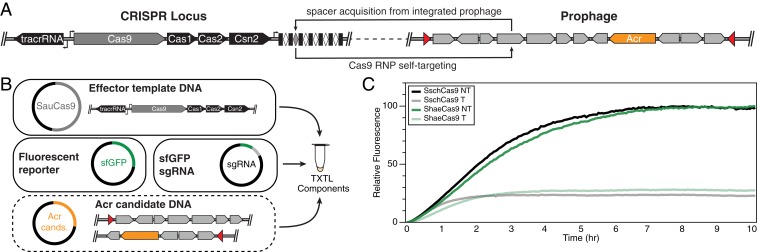
Fig. 1.
Identification of self-targeting Staphylococcus strains that contain active type II CRISPR-Cas systems. (A) Acquisition of protospacers from MGEs can result in self-targeting if the MGE is capable of stably integrating (e.g., prophages) or associating (e.g., plasmid) with the genome. (B) Overview of TXTL assay to test for Acr activity. Cas9 template DNA is combined with an sfGFP reporter plasmid and a plasmid expressing a sgRNA targeting the reporter. In the absence of Acr activity, expression of the Cas9 RNP suppresses sfGFP expression. In the presence of Acrs, the Cas9 RNP is inhibited, resulting in increased fluorescence. (C) Genomic amplicons from S. schleiferi and S. haemolyticus containing Cas9 lower sfGFP expression with an sfGFP-targeting sgRNA, demonstrating that the natural CRISPR loci are active.
Multiple anti-CRISPR (Acr) families inhibit Streptococcus pyogenes Cas9 (SpyCas9) and various Cas12a proteins and can be used in cell-based experiments to control genome editing outcomes (7, 10, 11, 13, 14, 21, 22). Although weak cross-reactivity with other noncognate Cas9 orthologs has been detected for a subset of these (10, 11, 23), we wondered whether more potent inhibitors for a wider selection of particular Cas9 variants might exist in nature. To address this question, we focused on Staphylococcus genomes that might encode inhibitors of Staphylococcus aureus Cas9 (SauCas9), a genome editing alternative to SpyCas9 whose smaller size could offer advantages for delivery into mammalian cells (24, 25). We used a combination of self-targeting CRISPR screening and guilt-by-association genomic searches to discover three peptide inhibitors of SauCas9. We show that these SauCas9 Acrs, AcrIIA13, AcrIIA14, and AcrIIA15, limit or prevent RNA-guided DNA cleavage in vitro and genome editing in human cells. These three inhibitors share a common N-terminal domain with a predicted helix-turn-helix (HTH) structure that is dispensable for DNA cleavage inhibition but can bind specifically to the inverted repeat (IR) sequence in the promoter of these Acr genes. The C terminus of each Acr is distinct and is responsible for SauCas9 inhibition in each case, likely by differing mechanisms. These SauCas9 inhibitors provide tools for the selective control of genome editing outcomes and validate a multipronged strategy for discovering diverse Acrs in nature.
Results
Bioinformatic Identification of Self-Targeting Type II-A CRISPR Systems.
To identify potential Acrs that inhibit SauCas9, we first used the Self-Target Spacer Searcher (STSS) (19) to query all Staphylococcus species deposited in the National Center for Biotechnology Information (NCBI) database for instances of CRISPR self-targeting. We observed 99 total instances of self-targeting in Staphylococcus CRISPR systems across 43 different strains out of a potential 11,910 assemblies searched (Dataset S1). Of the 99 self-targeting instances predicted, 50 could not be attributed to any particular CRISPR subtype, 48 were associated with a type II-A system, and 1 occurred as part of a type III-A system. We did not observe any self-targeting CRISPR type I-C systems that are occasionally found in Staphylococcus (26). It should also be noted that 29 of the predicted CRISPR self-targeting systems occurred in eight species whose CRISPR loci were manually annotated as type II-A based on identity to other type II-A Cas9-encoding genes.
To select the genomes most likely to contain Acrs, we filtered the list of 48 self-targets to exclude those with target protospacer-adjacent motifs (PAMs) that were more than one indel/mutation away from the known 3′-NNGRR(T) PAM for SauCas9 (25). This step eliminated genomes in which an incorrect PAM sequence could explain survival without the need for Acrs. The remaining 14 self-targeting instances, belonging to 12 different strains (Dataset S1), were ranked according to similarity of their encoded Cas9 and SauCas9 (SI Appendix, Table S1). Of the seven Cas9s displaying high similarity to SauCas9 (>50% identity with blastp), two species, Staphylococcus haemolyticus and Staphylococcus schleiferi, were selected for further screening for Acrs active against SauCas9 based on strain availability.
Self-Targeting Screen for Acr Activity.
To reduce the sequence space to search for Acr genes, we selected only one of the two self-targeting strains identified for each species (S. schleiferi and S. haemolyticus), since the two strains of each respective species contained highly similar genomic sequences and MGE content. The S. schleiferi strain 5909-02 was chosen for having three self-targets vs. the two self-targets present in strain 2713-03; and S. haemolyticus strain W_75, containing a single self-target, was selected arbitrarily over strain W_139 (SI Appendix, Fig. S1).
To first assess whether the CRISPR systems in S. schleiferi and S. haemolyticus are active, we generated amplicons encoding the Cas9, Cas1, Cas2, and Csn2 of each system under their natural promoters and a plasmid expressing a single-guide RNA (sgRNA) for SauCas9 (27). Two versions of the sgRNA were designed, one targeting a superfolder GFP reporter (sfGFP) and the other containing a nontargeting sequence. We coexpressed each amplicon with the targeting or control sgRNA-encoding plasmid, and a plasmid encoding sfGFP, in a cell-free transcription–translation extract (TXTL; Fig. 1B) (19, 23). We observed a strong reduction in sfGFP expression in the TXTL reaction mixtures expressing the targeting, but not the nontargeting, sgRNAs (Fig. 1C). This result suggests that the native type II-A CRISPR systems in both S. schleiferi strain 5909-02 and S. haemolyticus strain W_75 are active.
Nearly all currently known Acrs can be found in MGE sequences, especially phages, highlighting their use as a CRISPR counterdefense mechanism (3). Therefore, we examined the genomes of both S. schleiferi and S. haemolyticus for integrated MGEs using PHASTER (28) and Islander (29) to identify potential hotspots that might harbor Acrs. We found four predicted MGEs in S. schleiferi strain 5909-02 and eight in the contigs of the S. haemolyticus strain W_75 assembly. We then designed 27 genome fragment (GF) amplicons in total, covering all of the predicted sequences, each up to ∼10 kb, with the goal of best capturing complete operons wherever possible (SI Appendix, Table S2). We then prepared TXTL reactions containing DNA encoding the sfGFP reporter, the sgRNA-encoding plasmid, a GF amplicon, and the associated Cas protein-encoding amplicon from the GF’s source organism. By comparing GFP expression over time, we found that the presence of GF5 from S. schleiferi corresponded to nearly complete inhibition of Cas9-mediated sfGFP knockdown (Fig. 2A). We observed no inhibitory activity in the presence of any GFs derived from S. haemolyticus.
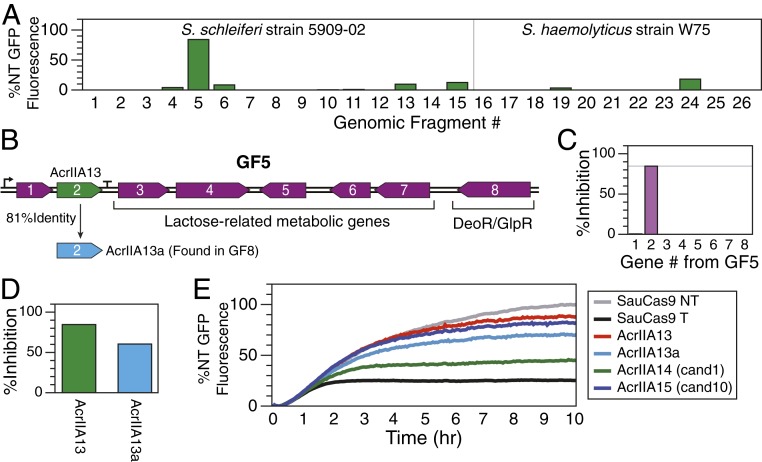
Fig. 2.
Identification of three Acrs using amplicon screening and guilt by association. (A) The relative level of S. schleiferi (Left) or S. haemolyticus (Right) Cas9 DNA cleavage inhibition for each fragment was measured as percentage of the GFP expression for the nontargeting (NT) control after subtracting the fluorescence level observed in the targeting (T) control with no Acr present. Thus, 0% inhibition is equivalent to the GFP expression level measured with the targeting sgRNA, while 100% inhibition represents no reduction in the maximum GFP expression. GF5 is the only amplicon that exhibits Acr activity. (B) Genes found in GF5. (C) Each gene in GF5 was individually cloned and tested for Acr activity with TXTL and SauCas9. The second gene of GF5 (AcrIIA13) inhibits SauCas9. (D) AcrIIA13a, a homolog of AcrIIA13, is found in GF8 and is also able to inhibit SauCas9 in a TXTL assay. GF8, which contains AcrIIA13a, did not exhibit Acr activity, however. (E) Of 10 candidates chosen from a guilt-by-association search seeded by GF5 gene 1, candidates 1 (AcrIIA14) and 10 (AcrIIA15) were found to inhibit SauCas9 using the TXTL assay. All data in this figure are from single TXTL runs.
S. schleiferi GF5 contains eight genes (Fig. 2B), most of which are predicted to be part of the lactose metabolic pathway. Genes 1 and 2, however, have no predicted function. We cloned each gene from GF5 into a heterologous plasmid under the control of a pTet promoter and then used the TXTL assay to test each individual gene for its ability to inhibit SauCas9 expressed from a plasmid. We observed that gene 2 alone produced the inhibitory effect previously observed with the complete GF5 (Fig. 2C). This gene was named AcrIIA13 to indicate its inhibition of a CRISPR type II-A system, in accord with standard Acr nomenclature (22). There is also a homologous protein, AcrIIA13a, found within GF8, and although no inhibitory activity from GF8 was ever observed, when AcrIIA13a is expressed separately using the pTet expression plasmid, it does exhibit inhibitory activity against SauCas9 (Fig. 2D).
Guilt-By-Association Identification of SauCas9 Acrs.
We wondered whether AcrIIA13 or the neighboring gene in its operon (GF5 gene 1) might be associated with other Acrs, and whether these inhibitors might have the ability to block activity of the SauCas9 protein. We first searched AcrIIA13 homologs to find potential Acr candidates co-occurring with it (SI Appendix, Fig. S2A). While we did find a truncated homolog missing the AcrIIA13 N terminus, AcrIIA13b, we did not observe any new genes commonly found near AcrIIA13. However, when we searched for homologs of GF5 gene 1, we found that they occur widely in bacteria (SI Appendix, Fig. S2B), a typical hallmark of aca genes (6, 9, 10, 15). We identified genes that frequently co-occur with GF5 gene 1 and selected them as an initial set of Acr candidates. Then, because many Acrs are clustered together, we also added genes that co-occurred with the initially chosen Acr candidates to create the final test set. This set of 10 candidate Acr genes was cloned into the pTet expression plasmid for further analysis (SI Appendix, Table S3).
Each of the 10 new candidate Acr genes was tested in the TXTL sfGFP depletion experiment, using SauCas9 expressed from a recombinant plasmid. After 10 h, we observed that TXTL reaction mixtures for two of the new Acr candidates, candidate 1 and candidate 10, expressed GFP levels that were significantly higher than the GFP levels observed when no Acr candidate was present (Fig. 2E and SI Appendix, Fig. S3). Candidate 1, hereafter referred to as AcrIIA14, is phylogenetically associated with a close homolog of GF5 gene 1 and occurs in Staphylococcus simulans strain 19, which had previously passed the PAM filtering step for self-targeting Staphylococcus genomes and has a Cas9 closely related to SauCas9 (86% identical via blastp) (SI Appendix, Table S1). Although candidate 10, hereafter referred to as AcrIIA15, does not occur in a CRISPR self-targeting strain, a close homolog (80% identical via blastp) can be found in Staphylococcus pseudintermedius strain 104N, which is self-targeting (SI Appendix, Fig. S2 and Dataset S1).
Reconstitution of Acr Activity against SauCas9.
To examine the inhibitory activity detected in the TXTL experiments with AcrIIA13–AcrIIA15, we purified each Acr protein and tested its ability to inhibit SauCas9-mediated double-stranded DNA (dsDNA) cleavage in vitro. We began by first assembling the SauCas9-sgRNA RNP complex, then added the individually purified Acr proteins and dsDNA target. Under these conditions, we observed near total inhibition of SauCas9 dsDNA cleavage at 20-fold molar excess of Acr relative to Cas9 RNP (Fig. 3 A, Left and SI Appendix, Fig. S4 A, Top). To verify that the inhibition we observed was not an artifact due to nonspecific binding at high Acr protein concentrations, we also performed the cleavage assays in the presence of AcrVA4 and bovine serum albumin (BSA), and observed no difference in cleavage activity of SauCas9 in the presence of either BSA or AcrVA4 (SI Appendix, Fig. S4B). To determine the stage at which the Acrs are able to inhibit SauCas9 activity, we performed the SauCas9 in vitro cleavage experiments again, but added the Acr proteins before the addition of sgRNA. Adding the Acr protein before the sgRNA led to an approximately fourfold increase in inhibition by the Acr proteins, on par with the only currently known broad spectrum Acr for type II-A Cas9, AcrIIA5 (11, 30) (Fig. 3 A, Right and SI Appendix, Fig. S4 A, Bottom). The addition of BSA or AcrVA4 before RNP formation had no effect on cleavage of the target (SI Appendix, Fig. S4C).
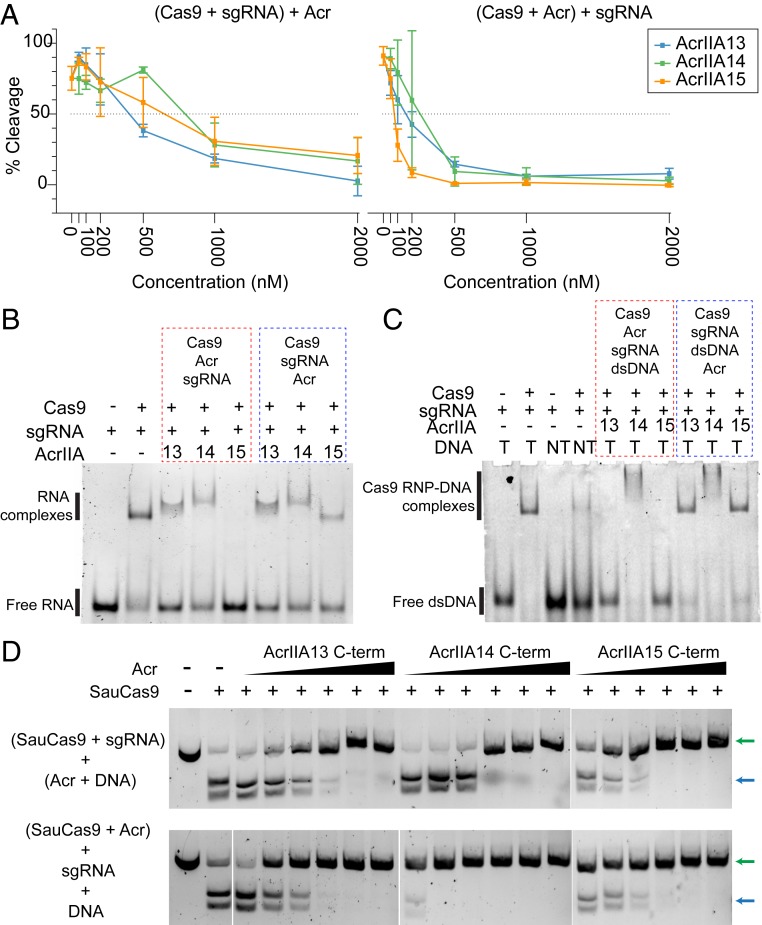
Fig. 3.
Three SauCas9 inhibitors with distinct features and activities. (A) Plot showing inhibition of SauCas9 cleavage activity by AcrIIA13, AcrIIA14, and AcrIIA15 with %cleavage plotted on y axis and Acr concentration (nanomolar) on the x axis. (Left) Plot corresponding to the cleavage assays performed by first complexing Cas9 and sgRNA followed by addition of Acr and DNA. (Right) Plot corresponding to the cleavage assays performed by first complexing Cas9 with Acrs followed by the addition of sgRNA and target dsDNA. The plotted data are the average %cleavage activity from three independent replicates. Error bars represent average ± SD. (B) Six percent polyacrylamide gel showing formation of Cas9-sgRNA RNP in the presence and absence of different Acrs. Lanes boxed in red represent reactions where Acrs were added before the addition of sgRNA. Lanes boxed in blue represent reactions where Acrs were added after the addition of sgRNA. Order of the addition of different reaction components are shown above the boxes. (C) Six percent polyacrylamide gel showing binding of Cas9-sgRNA RNP to either target DNA (T) or nontarget DNA (NT) in the presence and absence of different Acrs. Lanes boxed in red represent reactions where Acrs were added to Cas9 before the addition of sgRNA and dsDNA. Lanes boxed in blue represent reactions where Acrs were added after the addition of dsDNA to Cas9-sgRNA RNP. Order of the addition of different reaction components are shown above the boxes. (D) Agarose gels showing cleavage of dsDNA target by SauCas9 in the presence of increasing amounts of each Acr with the N terminus removed. Reactions were performed by adding Acrs after complexing Cas9 and sgRNA (Top) or prior to the addition of sgRNA (Bottom), as indicated to the Left of each panel. Each reaction contains 5 nM dsDNA, 100 nM SauCas9, 100 nM sgRNA, and the following Acr concentrations: 50 nM, 100 nM, 200 nM, 500 nM, 1,000 nM, and 2,000 nM. Uncleaved and cleaved dsDNA products are indicated by green and blue arrows, respectively.
We then performed RNA electrophoretic mobility shift assays (EMSAs) to determine if the Acrs were able to prevent, or reverse, Cas9-sgRNA RNP formation. While AcrIIA13 and AcrIIA14 did not prevent RNP formation, the addition of AcrIIA15 before the sgRNA during complex formation eliminated the presence of the slow-migrating species corresponding to Cas9 RNP that is visible when the Acr is added after RNP complex formation (Fig. 3B). The lack of a mobility-shifted species when AcrIIA15 is added before sgRNA suggests that AcrIIA15 binds directly to Cas9 to prevent RNP formation rather than causing RNP disassembly. Next, we performed EMSAs to evaluate the ability of the Acrs to prevent binding of the Cas9-sgRNA RNP to its target dsDNA. We found that both AcrIIA13 and AcrIIA15 inhibited dsDNA binding of Cas9, but only when added prior to the addition of target dsDNA. Interestingly, although AcrIIA14 completely inhibited Cas9-cleavage activity (Fig. 3A), it did not prevent Cas9 RNP from binding to its target. However, a supershifted species was observed after the addition of AcrIIA14, either before or after addition of dsDNA. This suggests that AcrIIA14 may act by binding the SauCas9 active site without impacting DNA binding, similar to the behavior of AcrIIC1 (31). AcrIIA14 might also trigger complex dimerization to form an inactive conformation, similar to the mechanisms of AcrIIC3 (31, 32) or AcrVA4 (33) (Fig. 3C). Altogether, these biochemical data suggest that each of the Acrs inhibits SauCas9-mediated DNA cleavage by a different method.
AcrIIA13, AcrIIA14, and AcrIIA15 Share an N-Terminal Domain That Is Dispensable for Cleavage Inhibition.
To determine the level of sequence similarity among AcrIIA13, AcrIIA14, and AcrIIA15, we performed a multiple sequence alignment with MUSCLE and observed a shared N-terminal domain across all three Acrs, which was absent in AcrIIA13b, a truncated homolog of AcrIIA13 found during the guilt-by-association search (SI Appendix, Fig. S2B). The remaining C terminus, however, was not closely related among AcrIIA13, AcrIIA14, and AcrIIA15 (SI Appendix, Fig. S5A). Given the apparently distinct mechanisms of inhibition by each Acr, we suspected that the residues downstream of the shared N terminus were responsible for the Acr activity. To test this, we cloned, expressed, and purified truncated versions of all three Acrs with the shared N termini removed (SI Appendix, Fig. S5A). In DNA cleavage assays with the purified C-terminal Acr proteins, all showed complete inhibition of Cas9 cleavage activity at approximately fivefold molar excess whether added before or after Cas9-sgRNA RNP formation (Fig. 3D). We also observed strong inhibition with the C-terminal portion of AcrIIA13 in the TXTL sfGFP expression assay, while no inhibition was detected using the N-terminal domain alone (SI Appendix, Fig. S5B). Overall, these data support the conclusion that the variable C-terminal domains of AcrIIA13, AcrIIA14, and AcrIIA15 constitute the Acr activity of each protein. We also tested the truncated Acr proteins for inhibition of SpyCas9 in an RNA-guided DNA cleavage assay, but observed no significant reduction in DNA cutting (SI Appendix, Fig. S5C).
The Shared Acr N-Terminal Domain Binds an Inverted-Repeat DNA Sequence.
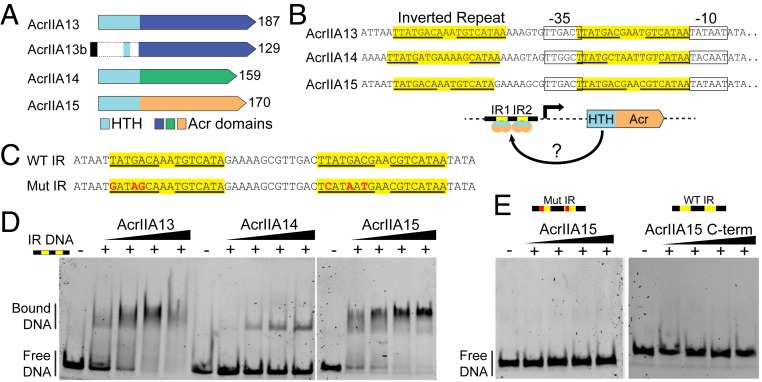
On further sequence analysis of the homologous N-terminal section of AcrIIA13–AcrIIA15 using HHpred (34), we found they contain a conserved HTH motif (Fig. 4A and SI Appendix, Fig. S5A). HTH domains have been found in aca genes and were recently shown to repress their own transcription along with other Acrs in the same operon (16, 35). These aca genes encode proteins that bind to promoter-proximal sequences containing two IRs to block polymerase access. Upstream of the operons containing AcrIIA13–AcrIIA15, we observed IRs proximal to the expected promoter sequences for these operons based on sequence similarity to the Escherichia coli σ70 consensus promoter (TTGACA-N17-TATAAT) (Fig. 4B). To determine if AcrIIA13–AcrIIA15 are capable of binding to the promoter-proximal IR sequences, we performed DNA EMSAs, combining each purified Acr protein with a fluorescein amidite (FAM)-labeled dsDNA sequence spanning the IRs in the promoter-proximal region of AcrIIA15 (Fig. 4C). We found that AcrIIA13 and AcrIIA15 strongly interact with the promoter-proximal IR sequence, while AcrIIA14 only weakly interacted with it (Fig. 4D). Mutating the IR sequence of the dsDNA substrate completely abolished the Acr-IR DNA interaction (SI Appendix, Fig. S4E). To further assess if this interaction was mediated by the HTH motif, we repeated the DNA EMSA using the C-terminal AcrIIA15 and failed to detect any interaction with the dsDNA IR-DNA substrate. Together, these data suggest that, similar to AcrIIA1 (16), AcrIIA13–AcrIIA15 also are fusion proteins that link an anti-SauCas9 Acr with a DNA binding domain that is likely to regulate the expression of its own operon.
Fig. 4.
AcrIIA13, AcrIIA14, and AcrIIA15 share an N-terminal domain that binds a set of inverted repeats proximal to their promoters. (A) Schematic showing the gene structure of AcrIIA13, AcrIIA14, and AcrIIA15. The gene section highlighted in blue is highly conserved across the different Acrs and contains the conserved HTH motif. The C-terminal region shows high variability across the three Acrs. (B) Promoter-proximal sequences for AcrIIA13–AcrIIA15 showing the presence of two sets of IRs. IR segments are highlighted in yellow with the inverted repeat sequence underlined. Schematic outlines hypothesis that the HTH motif within the Acrs binds to the promoter-proximal sequence spanning inverted repeats IR1 and IR2 to self-regulate Acr transcription. (C) Sequence of the dsDNA substrates used for EMSAs in D and E. These sequences are encoded within the promoter-proximal region of AcrIIA15 that were either synthesized in its wild-type form or mutated to serve as negative control. (D) Individually purified Acr proteins bind to the AcrIIA15 promoter-proximal sequence containing inverted repeats, causing an upshift. (E) AcrIIA15 does not bind to a mutated form of the AcrIIA15 IR dsDNA substrate (Left). Removing the N-terminal domain from AcrIIA15 prevents binding to the wild-type (WT) IR DNA (Right).
Inhibition of SauCas9-Mediated Genome Editing in Human Cells.
To assess the ability of AcrIIA13–AcrIIA15 to inhibit genome editing in the context of human cells, we designed a lentiviral validation system to express Cas9, an sgRNA, and an Acr protein from three different, stably integrated constructs (Fig. 5A). Using HEK-RT1 genome editing reporter cells (36, 37)—a monoclonal HEK293T-based cell line expressing a doxycycline-inducible GFP reporter—lentiviral constructs encoding the AcrIIA proteins, or mTagBFP2, and SauCas9 or SpyCas9, were sequentially transduced and stable integrants selected. The resulting cell lines were then further transduced with a vector expressing a GFP-targeting or negative control sgRNA and an associated mCherry fluorescence marker. In this system, we observed a baseline of ∼75% GFP-positive cells after induction in the presence of a negative control sgRNA, which was reduced to near 0% by an on-target sgRNA disrupting GFP expression (SI Appendix, Fig. S6).
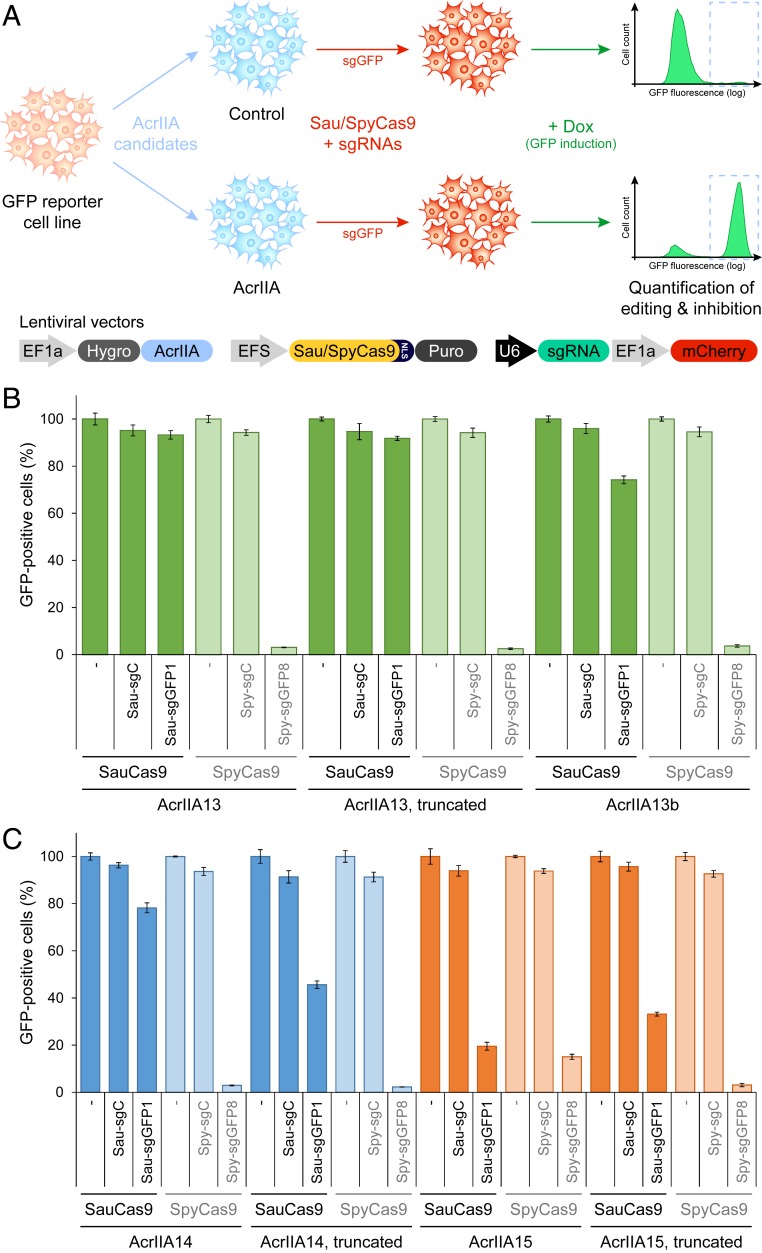
Fig. 5.
AcrIIA13 is a potent and selective inhibitor of SauCas9 in mammalian cells. (A) Schematic of a mammalian validation platform to express Cas9, sgRNAs, and an Acr protein from different stably integrated lentiviral vectors in human cells, to quantify the CRISPR-Cas genome editing inhibitory potential of the Acr candidates by flow cytometry. HEK-RT1 cells, a human HEK293T-based genome editing reporter cell line with a doxycycline-inducible GFP, were sequentially transduced with lentiviral vectors expressing Acr candidates, SauCas9 or SpyCas9, and guide RNAs targeting GFP or a negative control. Three days posttransduction of the guide RNAs, GFP expression was induced by treatment with doxycycline for 24 h. Genome editing inhibition efficiency for each Acr candidate was measured as the increase in percentage GFP-positive cells expressing on-target guide RNAs (sgGFP, mCherry-positive), compared to controls. (B) Quantification of SauCas9 and SpyCas9 inhibition efficiency of AcrIIA13, an N-terminal truncation of AcrIIA13, and an AcrIIA13 homolog (AcrIIA13b) in human cells. Error bars represent the SD of triplicates. sgC, sgGFP1/sgGFP8: negative control and GFP-targeting guide RNAs for the respective Cas9s. The data shown for AcrIIA13 are partial quantification of raw flow cytometry data shown in SI Appendix, Fig. S8. (C) Assessment of SauCas9 and SpyCas9 inhibition by AcrIIA14, an N-terminal truncation of AcrIIA14, AcrIIA15, and an N-terminal truncation of AcrIIA15, in human cells. The assay was run as described above.
We next tested three known Acrs for their ability to inhibit SauCas9-mediated genome editing in mammalian cells, including the robust SpyCas9 inhibitor AcrIIA4 (10), the broad-spectrum type II-A Cas9 inhibitor AcrIIA5 (12, 30), and the species-specific Streptococcus thermophilus Cas9 inhibitor AcrIIA6 (12). While AcrIIA4 prevented genome editing by SpyCas9 as expected (10), we found that it had no effect against SauCas9 (SI Appendix, Fig. S7). Conversely, AcrIIA5 displayed strong inhibitory activity toward both SauCas9 and SpyCas9, as recently shown (30), while AcrIIA6 did not show any inhibition of either of the two Cas9s.
Similar genome editing inhibition experiments using AcrIIA13, AcrIIA14, and AcrIIA15 showed that AcrIIA13 is a potent inhibitor of SauCas9, with near complete inhibition of GFP gene editing activity (Fig. 5B and SI Appendix, Fig. S8). However, in contrast to AcrIIA5, AcrIIA13 did not inhibit SpyCas9, making it to our knowledge the only Acr selective for SauCas9, and further expanding the CRISPR-Cas modulation toolkit. Interestingly, the truncated form of AcrIIA13, which is substantially smaller, was equally effective at inhibiting SauCas9-mediated genome editing in human cells. The AcrIIA13b homolog that lacks the N-terminal DNA binding domain showed a comparable trend of SauCas9 selective inhibition. Full-length AcrIIA14 also proved to be a potent inhibitor of SauCas9, although less robust than AcrIIA13, and removing its N-terminal domain had a larger impact on its inhibitory efficacy (Fig. 5C). SpyCas9 was not affected by AcrIIA14. Finally, only AcrIIA15 demonstrated any activity toward SpyCas9-mediated genome editing, but its inhibition was minimal (Fig. 5C), showing that AcrIIA13, AcrIIA14, and AcrIIA15 are potent and selective inhibitors for SauCas9.
Discussion
The potential role of anti-CRISPRs in regulating CRISPR-Cas gene editing in therapeutic applications has helped spur the discovery of over 40 different families of Acrs (3). However, creating “designer” inhibitors for specific gene editors and editing scenarios is very challenging, as Acr proteins are small and lack any sequence and structural conservation to use for direct identification of desired modalities. One method to overcome these limitations is to cast a wide net in the genomic space, such as through metagenomic DNA screening (13, 14). However, coverage is a challenge in these screens, as megabases of DNA need to be sifted through to find rare Acr genes. The selection of self-targeting genomes (7, 10, 19) is meant to address this issue by narrowing screening efforts to only a few candidate genomes. We further focus our search efforts to MGEs only, where nearly all Acrs have been discovered (2), reducing the search space to a few hundred kilobases or less. Identifying self-targeting events is particularly powerful when searching for specific Acrs against a Cas protein of interest (especially in organisms with multiple CRISPR systems), because the search inherently identifies which Cas system is likely being inhibited. Yet, an even more direct method is to use a guilt-by-association search approach, which leverages the repeated observations that Acrs tend to coexist in similar operons and that the genomic neighborhood of several Acrs contain anti-CRISPR-associated (aca) genes that are closely coupled with Acrs and widespread in bacteria and MGEs.
In this work, we sought to identify potent inhibitors of SauCas9, a common SpyCas9 alternative. Using a combination of the methods described above, we identified three families of Acrs (AcrIIA13, AcrIIA14, and AcrIIA15) that are potent inhibitors of SauCas9. Using biochemical approaches, we find that all three Acrs possess different mechanisms of Cas9 inhibition. AcrIIA13 prevents SauCas9-mediated dsDNA cleavage activity by blocking DNA binding, while AcrIIA15 binds to SauCas9 to block RNP assembly (Fig. 3 B and C). Our data also show that AcrIIA14 does not interfere with SauCas9 binding to either RNA or dsDNA, but inhibits dsDNA cleavage activity at a potency comparable to AcrIIA13 and AcrIIA15. It is possible that AcrIIA14 either causes the SauCas9-sgRNA RNP to form higher order structures like AcrVA4 (33) or AcrIIC3 (31, 32), or interacts with RNP to prevent cleavage but not binding like AcrIIC1 (31). We also observed potent inhibition of SauCas9, but not SpyCas9, by AcrIIA13 and AcrIIA14 in mammalian cells (Fig. 5), suggesting both have a narrow range of inhibition. AcrIIA15 was able to inhibit both SpyCas9 and SauCas9 in human cells, but to a much lesser extent. Of the three Acrs, AcrIIA13 was the most potent inhibitor of SauCas9 in human cells.
In comparing the protein sequences among AcrIIA13–AcrIIA15, we observe that the N termini of the Acrs possessed a highly conserved HTH motif (Fig. 4A and SI Appendix, Fig. S5A). This motif is a prominent feature of aca genes, which were recently shown to negatively regulate the transcription of their own operon (16, 35). The self-regulating activity of aca proteins was shown to be mediated by a direct interaction between the HTH motif and promoter-proximal sequences containing two IRs. Examining the promoter regions of AcrIIA13–AcrIIA15, we found that all of them also contain similar promoter-proximal IR sequences (Fig. 4B), and that all three Acrs are able to bind to the IR sequences specifically and only when the N-terminal domain is present (Fig. 4E). The DNA binding activity of the N-terminal region also has little to no effect on Acr potency when removed from the protein in in vitro cleavage assays (Fig. 3D and SI Appendix, Fig. S5B) and only a minor effect, or none, in human cells depending on the Acr (Fig. 5 B and C). AcrIIA13 in particular is unaffected by the removal of its N terminus, and represents a highly potent, SauCas9-specific inhibitor for use in mammalian contexts for regulating Cas9 activity.
Based on our data, the AcrIIA13, AcrIIA14, and AcrIIA15 proteins we identified are likely fusions between each of three unique Acr proteins and a shared aca gene in the form of an Acr gene that inhibits Cas9 with an aca gene fused to the N terminus that is dispensable for DNA cleavage inhibition. It is unclear how such an aca gene would have become fused with different Acr proteins, or what benefit a single protein would have over the split alternative. Regardless, the aca gene sequence provides another seed to search for further discoveries using the guilt-by-association approach. With the addition of yet more proteins, we increase the chances of finding new activities that are particularly attractive to biotechnology and therapeutic applications, such as catalytic Acrs that would have low delivery requirements for potent inhibition (38, 39). We believe that the search for more Acrs with potent and novel mechanisms of inhibition will continue to rapidly add new dimensions of control into the CRISPR toolbox.
Materials and Methods
Self-Targeting Strain Selection.
The bioinformatics search for self-targeting Staphylococcus strains was carried out in a similar manner as described in Watters et al. (19). Briefly, the STSS was used in the NCBI database for all assemblies returned using a search of “Staphylococcus[organism] NOT phage NOT virus.” Each assembly was checked for instances of self-targeting, which were reported along with details about the instance including: target site mutations, up/downstream target sequences, Cas genes found near the array of the self-targeting spacer, predicted CRISPR subtype based on repeats/Cas proteins, repeat sequences, and conservation, etc. Systems that were output as ambiguous type II CRISPR systems were manually checked for 1) repeat sequence similarity to other known loci; 2) presence of defining Cas proteins (i.e., Csn2 for II-A); and 3) alignment of Cas9 of the system in question to a set of type II-A, II-B, and II-C Cas9s to determine similarity. The final set of self-targeting instances found in Staphylococcus genomes in the NCBI database can be found in Dataset S1. Strains were chosen for further study based on similarity of the downstream target site to the SauCas9 PAM [3′-NNGRR(T)] and similarity of the self-targeting system’s Cas9 to SauCas9 by blastp.
Guilt-By-Association Search.
GF5 genes 1 and 2 (AcrIIA13) were used to query the NCBI protein database with blastp to identify homologs. Each homolog found was then manually surveyed for neighboring genes that were not found within the genome fragment amplicons tested with the TXTL sfGFP assay. This approach was repeated using any neighboring proteins to further locate their neighbors. Only alignments with an E-value <0.001 were considered. The search was ended after 10 candidate proteins were identified.
gDNA Extraction.
To extract gDNA, 4 mL of a culture containing S. haemolyticus (strains W_75 and W_139) or S. schleiferi (strains 5909-02 and 2713-03) cells were grown overnight in tryptic soy broth. The cultures were then pelleted and resuspended in 500 μL of phosphate buffered saline (PBS). Cells were then treated with lysostaphin for 1 h at 37 °C. gDNA was then extracted using the Monarch gDNA Purification Kit from NEB following the manufacturer’s instructions.
Template Preparation for TXTL.
Each TXTL reaction contained three template DNA sources: a sfGFP constitutive reporter plasmid (pUC), a plasmid expressing a sgRNA targeting GFP (pUC) using the SauCas9 sgRNA scaffold sequence from Strutt et al. (27), an either a plasmid expressing SauCas9 under an arabinose promoter (p15A) or a genomic amplicon containing the Cas9 from S. haemolyticus or S. schleiferi. Reactions may also contain an Acr or Acr candidate expressed from a Tet-repressible promoter or a genomic fragment amplicon with the naturally occurring sequence. The plasmid preparation for the reporter, sgRNA plasmid, and SauCas9 were as described in Watters et al. (19). To prepare the Cas9 genomic amplicons, PCR of purified gDNA with Q5 polymerase (New England Biolabs) was performed with primers that spanned all of the Cas genes (SI Appendix, Table S2). Similarly, genomic fragment amplicons were prepared with PCR using the gDNA and primers listed in SI Appendix, Table S2. Acr candidates that were to be tested individually were cloned into a plasmid for use in TXTL.
To prepare the plasmids for TXTL, a 20-mL culture of E. coli containing one of the plasmids was grown to high density, then isolated across five preparations using the Monarch Plasmid Miniprep Kit (New England Biolabs), eluting in a total of 200 μL nuclease-free H2O. A total of 200 μL of AMPure XP beads (Beckman Coulter) were then added to each combined miniprep and purified according to the manufacturer’s instructions, eluting in a final volume of 20 μL in nuclease-free H2O.
All anti-CRISPR candidate amplicons and subfragments were prepared using 100-μL PCRs with Q5 or Phusion polymerase (New England Biolabs), under various conditions to yield a clear single band on an agarose gel such that the correct fragment length was greater than 95% of the fluorescence intensity of the lane on the gel. A total of 100 μL of AMPure XP beads (Beckman Coulter) were then added to each reaction and purified according to the manufacturer’s instructions, eluting in a final volume of 10 μL in nuclease-free H2O. The PCRs and purification of the Cas9 genomic amplicons were scaled up fivefold. A listing of the plasmids used in the TXTL experiments can be found in SI Appendix, Table S4.
TXTL Reactions.
The reactions were carried out as described in Watters et al. (19). Briefly, each reaction was mixed from 9 μL of TXTL master mix, 0.125 nM of each reporter plasmid, 1 nM of Cas9 amplicon or plasmid, 2 nM of sgRNA plasmid, 1 nM of genomic fragment amplicon or Acr candidate plasmid, 1 μM of isopropyl β-d-1-thiogalactopyranoside (IPTG), 0.5 μM of anhydrotetracycline, 0.1% arabinose, and 2 μM of annealed oligos containing six χ sites (40).
The reactions were run at 29 °C in a TECAN Infinite Pro F200, measuring GFP (λex: 485 nm, λem: 535 nm) fluorescence levels every 3 min for 10 h. To plot kinetic data, the minimum measured fluorescence intensity was subtracted from each point on the curve (to compensate for early variations due to condensation on the sealing film), then the overall curve was normalized by the fluorescence level measured for the nontargeting negative control after 10 h of reporter expression.
Protein Purification.
DNA sequences corresponding to SauCas9 and variants of AcrIIA13, AcrIIA14, AcrIIA15 were cloned downstream of 10×His tag, MBP tag, and a tobacco etch virus (TEV) cleavage site. Each of these constructs was grown in Rosetta2 cells overnight in lysogeny broth and subcultured in 750 mL of terrific broth. After the cultures reached an OD600 of 0.6 to 0.8, proteins were induced by treatment with 0.5 mM IPTG for ∼20 h at 16 °C. Cells were then resuspended in lysis buffer containing 50 mM Hepes pH 7.0, 500 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP), and 5% (vol/vol) glycerol and 1× Roche cOmplete protease inhibitor mixture and lysed by sonication, followed by purification using Ni-NTA Superflow resin (Qiagen). The eluted proteins were then cleaved with TEV protease overnight at 4 °C while performing buffer exchange in buffer containing 250 mM NaCl. The cleaved proteins were further purified using ion exchange chromatography with a linear NaCl gradient using Heparin HiTrap column (GE) or HiTrap Q column (GE). The protein samples were then concentrated and subjected to size exclusion chromatography with Superdex 200 size exclusion column (GE). The proteins were eluted in final storage buffer containing 20 mM Hepes pH 7.0, 200 mM KCl, 1 mM TCEP, and 5% glycerol. Plasmids for expressing the Acrs are available on Addgene with numbers between 138281 and 138286.
In Vitro Cleavage Assays.
All dsDNA cleavage assays were performed in a 1× cleavage buffer containing 20 mM Hepes-HCl, pH 7.5, 150 mM KCl, 10 mM MgCl2, 0.5 mM TCEP. sgRNA sequences synthesized by IDT were first refolded in 1× cleavage buffer by heating at 70 °C for 5 min then cooling to room temperature. For all of the cleavage assays involving SauCas9, reaction components were added in the following two ways: 1) SauCas9 and Acrs were complexed together, followed by sequential addition of sgRNA and dsDNA; and 2) SauCas9 and sgRNA were complexed together, followed by addition of Acrs and dsDNA target. Each step of sequential additions in both reactions were preceded by incubation at 37 °C for 10 min. All cleavage reactions were then quenched on completion with 2 µL of 6× quenching buffer containing 30% glycerol, 1.2% SDS, 250 mM EDTA. Cleavage products were read using a 1% agarose gel prestained with SYBRGold. All DNA/RNA sequences used for biochemical assays can be found in SI Appendix, Table S5.
Electrophoretic Mobility Shift Assays.
RNA EMSAs were performed by incubating SauCas9 and sgRNA in 1× cleavage buffer lacking MgCl2 in the presence or absence of 10-fold molar excess of different Acrs relative to SauCas9. The reactions were then incubated at 37 °C for 10 min and run on 6% Tris-borate-EDTA polyacrylamide gels for 1 h at 50V. The gels were then visualized using SYBRGold stain. DNA EMSAs to evaluate binding of SauCas9 RNP to dsDNA target were performed by incubating the reaction components described in their respective figures in 1× cleavage buffer lacking MgCl2 at 37 °C for 10 min. The dsDNA substrates used for this assay were synthesized as 5′ FAM-labeled 34 bp DNA oligos that were annealed prior to their addition to the EMSA reaction. The reactions were run on 6% TBE polyacrylamide gels and visualized on a Typhoon imager (GE). DNA EMSAs for the binding of AcrIIA13, AcrIIA14, and AcrIIA15 to IR DNA were performed by incubating 50 nM IR DNA with 0, 50, 100, 151.5, or 200 nM of Acr in 1× cleavage buffer lacking MgCl2 at 37 °C for 10 min. The completed reactions were then run on 6% TBE polyacrylamide gels for 1 h at 50V and visualized with SYBRGold stain.
Lentiviral Vectors.
The lentiviral vector pCF525-mTagBFP2, expressing an EF1a-driven polycistronic construct containing a hygromycin B resistance marker, P2A ribosomal skipping element, and an mTagBFP2 fluorescence marker had been cloned before (19). For stable lentiviral expression of AcrIIA candidates, mTagBFP2 was replaced by the gene-of-interest open reading frame (ORF) using custom oligonucleotide gBlocks (IDT), Gibson assembly reagents (NEB), and standard molecular cloning techniques. The SpyCas9 guide RNA-only lentiviral vector pCF820, encoding a SpyCas9 U6-sgRNA cassette and an EF1a-driven, human codon optimized mCherry2 marker (41), was based on the pCF221 vector (36). To make the backbone more efficient and increase viral titers, the f1 bacteriophage origin of replication and bleomycin resistance marker were removed, as has previously been done for pCF525 (19). The SauCas9 guide RNA-only lentiviral vector pCF824, encoding an SauCas9 U6-sgRNA cassette and an EF1a-driven, human codon optimized mCherry2 marker, was based on the pCF820 vector. The SauCas9 guide RNA scaffold was designed as previously reported (42), synthesized as custom oligonucleotides (IDT), and assembled using Gibson techniques and standard molecular cloning methods. A lentiviral vector referred to as pCF823, expressing an EFS-driven SpyCas9-P2A-PuroR cassette, was based on the vector pCF204 (36) by removing the f1 bacteriophage origin of replication and bleomycin resistance marker to increase viral titers. A lentiviral vector referred to as pCF825, expressing an EFS-driven SauCas9-P2A-PuroR cassette, was based on the vector pCF823 by replacing the SpyCas9 ORF with the SauCas9 ORF from pX601 (Addgene no. 61591) (25). Vector sequences are provided in a separate file (Dataset S2). Gene editing plasmid vectors are available on Addgene with nos. 138305 to 138319.
Design of sgRNAs.
The following spacer sequences were used for SpyCas9 sgRNAs: sgC (GGAGACGGAGGACGACGAACGTCTCT) and sgGFP8 (GCAGGGTCAGCTTGCCGTAGG). The following sequences were used for SauCas9 sgRNAs: sgC (GGAGACGGAGGACGACGAACGTCTCT) and sgGFP1 (GCAAGGGCGAGGAGCTGTTCAC).
Mammalian Cell Culture.
All mammalian cell cultures were maintained in a 37 °C incubator at 5% CO2. HEK293T (293FT; Thermo Fisher Scientific) human kidney cells and derivatives thereof were grown in Dulbecco’s modified Eagle medium (DMEM; Corning Cellgro, no. 10-013-CV) supplemented with 10% FBS (Seradigm no. 1500-500), and 100 units/mL penicillin (Pen) and 100 μg/mL streptomycin (Strep) (100-Pen-Strep; Gibco no. 15140-122). HEK293T and HEK-RT1 cells were tested for absence of mycoplasma contamination (University of California Berkeley Cell Culture facility) by fluorescence microscopy of methanol fixed and Hoechst 33258 (Polysciences no. 09460)-stained samples.
Lentiviral Transduction.
Lentiviral particles were produced in HEK293T cells using polyethylenimine (PEI; Polysciences no. 23966)-based transfection of plasmids, as previously described (36). In brief, lentiviral vectors were cotransfected with the lentiviral packaging plasmid psPAX2 (Addgene no. 12260) and the VSV-G envelope plasmid pMD2.G (Addgene no. 12259). Transfection reactions were assembled in reduced serum media (Opti-MEM; Gibco no. 31985-070). For lentiviral particle production on six-well plates, 1 µg lentiviral vector, 0.5 µg psPAX2 and 0.25 µg pMD2.G were mixed in 0.4 mL Opti-MEM, followed by addition of 5.25 µg PEI. After a 20- to 30-min incubation at room temperature, the transfection reactions were dispersed over the HEK293T cells. Media were changed 12 h posttransfection, and virus was harvested at 36 to 48 h posttransfection. Viral supernatants were filtered using 0.45 µm polyethersulfone (PES) membrane filters, diluted in cell culture media if appropriate, and added to target cells. Polybrene (5 µg/mL; Sigma-Aldrich) was supplemented to enhance transduction efficiency, if necessary.
Gene Editing Inhibition Assays in Human Cells.
Candidate Acr proteins were tested in a HEK293T-based monoclonal reporter cell line called HEK-RT1 (36, 37), which features a doxycycline-inducible GFP reporter (43) that can be edited before doxycycline-based induction of its expression. To test the effect of genomic integration and expression of AcrIIA candidate proteins in mammalian cells, HEK-RT1 cells were stably transduced with the lentiviral vector pCF525 (19) encoding the various candidate proteins or mTagBFP2, and selected on hygromycin B as previously described (19). The resulting HEK-RT1-AcrIIA candidates and HEK-RT1-mTagBFP2 genome protection and editing reporter cell lines were then used to quantify genome editing inhibition by flow cytometry. For this, all reporter cell lines were stably transduced with the lentiviral vector pCF823-SpyCas9 (expressing SpyCas9-P2A-PuroR from an EFS promoter) or pCF825-SauCas9 (expressing SauCas9-P2A-PuroR from an EFS promoter) and selected on puromycin (1.0 µg/mL). AcrIIA/mTagBFP2 and SpyCas9/SauCas9 expressing HEK-RT1 reporter cell lines were then stably transduced with lentiviral vectors pCF820-Spy-sgGFP8/sgC (U6 promoter-driven expression of SpyCas9-specific guide RNAs targeting GFP or a nontargeting recipient control, along with an EF1a-driven humanized mCherry2 marker) or pCF824-Sau-sgGFP1/sgC (U6 promoter-driven expression of SauCas9-specific guide RNAs targeting GFP or a nontargeting recipient control, along with an EF1a-driven humanized mCherry2 marker). At day 3 posttransduction of the guide RNA vectors, anti-CRISPR (AcrIIA candidates, mTagBFP2) + Cas9 (SpyCas9, SauCas9) + guide RNA (Spy-sgC/sgGFP8, Sau-sgC/sgGFP1) expressing HEK-RT1 reporter cells were treated with doxycycline (1 µg/mL) for GFP reporter induction. At 24 h postinduction, transgenic GFP genome editing efficiency and inhibition thereof were quantified by flow cytometry (Attune NxT, Thermo Fisher Scientific). For guide RNA-expressing samples, quantification was gated on the mCherry-positive (lentiviral guide RNA transduced) population. Data were normalized on samples without guide RNA expression treated ±doxycycline (1 µg/mL).
Data Availability.
The code for Self-Target Spacer Searcher can be found on GitHub at: https://github.com/kew222/Self-Targeting-Spacer-Searcher. All other data discussed in the paper are available in the main text, SI Appendix, and Datasets S1 and S2.
Supplementary Material
Acknowledgments
We thank Hua B. Bai for cloning plasmids used in the TXTL assay, Shawn M. Ren for help with preliminary cell line construction, Fuguo Jiang for a gift of AcrIIA4 protein, Gavin Knott for a gift of AcrVA4 protein, the Bondy-Denomy laboratory for sharing their lab space, and the J.A.D. laboratory for thoughtful comments and feedback. We also thank Kathleen Boyajian and Shelley Rankin at the University of Pennsylvania for the generous gift of the S. schleiferi strains, and Jorunn Pauline Cavanagh from the University of Norway, Tromsø for the generous gift of the S. haemolyticus strains. We acknowledge financial support from the Defense Advanced Research Projects Agency (award HR0011 17-2-0043 to J.A.D.), the Paul G. Allen Frontiers Group, and a grant from the National Science Foundation (MCB-1244557 to J.A.D.). C.F. is supported by a National Institutes of Health K99/R00 Pathway to Independence Award (K99GM118909, R00GM118909) from the National Institute of General Medical Sciences. J.A.D. is an investigator of the Howard Hughes Medical Institute (HHMI), and this study was supported in part by the HHMI; J.A.D. is also a Paul Allen Distinguished Investigator.
Footnotes
Competing interest statement: The Regents of the University of California have patents pending for CRISPR technologies on which the authors are inventors. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Felix Biosciences, Mammoth Biosciences, and Inari. J.A.D. is a Director at Johnson & Johnson and has sponsored research projects supported by Pfizer and Biogen.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917668117/-/DCSupplemental.
References
- 1.Knott G. J., Doudna J. A., CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F., Song G., Tian Y., Anti-CRISPRs: The natural inhibitors for CRISPR-Cas systems. Animal Model Exp. Med. 2, 69–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang S., Maxwell K. L., Meet the anti-CRISPRs: Widespread protein inhibitors of CRISPR-Cas systems. CRISPR J. 2, 23–30 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Bondy-Denomy J., Pawluk A., Maxwell K. L., Davidson A. R., Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawluk A., Bondy-Denomy J., Cheung V. H. W., Maxwell K. L., Davidson A. R., A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio 5, e00896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pawluk A., et al. , Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 1, 16085 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Marino N. D., et al. , Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362, 240–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He F., et al. , Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat. Microbiol. 3, 461–469 (2018). Correction in: Nat. Microbiol.3, 1076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pawluk A., et al. , Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauch B. J., et al. , Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168, 150–158.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hynes A. P., et al. , An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat. Microbiol. 2, 1374–1380 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Hynes A. P., et al. , Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun. 9, 2919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uribe R. V., et al. , Discovery and characterization of Cas9 inhibitors disseminated across seven bacterial phyla. Cell Host Microbe 25, 233–241.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Forsberg K. J., et al. , Functional metagenomics-guided discovery of potent Cas9 inhibitors in the human microbiome. elife 8, e46540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J., et al. , Potent Cas9 inhibition in bacterial and human cells by AcrIIC4 and AcrIIC5 anti-CRISPR proteins. MBio 9, e02321-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osuna B. A., et al. , Listeria phages induce Cas9 degradation to protect lysogenic genomes. bioRxiv:10.1101/787200 (30 September 2019). [DOI] [PMC free article] [PubMed]
- 17.Bhoobalan-Chitty Y., Johansen T. B., Di Cianni N., Peng X., Inhibition of type III CRISPR-cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell 179, 448–458.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Athukoralage J. S., Graham S., Grüschow S., Rouillon C., White M. F., A type III CRISPR ancillary ribonuclease degrades its cyclic oligoadenylate activator. J. Mol. Biol. 431, 2894–2899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watters K. E., Fellmann C., Bai H. B., Ren S. M., Doudna J. A., Systematic discovery of natural CRISPR-Cas12a inhibitors. Science 9, eaau5138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heussler G. E., O’Toole G. A., Friendly fire: Biological functions and consequences of chromosomal targeting by CRISPR-cas systems. J. Bacteriol. 198, 1481–1486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondy-Denomy J., et al. , A unified resource for tracking anti-CRISPR names. CRISPR J. 1, 304–305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin J., et al. , Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 3, e1701620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall R., et al. , Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell 69, 146–157.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., et al. , Genome scale screening identification of SaCas9/gRNAs for targeting HIV-1 provirus and suppression of HIV-1 infection. Virus Res. 250, 21–30 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Ran F. A., et al. , In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X., Yu Z., Xu Z., Study the features of 57 confirmed CRISPR loci in 38 strains of Staphylococcus aureus. Front. Microbiol. 9, 1591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strutt S. C., Torrez R. M., Kaya E., Negrete O. A., Doudna J. A., RNA-dependent RNA targeting by CRISPR-Cas9. elife 7, e32724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arndt D., et al. , PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hudson C. M., Lau B. Y., Williams K. P., Islander: A database of precisely mapped genomic islands in tRNA and tmRNA genes. Nucleic Acids Res. 43, D48–D53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia B., et al. , Anti-CRISPR AcrIIA5 potently inhibits all Cas9 homologs used for genome editing. Cell Rep. 29, 1739–1746.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrington L. B., et al. , A broad-spectrum inhibitor of CRISPR-cas9. Cell 170, 1224–1233.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y., et al. , Diverse mechanisms of CRISPR-cas9 inhibition by type IIC anti-CRISPR proteins. Mol. Cell 74, 296–309.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knott G. J., et al. , Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a. elife 8, e49110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann L., et al. , A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Stanley S. Y., et al. , Anti-CRISPR-associated proteins are crucial repressors of anti-CRISPR transcription. Cell 178, 1452–1464.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oakes B. L., et al. , CRISPR-Cas9 circular permutants as programmable scaffolds for genome modification. Cell 176, 254–267.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park H. M., et al. , Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo. Nat. Commun. 9, 3313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knott G. J., et al. , Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat. Struct. Mol. Biol. 26, 315–321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong L., et al. , An anti-CRISPR protein disables type V Cas12a by acetylation. Nat. Struct. Mol. Biol. 26, 308–314 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Marshall R., Maxwell C. S., Collins S. P., Beisel C. L., Noireaux V., Short DNA containing χ sites enhances DNA stability and gene expression in E. coli cell-free transcription-translation systems. Biotechnol. Bioeng. 114, 2137–2141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ai H.-W., Shaner N. C., Cheng Z., Tsien R. Y., Campbell R. E., Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry 46, 5904–5910 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Chen B., et al. , Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 44, e75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fellmann C., et al. , An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code for Self-Target Spacer Searcher can be found on GitHub at: https://github.com/kew222/Self-Targeting-Spacer-Searcher. All other data discussed in the paper are available in the main text, SI Appendix, and Datasets S1 and S2.