Significance
Pseudomonas aeruginosa is an important bacterial pathogen of clinical concern. Evidence suggests that highly pathogenic isolates contain novel virulence genes that other less virulent isolates lack. We utilized a comparative genomics approach to identify such virulence genes from the accessory genomes of 100 clinical isolates. From this work, we identified a contact-dependent growth inhibition (CDI) system known for its ability to mediate direct competition between bacteria. We demonstrate that this CDI system has toxic effects on mammalian cells and is required for virulence in murine infection models. This work highlights the potential of CDI systems to act as virulence factors during invasive P. aeruginosa infection.
Keywords: Pseudomonas aeruginosa, whole-genome sequencing, comparative genomics, virulence, contact-dependent growth inhibition
Abstract
Emerging evidence suggests the Pseudomonas aeruginosa accessory genome is enriched with uncharacterized virulence genes. Identification and characterization of such genes may reveal novel pathogenic mechanisms used by particularly virulent isolates. Here, we utilized a mouse bacteremia model to quantify the virulence of 100 individual P. aeruginosa bloodstream isolates and performed whole-genome sequencing to identify accessory genomic elements correlated with increased bacterial virulence. From this work, we identified a specific contact-dependent growth inhibition (CDI) system enriched among highly virulent P. aeruginosa isolates. CDI systems contain a large exoprotein (CdiA) with a C-terminal toxin (CT) domain that can vary between different isolates within a species. Prior work has revealed that delivery of a CdiA-CT domain upon direct cell-to-cell contact can inhibit replication of a susceptible target bacterium. Aside from mediating interbacterial competition, we observed our virulence-associated CdiA-CT domain to promote toxicity against mammalian cells in culture and lethality during mouse bacteremia. Structural and functional studies revealed this CdiA-CT domain to have in vitro tRNase activity, and mutations that abrogated this tRNAse activity in vitro also attenuated virulence. Furthermore, CdiA contributed to virulence in mice even in the absence of contact-dependent signaling. Overall, our findings indicate that this P. aeruginosa CDI system functions as both an interbacterial inhibition system and a bacterial virulence factor against a mammalian host. These findings provide an impetus for continued studies into the complex role of CDI systems in P. aeruginosa pathogenesis.
Bacterial virulence is a complex trait dependent upon numerous attributes (1), each of which contributes in a cumulative way to the overall pathogenic potential of a given microbe (2). The formula by which a microbe may achieve a high level of virulence varies greatly and is influenced by genes in both the core and accessory genomes (3–5). The bacterial core genome refers to those sequences shared among almost all strains within the species while the accessory genome consists of the remaining variable genetic content that accounts for many strain-specific attributes (6, 7). Emerging evidence suggests that the accessory genome is enriched for virulence genes with the potential to confer distinct pathogenic traits on some members of a bacterial species (8).
Pseudomonas aeruginosa is a widespread and highly diverse species of bacteria (9, 10). It is responsible for many types of severe acute infections (11) and continues to be one of the most common bacteria cultured from the chronically infected lungs of individuals with cystic fibrosis (12). P. aeruginosa exhibits a nonclonal epidemic population structure in which a limited number of widespread clones exist among a much larger cloud of diverse genotypes recombining at high frequencies (13). These complex evolutionary dynamics can generate accessory genomes of extensive diversity through the lateral transfer of different mobile genetic elements (14–17).
Individual P. aeruginosa isolates differ in their intrinsic virulence (18), yet few studies have investigated the role the accessory genome plays in this variability. In this study, our goal was to interrogate the accessory genomes of highly virulent isolates to identify novel virulence genes. To this end, we developed a comparative genomics approach to identify genes that were present in high-virulence P. aeruginosa isolates but absent in low-virulence isolates. We established a quantifiable virulence phenotype using a murine infection model and investigated a large collection of 100 clinical P. aeruginosa isolates to increase our analytical power. Using this approach, we identified several loci that contributed to the pathogenesis of high-virulence P. aeruginosa isolates. We further characterized one of these loci and demonstrated that it encodes a portion of a contact-dependent growth inhibition (CDI) system that functions in both interbacterial competition and virulence in animal infection models. This work provides insight into the potential of CDI systems to act as virulence factors during invasive P. aeruginosa infection.
Results
P. aeruginosa Clinical Isolates Differ in Their Intrinsic Virulence.
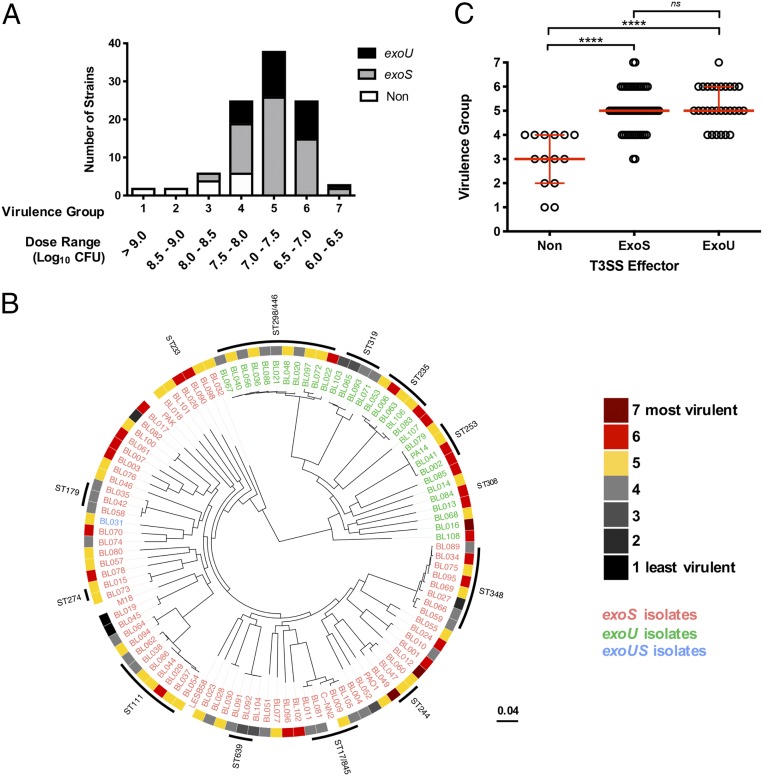
It is understood that individual isolates of P. aeruginosa can substantially differ in virulence, with some isolates exhibiting a significantly heightened pathogenicity (18). We hypothesized that the accessory genomes of such highly pathogenic P. aeruginosa isolates would be enriched for novel virulence genes. As a first step in identifying such genes, we quantified the virulence of a large population of P. aeruginosa isolates. One-hundred P. aeruginosa isolates obtained from the blood cultures of hospitalized patients at Northwestern Memorial Hospital in Chicago (“PABL collection”) (SI Appendix, Table S1) (19) were individually assessed for virulence in mice. A modified LD50 (mLD50) was estimated based upon the development of prelethal illness following tail vein injection at specific half-logarithmic infectious doses. These results were then used to bin the isolates into virulence groups from 7 (high virulence) to 1 (low virulence) based upon these dosing breakpoints (Fig. 1A). The complete virulence distribution as measured by mLD50 spanned over three logarithms, highlighting major differences in virulence between individual P. aeruginosa isolates, even those obtained from a single type of infection.
Fig. 1.
P. aeruginosa clinical isolates differ in virulence. (A) BALB/c mice were infected by intravenous injection with a collection of P. aeruginosa bloodstream (PABL) isolates. The isolates were ranked into virulence groups (7 = most virulent, 1 = least virulent) based upon the morbidity and mortality at several infectious doses. All strains were screened by Western blot for in vitro secretion of the known T3SS effectors ExoS (gray) and ExoU (black). Nonsecreting strains (“non”) are represented in white. (B) A maximum-likelihood tree of the PABL isolates was generated using kSNP v3.0.21. Strains that contain the T3SS effector gene exoU are labeled in green and those that contain exoS are labeled in red. Isolates in blue contain both exoS and exoU genes. The virulence rank for each strain is represented in the outer ring. Several well-documented MLST types are also highlighted. Reference strains UCBPP-PA14, PAO1, PAK, C-NN2, LESB58, and M18 were included in the phylogenetic analysis. (C) The strain virulence data were plotted against the respective type III secretion profile. The red bars indicate the median ± 95% CI of each group (Kruskal–Wallis test with Dunn’s multiple comparisons, ****P < 0.0001, ns = not significant).
To rule out that the observed virulence distribution was driven solely by a clonally related group of P. aeruginosa isolates, whole-genome sequencing was performed on all isolates (20) and phylogenetic relationships were analyzed based upon single nucleotide variations in the core genome. While isolated instances of sequence types (STs) with either relatively high or low virulence could be observed (Fig. 1B), the overall P. aeruginosa bloodstream (PABL) population displayed no evidence of any phylogenetic signal associated with virulence (Pagel’s λ = 6.67 × 10−5, P = 1) (21), suggesting that virulence is not lineage-specific.
We also examined secretion of the type III secretion system (T3SS) effectors ExoS and ExoU, as differences in their secretion are known to impact the virulence of a given isolate (22–25). T3SS effector-secreting isolates were significantly more virulent than nonsecreting isolates (P < 0.0001), but ExoS and ExoU secreting isolates were not statistically different from each other (Fig. 1C). This suggests the observed virulence disparity among the P. aeruginosa PABL isolates cannot be explained simply by production of the different T3SS effectors. Altogether, these data highlight the intrinsic virulence disparity that exists among P. aeruginosa clinical isolates and underscore the limited understanding of what factors account for these differences.
Identification of Novel Virulence Determinants through a Comparative Genomics Approach.
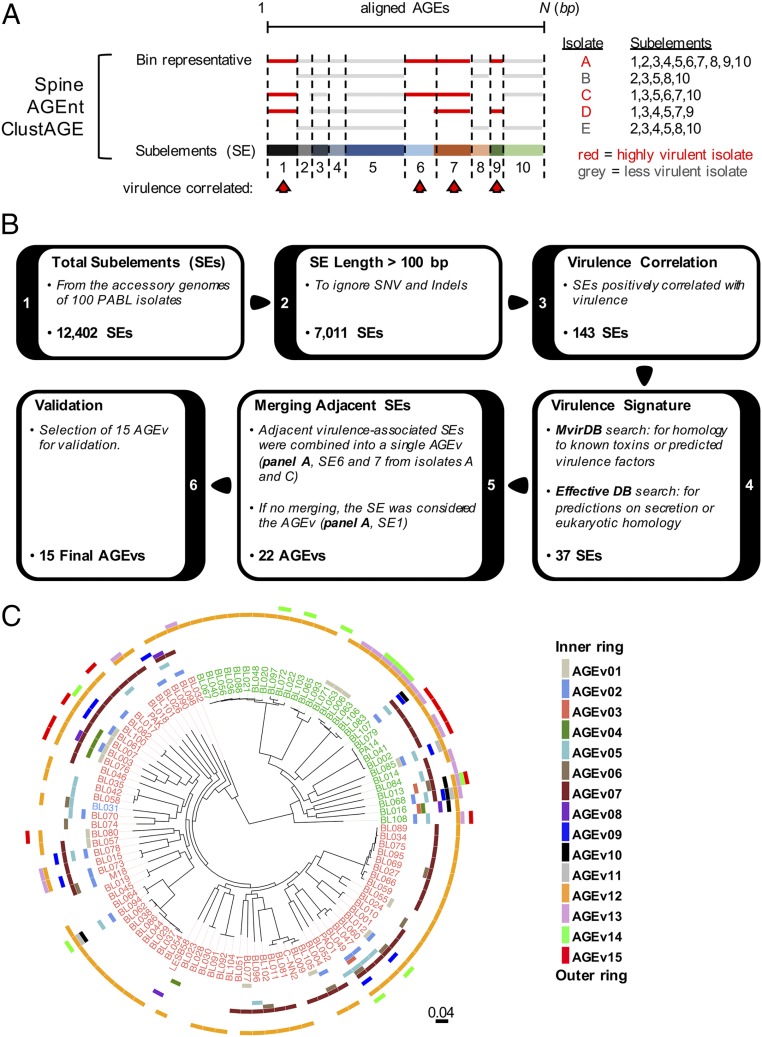
We hypothesized that highly virulent PABL isolates harbored certain accessory genomic elements (AGEs) that contribute to their increased virulence. Although single nucleotide variations and small insertions/deletions (indels) that alter the expression or function of core genes may partially explain these differences, we chose to focus on identifying novel accessory virulence genes. A comparative genomics pipeline was developed to identify accessory genome sequences that were significantly associated with highly virulent isolates (Fig. 2). The core genome of the PABL isolates was first determined using the program Spine. Accessory sequences were then identified by subtracting the core genome from the total genome of each isolate using the program AGEnt (26). These accessory sequences were analyzed using ClustAGE to determine patterns of shared sequences among different isolates (27). An advantage of this approach is that it is independent of gene annotation and is capable of identifying intergenic and intragenic sequences that differ between isolates.
Fig. 2.
Overview of the comparative genomics pipeline. (A) Schematic representation of the process by which accessory sequences were divided into subelements to be analyzed for virulence association. The core and accessory genomes of each PABL isolate were defined using Spine and AGEnt (26). Common contiguous nucleotide sequences among all PABL isolates were identified and organized using ClustAGE (27). The cartoon representation depicts the alignment of sequences in a bin as described in the main text. Alignment breakpoints within an individual bin determine shared contiguous sequences referred to as genetic SEs (numbered 1 through 10). With this organization, SEs can be easily analyzed for their association with highly virulent isolates (red arrowheads). (B) SEs were analyzed for their virulence association using a series of filtering steps. SEs ≤ 100 bp were filtered to remove small insertions and deletions. From this filtered list, SEs associated with PABL isolates in higher virulence groups were statistically determined (positive Spearman correlation, P < 0.05). These virulence-associated SEs were screened through the databases MvirDB and EffectiveDB to identify potential virulence signatures and further filtered to generate a focused list for further study (see main text for details). The ultimate 15 targets were referred to as AGEv. (C) The distribution of each AGEv was overlaid as a separate ring onto the maximum-likelihood tree of the PABL isolates. Strains that encode the T3SS effector exoU are labeled in green and those that encode exoS are labeled in red. Isolates in blue are encode both exoS and exoU.
Subelements (SEs) were examined over a series of filtering steps to identify those with the highest likelihood of impacting virulence (Fig. 2B). SEs were first size selected to be greater than 100 bp, so as to ignore small insertions and deletions. Filtered SEs were then analyzed for a significant positive correlation with virulence rank (e.g., overrepresentation in high-virulence groups). Because our final goal was to identify virulence determinants that may function by a novel mechanism, we chose to narrow this extensive list down to a focused set of elements for further study. Candidate SEs were screened against the MvirDB and EffectiveDB databases for coding regions with sequence homology to known or predicted bacterial toxins, potential secretion signals, or regions with eukaryotic homology. Finally, if SEs passing these filters were located adjacent to each other in highly virulent strains, they were merged together as a single virulence-associated AGE (AGEv). If this were not the case, the individual SE was referred to as an AGEv (Fig. 2 A and B). Twenty-two AGEv passed filtering and represented regions that were predicted to have a high likelihood of encoding novel virulence factors.
To determine whether the computationally identified AGEv actually contributed to the virulence of P. aeruginosa, deletion mutants were constructed for a subset of 15 AGEv in highly virulent isolates and tested for attenuation in vivo. These 15 AGEv were selected to obtain a representative sampling over the range of statistical correlations observed within the group of 22 AGEv. Ultimately, 11 of the 15 AGEv mutants were significantly attenuated (i.e., an increased survival of infected mice relative to wild-type (WT) infected controls) (Table 1 and SI Appendix, Fig. S1). To ensure that the high proportion of attenuated AGEv mutants was not due to chance, we also investigated five AGEs that had no virulence association (AGEx) but were of comparable size and in similar strain backgrounds as AGEv. The AGEx mutants had no statistically significant impact on survival in the mouse model (Table 1 and SI Appendix, Fig. S1), supporting our hypothesis that AGEv have a greater likelihood of encoding virulence factors.
Table 1.
Summary of investigated AGEs
| AGE | Isolates* | Features | Context† | Mutant background‡ | Dose§ | n¶ | P level# |
| AGEv | |||||||
| AGEv1 | 14 | Glucan synthesis, fatty acid metabolism | GI | PABL012 | 6.5 | 10 | <0.0001 |
| PABL057 | 6.2 | 10 | 0.0010 | ||||
| AGEv2 | 16 | Phage-related, hypothetical | Phage | PABL026 | 7.0 | 10 | <0.0001 |
| PABL049 | 6.7 | 10 | <0.0001 | ||||
| AGEv3 | 8 | Transposon, hypothetical | Tn | PABL016 | 6.6 | 15 | 0.5755 |
| AGEv4 | 5 | Restriction modification | Tn | PABL007 | 7.0 | 10 | 0.0017 |
| AGEv5 | 15 | Substrate transport & metabolism | PABL049 | 6.6 | 10 | 0.0002 | |
| PABL070 | 6.8 | 10 | 0.0002 | ||||
| AGEv6 | 9 | Phage-related, hypothetical | Phage | PABL012 | 6.6 | 20 | <0.0001 |
| PABL049 | 6.5 | 10 | 0.0085 | ||||
| AGEv7 | 49 | Phage-related, hypothetical | Phage | PABL012 | 6.5 | 15 | <0.0001 |
| PABL049 | 6.9 | 10 | <0.0001 | ||||
| AGEv8 | 3 | RHS-containing gene | PMT | PABL016 | 6.6 | 15 | 0.0008 |
| AGEv9 | 10 | Phage-related | Phage | PABL012 | 6.6 | 15 | 0.0164 |
| PABL049 | 6.7 | 10 | 0.0139 | ||||
| AGEv10 | 4 | RND family heavy metal efflux proteins | PABL016 | 6.8 | 15 | 0.4811 | |
| PABL068 | 5.8 | 10 | 0.3933 | ||||
| AGEv11 | 2 | Phage-related | Phage | PABL012 | 6.5 | 30 | 0.0001 |
| AGEv12 | 82 | ICE proteins | GI | PABL012 | 6.6 | 15 | 0.0008 |
| PABL049 | 6.7 | 10 | <0.0001 | ||||
| AGEv13 | 18 | Autotransporter protein | PABL083 | 6.5 | 20 | 0.5694 | |
| PABL107 | 6.5 | 30 | 0.0526 | ||||
| AGEx | |||||||
| AGEx1 | 14 | Phage related | Phage | PABL012 | 6.5 | 10 | 0.6789 |
| AGEx2 | 69 | Helicase, hypothetical | Phage | PABL012 | 6.5 | 10 | 0.2059 |
| AGEx3 | 48 | Peptide synthase, hypothetical | PABL016 | 6.7 | 10 | 0.2247 | |
| AGEx4 | 35 | Hypothetical | PABL012 | 6.5 | 10 | 0.4328 | |
| AGEx5 | 32 | Hypothetical | PABL049 | 6.7 | 10 | 0.2402 |
Number of isolates in which the AGEv was observed.
Genomic context in which the AGEs were observed. Abbreviations include pKLC102-like genomic island (GI), transposon (Tn), polymorphic toxin (PMT), or predicted prophage (Phage).
PABL isolate in which the AGE mutation was constructed.
Dose for mouse bacteremia infection (log10 CFU).
Number of mice infected.
Log-rank test.
The AGEv with confirmed virulence phenotypes contained genes with homology to polymorphic toxins, substrate transport, and metabolism systems, and various phage-related and hypothetical proteins (Table 1 and SI Appendix, Fig. S2). Many of the AGEv were located within predicted mobile genetic elements (e.g., pKLC2-like genomic islands, transposons, phages), suggesting acquisition through horizontal transfer. No single AGEv occurred in all highly virulent isolates (Fig. 2C). Rather, the highly virulent isolates often had unique combinations of different AGEvs, suggesting that P. aeruginosa strains utilize different pathogenic mechanisms to increase virulence in a mammalian host. Moreover, this work demonstrates the power of using comparative genomic approaches to identify novel virulence determinants.
AGEv15 Is Part of a Functional CDI System.
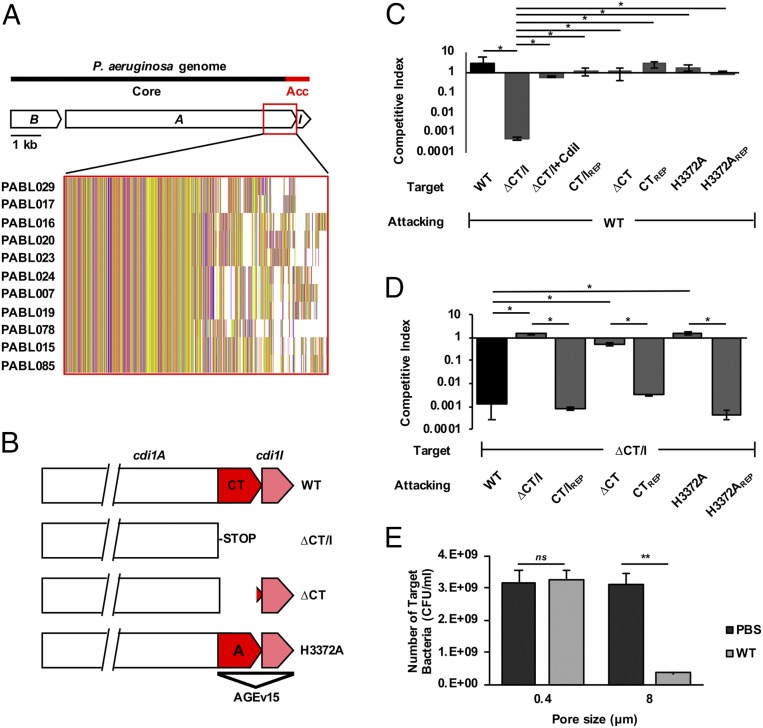
As an extension of our computational studies, we sought to elucidate how a novel AGEv may function to enhance the virulence of P. aeruginosa. While many AGEv were of great interest, we chose to further investigate the function of AGEv15 because of the novelty, system diversity, and potential impact it may have on P. aeruginosa pathogenesis. AGEv15 was found to encompass a portion of a CDI system (Fig. 3A) (28, 29). These systems were first recognized for their ability to mediate interbacterial competition (30). CDI systems utilize a type Vb secretion mechanism to display a large exoprotein (CdiA) at the bacterial surface. Upon direct cell-to-cell contact, CdiA transfers its C-terminal toxin (CT) domain into a susceptible target bacterium (31). Siblings are protected from self-intoxication by production of a cognate CdiI immunity protein. Similar to CDI systems from Escherichia coli (32) and Burkholderia species (33, 34), the amino-half of the CdiA protein is highly conserved while the CT domains and cognate CdiI proteins vary between different P. aeruginosa isolates (35). Our computational analysis had designated the 3′ end of the cdi1A gene, encoding the CT domain, and corresponding cdiI gene, as part of the accessory genome because of this allelic diversity (Fig. 3A). Thus, we had identified a specific cdi1A allele encoding a CT domain that was associated with and contributed to P. aeruginosa virulence (Table 1).
Fig. 3.
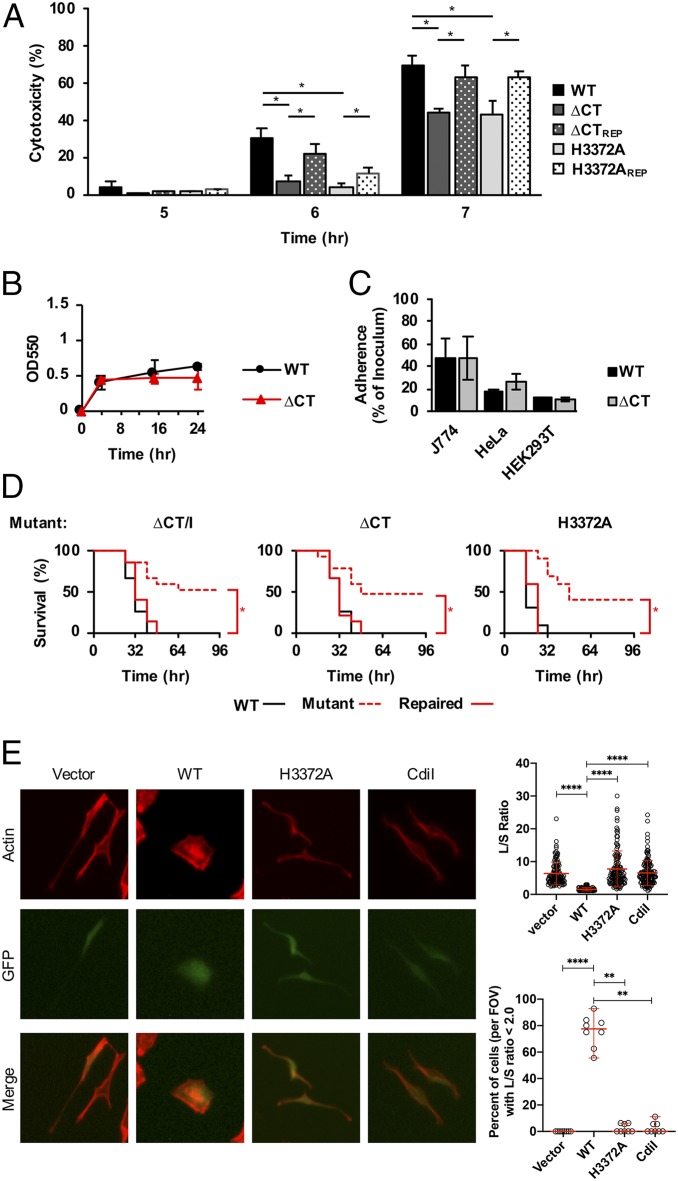
AGEv15 is part of a CDI system. (A) A diagram of the cdi1BAIPABL017 locus denotes the distribution of cdi1B (“B”), cdi1A (“A”) and cdi1I (“I”) within the core and accessory (“Acc”) genome. Comparative sequence analysis uncovered 11 different cdi1A alleles distributed among the PABL isolates (35) (Inset). Representative isolates for each of these 11 cdi1A alleles are listed. Translated sequences from the representative alleles were aligned using Clustal Omega and visualized with Jalview to highlight variability in the C-terminal region. (B) Targeted mutations within the cdi1APABL017 gene were generated for specific functional studies. WT cdi1APABL017 and three specific mutations are depicted: 1) The nucleotide sequence encoding the Cdi1APABL017-CT domain and cognate cdi1I immunity gene were deleted as a pair (∆CT/I), 2) the nucleotide sequence encoding the Cdi1A-CTPABL017 domain was deleted up to the putative cdi1I Shine-Dalgarno site (∆CT), 3) a point mutation resulting in a single amino acid substitution from histidine to alanine at position 3372 (H3372A). (C and D) Competition experiments were conducted using attacking cells cocultured at a 10:1 ratio with target cells as described in Methods. Mutations in the target cells (C) and the attacking cells (D) were tested to define the roles of cdi1A and cdi1I in bacterial inhibition. Target cell growth is represented as the competitive index relative to the attacking cell. Strain notations are the same as B. Mutation repaired to WT (REP), ∆CT/I+Cdi1I (intact copy of the cdi1I gene under pBAD control located at the attB site, strain JPA043). Results indicate means ± SD (Kruskal–Wallis test with Dunn’s multiple comparisons, n = 6, *P < 0.05). (E) To assay for the dependence of cell contact during competition, attacking and target cells were cultured within individual chambers using transwell plates separated by membranes containing either 0.4 µM or 8 µM pores for 2 h in LB broth. Growth of the CDI-susceptible ∆CT/I target strain was enumerated in the presence of the WT attacking strain or PBS control. Results indicate mean ± SD (two-way ANOVA with Sidak’s multiple comparisons compared to PBS control, n = 6, **P < 0.01, ns = not significant).
We first questioned whether this specific cdi1A allele retained its ability to function in bacterial competition by performing competition assays on a series of specific mutant strains. The genetic region encoding the Cdi1A-CT domain was deleted from the representative highly virulent strain PABL017, leaving the immunity gene intact (Cdi1A∆CTPABL017). Alternatively, the genetic region encoding the Cdi1A-CTPABL017 domain and its cognate immunity gene were both deleted (Cdi1A∆CT/IPABL017) (Fig. 3B). Disruption of the immunity gene rendered the mutant susceptible to killing by the parental WT strain, and resistance to CDI-mediated killing was restored by supplying a copy of the immunity gene in trans (Fig. 3C). Moreover, a functional CT-domain was required for CDI-mediated killing of a susceptible target strain (Fig. 3D), and this inhibition was contact-dependent (Fig. 3E). These results demonstrate that the AGEv15-associated cdi1APABL017 encodes a functional CDI system capable of mediating interbacterial competition.
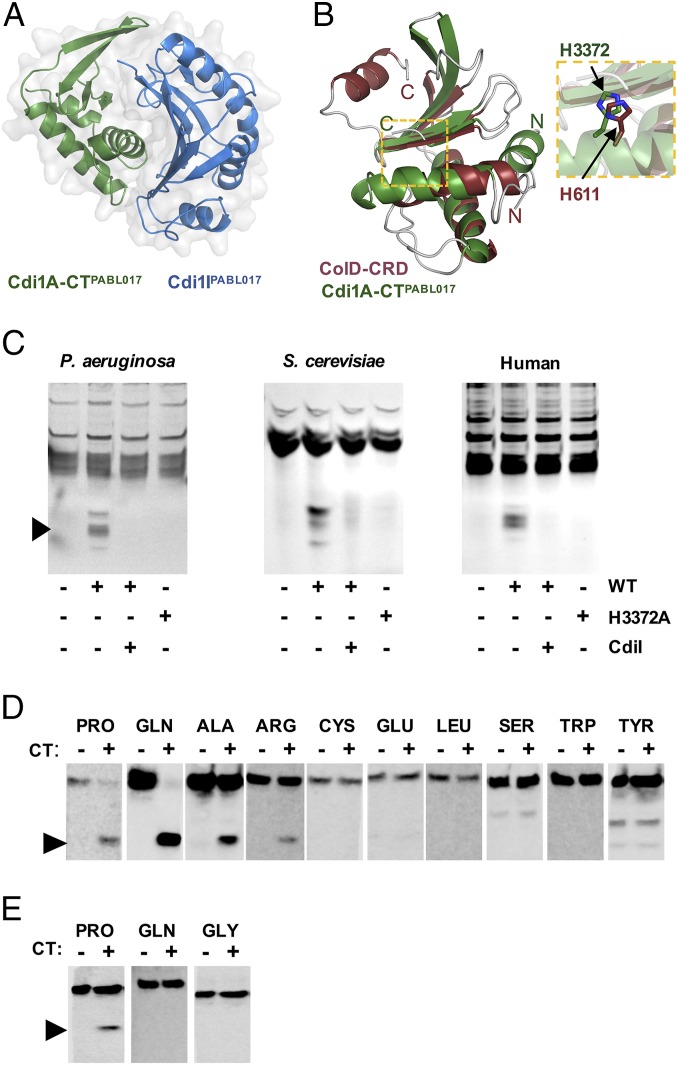
The AGEv15-Associated Cdi1A-CTPABL017 Is a Specific tRNase.
We sought to gain insight into how this CDI system could play a dual role in bacterial virulence and interbacterial competition by investigating the function of the CT domain. The primary sequence of Cdi1A-CTPABL017 did not provide any insight into its activity. To help elucidate the enzymatic function of this CT domain, we determined its three-dimensional structure in complex with Cdi1IPABL017 by X-ray crystallography (Fig. 4A) (36). Cdi1A-CTPABL017 adopts a crescent shape with the second of three α-helices acting as a major rim opposite a four-stranded antiparallel β-sheet that forms the minor rim and central groove of a putative enzymatic cleft. The immunity protein binds tightly over the major rim and central groove of the CT through opposite electrostatic charges on the interacting surfaces. Structural comparison with the DALI server revealed Cdi1A-CTPABL017 shares significant structural homology with the C-terminal RNase domain (CRD) of Colicin D (ColD) from E. coli (Fig. 4B). ColD is a known tRNase, and a histidine residue in CdiAPABL017 at position 3372 occurred in the same orientation as the known catalytic histidine 611 of ColD (37, 38). Substitution of this single His3372 residue to alanine (H3372A) was sufficient to abrogate CdiAPABL017-mediated bacterial killing (Fig. 3D). Incubation of purified Cdi1A-CTPABL017 with nucleic acid preps from prokaryotic or eukaryotic (including human) sources resulted in a pattern consistent with tRNA cleavage that required the His3372 residue (Fig. 4C). Northern blots confirmed the cleavage products were tRNA halves, with apparent preference for proline and glutamine tRNA molecules (Fig. 4 D and E). These data indicate that Cdi1A-CTPABL017 is a specific tRNase that is functional against tRNA species from both prokaryotic and eukaryotic organisms and suggest that this tRNAse activity is responsible for CDI.
Fig. 4.
Cdi1A-CTPABL017 is a tRNase. (A) The crystal structure of recombinant Cdi1A-CTPABL017 was solved in complex with recombinant Cdi1IPABL017. (B) Cdi1A-CTPABL017 structure is highly similar to the ColD-CRD. The individual structures are overlaid to reveal structural similarity and highlight the position and orientation of the catalytic His611 from ColD with the putative catalytic His3372 of Cdi1APABL017 (Inset). (C) Purified Cdi1A-CTPABL017 (WT) or Cdi1A-CT[H3372A]PABL017 (H3372A) was incubated with tRNA preparations from Saccharomyces cerevisiae (Sigma) or total nucleic extracts from P. aeruginosa or HeLa cells at 37 °C for 60 min. The preparations were separated on a denaturing polyacrylamide gel, stained with ethidium bromide and visualized with UV light. Cdi1A-CTPABL017 protein was preincubated with purified Cdi1IPABL017 protein for 30 min where indicated (CdiI). Black arrowhead denotes cleaved products. Northern blot analysis of (D) P. aeruginosa or (E) HeLa cell total nucleic acid preparations treated with WT Cdi1A-CTPABL017 as above and hybridized with tRNA-specific probes. Black arrowheads indicate cleaved tRNA molecules.
The Cdi1A-CTPABL017 tRNase Activity Is Required for P. aeruginosa Virulence.
Because the AGEv15 mutant was attenuated for virulence in mice, we reasoned that this CDI system may have cytotoxic effects on eukaryotic cells. Incubation of HeLa cells with WT PABL017 resulted in the majority of cells rounding by 5 h postinfection (SI Appendix, Fig. S4). In contrast, incubation with the Cdi1A∆CTPABL017 mutant resulted in a modest but significant reduction in cell rounding. Moreover, the Cdi1A∆CTPABL017 mutant displayed a significant reduction in cytotoxicity (as measured by LDH release) relative to the parental strain (Fig. 5A). This reduction in cytotoxicity was also observed with the Cdi1A(H3372A)PABL017 mutant (Fig. 5A), suggesting the Cdi1APABL017 tRNase activity may be important for this process. This attenuation in cytotoxicity was not caused by an adherence defect, as deletion of the Cdi1A-CTPABL017 domain had little impact on adherence to biotic or abiotic surfaces (Fig. 5 B and C). Overall, these findings suggest that Cdi1APABL017 enhances cytotoxic effects against eukaryotic cells.
Fig. 5.
The Cdi1APABL017 tRNase activity contributes to P. aeruginosa virulence. (A) HeLa cells were infected at a multiplicity of infection of 10 with PABL017 (WT), the previously described mutant strains Cdi1A-∆CTPABL017 (∆CT) and Cdi1A-CT[H3372A]PABL017 (H3372A), or strains in which the respective cdi1A mutations were repaired to WT PABL017 (REP). Cytotoxicity (percent relative to Triton X-100 treatment) was monitored by the release of lactate dehydrogenase (LDH) over time. Results indicate mean ± SD (two-way ANOVA with Tukey’s multiple comparisons, n = 6. *P < 0.05; all other comparisons within a single time point were not significant). PABL017 WT and Cdi1A∆CTPABL017 (∆CT) strains were assayed for adherence and biomass formation on polystyrene plates using a Crystal violet staining assay (B) or adherence (percent relative to inoculum) to J774 murine-like macrophages, HeLa human cervical epithelial cells or HEK 293T cells (C). Results indicate mean ± SD (n = 3, differences were not statistically significant). (D) BALB/c mice were infected by intravenous injection with a lethal dose of PABL017 WT or previously described mutant or repaired strains and monitored for disease progression over 96 h. Data were plotted on Kaplan–Meier curves and analyzed for significance by the Mantel–Cox (log-rank) test (n = 10 mice, *P < 0.05). (E) HeLa cells were transfected with empty pEF1α-IRES-AcGFP1A (Vector), or vector containing the coding regions for Cdi1A-CTPABL017 (WT), Cdi1A-CT[H3372A]PABL017 (H3372A) or Cdi1IPABL017 (Cdil). Cells were fixed, stained for actin, and visualized by fluorescence microscopy (magnification: 40×). Cell rounding was quantified for transfected (GFP+) cells by determining the ratio of length of the long cell axis divided by the length of the short cell axis (L/S ratio). For each construct, over 100 transfected cells were analyzed from eight fields-of-view (FOV) obtained from two independent experiments. Individual data points are presented with means ± SD (Kruskal–Wallis test with Dunn’s multiple comparisons, ****P < 0.0001). The data were also analyzed for the percentage of transfected cells (per FOV) with an L/S ratio less than 2, indicative of increased cell rounding. Individual data points are presented as medians ± 95% CI (Kruskal–Wallis test with Dunn’s multiple comparisons, n = 8, ****P < 0.0001, **P < 0.01).
We also questioned whether the catalytic activity of Cdi1APABL017 was required for the observed virulence defects in mice. Mice intravenously infected with the Cdi1A∆CTPABL017 or Cdi1A(H3372A)PABL017 mutants showed increased survival that was reversed when the mutations were repaired (Fig. 5D). Similarly, Cdi1A∆CTPABL017 mutants were attenuated in mouse pneumonia and subcutaneous abscess infection models (SI Appendix, Fig. S3), suggesting the route of infection is not critical. Collectively, these findings suggest the tRNAse activity of Cdi1APABL017 broadly enhances the virulence of P. aeruginosa.
Interbacterial Contact-Dependent Signaling through CdiA Is Not Required for P. aeruginosa Virulence.
We next examined the mechanism by which Cdi1APABL017 acted as a virulence factor. Two virulence mechanisms presented themselves: 1) As with CDI and bacteria, Cdi1APABL017 may gain access to the cytosol of mammalian cells, where its nuclease activity leads to pathogenic changes; or 2) Cdi1APABL017 may be delivered into immune bacterial siblings, resulting in physiological changes to the “injected” siblings that lead to enhanced virulence. The latter mechanism, referred to as contact-dependent signaling (CDS) (39), is responsible for enhanced biofilm formation and broad gene-expression changes in Burkholderia thailandensis E264 (40).
The first possibility is supported by the fact that the tRNA substrates of Cdi1A-CTPABL017 are present in mammalian cells. We reasoned that if Cdi1A-CTPABL017 was injected into mammalian cells, then transfection of a construct expressing Cdi1A-CTPABL017 should mimic coincubation with P. aeruginosa. Transfected HeLa cells expressing a functional Cdi1A-CTPABL017 domain displayed cell rounding when visualized by immunofluorescent staining (Fig. 5E and SI Appendix, Fig. S5). In contrast, transfected cells expressing a Cdi1A-CT(H3372A)PABL017 tRNase mutant domain, Cdi1IPABL017 immunity protein, or vector control demonstrated little cell rounding. Overall, these results suggest an enzymatically active Cdi1A-CTPABL017 can have direct pathogenic effects on eukaryotic cells.
Next, we attempted to directly demonstrate injection of Cdi1A-CTPABL017 into HeLa cells. Despite multiple attempts, we were unsuccessful in introducing various epitope tags and reporter fusions, including both internal and external placement of β-lactamase, polyhistidine, and glycogen synthase kinase tags, into Cdi1A-CTPABL017. In vivo detection of tRNA halves in both prokaryotic and eukaryotic cells was also unsuccessful. Since attempts to demonstrate direct injection of Cdi1A-CTPABL017 into mammalian cells were unsuccessful, we focused on testing whether CDS accounted for the virulence associated with Cdi1A-CTPABL017. CDS requires proper delivery of Cdi1A-CT into the cytosol of the target bacterium (39, 40), which in turn requires the presence of specific inner and outer membrane receptors in the target bacterium (SI Appendix, Fig. S6A) (41). Mutation of these specific receptors would abrogate CdiA-CT uptake (e.g., CDS) while maintaining the ability of the bacteria to present Cdi1A-CT to susceptible target bacteria and mammalian cells (SI Appendix, Fig. S6B). Thus, if Cdi1A-dependent CDS drives virulence, then a receptor mutant would be attenuated in mouse infection models (SI Appendix, Fig. S6C). Alternatively, if the impact on P. aeruginosa virulence is through a direct effect of Cdi1A on mammalian cells, then a CDI-receptor mutant, which still produces a functional Cdi1A exoprotein, would display no virulence defect.
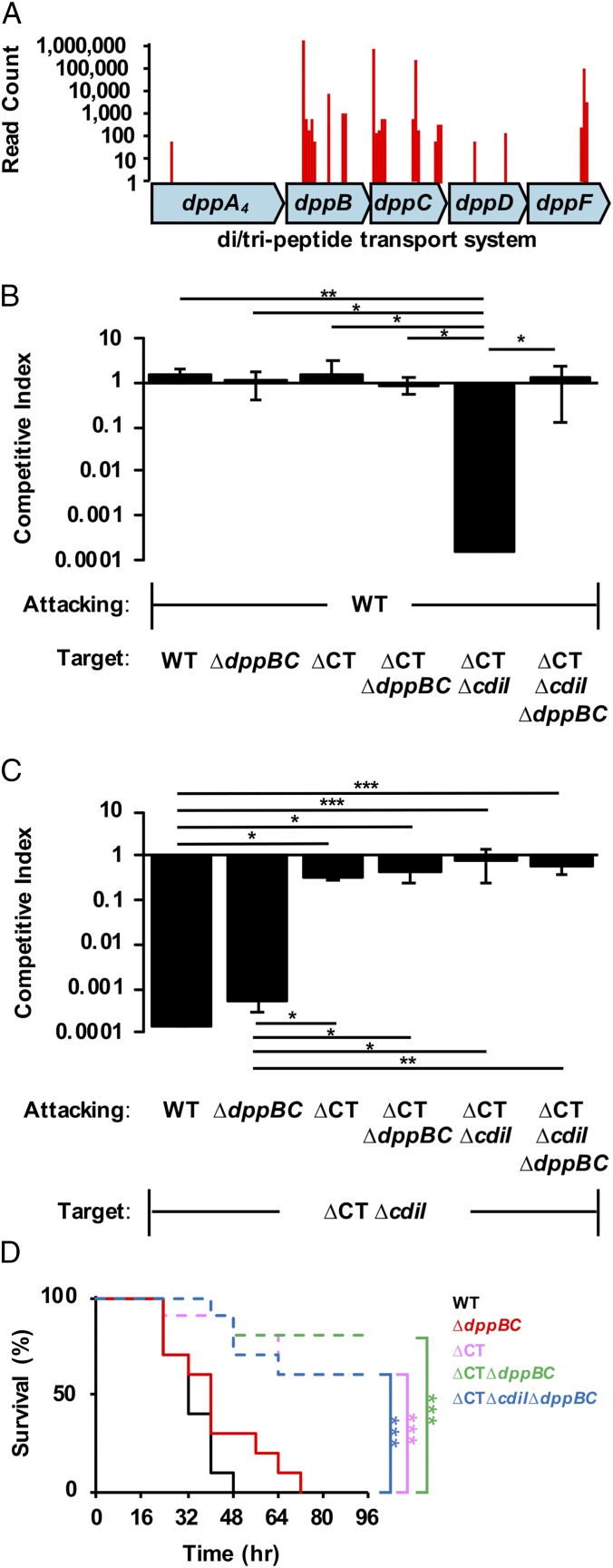
To identify such a receptor in P. aeruginosa, we took advantage of the fact that a CDI-sensitive strain would become resistant to CDI-mediated killing upon mutation of the cognate receptor. Transposon mutagenesis was performed on the CDI-sensitive Cdi1A∆CT/IPABL017 strain, and selection was performed for CDI-resistant mutants. These mutants contained an abundance of transposon hits in dppB and dppC (Fig. 6A), part of an ABC import system of small oligopeptides predicted to be localized to the inner membrane (42). As hypothesized, deletion of both dppB and dppC (∆dppBC) in the CDI-sensitive Cdi1A∆CT/IPABL017 strain rescued growth in competition with parental PABL017 (Fig. 6B), suggesting that DppB and DppC act as the inner membrane receptor for Cdi1APABL017. The WT and Cdi1A∆CTPABL017 strains displayed no defect in competition against a WT attacking strain as before, nor did their respective ∆dppBC mutant constructs, indicating that the ∆dppBC mutation has no inherent fitness defect. Furthermore, deletion of dppBC in WT PABL017 did not abrogate the ability to inhibit the growth of a CDI-sensitive strain (Fig. 6C). Loss of CDI capability was only observed in attacking strains when a functional Cdi1A-CT domain was absent, independent of dppB and dppC. These results indicate that the PABL017∆dppBC deletion strain is capable of delivering a functional Cdi1A exoprotein but cannot receive a Cdi1A-CT domain into its cytosol and is therefore CDI+ but CDS− (SI Appendix, Fig. S6C).
Fig. 6.
Cdi1APABL017-dependent virulence effects are independent of CDS. (A) Transposon mutagenesis of a CDI-susceptible strain Cdi1A∆CT/IPABL017 was performed to identify genes encoding an inner membrane receptor for CT uptake into a target cell. The data represent the read counts at specific locations within the dppABCDF operon generated from Illumina sequencing of the CDI-resistant transposon pool. (B and C) Competition experiments were conducted using attacking cells cocultured at a 10:1 ratio with target cells as described in Methods. Target cell growth is represented as the competitive index relative to the attacking cell after 2 h. Results indicate means ± SD (Kruskal–Wallis test with Dunn’s multiple comparisons, n = 6, ***P < 0.001, **P < 0.01, *P < 0.05). Strains used include PABL017 (WT), PABL017∆dppBC (∆dppBC), CdiA∆CTPABL017 (∆CT), CdiA∆CTPABL017 + ∆dppBC double mutant (∆CT∆dppBC), CdiA∆CTPABL017 + ∆cdi1I double mutant (∆CT∆cdiI), or a CdiA∆CTPABL017 + ∆cdi1I + ∆dppBC triple mutant (∆CT∆cdiI∆dppBC). (D) BALB/c mice were infected by intravenous injection with a lethal dose of PABL017 (WT) or mutant strains listed above. Infected mice were monitored for disease progression over 96 h. Data were plotted on a Kaplan–Meier curve. Differences were analyzed for significance by the Mantel–Cox (log-rank) test (n = 10 mice, ***P < 0.001).
We next tested whether this CDI+ CDS− strain maintained virulence in mice. The PABL017∆dppBC mutant demonstrated no significant difference in virulence from WT PABL017 (Fig. 6D). Only when the Cdi1A-CTPABL017 domain was disrupted was virulence attenuation observed, independent of the ∆dppBC mutation. These results suggest that the influence of Cdi1APABL017 on P. aeruginosa virulence is not through CDS but rather suggest a direct effect of the Cdi1A molecule.
Discussion
In the present study, we show that a P. aeruginosa CDI system can function as both an interbacterial inhibition system and a bacterial virulence factor against a mammalian host. We demonstrate that the Cdi1A-CTPABL017 domain functions as a specific tRNAse and that a catalytic histidine residue important for this enzymatic activity in vitro was also required for CdiA-mediated toxicity against mammalian cells and lethality during mouse bacteremia infections. Moreover, Cdi1A-CTPABL017 appears to have a broad pathogenic role, as it was also required for virulence in both mouse intranasal and subcutaneous infection models. Overall, these findings support the idea that there may be a dual role for the CDI systems of P. aeruginosa (28).
We have also demonstrated the utility of using a comparative genomics approach to identify accessory virulence factors that act by novel mechanisms. By quantifying the virulence of 100 P. aeruginosa isolates in mice, we were able to combine genotypic and phenotypic data to identify AGEs overrepresented in highly virulent isolates and subsequently determine those that played a causal role in pathogenicity. Because the genomic diversity of P. aeruginosa is so great (an average of 11% of the genome differs from isolate to isolate), population approaches can be a powerful tool to identify novel accessory genes that enhance the pathogenicity of isolates within the species.
While our findings shed light on the pathogenic mechanism of Cdi1APABL017, there is still much to uncover regarding how this protein functions as a virulence factor within an infected host. One possibility is that the enzymatically active CT of Cdi1APABL017 is translocated into mammalian cells and cleaves tRNA molecules similar to the model of CDI-dependent intoxication of bacteria. The following results are consistent with this proposed mechanism: 1) The substrates of Cdi1A-CTPABL017, tRNAs, are present in both bacteria and mammalian cells; 2) Cdi1A-CTPABL017 is capable of cleaving mammalian tRNAs (Fig. 4C); 3) Cdi1A-CTPABL017 is sufficient to cause injury to mammalian cells (Fig. 5E); 4) the enzymatic activity of Cdi1A-CTPABL017 is necessary for its virulence effect in mice (Fig. 5D); 5) the virulence effect of Cdi1A-CTPABL017 is independent of CDS (Fig. 6D), which is consistent with previous work demonstrating that Cdi1A does not influence the expression of virulence genes in P. aeruginosa strain PAO1 (28); and 6) biofilm formation or adherence to eukaryotic cells did not differ between WT PABL017 and the Cdi1A∆CTPABL017 mutant (Fig. 5 B and C). In contrast, several inconsistencies remain with this hypothesis: 1) Studies (including this one) have failed to demonstrate transfer of CdiA proteins across eukaryotic membranes; 2) prokaryotic and eukaryotic cell toxicity has not been directly attributed to degradation of tRNAs; and 3) HeLa cell rounding observed in our transfection and infection experiments has not been directly related to the observed in vivo virulence effects. Nonetheless, these findings provide an impetus for continued studies into the complex role of CDI systems in P. aeruginosa pathogenesis.
To our knowledge, CDI systems have not previously been shown to play a direct role in virulence in mouse infection models; however, findings from other groups have suggested roles for CdiA beyond bacterial competition. The CDI system of P. aeruginosa strain PAO1 was important for tissue maceration of lettuce leaves, for bacterial burden in Galleria mellonella larva, and in the paralytic killing of Caenorhabditis elegans (28, 43). The CdiA homolog XacFhaB of Xanthomonas axonopodis pv. citri was necessary for increased bacterial burden and formation of cancer lesions on citrus leaves (44), and the Xylella fastidosa CdiA homolog was required for efficient colonization of insect vectors (45). Furthermore, the CdiA homolog HrpA of the human pathogen Neisseria meningitidis facilitated escape from intracellular vacuoles in HeLa cells (46, 47). Because our findings reveal that CDI systems can impact host survival outcomes in mouse infection models, this aspect of CDI biology will be considered in future studies on P. aeruginosa pathogenesis.
Ultimately, this work supports the evolving narrative that CDI systems likely function on multiple levels of host–pathogen interactions (28). For example, Cdi1A-CTPABL017 may target other P. aeruginosa bacteria to provide PABL017 with a competitive advantage during colonization of the mammalian upper airway, and following subsequent aspiration into the lung it may target mammalian cells to cause severe pneumonia. With such astonishing diversity, CDI systems and other polymorphic toxins will likely provide a fertile field for future studies (48).
Methods
Reagents and Strains and Culture Conditions.
All bacterial strains used in this study are listed in SI Appendix, Table S1. P. aeruginosa clinical isolates were cultured from patient blood samples collected at Northwestern Memorial Hospital between August 1999 and September 2003 (19). Bacteria were routinely cultured in Luria-Bertani (LB) medium or MINS (49) at 37 °C with shaking at 250 rpm or on agar plates. Antibiotics were supplemented at the following concentrations for E. coli and P. aeruginosa, respectively: Gentamicin (gm), 15 or 100 μg/mL; carbenicillin (carb), 50 or 500 μg/mL; tetracycline (tet) 10 or 100 μg/mL Irgasan (irg) was used to select for P. aeruginosa at 5 μg/mL. Common chemicals, antibiotics, and growth media components were purchased from Sigma-Aldrich, VWR, Thermo Fisher, or are specified where appropriate.
Experimental Animals.
All procedures were performed in accordance with the guidelines approved by the Northwestern University Animal Care and Use Committee as described in protocol IS00002172. Female BALB/c mice aged 6 to 8 wk were purchased from Envigo and female C57BL/6J mice aged 6 to 8 wk were purchased from The Jackson Laboratory. Mice were housed under ABSL-2 containment at the Northwestern University Center for Comparative Medicine research facility. Mice were infected by either tail-vein injection, intranasal inoculation, or subcutaneous injection. During infection experiments, mice were checked every 8 h by research staff for the first 72 h then twice per day through 96 h for signs of disease progression. Detailed infection procedures are provided in SI Appendix.
Whole-Genome Sequencing and Bioinformatic Analyses.
Genomic DNA was purified from overnight bacterial growth in LB broth using a Maxwell 16 Cell DNA Purification Kit (Promega). Samples were sent for library preparation and Illumina sequencing on a HiSeq-2000 platform at the University of Maryland Institute for Bioscience and Biotechnology Research. Raw reads were randomly downsampled to at most 100× coverage assuming an average genome size of 6.6 Mb. No trimming was performed. Reads were de novo-assembled using Ray (v2.0.0-rc8). Assemblies were annotated using Prokka v1.8. The whole-genome sequencing project was deposited in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) under the accession number PRJNA485889, and whole-genome sequences were deposited in the Assembly database under the accession numbers listed in SI Appendix, Table S1. P. aeruginosa accessory genomes were identified and grouped using SPINE, AGEnt, and ClustAGE programs (26, 27). A detailed description of these programs as well as the methods for determining virulence-associated accessory genomic elements are contained within SI Appendix.
kSNP3 was used to determine phylogenetic relationships between isolates and construct maximum-likelihood core genome trees (50). Trees were visualized in R using the ggtree package (51). Determination of phylogenetic signal with virulence was estimated in R using the Phylosignal package (52). Multilocus sequence typing (MLST) of the strain collection was performed using the Genome Comparator tool at the P. aeruginosa MLST website (https://pubmlst.org/paeruginosa/) developed by Keith Jolley and sited at the University of Oxford (53). Amino acid variation in the Cdi1A exoprotein was analyzed as previously described (35). Amino acid alignments were generated using Clustal Omega and visualized in Jalview (54).
Cloning and Strain Construction.
Oligonucleotides were purchased from Integrated DNA Technologies. Polymerase chain reactions (PCR) were performed using Phusion High-Fidelity DNA Polymerase (New England Biolabs), and gel-purified products were cloned into appropriate vectors using the Gibson Assembly Cloning Kit (New England Biolabs) or T4-DNA ligase (New England Biolabs) per the manufacturer’s instructions. All strains used in this study are listed in SI Appendix, Table S1, all plasmids with corresponding primers and templates used in plasmid construction are listed in SI Appendix, Table S2, and all primer sequences are listed in SI Appendix, Table S3. P. aeruginosa chromosomal deletions were generated using the procedure of Schweizer and colleagues (55). Detailed procedures for mutant generation as well as protein purification and crystallization are described in SI Appendix.
Transposon Mutagenesis.
Transposon mutagenesis was performed with the Himar1 mariner transposon carried on the mini transposon vector, pBT20, as previously described (56). Mutant pools were subjected to three rounds of bacterial competition with WT PABL017, as described below. After each round, CDI-resistant colonies were collected on selective media (LB-gm) and pooled to initiate growth for the next round of competition. After three rounds, the resulting mutant pool was stored at −80 °C and analyzed for identification of transposon insertion sites as described in the SI Appendix.
Bacterial Competition Assays.
Bacterial competition assays were performed on solid media with a modified protocol from Basler et al. (57). Briefly, P. aeruginosa attacking and target strains were passaged separately at a 1:50 dilution in fresh LB from overnight growth and cultured to an OD600 = 1. Cells were then separately pelleted and suspended in PBS to an OD600 = 10. Attacking and target cells were then combined at a 10:1 ratio respectively, mixed, and spotted as 5-µL drops in triplicate onto dry competition plates (LB agar). At indicated times, the bacterial spots were cut out and suspended in 1 mL LB. Serial dilutions were plated on LB or selective plates (LB-gm or LB-5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal]) for bacterial enumeration of attacking and target strains. For strain JPA040, expression of Cdi1I was induced by including 0.2% arabinose during growth in both liquid media and competition plates. The competitive index was calculated as follows: C.I. = (Target CFUtime = 2 h/Attacking CFUtime = 2 h) ÷ (Target CFUtime = 0 h/Attacking CFUtime = 0 h).
Bacterial Contact-Dependent Competition Assay.
Bacterial contact-dependent competition assays were performed using a modified protocol from Aoki et al. (30). P. aeruginosa strains were cultured overnight in LB, washed in PBS and normalized to 109 CFU/mL as determined by OD600. Next, 24-well polystyrene plates (Fisher Scientific) were fitted with polyethylene terephthalate (PET) track-etched membrane inserts with a 0.4-µm or 8-µm pore size (Corning). Bacterial strains were inoculated into LB broth at a 10:1 ratio of attacking to target cells into the upper and lower chambers, respectively. Cells were placed at 37 °C in a shaking incubator (120 rpm) for 6 h. Serial dilutions from the upper and lower chambers were plated on LB or selective plates (LB-gm or LB–X-Gal) for bacterial enumeration of attacking and target strains. Assays were performed a minimum of three times in triplicate.
Biofilm Assay.
P. aeruginosa strains were cultured overnight in LB, washed in PBS, and normalized to 109 CFU/mL as determined by OD600. Falcon 96-well polystyrene plates (Fisher Scientific) were filled with 180 µL of LB, inoculated with 20 µL of prepared cells and stored at 37 °C with no shaking. At specific times plates were inverted to decant planktonic cells. Two-hundred microliters of Crystal violet (0.1% in PBS) was added to each well for 10 min followed by three PBS washes. Biofilms were de-stained with 95% ethanol for 10 min at room temperature, and 100 µL of the de-stained ethanol mix was transferred to a fresh 96-well polystyrene assay plate. Absorbance at 550 nm was measured on a SpectraMax M3 plate reader (Molecular Devices).
Mammalian Cell Culture.
HeLa (female, human epithelioid cervix carcinoma) cells, HEK293T (female, human embryonic kidney) cells, and J774 (female, BALB/cN mouse macrophage) cells were obtained from ATCC. Cells were grown at 37 °C in DMEM (Gibco) supplemented with 10% FBS (GE Healthcare Life Sciences) under 5% CO2 conditions. Cell infection and transfection experimental details are described in the SI Appendix.
Immunoblot Analysis.
For detection of secreted (extracellular) effectors of the P. aeruginosa T3SS, culture supernatants were prepared from 5 mL of overnight growth in MINS. Proteins were precipitated from solution at 4 °C by overnight incubation with 50% (wt/vol) ammonium sulfate and collected by centrifugation at 13,000 × g for 30 min. Protein pellets were resuspended in 1× sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis sample buffer and separated on a 10% SDS-polyacrylamide gel. Proteins were transferred to a PVDF Immobilon-FL membrane (Millipore Sigma) by electro-transfer and blocked in 5% milk powder (1× PBS) solution. Proteins were probed by immunoblot with a mixture of rabbit antibodies against the P. aeruginosa type III effectors ExoU and ExoS/T (24). Effectors were detected following a triplicate wash step using the IRDye680RD goat anti-rabbit IgG antibody (LI-COR Biosciences) and visualization with the Odyssey FC imaging system (LI-COR Biosciences). Images were captured and processed with the Image Studio software package (LI-COR Biosciences).
Detection of tRNA Cleavage In Vitro.
Total RNA was collected from overnight cultures of P. aeruginosa or HeLa cells grown to confluency. Harvested cell pellets were frozen overnight at −80 °C and subsequently suspended in acid guanidinium thiocyanate-phenol-chloroform solution for total RNA purification as previously described (58). Three micrograms of mixed tRNA preparations from Baker’s Yeast (Roche) or total RNA preparations from P. aeruginosa or HeLa cells were combined with a 10 µM final concentration of purified Cdi1A-CTPABL017 or Cdi1A-CT(H3372A)PABL017 for 60 min at 37 °C. For reactions with Cdi1IPABL017, Cdi1A-CTPABL017 was incubated on ice for 30 min with 9.5 µM Cdi1IPABL017 prior to the addition of tRNA. Samples were suspended in 2× RNA loading dye (New England Biolabs) and separated on a 12.5% polyacrylamide TBE gel containing 50% urea. For visualization of general tRNA cleavage, gels were stained with ethidium bromide and imaged on an Axygen gel documentation system (Corning). Direct detection of specific tRNA cleavage was determined by northern analysis as described in SI Appendix.
Quantification and Statistical Analysis.
Spearman correlation analysis was performed in R (v3.6.1). When appropriate, data were analyzed for normality using Prism (GraphPad Software, v8.2.1). ANOVA, area under the curve (AUC) measurements, Kaplan–Meier plots with log-rank tests and Student’s t tests were also performed using Prism (GraphPad Software, v8.2.1). Specific statistical tests used, the number of subjects tested in each experiment and notations of significance are described in the figure legends or within the individual figure panel. A P value of 0.05 or less was considered significant for all analyses.
Data and Code Availability.
All PABL genome assemblies were deposited through the NCBI (https://www.ncbi.nlm.nih.gov/assembly) and the respective accession numbers are listed in SI Appendix, Table S1. Structural information for the CdiA-CTPABL017 + CdiIPABL017 complex was deposited in the Protein Data Bank (https://www.rcsb.org/) with the assigned PDB ID code 6D7Y. All software packages used in this study are freely available for download.
Supplementary Material
Acknowledgments
We thank I. Dubrovska for help with crystallization; Z. Wawrzak for help in structure solution; and Peter Agron for discussions that were the inspiration for this study. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by the Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contracts HHSN272201200026C (to Wayne F. Anderson) and HHSN272201700060C (to K.J.F.S.), F32 AI108247 (to J.P.A.), and R01 AI118257, R01 AI053674, U19 AI135964, K24 AI04831, and R21 AI129167 (all to A.R.H.). Financial support was also provided by the American Heart Association under Contract 15POST25830019 (to J.P.A.), and the American Cancer Society under Contract MRSG-13-220-01 (to E.A.O.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Competing interest statement: A.R.H. serves on an advisory board and as a consultant for Microbiotix, Inc. to develop type III secretion inhibitors. K.J.F.S. has a significant financial interest in Situ Biosciences, which conducts research unrelated to this work.
This article is a PNAS Direct Submission.
Data deposition: Crystal structures were deposited in the Protein Data Bank, https://www.rcsb.org (PDB ID code 6D7Y). Genome assemblies were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive, https://www.ncbi.nlm.nih.gov/bioproject/ (Bioproject code PRJNA485889).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919198117/-/DCSupplemental.
References
- 1.Casadevall A., Cards of virulence and the global virulome for humans. Microbe 1, 359–364 (2006). [Google Scholar]
- 2.Webb S. A., Kahler C. M., Bench-to-bedside review: Bacterial virulence and subversion of host defences. Crit. Care 12, 234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouli L., Merhej V., Fournier P. E., Raoult D., The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 7, 72–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis B. B., et al. , Pathogenicity locus, core genome, and accessory gene contributions to Clostridium difficile virulence. MBio 8, e00885-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson R. W., Vinatzer B., Arnold D. L., Dorus S., Murillo J., The influence of the accessory genome on bacterial pathogen evolution. Mob. Genet. Elements 1, 55–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segerman B., The genetic integrity of bacterial species: The core genome and the accessory genome, two different stories. Front. Cell. Infect. Microbiol. 2, 116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan R., Reeves P. R., Intraspecies variation in bacterial genomes: The need for a species genome concept. Trends Microbiol. 8, 396–401 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Ho Sui S. J., Fedynak A., Hsiao W. W., Langille M. G., Brinkman F. S., The association of virulence factors with genomic islands. PLoS One 4, e8094 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green S. K., Schroth M. N., Cho J. J., Kominos S. K., Vitanza-jack V. B., Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl. Microbiol. 28, 987–991 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellett S., Bigley D. V., Grimes D. J., Distribution of Pseudomonas aeruginosa in a riverine ecosystem. Appl. Environ. Microbiol. 45, 328–332 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauser A. R., Rello J., Eds. Severe Infections Caused by Pseudomonas aeruginosa. (Kluwer Academic Publishers, Boston, 2003). [Google Scholar]
- 12.Granchelli A. M., et al. , Microbial interactions in the cystic fibrosis airway. J. Clin. Microbiol. 56, e00354-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiehlmann L., et al. , Functional genomics of Pseudomonas aeruginosa to identify habitat-specific determinants of pathogenicity. Int. J. Med. Microbiol. 297, 615–623 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Wiehlmann L., Cramer N., Tümmler B., Habitat-associated skew of clone abundance in the Pseudomonas aeruginosa population. Environ. Microbiol. Rep. 7, 955–960 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Fischer S., et al. , Intraclonal genome diversity of the major Pseudomonas aeruginosa clones C and PA14. Environ. Microbiol. Rep. 8, 227–234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stover C. K., et al. , Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Mathee K., et al. , Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 3100–3105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battle S. E., Meyer F., Rello J., Kung V. L., Hauser A. R., Hybrid pathogenicity island PAGI-5 contributes to the highly virulent phenotype of a Pseudomonas aeruginosa isolate in mammals. J. Bacteriol. 190, 7130–7140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheetz M. H., et al. , Morbidity associated with Pseudomonas aeruginosa bloodstream infections. Diagn. Microbiol. Infect. Dis. 64, 311–319 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oze E. A., Hauser A. R., Pseudomonas aeruginosa bloodstream isolate sequencing. National Center for Biotechnology Information Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA485889. Deposited 15 August 2018.
- 21.Pagel M., Inferring the historical patterns of biological evolution. Nature 401, 877–884 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Mikkelsen H., McMullan R., Filloux A., The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One 6, e29113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Ledizet M., Kar K., Koski R. A., Kazmierczak B. I., An indirect enzyme-linked immunosorbent assay for rapid and quantitative assessment of type III virulence phenotypes of Pseudomonas aeruginosa isolates. Ann. Clin. Microbiol. Antimicrob. 4, 22 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaver C. M., Hauser A. R., Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 72, 6969–6977 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulert G. S., et al. , Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188, 1695–1706 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Ozer E. A., Allen J. P., Hauser A. R., Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics 15, 737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozer E. A., ClustAGE: A tool for clustering and distribution analysis of bacterial accessory genomic elements. BMC Bioinformatics 19, 150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melvin J. A., et al. , Pseudomonas aeruginosa contact-dependent growth inhibition plays dual role in host-pathogen interactions. MSphere 2, e00336-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercy C., Ize B., Salcedo S. P., de Bentzmann S., Bigot S., Functional characterization of Pseudomonas contact dependent growth inhibition (CDI) systems. PLoS One 11, e0147435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aoki S. K., et al. , Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Ruhe Z. C., et al. , Programmed secretion arrest and receptor-triggered toxin export during antibacterial contact-dependent growth inhibition. Cell 175, 921–933.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aoki S. K., et al. , A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468, 439–442 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson M. S., Garcia E. C., Cotter P. A., The Burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet. 8, e1002877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolakakis K., et al. , The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol. Microbiol. 84, 516–529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen J. P., Hauser A. R., Diversity of Pseudomonas aeruginosa contact-dependent growth inhibition systems. J. Bacteriol., JB.00776-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minasov G., et al. , 1.75 angstrom resolution crystal structure of the toxic C-terminal tip of CdiA from Pseudomonas aeruginosa in complex with immune protein. Research Collaboratory for Structural Bioinformatics Protein Data Bank. https://www.rcsb.org/structure/6D7Y. Deposited 25 April 2018.
- 37.Tomita K., Ogawa T., Uozumi T., Watanabe K., Masaki H., A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl. Acad. Sci. U.S.A. 97, 8278–8283 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graille M., Mora L., Buckingham R. H., van Tilbeurgh H., de Zamaroczy M., Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein. EMBO J. 23, 1474–1482 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia E. C., Perault A. I., Marlatt S. A., Cotter P. A., Interbacterial signaling via Burkholderia contact-dependent growth inhibition system proteins. Proc. Natl. Acad. Sci. U.S.A. 113, 8296–8301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ocasio A. B., Cotter P. A., CDI/CDS system-encoding genes of Burkholderia thailandensis are located in a mobile genetic element that defines a new class of transposon. PLoS Genet. 15, e1007883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willett J. L., Gucinski G. C., Fatherree J. P., Low D. A., Hayes C. S., Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc. Natl. Acad. Sci. U.S.A. 112, 11341–11346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abouhamad W. N., Manson M. D., The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 14, 1077–1092 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Gallagher L. A., Manoil C., Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183, 6207–6214 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gottig N., Garavaglia B. S., Garofalo C. G., Orellano E. G., Ottado J., A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One 4, e4358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Killiny N., Almeida R. P., Factors affecting the initial adhesion and retention of the plant pathogen Xylella fastidiosa in the foregut of an insect vector. Appl. Environ. Microbiol. 80, 420–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arenas J., Schipper K., van Ulsen P., van der Ende A., Tommassen J., Domain exchange at the 3′ end of the gene encoding the fratricide meningococcal two-partner secretion protein A. BMC Genomics 14, 622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talà A., et al. , The HrpB-HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell. Microbiol. 10, 2461–2482 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Zhang D., de Souza R. F., Anantharaman V., Iyer L. M., Aravind L., Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicas T. I., Iglewski B. H., Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect. Immun. 45, 470–474 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardner S. N., Slezak T., Hall B. G., kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T. Y., GGTree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 52.Keck F., Rimet F., Bouchez A., Franc A., phylosignal: An R package to measure, test, and explore the phylogenetic signal. Ecol. Evol. 6, 2774–2780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jolley K. A., Maiden M. C., BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P., A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Kulasekara H. D., et al. , A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55, 368–380 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Basler M., Ho B. T., Mekalanos J. J., Tit-for-tat: Type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chomczynski P., Sacchi N., The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 1, 581–585 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All PABL genome assemblies were deposited through the NCBI (https://www.ncbi.nlm.nih.gov/assembly) and the respective accession numbers are listed in SI Appendix, Table S1. Structural information for the CdiA-CTPABL017 + CdiIPABL017 complex was deposited in the Protein Data Bank (https://www.rcsb.org/) with the assigned PDB ID code 6D7Y. All software packages used in this study are freely available for download.