Vaccinia virus (VACV), the prototype orthopoxvirus, is the first live viral vaccine used to protect against smallpox, one of the most feared infectious diseases of humans (1). This outstanding achievement was possible because even as the various orthopoxviruses evolved and diversified they often retained major parts of their virion composition in common. In consequence, productive infection with one orthopoxvirus, VACV, effectively immunizes the host against infection by other orthopoxviruses including variola virus, the closely related agent of smallpox. Successful vaccination against smallpox positively correlated with the productive replication of VACV in the skin of vaccinated individuals. However, on occasion the inoculation of live VACV had risk because the vaccine virus could develop generalized infections and cause other severe adverse events following vaccination (2). Ever since, research with VACV seeks to elucidate the many secrets of this unique virus–human host interaction. What makes VACV efficiently grow in human cells? This might sound like a simple question since the growth tropism of many other viruses is largely determined by the binding of a specific viral protein (ligand) to its host cell counterpart (receptor) for successful entry. In contrast, the host tropism of VACV and poxviruses, in general, is far more complex and wonderful with numerous, postentry, intracellular events dictating whether productive virus replication will occur at the level of the host cell, the tissue, or the whole organism (3). VACV can readily bind to and enter many different cells from diverse animal species. After entry, however, successful VACV replication in a specific host cell depends on the functional activity of an appropriate subset of viral genes, the so-called host-range genes, predicted to control the intracellular host defenses at various levels.
In PNAS, Peng and Moss (4) identify and explore a previously unknown viral protein as a major facilitator of VACV growth in human cells. As highlighted by their study, this viral regulatory protein turns out to be the last missing host-range factor to repair the severe replication deficiency of modified vaccinia virus Ankara (MVA). The MVA infection of human cells is abortive and fails to produce new infectious progeny. Importantly, this nonpermissive infection still allows for an unimpaired synthesis of MVA-encoded proteins, and this unusual trait for a VACV host-range mutant allowed for the advancement of MVA as an exceptionally safe vaccine and vector virus (5). Today, MVA serves as the next-generation smallpox vaccines licensed in Europe, Canada, and the United States. Moreover, its exceptional safety profile has allowed MVA to become a versatile vector platform for the rapid generation and clinical testing of recombinant vaccines against a host of infectious diseases and cancer (6). After almost three decades of research to elucidate its particular host-range phenotype in human cells, here Peng and Moss finally reveal the genetic basis of the late MVA growth restriction (4).
MVA obtained its highly attenuated phenotype via serial tissue culture propagation on primary chicken embryo fibroblasts (CEF) (7). Due to its growth adaptation to avian cells, MVA lost the ability to replicate in most mammalian host cells along with its ability to cause infectious disease when inoculated into animals or humans. MVA abandoned many of the nonessential gene functions VACV uses to manipulate its host (cell) environment (6). Normal VACV has a broad cellular host range and can productively grow in a variety of mammalian and avian cells. Early studies with mutant viruses of VACV or the closely related rabbitpox virus revealed that growth restrictions in specific host cells were associated with large deletions in the viral genomes (8, 9). The first viral genes identified to control VACV replication in human cells were the ORFs K1L and C7L (10–12). In the absence of functional K1L and C7L, VACV infection is arrested at the level of viral early gene expression with a striking inhibition of postreplicative protein synthesis, and either gene is necessary and sufficient to allow standard VACV replication on human cells (11, 13). Yet, the C7L gene sequence is conserved in the MVA genome, and even when the virus is engineered to contain functional copies of both K1L and C7L, the late growth defect of MVA in human cells remained unrescued (6). These observations suggested the existence of an unknown VACV factor(s) to counteract other intracellular mechanisms mediating the MVA host restriction. However, the large deletions and the many other minor genetic changes in the MVA genome complicated the search for the important viral gene products. A laborious approach was chosen by the Moss laboratory in marker rescue experiments to select for growth-competent recombinant MVA viruses following the transfection of large genomic DNA fragments from a parental VACV Ankara (14). Indeed, recombinant viruses with repaired gene sequences at the left end of the viral genome showed a promising growth phenotype, efficiently replicated in cell lines of human origin, and were referred to as host-range extended (HRE) MVA. Whole-genome sequencing of these HRE MVAs in comparison to MVA allowed researchers to identify the gene C12L encoding VACV serine protease inhibitor 1 (SPI-1) as an important host-range factor to support MVA growth in human cells (15). In comparison to HRE MVA, however, reinsertion of C12L rescues MVA growth only in human MRC-5 cells but it does not substantially facilitate productive replication in human A549 cells (15), thus suggesting there must be (an)other VACV host-range gene(s).
In PNAS, Peng and Moss (4) extend the search for the missing factor(s) and targeted selected gene sequences of HRE MVA for deletion through homologous recombination and tested the deletion mutants for replication on A549 and MRC-5 cells. Surprisingly, the desired growth defect in human A549 cells was observed with the inactivation of selected genes that are duplicated both in the left (C16L-C17L) and in the right (B22R-B23R) end of the VACV genome. Separate insertions of single ORFs into the MVA genome and comparative growth analysis in A549 cells clearly revealed the previously unexplored ORF C16L/B22R to encode for the missing host-range factor. As with most of the genes expressed from the terminal ends of the VACV genome, C16L/B22R encodes an early virus protein that enables a crucial step in the virus life cycle influencing synthesis and processing of late structural virus proteins.
The work of Peng and Moss (4) presents a solution to the longstanding enigma of the elusive genetic basis for the MVA growth restriction in human cells and defines the reinsertion or repair of the C12L and C16L/B22R ORFs in the MVA genome as sufficient to rescue replicative capacity.
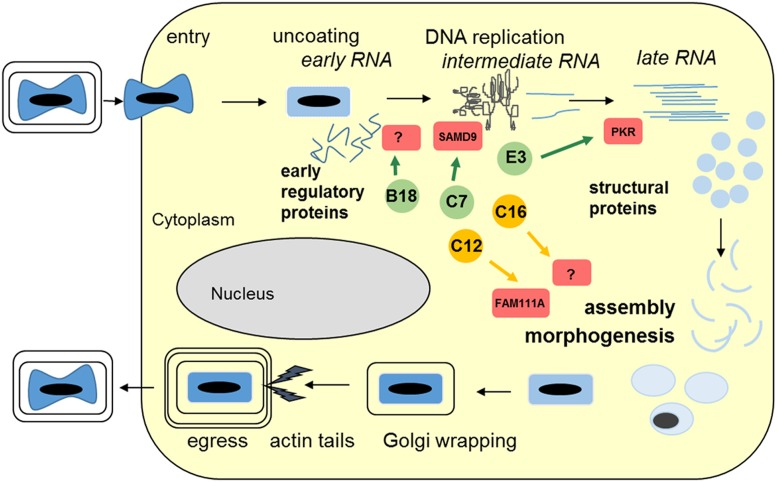
With this breakthrough we understand the requirements of a minimal set of five VACV early regulatory proteins needed for the completion of the life cycle of MVA in human cells (Fig. 1). The polypeptides C16 and SPI-1 (encoded by C12L) represent the sought viral factors to overcome the host restrictions disabling
Fig. 1.
Schematic overview of the MVA life cycle and important virus proteins to overcome intracellular host restrictions in human cells. As all poxviruses, MVA replicates in the cytoplasm of infected cells. Following entry, the virus replication comprises three steps of viral RNA and protein synthesis defined as expression of early, intermediate, and late viral genes. Production and processing of late structural proteins result in the morphogenesis of new infectious virions enveloped by one or two additional lipid membranes. Depicted are five early viral proteins needed to complete the MVA life cycle. The VACV proteins B18, C7, and E3 are conserved in MVA; interfere with different host restriction targets (including SAMD9, PKR, and unknown); and are important to maintain viral uncoating, DNA replication, and postreplicative gene expression in human cells. VACV C12 and C16 are the additional host-range proteins needed to rescue full MVA replicative capacity in human cells through association with host restrictions interfering with viral late protein processing, assembly, and morphogenesis (targets include FAM111A and unknown).
In PNAS, Peng and Moss identify and explore a previously unknown viral protein as a major facilitator of VACV growth in human cells.
MVA virion assembly and morphogenesis at the late stages of MVA infection. While the direct target of SPI-1 remains elusive, the protein is believed to interact with a host complex including the cellular DNA replication proteins FAM111A and RFC to modulate the replication of poxviruses in human cells (16). In addition to C12 and C16, some eminent regulatory proteins are functional in MVA infection and include the early proteins C7, E3, and B18 [following the nomenclature established for the genome of VACV strain Copenhagen (12)]. Inactivation of either of the encoding genes results in an early inhibition of the MVA molecular life cycle in human cells (6). The pleiotropic VACV immunomodulator E3, well known for its role to sequester viral dsRNA to inhibit the activation of antiviral IFN response proteins PKR, 2′-5′ oligoadenylate synthase, and RNase L, is also needed to support MVA replication in primary chicken embryo cells, whereas C7 and B18 are not essential for MVA replication in this avian cell culture. The C7 protein serves to counteract a host restriction in which the human protein SAMD9 has been identified as a major restriction factor (17) triggering a block early in infection and involving inhibition of viral mRNA translation. B18, also called 68k-ank, is the only MVA protein to contain ankyrin repeats and an F-box–like structure as obvious protein-interacting domains and serves to overcome an early intracellular restriction event interfering with genome uncoating and viral DNA replication (18). The molecular target(s) of B18/68k-ank within the host cell remain to be identified, and this is an exciting question for important future research on the functional activity of the C16L/B22R gene product. In this respect, one needs to keep in mind that the definition of essential genes in VACV infection is complicated by the phenomenon that inactivation of two or more host regulatory genes may be required to reveal the detrimental host restriction and the lethal interruption of the viral life cycle. Good examples for such so-called synthetic lethality in VACV replication in human cells are the redundant activities of the viral regulatory proteins C7/K1 (11) and B18/M2/C5 (18). Indeed, the fact that the genome of fully replication-competent VACV strain Western Reserve lacks a copy of the newly identified host-range gene C16L/B22R strongly suggests the existence of at least another VACV protein with a complementary activity (4). A particular aspect of this pioneering work is that we are only just starting to learn about the various molecular roles of all these important regulatory VACV proteins and their counterparts in the human host cell. Even with the very little we now know, it seems clear that with these viral proteins as guides we will discover whole networks of host factors that serve as potent antiviral response pathways that until now have escaped our view.
Of note, the discovery of the host-range genes also has intriguing practical implications in translational vaccine research. The results are an important step toward a better understanding of the basis for MVA attenuation, as the replication deficiency of the virus serves as a key determinant of its clinical and biological safety in compliance with regulatory requirements for human health and environmental release (19). In addition, the presented evidence here, that human cell lines expressing C16L and C12L allow for productive MVA replication, may enable the development of additional mammalian cell substrates for improved propagation of safety-tested nonreplicating MVA vaccines at an industrial scale. Finally, the reinsertion or repair of these host-range genes in the MVA genome will allow us to test the performance of replicating MVA recombinant vaccines or oncolytic vectors, which may further propel the usefulness of these viruses in our struggle against infectious diseases and cancers.
Footnotes
The author declares no competing interest.
See companion article, “Repair of a previously uncharacterized second host-range gene contributes to full replication of modified vaccinia virus Ankara (MVA) in human cells,” 10.1073/pnas.1921098117.
References
- 1.Fenner F., The eradication of smallpox. Prog. Med. Virol. 23, 1–21 (1977). [PubMed] [Google Scholar]
- 2.Lane J. M., Ruben F. L., Neff J. M., Millar J. D., Complications of smallpox vaccination, 1968. N. Engl. J. Med. 281, 1201–1208 (1969). [DOI] [PubMed] [Google Scholar]
- 3.McFadden G., Poxvirus tropism. Nat. Rev. Microbiol. 3, 201–213 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng C., Moss B., Repair of a previously uncharacterized second host-range gene contributes to full replication of modified vaccinia virus Ankara (MVA) in human cells. Proc. Natl. Acad. Sci. U.S.A. 117, 3759–3767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutter G., Moss B., Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. U.S.A. 89, 10847–10851 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volz A., Sutter G., Modified vaccinia virus Ankara: History, value in basic research, and current perspectives for vaccine development. Adv. Virus Res. 97, 187–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayr A., Munz E., [Changes in the vaccinia virus through continuing passages in chick embryo fibroblast cultures]. Zentralbl. Bakteriol. Orig. 195, 24–35 (1964). [PubMed] [Google Scholar]
- 8.Drillien R., Koehren F., Kirn A., Host range deletion mutant of vaccinia virus defective in human cells. Virology 111, 488–499 (1981). [DOI] [PubMed] [Google Scholar]
- 9.Moyer R. W., Rothe C. T., The white pock mutants of rabbit poxvirus. I. Spontaneous host range mutants contain deletions. Virology 102, 119–132 (1980). [DOI] [PubMed] [Google Scholar]
- 10.Gillard S., Spehner D., Drillien R., Kirn A., Localization and sequence of a vaccinia virus gene required for multiplication in human cells. Proc. Natl. Acad. Sci. U.S.A. 83, 5573–5577 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkus M. E., et al. , Vaccinia virus host range genes. Virology 179, 276–286 (1990). [DOI] [PubMed] [Google Scholar]
- 12.Goebel S. J., et al. , The complete DNA sequence of vaccinia virus. Virology 179, 247–266, 517–563 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Meng X., Chao J., Xiang Y., Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology 372, 372–383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyatt L. S., et al. , Marker rescue of the host range restriction defects of modified vaccinia virus Ankara. Virology 251, 334–342 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Liu R., et al. , SPI-1 is a missing host-range factor required for replication of the attenuated modified vaccinia Ankara (MVA) vaccine vector in human cells. PLoS Pathog. 15, e1007710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panda D., Fernandez D. J., Lal M., Buehler E., Moss B., Triad of human cellular proteins, IRF2, FAM111A, and RFC3, restrict replication of orthopoxvirus SPI-1 host-range mutants. Proc. Natl. Acad. Sci. U.S.A. 114, 3720–3725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivan G., Ormanoglu P., Buehler E. C., Martin S. E., Moss B., Identification of restriction factors by human genome-wide RNA interference screening of viral host range mutants exemplified by discovery of SAMD9 and WDR6 as inhibitors of the vaccinia virus K1L-C7L- mutant. MBio 6, e01122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B., et al. , Identification of poxvirus genome uncoating and DNA replication factors with mutually redundant roles. J. Virol. 92, e02152-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verheust C., Goossens M., Pauwels K., Breyer D., Biosafety aspects of modified vaccinia virus Ankara (MVA)-based vectors used for gene therapy or vaccination. Vaccine 30, 2623–2632 (2012). [DOI] [PubMed] [Google Scholar]