Abstract
Misregulation of Wnt signaling is common in human cancer. The development of small molecule inhibitors against the Wnt receptor, frizzled (FZD), may have potential in cancer therapy. During small molecule screens, we observed binding of carbamazepine to the cysteine-rich domain (CRD) of the Wnt receptor FZD8 using surface plasmon resonance (SPR). Cellular functional assays demonstrated that carbamazepine can suppress FZD8-mediated Wnt/β-catenin signaling. We determined the crystal structure of the complex at 1.7 Å resolution, which reveals that carbamazepine binds at a novel pocket on the FZD8 CRD. The unique residue Tyr52 discriminates FZD8 from the closely related FZD5 and other FZDs for carbamazepine binding. The first small molecule-bound FZD structure provides a basis for anti-FZD drug development. Furthermore, the observed carbamazepine-mediated Wnt signaling inhibition may help to explain the phenomenon of bone loss and increased adipogenesis in some patients during long-term carbamazepine treatment.
Introduction
Ligands belonging to the Wnt family of secreted lipoproteins play central roles in tissue morphogenesis and homeostasis through binding to members of the frizzled (FZD) family of cell surface receptors.1 Overexpression of FZD proteins has been observed in cancers,2,3 and FZD8 has been proposed as a therapeutic target in human lung cancer4 and renal cell carcinoma.5 The anti-FZD antibodies vantictumab (OMP-18R5), IgG-2919, and IgG-2921 have been taken into phase 1 (ClinicalTrials.gov: NCT01345201) or preclinical trials for cancers.6 These antibodies usually target multiple FZDs due to the high level of sequence conservation between FZD proteins. For example, vantictumab interacts with FZD1, 2, 5, 7, and 8.7 However, structural information on antibody–FZD complexes can guide engineering to improve specificity and avoid unwanted toxicity.8 The development of small molecules against specific FZD receptors has proved challenging. FZD proteins comprise an extracellular Wnt-binding cysteine-rich domain (CRD), a seven helix transmembrane domain (TMD), and a cytoplasmic tail. The small molecule FZM1 has been identified as an FZD4 misfolding chaperone, which is likely to bind to the intracellular loops of FZD4,9 and its derivatives can act as allosteric agonists of noncanonical Wnt signaling.10 The recent FZD4 TMD structure suggests that FZDs are not amenable to targeting with traditional G protein-coupled receptor (GPCR) small molecule ligands.11 Although computer modeling can potentially be used for anti-FZD drug development,12,13 experimentally determined high-resolution crystal structures of a target protein in complex with small molecule inhibitors remain the gold standard for structure-guided drug design. To date, no structural information about small molecules binding to FZD receptors has been published. We have used surface plasmon resonance (SPR) to screen a set of small molecules and report here the high-resolution crystal structure of the extracellular FZD8 cysteine-rich domain (FZD8CRD) bound with carbamazepine.
Results
Screening of Small Molecules for Binding to FZD8CRD
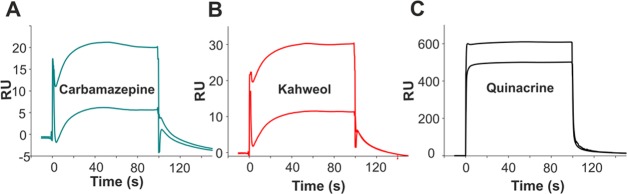
Wnt ligands bind to the FZD receptor CRD to initiate Wnt signaling. Small molecule antagonists that bind to the FZD CRD could therefore have therapeutic potential in cancers with upregulated Wnt signaling.1 We used SPR to screen for small molecules that bind to FZD8CRD. Small molecules from the Maybridge fragments library (58 compounds) and our own laboratory collection (44 compounds) were screened. All of the compounds screened are listed in Supporting Information, Table S1. Initial screening identified three hits showing SPR responses (Figure 1): carbamazepine, kahweol, and quinacrine (number 79, 87, and 99 in Supporting Information, Table S1). Further analysis of these hits with other irrelevant protein controls eliminated quinacrine as a nonspecific binder. Kahweol and carbamazepine were then carried forward to co-crystallization with FZD8CRD. While kahweol did not yield a complex structure, the structure of carbamazepine in complex with FZD8CRD was determined (Figure 2). In the absence of further evidence to support kahweol binding, we focused all further work on carbamazepine.
Figure 1.
SPR sensorgrams from FZD8CRD-positive response compounds. The biotinylated FZD8CRD protein (1000 response units) was immobilized on a streptavidin (SA) sensor chip. Analyte compounds carbamazepine (A), kahweol (B), and quinacrine (C) are in two concentrations (50 and 5 μM).
Figure 2.
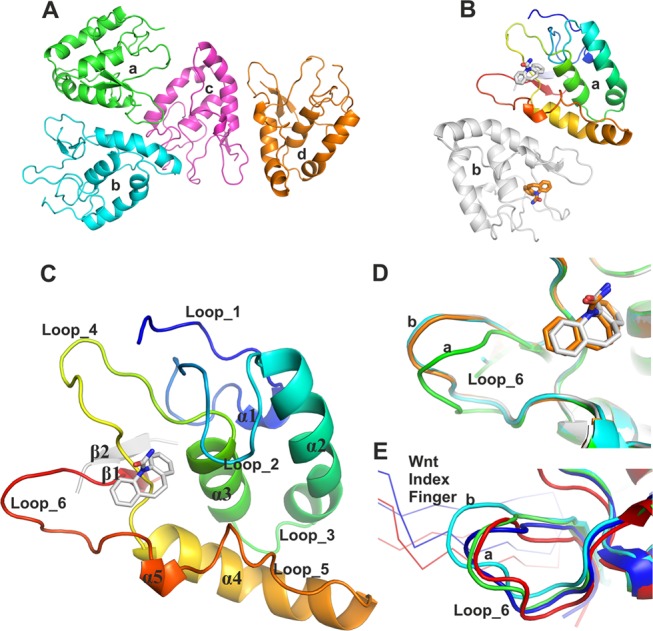
Overall structure of apo and carbamazepine-bound FZD8CRD and structural comparison. (A) Four molecules of the apo structure of FZD8CRD in the asymmetric unit (ASU) (PDB code 6TFM). (B) Two molecules of FZD8CRD in complex with carbamazepine in the ASU (PDB code 6TFB). (C) Cartoon representation of FZD8CRD, rainbow-colored from N-(blue) to C-(red) terminus. Residues originating from the Rhinovirus 3C cleavage site are colored in gray. In both our apo and carbamazepine-bound crystal structures, these additional residues contribute to an antiparallel β-strand (β2), which stabilizes β1. The bound carbamazepine is shown as gray sticks (PDB code 6TFB). (D) Close-up view of loop_6 from the two aligned carbamazepine-bound FZD8CRD molecules (gray and brown) superimposed on two representative apo FZD8CRD molecules from the ASU (green and cyan, PDB code 6TFM). (E) Alignment of two FZD8CRD copies from previously published complex structures with Wnt8 (blue, PDB code 4F0A) and Wnt3 (red, PDB code 6AHY). The Wnt index fingers are shown as Cα traces.
Structure of FZD8CRD and Its Complex with Carbamazepine
The amino acid sequence of FZD8CRD is fully conserved between human and mouse. Therefore, although we used a mouse cDNA sequence, the resulting structure is identical to human FZD8CRD. Thus, we refer to it simply as FZD8CRD hereafter. The apo FZD8CRD structure was determined at 2.3 Å resolution in space group P21 with four molecules in the asymmetric unit (ASU, Figure 2A). The carbamazepine complex structure (1.7 Å resolution) crystallized in the same condition (see the Experimental Section) with similar lattice contacts to the apo structure, but with higher crystallographic symmetry resulting in space group P212121, with two molecules in each ASU (Figure 2B).
The overall structure of FZD8CRD, either apo or in complex with carbamazepine, is almost identical to those of previously reported apo or Wnt-bound FZD8CRD14−16 except at the CRD C-terminus, which forms a β-hairpin with the remaining residues of a Rhinovirus 3C protease cleavage site (used to remove purification tags; Figure 2C and the Experimental Section). In both our structures, the ASUs contain dimers resulting from two-fold noncrystallographic symmetry (Figure 2A,B). This dimeric arrangement, as well as the other lattice packing interactions, differs from the distinctive dimer, mediated by unsaturated fatty acyl–FZD CRD binding, that has been observed for a number of FZD CRD structures, including FZD8.15,17−19
In the apo structure, the four molecules of FZD8CRD in the ASU fall into two conformations (“a” and “b”, Figure 2D) regarding loop_6 (residues R137 to L147). However, both copies of the carbamazepine-bound FZD8CRD are in conformation “b” (Figure 2D). This suggests that carbamazepine may stabilize conformation “b” upon binding. The Xenopus Wnt8-15 or human Wnt3-16 bound FZD8CRD loop_6 corresponds most closely to conformation “a” (Figure 3E). This suggests that Wnt binding may prefer the loop_6 conformation “a”.
Figure 3.
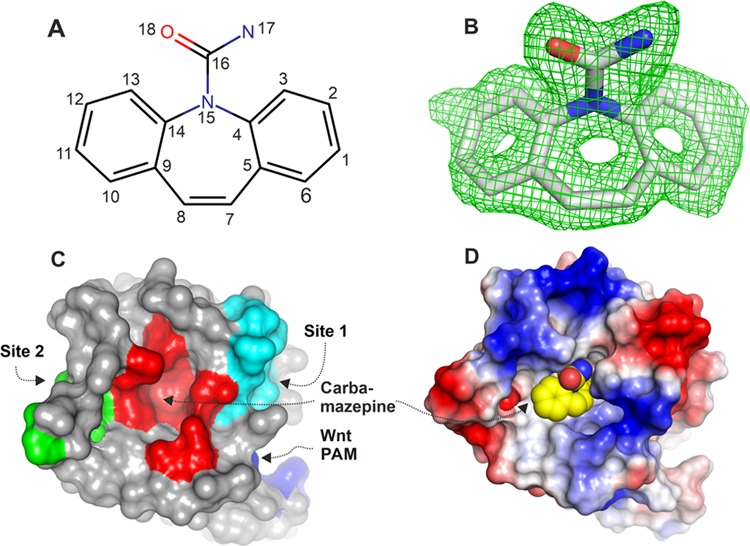
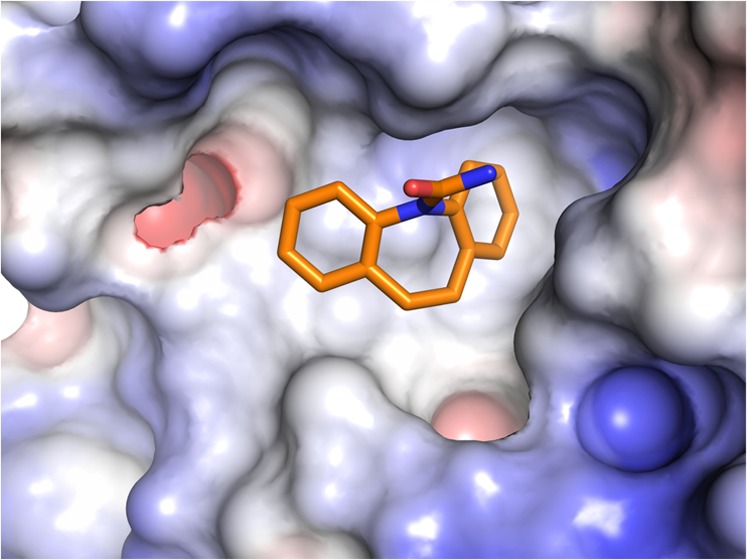
Carbamazepine chemical structure, electron density, and binding pocket. (A) Chemical structure of carbamazepine with atom numbers indicated. (B) Simulated annealing |Fo – Fc| omit electron density map for carbamazepine contoured at 2.5σ from one molecule from the ASU (the other has similar electron density and is not shown, PDB code 6TFB). (C) Surface representation of FZD8CRD with areas interacting with carbamazepine (red), Wnt index finger (site 2, green), Wnt thumb (site 1, cyan), and lipid palmitoleic acid (PAM, blue). (D) Electrostatic properties of FZD8CRD. The protein surface is colored by the electrostatic potential at ±5 kT/e (red, acidic; blue, basic; carbamazepine shown as spheres: yellow, carbon; blue, nitrogen; and red, oxygen).
Carbamazepine Binds FZD8CRD at a Novel Pocket
Carbamazepine (5H-dibenzo[b,f]azepine-5-carboxamide), sold under the trade names Tegretol, Equetro, Carbatrol, Epitol, and Orteril, is a tricyclic compound (Figure 3A). The 1.7 Å resolution complex structure unambiguously showed carbamazepine binding to the FZD8CRD. The simulated annealing omit electron density map showed clear electron density for all atoms of carbamazepine (Figure 3B) and both molecules in the ASU show similar quality density. FZD8CRD possesses two well-documented Wnt binding sites: a hydrophobic groove that binds the palmitoleic acid moiety (PAM) that is appended to the Wnt thumb (site 1) and the Wnt index finger binding site (site 2).15,16 The carbamazepine binding pocket is sandwiched by the two Wnt binding sites (Figures 3C and 4A). This pocket is largely hydrophobic and neutral in surface charge but is surrounded by positively and negatively charged patches (Figure 3D). Aside from Wnt, there are many FZD CRD binding proteins that have been reported, but none of them bind at this pocket. The Wnt mimic Norrin protein binds at site 2 on FZD4CRD (Figure 4A),20,21 while all other reported binders bind at or near site 1. These include the Clostridium difficile toxin B (TcdB22) that binds FZD2CRD (Figure 4B), the Wnt surrogate B12 module that binds FZD8CRD23 and the peptide FZ7-21 that binds FZD7CRD18 (Figure 4C), the DARPin module DRPB that binds FZD8CRD,24 and the antibody Fab F2.I that binds FZD5CRD8 (Figure 4D). Carbamazepine does not overlap with any of the reported FZD CRD ligands when the various structures are superimposed (Figure 4) and instead binds to residues located between the two Wnt binding sites in a novel binding pocket (Figures 3 and 4).
Figure 4.
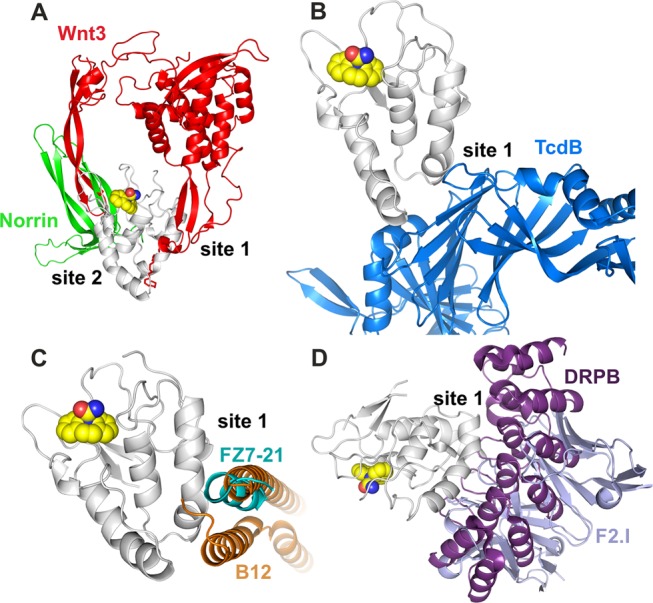
Carbamazepine binding site (PDB code 6TFB) relative to other FZD ligand binding sites. (A) Wnt3 (PDB code 6AHY(15)) and Norrin (PDB code 5BQE(20)); (B) C. difficile toxin B (TcdB, PDB code 6C0B(22)); (C) peptide FZ7-21(PDB code 5WBS(18)) and Wnt surrogate B12 module (PDB code 5UN5(23)); and (D) DARPin B (PDB code 6NDZ(24)) and the Fab fragment of antibody F2.I (PDB code 6O39(8)). The carbamazepine-bound FZD8CRD structure is shown as gray cartoon and spheres as in Figure 3D. For clarity, in all other ligand-bound structures, only our FZD8CRD–carbamazepine structure (gray ribbon and spheres) is shown. All other FZD CRDs are hidden from view.
The carbamazepine binding pocket comprises residues from helix α3 (S90, M91), loop_2 (Y52, Q56), loop_4 (P94, P103, L104, P105, P106), and loop_6 (R137). The residues L104 and R137 use their main chain atoms to interact with carbamazepine, while the other interacting residues use their side chains (Figure 5A). A distinctive feature of this pocket is a cluster of four hydrophobic prolines. The interacting residues from the two copies of FZD8CRD in the crystallographic ASU (“a” and “b”) show similar interactions with carbamazepine, except for the residue Q56. Q56 of molecule “a” interacts with a carbamazepine nitrogen (N17) atom and azepine ring (C7, Figure 5B), while in the other copy, “b”, Q56 interacts with the carbamazepine azepine and benzyl rings (Figure 5C). The difference in the Q56 side-chain conformation may be due to crystal packing, as this area of molecule “a” contacts a neighbor packing molecule. When two representative copies of apo structures are aligned with carbamazepine-bound structures, the Q56 side chain from the apo structure would sterically hinder carbamazepine binding (Figure 5D). This suggests that Q56 may undergo a conformational change upon carbamazepine binding. The carbamazepine–protein interactions are mainly hydrophobic, especially those involving P105 and P103, which form extensive hydrophobic interactions with the azepine ring and two benzyl rings of carbamazepine (Figure 5E). Y52 forms a hydrogen bond through a water molecule to the nitrogen (N17) of the carboxamide head of carbamazepine (Figure 5A). There are three additional ordered water molecules within the pocket that are conserved between the two molecules in the ASU. The positions of water molecules could guide carbamazepine modification to develop more potent FZD8 inhibitors.
Figure 5.
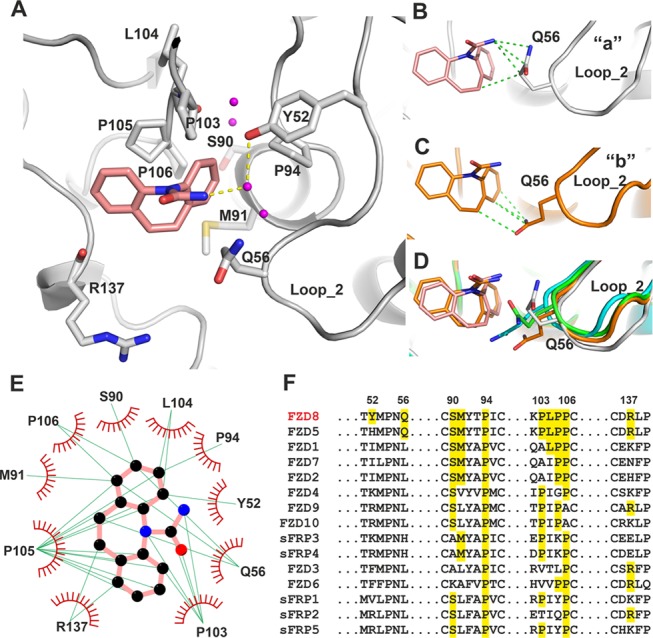
Interactions between carbamazepine and FZD8CRD. (A) Details of the carbamazepine binding site (gray cartoon, from molecule “a” of the ASU, PDB code 6TFB). The carbamazepine interacting amino acid side chains are shown as gray sticks. Water molecules within the pocket are shown as magenta balls, carbamazepine as salmon sticks, and hydrogen bonds as yellow dashed lines. (B–D) Conformations of Q56 from two carbamazepine binding molecules of the ASU, “a” (gray, B) and “b” (brown, C), and superimposed with two representative molecules from apo structures, (green and cyan; D). Interactions are defined as distances between protein and carbamazepine of less than 3.9 Å. The hydrophobic interactions are shown as green dashed lines. (E) LIGPLOT25 of FZD8CRD–carbamazepine interactions. The green lines indicate hydrophobic interactions. Carbamazepine carbon atoms are shown as black, nitrogen as blue, and oxygen as red spheres. Only molecule “a” from the ASU is shown. (F) Sequence alignment of human FZDs and sFRPs, with carbamazepine interacting FZD8 residues and all matching residues highlighted. The residue numbers indicated are for mouse FZD8.
Binding Specificity and Affinity of Carbamazepine
In addition to the 10 FZDs, the human genome also encodes 5 secreted FZD-related proteins (sFRPs), important Wnt regulatory proteins,26 and all FZDs and sFRPs possess a conserved FZD CRD. Sequence alignment of all FZD and sFRP CRDs shows that only one proline residue (P94) is fully conserved among the residues interacting with carbamazepine (Figure 5F). Another proline, P106, is conserved in all FZDs/sFRPs, except for FZD9/10. The residue S90 is highly conserved but absent in FZD3/6 and sFRP3/4. Other carbamazepine interacting residues are only partially conserved among FZDs and sFRPs. FZD8 and FZD5 are closely related (>80% sequence identity within the CRD), sharing most of the carbamazepine interacting residues, including Q56, which is unique to these FZDs. However, the residue Y52 is found only in FZD8 (Figure 5F). The sequence alignment suggests that of all of the other FZDs/sFRPs, FZD5 is the most likely candidate to bind to carbamazepine, followed by FZD1, 2, and 7.
We then used biophysical methods to measure the affinity of the interaction between carbamazepine and FZD8CRD, as well as the cross-reactivity of carbamazepine with the CRDs of FZD5 and 7. We have previously used a thermal shift assay (also known as differential scanning fluorimetry) to investigate small molecule–protein interactions.27,28 However, we found that FZD CRDs are highly thermostable (remaining folded at 95 °C), which precluded the measurement of melting curves. We therefore turned to SPR as an alternative method for the detection of small molecule–protein interactions.29,30 All three (FZD8, 5, and 7) CRD constructs yielded correctly folded protein samples as evidenced by their gel filtration profiles (Supporting Information, Figure S1). Biotinylated samples of each CRD were immobilized on a streptavidin (SA) chip in separate flow chambers with carbamazepine as an analyte. FZD8CRD has a clear concentration-dependent response to carbamazepine (Figure 6), with a calculated Kd of 17 μM. Surprisingly, carbamazepine does not interact with the closely related FZD5CRD. This is despite FZD5CRD differing from FZD8CRD at only one carbamazepine interacting residue, Y52, which corresponds to FZD5 H50. In fact, Y52 is unique among all FZDs and sFRPs (Figure 5F), suggesting that this residue is a key determinant of the specificity of the interaction between carbamazepine and FZD8. As FZD7CRD shares only half of the interacting residues, it is less surprising that it also does not bind detectably to carbamazepine (Figure 6).
Figure 6.
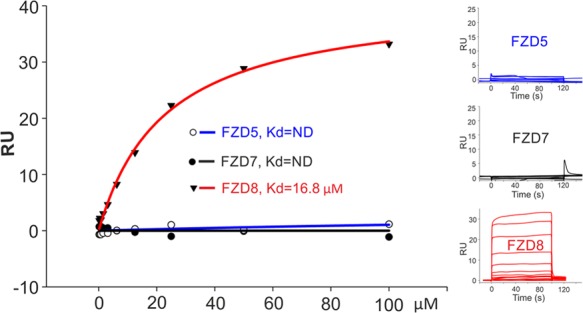
SPR analysis of carbamazepine interaction with FZD5, 7, 8 CRD. Biotinylated mouse FZD5, human FZD7, and mouse FZD8 CRD were immobilized on individual flow cells of a SA chip, respectively. A carbamazepine concentration series was used as an analyte. The SPR sensorgrams are shown in the right panels. ND, not detectable; RU, resonance units.
Carbamazepine Inhibits Wnt Signaling in a Cellular Reporter Gene Assay
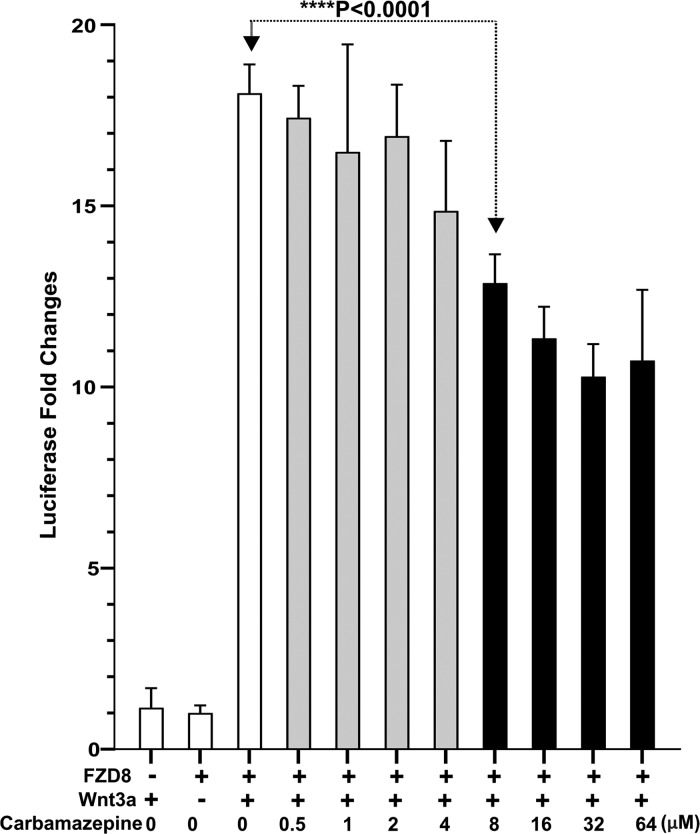
We used cellular luciferase reporter assays to assess the effects of carbamazepine on Wnt signaling. Carbamazepine only binds to FZD8; however, the commonly used cell line (HEK293T) expresses FZD8 at a very low level, and the HEK293T TOPFlash response to canonical Wnt ligands is dependent mainly on FZD1, 2, and 7.31 To observe the specific response from FZD8, we therefore used an FZD1, 2 and 7 knockout HEK293T cell line31 and introduced the full-length mouse FZD8 expression cassette by lentiviral transduction.32 The cell line was further stably transformed with a T-cell factor/lymphoid enhancer-binding factor 1 luciferase (TCF/LEF, TOPFlash) plasmid to minimize reporter plasmid transfection variations. The cells were then stimulated with a conditioned medium from mouse L-cells expressing Wnt3a, in the presence of a carbamazepine concentration series (Figure 7). Carbamazepine starts to inhibit Wnt3a-induced TOPFlash luciferase activity at a concentration of 8 μM (unpaired t-test, P < 0.0001), with greater inhibition seen at higher concentrations. However, we found that carbamazepine can only partially suppress Wnt3a-induced luciferase activity and even at the high concentration of 64 μM, luciferase activity remained around 60% (Figure 7).
Figure 7.
Carbamazepine inhibits FZD8-mediated TOPFLASH luciferase activity. HEK293T cells with FZD1, 2, and 7 knocked out and with mouse FZD8 and TOPFLASH expression cassettes introduced were transfected with tk-Renilla luciferase and treated with carbamazepine at the indicated concentrations and Wnt3a for luciferase induction. Each data point represents the Renilla luciferase-normalized firefly luciferase fold changes in quadruplicate. Error bars represent the standard deviation of the mean and the Student’s t-test was used to calculate statistical significance. Gray columns indicate no statistical difference and black columns indicate a significant difference.
Discussion and Conclusions
FZD proteins, as essential Wnt receptors, are a central point for Wnt signaling intervention in diseases such as cancer. While macromolecules like antibodies, peptides, the FZD5/8-binding B12 protein, and DARPin molecules targeting FZD CRDs have been described,8,18,23,24 small molecules may offer advantages such as being easier to manufacture, more stable, less expensive, and having the potential to be administered orally. Among the methods of screening small molecules for binding to target proteins, SPR provides a measure of direct molecular interactions, allowing an effective triage for candidates with the highest potential for generating complex crystal structures. Structural information forms the basis for rational drug design. We have obtained the first FZD-small molecule structure and discovered that carbamazepine specifically binds to the FZD8CRD and not to the closely related FZD5. To date, no antibodies or synthetic FZD binders have been reported that can distinguish between FZD8 and FZD5. Conceivably, carbamazepine may offer this potential.
We noticed that the carbamazepine can only partially suppress Wnt3a-induced luciferase activity, even at a high concentration of 64 μM (Figure 7), which is in agreement with the recently reported weak Wnt inhibitory effects in mouse adipose cells.33 The binding pocket that we have identified in FZD8CRD does not overlap with the known Wnt binding sites. Thus, the mechanism of action is allosteric. However, the carbamazepine binding pocket may still be important for Wnt signaling. It was previously demonstrated that the disruption of the carbamazepine binding region (insertion of a tripeptide, GSG, before the carbamazepine interacting residues Y52 or R137) abolished Wnt8 binding to FZD8.14 The carbamazepine binding affinity Kd value of 17 μM and starting inhibitory concentration of 8 μM are comparable to the reported Wnt/β-catenin inhibitory concentration of 5–10 μM in adipose cells33 and colon cancer cells.34 Carbamazepine is the primary drug used to treat epilepsy. Typical doses are between 400 and 1000 mg per day and the carbamazepine plasma concentration in patients during treatment reaches 20–40 μM,35 which is higher than the Kd value and inhibitory concentration we observed.
Epilepsy is a common serious neurological disorder, affecting 1–2% of the population (over 50 million people) worldwide.36 The etiology of epilepsy is multifactorial and the involvement of Wnt signaling misregulation has recently received attention.37 Wnt/β-catenin signaling may play a role in the development of temporal lobe epilepsy.38 Conditional knockout of the major Wnt/β-catenin negative regulator adenomatous polyposis coli (APC) gene in mice, which results in elevated Wnt/β-catenin signaling, causes chronic seizures39 with similar features to those seen in humans with infantile spasms, a common syndrome in childhood epilepsy.40 In a mouse model, both deletion and overexpression of β-catenin have a significant impact on seizure susceptibility.41 The kainic acid-induced epilepsy animal model shows upregulated Wnt/β-catenin signaling.42 Increased Wnt/β-catenin signaling is associated with the increased number of neuronal stimuli,43 while decreased Wnt/β-catenin signaling by the Wnt antagonist Dickkopf-related protein 1 (DKK1) has been shown to be able to protect against the development of hippocampal sclerosis, which is a hallmark of temporal lobe epilepsy.44 The antiepileptic drug carbamazepine (an inhibitor of voltage-gated sodium channels45) has been shown to decrease Wnt/β-catenin signaling in the human colon adenocarcinoma SW480 cell line34 and mouse adipocyte 3T3-L1 cells.33 The results presented here contribute further evidence that Wnt signaling modulation may be involved in carbamazepine treatment of epilepsy. This warrants further studies of the role of Wnt-FZD8 signaling modulation in the therapeutic mechanism of carbamazepine against epilepsy.
It is noteworthy that long-term antiepileptic drug (AED) treatment can have side effects, including disorders of bone metabolism leading to bone loss.46−50 The exact mechanism of this pathological change is not completely understood. It is commonly accepted that intact Wnt signaling is essential for proper bone formation and remodeling.51 The strength and integrity of the human skeleton depend on a delicate equilibrium between bone resorption by osteoclasts and bone formation by osteoblasts. Wnt/β-catenin signaling can inhibit osteoclastogenesis52 and increase bone mass. On the other hand, inhibition of Wnt signaling can lead to bone loss. Among the 10 FZD receptors, FZD8 and FZD9 are particularly relevant to bone metabolism. While FZD9 regulates osteoblast function through noncanonical Wnt signaling,53,54 FZD8 mediates canonical Wnt/β-catenin signaling and inhibits osteoclastogenesis.52 The discovery that carbamazepine can inhibit FZD8-mediated Wnt signaling may help to explain the loss of bone density associated with long-term treatment with carbamazepine. In addition, weight gain affecting patients treated with carbamazepine has also been linked to Wnt signaling inhibition.33 Carbamazepine use has also been shown to reduce the risk of prostate cancer55,56 and synergistically inhibits breast cancer cell proliferation when combined with other anticancer treatment,57 although the exact mechanisms remain to be investigated.
In summary, we have identified carbamazepine as a specific ligand for the Wnt receptor FZD8 using an SPR screen of small molecules. The high-resolution crystal structure of the complex reveals a novel binding site in FZD8CRD that allows small molecule interactions to discriminate between closely related FZDs. As well as potentially explaining the loss of bone density observed in patients following long-term treatment with carbamazepine, our carbamazepine–FZD8CRD structure also provides a new avenue to explore the design and development of FZD specific inhibitors.
Experimental Section
Protein Production and Crystallization
Mouse FZD8 (UniProtKB Q61091) residues A28-R153 (identical amino acid sequence to human FZD8CRD) with glycosylation site mutations (N49E and N152E) were PCR amplified from the template cDNA (Source Bioscience, Nottingham U.K.; clone ID 40130820) and cloned into a stable cell line vector pNeo-sec.58 with a Rhinovirus 3C cleavage site, a monoVenus fluorescent protein, and 6xHis tags. The resulting 3C protease treated protein contains two additional amino acids (GT) from KpnI cloning site and six amino acids (LEVLFQ) from the 3C cleavage site at the C-terminus. HEK293S GnTI– cells (ATCC CRL-3022) were co-transfected with this plasmid and a PhiC31 integrase expression vector (pCB92/pgk-φC31).59 The polyclonal population of cells following G418 (1 mg/mL) selection was cultured in a CompacT automated cell culture system.60 The secreted proteins were captured by Talon Co2+ affinity resin (Takara Bio Europe SAS) after the conditional media were dialyzed against PBS. The protein-bound Talon beads were washed with 20 mM imidazole in PBS and eluted with 300 mM imidazole in PBS. The eluted fusion protein was treated with Rhinovirus 3C protease. After reverse tag capture with Talon beads (to remove the monoVenus and His tags), the protein was further purified by size-exclusion chromatography using a Superdex 200 16/60 column (GE Healthcare) and concentrated to 10 mg·mL–1 in 10 mM HEPES, pH 7.4, 50 mM NaCl buffer. Crystallization screening was carried out using the sitting-drop vapor diffusion method in 96-well plates, and crystals from both FZD8CRD protein alone or mixed with carbamazepine (∼10 μg.ML–1) grew in a condition containing 0.1 M sodium acetate, pH 5, and 1 M ammonium sulfate.
Data Collection and Structure Determination
Crystals were flash-frozen by immersion in a reservoir solution supplemented with 25% v/v glycerol followed by transferring to liquid nitrogen. Data sets were recorded from crystals at 100 K at the Diamond Light source (Didcot U.K., beamline I04-1) and processed with Xia2.61 Structures were determined by molecular replacement using Molrep (CCP4) using the previously reported FZD8CRD structure (PDB code 1IJY(14)) as a search model. The model was then manually built with COOT62 and refined with Phenix. Data collection and refinement statistics are shown in Table 1. We noticed that the Rfree for both of the structures are relatively high, and we then used Zanuda63 to check if the space group for each structure was misassigned; refinement in lower symmetry space groups (P1 and P21 for the apo and complex structures, respectively) did not result in significantly lower R factors and both structures passed Zanuda tests. The PyMOL Molecular Graphics System (Schrödinger, LLC.) was used for preparing figures.
Table 1. Data Collection and Refinement Statisticsa.
| data collection | ||
|---|---|---|
| data set | FZDCRD | FZDCRD-carbamazepine |
| PDB code | 6TFM | 6TFB |
| wavelength (Å) | 0.916 | 0.916 |
| space group | P21 | P21 21 21 |
| unit cell dimensions (Å) | a = 52.3, b = 66.0, c = 72.7; α = γ = 90°, β = 90.1° | a = 52.0, b = 68.1, c = 73.8; α = β = γ = 90° |
| resolution (Å) | 72.7–2.34 (2.38–2.34) | 52–1.68 (1.71–1.68) |
| unique reflections | 20741 (1024) | 25589 (1212) |
| Rmerge | 0.19 (0.83) | 0.13 (---) |
| < I > / < σI> | 7.3 (2.3) | 9.4 (1.0) |
| CC half | 0.99 (0.89) | 0.99 (0.84) |
| completeness (%) | 98.9 (98.5) | 99.2 (95.3) |
| redundancy | 6.5 (5.9) | 11.5 (8.5) |
| refinement | ||
|---|---|---|
| resolution (Å) | 72.7–2.34 | 50–1.68 |
| no. reflections | 19675 | 24108 |
| Rwork /Rfree (%) | 24.3/30.0 | 22.3/26.9 |
| no. atoms | 4236 | 2179 |
| average B-factor (Å2) | 20 | 16 |
| r.m.s. deviations | ||
| bond lengths (Å) | 0.004 | 0.009 |
| bond angles (deg) | 1.6 | 1.2 |
Numbers in parentheses refer to the highest resolution shell of data.
SPR Equilibrium Binding Studies
Human FZD5 (UniProtKB - Q13467) CRD (P31- Y150), human FZD7 (UniProtKB - O75084) CRD (Q33-G173), and mouse FZD8CRD (A28-R153) were cloned into a pHL-Avi3 vector.64 To produce biotinylated proteins, these plasmids were co-transfected with the pDisplay_BirA-ER plasmid65 into HEK293T cells with the media supplemented with 20 μM biotin. This procedure allows in vivo biotinylation to occur.65 The dialyzed conditioned media were directly used for the immobilization of ligands. The affinity was measured at 25 °C in 10 mM HEPES, pH 7.4, 150 mM NaCl, 0.005% Tween20, and 2% DMSO using a Biacore S200 machine (GE Healthcare). The biotinylated ligands (1000 RU each) were coupled to a SA sensor chip (GE Healthcare), and the analyte carbamazepine was tested using a two-fold serial dilution. The response was plotted against the analyte concentration and fitted by nonlinear regression to a one-site saturation binding model (Sigma Plot, Systat software, Inc.)
Cellular Wnt Signaling TOPFlash Luciferase Assay
The HEK293T cell line with FZD1/2/7 knocked out31 was a kind gift from Professor Michel Boutrous (DFKZ, Germany). The mouse full-length FZD8 was introduced into this cell line using a lentiviral vector with an IRES-GFP sequence after the expression cassette.32 The resulting cell line was further transformed using a Super TOPFlash firefly luciferase66 gene expression cassette in a stable cell line vector, followed by blasticidine selection. The polyclonal population from the blasticidine selection (20 μg mL–1) was seeded in a 96-well plate (105 cells/well) and transfected with a constitutive Renilla luciferase plasmid (pRL-tk; Promega) at a concentration of 10 ng mL–1 with lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, the media were replaced with carbamazepine dilution series diluted in Wnt3a conditional media from the Wnt3a producing mouse L Wnt-3A cell line (ATCC CRL-2647, ref (67)). The culture medium from L-cells served as the Wnt3a control medium. The firefly and Renilla luciferase activities were measured 24 h later using the Dual-Glo luciferase reporter assay system (Promega) with an Ascent Lunimoskan luminometer (Labsystems). The firefly luciferase activity was normalized with the constitutive Renilla luciferase activity.
Acknowledgments
We thank Diamond Light Source for beamtime and the staff of beamline I04-1 for assistance with crystal testing and data collection (under BAG application MX14744). We thank Thomas Walter for assistance with crystallization. We are grateful to Professor Michel Boutrous (DFKZ, Germany) for the HEK293T FZD1, 2, 7 knockout cell line. We thank Drs Laura Diaz Saez and Oleg Fedorov (TDI, Oxford) for assistance with using the Biacore S200 machine. This work was funded by Cancer Research UK, the UK Medical Research Council (to EYJ, C375/A17721 and MR/M000141/1), and the Wellcome Trust (grant 203141/Z/16/Z supporting the Wellcome Centre for Human Genetics).
Glossary
Abbreviations
- FZDCRD
frizzled cysteine-rich domain
- ASU
asymmetric unit
- SPR
surface plasmon resonance
- TCF/LEF
T-cell factor/lymphoid enhancer-binding factor 1
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.9b02020.
Accession Codes
PDB codes for structures of FZD8CRD and its complex with carbamazepine are 6TFM and 6TFB, respectively. The authors will release the atomic coordinates and experimental data upon article publication.
Author Contributions
Y.Z. and E.Y.J. designed the project and wrote the manuscript together with J.H. and J.R., Y.Z. performed experiments and analyzed data with J.R. and J.H. W.L. helped with tissue culture.
The authors declare no competing financial interest.
Supplementary Material
References
- Nusse R.; Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Merle P.; Kim M.; Herrmann M.; Gupte A.; Lefrancois L.; Califano S.; Trepo C.; Tanaka S.; Vitvitski L.; de la Monte S.; Wands J. R. Oncogenic Role of the Frizzled-7/beta-Catenin Pathway in Hepatocellular Carcinoma. J. Hepatol. 2005, 43, 854–862. 10.1016/j.jhep.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Simmons G. E. Jr; Pandey S.; Nedeljkovic-Kurepa A.; Saxena M.; Wang A.; Pruitt K. Frizzled 7 Expression is Positively Regulated by SIRT1 and Beta-Catenin in Breast Cancer Cells. PLoS One 2014, 9, e98861 10.1371/journal.pone.0098861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Q.; Xu M. L.; Ma J.; Zhang Y.; Xie C. H. Frizzled-8 as a Putative Therapeutic Target in Human Lung Cancer. Biochem. Biophys. Res. Commun. 2012, 417, 62–66. 10.1016/j.bbrc.2011.11.055. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Wang Y.; Pan X.; Ye J.; Gan S.; Qu F.; Chen L.; Chu C.; Gao Y.; Cui X. Frizzled 8 Promotes the Cell Proliferation and Metastasis of Renal Cell Carcinoma. Oncotarget 2017, 8, 78989–79002. 10.18632/oncotarget.20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhart Z.; Pavlovic Z.; Chandrashekhar M.; Hart T.; Wang X.; Zhang X.; Robitaille M.; Brown K. R.; Jaksani S.; Overmeer R.; Boj S. F.; Adams J.; Pan J.; Clevers H.; Sidhu S.; Moffat J.; Angers S. Genome-Wide CRISPR Screens Reveal a Wnt-FZD5 Signaling Circuit as a Druggable Vulnerability of RNF43-mutant Pancreatic Tumors. Nat. Med. 2017, 23, 60–68. 10.1038/nm.4219. [DOI] [PubMed] [Google Scholar]
- Gurney A.; Axelrod F.; Bond C. J.; Cain J.; Chartier C.; Donigan L.; Fischer M.; Chaudhari A.; Ji M.; Kapoun A. M.; Lam A.; Lazetic S.; Ma S.; Mitra S.; Park I. K.; Pickell K.; Sato A.; Satyal S.; Stroud M.; Tran H.; Yen W. C.; Lewicki J.; Hoey T. Wnt Pathway Inhibition via the Targeting of Frizzled Receptors Results in Decreased Growth and Tumorigenicity of Human Tumors. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 11717–11722. 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S.; Beilschmidt M.; To M.; Lin K.; Lui F.; Jmeian Y.; Ng M.; Fernandez M.; Fu Y.; Mascall K.; Duque A.; Wang X.; Pan G.; Angers S.; Moffat J.; Sidhu S. S.; Magram J.; Sinclair A. M.; Fransson J.; Julien J. P. Structure-guided Design Fine-tunes Pharmacokinetics, Tolerability, and Antitumor Profile of Multispecific Frizzled Antibodies. Proc. Natl. Acad.Sci. U.S.A. 2019, 116, 6812–6817. 10.1073/pnas.1817246116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generoso S. F.; Giustiniano M.; La Regina G.; Bottone S.; Passacantilli S.; Di Maro S.; Cassese H.; Bruno A.; Mallardo M.; Dentice M.; Silvestri R.; Marinelli L.; Sarnataro D.; Bonatti S.; Novellino E.; Stornaiuolo M. Pharmacological Folding Chaperones Act as Allosteric Ligands of Frizzled4. Nat. Chem. Biol. 2015, 11, 280–286. 10.1038/nchembio.1770. [DOI] [PubMed] [Google Scholar]
- Riccio G.; Bottone S.; La Regina G.; Badolati N.; Passacantilli S.; Rossi G. B.; Accardo A.; Dentice M.; Silvestri R.; Novellino E.; Stornaiuolo M. A Negative Allosteric Modulator of WNT Receptor Frizzled 4 Switches into an Allosteric Agonist. Biochemistry 2018, 57, 839–851. 10.1021/acs.biochem.7b01087. [DOI] [PubMed] [Google Scholar]
- Yang S.; Wu Y.; Xu T. H.; de Waal P. W.; He Y.; Pu M.; Chen Y.; DeBruine Z. J.; Zhang B.; Zaidi S. A.; Popov P.; Guo Y.; Han G. W.; Lu Y.; Suino-Powell K.; Dong S.; Harikumar K. G.; Miller L. J.; Katritch V.; Xu H. E.; Shui W.; Stevens R. C.; Melcher K.; Zhao S.; Xu F. Crystal Structure of the Frizzled 4 Receptor in a Ligand-Free State. Nature 2018, 560, 666–670. 10.1038/s41586-018-0447-x. [DOI] [PubMed] [Google Scholar]
- Hamdoun S.; Fleischer E.; Klinger A.; Efferth T. Lawsone Derivatives Target the Wnt/Beta-Catenin Signaling Pathway in Multidrug-resistant Acute Lymphoblastic Leukemia Cells. Biochem. Pharmacol. 2017, 146, 63–73. 10.1016/j.bcp.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Lee H. J.; Bao J.; Miller A.; Zhang C.; Wu J.; Baday Y. C.; Guibao C.; Li L.; Wu D.; Zheng J. J. Structure-based Discovery of Novel Small Molecule Wnt Signaling Inhibitors by Targeting the Cysteine-rich Domain of Frizzled. J. Biol. Chem. 2015, 290, 30596–30606. 10.1074/jbc.M115.673202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann C. E.; Hsieh J. C.; Rattner A.; Sharma D.; Nathans J.; Leahy D. J. Insights into Wnt Binding and Signalling from the Structures of Two Frizzled Cysteine-Rich Domains. Nature 2001, 412, 86–90. 10.1038/35083601. [DOI] [PubMed] [Google Scholar]
- Hirai H.; Matoba K.; Mihara E.; Arimori T.; Takagi J. Crystal Structure of a Mammalian Wnt-Frizzled Complex. Nat. Struct. Mol. Biol. 2019, 26, 372–379. 10.1038/s41594-019-0216-z. [DOI] [PubMed] [Google Scholar]
- Janda C. Y.; Waghray D.; Levin A. M.; Thomas C.; Garcia K. C. Structural Basis of Wnt Recognition by Frizzled. Science 2012, 337, 59–64. 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruine Z. J.; Ke J.; Harikumar K. G.; Gu X.; Borowsky P.; Williams B. O.; Xu W.; Miller L. J.; Xu H. E.; Melcher K. Wnt5a Promotes Frizzled-4 Signalosome Assembly by Stabilizing Cysteine-Rich Domain Dimerization. Genes Dev. 2017, 31, 916–926. 10.1101/gad.298331.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nile A. H.; de Sousa E. M. F.; Mukund S.; Piskol R.; Hansen S.; Zhou L.; Zhang Y.; Fu Y.; Gogol E. B.; Komuves L. G.; Modrusan Z.; Angers S.; Franke Y.; Koth C.; Fairbrother W. J.; Wang W.; de Sauvage F. J.; Hannoush R. N. A Selective Peptide Inhibitor of Frizzled 7 Receptors Disrupts Intestinal Stem Cells. Nat. Chem. Biol. 2018, 14, 582–590. 10.1038/s41589-018-0035-2. [DOI] [PubMed] [Google Scholar]
- Nile A. H.; Mukund S.; Stanger K.; Wang W.; Hannoush R. N. Unsaturated Fatty Acyl Recognition by Frizzled Receptors Mediates Dimerization upon Wnt Ligand Binding. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 4147–4152. 10.1073/pnas.1618293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. H.; Hsieh F. L.; Zebisch M.; Harlos K.; Elegheert J.; Jones E. Y. Structure and Functional Properties of Norrin Mimic Wnt for Signalling With Frizzled4, Lrp5/6, and Proteoglycan. Elife 2015, 4, e06554 10.7554/eLife.06554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G.; Ke J.; Wang Z.; Cheng Z.; Gu X.; Wei Y.; Melcher K.; Xu H. E.; Xu W. Structural Basis of the Norrin-Frizzled 4 Interaction. Cell Res. 2015, 25, 1078–1081. 10.1038/cr.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.; Tao L.; Wang T.; Zhang J.; He A.; Lam K. H.; Liu Z.; He X.; Perry K.; Dong M.; Jin R. Structural Basis for Recognition of Frizzled Proteins by Clostridium Difficile Toxin B. Science 2018, 360, 664–669. 10.1126/science.aar1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda C. Y.; Dang L. T.; You C.; Chang J.; de Lau W.; Zhong Z. A.; Yan K. S.; Marecic O.; Siepe D.; Li X.; Moody J. D.; Williams B. O.; Clevers H.; Piehler J.; Baker D.; Kuo C. J.; Garcia K. C. Surrogate Wnt Agonists that Phenocopy Canonical Wnt and Beta-Catenin Signalling. Nature 2017, 545, 234–237. 10.1038/nature22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L. T.; Miao Y.; Ha A.; Yuki K.; Park K.; Janda C. Y.; Jude K. M.; Mohan K.; Ha N.; Vallon M.; Yuan J.; Vilches-Moure J. G.; Kuo C. J.; Garcia K. C.; Baker D. Receptor Subtype Discrimination using Extensive Shape Complementary Designed Interfaces. Nat. Struct. Mol. Biol. 2019, 26, 407–414. 10.1038/s41594-019-0224-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A.; Swindells M. B. LigPlot +: Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Mii Y.; Taira M. Secreted Wnt ″Inhibitors″ are not Just Inhibitors: Regulation of Extracellular Wnt by Secreted Frizzled-Related Proteins. Dev. Growth Differ. 2011, 53, 911–923. 10.1111/j.1440-169X.2011.01299.x. [DOI] [PubMed] [Google Scholar]
- Ren J.; Zhao Y.; Fry E. E.; Stuart D. I. Target Identification and Mode of Action of Four Chemically Divergent Drugs against Ebolavirus Infection. J. Med. Chem. 2018, 61, 724–733. 10.1021/acs.jmedchem.7b01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Ren J.; Harlos K.; Jones D. M.; Zeltina A.; Bowden T. A.; Padilla-Parra S.; Fry E. E.; Stuart D. I. Toremifene Interacts with and Destabilizes the Ebola Virus Glycoprotein. Nature 2016, 535, 169–172. 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaru A.; Bala C.; Jaffrezic-Renault N.; Aboul-Enein H. Y. Surface Plasmon Resonance (SPR) Biosensors in Pharmaceutical Analysis. Crit Rev Anal Chem 2015, 45, 97–105. 10.1080/10408347.2014.881250. [DOI] [PubMed] [Google Scholar]
- Yadav S. P.; Bergqvist S.; Doyle M. L.; Neubert T. A.; Yamniuk A. P. MIRG Survey 2011: Snapshot of Rapidly Evolving Label-free Technologies used for Characterizing Molecular Interactions. J. Biomol. Tech. 2012, 23, 94–100. 10.7171/jbt.12-2303-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshanenko O.; Gmach P.; Winter J.; Kranz D.; Boutros M. Mapping of Wnt-Frizzled Interactions by Multiplex CRISPR Targeting of Receptor Gene Families. FASEB J. 2017, 31, 4832–4844. 10.1096/fj.201700144R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elegheert J.; Behiels E.; Bishop B.; Scott S.; Woolley R. E.; Griffiths S. C.; Byrne E. F. X.; Chang V. T.; Stuart D. I.; Jones E. Y.; Siebold C.; Aricescu A. R. Lentiviral Transduction of Mammalian Cells for Fast, Scalable and High-Level Production of Soluble and Membrane Proteins. Nat. Protoc. 2018, 13, 2991–3017. 10.1038/s41596-018-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im D. U.; Kim S. C.; Chau G. C.; Um S. H. Carbamazepine Enhances Adipogenesis by Inhibiting Wnt/beta-Catenin Expression. Cells 2019, 8, e1460 10.3390/cells8111460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarzadeh L.; Moini Zanjani T.; Sabetkasaei M. Comparison of Anticancer Effects of Carbamazepine and Valproic Acid. Iran Red Crescent Med. J. 2016, 18, e37230 10.5812/ircmj.37230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertilsson L. Clinical Pharmacokinetics of Carbamazepine. Clin. Pharmacokinet. 1978, 3, 128–143. 10.2165/00003088-197803020-00003. [DOI] [PubMed] [Google Scholar]
- Behr C.; Goltzene M. A.; Kosmalski G.; Hirsch E.; Ryvlin P. Epidemiology of Epilepsy. Rev. Neurol. 2016, 172, 27–36. 10.1016/j.neurol.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Hodges S. L.; Lugo J. N. Wnt/beta-Catenin Signaling as a Potential Target for Novel Epilepsy Therapies. Epilepsy Res. 2018, 146, 9–16. 10.1016/j.eplepsyres.2018.07.002. [DOI] [PubMed] [Google Scholar]
- Huang C.; Fu X. H.; Zhou D.; Li J. M. The Role of Wnt/Beta-Catenin Signaling Pathway in Disrupted Hippocampal Neurogenesis of Temporal Lobe Epilepsy: a Potential Therapeutic Target?. Neurochem Res. 2015, 40, 1319–1332. 10.1007/s11064-015-1614-1. [DOI] [PubMed] [Google Scholar]
- Pirone A.; Alexander J.; Lau L. A.; Hampton D.; Zayachkivsky A.; Yee A.; Yee A.; Jacob M. H.; Dulla C. G. APC Conditional Knock-Out Mouse Is a Model of Infantile Spasms with Elevated Neuronal Beta-Catenin Levels, Neonatal Spasms, and Chronic Seizures. Neurobiol. Dis. 2017, 98, 149–157. 10.1016/j.nbd.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorkowski A. R.; Thio L. L.; Dobyns W. B. Genetic and Biologic Classification of Infantile Spasms. Pediatr. Neurol. 2011, 45, 355–367. 10.1016/j.pediatrneurol.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Zhang X.; Wu Y.; Zhao B.; Liu X.; Pan Y.; Liu Y.; Ding Y.; Qiu M.; Wang Y. Z.; Zhao G. Wnt/beta-Catenin Signaling Mediates the Seizure-Facilitating Effect of Postischemic Reactive Astrocytes after Pentylenetetrazole-Kindling. Glia 2016, 64, 1083–1091. 10.1002/glia.23073. [DOI] [PubMed] [Google Scholar]
- Qu Z.; Su F.; Qi X.; Sun J.; Wang H.; Qiao Z.; Zhao H.; Zhu Y. Wnt/beta-Catenin Signalling Pathway Mediated Aberrant Hippocampal Neurogenesis in Kainic Acid-Induced Epilepsy. Cell Biochem. Funct. 2017, 35, 472–476. 10.1002/cbf.3306. [DOI] [PubMed] [Google Scholar]
- Rubio C.; Rosiles-Abonce A.; Trejo-Solis C.; Rubio-Osornio M.; Mendoza C.; Custodio V.; Martinez-Lazcano J. C.; Gonzalez E.; Paz C. Increase Signaling of Wnt/beta-Catenin Pathway and Presence of Apoptosis in Cerebellum of Kindled Rats. CNS Neurol. Disord. Drug Targets 2017, 16, 772–780. 10.2174/1871527316666170117114513. [DOI] [PubMed] [Google Scholar]
- Busceti C. L.; Biagioni F.; Aronica E.; Riozzi B.; Storto M.; Battaglia G.; Giorgi F. S.; Gradini R.; Fornai F.; Caricasole A.; Nicoletti F.; Bruno V. Induction of the Wnt Inhibitor, Dickkopf-1, Is Associated with Neurodegeneration Related to Temporal Lobe Epilepsy. Epilepsia 2007, 48, 694–705. 10.1111/j.1528-1167.2007.01055.x. [DOI] [PubMed] [Google Scholar]
- Catterall W. A. Molecular Properties of Brain Sodium Channels: An Important Target for Anticonvulsant Drugs. Adv. Neurol. 1999, 79, 441–456. [PubMed] [Google Scholar]
- Akhoundi M. S. A.; Sheikhzadeh S.; Mirhashemi A.; Ansari E.; Kheirandish Y.; Allaedini M.; Dehpour A. Decreased Bone Density Induced by Antiepileptic Drugs can Cause Accelerated Orthodontic Tooth Movement in Male Wistar Rats. Int. Orthod. 2018, 16, 73–81. 10.1016/j.ortho.2018.01.021. [DOI] [PubMed] [Google Scholar]
- Coppola G.; Fortunato D.; Auricchio G.; Mainolfi C.; Operto F. F.; Signoriello G.; Pascotto A.; Salvatore M. Bone Mineral Density in Children, Adolescents, and Young Adults with Epilepsy. Epilepsia 2009, 50, 2140–2146. 10.1111/j.1528-1167.2009.02082.x. [DOI] [PubMed] [Google Scholar]
- Feldkamp J.; Becker A.; Witte O. W.; Scharff D.; Scherbaum W. A. Long-Term Anticonvulsant Therapy Leads to Low Bone Mineral Density--Evidence for Direct Drug Effects of Phenytoin and Carbamazepine on Human Osteoblast-Like Cells. Exp. Clin. Endocrinol. Diabetes 2000, 108, 37–43. 10.1055/s-0032-1329213. [DOI] [PubMed] [Google Scholar]
- Hoikka V.; Alhava E. M.; Karjalainen P.; Keranen T.; Savolainen K. E.; Riekkinen P.; Korhonen R. Carbamazepine and Bone Mineral Metabolism. Acta Neurol. Scand. 1984, 70, 77–80. 10.1111/j.1600-0404.1984.tb00806.x. [DOI] [PubMed] [Google Scholar]
- Kumandas S.; Koklu E.; Gumus H.; Koklu S.; Kurtoglu S.; Karakukcu M.; Keskin M. Effect of Carbamezapine and Valproic Acid on Bone Mineral Density, IGF-I and IGFBP-3. J. Pediatr. Endocrinol. Metab 2006, 19, 529–534. [PubMed] [Google Scholar]
- Zhong Z.; Ethen N. J.; Williams B. O. WNT Signaling in Bone Development and Homeostasis. Wiley Interdiscip. Rev. Dev. Biol. 2014, 3, 489–500. 10.1002/wdev.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers J.; Keller J.; Baranowsky A.; Beil F. T.; Catala-Lehnen P.; Schulze J.; Amling M.; Schinke T. Canonical Wnt Signaling Inhibits Osteoclastogenesis Independent of Osteoprotegerin. J. Cell Biol. 2013, 200, 537–549. 10.1083/jcb.201207142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers J.; Schulze J.; Beil F. T.; Gebauer M.; Baranowsky A.; Keller J.; Marshall R. P.; Wintges K.; Friedrich F. W.; Priemel M.; Schilling A. F.; Rueger J. M.; Cornils K.; Fehse B.; Streichert T.; Sauter G.; Jakob F.; Insogna K. L.; Pober B.; Knobeloch K. P.; Francke U.; Amling M.; Schinke T. Control of Bone Formation by the Serpentine Receptor Frizzled-9. J. Cell Biol. 2011, 192, 1057–1072. 10.1083/jcb.201008012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann A.; Schinke T.; Bindl R.; Wehner T.; Rapp A.; Haffner-Luntzer M.; Nemitz C.; Liedert A.; Amling M.; Ignatius A. The Wnt Serpentine Receptor Frizzled-9 Regulates New Bone Formation in Fracture Healing. PLoS One 2013, 8, e84232 10.1371/journal.pone.0084232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen J. K.; Tammela T. L.; Auvinen A.; Murtola T. J. Antiepileptic Drugs with Histone Deacetylase Inhibition Activity and Prostate Cancer Risk: a Population-based Case-Control Study. Cancer Causes Control 2016, 27, 637–645. 10.1007/s10552-016-0737-2. [DOI] [PubMed] [Google Scholar]
- Stettner M.; Kramer G.; Strauss A.; Kvitkina T.; Ohle S.; Kieseier B. C.; Thelen P. Long-term Antiepileptic Treatment with Histone Deacetylase Inhibitors may Reduce the Risk of Prostate Cancer. Eur. J. Cancer Prev. 2012, 21, 55–64. 10.1097/CEJ.0b013e32834a7e6f. [DOI] [PubMed] [Google Scholar]
- Meng Q.; Chen X.; Sun L.; Zhao C.; Sui G.; Cai L. Carbamazepine Promotes Her-2 Protein Degradation in Breast Cancer Cells by Modulating HDAC6 Activity and Acetylation of Hsp90. Mol. Cell Biochem. 2011, 348, 165–171. 10.1007/s11010-010-0651-y. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Ren J.; Padilla-Parra S.; Fry E. E.; Stuart D. I. Lysosome Sorting of Beta-Glucocerebrosidase by LIMP-2 Is Targeted by the Mannose 6-Phosphate Receptor. Nat. Commun. 2014, 5, 4321 10.1038/ncomms5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. M.; Krohn J.; Bhattacharya S.; Davies B. A Comparison of Exogenous Promoter Activity at the ROSA26 Locus Using a PhiC31 Integrase Mediated Cassette Exchange Approach in Mouse ES Cells. PLoS One 2011, 6, e23376 10.1371/journal.pone.0023376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Bishop B.; Clay J. E.; Lu W.; Jones M.; Daenke S.; Siebold C.; Stuart D. I.; Jones E. Y.; Aricescu A. R. Automation of Large Scale Transient Protein Expression in Mammalian Cells. J. Struct. Biol. 2011, 175, 209–215. 10.1016/j.jsb.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G.; Lobley C. M.; Prince S. M. Decision Making in Xia2. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 1260–1273. 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Lebedev A. A.; Isupov M. N. Space-Group and Origin Ambiguity in Macromolecular Structures with Pseudo-Symmetry and its Treatment with the Program Zanuda. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2014, 70, 2430–2443. 10.1107/S1399004714014795. [DOI] [PubMed] [Google Scholar]
- Aricescu A. R.; Lu W.; Jones E. Y. A Time- and Cost-Efficient System for High-Level Protein Production in Mammalian Cells. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006, 62, 1243–1250. 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Howarth M.; Liu W.; Puthenveetil S.; Zheng Y.; Marshall L. F.; Schmidt M. M.; Wittrup K. D.; Bawendi M. G.; Ting A. Y. Monovalent, Reduced-size Quantum Dots for Imaging Receptors on Living Cells. Nat. Methods 2008, 5, 397–399. 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R.; Kaykas A.; Moon R. T.; Perrimon N. Functional Genomic Analysis of the Wnt-Wingless Signaling Pathway. Science 2005, 308, 826–833. 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- Willert K.; Brown J. D.; Danenberg E.; Duncan A. W.; Weissman I. L.; Reya T.; Yates J. R 3rd.; Nusse R. Wnt Proteins are Lipid-modified and can Act as Stem Cell Growth Factors. Nature 2003, 423, 448–452. 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.