Highlights
-
•
The most common foodborne viral syndromes are gastroenteritis and hepatitis.
-
•
Riskiest foods include bivalve mollusks, soft fruits and ready-to-eat products.
-
•
Reference methods are now ready for hepatitis A and norovirus detection in food.
-
•
Hepatitis E virus is regarded as an emerging zoonotic foodborne threat.
-
•
NGS approaches are technically challenging but highly promising for outbreak biotracing.
Abstract
Among the wide variety of viral agents liable to be found as food contaminants, noroviruses and hepatitis A virus are responsible for most well characterized foodborne virus outbreaks. Additionally, hepatitis E virus has emerged as a potential zoonotic threat.
Molecular methods, including an ISO standard, are available for norovirus and hepatitis A virus detection in foodstuffs, although the significance of genome copy detection with regard to the associated health risk is yet to be determined through viability assays.
More precise and rapid methods for early foodborne outbreak investigation are being developed and they will need to be validated versus the ISO standard. In addition, protocols for next-generation sequencing characterization of outbreak-related samples must be developed, harmonized and validated as well.
Current Opinion in Food Science 2016, 8:110–119
This review comes from a themed issue on Food microbiology
Edited by Luca Cocolin and Kalliopi Rantsiou
For a complete overview see the Issue and the Editorial
Available online 16th April 2016
http://dx.doi.org/10.1016/j.cofs.2016.04.002
2214-7993/© 2016 Elsevier Ltd. All rights reserved.
Introduction
A wide variety of viruses may be foodborne transmitted (Table 1 ). These viruses belong to numerous different families and the diseases associated with their infection may range from mild diarrhea to severe neural diseases, flaccid paralysis, with even rare events of myocarditis, respiratory disease or hemorrhagic fever. Nevertheless, the most frequently reported foodborne syndromes are gastroenteritis and hepatitis. This review will be focused on the viruses most commonly found as food contaminants, noroviruses (NoV); the virus causing the most abundant type of hepatitis, hepatitis A virus (HAV); and on another hepatitis virus that represents an emerging foodborne threat, hepatitis E virus (HEV).
Table 1.
Viruses that may be foodborne transmitted.
| Primary tissue tropism | Common name | Particle/genome | Genus | Family | Associated disease(s) |
|---|---|---|---|---|---|
| Enterotropic | Human norovirus | Nonenveloped/ssRNA | Norovirus | Caliciviridae | Gastroenteritis |
| Human sapovirus | Nonenveloped/ssRNA | Sapovirus | Caliciviridae | Gastroenteritis | |
| Aichi virus | Nonenveloped/ssRNA | Kobuvirus | Picornaviridae | Gastroenteritis | |
| Human astrovirus | Nonenveloped/ssRNA | Mamastrovirus | Astroviridae | Gastroenteritis | |
| Human rotavirus | Nonenveloped/segmented dsRNA | Rotavirus | Reoviridae | Gastroenteritis | |
| Human reovirus | Nonenveloped/segmented dsRNA | Orthoreovirus | Reoviridae | Unknown | |
| Human enteric adenovirus | Nonenveloped/dsDNA | Mastadenovirus | Adenoviridae | Gastroenteritis, fever, respiratory disease | |
| Human parvovirus | Nonenveloped/ssDNA | Parvovirus | Parvoviridae | Gastroenteritis | |
| Human picorbirnavirus | Nonenveloped/segmented dsRNA | Picobirnavirus | Picobirnaviridae | Gastroenteritis? | |
| Hepatotropic | Hepatitis A virus | Nonenveloped/ssRNA | Hepatovirus | Picornaviridae | Hepatitis |
| Hepatitis E virus | Nonenveloped/ssRNA | Orthohepevirus | Hepeviridae | Hepatitis | |
| Neurotropic | Poliovirus | Nonenveloped/ssRNA | Enterovirus | Picornaviridae | Flaccid paralysis, meningitis, fever |
| Non-polio enteroviruses (incl. Coxsackie A and B virus, Echovirus, and Enterovirus D68 and 71) | Nonenveloped/ssRNA | Enterovirus | Picornaviridae | Meningitis, herpangina, flaccid paralysis, cranial nerve dysfunction, hand-foot-and-mouth disease, myocarditis, heart anomalies, respiratory illness, rush, pleurodynia | |
| Human parechovirus | Nonenveloped/ssRNA | Parechovirus | Picornaviridae | Meningitis, respiratory disease, gastroenteritis | |
| Nipah virus | Enveloped/ssRNA | Henipavirus | Paramyxoviridae | Encephalitis, respiratory disease | |
| Polyoma virus (JC, BK) | Nonenveloped/circular dsDNA | Polyomavirus | Polyomaviridae | Persistent infections, progressive multifocal leukoencephalopathy, urinary track diseases | |
| Tick-borne encephalitis virus | Nonenveloped/ssRNA | Flavivirus | Flaviviridae | Encephalitis, meningitis | |
| Pneumotropic | Human coronavirus (incl. SARS and MERS CoV) | Enveloped/ssRNA | Betacoronavirus | Coronaviridae | Respiratory disease, SARS, MERS, gastroenteritis |
| Avian influenza virus | Enveloped/segmented ssRNA | Influenzavirus A | Orthomyxoviridae | Influenza, respiratory disease | |
| Multitropic | Ebola virus | Enveloped/ssRNA | Ebolavirus | Filoviridae | Gastroenteritis, hemorrhagic fever |
Despite food is nowadays safer than ever, foodborne diseases are still an important cause of morbidity and mortality, although the actual global burden of unsafe food consumption remains hard to estimate [1]. Several factors, among them the increasing population and the demand for continuous availability of seasonal products all year-around, lead to global food trade among regions with different hygienic standards and the vulnerability of the food supply.
The World Health Organization (WHO) Foodborne Disease Burden Epidemiology Reference Group provided in 2015 the first estimates of global foodborne disease incidence, mortality, and disease burden in terms of Disability Adjusted Life Years (DALYs) [1]. The global burden of foodborne hazards was 33 million DALYs in 2010 (95% uncertainty interval [UI] 25–46); 40% affecting children under 5 years of age. The US Centers for Disease Control and Prevention (CDC) estimates that each year roughly 48 million people in the US gets sick, 128 000 are hospitalized, and 3000 die from foodborne diseases (http://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html).
Foodborne virus transmission
Figure 1 depicts the routes of enteric virus transmission, which essentially is through the fecal–oral route. Patients suffering from viral gastroenteritis may shed very high numbers of viruses in their feces, for example, may reach over 1010 NoV genome copies per gram (gc/g) of stool [2], while it is estimated that as many as 3 × 107 virus particles are released in a single episode of vomiting [3]. Fecal shedding of HAV reaches its maximum, up to 1011 gc/g just before the onset of symptoms, at which point there is the maximum risk of fecal–oral transmission [4•]. For HEV, peak shedding of the virus (around 108 gc/g) occurs during the incubation period and early acute phase of disease [5].
Figure 1.
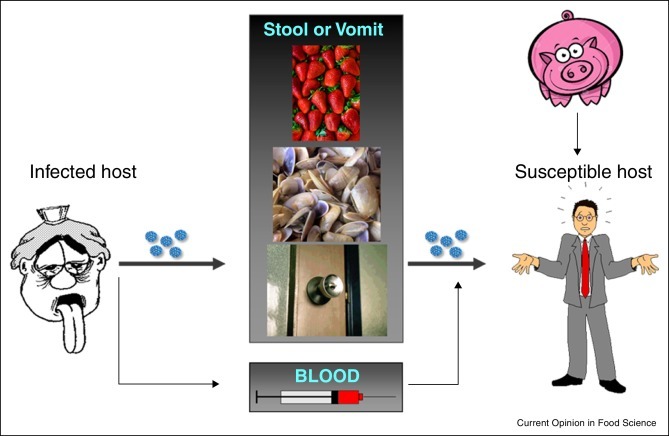
Routes of enteric virus transmission (see text for details).
Viruses may contaminate a wide variety of food products at pre-harvest or post-harvest stages. Among those foods at risk of pre-harvest contamination, bivalve molluscan shellfish and fruits are most commonly associated with foodborne outbreaks. The 2014 report on virus alerts in Europe of the Rapid Alert System for Food and Feed (RASFF, http://ec.europa.eu/food/safety/rasff/), bivalve mollusks were involved in 85% of the alerts, while fruits accounted for 15% of the alerts. Among bivalves, clams, usually imported frozen, caused 57% of all foodborne alerts, followed by oysters (15%) and mussels (11%). Among fruits, frozen strawberries and raspberries were involved each in 5% of all foodborne alerts, while 3% of the alerts involved frozen berry mix.
Post-harvest contamination results most likely from poor hygiene practices during food handling, and hence the foods most at risk are uncooked or lightly cooked products. Surfaces employed for food preparation, as well as other types of fomites, may act as vehicles for foodborne virus transmission. Some enteric viruses, for example, HAV and HEV, may also be parentally transmitted. Foodborne infection can also be acquired, although much more rarely, through ingestion of products from an animal infected with a zoonotic virus, as has been documented for HEV after consumption of pork, wild boar or deer [6, 7•].
Norovirus gastroenteritis
NoV gastroenteritis by itself contributes to an estimated 7.6% of the total DALYs global burden of foodborne disease [1]. Gastroenteritis refers to any inflammatory process of the enteric track although the term is mostly employed to describe acute diarrhea, frequently accompanied by vomiting, nausea and abdominal pain [8]. Gastroenteritis may be non-invasive, inflammatory or invasive. Although bacteria usually cause the most severe cases of invasive gastroenteritis, viruses, more precisely NoV, are responsible for the largest number of cases, usually non-inflammatory episodes of gastroenteritis (http://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html) [3]. More severe disease and cases of chronic gastroenteritis may also occur among the elderly and immunosuppressed [9••].
Out of all foodborne alerts reported in 2014 by the RASFF (http://ec.europa.eu/food/safety/rasff/), NoV accounted for 92% of these alerts. In addition, NoV is nowadays the leading cause of acute gastroenteritis among children less than 5 years of age who seek medical care [10]. In the US, NoV are the foremost cause of domestically acquired foodborne infections in general, and the second cause of domestically acquired foodborne illness resulting in hospitalization (http://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html).
NoV are genetically and antigenically highly diverse agents distributed into seven genogroups (GI to GVII) with altogether more than 30 genotypes distributed worldwide, and with GI, GII and GIV infecting humans [11, 12]. In the last decade, strains belonging to GII, genotype 4 (GII.4) accounted for the majority of cases worldwide, with pandemic GII.4 strains periodically emerging and replacing the previous predominant strain [13, 14]. A new variant (GII.17) causing outbreaks has recently emerged in China and Japan replacing the previously dominant GII.4 genotype Sydney 2012 variant in some areas in Asia, although it has been reported in only a limited number of cases on other continents [15].
Susceptibility to NoV infection is related to histo-blood group antigens (HBGAs), which act as co-receptor factors for these viruses [16]. Expression of HBGAs is determined by the FUT2 gene and resistance to NoV infection follows a Mendelian pattern. NoV fulfill the following axioms: (i) different specific glycans are employed by different NoV strains, (ii) everyone may be infected by a specific NoV strain, and (iii) no NoV strain is able to infect all the human population.
Asymptomatic NoV shedding by healthy individuals is common, with a reported prevalence ranging from 1 to 16% in different parts of the world [2]. In a screening performed in food handlers and healthcare workers related with outbreaks, around 60% shed NoV and 70% of them were asymptomatic shedders (Sabrià et al., submitted).
Nevertheless, irrigation with sewage contaminated water rather than from a single infected food handler seems to be the main cause of large outbreaks of NoV gastroenteritis linked to soft fruit consumption, such as the large outbreak affecting 11,000 individuals in Germany caused by imported frozen strawberries [17]. The detection of several different genotypes in the strawberries reinforce the idea that sewage contamination originated the outbreak. Bivalve shellfish grown and harvested from sewage-contaminated waters have also been recognized as frequent vehicles for NoV transmission [18]. In vitro, in vivo and environmental studies have demonstrated that some bivalve mollusks may selectively accumulate NoV strains due to the presence in bivalve tissues of HBGAs shared with humans [19].
The fecal virus titers after natural and experimental infection is around 105–1010 gc/g without significant differences between symptomatic and asymptomatic individuals [2] (Sabrià et al., submitted). Besides being transmitted by the fecal–oral route, NoV are readily spread through the release of enormous amounts in episodes of projectile vomiting. Having in mind their low infectious dose, reported to be between 20 and 1300 particles [20•], it is obvious that vomiting greatly contributes to the spread of NoV gastroenteritis in closed settings such as restaurants, hotels, health care facilities and cruise ships [3]. In fact, NoV genomes have been detected in the air of healthcare facilities during outbreaks [21].
There are presently several on-going efforts to develop vaccines to prevent NoV infections. Besides generating systemic and mucosal immune responses, intranasal vaccination with virus-like particles (VLP) corresponding to NoV GI.1 (Norwalk virus) reduces the symptoms of illness by more than 50%. However, despite the substantial impact on morbidity that NoV vaccines could have, extensive work is required to target multiple genotypes of interest.
Hepatitis A
Hepatitis A infection is highly endemic in developing regions while is much less frequent in developed regions. This epidemiological pattern has important implications on the average age of exposure and on the severity of the clinical disease. The infection is mostly asymptomatic in children younger than six while the severity increases thereafter, being the illness very severe in those older than sixty (http://www.who.int/mediacentre/factsheets/fs328/en/). Since the infection induces a life-long immunity, severe infections among adults are rare in endemic regions where most children are infected early in life. By contrast, in non-endemic developed countries the disease occurs mostly in adulthood, mainly as a consequence of consuming contaminated water or food, traveling to endemic regions or having risky sexual practices and hence the likelihood of developing severe symptomatic illness is high [22].
Despite a nucleotide diversity similar to that of other picornaviruses, capsid structural constraints limit its amino acid variability, and thus HAV exists as a single serotype, with human strains distributed into three genotypes (I, II and III) and seven subgenotypes (IA, IB, IC, IIA, IIB, IIIA and IIIB) [23]. Genotypic characterization is highly relevant to trace the origin of an outbreak [24, 25•], but also to anticipate the severity of cases. Hepatitis cases associated to subgenotype IIIA have been reported to be more severe, with higher alteration of clinical parameters and requiring longer hospitalization. In a study conducted in Catalonia, covering the decade 2004–2013, a significant increase of subgenotype IIIA was detected in 2012 and associated cases were all in toddlers younger than 4 years [26•]. This is an unexpected result since under the age of six years hepatitis A is mostly asymptomatic and thus indicates a more severe outcome compared to other genotypes.
Some studies suggested the association of a given subgenotype to fulminant cases but different conclusions were drawn from studies conducted in different countries, or even in the same country but in different years [27]. We have performed for this revision a meta-analysis with all data included in previous studies to balance the worldwide prevalence of subgenotypes in different years. This meta-analysis has revealed that the prevalence of subgenotypes IA, IB and IIIA is of 66%, 14% and 21%, respectively, while their association to fulminant cases is of 30%, 30% and 41%, respectively. Thus, it can be concluded that fulminant outcomes associate with infections with viruses belonging to subgenotypes IB and IIIA. From this meta-analysis it can also be inferred that subgenotypes IA, IIIA and IB are the most abundant worldwide. Particularly, IA is the most prevalent genotype followed by IB (particularly in Africa) with the exception of the South Asian continent, including India and Pakistan, where IIIA is the most abundant type. However, it should be pointed out that subgenotype IIIA is rapidly spreading to other parts of the world [27, 28].
Several highly effective inactivated vaccines exist, thanks to the occurrence of a single serotype [4•, 23], however, vaccine-escape mutants have been isolated from HIV+ patients [29] who underwent incomplete vaccination. These are optimal conditions for the selection of mutants present in the quasispecies able to escape the neutralization action of antibodies [23]. Fortunately, these vaccine-escape mutants circulated for a short period of time [26•] likely due to a lower fitness than the wild-type viruses [29]. Recently, several HAV-related viruses have been found in seals [30] and small mammals [31•] which indicate that the possibility of emergence of a new serotype through a zoonotic origin cannot be ruled out.
Hepatitis E
HEV infects a wide range of mammalian species, as well as chickens and trouts [32]. HEV infection usually leads to acute hepatitis that can become fulminant, particularly among pregnant women and in patients with preexisting liver disease, or may even evolve to a chronic state, especially in immunosuppressed individuals [33••]. HEV has been shown to produce a range of extra-hepatic manifestations including aplastic anemia, acute thyroiditis, glomerulonephritis as well as neurological disorders such as Guillain-Barré syndrome, neuralgic amyotrophy and encephalitis [34, 35•].
Through whole genome analysis of existing sequences, seven genotypes of mammalian HEV have been established within subgenus Orthohepevirus A [32]. The predominant host species for genotypes in this subgenus are: human for HEV-1 and HEV-2; human, pig, wild boar, rabbit, deer and mongoose for HEV-3; human and pig for HEV-4; wild boar for HEV-5 and HEV-6; and camel for HEV-7. In addition, Orthohepevirus B predominantly infects chickens, Orthohepevirus C rats and ferrets, and Orthohepevirus D bats [32].
Until recently, hepatitis E was considered endemic in developing countries and rare in developed countries, essentially linked to travelers returning from endemic regions. However, recent evidence points that autochthonous HEV infection in developed areas is much more prevalent than previously acknowledged [36]. For instance, in some industrialized parts of Europe, seroprevalence rates higher than 50% of the population are reported [37, 38, 39]. The source and route of these silent infections remain unclear but evidence points to a porcine zoonosis with HEV-3 circulating among European pigs [6, 40].
Sporadic cases of hepatitis E have been clearly linked to the consumption of raw or undercooked animal meats such as pig livers, wild boar, sausages, and deer meats [7•, 41]. HEV is also present in porcine muscle. In addition, since large amounts of viruses excreted in feces, animal manure land application and runoffs can contaminate irrigation and drinking water with concomitant contamination of fresh produce or shellfish. HEV RNA of swine origin has been detected in swine manure, sewage water and oysters, and consumption of contaminated shellfish has been as well implicated in sporadic cases of hepatitis E [42, 43•]. Therefore, the animal strains of HEV pose not only a zoonotic risk but also food and environmental safety concerns.
A recombinant hepatitis E vaccine, HEV 239, has been licensed in China although so far it is not globally available [44].
The tools to control viral contamination
Molecular methods such as quantitative real time RT-PCR (RTqPCR) have been the methods of choice for virological analysis of food due to the low concentration of viruses normally present on contaminated foodstuffs. In addition, the low number of contaminating virus particles may not be uniformly distributed and some of the components in the food matrix may be potent inhibitors of molecular assays. Two International Organization for Standardization (ISO) procedures for quantitative and qualitative detection of NoV (GI and GII) and HAV in selected foodstuffs (soft fruits, salad vegetables and bivalve molluscan shellfish), bottled water and food surfaces were published in 2013 (ISO/TS 15216-1 and ISO/TS 15216-2) [45••]. The availability of these standard methods may set the basis for the formulation of regulatory standards for viruses in food and water in the near future. The following sections describe these validated methods and other published efforts channeled toward the optimization of the virus concentration procedure, as well as approaches aiming at providing added value to the basic viral detection and assisting in the interpretation of a positive result. A general flow chart for the recently developed analytical options for the detection and characterization of viruses in food is shown in Figure 2 .
Figure 2.
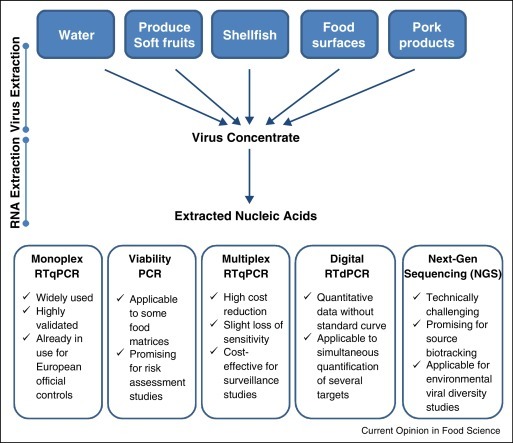
General flow diagram for the analytical options available and under development for the detection and characterization of viruses in food and water. Most protocols involve two steps for virus purification from food or water. The first step extracts and/or concentrates viruses from food or water samples and the second step further purifies and concentrates the viral genomes. Several molecular approaches allowing virus detection, quantification and/or characterization, as well as the most outstanding traits of each methodological approach are summarized.
Validated method for screening NoV and HAV
Box 1 summarizes the key features of the methods and the minimum quality control requirements. With slight modifications in some instances, the ISO methods have been widely used to screen naturally contaminated sample matrices [46, 47], and several companies also offer commercial kits based on RTqPCR assays closely related to the one used in the ISO validation. However, despite the wide acceptance of the standardized method and its variations among research labs, most studies do not report the limit of detection (LOD) and limit of quantification (LOQ) of the assay and this complicates comparisons and drawing of general conclusions. Regarding the process control virus, the use of Mengo virus strain MCo or murine norovirus (MNV) is widespread in most studies [48]. Due to the complexity of food matrices, an extraction efficiency above 1% is considered satisfactory. An assay multiplexing HAV, NoV GI, NoV GII and Mengo virus as a process control was optimized and validated on naturally contaminated bivalve mollusks and water samples [49••]. The quadruplex assay fulfills the ISO requirements, showing in the worst-case scenario an average sensitivity loss of 0.4 logs. Most virus concentration protocols are time-consuming and laborious, and the great variety of food matrices makes selection of a single method not straightforward. For berries, baby spinaches, lettuce and sliced tomatoes for example, alternative methods of extraction in which RNAs are directly extracted from food have shown good performance and shorter times [50, 51]. However, all alternative methods must be validated versus the ISO standard.
Box 1. Key features of the validated ISO/TS 15216-1 and ISO/TS 15216-2 procedures.
| Step 1: Virus extraction and concentration | ||
| Sample size | Method | |
| Soft fruits and salad vegetables | 25 g/chopped | Elution with agitation followed by precipitation with PEG/NaCl |
| Bivalve molluscan shellfish | 2 g digestive gland from 10 animals | Treatment with a proteinase K solution |
| Bottled water | Up to 5 L | Adsorption and elution using positively charged membranes followed by concentration by ultrafiltration |
| Food surfaces | Maximum area 100 cm2 | Swabbing |
| Step 2: RNA extraction | ||
| ✓ Common to all samples | ||
| ✓ Reagents should enable processing of 500 μl of extracted virus | ||
| ✓ Addition of a process control virus | ||
| ✓ Based on virus capsid disruption with chaotropic reagents and adsorption of RNA to silica particles | ||
| Step 3: RTq-PCR | ||
| ✓ One-step RT-qPCR assay | ||
| ✓ Reagents should allow processing of 5 μl RNA in 25 μl total volume | ||
| ✓ Simultaneous monoplex assays for each specific target (NoV GI, NoV GII, HAV and process control virus) | ||
| ✓ Use of hydrolysis probes | ||
| ✓ Addition of an external control RNA (purified single-stranded RNA carrying the target sequence for each target virus) | ||
| ✓ Use of double-stranded DNA control material to make a standard curve | ||
| Step 4: Quality control | ||
| ✓ Virus extraction efficiency should be ≥1% | ||
| ✓ RT-PCR inhibition should be ≤75% | ||
| ✓ Amplification efficiencies should range between 90 and 110% | ||
Special case: in-house protocols to detect HEV in pork products
HEV contamination of meat products is not only restricted to the product surface and hence virus extraction requires other experimental approaches. Although not standardized yet, there are several methods available for detection of HEV in meat and meat products that have been applied to screen retail products in several countries, finding a broad distribution in most cases [41, 52, 53]. There is the need for an ISO standard for HEV detection in food products.
Digital RT-PCR (RTdPCR)
Digital RT-PCR (RTdPCR) is an endpoint quantitative approach that accurately estimates genome copies based on the Poisson distribution. Sensitivity of RTdPCR is comparable to RTqPCR for most targets with increased accuracy since RTqPCR tends to overestimate the number of genome copies in a given sample [54•].
Microfluidic and nanofluidic assays are novel high-throughput methods for simultaneous qualitative detection of numerous pathogens in the same sample, but due to the small volumes of reactions, all reported developments require a pre-amplification step between the RT and the PCR amplification [54•, 55•].
Interpretation of the public health significance of PCR positive results
One of the main pitfalls of the use of molecular methods in food safety is that they do not discern between infectious and noninfectious viruses. The proportion of inactivated viruses in food and water samples may vary depending on how long these viruses have persisted in the environment and/or in the samples. On the other hand, since viral nucleic acids (intact or fragmented) may stand some of the inactivation treatments applied in the food industry, the lack of correlation between genome copies and infectious titers also hampers the assessment of the efficacy of inactivation procedures (extensively reviewed by Ceupens et al., [56•]). Improper interpretation of molecular data can easily lead to wrong decision-making in the food industry and misconceptions of the associated public health risk.
During the last decade, adaptations to the conventional RTqPCR protocols (also denominated ‘viability PCR’ assays) have been developed to reduce and minimize the detection of non-infectious viruses and free nucleic acids (detailed reviews have been published elsewhere [57, 58•]). However, validation of these ‘viability PCR assays’ depends on the use of cell-adapted viral strains, when available, or virus surrogates since none of main foodborne virus threats may be detected by infectivity assays.
Briefly, molecular approaches for predicting virus infectivity examine two major virus characteristics: integrity of the virus genome, and capsid structural stability. Viability dyes such as ethidium monoazide (EMA) or propidium monoazide (PMA) in combination with RTqPCR have been employed to examine capsid integrity and the validity of the approach depends on the target virus and the applied disinfection procedures [59, 60•]. Factors such as the degree of secondary structure present within the target RTqPCR region, its level of interaction and protection by capsid proteins, the mechanical stability or plasticity of the viral capsid, or the level of viral aggregation within the sample, may cause bias in the application of EMA/PMA-RTqPCR methods. Finally, some of the most promising alternative approaches, developed for NoV, are based on integrated in situ capture assays, combining virus binding to cells or to porcine gastric mucin with RTqPCR [61, 62]. Whether ‘viability PCR’ protocols could be used in combination with the ISO extraction/concentration methods requires further investigation.
Biotracing foodborne outbreaks
Genotype and subtype information from food contaminant strains is required to trace the transmission source, and to characterize and compare strains circulating in the environment with strains causing clinical burden, which may help to infer virulence properties of specific strains. Next-generation sequencing (NGS) approaches are progressively replacing standard Sanger sequencing, opening a new era of public health microbiology and outbreak investigations of foodborne pathogens. Thousands of sequence reads are obtained which may belong to different genomic regions enabling multiple genetic characterizations. Depending on virus and regions employed, this approach may provide information on the genetic variability, recombination events, virulence traits and, even for some viruses, geographic tracing. Excellent reviews have been published on the use of NGS platforms for viral diagnostics and pathogen discovery [63••, 64, 65] although it is still unclear whether they could be cost-effective and sensitive enough to be employed in food samples with low levels of contaminant viruses.
Conclusions
The multinational hepatitis A outbreaks occurring in Europe in 2013 and 2014 with over 1400 cases linked to fresh and frozen strawberries and berry mix evidenced the usefulness of virus sequencing to link sporadic cases reported in different EU/EEA countries in outbreaks [66, 67]. However, due to different sequencing practices and protocols in EU/EEA countries, the interpretation of the sequencing results was often challenging and untimely.
Molecular data based on NGS are increasingly replacing the numerous different methodologies currently in use in human and veterinary reference laboratories for outbreak investigation and attribution modeling. These methods have the potential for early identification of foodborne organisms with epidemic character and the resulting data is beginning to be integrated into risk assessment studies. The epidemic potential of a virus genotype or even a subtype, may vary considerably, and is a function of its inherent genetic characteristics and their capacity to mutate, survive and spread through the food chain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported in part by projects Food-FP7-311846 (European Union), BIO2014-53285 (Ministry of Economy and Competitiveness, Spain), 2014SGR00174 and XRB-Biotechnology Reference Network (Generalitat de Catalunya). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- 1.WHO . Geneva; 2015. WHO estimates of the global burden of foodborne diseases. [Google Scholar]
- 2.Teunis P.F., Sukhrie F.H., Vennema H., Bogerman J., Beersma M.F., Koopmans M.P. Shedding of norovirus in symptomatic and asymptomatic infections. Epidemiol Infect. 2015;143:1710–1717. doi: 10.1017/S095026881400274X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall A.J., Wikswo M.E., Pringle K., Gould L.H., Parashar U.D. Vital signs: foodborne norovirus outbreaks — United States, 2009–2012. MMWR Morb Mortal Wkly Rep. 2014;63:491–495. [PMC free article] [PubMed] [Google Scholar]
- 4•.Pintó R., Bosch A., Kaplan G. Hepatitis A: immune response and virus evolution. In: Gershwin M.E., Vierling J.M., Manns M.P., editors. Liver Immunology. Springer International Publishing; 2014. pp. 173–189. [Google Scholar]; A comprehensive overview of HAV evolution and the potential for emergence of vaccine-escape variant.
- 5.Takahashi M., Okamoto H. Features of hepatitis E virus infection in humans and animals in Japan. Hepatol Res. 2014;44:43–58. doi: 10.1111/hepr.12175. [DOI] [PubMed] [Google Scholar]
- 6.Ruggeri F.M., Di Bartolo I., Ponterio E., Angeloni G., Trevisani M., Ostanello F. Zoonotic transmission of hepatitis E virus in industrialized countries. New Microbiol. 2013;36:331–344. [PubMed] [Google Scholar]
- 7•.Van der Poel W.H.M. Food and environmental routes of Hepatitis E virus transmission. Curr Opin Virol. 2014;4:91–96. doi: 10.1016/j.coviro.2014.01.006. [DOI] [PubMed] [Google Scholar]; A good synopsis on the transmission routes of a relevant food zoonosis.
- 8.Morgan D.R., Chidi V., Owen R.L. Gastroenteritis. In: Schlossberg D., editor. Clinical infectious disease. 2nd edn. Cambridge University Press; 2015. pp. 334–341. [Google Scholar]
- 9••.Robilotti E., Deresinski S., Pinsky B.A. Norovirus. Clin Microbiol Rev. 2015;28:134–164. doi: 10.1128/CMR.00075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; A detailed overview of the current knowledge of Norovirus epidemiology, clinical diagnostics and environmental detection.
- 10.Payne D.C., Vinjé J., Szilagyi P.G., Edwards K.M., Staat M.A., Weinberg G.A., Hall C.B., Chappell J., Bernstein D.I., Curns A.T. Norovirus and medically attended gastroenteritis in U.S. Children. New Engl J Med. 2013;368:1121–1130. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroneman A., Vega E., Vennema H., Vinje J., White P.A., Hansman G., Green K., Martella V., Katayama K., Koopmans M. Proposal for a unified norovirus nomenclature and genotyping. Arch Virol. 2013;158:2059–2068. doi: 10.1007/s00705-013-1708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol. 2015;53:373–381. doi: 10.1128/JCM.01535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed S.M., Hall A.J., Robinson A.E., Verhoef L., Premkumar P., Parashar U.D., Koopmans M., Lopman B.A. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabria A., Pintó R.M., Bosch A., Bartolome R., Cornejo T., Torner N., Martinez A., de Simon M., Dominguez A., Guix S. Molecular and clinical epidemiology of norovirus outbreaks in Spain during the emergence of GII.4 2012 variant. J Clin Virol. 2014;60:96–104. doi: 10.1016/j.jcv.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Matsushima Y., Ishikawa M., Shimizu T., Komane A., Kasuo S., Shinohara M., Nagasawa K., Kimura H., Ryo A., Okabe N. Genetic analyses of GII.17 norovirus strains in diarrheal disease outbreaks from December 2014 to March 2015 in Japan reveal a novel polymerase sequence and amino acid substitutions in the capsid region. Euro Surveill. 2015:20. doi: 10.2807/1560-7917.es2015.20.26.21173. [DOI] [PubMed] [Google Scholar]
- 16.Singh B.K., Leuthold M.M., Hansman G.S. Human noroviruses’ fondness for histo-blood group antigens. J Virol. 2015;89:2024–2040. doi: 10.1128/JVI.02968-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernard H., Faber M., Wilking H., Haller S., Hohle M., Schielke A., Ducomble T., Siffczyk C., Merbecks S.S., Fricke G. Large multistate outbreak of norovirus gastroenteritis associated with frozen strawberries, Germany, 2012. Euro Surveill. 2014;19:20719. doi: 10.2807/1560-7917.es2014.19.8.20719. [DOI] [PubMed] [Google Scholar]
- 18.Lowther J.A., Gustar N.E., Hartnell R.E., Lees D.N. Comparison of norovirus RNA levels in outbreak-related oysters with background environmental levels. J Food Prot. 2012;75:389–393. doi: 10.4315/0362-028X.JFP-11-360. [DOI] [PubMed] [Google Scholar]
- 19.Le Guyader F.S., Atmar R.L., Le Pendu J. Transmission of viruses through shellfish: when specific ligands come into play. Curr Opin Virol. 2012;2:103–110. doi: 10.1016/j.coviro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Atmar R.L., Opekun A.R., Gilger M.A., Estes M.K., Crawford S.E., Neill F.H., Ramani S., Hill H., Ferreira J., Graham D.Y. Determination of the 50% human infectious dose for Norwalk virus. J Infect Dis. 2014;209:1016–1022. doi: 10.1093/infdis/jit620. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study provides evidence of the infectious dose for Noroviruses calculated from volunteer studies, which is valuable for future assessing the limit of viral contamination acceptable on foodstufs.
- 21.Bonifait L., Charlebois R., Vimont A., Turgeon N., Veillette M., Longtin Y., Jean J., Duchaine C. Detection and quantification of airborne Norovirus during outbreaks in healthcare facilities. Clin Infect Dis. 2015;61:299–304. doi: 10.1093/cid/civ321. [DOI] [PubMed] [Google Scholar]
- 22.Pérez-Sautu U., Costafreda M.I., Lite J., Sala R., Barrabeig I., Bosch A., Pintó R.M. Molecular epidemiology of hepatitis A virus infections in Catalonia, Spain, 2005–2009: circulation of newly emerging strains. J Clin Virol. 2011;52:98–102. doi: 10.1016/j.jcv.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Pintó R.M., D’Andrea L., Perez-Rodriguez F.J., Costafreda M.I., Ribes E., Guix S., Bosch A. Hepatitis A virus evolution and the potential emergence of new variants escaping the presently available vaccines. Future Microbiol. 2012;7:331–346. doi: 10.2217/fmb.12.5. [DOI] [PubMed] [Google Scholar]
- 24.Pintó R.M., Costafreda M.I., Bosch A. Risk assessment in shellfish-borne outbreaks of hepatitis A. Appl Environ Microbiol. 2009;75:7350–7355. doi: 10.1128/AEM.01177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Severi E., Vennema H., Takkinen J., Lopalco P.L., Coulombier D. Hepatitis A outbreaks. Lancet Infect Dis. 2015;15:632–634. doi: 10.1016/S1473-3099(15)00021-3. [DOI] [PubMed] [Google Scholar]; Lessons learnt from data from large hepatitis A outbreaks.
- 26•.D’Andrea L., Perez-Rodriguez F.J., de Castellarnau M., Manzanares S., Lite J., Guix S., Bosch A., Pintó R.M. Hepatitis A virus genotype distribution during a decade of universal vaccination of preadolescents. Int J Mol Sci. 2015;16:6842–6854. doi: 10.3390/ijms16046842. [DOI] [PMC free article] [PubMed] [Google Scholar]; An analysis of the distribution of HAV genotypes in an area covered by vaccination.
- 27.Miyamura T., Ishii K., Kanda T., Tawada A., Sekimoto T., Wu S., Nakamoto S., Arai M., Fujiwara K., Imazeki F. Possible widespread presence of hepatitis A virus subgenotype IIIA in Japan: recent trend of hepatitis A causing acute liver failure. Hepatol Res. 2012;42:248–253. doi: 10.1111/j.1872-034X.2011.00919.x. [DOI] [PubMed] [Google Scholar]
- 28.Mukomolov S., Kontio M., Zheleznova N., Jokinen S., Sinayskaya E., Stalevskaya A., Davidkin I. Increased circulation of hepatitis A virus genotype IIIA over the last decade in St Petersburg, Russia. J Med Virol. 2012;84:1528–1534. doi: 10.1002/jmv.23378. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Sautu U., Costafreda M.I., Cayla J., Tortajada C., Lite J., Bosch A., Pintó R.M. Hepatitis A virus vaccine escape variants and potential new serotype emergence. Emerg Infect Dis. 2011;17:734–738. doi: 10.3201/eid1704.101169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anthony S.J., Leger St., Liang J.A., Hicks E., Sánchez-León A.L., Jain M.D., Lefkowitch K., Navarrete-Macias J.H., Knowles I., Goldstein N. Discovery of a novel hepatovirus (phopivirus of seals) related to human hepatitis A virus. mBio. 2015:6. doi: 10.1128/mBio.01180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Drexler J.F., Corman V.M., Lukashev A.N., van den Brand J.M.A., Gmyl A.P., Brünink S., Rasche A., Seggewiβ N., Feng H., Leijten L.M. Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci USA. 2015;112:15190–15195. doi: 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Novel insight into the origins of HAV, a human pathogen recovered previously only from primates, by analyzing unpredicted animal reservoirs of viruses closely related with HAV.
- 32.Smith D.B., Simmonds P., Group motICotToVHS, Jameel S., Emerson S.U., Harrison T.J., Meng X.-J., Okamoto H., Van der Poel W.H.M., Purdy M.A. Consensus proposals for classification of the family Hepeviridae. J Gen Virol. 2014;95:2223–2232. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Kamar N., Dalton H.R., Abravanel F., Izopet J. Hepatitis E virus infection. Clin Microbiol Rev. 2014;27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; detailed overview of the current knowledge of HEV epidemiology, clinical diagnostics and environmental detection.
- 34.Kamar N., Abravanel F., Lhomme S., Rostaing L., Izopet J. Hepatitis E virus: chronic infection, extra-hepatic manifestations, and treatment. Clin Res Hepatol Gastroenterol. 2015;39:20–27. doi: 10.1016/j.clinre.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 35•.Dalton H.R., Kamar N., van Eijk J.J., McLean B.N., Cintas P., Bendall R.P., Jacobs B.C. Hepatitis E virus and neurological injury. Nat Rev Neurol. 2016;12:77–85. doi: 10.1038/nrneurol.2015.234. [DOI] [PubMed] [Google Scholar]; An overview of studies on a number of extrahepatic manifestations of HEV infection, including a range of neurological injuries.
- 36.Petrik J., Lozano M., Seed C.R., Faddy H.M., Keller A.J., Prado Scuracchio P.S., Wendel S., Andonov A., Fearon M., Delage G. Hepatitis E. Vox Sang. 2016;110:93–103. doi: 10.1111/vox.12285. [DOI] [PubMed] [Google Scholar]
- 37.Izopet J., Labrique A.B., Basnyat B., Dalton H.R., Kmush B., Heaney C.D., Nelson K.E., Ahmed Z.B., Zaman K., Mansuy J.M. Hepatitis E virus seroprevalence in three hyperendemic areas: Nepal, Bangladesh and southwest France. J Clin Virol. 2015;70:39–42. doi: 10.1016/j.jcv.2015.06.103. [DOI] [PubMed] [Google Scholar]
- 38.Slot E., Hogema B.M., Riezebos-Brilman A., Kok T.M., Molier M., Zaaijer H.L. Silent hepatitis E virus infection in Dutch blood donors, 2011 to 2012. Euro Surveill. 2013:18. doi: 10.2807/1560-7917.es2013.18.31.20550. [DOI] [PubMed] [Google Scholar]
- 39.Hewitt P.E., Ijaz S., Brailsford S.R., Brett R., Dicks S., Haywood B., Kennedy I.T.R., Kitchen A., Patel P., Poh J. Hepatitis E virus in blood components: a prevalence and transmission study in southeast England. Lancet. 2014;384:1766–1773. doi: 10.1016/S0140-6736(14)61034-5. [DOI] [PubMed] [Google Scholar]
- 40.Lhomme S., Top S., Bertagnoli S., Dubois M., Guerin J.L., Izopet J. Wildlife reservoir for hepatitis E virus, Southwestern France. Emerg Infect Dis. 2015;21:1224–1226. doi: 10.3201/eid2107.141909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Bartolo I., Diez-Valcarce M., Vasickova P., Kralik P., Hernandez M., Angeloni G., Ostanello F., Bouwknegt M., Rodriguez-Lazaro D., Pavlik I. Hepatitis E virus in pork production chain in Czech Republic, Italy, and Spain, 2010. Emerg Infect Dis. 2012;18:1282–1289. doi: 10.3201/eid1808.111783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao S., Li D., Zha E., Zhou T., Wang S., Yue X. Surveillance of hepatitis E virus contamination in shellfish in China. Int J Environ Res Public Health. 2015;12:2026–2036. doi: 10.3390/ijerph120202026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Grodzki M., Schaeffer J., Piquet J.-C., Le Saux J.-C., Chevé J., Ollivier J., Le Pendu J., Le Guyader F.S. Bioaccumulation efficiency, tissue distribution, and environmental occurrence of hepatitis E virus in bivalve shellfish from France. Appl Environ Microbiol. 2014;80:4269–4276. doi: 10.1128/AEM.00978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; This surveillance work on hepatitis E virus contamination on shellfish suggests that the role of shellfish as vehicle of transmission of hepatitis E is minor.
- 44.Li S.W., Zhao Q., Wu T., Chen S., Zhang J., Xia N.S. The development of a recombinant hepatitis E vaccine HEV 239. Hum Vaccin Immunother. 2015;11:908–914. doi: 10.1080/21645515.2015.1008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.ISO – International Organization for Standardization: ISO/TS 15216. Microbiology of food and animal feed – Horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR. 2013.; This is the first standardized procedure for qualitative and quantitative virological analysis of certain foodstuffs.
- 46.El-Senousy W.M., Costafreda M.I., Pintó R.M., Bosch A. Method validation for norovirus detection in naturally contaminated irrigation water and fresh produce. Int J Food Microbiol. 2013;167:74–79. doi: 10.1016/j.ijfoodmicro.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 47.Polo D., Varela M.F., Romalde J.L. Detection and quantification of hepatitis A virus and norovirus in Spanish authorized shellfish harvesting areas. Int J Food Microbiol. 2015;193:43–50. doi: 10.1016/j.ijfoodmicro.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Hennechart-Collette C., Martin-Latil S., Guillier L., Perelle S. Determination of which virus to use as a process control when testing for the presence of hepatitis A virus and norovirus in food and water. Int J Food Microbiol. 2015;202:57–65. doi: 10.1016/j.ijfoodmicro.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 49••.Fuentes C., Guix S., Pérez-Rodriguez F.J., Fuster N., Carol M., Pintó R.M., Bosch A. Standardized multiplex one-step qRT-PCR for hepatitis A virus, norovirus GI and GII quantification in bivalve mollusks and water. Food Microbiol. 2014;40:55–63. doi: 10.1016/j.fm.2013.12.003. [DOI] [PubMed] [Google Scholar]; This work describes a multiplex protocol based on the current ISO procedure aiming at reducing the cost of the analysis with minimal losses of sensitivity.
- 50.Perrin A., Loutreul J., Boudaud N., Bertrand I., Gantzer C. Rapid, simple and efficient method for detection of viral genomes on raspberries. J Virol Methods. 2015;224:95–101. doi: 10.1016/j.jviromet.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Hida K., Kulka M., Papafragkou E. Development of a rapid total nucleic acid extraction method for the isolation of hepatitis A virus from fresh produce. Int J Food Microbiol. 2013;161:143–150. doi: 10.1016/j.ijfoodmicro.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 52.Szabo K., Trojnar E., Anheyer-Behmenburg H., Binder A., Schotte U., Ellerbroek L., Klein G., Johne R. Detection of hepatitis E virus RNA in raw sausages and liver sausages from retail in Germany using an optimized method. Int J Food Microbiol. 2015;215:149–156. doi: 10.1016/j.ijfoodmicro.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 53.Martin-Latil S., Hennechart-Collette C., Guillier L., Perelle S. Method for HEV detection in raw pig liver products and its implementation for naturally contaminated food. Int J Food Microbiol. 2014;176:1–8. doi: 10.1016/j.ijfoodmicro.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 54•.Coudray-Meunier C., Fraisse A., Martin-Latil S., Guillier L., Delannoy S., Fach P., Perelle S. A comparative study of digital RT-PCR and RT-qPCR for quantification of hepatitis A virus and Norovirus in lettuce and water samples. Int J Food Microbiol. 2015;201:17–26. doi: 10.1016/j.ijfoodmicro.2015.02.006. [DOI] [PubMed] [Google Scholar]; This work introduces for the first time the use of digital RT-PCR technology for the analysis of viral contamination in foods.
- 55•.Ishii S., Kitamura G., Segawa T., Kobayashi A., Miura T., Sano D., Okabe S. Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples. Appl Environ Microbiol. 2014;80:7505–7511. doi: 10.1128/AEM.02578-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work introduces for the first time the use of microfluidic RT-PCR technology for the qualitative cost-effective analysis of viral contamination targeting over 10 pathogens at the same time.
- 56•.Ceuppens S., Li D., Uyttendaele M., Renault P., Ross P., Ranst M.V., Cocolin L., Donaghy J. Molecular methods in food safety microbiology: interpretation and implications of nucleic acid detection. Compr Rev Food Sci Food Saf. 2014;13:551–577. doi: 10.1111/1541-4337.12072. [DOI] [PubMed] [Google Scholar]; A review highlighting the caveats of molecular procedures for pathogen detection in food and estimation of the associated risk.
- 57.Elizaquível P., Aznar R., Sánchez G. Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J Appl Microbiol. 2014;116:1–13. doi: 10.1111/jam.12365. [DOI] [PubMed] [Google Scholar]
- 58•.Knight A., Li D., Uyttendaele M., Jaykus L.-A. A critical review of methods for detecting human noroviruses and predicting their infectivity. Crit Rev Microbiol. 2013;39:295–309. doi: 10.3109/1040841X.2012.709820. [DOI] [PubMed] [Google Scholar]; Using norovirus as an example of a non-culturable virus, this review broadly covers developed approaches for a better assessment of the risk of infection from foods and the environment.
- 59.Leifels M., Jurzik L., Wilhelm M., Hamza I.A. Use of ethidium monoazide and propidium monoazide to determine viral infectivity upon inactivation by heat, UV-exposure and chlorine. Int J Hyg Environ Health. 2015;218:686–693. doi: 10.1016/j.ijheh.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 60•.Karim M.R., Fout G.S., Johnson C.H., White K.M., Parshionikar S.U. Propidium monoazide reverse transcriptase PCR and RT-qPCR for detecting infectious enterovirus and norovirus. J Virol Methods. 2015;219:51–61. doi: 10.1016/j.jviromet.2015.02.020. [DOI] [PubMed] [Google Scholar]; This study represents a good example of the use of viability PCR to predict infectivity of viruses and for assessing the damage of viral capsid caused by different inactivation mechanisms.
- 61.Dancho B.A., Chen H., Kingsley D.H. Discrimination between infectious and non-infectious human norovirus using porcine gastric mucin. Int J Food Microbiol. 2012;155:222–226. doi: 10.1016/j.ijfoodmicro.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 62.Li D., Baert L., Van Coillie E., Uyttendaele M. Critical studies on binding-based RT-PCR detection of infectious Noroviruses. J Virol Methods. 2011;177:153–159. doi: 10.1016/j.jviromet.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 63••.Gilchrist C.A., Turner S.D., Riley M.F., Petri W.A., Hewlett E.L. Whole-genome sequencing in outbreak analysis. Clin Microbiol Rev. 2015;28:541–563. doi: 10.1128/CMR.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; An overview of the current application of next-generation sequencing technology to microbial whole-genome sequencing and microbial forensics.
- 64.Barzon L., Lavezzo E., Militello V., Toppo S., Palu G. Applications of next-generation sequencing technologies to diagnostic virology. Int J Mol Sci. 2011;12:7861–7884. doi: 10.3390/ijms12117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiapponi C., Pavoni E., Bertasi B., Baioni L., Scaltriti E., Chiesa E., Cianti L., Losio M.N., Pongolini S: Isolation and genomic sequence of hepatitis A virus from mixed frozen berries in Italy. Food Environ Virol. 2014;6:202–206. doi: 10.1007/s12560-014-9149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montano-Remacha C., Ricotta L., Alfonsi V., Bella A., Tosti M., Ciccaglione A., Bruni R., Taffon S., Equestre M., Losio M. Hepatitis A outbreak in Italy. 2013: a matched case-control study. Euro Surveill. 2014:19. [PubMed] [Google Scholar]


