Significance
The SNARE complex plays a central role in exocytosis. Secretagogin (SCGN) modulates exocytosis in multiple cell lines and tissues, and its deficiency has been linked with several neurodevelopmental and neurodegenerative diseases. To precisely define the mechanisms by which SCGN regulates exocytosis, we utilized an array of approaches, including X-ray crystallography, cell biology, and a zebrafish model. We show that SNAP-25 binds SCGN, in a conformation incompatible with assembly with other SNARE proteins. SCGN promotes proper cellular localization of SNAP-25 in multiple SCGN-expressing cell lines. Furthermore, the interaction between SCGN and SNAP-25 (and/or SNAP-23) is critical for neuronal growth and brain development in zebrafish. Altogether, our results highlight the importance of precise regulation of SNARE functions for human development and diseases.
Keywords: regulated exocytosis, SNARE, neurological disorder, SNAP-25, neuronal development
Abstract
Secretagogin (SCGN) is a hexa–EF-hand protein that is highly expressed in the pancreas, brain, and gastrointestinal tract. SCGN is known to modulate regulated exocytosis in multiple cell lines and tissues; however, its exact functions and underlying mechanisms remain unclear. Here, we report that SCGN interacts with the plasma membrane SNARE SNAP-25, but not the assembled SNARE complex, in a Ca2+-dependent manner. The crystal structure of SCGN in complex with a SNAP-25 fragment reveals that SNAP-25 adopts a helical structure and binds to EF-hands 5 and 6 of SCGN. SCGN strongly inhibits SNARE-mediated vesicle fusion in vitro by binding to SNAP-25. SCGN promotes the plasma membrane localization of SNAP-25, but not Syntaxin-1a, in SCGN-expressing cells. Finally, SCGN controls neuronal growth and brain development in zebrafish, likely via interacting with SNAP-25 or its close homolog, SNAP-23. Our results thus provide insights into the regulation of SNAREs and suggest that aberrant synapse functions underlie multiple neurological disorders caused by SCGN deficiency.
Regulated exocytosis is an essential process that secretory cells use to deliver molecules to the extracellular environment. During this process, vesicles carrying diverse bioactive molecules dock and fuse with the plasma membrane (PM) to release their cargoes in response to cellular stimuli. This process is fundamental to many cellular and physiological processes, such as neurotransmitter release, insulin secretion, immune response, and organism development. A central role in neuronal exocytosis is played by the conserved SNARE complex formed by syntaxin-1, SNAP-25, and synaptobrevin, which forms a four-helix bundle that helps to bring the vesicle and plasma membranes into proximity and is crucial for membrane fusion (1–4). N-ethylmaleimide–sensitive factor (NSF) and soluble NSF adaptor proteins (SNAPs) disassemble this complex to recycle the SNAREs for another round of fusion (1, 5), whereas at synapses SNARE complex assembly is orchestrated in an NSF-SNAP–resistant manner by Munc18-1 and Munc13-1 (6, 7). Interestingly, Munc18-1 plays inhibitory (8, 9) and active roles (10–12) in the process. Similar to Munc18-1, other key components of the neurotransmitter release machinery, such as synaptotagmin-1 and complexin-1, have dual roles in exocytosis, suggesting that a balance between inhibition and stimulation is crucial for the exquisite regulation of neurotransmitter release (reviewed in refs. 13 and 14).
In vertebrates, the SNAP-25 family also includes SNAP-23, SNAP-29, and SNAP-47 (15). Whereas both SNAP-25 and SNAP-23 function in driving regulated exocytosis, SNAP-29 and SNAP-47 regulate other cellular events. As the closest homolog of SNAP-25, SNAP-23, is nearly 60% identical with SNAP-25 at the amino acid level. Whereas SNAP-25 is a neuron-specific SNARE protein, SNAP-23 is ubiquitously expressed. In neurons, SNAP-23 is found mainly postsynaptically and functions to regulate trafficking of NMDA and GABAA receptors (16–18).
Although past research has significantly advanced our understanding of SNARE function and regulation, many regulators of exocytosis are still poorly characterized. Particularly important among these regulators is secretagogin (SCGN), a protein highly conserved from fruit flies to humans (19). In humans, SCGN is highly expressed and enriched in pancreatic β-cells, and can also be found in the central nervous systems and enteroendocrine cells (EECs) of the gastrointestinal tract (20). SCGN encodes an EF-hand Ca2+-sensor protein, and is known to interact with cytoskeletal proteins, SNAREs, and cargo proteins (21, 22). Recent studies have revealed that SCGN controls the secretion of multiple hormones in diverse tissues or cells. For example, SCGN suppression in pancreatic β-cells significantly impairs glucose-stimulated insulin secretion (23). Accordingly, SCGN knockout (KO) mice display glucose intolerance at an early life-stage and age-associated progressive hyperglycemia (24). SCGN is also critical for the secretion of corticotropin-releasing hormone and matrix metalloprotease-2 in different types of neurons (25, 26). In addition to its role in exocytosis, SCGN may have a role in diverse cellular processes, including insulin synthesis and function, endoplasmic reticulum stress, and protein folding (27). Emphasizing the importance of SCGN in human physiology is the observation that SCGN deficiency has been linked to the pathogenesis of multiple diseases, such as neurodegeneration, autism, and schizophrenia (19, 28–30). More recently, deficiency in SCGN was shown to contribute to inflammatory bowel disease (31). In contrast with the wealth of information regarding the cellular and physiological functions of SCGN, little is known about the mechanisms by which SCGN regulates exocytosis in diverse cellular events. Similarly, it is unclear how the roles of SCGN in exocytosis are linked with its functions in physiological and developmental processes.
To bridge the gap between the understanding of the functional properties of SCGN and its molecular features, we performed a combination of biochemical, structural, cellular, and zebrafish studies. We show that SCGN binds to SNAP-25, which is mutually exclusive with incorporation of SNAP-25 into the SNARE complex. The crystal structure of SCGN in complex with SNAP-25 in the presence of Ca2+ reveals that SCGN mainly binds to hydrophobic residues of SNAP-25 that are also used to contact other SNAREs in the SNARE complex. In EECs, the interaction between SCGN and SNAP-25 is important for the cellular localization of both proteins, and for their exocytotic functions. Finally, we demonstrate that SCGN is important for brain development in zebrafish, via interaction with SNAP-25 or SNAP-23.
Results
SCGN Interacts with SNAP-25, but Not with the Assembled SNARE Complex.
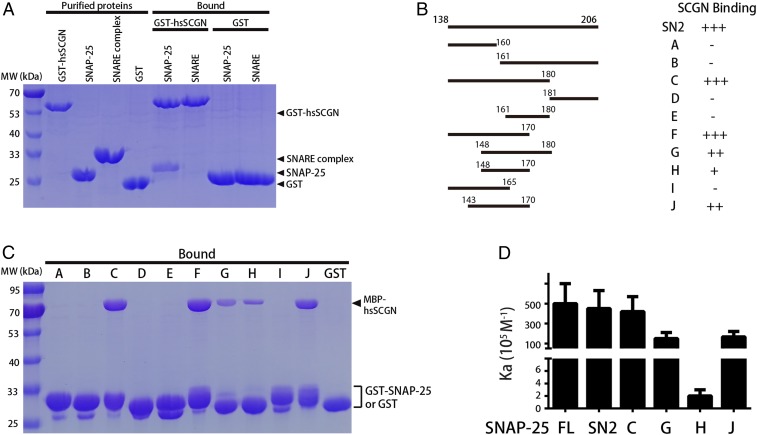
Previous studies have shown that SCGN binds to SNAP-25, among other proteins (21, 22). Since SNAP-25 forms the SNARE complex together with syntaxin-1 and Synaptobrevin, we tested whether SCGN interacted with isolated SNAP-25 or with the SNARE complex. As shown in the pull-down assays of Fig. 1A, GST-human SCGN (hsSCGN) specifically retained SNAP-25, but not the SNARE complex, in a Ca2+-dependent manner. To address which region of SNAP-25 interacts with SCGN, we engineered two SNAP-25 fragments (SN1 and SN2), each of which harbors one SNARE motif. GST pull-down assays revealed that hsSCGN bound to the SN2 fragment, which encompasses the C-terminal SNARE motif, but not the SN1 fragment that spans the N-terminal SNARE motif (SI Appendix, Fig. S1A).
Fig. 1.
Interaction between SCGN and SNAP-25. (A) GST pull-down assays performed with GST-human SCGN (hsSCGN) or GST, and purified full-length SNAP-25 and the assembled SNARE complex (SNARE), in the presence of 2 mM CaCl2 and 0.005% Triton X-100. After incubation with soluble proteins, the resin was extensively washed. The resin-bound proteins were then subjected to SDS/PAGE and Coomassie blue staining. The SNARE complex was assembled using full-length SNAP-25, synaptobrevin (amino acids 29 to 94), and syntaxin 1A (amino acids 191 to 253). (B) Schematic representation of the SNAP-25 fragments used in C and D, and of the amount of binding to SCGN observed: +++, strong affinity; ++,medium affinity; +, weak affinity; −, no interaction detected by pull-down. (C) GST pull-down assays performed with GST–SNAP-25 fragments or GST, and purified MBP-hsSCGN, in the presence of 2 mM CaCl2 and 0.005% Triton X-100. After incubation with soluble proteins, the resin was extensively washed. The resin-bound proteins were then subjected to SDS/PAGE and Coomassie blue staining. (D) Affinity between SNAP-25 fragments and human SCGN (hsSCGN) in the presence of 2 mM CaCl2, determined by ITC. Association constants (Ka) were measured from three independent titrations and shown as mean ± SD.
To further define the region of SN2 that associates with SCGN, we used additional GST pull-down assays and isothermal titration calorimetry (ITC), and examined the binding of hsSCGN to a series of SN2 fragments in the presence of Ca2+ (Fig. 1 B–D). These experiments demonstrated that: 1) Fragment H (amino acids 148 to 170) was the minimal fragment that associates with SCGN; 2) inclusion of additional N-terminal residues (J: amino acids 143 to 170) or C-terminal residues (G: amino acids 148 to 180) could dramatically increase the affinity for hsSCGN about 80-fold (dissociation constant ∼60 nM for G and J, 5 μM for H); 3) fragments F and H showed significantly higher affinity than fragment I, indicating that amino acids 166 to 170 are critical for the binding to SCGN. Finally, fragment J bound to hsSCGN in the presence or absence of Ca2+ with an apparent affinity of 60 nM and 8 μM, respectively, comparable to the measurement with full-length SNAP-25 (21) (SI Appendix, Fig. S1B). Taken together, our biochemical experiments indicate that SNAP-25 interacts with SCGN through a relatively small fragment involving residues 166 to 170 and surrounding residues, which could be buried upon formation of the SNARE complex.
Crystal Structure of the SCGN–SNAP-25 Complex.
To understand the binding mechanism between SCGN and SNAP-25, we performed crystallization trials between human or zebrafish SCGN (drSCGN), and full-length SNAP-25 or its multiple fragments, in the presence of Ca2+. We were able to produce diffraction-quality crystals using drSCGN together with one of the SNAP-25 fragments that we tried, fragment J. The structure of the complex of drSCGN with fragment J was determined by selenium single-wavelength anomalous diffraction (Se-SAD), and refined to a resolution of 2.37 Å (SI Appendix, Fig. S2 and Table S1).
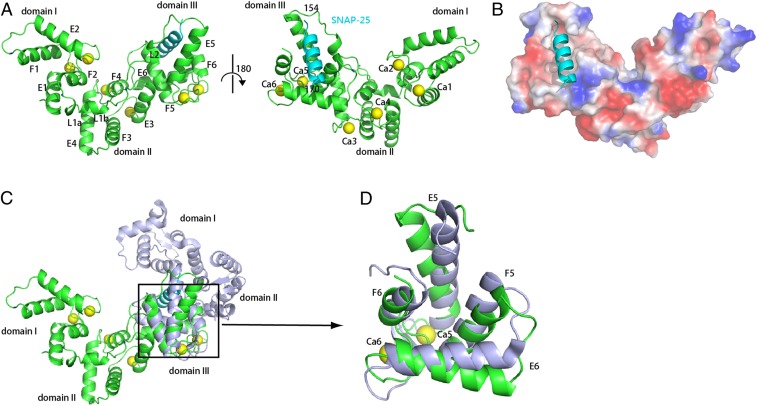
Each asymmetric unit contains two copies of drSCGN and SNAP-25 residues 154 to 170, and 12 calcium ions, with each drSCGN molecule binding to one SNAP-25 fragment and 6 calcium ions (Fig. 2 A and B). Since SCGN predominately exists as a monomer in solution (32), we will focus on the drSCGN monomer and its interaction with SNAP-25 and calcium (the Holo form) in the following discussion.
Fig. 2.
Crystal structure of drSCGN in complex with a SNAP-25 fragment and Ca2+. (A) Ribbon diagrams of drSCGN in complex with a SNAP-25 fragment, shown in two orientations rotated 180° with respect of each other. Green: drSCGN; cyan: SNAP-25 peptide; yellow balls: calcium ions. (B) Electrostatic potential surface of the complex of drSCGN with the SNAP-25 peptide (ribbon diagram). Blue: positive potential; red: negative potential. The complex is shown in the same orientation as that of the right molecule in A. (C) SCGN has different domain arrangement in the Apo (light blue) and Holo forms (green). The two structures were aligned by superimposing their domain III. (D) Overlay of domain III of SCGN in the Apo (light blue) and Holo forms (green).
Previous structural studies revealed that drSCGN consists of three globular domains, and each contains a pair of apposed EF-hand motifs (32). In the absence of Ca2+ (the Apo form), the three domains are arranged into a V-shaped structure (32). The Holo form of drSCGN still adopts a similar V-shape, although the protein undergoes significant structural arrangement throughout its entire protein (Fig. 2C). The most dramatic change is the spatial arrangement of the three domains in the two structures. When domains III of both structures are aligned, domains I and II are rotated nearly 180° with respect to domain III (Fig. 2C).
SCGN contains six EF-hand motifs, and our structure confirms that all of the motifs can bind to a calcium ion. Each calcium ion is coordinated by residues from a 12-residue loop connecting the E helix and F helix, including positions 1, 3, 5, 7, 9, and 12, analogous to the canonical EF-hands (33). The SNAP-25 peptide specifically binds to domain III, which consists of EF5 and EF6 (Fig. 2 A, B, and D). Although we used SNAP-25 J fragment (amino acids 143 to 170) for crystallization, only residues 154 to 170 show clear electron density. Binding to calcium ions and the SNAP-25 peptide leads to significant conformation rearrangements in domain III (Fig. 2D). The root mean square deviation of the C-α carbons of domain III between the Apo and Holo structures is about 4 Å. All four helices from EF5 and EF6 undergo substantial movement, creating a continuous hydrophobic patch to accommodate the SNAP-25 peptide (SI Appendix, Fig. S3 A and B). In addition to EF5 and EF6, the conformations of other EF-hand motifs also change dramatically (SI Appendix, Fig. S3 C–E). In the Apo form, the metal binding loops form a two-stranded antiparallel β-sheet in each domain (32); in the Holo form, all metal binding loops—including those from EF1, EF2, EF3, and EF4—lose the secondary structure, suggesting a cooperativity among different EF-hand motifs.
Interaction between SCGN and SNAP-25.
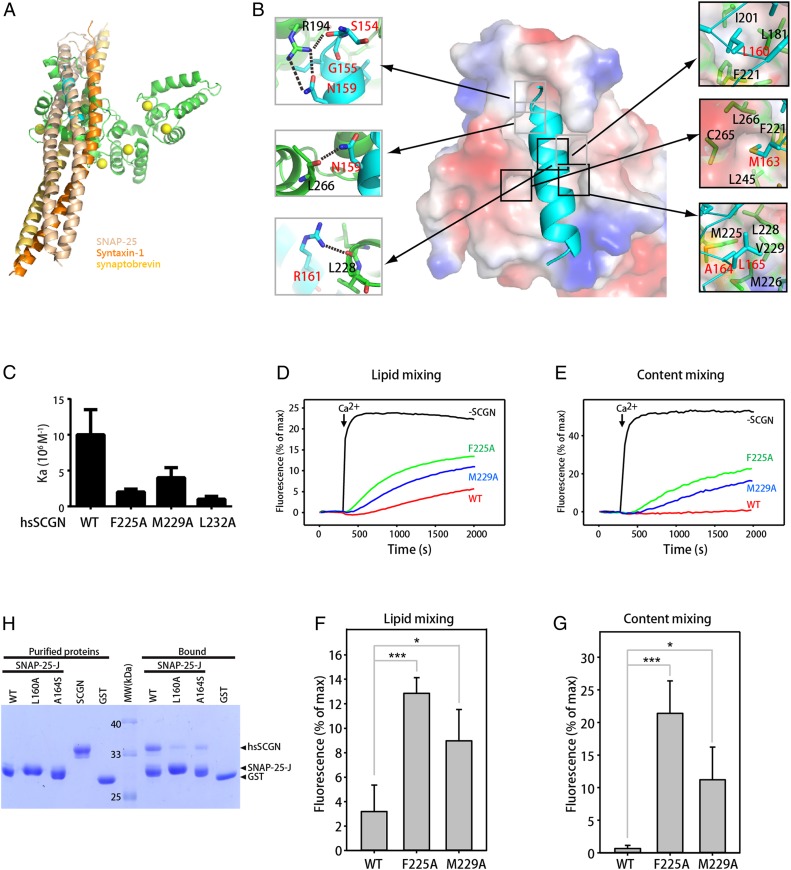
The SNARE complex is a four α-helix bundle, and is composed of layers of interactions, dominated by hydrophobic interactions (4). Most hydrophobic residues of SNAP-25 are buried in the SNARE complex, whereas most of hydrophilic and charged residues face the solvent. Superposition of the crystal structure of the SNARE complex and that of the complex between drSCGN and SNAP-25 fragment J reveals that this segment of SNAP-25 adopts similar conformations in the two structures. The hydrophobic residues of SNAP-25 involved in contacts with SCGN are also involved in interactions with other SNAREs in the SNARE complex (Fig. 3A).
Fig. 3.
Residues of SNAP-25 that mediate SCGN binding are also involved in interactions with other SNAREs in the SNARE complex. (A) Overlay of the structure of the SCGN–SNAP-25 complex with that of the SNARE complex by superimposing the SNAP-25 peptide. Green: drSCGN; cyan: SNAP-25 peptide; wheat: SNAP-25 within the SNARE complex; yellow orange: synaptobrevin; orange: Syntaxin-1; yellow balls: calcium ions. (B) Detailed view of the interactions between domain III of drSCGN and the SNAP-25 peptide. drSCGN is shown in electrostatic potential surface representation (orientation identical to that of Fig. 2B), and SNAP-25 is shown in ribbon representation. Residues forming intramolecular hydrogen bonds (dashed line) and van der Waals’ interactions are shown on the left and right of the main figure, respectively. (C) Affinity between SNAP-25 J and human SCGN (hsSCGN) wild-type or mutants in the presence of 2 mM CaCl2, determined by ITC. Association constants (Ka) were measured from three independent titrations, and shown as mean ± SD. (D–E) Lipid mixing (D) between V- and T-liposomes was monitored from the fluorescence de-quenching of Marina blue lipids and content mixing (E) was monitored from the increase in the fluorescence signal of Cy5-streptavidin trapped in the V-liposomes caused by FRET with PhycoE-biotin trapped in the T-liposomes upon liposome fusion. Assays were performed with V- and T-liposomes in the presence of Munc18-1, M13C1C2BMUNC2C, NSF, αSNAP, and 1.5 μM wild-type, F225A or M229A SCGN. Experiments were started in the presence of 100 μM EGTA and 5 μM streptavidin, and Ca2+ (600 μM) was added at 300 s. (F and G) Quantification of the fusion assays shown in the figure. (D and E) Bars represent averages of the normalized fluorescence intensities observed in lipid mixing and content mixing assays at 2,000 s, performed in triplicates. Error bars represent SDs. Statistical significance and P values were determined by one-way analysis of variance (ANOVA) with Holm-Sidak test (*P < 0.05; ***P < 0.001). (H) GST pull-down assays performed with GST–SNAP-25 J wild-type or mutants, or GST, and purified hsSCGN, in the presence of 2 mM CaCl2 and 0.005% Triton X-100. After incubation with soluble proteins, the resin was extensively washed. The resin-bound proteins were then subjected to SDS/PAGE and Coomassie blue staining.
In the Holo structure, the SNAP-25 peptide is wrapped around and engulfed by domain III of drSCGN (Fig. 2A). Similar to classic EF-hands proteins, drSCGN interacts with SNAP-25 mainly through hydrophobic interactions made by shallow hydrophobic pockets of drSCGN and certain hydrophobic residues of SNAP-25 (Fig. 3B). For example, L160 of SNAP-25 inserts into a hydrophobic pocket made by L181, I201, and F221 of drSCGN. Next to this pocket, F221, L245, C265, and L266 of drSCGN form a second pocket to accommodate M163 of SNAP-25. Furthermore, A164 and L165 of SNAP-25 form a hydrophobic cluster together with M225, M226, L228, and V229 of drSCGN.
In addition to hydrophobic interactions, the interaction between drSCGN and SNAP-25 also involves multiple hydrogen bonds, particularly at the N terminus of the SNAP-25 peptide (Fig. 3B). R194 of drSCGN forms multiple hydrogen bonds with the side chain of N159 and the carboxyl oxygens of S154 of SNAP-25. N159 of SNAP-25 forms another hydrogen bond with the carboxyl oxygen of L266 of drSCGN, in addition to R194. Furthermore, R161 of SNAP-25 also engages with L228 of drSCGN via a hydrogen bond.
To verify our structure, we mutated several SNAP-25–contacting residues in human SCGN and examined their binding to SNAP-25, using GST pull-down and ITC assays (Fig. 3C and SI Appendix, Fig. S4). Human and zebrafish SCGN show 86% amino acid sequence identity within their domain III, with the numbering of human amino acids being four larger than that of zebrafish. All single alanine substitution in hsSCGN, except for V233A, dramatically reduced the binding to SNAP-25 (SI Appendix, Fig. S4). Among them, R198A, F225A, M230A, and L232A showed the most dramatic reduction, with nearly complete loss of the interaction with SNAP-25 when the binding buffer included 0.5% Triton X-100; in contrast, M229A showed reduced affinity while retaining some binding to SNAP-25. Quantitative measurements using ITC under conditions without Triton X-100 revealed a similar trend: hsSCGN L232A, F225A, and M229A reduced the affinity to SNAP-25 about 10-, 5-, and 2.5-fold, respectively, relative to the wild-type protein (Fig. 3C).
To further validate our structural and biochemical observations, we examined how SCGN regulates SNARE-mediated vesicle fusion using reconstitution assays that allow to simultaneously monitor lipid and content mixing, and that incorporate the three neuronal SNAREs together with NSF, αSNAP, Munc18-1, and a fragment spanning the conserved C-terminal region of Munc13-1 (Munc13-1 C1C2BMUNC2C) (34). SCGN wild-type inhibits both SNARE-mediated lipid and content mixing in a concentration-dependent manner (SI Appendix, Fig. S5). Interestingly, hsSCGN showed a dramatic cooperativity in the inhibitory action, as the curves changed drastically when the concentration of hsSCGN was increased from 1 to 1.5 μM (SI Appendix, Fig. S5). The inhibition was consistent with the notion that SCGN competes with synaptobrevin and syntaxin-1 for binding to SNAP-25, thus inhibiting SNARE complex assembly. Critically, both the F225A and M229A mutants showed a diminished inhibition relative to wild-type hsSCGN (Fig. 3 D–G). M229A displayed slightly more inhibition than F225A, in agreement with the observation that M229A bind to SNAP-25 more tightly than F225A (Fig. 3 D–G). Thus, SCGN inhibits SNARE-dependent liposome fusion in vitro through its interaction with SNAP-25.
To test the importance of calcium for SNAP-25 binding, we mutated each of the EF-hand motifs of hsSCGN by converting the first and third positions of the loop to alanines, and tested SNAP-25 binding (SI Appendix, Fig. S6A). Residues at the first and third positions are involved in coordinating calcium, and are highly conserved among the EF-hand motifs (21). Position 1 is especially critical, and harbors an invariant aspartic acid residue (21). The alanine substitutions were thus expected to disrupt calcium binding. Indeed, mutations at loops of EF5 and EF6 completely disrupted the binding between SCGN and SNAP-25, consistent with the notion that both EF5 and EF6 are necessary to contact SNAP-25. Interestingly, mutations at EF1 and EF3, but not EF2 and EF4, also dramatically decreased the amount of SNAP-25 retained, suggesting that different EF-hand motifs of SCGN may coordinate to achieve optimal binding to target proteins. Conversely, alanine substitution of SCGN-interacting residues of SNAP-25, L160A, and A164S, also significantly reduced the binding between SNAP-25 and hsSCGN (Fig. 3H).
Cbp53E, also known as CALB2, is the only hexa–EF-hand protein in Drosophila and shares only 44% amino acid sequence identity with human SCGN (35). It also shares nearly the same identity with two other mammalian hexa–EF-hand proteins, calbindin-D28k and calretinin (35). Thus, it is unclear whether Cbp53E is the genuine ortholog of mammalian SCGN. Cbp53E functions to regulate axonal growth at the neuromuscular junction, and also plays a role in the regeneration of axons and synapses (35, 36); however, it remains to be determined whether Cbp53E functions via SNAP-25 interaction. Utilizing GST pull-down assays, we found that both GST-Cbp53E and GST-hsSCGN proteins, but not GST alone, could retain Drosophila SNAP-25, in a calcium-dependent manner. (SI Appendix, Fig. S6B) Thus, the interaction between SCGN and SNAP-25 is likely to be conserved from fly to human. To probe the interaction between SCGN and other members of the SNAP-25 family, we engineered GST-fused proteins encoding a fragment from each protein equivalent to SNAP-25 J and tested their binding with SCGN in the presence of calcium. The fragment from SNAP-23, but not those from SNAP-29 or SNAP-47, could retain SCGN (SI Appendix, Fig. S6 C and D). Thus, it is likely that SCGN preferentially interacts with SNAP-25 and SNAP-23.
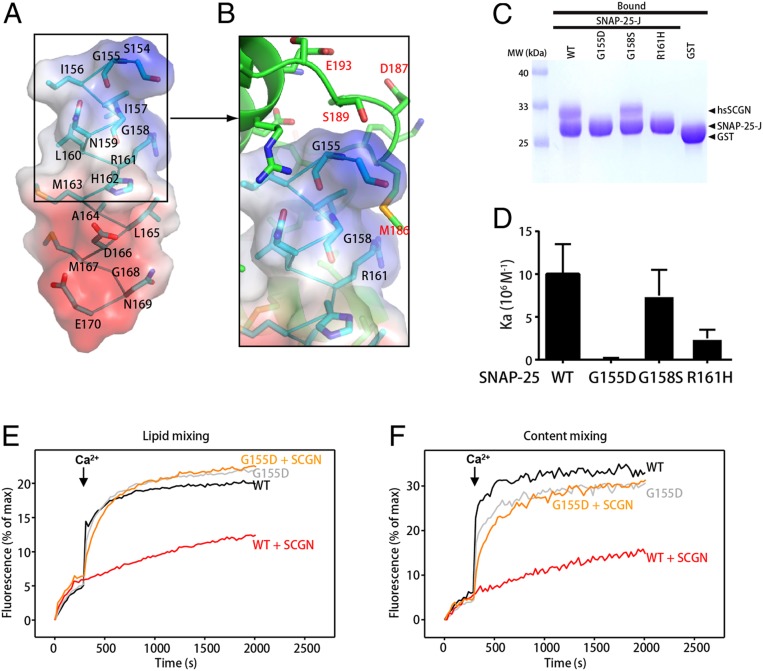
SNAP-25 G155D Specifically Disrupts the Interaction with SCGN without Impairing SNARE-Dependent Liposome Fusion.
Our structural and biochemical studies revealed that the surface of SNAP-25 that contacts SCGN also interacts with other SNARE proteins. To search for possible SNAP-25 mutations that only interfere with the binding to SCGN, we focused on the N terminus of the SNAP-25 J peptide, which harbors multiple residues that interact with SCGN through hydrogen bonds (Fig. 4 A and B). We introduced mutations that could weaken hydrogen bonding (R161H), or create steric clashing (G158S) or electric repulsion (G155D). Indeed, two of the mutants that we generated, G155D and R161H, strongly impaired the binding between SNAP-25 and hsSCGN based on pull-down assays (Fig. 4C). In agreement with these results, quantitative measurements using ITC indicated that R161H reduced the affinity toward hsSCGN about fourfold (Kd ∼ 100 nM for wild-type, Kd ∼ 400 nM for R161H), whereas no apparent binding was observed between hsSCGN and SNAP-25 G155D (Fig. 4D). On the other hand, SNAP-25 G158S appeared to display slightly decreased affinity toward hsSCGN.
Fig. 4.
Identification of SNAP-25 residues that specifically interact with SCGN. (A) Structure of the SNAP-25 peptide (stick presentation) observed in the SCGN–SNAP-25 complex, together with its electrostatic potential surface. Blue: positive potential; red: negative potential. (B) Detailed view of the interactions involving drSCGN and the N terminus of the SNAP-25 peptide. Three SNAP-25 residues (G155, G158, R161) that were mutated in later studies were highlighted. (C) GST pull-down assays performed with GST-SNAP-25 J wild-type or mutants, or GST, and purified hsSCGN, in the presence of 2 mM CaCl2 and 0.5% Triton X-100. After incubation with soluble proteins, the resin was extensively washed. The resin-bound proteins were then subjected to SDS/PAGE and Coomassie blue staining. (D) Affinity between SNAP-25 J fragment wild-type or mutants, and human SCGN (hsSCGN) in the presence of 2 mM CaCl2, determined by ITC. Association constants (Ka) were measured from three independent titrations, and shown as mean ± SD. (E and F) Lipid mixing (E) between V- and T-liposomes was monitored from the fluorescence de-quenching of Marina blue lipids, and content mixing (F) was monitored from the increase in the fluorescence signal of Cy5-streptavidin trapped in the V-liposomes caused by FRET with PhycoE-biotin trapped in the T-liposomes upon liposome fusion. Assays were performed with V- and T-liposomes in the presence of Munc18-1, M13C1C2BMUNC2C, NSF, and αSNAP, with or without 3 μM SCGN. The T-liposomes contained WT or G155D SNAP-25. Experiments were started in the presence of 100 μM EGTA and 5 μM streptavidin, and Ca2+ (600 μM) was added at 300 s.
Since SNAP-25 G155D showed the most dramatic effect in disrupting the interaction with SCGN, we tested whether it altered synaptic vesicle fusion. In the absence of SCGN, G155D displayed curves nearly identical to those of SNAP-25 wild-type for both lipid and content mixings, suggesting that the mutant did not alter normal SNARE complex assembly (Fig. 4 E and F). Importantly, when SCGN was included in the assays, hsSCGN inhibited SNAP-25 wild-type–mediated vesicle fusion but had little effect on SNAP-25 G155D-mediated fusion (Fig. 4 E and F). Thus, SNAP-25 G155D retains normal SNARE complex assembly activity, but specifically loses the SCGN interaction. We envisioned that this mutant would be useful to dissect SCGN functions in cells and in model organisms.
SCGN Regulates the PM Localization of SNAP-25.
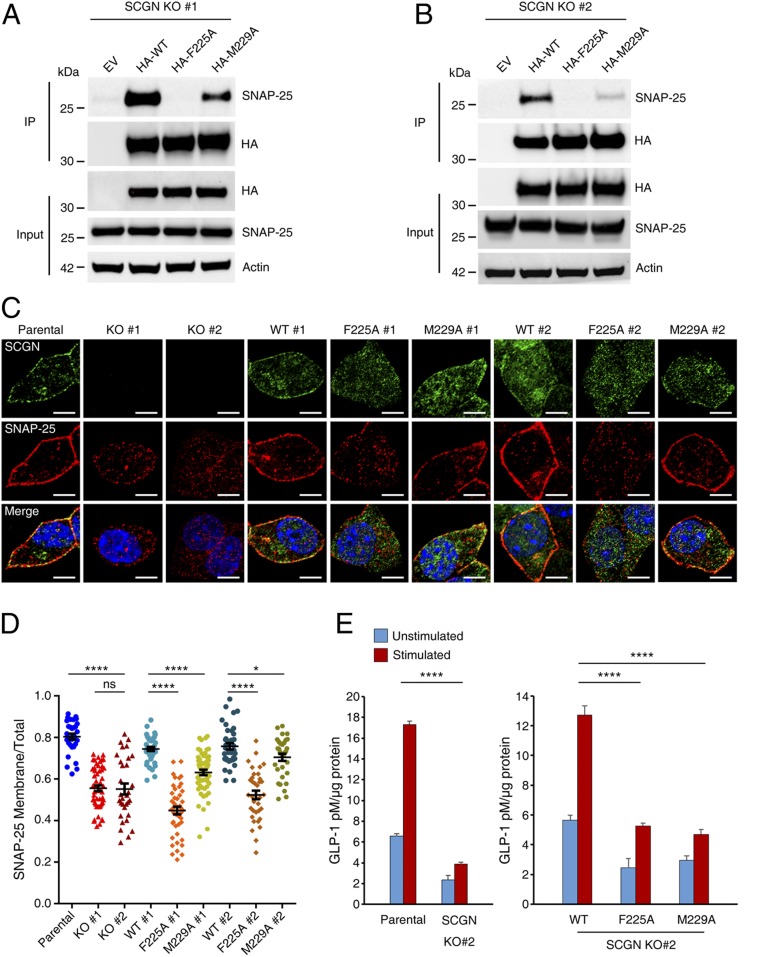
We chose the murine EEC line STC-1 and pancreatic β-cell line NIT-1 as cellular models to investigate the cellular functions of SCGN. STC-1 and NIT-1 cells secrete peptide hormones and insulin, respectively, in response to external signals, and both are known to express SCGN. Using CRISPR-Cas9 technology, we developed two individual STC-1 lines that lacked SCGN expression (SI Appendix, Fig. S7). We also generated rescue cell lines through lentiviral re-expression of HA-tagged wide-type human SCGN and two point-mutant variants (F225A, M229A) (SI Appendix, Fig. S8A). Immunoprecipitation against the HA tag revealed that the F225A mutant completely abolished the interaction with SNAP-25, while the M229A mutant showed reduced binding relative to wild-type SCGN (Fig. 5 A and B). These results are in nice agreement with our in vitro binding data.
Fig. 5.
SCGN controls SNAP-25 subcellular localization and optimal hormone secretion in EECs in a SNAP-25–dependent manner. (A and B) Coimmunoprecipitation (Co-IP) analyses of two SCGN KO STC-1 clones stably expressing N-terminal HA-tagged human wild-type SCGN, SCGN (F225A, M229A) point mutant variants, or empty vector (EV). Cell lysates were precipitated with anti-HA affinity matrix, and probed with anti–SNAP-25 antibody. (C) Parental, two SCGN KO clones, as well as their rescue STC-1 cells were costained with anti-SCGN and anti–SNAP-25 antibodies. Representative cells are shown. (Scale bars, 10 µm.) Blue: DAPI staining. (D) Quantification of SNAP-25 membranous and total cellular fluorescence intensity ratio was calculated from each of the cell lines used in C. On average ∼40 cells were used for analysis in each group. Bars, mean; error bars, SEM; differences among groups by unpaired Student t test. *P < 0.05; ****P < 0.0001; ns, not significant. (E) GLP-1 release assay. Parental, SCGN-KO clone #2 and rescue STC-1 cells seeded on 6-cm dishes were treated with 100 µM DHA or Hepes buffer for 15 min. Supernatants were collected for analysis and values were normalized to protein concentration. Error bar, SEM; differences among stimulated groups using unpaired Student t test. ****P < 0.0001.
In parental STC-1 cells, both SCGN and SNAP-25 showed vesicular and PM localization (Fig. 5 C and D). SCGN KO cells displayed a pronounced loss of accumulation of SNAP-25 at the PM, determined by quantitative immunofluorescence (Fig. 5 C and D). Lentiviral reexpression of SCGN wild-type restored the PM localization of SNAP-25. In contrast, the SCGN F225A mutant completely failed to rescue the SNAP-25 localization to the PM. Finally, the M229A mutant, which still retained some binding toward SNAP-25, could partially rescue the SNAP-25 localization. In contrast with SNAP-25, the PM localization of syntaxin-1 was not altered in SCGN KO or the SCGN mutant rescue cell lines (SI Appendix, Fig. S8 B and C). Thus, SCGN specifically promotes the PM localization of SNAP-25 via a direct interaction in STC-1 cells.
To determine whether SCGN regulate the membrane localization of SNAP-25 in other types of cells, we generated NIT-1 cell lines that displayed loss of SCGN expression (SI Appendix, Fig. S9A). In parental NIT-1 cells, both SCGN and SNAP-25 can be detected on vesicular and plasma membranes, similar to STC-1 cells (SI Appendix, Fig. S9B). The localization of SNAP-25 at the PM was dramatically diminished in the SCGN KO NIT-1 cells, with ∼40% loss of SNAP-25 plasma accumulation (∼75% of SNAP-25 localized at the plasma in parental Vs, 35% in SCGN-KO cells) (SI Appendix, Fig. S9 B and C). Taken together, our data suggest that directing the PM-localization of SNAP-25 is likely to be a general function of SCGN in SCGN-expressing cells.
The SCGN–SNAP-25 Interaction Promotes Hormone Secretion from EECs.
To further assess the cellular function of SCGN, we measured the amount of GLP-1 secreted by STC-1 cells in response to nutrient stimuli (37). Consistent with previous studies (37), about three times of GLP-1 was released into the medium when the STC-1 cells were stimulated by DHA, compared to the basal level (Fig. 5E). Loss of SCGN nearly completely abolished the DHA-mediated GLP-1 release. Critically, the loss of GLP-1 release could be rescued by the lentiviral reexpression of SCGN wild-type, but barely by that of the F225A or M229A mutants (Fig. 5E). Thus, the interaction between SCGN and SNAP-25 is critical for hormone secretion in cultured EECs.
The Association with SNAP-25 Is Critical for the Cellular Localization of SCGN.
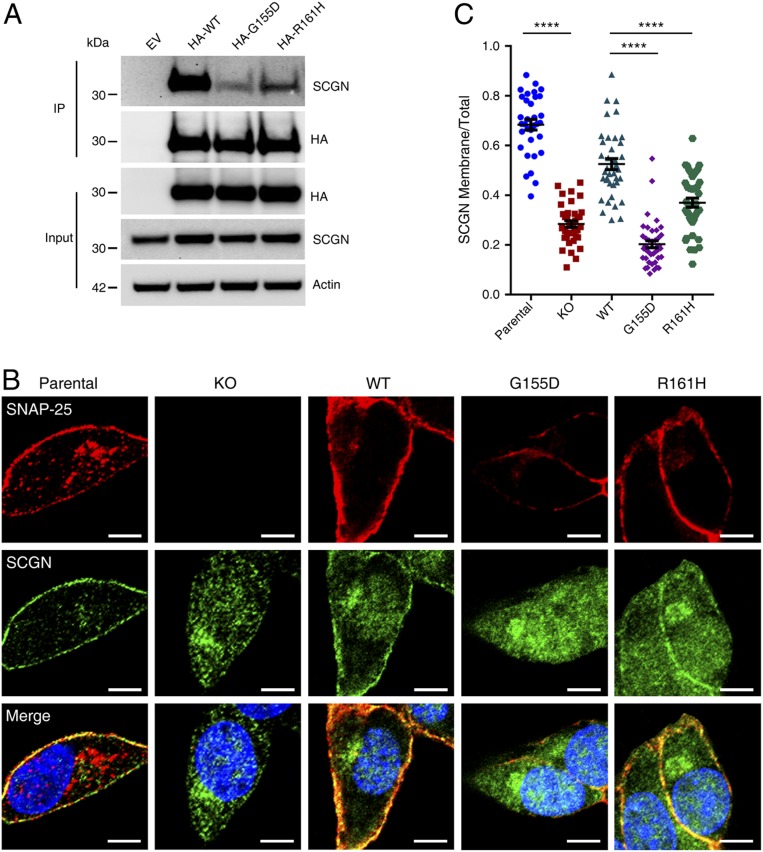
After demonstrating that SCGN promotes the proper subcellular distribution of SNAP-25, we next studied how SNAP-25 regulates the functions of SCGN. SNAP-25 is alternatively spliced, creating two isoforms, SNAP-25a and SNAP-25b (15). The two isoforms share most exons except for exon 5, encoding two proteins of same length but differing in nine amino acids. Using CRISPR/Cas9 targeting the first exon of SNAP-25, we were able to generate STC-1 cells that are deficient of both isoforms. We also generated rescue cell lines through lentiviral reexpression of HA-SNAP-25b (referred as SNAP-25) wild-type, G155D, and R161H. Immunoprecipitation assays against the HA tag revealed that the interaction of SCGN with both the G155D and the R161H mutant versions of SNAP-25 was impaired compared to that with wild-type SNAP-25 (Fig. 6A and SI Appendix, Fig. S10A). The interaction was more severely lost for the G155D mutant, consistent with our measurements with purified proteins.
Fig. 6.
The interaction with SNAP-25 is required for the plasma localization of SCGN. (A) Co-IP analyses of SNAP-25 knockout STC-1 clones stably expressing N-terminal HA tagged SNAP-25 (wild-type, G155D, R161H) or empty vector. Cell lysates were precipitated with anti-HA affinity matrix and probed for endogenous SCGN with anti-SCGN antibody. (B) Parental, SNAP-25 KO and SNAP-25 KO rescue STC-1 cells were costained with anti-SCGN and anti–SNAP-25 antibodies. (Scale bars, 10 µm.) (C) Quantification of SCGN membranous and total cellular fluorescence intensity ratio was calculated from each of the cell lines used in B. On average ∼40 cells were used for analysis in each group. Bars, mean; error bars, SEM; differences among groups by unpaired Student t test. ****P < 0.0001.
To test whether SNAP-25 regulates the subcellular distribution of SCGN, we performed immunofluorescence staining of SNAP-25 and SCGN in parental, SNAP-25 KO, and SNAP-25 rescue cells (Fig. 6B). Whereas SCGN displayed both vesicular and plasma membrane localization in parental cells, its PM distribution was lost in the SNAP-25–deficient cells (Fig. 6 B and C). The loss of PM distribution could be restored upon reexpression of SNAP-25 wild-type, but not the G155D mutant. Interestingly, SCGN PM localization was partially rescued in the SNAP-25 R161H cells, consistent with the finding that the R161H mutant retained some binding toward SCGN. In contrast with SCGN, the loss of SNAP-25 or the mutations in the rescues with SNAP-25 did not alter the cellular localization of syntaxin-1 (SI Appendix, Fig. S10 B and C). Thus, SNAP-25 and SCGN depend on each other to localize on the PM, and their association is necessary for their proper subcellular localization.
SCGN Controls Zebrafish Neuronal Development and Brain Growth Likely through Interaction with SNAP-25 or SNAP-23.
To define the role of SCGN in development, we chose zebrafish as a model organism and studied the functions of SCGN in early embryonic development. Semiquantitative RT-PCR (sqRT-PCR) revealed a highly dynamic temporal expression pattern (SI Appendix, Fig. S11A). No SCGN was detected for the first 10 h (one-cell stage to bud). The expression of SCGN peaked at 24 and 48 h postfertilization (hpf). At 72 hpf, the expression of SCGN started to decrease. Whole-mount in situ hybridization (WISH) showed that SCGN was mainly expressed in the pancreas, but can also be detected in the brain and spinal cord at 24 and 48 hpf (SI Appendix, Fig. S11B). The spatial expression of SCGN in zebrafish is highly similar to that of mammals (20).
Two different morpholinos (MO) were used to knockdown (KD) SCGN: A translation blocking morpholino (MO1) and a splicing blocking one (MO2) (29). To remove undesirable nonspecific effects, we coinjected p53 MO together with MO1 or MO2, which could effectively suppress off-target effects (38) (SI Appendix, Fig. S11C). Since MO1 was more effective in suppressing the expression of SCGN than MO2, most of our experiments were performed with MO1, although injection of MO1 and MO2 led to highly similar phenotypes (SI Appendix, Fig. S11 D and E). To further investigate the function of SCGN, we generated SCGN KO zebrafish lines using CRISPR/Cas9 technology (39). We obtained a mutant line (1-bp mutation in the exon 1 and +29-bp insertion in the exon 2 of SCGN), resulting in a frame shift and a premature stop at codon 105 (SI Appendix, Fig. S12).
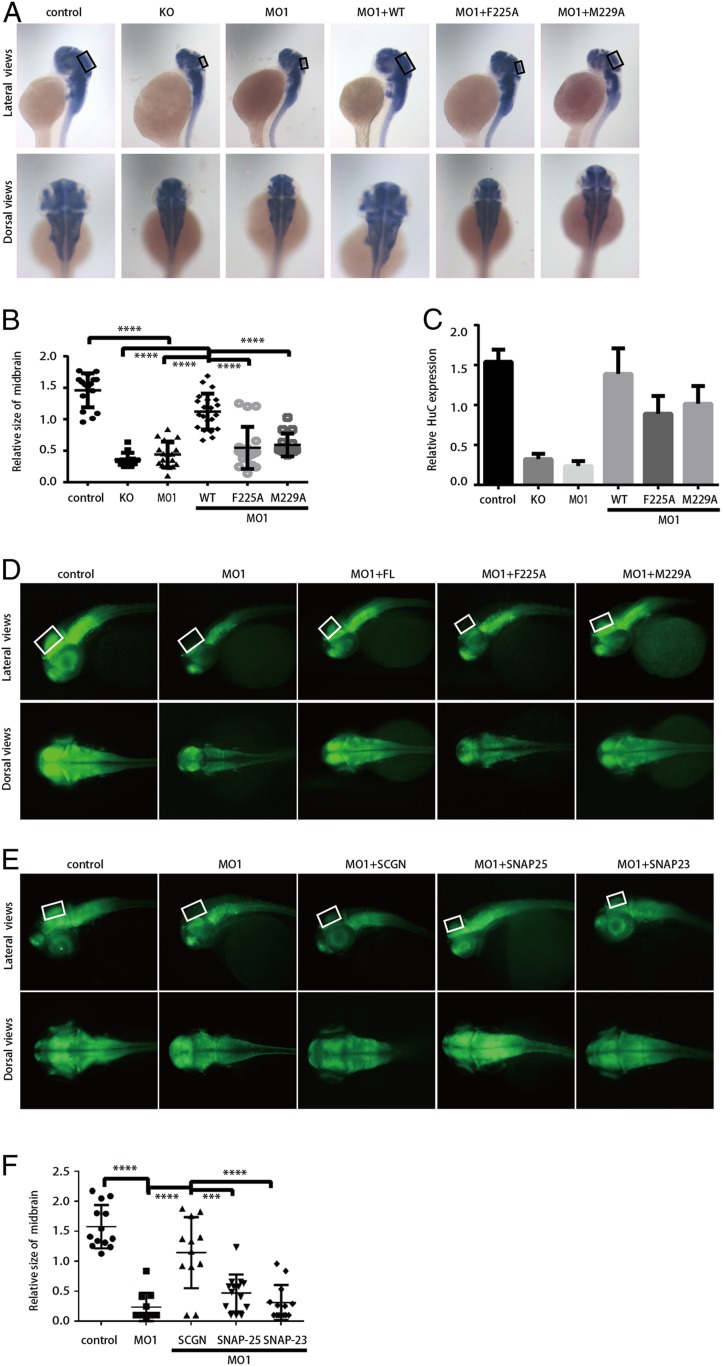
Remarkably, both KO and KD of SCGN using MO1 led to highly similar phenotypes (Fig. 7A). Both embryos were viable, but displayed significantly reduced midbrains, which are only about one-quarter to one-third the size of those of the control group (Fig. 7 A and B). To test whether this phenotype relies on the interaction between SCGN and SNAP-25, we coinjected MO1 together with mRNA encoding human SCGN wild-type, F225A, or M229A (Fig. 7 A and B). hsSCGN wild-type can effectively restore brain size, suggesting that human SCGN can compensate for the loss of zebrafish mRNA. Conversely, injection of mRNAs encoding F225A or M229A, which dramatically reduced the interaction with SNAP-25, failed to rescue the reduction of the midbrain size, confirming the importance of the interaction between SCGN and SNAP-25 for brain growth (Fig. 7 A and B). Furthermore, WISH analysis revealed that the amount of HuC (elavl3), an early marker of pan-neuronal cells, was dramatically decreased in both KO and MO1 embryos (40) (Fig. 7A). Human SCGN wild-type mRNA, but not those of the mutants, was able to restore the expression of HuC (Fig. 7A). sqRT-PCR further confirmed the WISH analysis and revealed a >fourfold reduction in the HuC mRNA level in both KO and MO1 samples (Fig. 7C). The reduction induced by the MO1 could be restored by coinjection of human SCGN WT mRNA, but not those of the mutants (Fig. 7C). In agreement with the WISH and RT-PCR analyses, immunofluorescence also revealed a similar trend in the HuC protein levels (Fig. 7D).
Fig. 7.
SCGN controls brain development in zebrafish through its interaction with SNAP-25 or SNAP-23. (A) WISH staining of HuC (elavl3) in zebrafish embryos at 48 hpf (80× magnification). KO: SCGN knockout; MO1: MO1 injection; MO1+WT, MO1 and hsSCGN wild-type mRNA coinjection; MO1+F225A: MO1 and hsSCGN F225A mutant mRNA coinjection; MO1+M229A: MO1 and hsSCGN M229A mutant mRNA coinjection; MO1+M230A/L232A: MO1 and hsSCGN M230A/L232A mutant mRNA coinjection. All injections were performed at one-cell stage of the development, together with p53 MO. The black rectangles label the position of midbrain. (A, Upper) Lateral view; (Lower) dorsal view. (B) Relative size of midbrain in zebrafish embryos, measured from A. Mean ± SD, ****P < 0.0001. P values were calculated using one-way ANOVA, Tukey's multiple comparisons test. (C) sqRT-PCR analysis of the relative transcription level of HuC. Mean ± SEM, n = 3. (D) HuC (green) expression in Tg[HuC: GFP] transgenic zebrafish (112× magnification). The white rectangles label the position of midbrain. (E) Tg[HuC: GFP] transgenic zebrafish was injected with control MO (control), or SCGN MO1 alone or together with mRNA encoding hsSCGN, SNAP-25, or SNAP-23. HuC expression was observed at 48 hpf (112× magnification). The white rectangles label the position of midbrain. (F) Relative size of midbrain in zebrafish embryos, measured from E. Mean ± SD, ****P < 0.0001; ***P < 0.001. P values were calculated using one-way ANOVA, Tukey’s multiple comparisons test.
To investigate the functions of SCGN in regulating neuronal development, we examined the CaP motor neurons in Tg[hb9: GFP]ml2 transgenic zebrafish (41, 42) (SI Appendix, Fig. S13). Localized in the middle of each spinal cord, the CaP motor neurons can be easily observed. KD of SCGN resulted in abnormally branched axons at 48 hpf. The averaged branch number of the CaP axon in MO1-injected embryo was 4.0, which was significantly more than that of the control group (0.39) (SI Appendix, Fig. S13 A and B). Furthermore, SCGN MO1 injection led to a shorter CaP axon outgrowth (relative length of control: 90; MO1: 37) (SI Appendix, Fig. S13 A and C). Importantly, SCGN wild-type, but not F225A, was able to rescue both the branch number and axonal length phenotypes (SI Appendix, Fig. S13). In contrast, M229A could restore the branch number, but not axonal length, consistent with in vitro measurement that F225A reduced the binding affinity to SNAP-25 more than M229A (SI Appendix, Fig. S13). Taken together, these findings demonstrate that SCGN is critical for zebrafish neuronal growth and brain development, likely by interacting with SNAP-25 or SNAP-23.
SCGN Is More than a Trafficking Factor of SNAP-25.
Our results so far have established that SCGN is critical for the membrane localization of SNAP-25, likely by functioning to chaperone SNAP-25 to the PM. If SCGN merely functions to promote the PM localization of SNAP-25, overexpression of a PM-localized SNAP-25 should bypass the requirement of SCGN. To test this hypothesis, we overexpressed SNAP-25 in SCGN KO STC-1 cells (SI Appendix, Fig. S14). As illustrated in SI Appendix, Fig. S14B, overexpression of SNAP-25 resulted in restored membrane localization of SNAP-25, implying that SCGN facilitates, but is not absolutely required, to deliver SNAP-25 to the PM. However, overexpression of SNAP-25 failed to rescue the GLP-1 secretion in SCGN KO cells, suggesting that SCGN is not a mere trafficking regulator of SNAP-25 (SI Appendix, Fig. S14C).
To examine whether overexpression of SNAP-25 could rescue the SCGN phenotype in zebrafish, we coinjected SCGN MO1 together with mRNA encoding human SNAP-25, or its close homolog, SNAP-23 (Fig. 7 E and F). Although expression of SCGN wild-type could effectively compensate for the loss of zebrafish SCGN, overexpression of SNAP-25 or SNAP-23 did not rescue the physiological defects (Fig. 7 E and F). Specifically, embryos injected with SNAP-25 or SNAP-23 together with SCGN MO1 had a much smaller midbrain, relative to the control or MO1+SCGN group (average of relative size, control: 1.57; MO1: 0.23; MO1+SCGN: 1.14; MO1+SNAP-25: 0.47; MO1+SNAP-23: 0.23) (Fig. 7 E and F). Altogether, our cellular and animal studies indicate that SCGN could have additional roles in regulating snare functions.
Discussion
SNARE activity needs to be precisely regulated, both spatially and temporally, to ensure the exquisite control of neurotransmitter release and other forms of regulated secretion. SCGN is a unique Ca2+ sensor with diverse functions in secretion in multiple tissues, but the mechanisms underlying these functions remained poorly understood. Here, we map the sequence of SNAP-25 that binds to SCGN and present the crystal structure of SCGN in complex with Ca2+ and this SNAP-25 sequence. Structure-based mutagenesis in combination with biochemical, functional, and localization studies lead to a model whereby SCGN regulates exocytosis in part by controlling the PM localization of SNAP-25, but has additional functions that remain to be determined.
It is well established that syntaxin-1 and SNAP-25 have a tendency to oligomerize, forming four-helix bundles with different combinations of their SNARE motifs that can potentially form large aggregates by participation of the two SNAP-25 SNARE motifs in distinct four-helix bundles (reviewed in ref. 43). This tendency may dictate the need for proteins that act as chaperones for maintenance of syntaxin-1 and SNAP-25 in fusion-competent states and for their transport from the endoplasmic reticulum through the Golgi and to the PM. Thus, Munc18-1 has been implicated in the correct transport of syntaxin-1 to the PM (44) and, although it is clear that Munc18-1 or Unc-18 plays crucial roles in neurotransmitter release beyond its syntaxin-1 chaperone function (13, 45), strong evidence indicates that Munc18-1 binding is critical to keep syntaxin-1 in the state that leads to release (46). Moreover, heat-shock cognate 70 (Hsc70), cysteine string protein α (CSPα), and small glutamine-rich protein (SGT) form a chaperone complex that is believed to keep SNAP-25 in a state that is competent for engagement in SNARE complex assembly in synapses (47). Our results now suggest that SCGN constitutes a key factor that chaperones SNAP-25 for proper transport to the PM. It is unclear whether this role is performed in neurons and secretory cells in general, or is restricted to particular cell types, but our data show that SCGN is important for SNAP-25 localization to the PM at least in two different types of SCGN-expressing cells that we have examined.
Our data also indicate that SCGN plays additional roles in exocytosis, in addition to facilitating the membrane localization of SNAP-25. It is unclear whether, once at the PM, SCGN is involved in maintenance of functional states of SNAP-25 and modulation of the ability of SNAP-25 to engage in SNARE complexes. Based on our results, it is tempting to speculate that SNAP-25 could exist in distinct pools where SNAP-25 is associated or not with SCGN (SI Appendix, Fig. S15). Non-SCGN–bound SNAP-25 could be readily incorporated into SNARE complexes and mediate exocytosis if monomeric, but could also form nonfunctional aggregates or be associated to the Hsc70–CSPα–SGT complex. SCGN-bound SNAP-25 needs to be released from this state prior to incorporation into SNARE complexes, which could mediate slow modes of vesicle exocytosis (SI Appendix, Fig. S15). In agreement with this notion, silencing SCGN in pancreatic β-cells specifically impaired the second phase of insulin secretion, but not the first phase (figure 2 of ref. 23). It is important to emphasize that the regulation of SNAP-25 availability and function by SCGN should depend on the relative levels of these proteins and of other SNAP-25 binding proteins, which is likely to be different in distinct forms of regulated secretion. For example, SNAP-25 is highly abundant at synaptic boutons purified from cortex and cerebellum of adult rats (48), where SCGN is known to have low expression levels (49). Therefore, it seems unlikely that SCGN can substantially alter the availability of functional SNAP-25 in these neurons unless it acts in a catalytic fashion, but SCGN might act in a more stoichiometric mode in other secretory cells. We note that, based on these considerations and our results, SCGN may act as both inhibitor and activator of SNARE activity and exocytosis, but this is not surprising, as many key regulators of exocytosis—such as Munc18-1, synaptotagmin-1, and complexin—also play inhibitory and stimulatory roles, which is an intrinsic, necessary feature of regulated secretion (reviewed in refs. 13 and 14). Future studies will be necessary to address how SCGN and other proteins function together to ensure the precise regulation of exocytosis.
Since synapses play a central role in brain functions, perturbations of synapse function can lead to both neuro-psychiatric disorders and neurodegenerative diseases (50). Although previous studies have linked SCGN with multiple neurodevelopmental disorders and neurodegeneration, the underlying mechanism remained unclear (19, 28–30). Using zebrafish as a model, we demonstrate that SCGN is critical for neuronal growth and brain development (Fig. 7 and SI Appendix, Figs. S11 and S13). Since both SNAP-25 and SNAP-23 are implicated in brain development (15, 18), and most SCGN-contacting residues in SNAP-25 are conserved in SNAP-23 (SI Appendix, Fig. S6C), SCGN functions likely by interacting with SNAP-25 and SNAP-23, and by modulating synapse activity. These results, together with the demonstration that fly Cbp53E is a homolog of SCGN and specifically interacts with SNAP-25, suggest that SCGN plays a conserved role in developing and maintaining neuronal networks in diverse organisms, likely via fine-tuning synapse activity.
Whereas our study addressed several important questions in the field, it also raised some interesting questions. First, how does SCGN control the PM localization of SNAP-25? Second, how is SCGN-binding to SNAP-25 activated or down-regulated? Last but not least, are the functions of SCGN in exocytosis and other nonexocytotic cellular processes related? Answering these questions will be interesting directions for the fields of exocytosis and for studying SCGN. Our studies lay a foundation for these future discoveries and provide insights into further understanding how SNARE function is regulated.
Methods
Antibodies and Plasmids.
DNA constructs and antibodies used in this paper are listed in SI Appendix, Tables S2 and S3, respectively.
Cell Culture and Generation of CRISPR KO and Rescued Cell Lines.
The murine enteroendocrine cell line, STC-1, and the pancreatic β-cell line, NIT-1, were purchased from the American Type Culture Collection. STC-1 cells were maintained in DMEM and NIT-1 cells were cultured in F-12K. Both were supplemented with 10% FBS. SCGN and SNAP-25 deficient STC-1 cell lines and SCGN-deficient NIT-1 cell line were generated using lentiviral CRISPR/Cas9 system targeting exon 1, as previously reported (31, 39). Transduced cells were puromycin-selected and clonally isolated using serial dilutions. Individual clones were expanded and screened for protein loss with immunoblotting. Two individual SCGN KO clones, a single SNAP-25 KO clone of STC-1 cells and a single SCGN KO clone of NIT-1 cells, were selected for experiments.
Protein expression in KO clones of STC-1 cells was restored by viral transduction of lentivirus carrying N-terminal HA-tagged human SCGN or SNAP-25b wide-type or mutants. Infected cells were selected under hygromycin and screened for protein re-expression using Western blot.
Immunofluorescence and Microscopy.
Immunofluorescence staining was performed as previously reported (51–53). Briefly, STC-1 or NIT-1 cell lines were seeded onto circular coverslips in 12-well plates. Cells were fixed for 30 min in cold 4% PFA, permeabilized with 0.15% Surfact-Amp (ThermoFisher) in PBS for 3 min, blocked for 30 min in blocking buffer (5% goat serum in PBS) at room temperature. Cells were then incubated with primary antibodies diluted in a blocking buffer overnight at 4 °C. Cells were washed four times with PBS prior to a second room-temperature, 30-min blocking step. Cells were incubated for 1 h at 4 °C in secondary antibodies. Cells were washed with PBS on four occasions and then incubated in Hoechst 33342 for 5 min at room temperature. Then coverslips were rinsed in PBS three times and affixed to slides with SlowFade Anti-fade reagent (Life Technologies).
Images were acquired with a confocal Nikon A1R microscope system. Membranous and total cellular fluorescence intensity was measured using ImageJ software. Approximately 40 cells per cell line were analyzed. A fluorescence intensity ratio of cell membrane to total cellular area was calculated and differences among groups were analyzed by unpaired Student t test.
Protein Extraction, Immunoblotting, and Immunoprecipitation.
Cultured STC-1 or NIT-1 cells were lysed using a prechilled Triton X-100 lysis buffer (25 mM Hepes, 160 mM NaCl, 10 mM DTT, 1mM EDTA, 10% glycerol, 1% Triton X-100) with 100 µM PMSF, 100 µM Na orthovanadate and 1% protease inhibitor mixture (ThermoFisher). Immunoprecipitation was performed using Anti-HA Affinity Matrix (Roche) following the manufacturer’s instructions, with minor modifications. Each immunoprecipitation solution contained 1 mg of total proteins, and 5 µL of anti-HA affinity matrix. After incubation for 2.5 h at 4 °C, the resin was washed by 1 mL of a cold completed lysis buffer five times. In each group, 30 µg of total proteins (3% of total) was used for immunoblotting.
GLP-1 Secretion Assays.
GLP-1secreation assays were performed as previously described (31). Briefly, DHA (Nu-chek, U-84-A) was dissolved in 100% ethanol at a concentration of 10 mg/mL, frozen with liquid N2, and stored at −20 °C. Fresh working DHA solution was prepared on the day of experiment by diluting stock solution in Hepes buffer (140 mM NaCl, 4.5 mM KCl, 1.2 mM CaCl2, 1.2 mM MgCl2, 20 mM Hepes, pH 7.4) at a concentration of 100 µM. DHA was sonicated for 3 min prior to adding to the cells. STC-1 cells were grown to 90% confluency on 6-cm dishes, and starved in Hepes buffer for 30 min before adding either 1 mL of working DHA solution or Hepes buffer alone. After 15-min incubation, supernatants were collected and analyzed with high sensitive GLP-1 active chemiluminescent ELISA kit (Millipore, EZGLPHS-35K) according to the manufacturer’s instructions. Cells were lysed as described above. Results were normalized to total protein concentration.
Cloning, Protein Expression, and Purification.
Human and zebrafish SCGN were cloned into PGEX 4T-1–based expression vector and transformed into BL21(DE3) cells for protein expression. Expression of proteins were induced by 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 37 °C for 4 h. Cells were harvested and resuspended with the lysis buffer (20 mM Tris⋅HCl, 200 mM NaCl and 1 mM PMSF, pH = 8.0). Cells were broken by high-pressure homogernizer, and subjected to centrifugation. Proteins were first captured with GST-affinity resin and eluted after tobacco etch virus protese cleavage. Proteins were further purified by anion exchange and Superdex 200 increase gel-filtration (GE Healthcare) chromatography. All SCGN mutants eluted off size-exclusion chromatography in the same position as the wild-type protein.
Bacterial expression and purification of full-length rat syntaxin-1A, a cysteine-free variant of full-length rat SNAP-25a, full-length rat synaptobrevin-2, full-length rat Munc18-1, full length Cricetulus griseus NSF, full-length Bos taurus αSNAP, and a rat Munc13-1 C1C2BMUNC2C fragment (residues 529 to 1725, Δ1408 to 1452) were described previously (1–5).
The SNARE complex was assembled using full-length rat SNAP-25, rat syntaxin-1A (residues 191 to 253), and rat synaptobrevin-2 (residues 29 to 94). The proteins were mixed at a ratio of 1:1.5:2 (syntaxin-1A: SNAP-25: synaptobrevin-2) in the presence of 1% triton X-100 and 1M NaCl. After incubation at room temperature for 1 h, the assembled SNARE complex was purified from unassembled proteins using gel-filtration chromatography.
Crystallization and Data Collection.
Zebrafish SCGN was concentrated to 10 mg/mL and mixed with human SNAP-25 peptide (amino acids 143 to 170) at 1:3 molar ratio, in a buffer (20 mM Tris⋅HCl, pH8.0, 200 mM NaCl, 2 mM CaCl2, 0.5 mM TCEP). The proteins were mixed with an equal volume of well solution containing 0.1 M glycine, 0.1 M Tris⋅HCl Bicine, pH 8.5, 50% (vol/vol) precipitant Mix4 (25% [vol/vol] MPD, 25% PEG 1000, 25% [vol/vol] PEG 3350), and crystal were grown at 16 °C. X-ray diffraction data were collected at Shanghai Synchrotron Radiation facility beamline BL17U1. The data collection statistics are given in SI Appendix, Table S1.
Structure Solution and Refinement.
Structure was solved by Se-SAD, using HySS and SHELXD. The resulting phase was used to allow model building through the program Autobuild (54), and manual building using the program COOT (55). Refinement was performed using the program Refmac5 (56). Figures were prepared using PyMOL.
ITC.
ITC experiments were performed at 25 °C using ITC 200 (Microcal) in ITC buffer (20 mM Tris⋅HCl, 200 mM NaCl, pH 8.0, 2 mM CaCl2 or EDTA), according to previous procedures (57). Full-length SNAP-25 or its fragments (100 to 200 μM) were titrated into the sample cell containing human or zebrafish SCGN or their mutants (10 to 20 μM). Data were analyzed with the Origin 7.0 software package (OriginLab) by fitting the “one set of sites” model.
Pull-Down Experiments.
GST pull-down assays using purified proteins were performed similar to previous studies (52, 57). Briefly, 20 μg of GST or GST-tagged bait protein were incubated with 100 μg of prey protein. The proteins were mixed with glutathione Sepharose 4B resin in 1 mL of PB buffer (20 mM Tris⋅HCl, pH 8.0, 200 mM NaCl, 2 mM CaCl2, 0.005% or 0.5% Triton-X100). After being washed with PB buffer five times, the bound proteins were separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) and visualized by Coomassie staining. Each experiment was repeated at least once and examined for consistency.
Simultaneous Lipid Mixing and Content Mixing Assays.
Assays that simultaneously measure lipid and content mixing were performed basically as previously described (2, 6), but including wild-type or mutant versions of SCGN and wild-type or mutant versions of SNAP-25. V-liposomes containing full length synaptobrevin-2 (protein-to-lipid ratio 1:500) were made with 39% POPC, 19% DOPS, 19% POPE, 20% cholesterol, 1.5% NBD-PE, and 1.5% Marina Blue DHPE. T-liposomes containing syntaxin-1 and wild-type or G155D SNAP-25 (syntaxin-1:lipid ratio 1:800) were made with 38% POPC, 18% DOPS, 20% POPE, 20% cholesterol, 2% PIP2, and 2% DAG. Dried lipid films were resuspended in 25 mM Hepes, pH 7.4, 150 mM KCl, 1mM TCEP, 2% bOG. Lipid solutions were then mixed with the respective proteins and with 4 μM Phycoerythrin-Biotin for T-liposomes or with 8 μM Cy5-Streptavidin for V- or VS-liposomes in 25 mM Hepes, pH 7.4, 150 mM KCl, 1 mM TCEP, 10% glycerol (vol/vol). Proteoliposomes were prepared by detergent removal using dialysis with 2 g/L Amberlite XAD-2 beads (Sigma) three times at 4 °C and subsequent cofloatation on a three-layer histodenz gradient (35%, 25%, and 0%), and harvested from the topmost layer. Acceptor T-liposomes were first incubated with 0.8 µM NSF, 2 µM αSNAP, 2.5 mM MgCl2, 2 mM ATP, 0.1 mM EGTA, and 1 µM Munc18-1 at 37 °C for 25 min. They were then mixed with donor V-liposomes, Munc13-1 C1C2BMUNC2C (0.1 μM for the experiments of Fig. 3 D–G and SI Appendix, Fig. S5; 0.5 µM for those of Fig. 4 E and F), 1 µM excess SNAP25 (wild-type or G155D), and the concentrations of wild-type or mutant SCGN indicated in the figures. All experiments were performed at 30 °C and 0.6 mM Ca2+ was added at 300 s. The fluorescence signal from Marina blue (excitation at 370 nm, emission at 465 nm) and Cy5 (excitation at 565 nm, emission at 670 nm) were recorded to monitor lipid and content mixing, respectively. At the end of the reaction, 1 to 2% β-OG was added to solubilize the liposomes and the lipid mixing data were normalized to the maximum-fluorescence signal. Most experiments were performed in the presence of 5 µM streptavidin, and control experiments without streptavidin were performed to measure the maximum Cy5 fluorescence after detergent addition for normalization of the content mixing data.
Animals.
All zebrafish (Danio rerio) experiments were performed according to standard procedures, and both adult fish and embryos were raised at 28.5 °C in the Aquatic Ecosystems. The following lines were used in this study: AB strain (wild-type), Tg[HuC:GFP] strain, and SCGN KO zebrafish. All experimental protocols were proved by the Animal Ethical Committee, West China Hospital of Sichuan University.
Generation of SCGN KO Zebrafish.
SCGN knockout zebrafish was obtained using the CISRP/Cas9 technology (39). Two different gRNA was designed to target the exon 1 and 2 of SCGN in zebrafish, respectively. These gRNA (75 pg) were injected at one-cell stage, together with mRNA encoding a zebrafish codon-optimized Cas9 (400 pg). Sequences of gRNA and of the primers used for genotyping are shown in SI Appendix, Table S4.
MO and mRNA Injections.
MO1 and MO2 (GeneTools) were used as previously described (29). MO1 binds to the start codon of zebrafish SCGN gene, and blocks translation. MO2 inhibits gene splicing. Control MO is a standard mismatched control. MO and mRNAs were injected into the yolk and the cell at the development of one cell stage, with 5 ng of MO and/or 200 pg of mRNA per injection, unless otherwise indicated.
WISH.
WISH analysis was performed as previously described (42). Briefly, samples at 24 hpf and 48 hpf were fixed, incubated with proteinase K (1mg/mL) at room temperature for 20 min, and then fixed with 4% paraformaldehyde. The samples were then incubated with digoxigenin (DIG)-labeled antisense RNAs (SCGN or HuC elavl3). After the probes were washed out, the samples were incubated with alkaline phosphatase (AP)-conjugated antidigoxigenin antibody (1:2,000) at 4 °C overnight. After washing, the samples were subjected to NBT/BCIP (Roche) staining according to the manufacturer’s instructions.
Zebrafish Total RNA Extraction and sqRT-PCR.
RNA isolation and sqRT-PCR was carried out in accordance with the published study (42). About 100 zebrafish embryos at 48 hpf were collected for each sample. The embryos were grounded, and tissue debris was removed by centrifugation. Total RNA was isolated by RNeasy Plant Mini Kit (FOREGRNE), and was then used to synthesize cDNA with the Prime Script Reverse-transcription PCR kit (TaKaRa DRR014A). sqRT-PCR was performed using CFX96 Touch Real-Time PCR Detection System (Bio-Rad). For each sample, 200 ng of cDNA were used as the templates for PCR with the Real Master Mix Kit (Roche). Sequences of the primers used for RT-PCR were listed in SI Appendix, Table S4.
Data Availability.
Structure factor and atomic coordinates were deposited to Protein Data Bank with ID code 6JLH.
Supplementary Material
Acknowledgments
We thank members of our laboratory for helpful discussions, and Mr. Chengxin Weng for help with making figures. This research is supported by Natural Science Foundation of China Grants 91854121, 31871429, and 31671477; National Key Research and Development Program of China (Grants 2018YFC1005004, 2015CB942800); Sichuan Science and Technology Program (Grant 2018RZ0128); Welch Foundation Grant I-1304 (to J.R.); and NIH Grant NS097333 (to J.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID code 6JLH).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919698117/-/DCSupplemental.
References
- 1.Söllner T., Bennett M. K., Whiteheart S. W., Scheller R. H., Rothman J. E., A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Hanson P. I., Roth R., Morisaki H., Jahn R., Heuser J. E., Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Poirier M. A., et al. , The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 5, 765–769 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T., Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Mayer A., Wickner W., Haas A., Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell 85, 83–94 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Ma C., Su L., Seven A. B., Xu Y., Rizo J., Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prinslow E. A., Stepien K. P., Pan Y. Z., Xu J., Rizo J., Multiple factors maintain assembled trans-SNARE complexes in the presence of NSF and αSNAP. eLife 8, e38880 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dulubova I., et al. , A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 18, 4372–4382 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misura K. M., Scheller R. H., Weis W. I., Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature 404, 355–362 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Parisotto D., et al. , An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J. Biol. Chem. 289, 9639–9650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker R. W., et al. , A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sitarska E., et al. , Autoinhibition of Munc18-1 modulates synaptobrevin binding and helps to enable Munc13-dependent regulation of membrane fusion. eLife 6, e24278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizo J., Mechanism of neurotransmitter release coming into focus. Protein Sci. 27, 1364–1391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunger A. T., Choi U. B., Lai Y., Leitz J., Zhou Q., Molecular mechanisms of fast neurotransmitter release. Annu. Rev. Biophys. 47, 469–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadkova A., Radecke J., Sorensen J. B., The SNAP-25 protein family. Neuroscience 420, 50–71 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Gu Y., et al. , Differential vesicular sorting of AMPA and GABAA receptors. Proc. Natl. Acad. Sci. U.S.A. 113, E922–E931 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imig C., et al. , The morphological and molecular nature of synaptic vesicle priming at presynaptic active zones. Neuron 84, 416–431 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Suh Y. H., et al. , A neuronal role for SNAP-23 in postsynaptic glutamate receptor trafficking. Nat. Neurosci. 13, 338–343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alhowikan A. M., -Ayadhi L. A., Halepoto D. M., Secretagogin (SCGN) plasma levels and their association with cognitive and social behavior in children with autism spectrum disorder ASD). J. Coll. Physicians Surg. Pak. 27, 222–226 (2017). [PubMed] [Google Scholar]
- 20.Wagner L., et al. , Cloning and expression of secretagogin, a novel neuroendocrine- and pancreatic islet of Langerhans-specific Ca2+-binding protein. J. Biol. Chem. 275, 24740–24751 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Rogstam A., et al. , Binding of calcium ions and SNAP-25 to the hexa EF-hand protein secretagogin. Biochem. J. 401, 353–363 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer M. C., et al. , Identification of a high-affinity network of secretagogin-binding proteins involved in vesicle secretion. Mol. Biosyst. 7, 2196–2204 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Yang S. Y., et al. , Secretagogin affects insulin secretion in pancreatic β-cells by regulating actin dynamics and focal adhesion. Biochem. J. 473, 1791–1803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malenczyk K., et al. , A TRPV1-to-secretagogin regulatory axis controls pancreatic β-cell survival by modulating protein turnover. EMBO J. 36, 2107–2125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romanov R. A., et al. , A secretagogin locus of the mammalian hypothalamus controls stress hormone release. EMBO J. 34, 36–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanics J., et al. , Secretagogin-dependent matrix metalloprotease-2 release from neurons regulates neuroblast migration. Proc. Natl. Acad. Sci. U.S.A. 114, E2006–E2015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma A. K., Khandelwal R., Sharma Y., Veiled potential of secretagogin in diabetes: Correlation or coincidence? Trends Endocrinol. Metab. 30, 234–243 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Krishnamurthy D., et al. , Metabolic, hormonal and stress-related molecular changes in post-mortem pituitary glands from schizophrenia subjects. World J. Biol. Psychiatry 14, 478–489 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Deciphering Developmental Disorders Study , Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahola P., et al. , Secretagogin expression in the vertebrate brainstem with focus on the noradrenergic system and implications for Alzheimer’s disease. Brain Struct. Funct. 224, 2061–2078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sifuentes-Dominguez L. F., et al. , SCGN deficiency results in colitis susceptibility. eLife 8, e49910 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitto E., et al. , X-ray structure of Danio rerio secretagogin: A hexa-EF-hand calcium sensor. Proteins 76, 477–483 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.da Silva A. C., Kendrick-Jones J., Reinach F. C., Determinants of ion specificity on EF-hands sites. Conversion of the Ca2+/Mg2+ site of smooth muscle myosin regulatory light chain into a Ca(2+)-specific site. J. Biol. Chem. 270, 6773–6778 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Liu X., et al. , Simultaneous lipid and content mixing assays for in vitro reconstitution studies of synaptic vesicle fusion. Nat. Protoc. 12, 2014–2028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagel K. R., Beriont J., Tessier C. R., Drosophila Cbp53E regulates axon growth at the neuromuscular junction. PLoS One 10, e0132636 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart T. M., et al. , Combining comparative proteomics and molecular genetics uncovers regulators of synaptic and axonal stability and degeneration in vivo. PLoS Genet. 8, e1002936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirasawa A., et al. , Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat. Med. 11, 90–94 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Robu M. E., et al. , p53 activation by knockdown technologies. PLoS Genet. 3, e78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim C. H., et al. , Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 216, 109–112 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Arkhipova V., et al. , Characterization and regulation of the hb9/mnx1 beta-cell progenitor specific enhancer in zebrafish. Dev. Biol. 365, 290–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang W., et al. , Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH. Proc. Natl. Acad. Sci. U.S.A. 116, 22598–22608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizo J., Südhof T. C., The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices—Guilty as charged? Annu. Rev. Cell Dev. Biol. 28, 279–308 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Han L., et al. , Rescue of Munc18-1 and -2 double knockdown reveals the essential functions of interaction between Munc18 and closed syntaxin in PC12 cells. Mol. Biol. Cell 20, 4962–4975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weimer R. M., et al. , Defects in synaptic vesicle docking in unc-18 mutants. Nat. Neurosci. 6, 1023–1030 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stepien K. P., Prinslow E. A., Rizo J., Munc18-1 is crucial to overcome the inhibition of synaptic vesicle fusion by αSNAP. Nat. Commun. 10, 4326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma M., Burré J., Südhof T. C., CSPα promotes SNARE-complex assembly by chaperoning SNAP-25 during synaptic activity. Nat. Cell Biol. 13, 30–39 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Wilhelm B. G., et al. , Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Maj M., et al. , Novel insights into the distribution and functional aspects of the calcium binding protein secretagogin from studies on rat brain and primary neuronal cell culture. Front. Mol. Neurosci. 5, 84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lepeta K., et al. , Synaptopathies: Synaptic dysfunction in neurological disorders—A review from students to students. J. Neurochem. 138, 785–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia D., et al. , Structural and mechanistic insights into regulation of the retromer coat by TBC1d5. Nat. Commun. 7, 13305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao J., et al. , Mechanism of inhibition of retromer transport by the bacterial effector RidL. Proc. Natl. Acad. Sci. U.S.A. 115, E1446–E1454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia D., Gomez T. S., Billadeau D. D., Rosen M. K., Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell 23, 2352–2361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terwilliger T. C., et al. , Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Sun Q., et al. , Structural and functional insights into sorting nexin 5/6 interaction with bacterial effector IncE. Signal Transduct. Target. Ther. 2, 17030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure factor and atomic coordinates were deposited to Protein Data Bank with ID code 6JLH.