Abstract
Accretionary prisms are thick masses of sedimentary material scraped from the oceanic crust and piled up at convergent plate boundaries found across large regions of the world. Large amounts of anoxic groundwater and natural gas, mainly methane (CH4), are contained in deep aquifers associated with these accretionary prisms. To identify the subsurface environments and potential for CH4 production by the microbial communities in deep aquifers, we performed chemical and microbiological assays on groundwater and natural gas derived from deep aquifers associated with an accretionary prism and its overlying sedimentary layers. Physicochemical analyses of groundwater and natural gas suggested wide variations in the features of the six deep aquifers tested. On the other hand, a stable carbon isotope analysis of dissolved inorganic carbon in the groundwater and CH4 in the natural gas showed that the deep aquifers contained CH4 of biogenic or mixed biogenic and thermogenic origins. Live/dead staining of microbial cells contained in the groundwater revealed that the cell density of live microbial cells was in the order of 104 to 106 cells mL–1, and cell viability ranged between 7.5 and 38.9%. A DNA analysis and anoxic culture of microorganisms in the groundwater suggested a high potential for CH4 production by a syntrophic consortium of hydrogen (H2)-producing fermentative bacteria and H2-utilizing methanogenic archaea. These results suggest that the biodegradation of organic matter in ancient sediments contributes to CH4 production in the deep aquifers associated with this accretionary prism as well as its overlying sedimentary layers.
Keywords: accretionary prism, deep aquifer, methane production, methanogenic archaea, fermentative bacteria
Accretionary prisms are thick layers of sedimentary material piled up at convergent plate boundaries. This material, which was originally deposited on a subducting ocean plate, is accreted onto a non-subducting continental plate during the subduction process (Taira et al., 1982). Accretionary prisms are found across large regions of the world, including Alaska and Washington in the United States, New Zealand, Chile, Peru, Indonesia, Taiwan, and Japan (Kano et al., 1991; Reed et al., 2002; Hervé et al., 2013; Lee et al., 2017).
The Shimanto Belt, an accretionary prism found in southwest Japan, was mainly formed during the Cretaceous and Paleogene periods and is traceable for 1,800 km in parallel with the Nankai Trough and Ryukyu Trench (Kano et al., 1991; Taira et al., 1992). The Shimanto Belt originated from ancient marine sediment deposited on the Philippine Sea Plate (Kano et al., 1991). This sediment is approximately 10 km thick and contains layers of water-bearing permeable sandstone and water-impermeable shale that are rich in complex organic matter (Maki et al., 1980; Taira et al., 1982). Rainwater and seawater recharge through faults or fracture zones formed by earthquakes that occur at the plate boundaries, and these waters then collect in the deep aquifers of accretionary prisms in which they become anoxic over time (Sakata et al., 2012; Baito et al., 2015). Deep aquifers contain large amounts of groundwater with dissolved and free phase natural gas, mainly methane (CH4) (Kimura et al., 2010; Matsushita et al., 2016; Matsushita et al., 2018).
The origin of CH4 in natural gas reserves in subsurface sedimentary deposits is biogenic (formed by methanogenic archaea) or thermogenic (formed by the thermal degradation of organic matter in sedimentary layers). Previous studies investigated CH4 production processes in the Shimanto Belt’s deep aquifers, which are affected by rainwater and seawater flowing down from surface environments, and reported that the syntrophic biodegradation of organic matter by hydrogen (H2)-producing fermentative bacteria and H2-utilizing methanogens, as well as a thermogenic reaction, contributes to CH4 production in these deep aquifers (Kimura et al., 2010; Matsushita et al., 2016; Imachi, 2017; Matsushita et al., 2018; Tamaki, 2019). On the other hand, Matsushita et al. (2016; 2018) demonstrated that groundwater in the deep aquifers associated with the Shimanto Belt had different chemical characteristics from site to site, based on measurements of temperature, salinity, chemical components, and oxygen and hydrogen stable isotope ratios. These findings suggest that the subsurface environments of the accretionary prisms are not constant and are affected by the geological and geochemical features of each region.
The anaerobic deep aquifers associated with accretionary prisms contain large amounts of CH4 (Kimura et al., 2010; Sakata et al., 2012). CH4 is a potential greenhouse gas and important energy resource. However, all of the research on CH4 production processes conducted to date has targeted deep aquifers that are affected by rainwater and seawater derived from surface environments. Accretionary prisms and their overlying sedimentary layers that contain deep aquifers are influenced by magmatic CO2 derived from the deep subsurface environments of active volcanoes or by ancient seawater containing high concentrations of iodine and bromine (Kato, 1985). Mayumi et al. (2013) previously reported that a high concentration of CO2 may change microbial CH4 production pathways in subsurface oil reservoirs. Additionally, iodine and bromine are generally known to exhibit antimicrobial activity (Odlaug, 1981). These factors may affect the microbial activity, community structure, and CH4 production pathway in deep aquifers associated with accretionary prisms; however, there is currently no experimental evidence to support this.
We herein investigated the deep aquifers associated with the accretionary prism and its overlying sedimentary layers in the southeastern part of Kyushu Island, Japan, near active volcanoes. To characterize each deep aquifer, we analyzed groundwater and natural gas samples with chemical and isotopic techniques. The aim of the present study was to clarify whether CH4 production by syntrophic communities of H2-producing fermentative bacteria and H2-utilizing methanogens occur universally in deep aquifers associated with accretionary prisms. Therefore, we investigated microbial community structures and their potential for CH4 production using microbial cell counts, a 16S rRNA gene analysis, and anaerobic culture methods. An understanding of the underlying mechanisms and potential for CH4 production in deep aquifers associated with accretionary prisms, which are distributed worldwide and at which geodynamic phenomena, such as huge earthquakes and fault formation, occur, may provide novel insights into greenhouse gas emissions and global warming.
Materials and Methods
Study sites
The present study was conducted in the southeastern part of Kyushu Island, Japan, near some of Japan’s most active volcanoes (e.g., Kirishima Volcano and Sakurajima Volcano). The accretionary prism known as the Nichinan Group (mainly Paleogene) in the Shimanto Belt is distributed across this region (Fig. 1). The Nichinan Group is composed principally of sandstone, siltstone, conglomerate, and basalt (Kato, 1985). Sedimentary layers that unconformably overlay the Nichinan Group are referred to as the Miyazaki Group (mainly Neogene). The Miyazaki Group is mainly composed of sandstone, mudstone, and conglomerate that were deposited in a forearc basin (Suzuki, 1987; Oda et al., 2011).
Fig. 1.
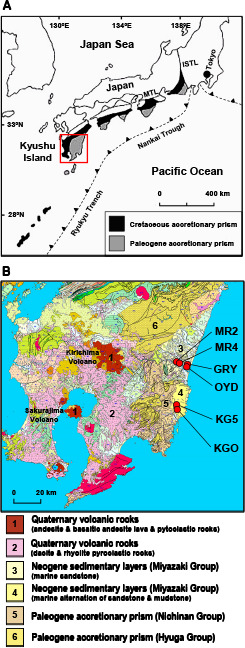
(A) The location of the accretionary prism in southwest Japan, known as the Shimanto Belt, was taken from Kano et al. (1991). The red square in map A is enlarged in the geological map in panel B. MTL, Median Tectonic Line; ISTL, Itoigawa-Shizuoka Tectonic Line. (B) The geological map modified from a 1:200,000 seamless digital geological map of Japan that was published by the Geological Survey of Japan, AIST (2014). The red circles indicate the location of the deep wells used for sampling.
Sampling of groundwater and natural gas
Anoxic groundwater and natural gas samples were collected from deep aquifers associated with the Nichinan and Miyazaki Groups through six deep wells: GRY, OYD, MR2, MR4, KGO, and KG5 (Fig. 1B and Table S1). These wells were drilled down to 810 to 1,301 m and constructed from tight steel-casing pipes including strainers (Table 1) (Kato et al., 2011; Sakata et al., 2012). Groundwater at these wells is anoxically drawn up to ground level by a water pump. In the present study, to prevent contamination by air and water remaining inside the pipe, 200 to 700 tons of groundwater was pumped before sampling. Groundwater samples were collected under anoxic conditions into autoclaved serum bottles and polycarbonate bottles using a sterile silicone tube. The concentrations of dissolved natural gas were so high that gas exsolved at ground level. Natural gas samples were collected into autoclaved 100-mL serum bottles underwater. Serum bottles were tightly sealed with sterile butyl-rubber stoppers and aluminum crimps to prevent contamination by air. Collected samples were transferred to the laboratory on ice and then stored at 4°C until later use.
Table 1.
Physical and chemical parameters of groundwater and components of natural gas.
| Site code | Well depth (Strainer depth) (m) |
Groundwater | Natural gas | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Temp. (ºC) |
pH | ORPa
(mV) |
ECb
(mS m–1) |
N2
(vol.%) |
CO2
(vol.%) |
CH4
(vol.%) |
C1/(C2+C3)c | |||
| GRY | 1,000 (591–983) | 24.9 | 7.1 | –231 | 5,000 | 0.6 | <0.01 | 99.2 | 813 | |
| OYD | 1,301 (947–1,240) | 44.5 | 7.4 | –197 | 2,220 | 3.0 | 2.90 | 94.0 | 666 | |
| MR2 | 847 (711–831) | 38.1 | 7.4 | –181 | 1,064 | 3.2 | 1.75 | 95.0 | 5,280 | |
| MR4 | 1,054 (774–1,054) | 44.7 | 7.3 | –169 | 1,409 | 3.2 | 2.10 | 94.7 | 6,312 | |
| KGO | 810 (581–779) | 50.0 | 7.2 | –199 | 1,009 | 2.2 | 34.80 | 62.9 | 571 | |
| KG5 | 1,070 (925–1,047) | 35.1 | 7.2 | –141 | 488 | 3.5 | 6.80 | 89.7 | 11,211 | |
a ORP, oxidation-reduction potential.
b EC, electrical conductivity.
c C1, CH4; C2, C2H6; C3, C3H8.
Physicochemical and stable isotope analyses
The temperature, pH, oxidation-reduction potential (ORP), and electrical conductivity (EC) of groundwater samples were measured at the outflows of the wells. Temperature was measured with a CT-460WR thermometer (Custom). pH was measured with an HM-20P portable meter (DKK-TOA). ORP and EC were measured with RM-20P and CM-21P portable meters (DKK-TOA), respectively. The concentrations of cations (Na+, Ca2+, Mg2+, K+, and NH4+) and anions (CI–, Br–, I–, F–, PO43–, NO3–, SO42–, acetate, and formate) in groundwater were analyzed using an ICS-1500 ion chromatography system (Dionex). The concentration of HCO3– was measured by the separation titration of groundwater with H2SO4 using the digital titrator model 16900 (Hach) according to the manufacturer’s instructions. Sulfide was analyzed using a No. 211 sulfide ion detector tube (Gastec). Dissolved organic carbon (DOC) in the groundwater, filtered through a pre-combusted Whatman GF/F glass microfiber filter (GE Healthcare), was measured on a TOC-V total organic carbon analyzer (Shimadzu).
H2, N2, O2, CO2, and CH4 concentrations in natural gas were measured with a GC-2014 gas chromatograph (GC) (Shimadzu) equipped with a thermal conductivity detector (TCD) and packed column (ShinCarbon ST, 6.0 m×3.0 mm i.d.; Shinwa Chemical Industries). The GC conditions used were as follows: injector temperature, 170°C; column oven temperature, 150°C; detector temperature, 170°C. Argon was used as a carrier gas at a constant flow mode of 50 mL min–1. CH4, C2H6, and C3H8 concentrations were measured with a GC-2014 GC equipped with a flame ionization detector and packed column (Sunpak-A, 2.0 m×3.0 mm i.d.; Shinwa Chemical Industries). The GC conditions used were as follows: injector temperature, 100°C; column oven temperature, 65°C; detector temperature, 100°C. N2 was used as a carrier gas at a constant flow rate of 50 mL min–1. Samples were analyzed in triplicate. The confidence limits of the measurement were 0.01 vol.% for H2, N2, O2, CO2, and CH4, and 0.001 vol.% for C2H6 and C3H8.
The stable hydrogen and oxygen isotope ratios of groundwater samples (D/H and 18O/16O) were measured with a DLT-100 liquid water isotope analyzer (Los Gatos Research) following the procedures described by Dawson et al. (2015). The stable carbon isotope ratio (13C/12C) of dissolved inorganic carbon (DIC; consisting mainly of HCO3–) was measured with a Trace GC Ultra GC (Thermo Fisher Scientific) connected to a Deltaplus XL IRMS (Thermo Fisher Scientific) according to the method described by Miyajima et al. (1995). The 13C/12C of CH4 in natural gas was measured with a Flash EA1112 elemental analyzer (Thermo Fisher Scientific) connected to a Delta V Advantage ConFlo IV system (Thermo Fisher Scientific) (Miyajima et al., 1995). All isotope ratios are reported relative to international standards: Vienna Standard Mean Ocean Water for δD and δ18O, and Vienna Pee Dee Belemnite for δ13C. The standard deviations of δD and δ18O in groundwater and δ13C of DIC and CH4 were ±0.5‰, ±0.1‰, ±1‰, and ±0.3‰, respectively.
Microbial cell counting and next-generation sequencing (NGS) analysis of 16S rRNA genes
Groundwater samples for microscopic observations were filtered using polycarbonate membrane filters (pore size, 0.2 μm; diameter, 25 mm) (Millipore). Total microbial cells trapped on the filters were stained with SYBR Green I (Thermo Fisher Scientific) (Yanagawa et al., 2016). A LIVE/DEAD BacLight bacterial viability kit (Thermo Fisher Scientific) was used to obtain the ratio of live to total microbial cells (Vezzulli et al., 2015). Stained cells were observed under a BX51 epifluorescence microscope equipped with a U-MNIB3 fluorescence filter (Olympus), and more than 20 microscopic fields (approximately 20 cells in each field) were counted for each sample. Cells were counted within 48 h of groundwater sampling.
To analyze the archaeal and bacterial communities in the groundwater, 10 L of groundwater samples was aseptically filtered using Sterivex-GV filter units (pore size, 0.22 μm) (Millipore). Bulk DNA was extracted from trapped cells using an ISOIL for Beads Beating kit (Nippon Gene). The V3–V4 region of the 16S rRNA gene was amplified using a primer set, Pro341F and Pro805R for prokaryotes (Takahashi et al., 2014). PCR products were purified through a MultiScreen PCR96 filter plate (Millipore) and analyzed using an Agilent DNA 1000 kit on an Agilent 2100 Bioanalyzer system (Agilent Technologies) to detect primer dimers and obtain the average molecular weight of each product. The Illumina sequencing library was generated according to the method described by Takahashi et al. (2014). Sequencing was conducted using a 600-cycle paired-end MiSeq reagent kit v3 (Illumina) on an Illumina MiSeq platform. After sequencing was complete, an image analysis, base calling, and error estimations were performed using Illumina Real-Time Analysis software version 1.17.28. Bioinformatic processing was performed using a combination of Quantitative Insights Into Microbial Ecology (QIIME) version 1.9.1 and USEARCH version 6.1 (Caporaso et al., 2010; Edgar, 2010). Only reads that had quality value scores of ≥20 for more than 98% of the sequence were extracted for further analyses. Potential chimeras were removed using USEARCH version 6.1. Quality-filtered reads were assigned to operational taxonomic units (OTUs) at a 97% similarity level, and the Good’s coverage, Chao 1, and Shannon indices were then calculated with QIIME version 1.9.1. Taxonomy was assigned using the Ribosomal Database Project (RDP) MultiClassifier version 2.11 with a confidence level of 80% (Wang et al., 2007). Sequences were deposited under GenBank/ENA/DDBJ accession numbers DRA004443 and DRA005244.
Measurement of potential microbial CH4 production
Thirty milliliters of each groundwater sample was anoxically injected into an autoclaved 70-mL serum bottle that was then tightly sealed with a sterile butyl-rubber stopper and aluminum crimp. To assess the potential for CH4 production by methanogenic archaea, groundwater samples were amended with acetate (20 mM), methanol (20 mM), formate (20 mM), or H2/CO2 (80:20 [v/v]; 150 kPa). Except for H2/CO2-amended bottles, the headspaces of serum bottles were filled with pure N2 at 150 kPa. These cultures were anoxically incubated without shaking at temperatures that were 10°C higher than those of the groundwater samples because the actual temperature in a deep aquifer is generally considered to be higher than that of a groundwater sample measured at the outflow of the well.
To assess the potential for CH4 production by a syntrophic microbial community of H2-producing fermentative bacteria and H2-utilizing methanogenic archaea, groundwater samples were amended with organic nutrients. The chemical components of organic matter contained in the deep aquifers of the Shimanto Belt currently remain unclear. Therefore, we used a mixture of yeast extract, peptone, and glucose (YPG) as organic substrates because this mixture is widely used for anaerobic cultures of microorganisms and may promote the rapid growth of microbial communities in groundwater. Groundwater samples were amended with 3 mL of YPG medium (10 g yeast extract, 10 g peptone, and 2 g glucose L–1 distilled water). As a killed control, groundwater samples were autoclaved and then supplemented with 3 mL of YPG medium. The headspaces of serum bottles were filled with pure N2 at 150 kPa. These cultures were anoxically incubated without shaking at the temperatures of the groundwater samples measured at the outflows of the wells and temperatures that were 10°C and 20°C higher than those measured at the outflows. As a control, cultures using groundwater samples without any amendment were also incubated.
All cultures were performed in triplicate. H2, N2, CH4, and CO2 concentrations in the headspaces were measured on a GC-2014 GC equipped with a TCD (Shimadzu) as described above.
Microorganisms that grew in cultures were identified according to the 16S rRNA gene clone library method. Briefly, cells in cultures were collected by centrifugation and lysed by lysozyme and proteinase K. Bulk DNAs were extracted using both phenol/chloroform/isoamyl alcohol and chloroform/isoamyl alcohol and purified with ethanol precipitation. Archaeal and bacterial 16S rRNA gene fragments were amplified by PCR from bulk DNA using the Archaea-specific primer set, 109aF and 915aR (Stahl and Amann, 1991; Großkopf et al., 1998), and Bacteria-specific primer set, 8bF and 1512uR (Eder et al., 1999). PCR products of the archaeal and bacterial 16S rRNA genes were cloned with a Zero Blunt TOPO PCR cloning kit (Invitrogen). The sequences of the inserted PCR products selected from recombinant colonies were elucidated with an Applied Biosystems 3730xl DNA analyzer (Life Technologies). A 3% distance level between sequences was considered to be the cut-off for distinguishing distinct OTUs. We identified the nearest relative of each OTU using the BLAST program (Altschul et al., 1990). Neighbor-joining phylogenetic trees were constructed using the CLUSTAL X version 2.1 program (Larkin et al., 2007). Bootstrap values were obtained from 1,000 replications and indicated at the branching points. Sequences were deposited under GenBank/ENA/DDBJ accession numbers LC123696 to LC123721 and LC194992 to LC194998.
Results
Physicochemical signatures of groundwater and natural gas
Anoxic groundwater and natural gas samples were collected from deep aquifers through six deep wells. The groundwater temperature measured at the outflows of the wells ranged between 24.9 and 50.0°C (Table 1). The pH of groundwater ranged between 7.1 and 7.4. The ORP was measured as an indicator of anoxic conditions in the deep aquifers and ranged between –231 and –141 mV. The EC, an indicator of salinity, ranged between 488 and 5,000 mS m–1. These parameters were measured several times between May 2014 and May 2019, and no temporal variations were detected. The concentration ranges of various chemical components were measured, including Br– and I– (0.04 to 1.38 mM and 0.05 to 0.95 mM, respectively; both were particularly high in samples from GRY), HCO3– (1.5 to 46 mM), and DOC (0.05 to 0.83 mM) (Table S2). PO43–, NO3–, SO42–, S2–, acetate, and formate were found at trace amounts or were below detection limits.
CH4 was the predominant component of natural gas (Table 1). The other principal components were CO2 and N2. In addition to CH4, natural gas collected from KGO contained a particularly large amount of CO2 (34.8 vol.%). C2H6 and C3H8 were found at trace amounts. The hydrocarbon gas composition C1/(C2+C3) of natural gas samples ranged between 571 and 11,211. H2, O2, and C4H10 concentrations were found at trace amounts or were below detection limits.
Stable isotopic signatures of groundwater and natural gas
The δD and δ18O of groundwater samples ranged between –33.9 and –5.8‰ and between –5.1 and 1.9‰, respectively (Fig. 2 and Table S3). To estimate the origins of groundwater in the deep aquifers, we plotted δD and δ18O values with respect to the global meteoric water line (Craig, 1961). Groundwater sampled from KG5 was plotted closer to the local surface water (Kato et al., 2011). The groundwater from OYD, MR2, MR4, and KGO fell in the upper right region of the local surface water. The groundwater from GRY was plotted closer to normal seawater and ancient seawater, as reported previously by Maekawa et al. (2006).
Fig. 2.
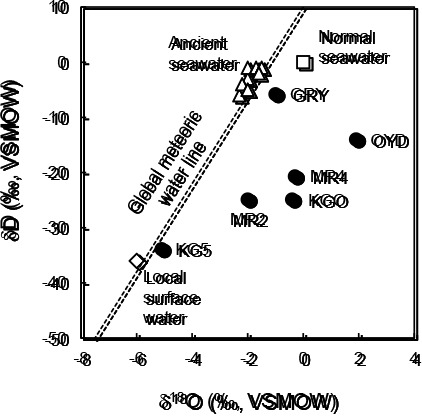
Stable hydrogen and oxygen isotope composition of groundwater samples (●) compared with those of local surface water (◇), normal seawater (□), and ancient seawater (△) (Maekawa et al., 2006; Kato et al., 2011). Dashed line: global meteoric water line (Craig, 1961). VSMOW, Vienna Standard Mean Ocean Water.
The δ13C of DIC (consisting mainly of HCO3–) in groundwater (δ13CDIC) ranged between 0.16 and 8.95‰ (Fig. 3A and Table S3). The δ13C of CH4 in natural gas (δ13CCH4) ranged between –57.8 and –39.6‰. To estimate the origin of CH4 in natural gas, we assessed carbon isotope fractionation (αc) between δ13CDIC and δ13CCH4. αc values were 1.041 to 1.065 (Fig. 3A and Table S3). In a plot of stable isotopic values on a δ13CDIC versus δ13CCH4 diagram (Smith and Pallasser, 1996), the sample from GRY fell within the region of biogenic origin (αc=1.06–1.08). In contrast, all other sample values fell within the boundary between biogenic and thermogenic origins (αc=1.04–1.06), indicating that natural gas samples contained CH4 of mixed biogenic and thermogenic origins. We also plotted the observed isotopic values in a δ13CCH4 versus hydrocarbon gas composition C1/(C2+C3) diagram according to Bernard et al. (1978). In this diagram, all sample values fell within the boundary between biogenic and thermogenic origins (Fig. 3B).
Fig. 3.
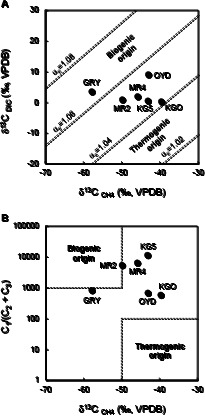
(A) Stable carbon isotope composition of CH4 in natural gas samples and dissolved inorganic carbon in groundwater samples. The categorization of CH4 origins was made according to Smith and Pallasser (1996). Dashed lines: equal carbon isotopic fractionation, αc=(δ13CDIC+103)/(δ13CCH4+103), for αc=1.02, 1.04, 1.06, and 1.08. (B) Stable carbon isotope composition of CH4 and hydrocarbon gas composition C1/(C2+C3) in natural gas samples. The categorization of CH4 origins was made according to Bernard et al. (1978). VPDB, Vienna Pee Dee Belemnite.
Abundance and diversity of microbial communities in groundwater
The abundance of total microbial cells stained with SYBR Green I ranged between 8.2×104 and 1.0×107 cells mL–1 (Table 2). Live/dead staining revealed that the density of live microbial cells ranged between 1.4×104 and 1.4×106 cells mL–1 (Fig. S1). The percent cell viability (% live microbial cells/total microbial cells) ranged between 7.5 and 38.9% (Table 2).
Table 2.
Microbial cell density and cell viability in groundwater.
| Site code | Total cell density (cells mL–1) |
Live cell density (cells mL–1) |
Cell viabilitya
(%) |
|---|---|---|---|
| GRY | 1.0×107 | 1.3×106 | 13.0 |
| OYD | 5.1×106 | 3.8×105 | 7.5 |
| MR2 | 3.6×106 | 1.4×106 | 38.9 |
| MR4 | 1.7×106 | 5.2×105 | 30.6 |
| KGO | 8.2×104 | 1.4×104 | 17.1 |
| KG5 | 5.2×105 | 1.2×105 | 23.1 |
a Cell viability=(% live cells/total cells).
To elucidate microbial community structures in anoxic groundwater derived from deep aquifers, we performed a NGS analysis targeting the archaeal and bacterial 16S rRNA genes. We obtained 12,678 to 54,714 reads and 234 to 804 OTUs (Table S4). Coverage reached >98.3%. The Chao1 and Shannon indices were 245 to 1,542 and 4.74 to 6.84, respectively.
Archaeal 16S rRNA genes, which accounted for between 1.0 and 23.9% of the total reads obtained from each sample (Fig. 4A), revealed the predominance of methanogenic archaea belonging to the order Methanobacteriales (Fig. 4B). We also detected minor amounts of Methanomicrobiales, Methanocellales, and Methanomassiliicoccales. These methanogenic archaea are generally known to use H2 and CO2 to produce CH4, while some are capable of methylotrophic methanogenesis (Sakai et al., 2008; Zhu et al., 2011; Dridi et al., 2012; Sakai et al., 2012). In groundwater samples from OYD, MR2, and KG5, we also identified archaeal 16S rRNA genes that are closely related to those of Methanosarcinales, an order of methanogenic archaea that uses acetate, methanol, or H2 and CO2 as a methanogenic substrate (Kamagata et al., 1992).
Fig. 4.
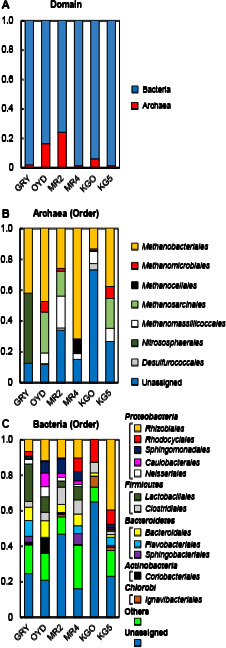
(A) Relative abundance of archaeal and bacterial 16S rRNA genes obtained from a NGS analysis. Taxonomic composition of microbial communities in natural groundwater samples based on the relative abundance of (B) archaeal and (C) bacterial 16S rRNA gene sequences. ‘Others’ indicate bacterial orders with relative abundance below 5% in all samples.
The analysis of bacterial 16S rRNA genes revealed the presence of bacterial groups that belong to the phyla Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Chlorobi (Fig. 4C). At the order level, 6–40% of bacterial 16S rRNA genes obtained from all sites, except KGO, were closely related to Rhizobiales, an order of Alphaproteobacteria. The bacterial 16S rRNA genes closely related to the orders Lactobacillales, Clostridiales, and Bacteroidales were also detected from all sites. The bacterial groups of the orders Coriobacteriales and Ignavibacteriales were identified mainly in OYD and KGO, respectively.
Potential for microbial CH4 production
To assess the potential for CH4 production by methanogenic archaea in the deep aquifers, we incubated groundwater samples amended with methanogenic substrates (acetate, methanol, formate, or H2/CO2) under anaerobic conditions. However, CH4 production was not observed in these cultures after incubations of longer than 60 d.
On the other hand, the high potential for CH4 production by microbial communities was confirmed in cultures using YPG medium-amended groundwater (Fig. 5 and S2). In cultures using groundwater from OYD, MR4, KGO, and KG5, the production of H2 and CO2 was observed within 3 d. In cultures using groundwater from GRY and MR2, H2 and CO2 were detected after 7 d. After the production of H2 and CO2, the concentration of H2 decreased to below the detection limit in all cultures. CH4 production was observed after H2 levels began to fall. In cultures using groundwater from GRY, MR2, MR4, KGO, and KG5, CH4 production was observed within 10 d. In cultures using groundwater from OYD, CH4 was detected after 17 d. In a killed control using the groundwater samples amended with YPG medium, abiotic H2 and CH4 production was not observed during an incubation of more than 120 d. Similarly, H2 and CH4 production was not observed in cultures using groundwater without organic substrates.
Fig. 5.
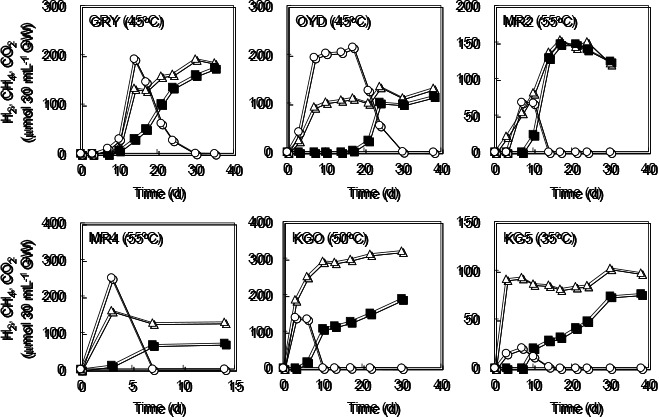
Biogas production from groundwater samples amended with YPG medium. The gas phase composition over time is shown as follows: H2 (○); CH4 (■); and CO2 (△). Incubation temperatures are shown in parentheses. Although representative results of cultures performed in triplicate are shown, all cultures showed similar potential for biogas production.
To identify microorganisms that generated biogases (i.e., H2, CO2, and CH4), we constructed archaeal and bacterial 16S rRNA gene clone libraries. The 16S rRNA gene analysis suggested that H2-utilizing methanogenic archaea and H2-producing fermentative bacteria were predominant in the microbial population (Table S5) and were related to the archaeal orders Methanobacteriales and Methanomicrobiales (Fig. S3) and the bacterial orders Bacillales, Bacteroidales, Clostridiales, Tissierellales, and Ignavibacteriales, respectively (Fig. S4) (Parshina et al., 2003; Watanapokasin et al., 2009). Additionally, bacterial 16S rRNA genes closely related to the order Desulfovibrionales, which is known to form syntrophic microbial communities with H2-utilizing methanogens, were also confirmed in the clone libraries associated with GRY and OYD (Morris et al., 2013).
Discussion
To characterize the subsurface environments and identify the potential for CH4 production in anoxic deep aquifers associated with the Shimanto Belt and its overlying sedimentary layers in the southeastern part of Kyushu Island, Japan, we herein revealed the geochemical and microbiological features of deep aquifer-derived anoxic groundwater and natural gas samples. Groundwater from the KG5 site had the lowest EC value (488 mS m–1), which was approximately 10% that of normal seawater (Table 1), and had similar δD and δ18O values to those of the local surface water (Fig. 2). Collectively, these characteristics suggest that the KG5 deep aquifer was affected by rainwater that flowed down from surface environments. In contrast, groundwater from GRY had almost the same EC value as normal seawater and ancient seawater (Table 1). The δD and δ18O values of groundwater sampled from the GRY site were also plotted closer to those of normal seawater and ancient seawater (Fig. 2). Additionally, the groundwater from GRY contained higher levels of Br– and I– than normal seawater (Table S2) (Millero et al., 2008). These chemical features are consistent with those of ancient seawater, which is comprised of groundwater that originated from seawater and was maintained for a long period in a low-temperature deep aquifer (Katayama et al., 2015). Therefore, the GRY deep aquifer was likely influenced by ancient seawater. This result differs from that previously reported in deep aquifers found in other regions of the Shimanto Belt that are affected by rainwater and normal seawater from surface environments (Matsushita et al., 2016; Matsushita et al., 2018).
The EC values of groundwater from OYD, MR2, MR4, and KGO were between approximately 20 and 44% that of normal seawater, suggesting that the deep aquifers at these sites hold groundwater derived from a mixture of rainwater and seawater that flowed down from the surface and the seafloor, respectively (Table 1). However, the δD and δ18O values of OYD, MR2, MR4, and KGO fell to the right of a line joining the plots of surface water and seawater, suggesting large δ18O enrichment (Fig. 2). The high δ18O values suggest that the groundwater in these deep aquifers was affected by water-rock interactions in thermal subsurface environments (Bowers and Taylor, 1985). Alternatively, these isotopic characteristics may be the result of groundwater evaporation due to geothermal heat (Connolly et al., 1990).
CH4 was the predominant component of all natural gas samples with the exception of KGO, which also contained particularly high amounts of CO2 (34.8 vol.%) and CH4 (62.9 vol.%) (Table 1). Kato et al. (2011) reported that CO2 in natural gas from the KGO site is of magmatic origin based on its δ13C value. The accumulation of magmatic CO2 in natural gas has frequently been found in tectonically active oil- and gas-bearing basins (e.g., Dai et al., 1999). The highest groundwater temperatures recorded in the present study were also from the KGO site (50°C). Thus, the deep aquifer at the KGO site may be affected by magmatic CO2 and geothermal groundwater that rose from deep subsurface environments of active volcanoes, e.g., the Kirishima Volcano and Sakurajima Volcano (Fig. 1). These results are distinct from previous findings on the deep aquifers of the Shimanto Belt, which is found in the mid-western part of Shizuoka Prefecture and the southern part of Okinawa Island, Japan (Matsushita et al., 2016; Matsushita et al., 2018).
The results of chemical and isotopic analyses of groundwater and natural gas samples suggested that the environmental features of the deep aquifers markedly differed among the sites studied herein. In contrast, stable carbon isotope analyses and microbiological assays suggested the presence of CH4 production by a syntrophic community of H2-producing fermentative bacteria and H2-utilizing methanogenic archaea in the deep aquifers. The δ13CCH4 versus δ13CDIC diagram and δ13CCH4 versus C1/(C2+C3) diagram both indicated that all natural gas samples contained CH4 of biogenic origin or a mixture of CH4 of biogenic and thermogenic origins (Fig. 3). The NGS analysis of target archaeal 16S rRNA genes revealed the predominance of H2-utilizing methanogenic archaea belonging to the orders Methanobacteriales, Methanomicrobiales, and Methanomassiliicoccales in the groundwater samples (Fig. 4B). The NGS analysis of target bacterial 16S rRNA genes indicated the presence of bacteria belonging to the orders Lactobacillales, Clostridiales, Bacteroidales, Coriobacteriales, and Ignavibacteriales (Fig. 4C). These bacterial groups have the ability to degrade organic matter to H2 and CO2 by fermentation (Benno et al., 1983; Kandler et al., 1983; Liu et al., 2008; Podosokorskaya et al., 2013). The presence of Rhizobiales in groundwater samples was also confirmed. Although Rhizobiales is generally known to comprise aerobic diazotrophs, some species that belong to this order may grow by fermentation and are present in deep subsurface environments (Kodama and Watanabe, 2011; Miettinen et al., 2015; Dutta et al., 2018). The presence of methanogenic archaea and fermentative bacteria identified in the present study has also been reported in deep aquifers of other regions associated with the Shimanto Belt (Matsushita et al., 2016; Matsushita et al., 2018). Additionally, these microorganisms have frequently been found in subsurface gas and oil reservoirs, coal deposits, and subseafloor sediment cores in which microbial hydrogenotrophic CH4 production has been confirmed (Colwell et al., 2004; Meslé et al., 2013; Katayama et al., 2015; Hirai et al., 2017). These results suggest that a syntrophic community of H2-producing fermentative bacteria and H2-utilizing methanogenic archaea contributes to CH4 production in the deep aquifers tested. The microbial CH4 production process may be flexible to environmental differences by changing the microbial community structure while maintaining similar metabolic functions.
The microbial cell densities observed in the present study were similar to or higher than those previously reported in deep aquifers of the accretionary prism and other geothermal aquifers (Table 2) (Katayama et al., 2015; Matsushita et al., 2016; Matsushita et al., 2018). These results also suggest the presence of microbial activity in the deep aquifers. Additionally, this is the first study to measure live cell density as well as total cell density in the groundwater of the Shimanto Belt. Groundwater from GRY showed relatively high live cell density (Table 2). Since CH4 from GRY was suggested to be of biogenic origin by the δ13CDIC versus δ13CCH4 diagram (Fig. 3A), the impact of high concentrations of Br– and I– on microbial CH4 production appears to be small. In contrast, the lowest live cell density was observed in KGO. This may be due to magmatic CO2 and geothermal groundwater from deep subsurface environments. The factors that influence microbial viability in deep aquifers may be more complex, such as the residence time of groundwater and the degradation rate of dead cells.
Although the potential for CH4 production was not confirmed in cultures using H2/CO2-amended groundwater, this may have been due to the growth inhibition of H2-utilizing methanogenic archaea caused by high concentrations of H2 and CO2 (Sakai et al., 2009). On the other hand, the addition of organic substrates to groundwater samples demonstrated a high potential for H2 and CO2 production by H2-producing fermentative bacteria as well as rapid H2 consumption and significant CH4 production by H2-utilizing methanogenic archaea (Fig. 5 and S2). Since we used high concentrations of organic substrates, the microbial activity observed in the cultures does not reflect the activity level in the actual subsurface environment. Changes in microbial community structures between natural groundwater and culture samples, particularly bacterial communities, are attributed to the strong selective pressure caused by using YPG medium (Fig. 4 and Table S5). On the other hand, the present results showed high live cell densities and a high potential for microbial CH4 production. Thus, previous and ongoing microbial CH4 production appears to contribute to the significant CH4 reserves in the deep aquifers. A previous study reported that the average contents of total organic carbon in the sedimentary layers of the Nichinan Group and Miyazaki Group were 0.86 and 0.58%, respectively (Maki et al., 1980). These contents are consistent with those of total organic carbon in the sediments of other accretionary prisms containing biogenic CH4 (Davie and Buffett, 2003; Oba et al., 2015). Therefore, this organic matter appears to support the activity of microbial communities that generate CH4 in the deep aquifers. Further clarification of the composition of this organic matter is of fundamental importance for future studies.
The NGS analysis of archaeal 16S rRNA genes in natural groundwater samples revealed the presence of Methanosarcinales, which generally uses acetate or methanol for CH4 production (Fig. 4B). Additionally, Methanobacteriales and Methanomassiliicoccales identified in the present study grow by methylotrophic methanogenesis. On the other hand, CH4 production was not confirmed in cultures using groundwater amended with acetate, methanol, and formate. These results suggest that the potential for CH4 production using acetate, methanol, and formate in the deep aquifers is lower than that of H2-utilizing methanogenesis. This is consistent with the low concentrations of these methanogenic substrates in the groundwater (Table S2). However, we cannot exclude the possibility that CH4 production from these methanogenic pathways occurs in the deep aquifers because the culture conditions used in the present study were significantly different from the actual subsurface environments and may not have been suitable for these methanogenic archaea.
Conclusion
In the present study, we demonstrated the potential for CH4 production by a syntrophic microbial community of H2-producing fermentative bacteria and H2-utilizing methanogens in deep aquifers that have widely varying geological and geochemical features. The production process of CH4 is similar to that previously reported in deep aquifers found in other regions of the Shimanto Belt that are affected by rainwater and normal seawater from surface environments (Matsushita et al., 2016; Matsushita et al., 2018). Since accretionary prisms are derived from ancient marine sediments scraped from the subducting ocean plate, they are rich in complex organic matter (Berner and Koch, 1993; Kaneko et al., 2010). The results of the present study support a model in which the biodegradation of organic matter in ancient sediments contributes to the generation of CH4 reserves in deep aquifers associated with accretionary prisms and their overlying sedimentary layers, which are distributed globally and have diverse geological and geochemical features.
Supplementary Material
Acknowledgements
We thank H. Harada, T. Yamagata, T. Kuroi, and K. Harada of Mingas Co., and I. Ueno, F. Yonee, M. Akiyama, and K. Hirashita of Nichinan City Hall for their help with groundwater and natural gas sampling. This work was partially supported by a Japan Society for the Promotion of Science (JSPS) KAKENHI Grant (No. 16H02968). This research was also supported by a Japan Science and Technology Agency (JST) MIRAI Project Grant (No. JPMJMI17EK).
References
- Altschul S.F., Gish W., Miller W., Myers E.W., and Lipman D.J. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Baito K., Imai S., Matsushita M., Otani M., Sato Y., and Kimura H. (2015) Biogas production using anaerobic groundwater containing a subterranean microbial community associated with the accretionary prism. Microb Biotechnol 8: 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benno Y., Watabe J., and Mitsuoka T. (1983) Bacteroides pyogenes sp. nov., Bacteroides suis sp. nov., and Bacteroides helcogenes sp. nov., new species from abscesses and feces of pigs. Syst Appl Microbiol 4: 396–407. [DOI] [PubMed] [Google Scholar]
- Bernard B.B., Brooks J.M., and Sackett W.M. (1978) Light hydrocarbons in recent Texas continental shelf and slope sediments. J Geophys Res 83: 4053–4061. [Google Scholar]
- Berner U., and Koch J. (1993) Organic matter in sediments of site 808, Nankai accretionary prism, Japan. Proc Ocean Drill Program: Sci Results 131: 379–385. [Google Scholar]
- Bowers T.S., and Taylor H.P. (1985) An integrated chemical and stable-isotope model of the origin of midocean ridge hot spring systems. J Geophys Res 90: 12583–12606. [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell F., Matsumoto R., and Reed D. (2004) A review of the gas hydrates, geology, and biology of the Nankai Trough. Chem Geol 205: 391–404. [Google Scholar]
- Connolly C.A., Walter L.M., and Baadsgaard H. (1990) Origin and evolution of formation waters, Alberta Basin, Western Canada Sedimentary Basin. II. Isotope systematics and water mixing. Appl Geochem 5: 397–413. [Google Scholar]
- Craig H. (1961) Isotopic variations in meteoric waters. Science 133: 1702–1703. [DOI] [PubMed] [Google Scholar]
- Dai J., Song Y., Dai C., and Wang D. (1999) Geochemistry and accumulation of carbon dioxide gases in China. AAPG Bull 80: 1615–1625. [Google Scholar]
- Davie M.K., and Buffett B.A. (2003) Sources of methane for marine gas hydrate: inferences from a comparison of observations and numerical models. Earth Planet Sci Lett 206: 51–63. [Google Scholar]
- Dawson K.S., Osburn M.R., Sessions A.L., and Orphan V.J. (2015) Metabolic associations with archaea drive shifts in hydrogen isotope fractionation in sulfate-reducing bacterial lipids in cocultures and methane seeps. Geobiology 13: 462–477. [DOI] [PubMed] [Google Scholar]
- Dridi B., Fardeau M.L., Ollivier B., Raoult D., and Drancourt M. (2012) Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62: 1902–1907. [DOI] [PubMed] [Google Scholar]
- Dutta A., Gupta S.D., Gupta A., Sarkar J., Roy S., Mukherjee A., and Sar P. (2018) Exploration of deep terrestrial subsurface microbiome in Late Cretaceous Deccan traps and underlying Archean basement, India. Sci Rep 8: 17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder W., Ludwig W., and Huber R. (1999) Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch Microbiol 172: 213–218. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Geological Survey of Japan, AIST. (2014) Seamless digital geological map of Japan 1: 200,000. Jan 14, 2014 version. Geological Survey of Japan. Tsukuba, Japan: National Institute of Advanced Industrial Science and Technology. URL https://gbank.gsj.jp/seamless/
- Großkopf R., Janssen P.H., and Liesack W. (1998) Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol 64: 960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé F., Calderón M., Fanning C.M., Pankhurst R.J., and Godoy E. (2013) Provenance variations in the Late Paleozoic accretionary complex of central Chile as indicated by detrital zircons. Gondwana Res 23: 1122–1135. [Google Scholar]
- Hirai M., Nishi S., Tsuda M., Sunamura M., Takai Y., and Nunoura T. (2017) Library construction from subnanogram DNA for pelagic sea water and deep-sea sediments. Microbes Environ 32: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imachi H. (2017) Topic of influence, methane and microbes. Microbes Environ 32: 297–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamagata Y., Kawasaki H., Oyaizu H., Nakamura K., Mikami E., Endo G., et al. (1992) Characterization of three thermophilic strains of Methanothrix (“Methanosaeta”) thermophila sp. nov. and rejection of Methanothrix (“Methanosaeta”) thermoacetophila. Int J Syst Bacteriol 42: 463–468. [DOI] [PubMed] [Google Scholar]
- Kandler O., Schillinger U., and Weiss N. (1983) Lactobacillus bifermentans sp. nov., nom. rev., an organism forming CO2 and H2 from lactic acid. Syst Appl Microbiol 4: 408–412. [DOI] [PubMed] [Google Scholar]
- Kaneko M., Shingai H., Pohlman J.W., and Naraoka H. (2010) Chemical and isotopic signature of bulk organic matter and hydrocarbon biomarkers within mid-slope accretionary sediments of the northern Cascadia margin gas hydrate system. Mar Geol 275: 166–177. [Google Scholar]
- Kano K., Nakaji M., and Takeuchi S. (1991) Asymmetrical melange fabrics as possible indicators of the convergent direction of plates: a case study from the Shimanto Belt of the Akaishi Mountains, central Japan. Tectonophysics 185: 375–388. [Google Scholar]
- Katayama T., Yoshioka H., Muramoto Y., Usami J., Fujiwara K., Yoshida S., et al. (2015) Physicochemical impacts associated with natural gas development on methanogenesis in deep sand aquifers. ISME J 9: 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S., Waseda A., and Iwano H. (2011) Geochemistry of natural gas and formation water from water-dissolved gas fields in Miyazaki Prefecture. J Jpn Assoc Pet Technol 76: 244–253 (in Japanese with an English abstract). [Google Scholar]
- Kato T. (1985) Stratigraphy of Nichinan Group in southeastern Kyushu, Japan. Contrib Inst Geol Paleontol, Tohoku Univ 87: 1–23 (in Japanese with an English abstract). [Google Scholar]
- Kimura H., Nashimoto H., Shimizu M., Hattori S., Yamada K., Koba K., et al. (2010) Microbial methane production in deep aquifer associated with the accretionary prism in Japan. ISME J 4: 531–541. [DOI] [PubMed] [Google Scholar]
- Kodama Y., and Watanabe K. (2011) Rhizomicrobium electricum sp. nov., a facultatively anaerobic, fermentative, prosthecate bacterium isolated from a cellulose-fed microbial fuel cell. Int J Syst Evol Microbiol 61: 1781–1785. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee H., Fischer T.P., Maarten de Moor J., Sharp Z.D., Takahata N., and Sano Y. (2017) Nitrogen recycling at the Costa Rican subduction zone: The role of incoming plate structure. Sci Rep 7: 13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Finegold S.M., Song Y., and Lawson P.A. (2008) Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 58: 1896–1902. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Igari S., and Kaneko N. (2006) Chemical and isotopic compositions of brines from dissolved-in-water type natural gas fields in Chiba, Japan. Geochem J 40: 475–484. [Google Scholar]
- Maki S., Nagata S., Fukuta O., and Furukawa S. (1980) Geochemical study on organic matter from sedimentary rocks in the Miyazaki Group and the Shimanto Supergroup of Miyazaki Prefecture, Japan. Bull Geol Surv Jpn 31: 1–24 (in Japanese with an English abstract). [Google Scholar]
- Matsushita M., Ishikawa S., Nagai K., Hirata Y., Ozawa K., Mitsunobu S., and Kimura H. (2016) Regional variation of CH4 and N2 production processes in the deep aquifers of an accretionary prism. Microbes Environ 31: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M., Magara K., Sato Y., Shinzato N., and Kimura H. (2018) Geochemical and microbiological evidence for microbial methane production in deep aquifers of the Cretaceous accretionary prism. Microbes Environ 33: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayumi D., Dolfing J., Sakata S., Maeda H., Miyagawa Y., Ikarashi M., et al. (2013) Carbon dioxide concentration dictates alternative methanogenic pathways in oil reservoirs. Nat Commun 4: 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslé M., Dromart G., and Oger P. (2013) Microbial methanogenesis in subsurface oil and coal. Res Microbiol 164: 959–972. [DOI] [PubMed] [Google Scholar]
- Miettinen H., Kietäväinen R., Sohlberg E., Numminen M., Ahonen L., and Itävaara M. (2015) Microbiome composition and geochemical characteristics of deep subsurface high-pressure environment, Pyhäsalmi mine Finland. Front Microbiol 6: 1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millero F.J., Feistel R., Wright D.G., and McDougall T.J. (2008) The composition of standard seawater and the definition of the reference-composition salinity scale. Deep Sea Res., Part I 55: 50–72. [Google Scholar]
- Miyajima T., Miyajima Y., Hanba Y.T., Yoshii K., Koitabashi T., and Wada E. (1995) Determining the stable isotope ratio of total dissolved inorganic carbon in lake water by GC/C/IRMS. Limnol Oceanogr 40: 994–1000. [Google Scholar]
- Morris B.E.L., Henneberger R., Huber H., and Moissl-Eichinger C. (2013) Microbial syntrophy: Interaction for the common good. FEMS Microbiol Rev 37: 384–406. [DOI] [PubMed] [Google Scholar]
- Oba M., Sakata S., and Fujii T. (2015) Archaeal polar lipids in subseafloor sediments from the Nankai Trough: Implications for the distribution of methanogens in the deep marine subsurface. Org Geochem 78: 153–160. [Google Scholar]
- Oda M., Chiyonobu S., Torii M., Otomo T., Morimoto J., Satou Y., et al. (2011) Integrated magnetobiochronology of the Pliocene–Pleistocene Miyazaki succession, southern Kyushu, southwest Japan: Implications for an Early Pleistocene hiatus and defining the base of the Gelasian (P/P boundary type section) in Japan. J Asian Earth Sci 40: 84–97. [Google Scholar]
- Odlaug T.E. (1981) Antimicrobial activity of halogens. J Food Prot 44: 608–613. [DOI] [PubMed] [Google Scholar]
- Parshina S.N., Kleerebezem R., Sanz J.L., Lettinga G., Nozhevnikova A.N., Kostrikina N.A., et al. (2003) Soehngenia saccharolytica gen. nov., sp. nov. and Clostridium amygdalinum sp. nov., two novel anaerobic, benzaldehyde-converting bacteria. Int J Syst Evol Microbiol 53: 1791–1799. [DOI] [PubMed] [Google Scholar]
- Podosokorskaya O.A., Kadnikov V.V., Gavrilov S.N., Mardanov A.V., Merkel A.Y., Karnachuk O.V., et al. (2013) Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ Microbiol 15: 1759–1771. [DOI] [PubMed] [Google Scholar]
- Reed D.W., Fujita Y., Delwiche M.E., Blackwelder D.B., Sheridan P.P., Uchida T., and Colwell F.S. (2002) Microbial communities from methane hydrate-bearing deep marine sediments in a Forearc Basin. Appl Environ Microbiol 68: 3759–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S., Imachi H., Hanada S., Ohashi A., Harada H., and Kamagata Y. (2008) Methanocella paludicolagen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage “Rice Cluster I”, and proposal of the new archaeal order Methanocellales ord. nov. Int J Syst Evol Microbiol 58: 929–936. [DOI] [PubMed] [Google Scholar]
- Sakai S., Imachi H., Sekiguchi Y., Tseng I.C., Ohashi A., Harada H., and Kamagata Y. (2009) Cultivation of methanogens under low-hydrogen conditions by using the coculture method. Appl Environ Microbiol 75: 4892–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S., Ehara M., Tseng I.C., Yamaguchi T., Bräuer S.L., and Cadillo-Quiroz H. (2012) Methanolinea mesophila sp. nov., a hydrogenotrophic methanogen isolated from rice field soil, and proposal of the archaeal family Methanoregulaceae fam. nov. within the order Methanomicrobiales. Int J Syst Evol Microbiol 62: 1389–1395. [DOI] [PubMed] [Google Scholar]
- Sakata S., Maekawa T., Igari S., and Sano Y. (2012) Geochemistry and origin of natural gases dissolved in brines from gas fields in southwest Japan. Geofluids 12: 327–335. [Google Scholar]
- Smith J.W., and Pallasser R.J. (1996) Microbial origin of Australian coalbed methane. AAPG Bull 80: 891–897. [Google Scholar]
- Stahl, D.A., and Amann, R. (1991) Development and application of nucleic acid probes in bacterial systematics. In Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt, E., and Goodfellow, M. (eds). New York, NY: John Wiley & Sons, pp. 205–248. [Google Scholar]
- Suzuki H. (1987) Stratigraphy of the Miyazaki Group in the southeastern part of Miyazaki Prefecture, Kyushu, Japan. Tohoku Univ. Inst Geol Pal Contr 90: 1–24 (in Japanese with an English abstract). [Google Scholar]
- Taira A., Okada H., Whitaker J.H., and Smith A.J. (1982) The Shimanto Belt of Japan: Cretaceous—Lower Miocene active margin sedimentation. Geol Soc Spec Publ 10: 5–26. [Google Scholar]
- Taira, A., Byrne, T., and Ashi, J. (1992) Photographic Atlas of an Accretionary Prism: Geologic Structures of the Shimanto Belt, Japan. Tokyo: University of Tokyo Press. [Google Scholar]
- Takahashi S., Tomita J., Nishioka K., Hisada T., and Nishijima M. (2014) Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS One 9: e105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki H. (2019) Cultivation renaissance in the post-metagenomics era: combining the new and old. Microbes Environ 34: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L., Pezzati E., Stauder M., Stagnaro L., Venier P., and Pruzzo C. (2015) Aquatic ecology of the oyster pathogens Vibrio splendidus and Vibrio aestuarianus. Environ Microbiol 17: 1065–1080. [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., and Cole J.R. (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanapokasin R.Y., Boonyakamol A., Sukseree S., Krajarng A., Sophonnithiprasert T., Kanso S., and Imai T. (2009) Hydrogen production and anaerobic decolorization of wastewater containing Reactive Blue 4 by a bacterial consortium of Salmonella subterranea and Paenibacillus polymyxa. Biodegradation 20: 411–418. [DOI] [PubMed] [Google Scholar]
- Yanagawa K., Tani A., Yamamoto N., Hachikubo A., Kano A., and Matsumoto R. (2016) Biogeochemical cycle of methanol in anoxic deep-sea sediments. Microbes Environ 31: 190–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Liu X., and Dong X. (2011) Methanobacterium movens sp. nov. and Methanobacterium flexile sp. nov., isolated from lake sediment. Int J Syst Evol Microbiol 61: 2974–2978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


