Significance
Auxin plays a crucial role in the life cycle of plants. In plant cells, auxin concentration must be closely controlled for optimal auxin responses. However, the underlying mechanism governing auxin homeostasis remains largely unknown. In this study, UDP-glycosyltransferase (UGT76F1) has been identified as involved in auxin homeostasis by glucosylation of the major auxin precursor (IPyA) to form IPyA glucose conjugates (IPyA-Glc). This glucosylation process is negatively regulated by the transcription factor PIF4 and plays a vital role in light- and temperature-dependent hypocotyl growth. This study suggests that IPyA-Glc formation is a key event in the control of auxin biosynthesis, thereby revealing a mechanism of auxin homeostasis that adapts to the plant environment through IPyA glucosylation.
Keywords: Arabidopsis thaliana, auxin, glycosylation
Abstract
Auxin is a class of plant hormone that plays a crucial role in the life cycle of plants, particularly in the growth response of plants to ever-changing environments. Since the auxin responses are concentration-dependent and higher auxin concentrations might often be inhibitory, the optimal endogenous auxin level must be closely controlled. However, the underlying mechanism governing auxin homeostasis remains largely unknown. In this study, a UDP-glycosyltransferase (UGT76F1) was identified from Arabidopsis thaliana, which participates in the regulation of auxin homeostasis by glucosylation of indole-3-pyruvic acid (IPyA), a major precursor of the auxin indole-3-acetic acid (IAA) biosynthesis, in the formation of IPyA glucose conjugates (IPyA-Glc). In addition, UGT76F1 was found to mediate hypocotyl growth by modulating active auxin levels in a light- and temperature-dependent manner. Moreover, the transcription of UGT76F1 was demonstrated to be directly and negatively regulated by PIF4, which is a key integrator of both light and temperature signaling pathways. This study sheds a light on the trade-off between IAA biosynthesis and IPyA-Glc formation in controlling auxin levels and reveals a regulatory mechanism for plant growth adaptation to environmental changes through glucosylation of IPyA.
In the evolutionary selection of higher plants, light is an important energy source for plant life (1). Other than this, with global climate change altering the availability of resources, light is also an important environmental cue for the plasticity response of growth to sustained climate change. For optimal growth and development, the perception and processing of information from the surrounding environment are crucial to plant performance. In addition to light, the ambient temperature also serves as another major stimulus that regulates plant development. The network of signaling pathways for these two key cues is the basis for adaptive plant growth.
Recently, it has been proven that the members of phytochrome-interacting factors (PIFs, such as PIF1, PIF3, PIF4, PIF5, and PIF7) play a central role in regulating multiple morphological responses in plants from seed germination to vegetation structure, with functional redundancy in some aspects, but also specificity in certain reactions (2–7). In addition, PIF transcription factors have also been identified as the core integrators or signaling hubs linking multiple environmental cues, including light, ambient temperature (8, 9), the circadian clock (10–12), and phytohormones (13–17).
PIF4 accumulates in the dark to promote skotomorphogenesis and plays a major role in sensing light and temperature to mediate plant growth and development (18–21). It is a key regulator not only in promoting elongation growth in response to external signals, but also in response to internal signals, such as the circadian clock (22–24) and hormones (25–29). Indole-3-acetic acid (IAA) is a type of the earliest discovered auxin hormones, which is essential for plant growth and development. It has been reported that, with the increase in ambient temperature, the content of free IAA increases, resulting in increased hypocotyl elongation (30). Moreover, PIF4 has been shown to activate the expression of auxin-related genes, such as YUCCA8 (YUC8) and several SAURs (9, 31). Indole-3-pyruvic acid (IPyA) is considered the predominant precursor for IAA biosynthesis. The YUC family has been shown to play a role in the conversion of IPyA to IAA in Arabidopsis thaliana, and it is believed that the conversion of IPyA to IAA is the main IAA biosynthesis pathway in plants (32, 33). However, the mechanisms that regulate auxin homeostasis for adaptive growth and development of plants under specific environmental cues remain largely unknown.
In the present work, we identify UGT76F1 as a glycosyltransferase which participates in the regulation of auxin homeostasis through the glucosylation of IPyA, the major precursor of IAA. In addition, we demonstrate that the transcription of UGT76F1 is directly regulated by PIF4. Moreover, we found that UGT76F1 regulates hypocotyl growth by modulating auxin levels in a light- and temperature-dependent manner. Thus, our findings reveal a mechanism for regulating auxin homeostasis for plant adaptive growth to changes in the external environment.
Results
UGT76F1 Is Transcriptionally Up-Regulated by Light Signal.
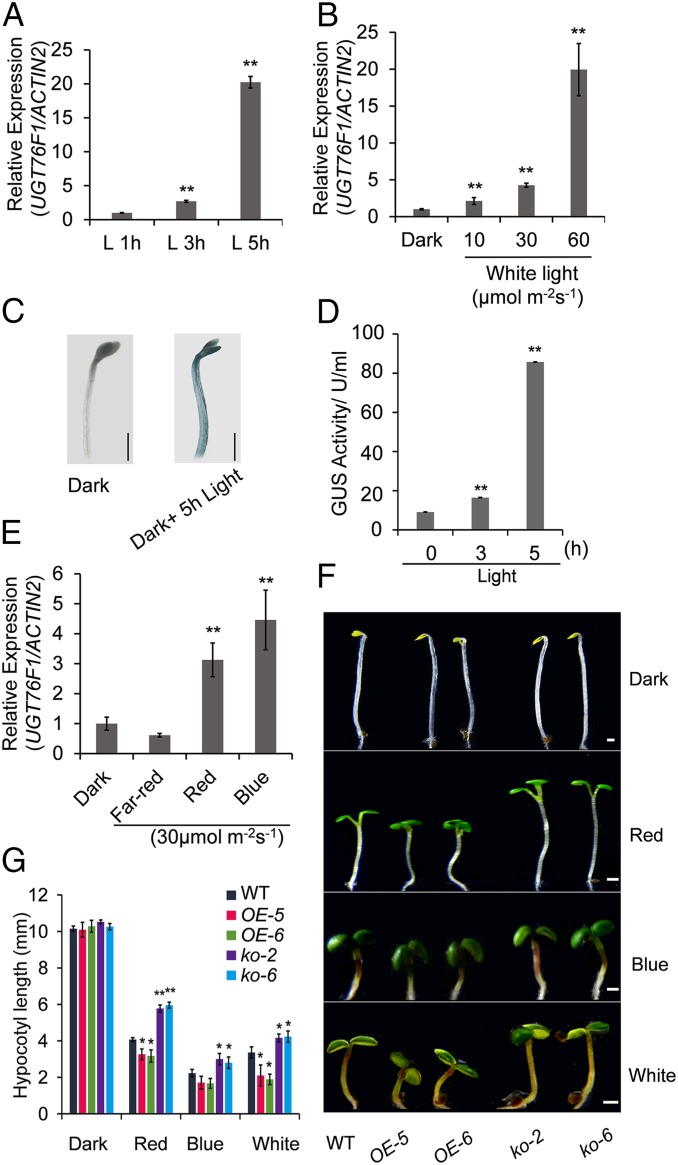
Upon screening possible transcriptionally induced members in response to natural light signals in the UDP-glycosyltransferase (UGT) family of Arabidopsis, the UGT76F1 gene was found to be evidently up-regulated by light (https://genevestigator.com/gv/). In order to verify whether the expression of UGT76F1 was altered by light, RT-qPCR analysis and the UGT76F1Pro::GUS transgenic plants were used in this study. UGT76F1 expression was found to be clearly induced at 3 and 5 h after exposure to light (Fig. 1A). In addition, continuous white light also induced UGT76F1 transcript accumulation in a fluence rate-dependent manner (Fig. 1B), indicating that high light intensity can more effectively induce the expression of UGT76F1. Consistent with RT-qPCR results, β-glucuronidase (GUS) staining and GUS activity analysis revealed that a 5-h white light treatment for dark-grown seedlings was sufficient to significantly induce UGT76F1 transcript accumulation (Fig. 1 C and D). Moreover, the inducibility of UGT76F1 transcription was retained in red and blue light (Fig. 1E). These results suggest that UGT76F1 expression is controlled by light signals.
Fig. 1.
The light induction of UGT76F1 expression and the phenotypes of UGT76F1 overexpression lines and mutant lines. (A) UGT76F1 transcript abundance was significantly induced in WT seedlings by 5 h of treatment with white light after growing in continuous darkness for 5 d. (B) Light-induced expression patterns of UGT76F1 in a light-intensity–dependent manner in 5-d-old WT seedlings. (C) GUS staining of pUGT76F1-GUS expressing seedlings grown in continuous darkness for 5 d and then shifted to light for 5 h. (Scale bars, 1 mm.) (D) Measurement of GUS activities for pUGT76F1-GUS–expressing seedlings grown in continuous darkness for 5 d and then shifted to light for the indicated time. Data are presented by means ± SD. **Significant difference at P < 0.01 relative to GUS activities in darkness (Student’s t test). (E) The induction of UGT76F1 expression by red and blue light. Data are presented by means ± SD. **Significant difference at P < 0.01 relative to GUS activities in darkness (Student’s t test). (F) Phenotypes of the UGT76F1 overexpression lines and mutants grown under continuous darkness or various light conditions for 5 d. (Scale bars, 1 mm.) (G) Hypocotyl length of seedlings shown in F. Data shown are means ± SD. n = 30. Student’s t test was performed (*P < 0.05, **P < 0.01). Experiments were conducted for three biological replicates, yielding similar results.
UGT76F1 Modulates Hypocotyl Growth in a Light-Dependent Manner.
To investigate whether the UGT76F1 gene is involved in photomorphogenesis or in skotomorphogenesis, we generated UGT76F1 overexpression lines (OE lines) and knockout mutants (ko lines). The ugt76f1ko-2 is an insertion mutant, and ugt76f1ko-6 is a deletion mutant created by the CRISPR/Cas9 system, both of which have the function loss of UGT76F1 (SI Appendix, Fig. S1 A–C). We have determined that these two mutants are Cas9-free (SI Appendix, Fig. S1D). OE-2, OE-3, OE-5, and OE-6 are overexpression lines with much higher transcript levels of UGT76F1 than those of wild-type (WT) plants (SI Appendix, Fig. S1E). Since hypocotyl growth is a good marker for the photomorphogenesis response, we compared the hypocotyls of WT, UGT76F1 overexpression lines, and ugt76f1 mutant lines after 5 d of growth under dark, red, blue, and white light with indicated light intensity. Although the hypocotyl lengths of different genotypes were comparable under the dark conditions, there were significant differences in the hypocotyl elongation between the WT and different transgenic lines of UGT76F1 under continuous red, blue, and white light conditions (Fig. 1F). In detail, ugt76f1 mutant lines displayed longer hypocotyls than WT under continuous white, red, and blue light. However, under these light conditions, UGT76F1 overexpression seedlings exhibited shorter hypocotyls than WT. These results indicate that UGT76F1 modulates hypocotyl growth in a light-dependent manner (Fig. 1 F and G). In addition, after 10 d of growth in the dark, UGT76F1 overexpression lines were more prone to cotyledon opening than WT (SI Appendix, Fig. S2 A and B). Typically, the expression levels of light-regulated genes increased during photomorphogenesis in young seedlings (34, 20). The study found that, in the dark and after light exposure, two light-regulated marker genes, namely the light-harvesting chlorophyll a/b-binding protein 2 (CAB2) and the photosystem I light-harvesting complex gene 6 (LHCA6), were up-regulated much more in UGT76F1OE lines than in WT (SI Appendix, Fig. S2 C and D). These observations suggest that UGT76F1 may be involved in the positive regulation pathway of photomorphogenesis.
PIF4 Negatively Regulates UGT76F1 Transcription by Physically Interacting with the UGT76F1 Promoter.
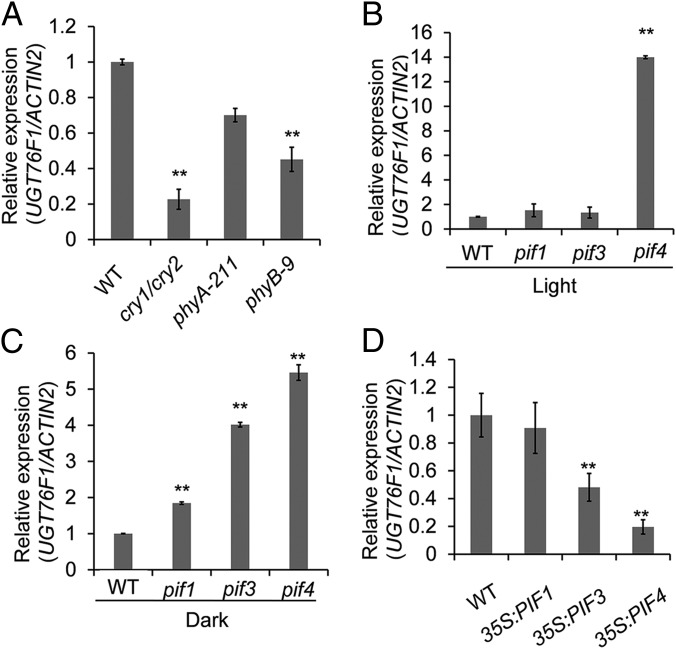
Previous work has shown that plants have evolved a series of photoreceptors to better sense light signals in the environment. To determine the contributions of photoreceptors to UGT76F1-mediated photomorphogenesis, the transcription levels of the UGT76F1 gene were examined in different light-signal receptor mutants. We found that once these photoreceptors (such as PHYA, PHYB, and CRYs) were removed, the light-induced accumulation of UGT76F1 transcripts reduced (Fig. 2A). This result suggests that UGT76F1 expression is affected by multiple photoreceptors in the light signal pathway.
Fig. 2.
Relative transcription abundance of UGT76F1 in different light signal mutants. (A) The light-induced up-regulation of UGT76F1 was abolished in the mutants of phyA, phyB, or crys under white light treatment for 5 h after growing in continuous darkness for 5 d. (B and C) The effects of removing PIF1, PIF3, and PIF4 on transcript accumulation of UGT76F1 under continuous white light (B) or darkness (C). (D) UGT76F1 transcript accumulation was significantly suppressed by the overexpression of PIF3 and PIF4. Data shown are means ± SD. n = 3. Student’s t test for each genotype was performed (**P < 0.01).
PIFs are negative regulators of photomorphogenic development mediated by photoreceptors and are critically relevant for the photoperiodic control of hypocotyl elongation. Based on this, we further clarified the relationship between UGT76F1 and PIFs and examined the expression levels of UGT76F1 in the mutants of PIF1, PIF3, and PIF4 under continuous white light or dark conditions. RT-qPCR analysis showed that UGT76F1 transcript accumulation in the pif4 mutant was most significantly up-regulated under continuous light or dark conditions (Fig. 2 B and C). In contrast, UGT76F1 transcription was most significantly repressed by PIF4 overexpression (Fig. 2D), suggesting that UGT76F1 may act downstream of PIF4.
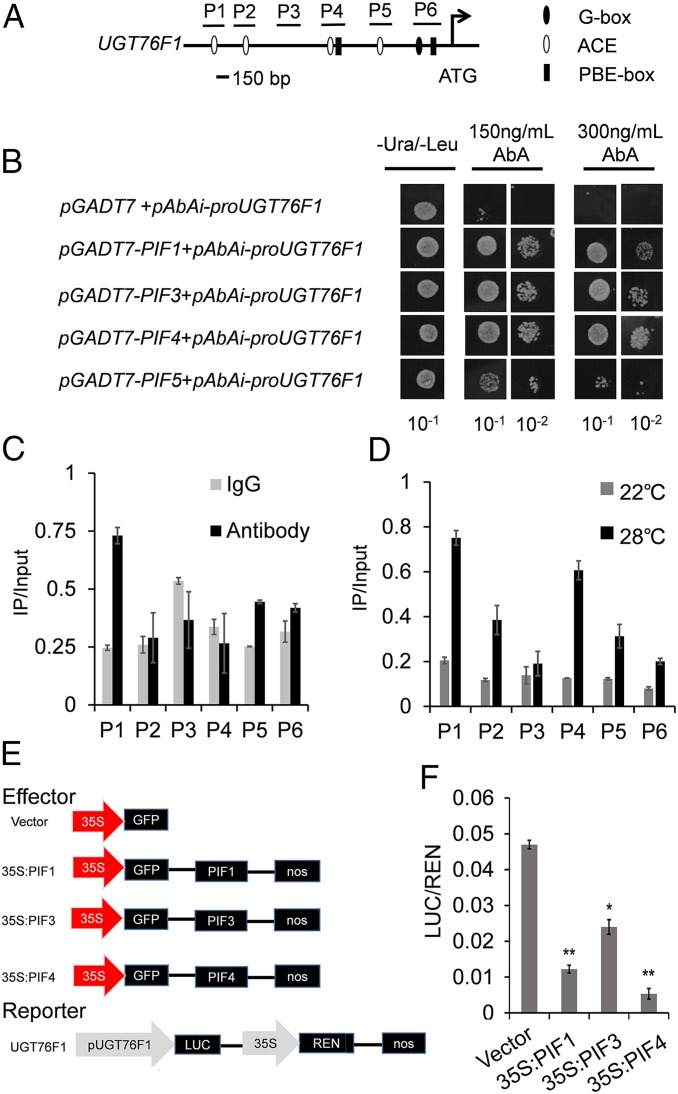
Next, we investigated whether PIF proteins, including PIF1, PIF3, PIF4, and PIF5, interact directly with the UGT76F1 promoter. PIF transcription factors are known to bind to variants of the E boxes (CANNTG), including the G box (CACGTG), the PIF-binding E (PBE)-box (CACATG/CATGTG), and the ACGT-containing element (ACE) (35–37). To identify the putative cis elements of UGT76F1 bound by PIFs, a 2,000-bp upstream DNA sequence before the translation initiation codon (ATG) was analyzed using the PlantCARE database (38). Sequence analysis indicated that multiple ACE, PBE-box, and G-box elements were found within the UGT76F1 promoter (Fig. 3A). Hereafter, yeast one-hybrid (Y1H) assays were performed to test the interaction between PIF proteins and these cis elements. As shown in Fig. 3B, the study found that PIF1, PIF3, and PIF4 had the possibility of binding to the UGT76F1 promoter in Y1H assays.
Fig. 3.
PIF4 is the major regulator for transcription activity of UGT76F1. (A) Promoter structure of the UGT76F1 gene and fragments used in the ChIP assay. The G box, ACE, and PBE box on sense strand of the UGT76F1 promoter are indicated by solid ovals, empty ovals and solid bars, respectively. P1–P6 indicate the promoter regions containing the above sequence or not, which were used in the ChIP/qPCR assay. (B) Yeast one-hybrid binding assay involving PIF1, PIF3, PIF4, PIF5, and UGT76F1 promoter. (C and D) ChIP-qPCR assay indicated that PIF4 is associated with the promoters of UGT76F1. Graphs show the ratio of bound promoter fragments (P1 to P6) versus total input detected by qPCR after immunoprecipitation. Mouse IgG was used as a mock control. Input sample was used to normalize the qPCR results in each ChIP. Data are means ± SD. n = 3. The experiments were repeated twice with similar results. (E and F) PIF4 transcriptionally activates the promoter of UGT76F1 in Arabidopsis protoplasts via transient dual-luciferase assays. The LUC-to-REN ratio indicates the activity of the transcription factors on the expression level of the promoters. LUC: firefly luciferase activity; REN: Renilla luciferase activity. Data are means ± SD. n = 30. Significant difference was compared to control. Student’s t test was performed (*P < 0.05, **P < 0.01).
The interaction between the PIF4 and UGT76F1 promoters was also confirmed by chromatin immunoprecipitation (ChIP) using an anti-GFP antibody followed by qPCR (ChIP-qPCR) assay, showing a stronger binding of PIF4 to the P1 and P5 regions of the UGT76F1 promoter (Fig. 3 A and C). PIF4 has a different affinity for these motifs. P2, P4, and P6 did not show a significant recovery compared to P3 (the negative control), indicating that there is no binding of PIF4 to these promoter elements.
Because PIF4 could mediate hypocotyl elongation at high temperature, we further examined the activity of PIF4 directly binding to the UGT76F1 promoter in planta in response to the elevated temperature. ChIP assays were performed at both 22 °C and 28 °C. Our results show that PIF4 has a universal binding with P1, P2, P4, P5, and P6 regions at a treatment of 28 °C (Fig. 3D). At 28 °C, P1 and P4 containing the ACE element are preferentially bound.
To further confirm that the transcription activity of UGT76F1 was affected by PIFs, a dual-luciferase (LUC) reporter plasmid was constructed, which encodes a firefly LUC gene driven by the UGT76F1 promoter (−1,812 to 0 bp) and a Renilla luciferase (REN) gene driven by the constitutive 35S promoter (Fig. 3E). The results show that all overexpression of PIF1, PIF3, and PIF4 can suppress the luciferase activity of the UGT76F1 reporter as compared to the empty vector control, especially the overexpression of PIF4, which suppresses ∼90% of the luciferase activity of the UGT76F1 reporter (Fig. 3F). All in all, these results provide strong evidence for the regulation activity of PIFs on UGT76F1 and indicate that PIF4 may play a major role in regulating UGT76F1 activity among the PIFs examined in this paper.
UGT76F1 Regulates High-Temperature–Mediated Hypocotyl Elongation.
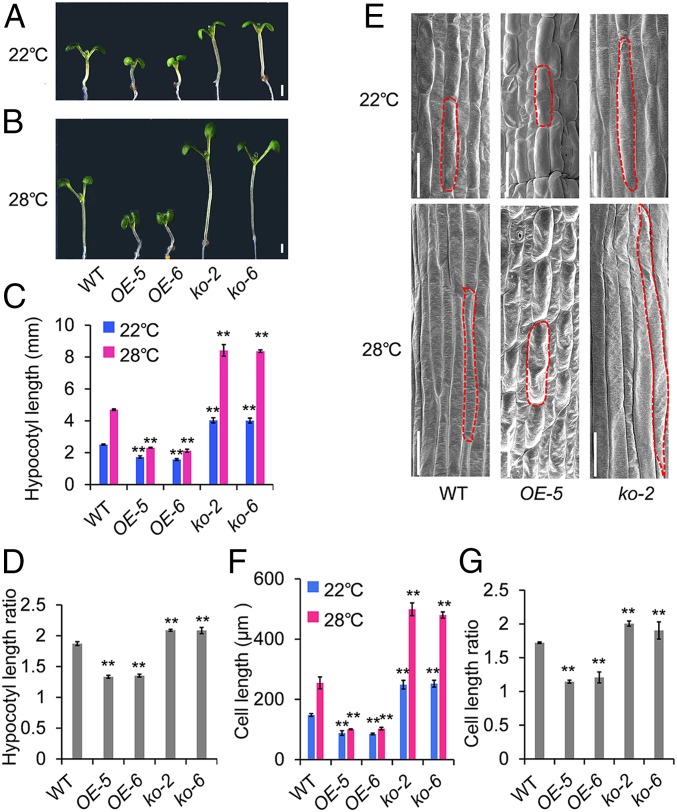
Given that PIF4 is involved in high-temperature–induced hypocotyl elongation in Arabidopsis (8, 39, 40) and transcriptionally regulates UGT76F1 expression, we investigated whether UGT76F1 is also involved in the plant response at an elevated temperature. An elevated temperature of 28 °C is known to promote significant hypocotyl elongation. Thus, hypocotyl elongation of WT, UGT76F1 overexpression lines, and ugt76f1 mutants was recorded after 6 d of growth in continuous white light at 22 °C or at 22 °C for 4 d and then transferred at 28 °C for another 48 h. The ugt76f1 mutants exhibited longer hypocotyls, whereas the UGT76F1 overexpression lines produced shorter hypocotyls at 22 °C (Fig. 4A). When transferred at 28 °C, both mutants and overexpressors further increased the differences in hypocotyl length with respect to WT (Fig. 4 B–D). We measured and quantified the cell length in the elongation zone of the hypocotyls. Similarly, both mutants and overexpressors increased the differences in the cell length of hypocotyls from WT at 28 °C compared with that at 22 °C (Fig. 4 E–G).
Fig. 4.
UGT76F1 inhibits high-temperature–promoted hypocotyl elongation. (A and B) Phenotypes of seedlings grown in continuous white light at 22 °C for 6 d (A) or at 22 °C for 4 d and then shifted to 28 °C for another 48 h (B). (Scale bars, 1 mm.) (C) Hypocotyl length of indicated seedlings shown in A and B. (D) Hypocotyl length ratios (28 °C/22 °C) of the quantified hypocotyl length in C. (E) Hypocotyl cell length observed by scanning electron microscope. (Scale bars, 100 µm.) (F) Cell-length quantification of indicated seedlings shown in E. (G) Cell-length ratios (28 °C/22 °C) of the quantified cell length in F. Data are means ± SD. n = 30. Significant difference was compared to respective WT. Student’s t test was performed (**P < 0.01).
Transcript analysis showed that the transcript abundance of the UGT76F1 gene was reduced in a 28 °C treatment (SI Appendix, Fig. S3A). As soon as PIF4 is mutated, the high temperature-induced suppression of UGT76F1 transcript accumulation attenuates (SI Appendix, Fig. S3B). These data indicate that PIF4 acts as a negative regulator of UGT76F1 transcription in high-temperature–induced hypocotyl elongation.
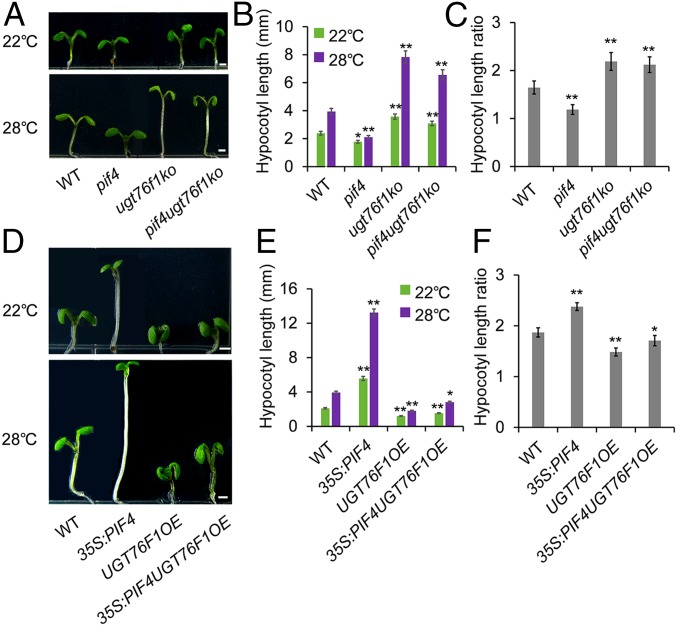
Furthermore, we compared the hypocotyl phenotypes of pif4, ugt76f1ko, and pif4 ugt76f1ko double mutants. We found that the removal of UGT76F1 dramatically increased the hypocotyl elongation at 28 °C. The pif4 ugt76f1ko double mutants significantly suppressed the phenotype of pif4 mutants (Fig. 5 A–C). In addition, we obtained the 35S:PIF4 UGT76F1OE hybrid plants (SI Appendix, Fig. S4). We found that hybrid plants clearly suppressed the long hypocotyl phenotype of the 35S:PIF4 plants at 22 °C and 28 °C (Fig. 5 D–F). In conclusion, the above data provide reliable evidence that UGT76F1 genetically acts downstream of PIF4 in regulating high-temperature–induced hypocotyl elongation.
Fig. 5.
UGT76F1 is involved in PIF4-mediated and temperature-promoted hypocotyl elongation. (A) Phenotypes of pif4, ugt76f1ko, and pif4 ugt76f1ko seedlings grown in continuous white light at 22 °C for 6 d or at 22 °C for 4 d and then transferred to 28 °C for another 48 h. (Scale bar, 1 mm.) (B) Hypocotyl length of seedlings shown in A. Data are means ± SD. n = 30. Significant difference was compared to respective WT at 22 °C or 28 °C. Student’s t test was performed (*P < 0.05, **P < 0.01). (C) Hypocotyl-length ratios (28 °C/22 °C) of the quantified hypocotyl length in B. (D) Phenotypes of 35S:PIF4, UGT76F1OE, and 35S:PIF4UGT76F1OE seedlings grown in continuous white light at 22 °C for 6 d or at 22 °C for 4 d and then transferred to 28 °C for another 48 h. (Scale bars, 1 mm.) (E) Hypocotyl length of seedlings shown in D. Data are means ± SD. n = 30. Significant difference was compared to respective WT at 22 °C or 28 °C. Student’s t test was performed (*P < 0.05, **P < 0.01). (F) Hypocotyl-length ratios (28 °C/22 °C) of the quantified hypocotyl length in E.
UGT76F1 Catalyzes IPyA Glucosylation In Vitro and In Vivo.
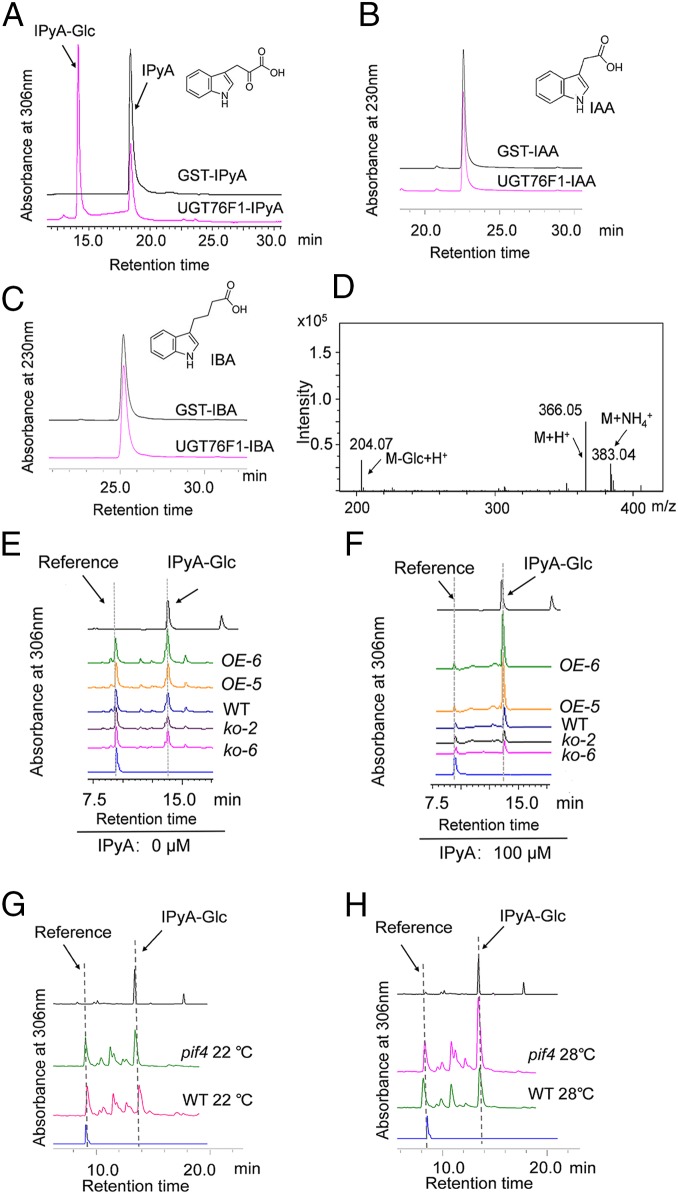
UGT76F1 is a member of the UDP-dependent glycosyltransferase family, which mainly uses UDP-glucose as a sugar donor to catalyze the glucosylation of plant secondary metabolites (41–43). To comprehend the mechanism by which UGT76F1 is involved in the regulation of hypocotyl growth, a series of natural plant secondary metabolites were screened to identify their possible substrates. UGT76F1 fused with GST was expressed in Escherichia coli, purified, and used in this enzyme assay. We found that UGT76F1 had a high and specific enzymatic activity only toward IPyA (SI Appendix, Table S1, and Fig. 6A). IPyA is known to be the major precursor of IAA in the auxin biosynthesis pathway. Interestingly, when IAA and indole-3-butytric acid (IBA), which exist widely in higher plants as active forms of auxin, were tested as potential substrates, no detectable enzyme activity was found (Fig. 6 B and C), suggesting that only IPyA, but not IAA and IBA, is the natural substrate of UGT76F1 in planta. Moreover, liquid chromatography-mass spectrometry (LC-MS) was used to verify the reaction products of IPyA. The molecular weight (M) of the IPyA-glucose conjugate (IPyA-Glc) is known to be 365. In the positive ionization mode, the reaction products exhibited dominant ion peaks at m/z 204 (M + H+− Glc), 366 (M + H+), and 383 (M + NH4+), which correspond well to the expected protonated molecular ions of IPyA-Glc (Fig. 6D). In addition to the end-point analysis, we also examined the kinetic data for IPyA glycosylation. As a result, the high glucosylating activity of UGT76F1 toward IPyA was demonstrated in vitro (SI Appendix, Table S2).
Fig. 6.
The in vitro and in vivo glucosylating activity of UGT76F1 toward IPyA. (A) Potential reaction product (IPyA-glucose conjugates, IPyA-Glc) from IPyA was found by HPLC analysis. UDP-glucose was used as the sugar donor. (B) No reaction product from IAA was found by HPLC analysis. (C) No reaction product from IBA was found by HPLC analysis. (D) Identity of reaction products from IPyA was further confirmed by LC-MS analysis under positive ion mode. (E and F) HPLC profiling of IPyA glucose conjugates (IPyA-Glc) from WT, overexpression lines (OE-5, OE-6), and mutant lines (ko-2, ko-6). The 7-d-old seedlings grown at 22 °C were incubated without (E) or with (F) 100 μM IPyA for 12 h before the extraction process. (G) HPLC profiling of IPyA glucose conjugates in WT and pif4 mutant lines grown at 22 °C for 7 d. (H) HPLC profiling of IPyA glucose conjugates in WT and pif4 mutants grown at 22 °C for 6 d and then shifted to 28 °C for 24 h. Caffeic acid was used as a reference in these assays to monitor the recovery rate. The IPyA glucose conjugates formed by UGT76F1 catalysis were used as the standards. Three biological replicates were conducted, yielding similar results.
To further investigate whether UGT76F1 contributes to IPyA glucosylation in vivo, the endogenous total IPyA-Glc from UGT76F1 overexpression lines and mutant lines was extracted and analyzed by high-performance liquid chromatography (HPLC). Even though the basal level of IPyA-Glc is very low, we can still find that the IPyA-Glc accumulation in UGT76F1 overexpression lines is much higher than that of WT. However, the IPyA-Glc level in ugt76f1 mutants was almost half that in WT (Fig. 6E). If the plant tissues were applied with exogenous IPyA for 12 h before the extraction process, then the UGT76F1 overexpression lines accumulated an even much higher level of IPyA-Glc than that in WT and that in overexpression lines without prior application. On the contrary, IPyA-Glc in the ugt76f1 mutants was much less than that in WT (Fig. 6F). These data indicate that UGT76F1 can catalyze IPyA glucosylation in vivo. The endogenous IPyA-Glc extracted from overexpression lines and mutant lines was further subjected to LC-MS analysis, and the IPyA-Glc identity was validated (SI Appendix, Fig. S5).
From the above-mentioned assays, we know that UGT76F1 is negatively regulated by PIF4. Thus, to figure out whether the IPyA-Glc formation is affected by PIF4, we compared the IPyA-Glc accumulation of pif4 mutants with WT under white light and different temperatures (22 °C and 28 °C). The experimental results indicated that IPyA-Glc contents were obviously increased upon PIF4 removal compared with the WT at either 22 °C or 28 °C under continuous white light (Fig. 6 G and H). This result not only shows the effect of PIF4 on IPyA-Glc formation, but also further proves that PIF4 negatively regulates UGT76F1 expression from a biochemical perspective.
In conclusion, our results demonstrate the biochemical function of UGT76F1 and indicate that UGT76F1 is responsible for IPyA glucosylation in plant cells.
UGT76F1 Functions in Regulating Endogenous Auxin Levels.
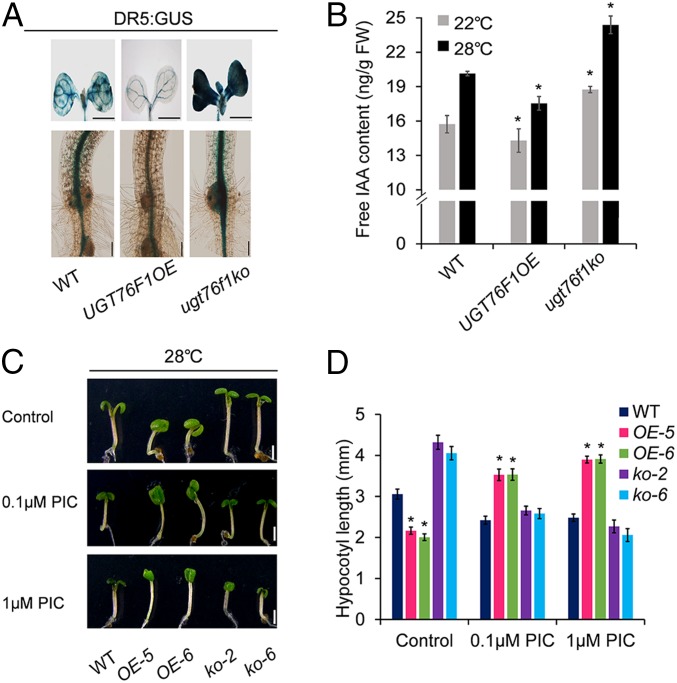
In the YUC flavin monooxygenase pathway of auxin biosynthesis, which is considered to be the main auxin biosynthesis pathway in Arabidopsis, IPyA is directly converted into IAA by the flavin monooxygenase-like proteins (YUC family). Given that IPyA can be glycosylated by UGT76F1, whether or not UGT76F1 affects endogenous auxin level and auxin response would be an interesting question. To this end, we introduced the auxin-responsive reporter construct DR5:GUS into UGT76F1 transgenic plants. After IAA treatment, the expression of DR5:GUS exhibited a dramatic decrease in UGT76F1OE plants compared to that of WT, whereas ugt76f1ko plants exhibited a dramatic increase in DR5:GUS expression (Fig. 7A). We further determined the content of free IAA in 5-d-old light-grown seedlings. UGT76F1 overexpression seedlings had lower levels of IAA, whereas ugt76f1 mutants accumulated much more IAA than WT at both 22 °C and 28 °C (Fig. 7B), indicating that UGT76F1 has an important role in regulating endogenous auxin levels through IPyA glucosylation. Our observation of root length supports this conclusion (SI Appendix, Fig. S6).
Fig. 7.
UGT76F1 functions in regulating IAA amount. (A) DR5:GUS expression of indicated seedlings showing auxin responses. (Scale bars: 200 µm, Upper; 100 µm, Lower.) (B) IAA content measurement of indicated seedlings grown in continuous white light (70 µmol m−2⋅s−1) conditions at 22 °C or 28 °C for 5 d. Data are means ± SD. n = 3. FW, fresh weight. Asterisks indicate significant differences relative to respective WT at 22 °C or 28 °C (Student’s t test, *P < 0.05). (C) Phenotypes of seedlings grown with or without the synthetic auxin picloram (PIC). Seedlings were grown in continuous light at 22 °C for 2 d and then transferred to plates containing picloram at 28 °C for another 2 d. (Scale bars, 1 mm.) (D) Hypocotyl length of seedlings shown in C. Data are means ± SD. n = 30. Significant difference was compared to WT (Student’s t test, *P < 0.05).
Picloram is a synthetic “proauxin” compound that can enter plant tissues and is subsequently hydrolyzed to substance with auxin activity, which can efficiently promote hypocotyl elongation (44). We used picloram to study the effect of UGT76F1 on endogenous auxin levels. We found that, at high concentrations of picloram capable of inhibiting hypocotyl elongation of WT at 28 °C, UGT76F1 overexpression reduced the inhibition efficiency, while loss of UGT76F1 function enhanced the inhibition efficiency (Fig. 7 C and D). This experiment suggests that UGT76F1 alleviates the inhibitory effect of auxin on hypocotyl elongation to some extent through glucosylation of endogenous IAA.
In the IPyA pathway of IAA biosynthesis, tryptophan aminotransferase (TAA/TAR1) converts Trp to IPyA, which is subsequently converted by the YUCs to IAA (32, 33, 45). L-kynurenine, a Trp analog, was identified as a competitive inhibitor of TAA1/TARs in Arabidopsis (46). To examine the relationship between UGT76F1 and TAA1, the phenotypes of kynurenine-treated WT (equivalent to taa mutant) and ugt76f1ko plants were studied. We discovered that UGT76F1 knockouts can suppress the phenotype of kynurenine-treated WT. In addition, the phenotype of UGT76F1 knockout mutants can also be suppressed by L-kynurenine (SI Appendix, Fig. S7). This experiment provides strong support for the role of UGT76F1 in the inactivation of endogenous IAA.
Furthermore, we studied the effects of UGT76F1 and PIF4 on the expression of several auxin-responsive genes in 5-d-old seedlings grown continuously at 22 °C or shifted to 28 °C for another 5 h. Experimental results show that the loss of UGT76F1 function results in significant up-regulation of these auxin-responsive genes, whereas UGT76F1 overexpression results in significant down-regulation (SI Appendix, Figs. S8 and S9). On the contrary, the loss of PIF4 function results in significant down-regulation of those auxin-responsive genes, whereas PIF4 overexpression results in significant up-regulation (with only a few We also examined gene expressions in pif4 ugt76f1ko double mutants and in 35S:PIF4 UGT76F1OE seedlings. It was revealed that double mutants have higher expression levels of those genes than pif4 single mutants. However, simultaneous overexpression of PIF4 and UGT76F1 resulted in significantly reduced gene expressions compared to only PIF4 overexpression (SI Appendix, Figs. S8 and S9). These data suggest that PIF4 regulates the expression of auxin-responsive genes in a UGT76F1-dependent and contrary manner.
UGT76F1 Antagonizes YUC in Regulating Auxin Biosynthesis and Hypocotyl Growth.
The direct conversion of IPyA to IAA by YUCs is considered to be the main auxin biosynthesis pathway in Arabidopsis (32, 47, 48). On the other hand, IPyA can also be directly converted to IPyA-Glc conjugates through UGT76F1. Thus, IPyA would be the shared substrate for both YUCs and UGT76F1. This means that there may be a competitive mechanism in plants to control different fluxes of IPyA. To investigate whether UGT76F1 and YUCs competitively affect auxin biosynthesis and auxin responses, yucasin [5-(4-chlorophenyl)-4H-1, 2, 4-triazole-3-thiol], an inhibitor of YUC activity (49), was introduced into our experiments. In comparison with the WT, when seedlings are grown with 20 or 200 µM yucasin under white light or dark conditions, UGT76F1 overexpression lines exhibit shorter hypocotyls, shorter roots, and severely disturbed gravitropism, indicating a reduced auxin biosynthesis and an auxin response due to UGT76F1 overexpression. In contrast, ugt76f1 mutants develop longer hypocotyls, longer roots and better gravitropism compared to the WT. These results suggest that UGT76F1 antagonizes YUCs in regulating auxin biosynthesis and auxin responses (SI Appendix, Figs. S10 and S11).
Furthermore, we also detected the levels of IPyA-Glc in WT and UGT76F1 overexpression lines before and after the addition of yucasin. We found that exogenous yucasin treatment did increase IPyA-Glc accumulation in UGT76F1 overexpression lines and in WT plants, implying that the inhibition of YUC activity would prevent IPyA from flowing into IAA, while UGT76F1 tends to catalyze more IPyA to form IPyA-Glc in the absence of YUC (SI Appendix, Fig. S11C). Therefore, how to balance the trade-off between UGT76F1 and YUC is very important for the normal growth of plants.
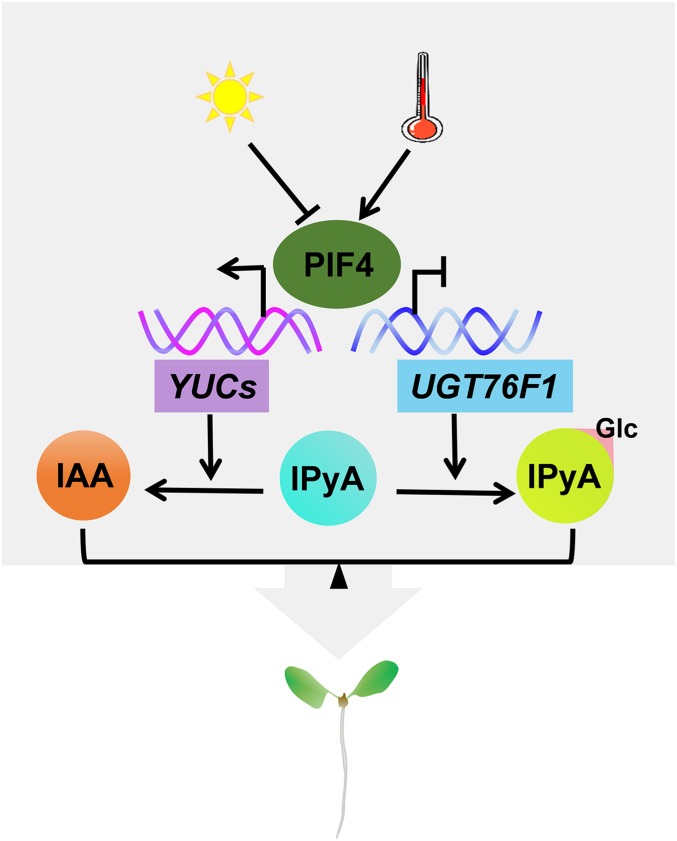
In summary, our results in this study reveal the crucial role of IPyA glucosyaltion in maintaining auxin homeostasis, which is a key watershed in auxin biosynthesis. Since PIF4 can simultaneously regulate the transcription of YUCs (28, 50) and UGT76F1 (this study), our data also suggest that PIF4 controls the contrary conversion of IPyA to IAA and IPyA-Glc to precisely modulate auxin homeostasis and plant growth in a light- and temperature-dependent manner (Fig. 8).
Fig. 8.
A proposed working model for UGT76F1-mediated auxin homeostasis in regulating light- and temperature-dependent seedling growth. IPyA is the major precursor for IAA biosynthesis in plants. YUCs promote the conversion from IPyA to IAA, whereas UGT76F1 antagonizes the role of YUCs through glucosylating IPyA to form its sugar conjugates. Acting as the pivotal integrator in light- and temperature-signaling pathways, PIF4 can transcriptionally regulate the expression of both YUCs (28, 50) and UGT76F1 (this study) through directly binding to their promoters, maintaining a trade-off between IAA biosynthesis and IPyA-Glc formation at the IPyA watershed and thereby fine-tuning the auxin homeostasis and the light- and temperature-dependent adaptation growth of seedlings.
Discussion
The glucosylation of auxins is a well-recognized modification that is believed to play an important role in hormonal homeostasis. Here, we demonstrate that the UDP-glucosyltransferase UGT76F1 strongly favors IPyA as a substrate (precursor of IAA biosynthesis), rather than auxin itself. So far, five Arabidopsis glycosyltransferases have been found to glucosylate auxin itself to form its glucose conjugate. UGT84B1 was identified to be an IAA preferring glucosyltransferase, whereas UGT74E2, UGT74D1, UGT75D1, and UGT84A2 were considered to be an IBA preferring glycosyltransferase (51–55). Previous researches have reported that overexpression of these auxin glycosyltransferase genes results in higher levels of IAA or IBA and related phenotypes. Unlike those reports, our study shows that overexpression of UGT76F1 displays phenotypes reminiscent of auxin-deficient mutants, such as short hypocotyls. In addition, UGT76F1 belongs to group H of the UGT family, whereas the five previously reported UGTs are members of group L (56). These differences suggest that UGT76F1 assumes a different responsibility from other auxin UGTs in the regulation of auxin homeostasis. In plant evolution, the development of multiple glycosyltransferases toward the same kind of phytohormones may be beneficial for plants to make good adaptive response to environmental changes. For example, in controlling auxin biosynthesis and metabolism, it may be necessary to glucosylate metabolites at different stages of auxin synthesis, with UGT76F1 for precursor and other UGTs for active auxin forms. This requirement not only reflects the importance of auxin homeostasis for plant growth and development, but also reflects the cooperation between the glycosyltransferase members of this large family.
Recent studies have shown that there exists a cross talk between light-signaling and temperature-signaling pathways in regulating plant growth and development (1, 57). PIF4 is identified as a vital player in sensing light and temperature to mediate plant growth and development by promoting the expression of YUCs. In this study, our results indicate that PIF4 directly binds to the promoter of UGT76F1, negatively regulates UGT76F1 expression, and causes adaptive changes in IAA levels and hypocotyl elongation in response to light and elevated temperature. Thus, this study proposes that UGT76F1 links PIF4 to the auxin pathway.
In this process, UGT76F1 antagonizes the role of YUCs through glucosylating IPyA to form its sugar conjugates. Acting as the pivotal integrator in light and temperature signaling pathways, PIF4 can transcriptionally control the expression of both YUCs and UGT76F1, maintaining a trade-off between IAA synthesis and IPyA-Glc formation at the IPyA watershed (Fig. 8). Thus, our findings reveal a mechanism for regulating auxin homeostasis for plant growth and development adaptive to external environmental changes.
Materials and Methods
Detailed description of plant materials, plant growth conditions, and methods for the preparation of the UGT76F1 promoter::GUS construct, GUS staining (58, 59), preparation for transgenic and knockout lines of UGT76F1 (60), phenotypic analyses, yeast one-hybrid assay, transient luciferase activity assay (61), ChIP assays, purification of UGT76F1 enzyme and assays of glucosyltransferase activity (41, 56, 62), free IAA measurement, RNA isolation, quantitative real-time PCR, and statistical analysis can be found in SI Appendix, SI Materials and Methods. The phyA and phyB mutants were ordered from The European Arabidopsis Stock Centre (NASC).
Data Availability.
All data and the associated protocols are included in the manuscript and SI Appendix. Plant materials presented in this article can be provided upon request.
Supplementary Material
Acknowledgments
We thank Prof. Ming-Yi Bai (Shandong University) for providing the mutant seeds of cry1/cry2 and the instruments for light treatments; Zhifeng Li, Jingyao Qu, and Jing Zhu (State Key Laboratory of Microbial Technology of Shandong University) for assistance in LC-MS/MS; and Sen Wang, Haiyan Yu, and Xiaomin Zhao (State Key Laboratory of Microbial Technology of Shandong University) for assistance in microimaging of SEM analysis. This research is supported by National Natural Science Foundation of China Grants 31770313, 91217301, and 31970290 (to B.-K.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000172117/-/DCSupplemental.
References
- 1.Kami C., Lorrain S., Hornitschek P., Fankhauser C., Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Lorrain S., Allen T., Duek P. D., Whitelam G. C., Fankhauser C., Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Brock M. T., Maloof J. N., Weinig C., Genes underlying quantitative variation in ecologically important traits: PIF4 (phytochrome interacting factor 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana. Mol. Ecol. 19, 1187–1199 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Li L., et al. , Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castillon A., Shen H., Huq E., Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12, 514–521 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Leivar P., Quail P. H., PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leivar P., Monte E., PIFs: Systems integrators in plant development. Plant Cell 26, 56–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koini M. A., et al. , High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Stavang J. A., et al. , Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60, 589–601 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Nozue K., et al. , Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Kidokoro S., et al. , The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151, 2046–2057 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashino T., et al. , A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44, 619–629 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Khanna R., et al. , The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19, 3915–3929 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng S., et al. , Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y., Lau O. S., Deng X. W., Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Lau O. S., Deng X. W., Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 13, 571–577 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Hornitschek P., et al. , Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Franklin K. A., Light and temperature signal crosstalk in plant development. Curr. Opin. Plant Biol. 12, 63–68 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Kumar S. V., et al. , Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma D., et al. , Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. U.S.A. 113, 224–229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quint M., et al. , Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2, 15190 (2016). [DOI] [PubMed] [Google Scholar]
- 22.de Lucas M., Prat S., PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 202, 1126–1141 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Zhu J.-Y., Oh E., Wang T., Wang Z. Y., TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7, 13692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Q., et al. , SHB1 and CCA1 interaction desensitizes light responses and enhances thermomorphogenesis. Nat. Commun. 10, 3110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lucas M., et al. , A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Bai M.-Y., et al. , Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh E., Zhu J.-Y., Wang Z.-Y., Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J., Qi L., Li Y., Chu J., Li C., PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 8, e1002594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh E., et al. , Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3, e03031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray W. M., Ostin A., Sandberg G., Romano C. P., Estelle M., High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 7197–7202 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franklin K. A., et al. , Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108, 20231–20235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mashiguchi K., et al. , The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y., Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma L., Zhao H., Deng X. W., Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130, 969–981 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Foster R., Izawa T., Chua N. H., Plant bZIP proteins gather at ACGT elements. FASEB J. 8, 192–200 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Izawa T., Foster R., Chua N. H., Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230, 1131–1144 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y., et al. , A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9, e1003244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rombauts S., Déhais P., Van Montagu M., Rouzé P., PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proveniers M. C., van Zanten M., High temperature acclimation through PIF4 signaling. Trends Plant Sci. 18, 59–64 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Huai J., et al. , SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Mol. Plant 11, 928–942 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Lim E. K., Bowles D. J., A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 23, 2915–2922 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gachon C. M., Langlois-Meurinne M., Saindrenan P., Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 10, 542–549 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Bowles D., Lim E. K., Poppenberger B., Vaistij F. E., Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57, 567–597 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Savaldi-Goldstein S., et al. , New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc. Natl. Acad. Sci. U.S.A. 105, 15190–15195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korasick D. A., Enders T. A., Strader L. C., Auxin biosynthesis and storage forms. J. Exp. Bot. 64, 2541–2555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He W., et al. , A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23, 3944–3960 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stepanova A. N., et al. , The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Won C., et al. , Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishimura T., et al. , Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 77, 352–366 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Liu G., et al. , Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PLoS Genet. 12, e1006360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson R. G., et al. , Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 276, 4350–4356 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Tognetti V. B., et al. , Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22, 2660–2679 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin S.-H., et al. , UGT74D1 is a novel auxin glycosyltransferase from Arabidopsis thaliana. PLoS One 8, e61705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang G.-Z., et al. , Ectopic expression of UGT75D1, a glycosyltransferase preferring indole-3-butyric acid, modulates cotyledon development and stress tolerance in seed germination of Arabidopsis thaliana. Plant Mol. Biol. 90, 77–93 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Zhang G.-Z., et al. , Ectopic expression of UGT84A2 delayed flowering by indole-3-butyric acid-mediated transcriptional repression of ARF6 and ARF8 genes in Arabidopsis. Plant Cell Rep. 36, 1995–2006 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Ross J., Li Y., Lim E., Bowles D. J., Higher plant glycosyltransferases. Genome Biol. 2, REVIEWS3004 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foreman J., et al. , Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 65, 441–452 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Stomp A. M., “Histochemical localization of β-glucuronidase” in GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression, Gallagher S. R., Ed. (Academic Press, London, 1992), pp. 103–113. [Google Scholar]
- 59.Jefferson R. A., Kavanagh T. A., Bevan M.-W., GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Hellens R. P., et al. , Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou B., Lim E. K., Higgins G. S., Bowles D. J., N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279, 47822–47832 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and the associated protocols are included in the manuscript and SI Appendix. Plant materials presented in this article can be provided upon request.