Significance
Human Vγ9Vδ2 T cells are important mediators of immunosurveillance and targets for cell-based immunotherapy. Activation by small molecular compounds called phosphoantigens is essential for their functions and involvement of butyrophilins (BTN3A) has been shown. However, it is not completely understood how the intracellular recognition of phosphoantigens by BTN3 is translated to the cell surface and finally to Vγ9Vδ2 T cell activation. In addition, phosphoantigen-reactive Vγ9Vδ2 T cells were only identified in primates but not in rodents. In this study, we present a phosphoantigen-reactive Vγ9Vδ2 T cell subset in Vicugna pacos (alpaca) and show similarities and instructive differences to the human system that can be applied for future structural and functional Vγ9Vδ2 T cell research and development of nonprimate animal models.
Keywords: Vicugna pacos, alpaca, Vγ9Vδ2 T cell, phosphoantigen, butyrophilin
Abstract
Vγ9Vδ2 T cells are a major γδ T cell population in the human blood expressing a characteristic Vγ9JP rearrangement paired with Vδ2. This cell subset is activated in a TCR-dependent and MHC-unrestricted fashion by so-called phosphoantigens (PAgs). PAgs can be microbial [(E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate, HMBPP] or endogenous (isopentenyl pyrophosphate, IPP) and PAg sensing depends on the expression of B7-like butyrophilin (BTN3A, CD277) molecules. IPP increases in some transformed or aminobisphosphonate-treated cells, rendering those cells a target for Vγ9Vδ2 T cells in immunotherapy. Yet, functional Vγ9Vδ2 T cells have only been described in humans and higher primates. Using a genome-based study, we showed in silico translatable genes encoding Vγ9, Vδ2, and BTN3 in a few nonprimate mammalian species. Here, with the help of new monoclonal antibodies, we directly identified a T cell population in the alpaca (Vicugna pacos), which responds to PAgs in a BTN3-dependent fashion and shows typical TRGV9- and TRDV2-like rearrangements. T cell receptor (TCR) transductants and BTN3-deficient human 293T cells reconstituted with alpaca or human BTN3 or alpaca/human BTN3 chimeras showed that alpaca Vγ9Vδ2 TCRs recognize PAg in the context of human and alpaca BTN3. Furthermore, alpaca BTN3 mediates PAg recognition much better than human BTN3A1 alone and this improved functionality mapped to the transmembrane/cytoplasmic part of alpaca BTN3. In summary, we found remarkable similarities but also instructive differences of PAg-recognition by human and alpaca, which help in better understanding the molecular mechanisms controlling the activation of this prominent population of γδ T cells.
All jawed vertebrates possess three lineages of rearranged antigen receptor-bearing lymphocytes: αβ T cells, γδ T cells, and Ig-expressing B cells. So far, a unifying concept on γδ T cell antigen recognition and function, analogous to αβ T cells and the association of MHC class restriction, is still pending, despite increasing knowledge about γδ T cell effector functions and the structure of γδ T cell receptors (TCRs) and γδ TCR–ligand interactions. Indeed, γδ T cells show features of adaptive and innate immune cells and a remarkable degree of diversification in function and anatomical localization, which is often linked to TCR usage that differs between species (1–3). This study focuses on Vγ9Vδ2 T cells, which are the major γδ T cell population in human blood and express a characteristic Vγ9JP (TRGV9/TRGJP) rearrangement paired with a Vδ2 chain using TRDV2 gene segments (4–7). Length restriction of the junctional region and a restricted repertoire of the Vγ9 chain are typical for Vγ9Vδ2 T cells, whereas the Vδ2 chain shows higher diversity (8, 9). The TCR-dependent activation of Vγ9Vδ2 T cells by so-called phosphoantigens (PAg), phosphorylated metabolites of isoprenoid synthesis pathways, is not MHC-restricted but requires cell–cell contact (10). For this activation, expression of B7-like butyrophilin (BTN3A1, CD277) molecules by the antigen-presenting or target cells is mandatory (11–13). Of note, the influence of other B7 receptor family-like butyrophilin (BTN) and butyrophilin-like (BTNL) molecules on the development of specific γδ T cell subsets has been shown in mice (Skint-1, Btnl1/Btnl6) and humans (BTNL3/BTNL8) (14–17).
Upon activation, human Vγ9Vδ2 T cells can rapidly release cytokines and kill transformed or infected cells by perforin and granzyme release, TCR-mediated and NKG2D-dependent mechanisms, as well as Fas–Fas ligand interactions (18–20). They proliferate upon activation in vitro and frequencies of up to 50% of circulating T cells have been observed in vivo after infection (21). Vγ9Vδ2 TCRs act in a more innate-like fashion (22), sensing accumulation of endogenous PAgs, such as isopentenyl pyrophosphate (IPP), as a consequence of changes in cell metabolism (23, 24) or treatment with drugs like aminobisphosphonates (25). Vγ9Vδ2 T cells are also able to sense microbial infection through the detection of higher potency PAgs deriving from these microorganisms (21). These features made Vγ9Vδ2 T cells the first clinically explored γδ T cell subset, for which objective antitumor responses have been found (22, 26–28). The most potent natural PAg is (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate (HMBPP), a precursor of IPP in the methylerythritol 4-phosphate pathway, which is found in many eubacteria, apicomplexan parasites (e.g., plasmodium and chloroplasts) and is responsible for the massive expansion or modulation of Vγ9Vδ2 T cells in infections like malaria or tuberculosis (7, 29).
Functional Vγ9Vδ2 T cells have so far only been described in humans and higher primates, but in recent years the longstanding belief that Vγ9Vδ2 T cells are only functionally conserved in primate species (12, 30–32) has been challenged by a genome-based approach of our group (3). In brief, we showed that the three genes, known so far to be essential for a PAg response—TRGV9, TRDV2, and BTN3—are conserved and in silico translatable in the genomes of a few nonprimate mammalian species and partially or completely lost in others, such as rodents. These genes appeared with the emergence of placental mammals as underlined by our analysis of the Xenarthra species armadillo (Dasypus novemcinctus), which possesses TRGV9, TRDV2, and BTN3 genes, although only partially functional, that show remarkably conserved features to their human counterparts (33). Furthermore, gene expression of productive TRGV9- and TRDV2-like rearrangements and a single BTN3-like gene was demonstrated in the cDNA of the New World camelid alpaca (Vicugna pacos) (2, 3). TCR rearrangements strongly resembling those of PAg-reactive primate TCRs could be shown for alpaca (3). Expression of cloned alpaca TCRs in TCR− murine T-cell hybridoma lines with endogenous CD3 expression resulted in the surface expression of murine CD3, suggesting formation of an alpaca TCR/murine CD3 complex. Culture with anti-mouse–CD3ε induced production of murine IL-2 (3) but a direct demonstration of PAg-reactivity of alpaca TCRs was still pending. Applying newly generated research tools, we now present alpaca as a nonprimate species with a Vγ9Vδ2-like T cell subset that recognizes PAgs in a BTN3-dependent manner. We found remarkable similarities but also instructive differences of PAg recognition by human and alpaca, which help us to better understand the molecular mechanisms controlling the activation of this prominent population of γδ T cells.
Results
Development of Monoclonal Antibodies Specific for Alpaca Vδ2 Chains and BTN3.
As previously published and highlighted in SI Appendix, Figs. S1 and S2, Vγ9, Vδ2, and BTN3 proteins of human and alpaca bear remarkable similarities, although differing in the number of BTN3 genes due to duplication events during primate evolution. Humans express three cooperatively acting BTN3 molecules, of which BTN3A1 possesses a PAg-binding groove in its intracellular B30.2 domain (34, 35). This is lost due to a H351R substitution in BTN3A3 and missing in BTN3A2 as a consequence of a lacking B30.2 domain. Alpaca expresses a single BTN3 (2, 3, 33), but its PAg-binding site is strikingly similar to that of BTN3A1, and both BTNs are identical in those amino acids (SI Appendix, Fig. S1) proposed to be pivotal for PAg-sensing or TCR recognition in different models of PAg-mediated activation (13, 34–36). The same identity is found for amino acids proposed to form a controversial extracellular binding site and for 3 amino acids of the Vγ9 located outside the classic CDRs recently shown to be critical for PAg-mediated activation (36). To directly identify PAg-reactive cells and analyze expression and function of the BTN3 molecule in the alpaca system, monoclonal antibodies were generated against murine cells expressing transduced alpaca Vγ9Vδ2 TCR or alpaca BTN3, as described in Methods.
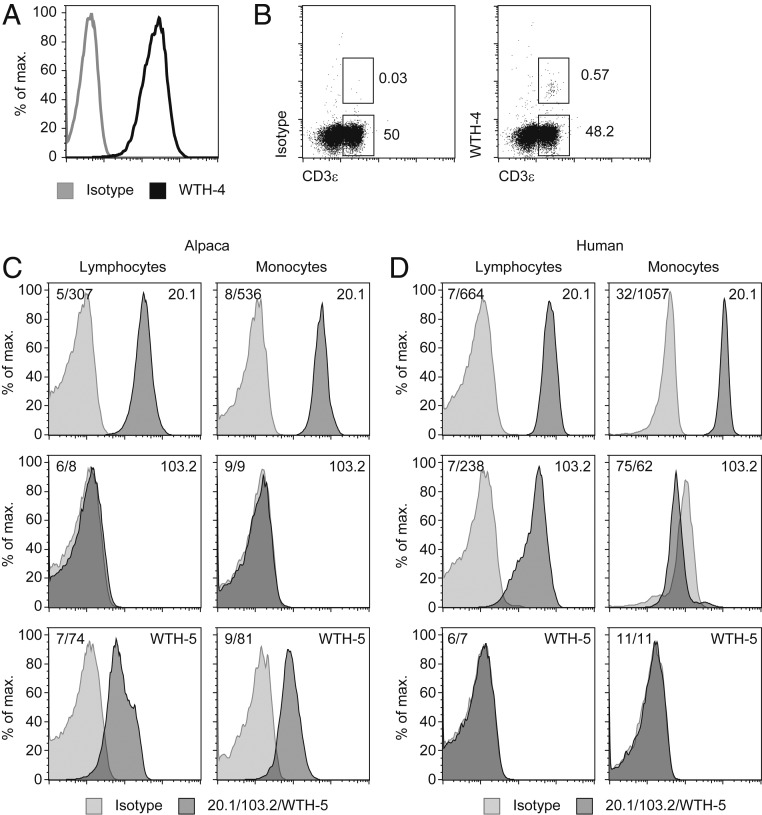
The specificity of the antibody WTH-4, which was raised against the alpaca Vγ9Vδ2 TCR, was verified by staining of TCRs expressed by different cell lines: For example, a mouse B cell lymphoma cotransduced with genes for the mouse CD3 complex and previously described alpaca Vγ9Vδ2 TCR genes (3) (Fig. 1A). Only a small population of CD3+ cells was stained by WTH-4 in peripheral blood mononuclear cells (PBMCs) of alpacas (Figs. 1B and 2A). Comparable to humans, frequencies of Vγ9Vδ2 T cells varied between individual animals and even for samples taken from the same individual (0.2 to 1.4% of alpaca T cells). Gating strategies for the identification of alpaca T cells are shown in SI Appendix, Fig. S3. The mAb LT97A (37) could be confirmed as CD3-specific and the identification of monocyte and lymphocyte populations by forward and side-scatter gates was verified (SI Appendix, Fig. S3). Binding of the WTH-4 mAb was dependent on the presence of an alpaca Vδ2 chain (SI Appendix, Fig. S4A), as indicated by cross-reactivity to heteromeric huVγ9/vpVδ2 TCRs and lack of binding to human TCR MOP (huTCR). Furthermore, lack of binding to an alpaca Vγ9Vδ2 TCR using a Jδ2 segment (SI Appendix, Fig. S4B) suggests that the WTH-4 epitope is most likely exclusive to alpaca Vδ2Jδ4 chains, the most widely used Jδ segment in alpacas (3).
Fig. 1.
Development and verification of monoclonal antibodies for alpaca surface molecules. (A) M12.4.1C3 cells expressing mouse CD3 and an alpaca γδ TCR (3) were stained with an mIgG1 isotype control or WTH-4 mAb (0.5 µg/mL) labeled with Fab mIgG1 R-PE. A representative staining is shown with the GM of 3.75 (isotype) and 203 (WTH-4). (B) Binding of an mIgG1 isotype control or WTH-4 mAb (0.5 µg/mL) labeled with Fab mIgG1 R-PE and CD3-FITC to lymphocytes within PBMC. Gating strategy is shown (SI Appendix, Fig. S3). Histograms depict binding of mAbs WTH5 or 103.2 (both 0.125 µg/mL) or of 1:50 dilution of mAb 20.1 PE and their isotype controls to alpaca (C) or human PBMCs (D) gated on cells with scatter characteristics of lymphocytes or monocytes (SI Appendix, Fig. S3).
Fig. 2.
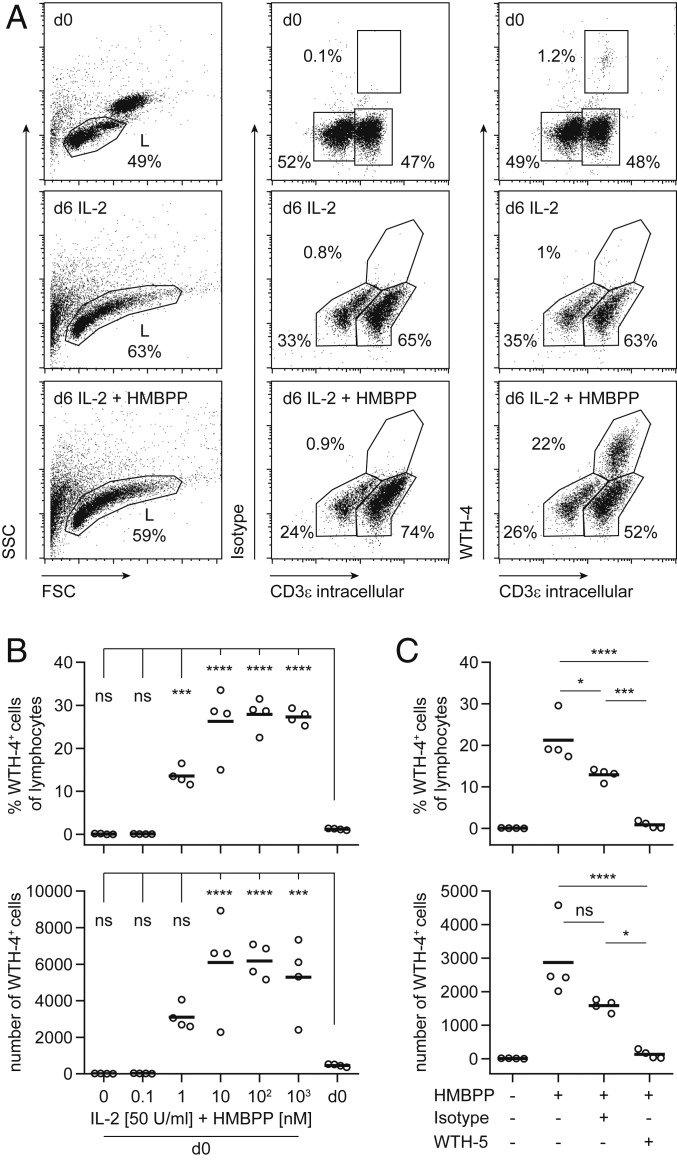
Alpaca PBMCs show BTN3-dependent PAg reactivity. PBMCs were cultured for 6 d in quadruplicates in the presence of 50 U/mL rhIL-2 with/without increasing doses of HMBPP. (A) Gating strategy of unstimulated and stimulated cells (50 U/Ml hIL-2, 100 nM HMBPP) with WTH-4 (0.5 µg/mL, labeled with Fab mIgG1 R-PE) and intracellular α-huCD3ε FITC (2 µg/mL). Cells were gated on live lymphocytes. Representative stainings of WTH-4 (y axis) and CD3ε (x axis) are shown. The frequencies and total cell numbers of Vδ2+/WTH-4+CD3+ cells in a HMBPP titration experiment (B) and in coculture (50 U/mL hIL-2, 1 µM HMBPP) with WTH-5 (1 µg/mL) or isotype (C) were calculated. One representative experiment of three is shown for B and C. Circles indicate single samples of quadruplicates. Results of a one-way ANOVA with Bonferroni post hoc test are shown (ns, not significant: P > 0.05, *P < 0.05, ***P < 0.001, and ****P < 0.0001). Adjusted P values of post hoc analysis of d0 compared to different HMBPP concentrations are plotted in B and of post hoc comparisons of HMBPP+WTH-5+, HMBPP+WTH-5− and HMBPP+Isotype+ with each other are plotted in C.
The monoclonal antibody WTH-5 was identified by a screening for vpBTN3-transduced vs. untransduced cells. As shown in Fig. 1 C and D, alpaca lymphocytes and monocytes were stained to a similar extent and lymphocytes showed a “shoulder” of cells with a slightly higher fluorescence. Human PBMCs were not stained by WTH-5 and the mAb 103.2 was specific for human PBMC but not alpaca PBMC. Human lymphocyte staining was about 20-fold higher than the isotype control with a geometric mean (GM) of 238 vs. 7. Furthermore, human monocyte binding of the IgG2a,κ isotype control was much higher than for IgG2b,κ, with the possible consequence that specific binding of 103.2 could not be observed. Nevertheless, since isotype staining was noticeably lower compared to staining of the same antibody to lymphocytes (GM 238 vs. 62), it can be stated that human monocytes expressed little if any 103.2 epitope. Moreover, PE-labeled mAb 20.1 stained human and alpaca PBMC rather well and binding to monocytes was somewhat higher in both species, as compared to lymphocytes. Curiously, titrations of the three mAbs on huBTN3A1- and vpBTN3-transduced Chinese hamster ovary (CHO) cells revealed species cross-reactivity of all three antibodies only at very high concentrations (SI Appendix, Fig. S5).
Both novel antibodies (WTH-4 and WTH-5) were subsequently used for the analysis of the BTN3/Vγ9Vδ2 T cell system in alpacas.
PAg-Dependent Activation of Alpaca Vγ9Vδ2 T Cells.
The hallmark of human Vγ9Vδ2 T cells is their reactivity to PAgs. Here, we report reactivity of alpaca PBMCs to the widely used PAg HMBPP as tested by in vitro cultures. Alpaca PBMCs were cultured with human recombinant IL-2 and different concentrations of HMBPP for 6 d and the frequency of Vδ2 cells was determined by flow cytometry using the novel marker WTH-4 and intracellular staining with anti-CD3 mAb (Fig. 2A). A significant increase in the frequency of WTH-4 and CD3 double-positive cells was observed compared to unstimulated PBMCs (Fig. 2B). This dose-dependent response was evident from 1 nM HMBPP but not detectable in control samples without HMBPP. The increase in numbers of WTH-4+ cells indicated proliferative activity. The significant down-modulation of the WTH-4 epitope in HMBPP-stimulated cells was most likely a consequence of cell culture and not of HMBPP-specific effects, since it could be observed at all HMBPP concentrations, in cultures with ConA or with medium plus IL-2 only.
Subsequently, the same cell culture approach was used to test whether alpaca BTN3 is involved in PAg recognition. To this end, the alpaca BTN3-specific antibody WTH-5 was added to PBMC stimulations. As shown in Fig. 2C, an HMBPP-dependent increase in frequency and cell numbers of WTH-4+ cells were significantly reduced in cultures with WTH-5 in comparison to cultures with an isotype-matched control antibody, similarly to that reported for the human BTN3-specific mAb BT3 103.2 (11, 31). Isotype-matched control antibody showed some inhibitory effect and this was statistically significant for frequencies but not cell numbers. However, differences between the isotype-matched control and WTH-5 were highly significant. The inhibiting effect of the isotype control may be due to other factors, like binding of the control mAb to Fc receptors of alpaca cells or minute amounts of residual sodium azide in the commercial mAb preparation. Importantly, WTH-5-mediated inhibition was complete and no unspecific effect by the isotype control was found in another experimental setting with TCR and BTN3 transductants (Fig. 3B). These findings imply that as for human Vγ9Vδ2 T cells, primary alpaca Vγ9Vδ2 T cells respond to PAg in a manner comparable to human Vγ9Vδ2 T cells and that BTN3 plays a pivotal role in PAg-induced activation of alpaca Vδ2+ cells.
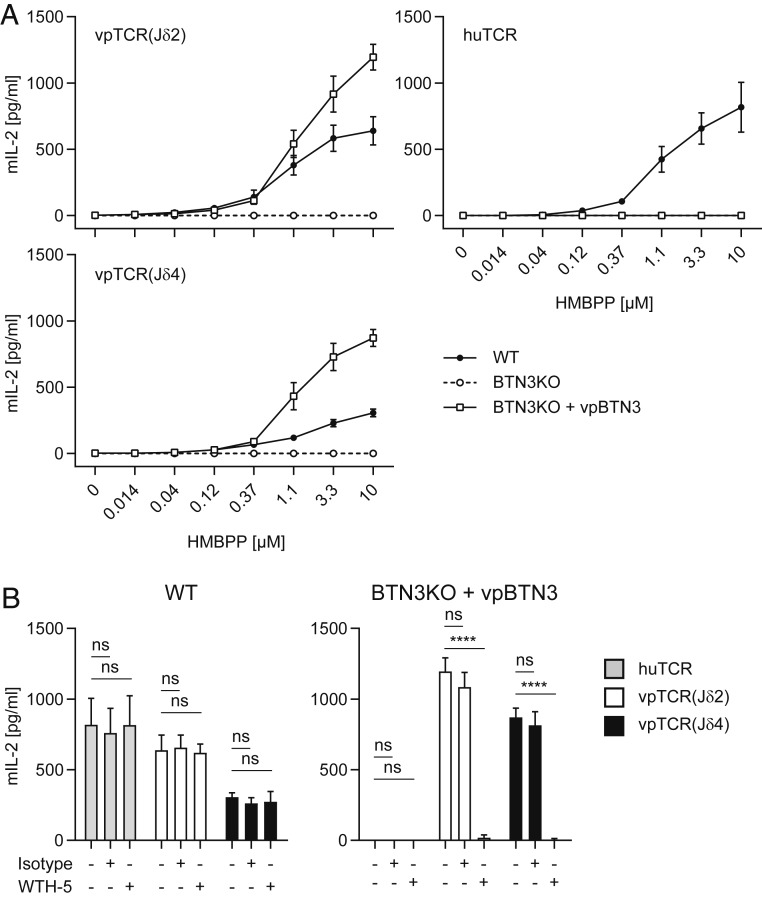
Fig. 3.
BTN3-dependent PAg reactivity of transduced alpaca Vγ9Vδ2 TCRs. (A) Murine responder cells (53/4) expressing vpTCR(Jδ2), vpTCR(Jδ4), or a human Vγ9Vδ2 TCR (huTCR) were cocultured with 293T cell lines (WT), 293T BTN3KO, or BTN3KO vpBTN3 (phNGFR mCherry) in the presence of titrated HMBPP doses. Supernatants were tested for mIL-2 after 22 h. (B) In addition, WTH-5 (1 µg/mL) or mIgG2b,κ isotype were added to cultures of TCR transductants with wild-type or BTN3KO vpBTN3 in the presence of 10 µM HMBPP. Duplicates were measured in three experiments and mean + SD are shown. Statistical analysis is shown in SI Appendix, Table S6 for A and a two-way ANOVA (B) was carried out with a Bonferroni post hoc test (ns, not significant: P > 0.05, ****P < 0.0001).
TCR Usage of Alpaca Vγ9Vδ2 T Cells.
The presence of transcripts encoding TRGV9 and TRDV2 chains in unstimulated alpaca PBMCs was previously shown by M. M. Karunakaran et al. (3, 38). In alpacas, TRGV9 was found to preferentially rearrange with different variants of TRGJP (JP-A, JP-B, or JP-C) (3). The TRDV2 chain rearranged with a TRDJ4 homolog in 16 of 17 clones and in one case with a TRDJ2 homolog (3). To investigate this further, the newly developed antibody WTH-4, most likely specific for alpaca Vδ2Jδ4 chains, was applied to sort alpaca PBMCs before and after HMBPP stimulation. Gene segment usage was determined by TRGV9/TRGC and TRDV2/TRDC amplicon generation from cDNA and TOPO TA cloning for sequencing. Single clonotypes were analyzed according to TRJ usage, CDR3 length, and frequency among analyzed sequences, and are summarized in SI Appendix, Tables S3 and S4. Certain rearrangements occurred in higher frequencies, indicating clonal expansion or better survival and we could verify the preferential rearrangement of TRGV9 with variants of TRGJP homologs and of TRDV2 with TRDJ4. Interestingly, in both animals two variants of TRGJP were used in rearrangements with TRGV9, hinting at the occurrence of allelic versions of TRGJP. As seen in the human system and earlier studies of alpaca TRGV9 rearrangements, CDR3 lengths of TRGV9 rearrangements showed homogenization to around 14 amino acids, whereas CDR3 lengths of Vδ2 chains varied substantially (SI Appendix, Fig. S6) (2, 3, 39). TRGV9 and TRDV2 rearrangements occurred in both sorted T cell populations (WTH-4+ and WTH-4−), indicating that WTH-4 is only specific for a subpopulation of Vδ2 T cells. Moreover, we observed the outgrowth of a single Vδ2Jδ2 clonotype in each animal in WTH-4− cells after HMBPP stimulation.
In addition, a formal proof of Vγ9 and Vδ2 chain pairing in alpacas was attempted adapting a previously published single-cell PCR approach to alpaca γδ TCR. The analysis of Vγ9Vδ2 TCR chain pairings in unstimulated and HMBPP-stimulated alpaca PBMCs permitted additional insights into the clonality and CDR3 usage of Vγ9Vδ TCRs (SI Appendix, Fig. S6). TRGJ usage of Vγ9 chains revealed a TRGJP preference for successfully sequenced TRGV9/TRGC PCR products and the use of one distinct TRGJP segment (JP-A or JP-C) in each animal. Moreover, CDR3 lengths were 14 amino acids in most of the clones. In the case of Vδ2, all chains uniformly used the TRDJ4 gene segment for rearrangement. In contrast to the CDR3 of Vγ9, CDR3 lengths of Vδ2 chains varied in length (11 to 17 amino acids). Some clones in stimulated PBMCs were detected at higher frequencies, and in all cases, unique Vγ9 chains were paired with a unique Vδ2 chain. Previously, hydrophobic amino acids (L, I, V) at position δ97 were reported to play a pivotal role in PAg recognition and that alanine substitutions abolish Vγ9Vδ2 activation (39). The use of L, I, or V at this position was also prevalent in alpaca Vδ2 chains; however, other amino acids (A, M, G) were observed as well (SI Appendix, Table S4).
This exploratory analysis of the Vγ9Vδ2 TCR repertoire in V. pacos provides conclusive evidence for PAg-reactive and clonally expanded Vγ9Vδ2 T cells and potentially highly reactive single clonotypes.
PAg Reactivity of Paired Alpaca Vγ9 and Vδ2.
Direct proof of BTN3-dependent PAg reactivity of specific alpaca Vγ9Vδ2 TCRs was demonstrated with TCR-transduced murine reporter cells. These cells lack BTN3 and have been previously used to compare different human Vγ9Vδ2 TCRs (11, 40). Stimulator cells were vpBTN3-expressing cell lines. To avoid interference of alpaca and human BTN3 (3), newly developed human 293T BTN3 knockout cells were used that lack either the BTN3A1 isoform (BTN3A1KO) or all human BTN3 isoforms (BTN3KO). A dose-dependent response of two alpaca TCRs to HMBPP in the context of alpaca BTN3 (measured by mIL-2 production) was shown in coculture with human BTN3KO cells transduced with alpaca BTN3 (vpBTN3) (Fig. 3A). This shows alpaca BTN3-dependent PAg reactivity and implies the conservation of other molecular interaction partners in the human and alpaca system. Cross-reactivity of alpaca TCRs to human BTN3A1 and wild-type expression of all BTN3 isoforms (293T) was evident; however, an approximately twofold lower IL-2 production was observed. Vice versa, no recognition of PAg in stimulation assays of alpaca BTN3-expressing BTN3KO cells with the human Vγ9Vδ2 (TCR MOP) was apparent; however, other human Vγ9Vδ2 TCRs might be reactive. Interestingly, the quantity of released IL-2 differed slightly between alpaca vpTCR(Jδ2) and vpTCR(Jδ4) with higher IL-2 production of vpTCR(Jδ2) transductants, implying differential responsiveness of alpaca TCR pairings. A summary of the magnitude of PAg responses of different alpaca TCR pairings is shown in SI Appendix, Table S5.
Next, the PAg-reactive alpaca Vγ9Vδ2 TCR transductants were used to test the antagonistic activity of the alpaca BTN3-specific antibody WTH-5 in stimulation assays with alpaca and human TCR transductants (Fig. 3B). Addition of this antibody completely abolished PAg-dependent IL-2 production by reporter cell lines cocultured with cells expressing alpaca BTN3, but not in coculture with BTN3-expressing human 293T cells, while no effects were found for isotype controls. This is in line with the mAb binding and inhibition data presented before (Figs. 1D and 2C). These experiments demonstrate the BTN3 dependence of PAg recognition of alpaca TCRs, as well as the impact of clonotypes on this response.
Cocultures of vpBTN3- or huBTN3A1-transduced CHO cells with WTH-5 or 20.1 induced no HMBPP-dependent IL-2 production of vpTCR transductants. This is in contrast to the massive activation in control cultures with TCR MOP transductants and CHO-huBTN3A1 cells plus mAb 20.1. (SI Appendix, Fig. S7). Tests of IL-2 production of vpTCR(Jδ2) transductants in culture with vpBTN3-transduced CHO cells plus HMBPP revealed no HMBPP-specific IL-2 production, while alpaca transductants responded well to human RAJI cells plus HMBPP in the same experiments (SI Appendix, Fig. S8).
BTN3 Function in the Alpaca.
Although the precise molecular mechanism of PAg recognition by human Vγ9Vδ2 T cells is not yet known, the human BTN3A1 molecule has been found to be essential for PAg reactivity of Vγ9Vδ2 T cells (11–13). As BTN3-dependent PAg reactivity seemed evident in our experiments, we investigated the contribution of BTN3 domains further. Stimulation assays similar to those in Fig. 3 were applied, and BTN3KO or BTN3A1KO cell lines were used as antigen presenting cells (APCs) (Fig. 4A). Those knockout cells were reconstituted by retroviral transduction with human BTN3A1 (huBTN3A1), alpaca BTN3 (vpBTN3), or the two chimeric BTN3 molecules, hu/vpBTN3 and vp/huBTN3, with an exchange of extracellular and intracellular domains of human and alpaca (Fig. 4B). To elucidate the influence of the C-terminally fused mCherry on HMBPP-stimulation, BTN3 molecules were expressed either as fusion proteins with mCherry (phNGFR mCherry vector) or with constructs expressing enhanced green fluorescent protein (EGFP) downstream of an internal ribosomal binding site. BTN3KO cells were transduced and sorted to a similar degree of BTN3 expression after staining with mAb 103.2 or WTH-5. As shown in SI Appendix, Fig. S9, the HMBPP response to cells expressing the mCherry fusion constructs was better compared to native molecules but the response pattern remained the same; poor activation by BTN3A1 plus HMBPP vs. much better activation by vpBTN3 plus HMBPP. In addition, for both types of huBTN3A1 transductants HMBPP-mediated activation was still far weaker than that induced by wild-type 293T cells, despite higher BTN3 expression, as measured by staining with mAb 103.2 (SI Appendix, Fig. S9). This is accordance with the reported need of cooperation of BTN3A1 with other BTN3-family members, especially BTN3A2, for efficient PAg stimulation (41). In the same experimental setting, we also tested the effect of a histidine 351 to arginine mutation (H351R) on PAg sensing of vpBTN3. H351 is part of a shallow basic PAg-binding pocket, which is located in the intracellular B30.2 domain of human BTN3A1 (34). The amino acids necessary for the formation of this pocket (H351, H378, K393, R412, R418, and R469) are identical among the vpBTN3 isoform, huBTN3A1 (SI Appendix, Fig. S1) and the hypothetical primordial BTN3 molecule of placental mammals (33). In accordance with published data on human BTN3A1 (34), no PAg-dependent activation by the vpBTN3-mutant was found (SI Appendix, Fig. S9).
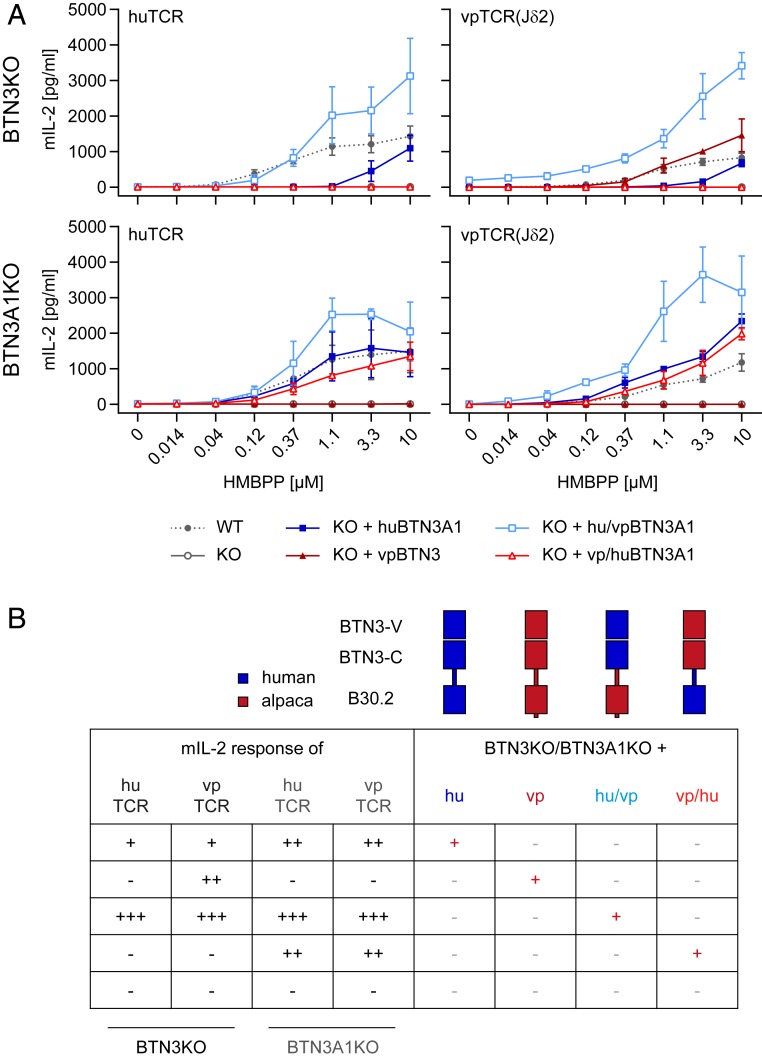
Fig. 4.
The alpaca B30.2 domain can sense PAgs. (A) A number of 1 × 104 293T BTN3KO or BTN3A1KO cells transduced with huBTN3A1, vpBTN3, hu/vpBTN3, or vp/huBTN3 (all in phNGFR mCherry) were plated overnight. Next, 5 × 104 Murine responder cells [53/4 huTCR or vpTCR(Jδ2)] were added as well as a dilution of HMBPP. Medium was used as a control, and the experiment was carried out in duplicates. Supernatants were tested for mIL-2 after 22 h with mIL-2 ELISA. Means of three independent experiments are shown with SDs. Statistical analysis is summarized in SI Appendix, Table S7. (B) Graphical summary of A with schematic representation of wild-type human (hu) and alpaca BTN3 (vp) and constructed chimeric molecules with an exchange of intracellular domains (hu/vp and vp/hu). The mIL-2 response of responder cells is indicated as high (+++), intermediate (++), low (+), or negative (−).
To test the ability of alpaca BTN3 and the chimeric molecules to reconstitute HMBPP reactivity, the mIL-2 production of murine responder cells expressing a human (huTCR) or alpaca [vpTCR(Jδ2)] Vγ9Vδ2 TCR were measured (Fig. 4A). Complete knockout of all BTN3 isoforms resulted in a total loss of IL-2 production of both responder cell lines upon HMBPP stimulation. Overexpression of human BTN3A1 failed to reach levels of mIL-2 observed upon HMBPP stimulation in coculture with 293T cells. HMBPP reactivity using huBTN3A1-transduced BTN3KO cells was only achieved at high levels of HMBPP (>3.3 µM) while fully reconstituting the activating capacity of BTN3A1KO cells. Interestingly, although huBTN3A1 expression of huBTN3A1-transduced BTN3A1 knockout cells was several log higher than expression of all BTN3 isoforms of wild-type 293T cells (SI Appendix, Fig. S10), it did not further increase the activating capacity, demonstrating that the extent of cell surface expression of BTN3A1 is not a limiting factor of the HMBPP response. This is reminiscent to previous observation made for BTN3 (39) and Skint1 (16). In contrast, a differential reactivity could be seen for vpBTN3-transduced knockout cells (BTN3KO and BTN3A1KO). These cells were not recognized by human responders although vpBTN3-transduced BTN3KO nicely activated the alpaca TCR transductants. This implies that at least this specific human Vγ9Vδ2 TCR (TCR MOP) does not respond to PAg in the context of alpaca BTN3, while alpaca Vγ9Vδ2 TCR show a more promiscuous response. Furthermore, unlike the human system, where BTN3A1 collaborates with BTN3A2 and BTN3A3 in eliciting a PAg response (41), vpBTN3 efficiently mediates PAg recognition on its own.
Somewhat surprising was the lack of a PAg-dependent response of alpaca TCR transductants in cocultures with vpBTN3-transduced BTN3A1KO cells, since staining with mAb WTH-5 revealed substantial amounts of alpaca BTN3 at the cell surface (SI Appendix, Fig. S10). This lack of activation might reflect negative effects of endogenous human BTN3A2 and A3 on acquiring a hypothetical PAg-induced activating conformation of vpBTN3. Vice versa, a chimeric molecule using human intracellular domains and alpaca extracellular domain (vp/huBTN3A1) induced activation of TCRs from both species, if expressed in BTN3A1KO cells. This suggests cooperation of this chimeric molecule with the non–PAg-binding BTN3A2 or BTN3A3 in the human system and interaction of the human TCR with the hypothetical human BTN3A2/3-containing complexes. The observation that expression of this chimera in BTN3KO cells did not allow PAg-induced activation of either human or alpaca TCR transductants could indicate an impaired capacity of this chimera to adapt an activating conformation in the absence of human BTN3A2 or BTN3A3. Finally, reconstitution of both BTN3 knockout cell lines with the hu/vp chimeric BTN3 molecule replenished PAg-dependent IL-2 production of both TCR transductants. This demonstrates that the alpaca transmembrane/intracellular domain can fully substitute for the “help” by BTN3A2 and/or BTN3A3. This “help” is needed for an efficient PAg response in the human system, as demonstrated by the substantially better PAg response of Vγ9Vδ2 TCR transductants to huBTN3A1-transduced BTN3A1KO cells compared to huBTN3A1-transduced BTN3KO cells.
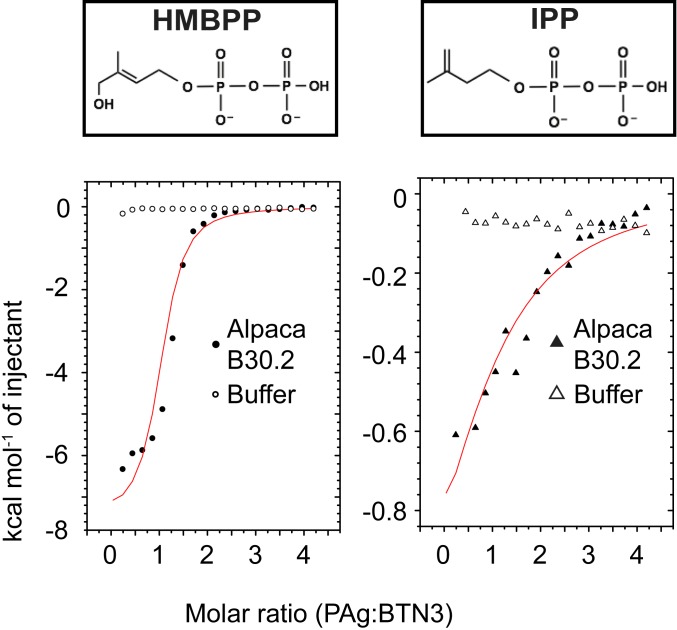
This was confirmed by isothermal titration calorimetry (ITC) experiments, which show interactions of two distinct PAgs (HMBPP and IPP) with the B30.2 domain of alpaca (Fig. 5 and SI Appendix, Fig. S11). Fitting of the ITC data revealed clear binding of both tested PAgs, HMBPP and IPP. As in humans, the affinities for IPP and HMBPP differed considerably and were ∼0.1 mM and 6 µM, respectively (SI Appendix, Table S8). These affinities are 5- to 10-fold lower, as reported for binding of HMBPP to the human BTN3A1-B30.2 domain and for IPP, 5- to 6-fold higher (34, 42). As expected, the prenyl ester of PAg is critical for binding to the B30.2 domain in alpacas, as ITC measurements with pyrophosphate showed no evidence of binding (SI Appendix, Fig. S12).
Fig. 5.
Alpaca B30.2 domain binds PAg similarly to human B30.2 domain. ITC binding isotherm traces of alpaca B30.2 domain with either HMBPP or IPP show interaction between the protein and PAgs. The binding measurements were all performed using 100 μM protein in the cell and 2 mM ligand in the syringe. The interaction with HMBPP is shown on the Left, whereas the interaction with IPP is shown on the Right. The buffer controls are shown in open circle (HMBPP) or open triangle (IPP); the trace for protein interaction is in filled circle (HMBPP) or filled triangle (IPP). The binding curves in red shown in both plots were fit assuming one-site binding. The reported molar ratio is the ratio of PAg:B30.2 domain.
Hence, alpaca BTN3 seems to retain the structural and functional features of its human counterpart with the important distinction that no other BTN3 isoforms seem to be involved in efficient PAg recognition. Further ITC experiments determined that the same residues necessary for coordinating PAg in humans (34, 35) are equally important for PAg binding in alpacas. Testing representative residues in the putative PAg-binding pocket (H351A, K393A, R412A) for their ability to bind HMBPP indicated that there are differential effects of each mutation (SI Appendix, Fig. S2). While mutation of R412A and H351A completely disrupted binding, mutation of K393A had a more modest effect. In the case of K393A, the sample never quite saturated, yet the curve fit to the data yields a binding affinity of 7.8 μM (SI Appendix, Fig. S13).
Discussion
In recent years, many groundbreaking studies contributed to a more in-depth understanding of the way Vγ9Vδ2 T cells recognize cellular stress or infection via phosphorylated metabolites. Yet, important pieces are still missing to resolve the jigsaw puzzle of Vγ9Vδ2 T cell activation. These include what exactly the Vγ9Vδ2 TCR interacts with, the identification of important key molecules besides BTN3A family members, and also how PAgs from an exogenous source can be sensed by intracellular domains of BTN3 molecules. In this study, we show a PAg-reactive subset of Vγ9Vδ2 T cells in a nonprimate species, the alpaca (V. pacos). The findings of this study are not only significant from an evolutionary perspective but provide a new point of view to understand the structural requirements of Vγ9Vδ2 T cell activation in humans.
Newly developed antibodies specific for alpaca Vδ2 chains and BTN3 were applied to test for surface expression of these essential markers. A small population of Vδ2+ cells was observed and we found evidence that WTH-4 is specific for a major subset of Vδ2 using Jδ4 segments. The BTN3-specific mAb WTH-5 allowed direct assessment of BTN3 expression on blood cells, where it was found to be differentially expressed on CD3+ and CD3− lymphocytes, and could also be detected on monocytes but not on granulocytes. The BTN3-specific antibody efficiently inhibited PAg-dependent activation of alpaca Vγ9Vδ2 T cells.
Unexpected were some of our results on species- and cell type-specificity of the BTN3-specific mAbs used in this study. While binding to transduced cells vs. primary cells by WTH-5 was largely consistent, mAb 103.2 and PE-labeled mAb 20.1 showed some surprising features. mAb 103.2 showed weak if any binding to human monocytes, which might indicate a masking of the epitope by translational modifications, conformational changes, or binding of associated molecules. Interestingly, Nerdal et al. (43) reported monocytes as much better “presenters” of PAg than lymphocytes. Thus, it is tempting to speculate that the loss of the 103.2 epitope might be related to this special capability. In any case, our observations on human cells warrant further analysis (15, 44), but are beyond the scope of this study. Another unexpected finding was the differential degree of species cross-reactivity of PE-labeled mAb 20.1 to primary cells vs. BTN3 transductants. The high cross-reactivity of PE-labeled mAb 20.1 might be especially useful when analyzing BTN3 in other species. The cross-reactivity of mAb 20.1 to human and alpaca PBMC also raises the question whether this mAb could act as an agonist of Vγ9Vδ2 T cells in the alpaca. We did not observe such reactivity so far, either with TCR transductants or in pilot experiments with primary alpaca cells. So far, we cannot exclude that these results were caused by a lack of reactivity due to the used TCR clonotypes (40) or by the fact that those TCRs are simply not detected by the mAb WTH-4.
Most importantly, the application of WTH-4 and WTH-5 antibodies allowed the measurement of a BTN3-dependent dose–response of alpaca PBMCs to HMBPP as measured by an increase in WTH-4+ cells. The magnitude of this response was around two-fold lower (in frequency) than in most human PBMC samples, which could be due to lower overall Vγ9Vδ2 frequencies or intrinsic mechanisms. Interestingly, the dose–response also varied between alpaca samples collected at different times and between individuals. Also, attempts of prolonged cultures (>7 d), easily performed with human Vγ9Vδ2 T cells, consistently failed despite testing various culture conditions with respect to cell density, culture media, and cytokine sources. Therefore, variability could be caused by a magnitude of other factors, including quality of sample material, nonideal culture conditions, or other effects, such as asymptomatic infections of animals or seasonal influences. In any case, the mAb WTH-4 provides a valuable tool to monitor PAg responses in vivo (e.g., during infections), and to study the physiology of PAg-reactive cells in a nonprimate species.
The analysis of TCR gene usage and TCR chain pairing, as well as the development of an in vitro system using TCR transductants and BTN3KO cells transduced with different BTN3 constructs, sheds new light on molecular parameters controlling the PAg response. As reported for humans (4), pairing of alpaca Vγ9 with Vδ2 was clearly predominant as shown by single-cell PCR. The results from this experiment and a pilot TCR repertoire analysis by TOPO TA cloning also showed restricted CDR3γ lengths and a dominance of Vγ9JP rearrangements. So far, no overt significance of usage of one of the presumed JP alleles was found. Combinations of Vδ2 and Vγ9 strongly affected the PAg response; however, PAg reactivity of random combinations was also evident. Contribution of the Vδ2 chain was indicated by a differential response of TCR transductants using Vδ2Jδ4 vs. Vδ2Jδ2 chains. We found no indication that certain CDR3δ lengths are favored per se, and although a tendency of hydrophobic amino acids was observed at δ97, we could not see a major effect of this position. The massive influence of TCR pairing on PAg reactivity warrants further studies on human γδ T cells and is reminiscent of the difference of certain human clonotypes in the response to stimulation by agonistic BTN3-specific mAbs (40).
Interestingly, the PAg-reactive transductants of either species showed similar dose–response. The lack of recognition of vpBTN3-transduced cells or cells expressing the extracellular domain of vpBTN3 by the human TCR MOP, while HMBPP-dependent stimulation of vpTCR transductants was observed, hints at interesting opportunities for defining functional domains of the BTN3 extracellular domain. The identification of differentially responding transductants will be used to define (species)-specific regions in the BTN3 extracellular domains controlling the TCR-dependent response as well as defining TCR regions controlling this response. This approach can be expected to be especially instructive, given that previously used alanine-scanning mutagenesis studies usually defined only loss-of-function but did not allow deciphering specific effects or a more general effect on the molecular integrity.
Another interesting finding is the differential response of human and alpaca TCR transductants to various types of BTN3-deficient cells and transductants. At first, we showed that the dose–response to endogenous human BTN3 (BTN3A1/A2/A3) of the human TCR was rather similar compared to that of the alpaca TCR to vpBTN3 expressed in BTN3KO cells. In addition, both alpaca TCRs showed a considerable cross-reactivity to human BTN3A1/A2/A3 and the Jδ2-containing TCR appeared to be especially reactive. The latter might reflect a special sensitivity of the rare Jδ2-containing TCRs to PAg, which would be consistent with the dominance of single Jδ2 TCRs seen in Vγ9Vδ2 rearrangements of WTH-4− HMBPP-stimulated cells. In either case, it is remarkable that alpaca TCR transductants showed a clear HMBPP response in cultures with human cells, indicating a high conservation of the key compounds of PAg “presentation” and recognition during evolution. The capacity of the vpBTN3-transduced 293T BTN3KO cells to stimulate in the presence of PAg also suggests a conservation of the interaction of alpaca BTN3 with the not yet defined human chromosome 6-encoded compound, which was previously demonstrated to be mandatory for PAg-induced activation of human Vγ9Vδ2 T cells (45).
As in the human system, a BTN3 homolog is essential for PAg recognition in alpaca and PAg interactions with B30.2 were shown. In a more detailed analysis of the contribution of alpaca BTN3 domains using chimeric molecules, we could show that the capacity of PAg-sensing is maintained or even improved if the region C-terminal to BTN3-IgC domain of human BTN3A1 is replaced by the corresponding alpaca BTN3 region. Most importantly, these chimeric molecules, as well as the wild-type alpaca BTN3, are not dependent on other BTN3 isoforms and we could see that they even outperform human BTN3 in our experimental set-up. This does not seem to be linked to a more efficient interaction of PAg with B30.2, since the affinity of the alpaca BTN3-B30.2 domain for HMBPP is even lower than for its human counterpart. The observed effects could be due to differential intracellular trafficking, subcellular localization, or surface expression, as shown by Vantourout et al. (41) for human BTN3 isoforms. Indeed, the alpaca BTN3 might represent an example of a primordial BTN3, which in its intracellular domain resembles the human BTN3A3 more than BTN3A1 but possesses a functional PAg-binding site. Such a primordial BTN3 has been predicted for lower primates that, after two successive gene-duplication events, gave rise to the three BTN3 isoforms found in humans (46). Furthermore, our analysis of armadillo BTN3 genes suggests the existence of a BTN3A3-like molecule with a functional PAg-binding site in a common ancestor of placental mammals (33). Apart from the PAg-binding B30.2 domain, the BTN3 JM domain has been shown to be crucial for inside-out signaling of PAg sensing (47). Interestingly, transplanting the JM of BTN3A3 into BTN3A1 has been reported to strongly increase PAg reactivity (48). Moreover, the coiled-coil domain of JM of alpaca and dolphin, which presumably also possesses functional Vγ9Vδ2 T cells (33, 49), clearly shows a higher degree of homology to the JM of BTN3A3 compared to BTN3A1 (SI Appendix, Table S2). Therefore, it is tempting to speculate that the BTN3A3-like nature of the alpaca JM and its cooperation with the nominal PAg-binding site of the B30.2 domain might enable the alpaca BTN3 to efficiently sense PAg, whereas formation of heteromers between the PAg-binding BTN3A1 and BTN3A2 and/or BTN3A3 are required in the human system (41). Whether alpaca BTN3 molecules act as homodimers as described for BTN3A1 (35, 50) needs to be elucidated. We hypothesized previously that in addition to BTN3A1, other human chromosome 6-encoded factors are required for PAg-induced but not for mAb 20.1-induced activation of human Vγ9Vδ2 TCR transductants or primary Vγ9Vδ2 T cells (45). This was concluded from a comparison of BTN3A1-transduced CHO cells with CHO cells containing human chromosome 6 and expressing BTN3A1. The fact that vpBTN3-transduced CHO cells failed to induce a PAg response while vpBTN3-transduced BTN3KO human 293T cells could do so, might indicate that the chromosome 6-encoded factors mandatory for PAg-sensing function of BTN3A1 support the same function for vpBTN3. Such interaction between partners of both species also suggests a high degree of conservation of the yet to be identified factors.
Given this remarkable conservation of TCRs and their putative ligands, the question arises as to why the BTN3/Vγ9Vδ2 T cell system is preserved in some species but not in others. Indeed, the case of primate and alpaca γδ TCR genes and BTN3 might reflect both the conservation of certain features needed to preserve type of function (probably the response to PAg) as well as the commonly observed “birth-and-death evolution” type, as it is found for many multigene families, including MHC and TCR genes (30, 51, 52). The fact that many mammalian species lack a functional BTN3/Vγ9Vδ2 T cell system suggests that other receptor–ligand interactions can overtake its function. Prominent examples for such convergent evolution of receptor–ligand systems are the NK cell receptors (53) and their ligands, as well as the variable lymphocyte receptor C-expressing γδ-like T like cells found in Agnatha (54). Another case might apply to divergent populations of γδ T cells in the response of mice and humans to Plasmodium species (55).
In summary, we could show that the participating surface molecules as well as the general mode of PAg recognition is strikingly conserved in human and alpaca, adding even more evidence to the importance of BTN3 for PAg recognition. It will be interesting to see whether PAg-reactive cells can also be found in other camelid species, including Old World camelids, which are of greater economic importance and serve as reservoir for zoonotic disease agents, such as Middle East respiratory syndrome corona virus, camel pox, or with respect to Vγ9Vδ2 T cells, most notably, Mycobacterium bovis (56).
Our finding that alpaca TCRs are able to recognize both human and alpaca BTN3 while human TCR recognizes PAg only in context of human BTN3, introduces new ways to map important regions of BTN3 using chimeric molecules or mutated versions of TCR and BTN3 of both species. The better knowledge on the molecular mechanism controlling the function of these molecules should also help targeting them in γδ T-cell based immunotherapy.
Methods
Animals.
BALB/cAnNCrl mice used for hybridoma development were bred at the Institute for Virology and Immunobiology (Julius-Maximilians University, Wuerzburg). Breeding pairs were provided by Charles River. Mice were kept and samples collected in accordance with the German regulations under protocols approved by the District of Lower Franconia (55.2-2531.01-81/14). Alpacas (V. pacos) were bred in the animal facility of the Ludwig-Maximilians University in Munich, Germany and provided by T.W.G. Blood samples were collected according to local regulations (Permission Regierung von Oberbayern: Az. 55.2-1-54-2532.3-11-11). Adult animals were between 1 and 8 y of age and female or male.
Cells.
HEK 293T (human embryonic kidney cells transduced with large T antigen of SV40) cells were purchased from the American Tissue Culture Collection and maintained in the laboratory of T.H. L929 mouse fibroblast cells and M12.4.1C3 mouse B cell lymphoma cells were used for hybridoma development. M12.4.1C3 cells were a kind gift of H. R. MacDonald, Ludwig Cancer Research Institute, Epalinges, Switzerland and originally purchased from Laurie Glimcher, Dana Farber Cancer Institute, Boston, MA (57). Both cell types were retrovirally transduced with pMIG II mCD3 δ-F2A-γ-T2A-ε-P2A-ζ, which contains all mouse CD3 subunits (pMIG II mCD3 δ-F2A-γ-T2A-ε-P2A-ζ), which was a gift from Dario Vignali, Department of Immunology, University of Pittsburgh, Pittsburgh, PA (Addgene plasmid # 52092; http://www.addgene.org/52092/; RRID:Addgene_52092) (58) and with alpaca γ and δ TCR chains [GenBank: KF734082/KF734084 (3)]. The mouse/rat T cell hybridoma cell line 53/4, a sister clone of 35/4, shows RT1B1-restricted specificity for guinea pig myelin basic protein and expresses endogenous αβ TCRs (59). Transduction with rat/mouse CD28 enhances antigen responsiveness in stimulation assays (40, 59–62). Expression of both, the endogenous αβ TCR and transduced γδ TCRs allows testing for αβ or γδ TCR-specific responses.
Cell Culture and Stimulation Assays.
Routine cell culture was carried out in RPMI 1640 (Gibco) supplemented with 10% FCS, 1 mM sodium pyruvate, 2.05 mM glutamine, 0.1 mM nonessential amino acids, 5 mM β‐mercaptoethanol, penicillin (100 U/mL), and streptomycin (100 μg/mL) or DMEM (Gibco) supplemented with 10% FCS. TCR transductants were stimulated using 293T cell lines and different stimulating or inhibiting agents for 22 h in 200 µL per well in 96-well flat-bottom cell culture plates at 37 °C, 5% CO2. The activation of TCR transductants was measured via mIL-2 ELISA (BD). One day prior to the stimulation, 293T cells or CHO cells were plated at 1 × 104 cells per well in 50-µL cell culture medium. Responders were added at a number of 1 × 105 cells per well.
Fresh or frozen PBMC were isolated from alpaca blood samples in EDTA with Histopaque-1077 centrifugation. For isolation of PBMCs, blood was layered over Histopaque-1077 (Sigma) in a 50-mL falcon tube and the gradient was centrifuged at 400 × g for 30 min. PBMCs were carefully aspirated from the interphase of the gradient and washed three times at 461 × g for 5 min with 15 mL PBS. Fresh or frozen PBMC were used. Cells were frozen in RPMI + 50% FCS + 10% DMSO. PBMC were plated at 1 × 105 cells per well in 200 µL per well in 96-well U-bottom suspension culture plates and cultured in the presence of hIL-2 50 U/mL (Novartis Pharma) for 6 to 7 d.
Monoclonal antibodies were raised against alpaca surface molecules using L929 cells retrovirally transduced with alpaca surface molecules (63). For immunization and screening by flow cytometry, transductants of L929 mouse fibroblasts and of M12.4.1C3 mouse B-cell lymphoma, were generated. vpBTN3 gene [GenBank: MG029164 (33, 38)] was expressed by applying gEGFP+ vector pMIG II using EcoRI/MfeI restrictions sites and the primers vpBTN3-MfeI-fwd and vpBTN3-MfeI-rev for amplification with Phusion Polymerase (Thermo Scientific). A vpTCR-mouse CD3 complex was expressed by cotransducing Vγ9Vδ2 TCR [GenBank: KF734082/KF734084 (3)] with vector pEGZ for Vγ9 and pIH for Vδ2 was and the vector pMIG II mCD3 δ-F2A-γ-T2A-ε-P2A-ζ, which encoded all chains of mouse CD3 (3, 58) and subsequent sorting of CD3+ cells. Screening was performed by staining transduced (Fig. 1A) vs. untransduced cells. Hybridomas were generated using PEG-mediated fusion of splenocytes from immunized mice with SP2/0 cells. Immunizations of female mice at 12 wk of age were performed intraperitoneally once a week for 5 successive weeks with 1 × 107 L929 cells overexpressing the selected surface molecule. Three weeks after the last immunization, an intravenous boost with 5 ×106 L929 cells was performed 3 d prior to organ removal. Following HAT-mediated selection, supernatants of hybridoma cells were tested for the presence of specific antibodies by flow cytometry. The Vδ2Jδ4-specific antibody WTH-4 (clone 118.7, mIgG1,κ) was purified by Protein G Sepharose Fast Flow (GE Healthcare). The antibody WTH-5 (clone 189.21.17, mIgG2b,κ) specific for alpaca BTN3 was purified using Protein A Sepharose 4B Conjugate (Invitrogen).
Flow Cytometry.
Flow cytometry samples were acquired on a FACSCalibur flow cytometer (BD) and analyzed using FlowJo (FlowJo LLC). The following antibodies were used to stain surface markers in flow cytometry assays: anti-vpVδ2Jδ4 clone WTH-4, anti-vpBTN3 clone WTH-5, anti-huBTN3/CD277 clone 103.2 was kindly provided by David Olive, Faculté de Médecine, Aix Marseille Université, Marseille, France (11, 31); PE-labeled anti-huBTN3/CD277 (11, 31) was from Milteny Biotech; anti-mouse CD3ε clone 145-2C-11 (BD Pharmingen), anti-alpaca CD3 (this paper) clone LT97A (37), and anti-alpaca CD14 clone CAM36A were kindly provided by William Davis and the Washington State University Monoclonal Antibody Center, Pullman, WA. Cell surface staining of human CD3 was performed with clone SK7 (Biolegend) and intracellular staining of CD3 of both species with CD3-12 FITC (Bio-Rad), which recognizes a highly conserved epitope of a CD3ε immunoreceptor tyrosine-based activation motif shared by mammals, birds, and amphibians. Isotype controls were mIgG2b,κ (BD Biosciences), isotype control mIgG1,κ clone P3.6.2.8.1 (Affymetrix), rat IgG1,κ RTK2071 (Biolegend). Purified antibodies were detected by F(ab’)2 Fragment Donkey α-Mouse IgG (H+L) R-PE (DαM R-PE, Jackson ImmunoResearch), biotinylated antibodies with Streptavidin-APC (BD Pharmingen), and WTH-4 was preincubated with AffiniPure Fab Fragment goat α-mouse IgG1 R-PE (Jackson ImmunoResearch). For instrument settings, fluorescence-minus-one controls were performed (64). Cell viability was assessed using Fixable Viability Dye eFluor 660 (eBioscience). Intracellular staining was carried out using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience).
TCR Clonotype Analysis and Single-Cell PCR.
The Vγ9Vδ2 TCR usage of alpaca PBMCs was assessed with two approaches. Unstimulated PBMCs and stimulated PBMCs (7 d, 1 µM HMBPP, 50 U/mL rhIL-2) were sorted on WTH-4+ and WTH-4− live cells. RNA was isolated using the RNeasy Micro Kit (Qiagen) and First Strand cDNA synthesis with Oligo dT primers was performed (Fermentas). Amplification of TRGV9/TRGC (TRGV9-fwd+TRGC-rev) and TRDV2/TRDC (TRDV2-fwd+TRDC-rev) sequences was performed in a Phusion PCR (Thermo Scientific) (primer sequences in SI Appendix, Table S1). The primers used for this experiment were TRGV9-fwd and TRGC-rev and TRDV2-fwd, as well as TRDC-rev. Single-cell PCR amplifications were performed according to a protocol described previouosly (65, 66). Unstimulated or stimulated PBMCs (7 d, 1 µM HMBPP, 50 U/mL rhIL-2) were single-cell sorted for WTH-4 expression on a FACSAria III cell sorter into 96-well plates. Cell lysis was mediated by freezing and heating to 65 °C for 2 min. The cells were placed on ice and combined cDNA synthesis, and amplification of Vγ9 and Vδ2 transcripts was performed with the OneStep RT-PCR Kit (Qiagen). Two separate PCR reactions were performed on this product (1:10 dilution) with the AmpliTaq Gold Polymerase (Applied Biosystems) resulting in TRGV9/TRGC (scTRGV9-fwd+scTRGC-rev) and TRDV2/TRDC (scTRDV2-fwd+scTRDC-rev) amplicons. PCR products were purified using agarose gel electrophoresis and QIAquick Gel Extraction Kit (Qiagen) and sequenced.
BTN3 Knockout Cell Lines and BTN3 Reconstitution and Mutation.
The 293T knockout cell lines were established using CRISPR/Cas9 (67). To achieve this, 293T cells (1.5 × 106) were seeded on day 1 in DMEM (10% FCS) without pyruvate. On day 2, cells were transfected with 5 µg of pFgH1t_BTN3A-CRISPR (GAGGGATCCATCTGGAGTGC) or pFgH1t-UTG_BTN3A1-CRISPR (GGGAGAGAACATCCCGACTG) and pFUCas9Cherry, which constitutively expresses the Cas9 endonuclease enzyme of Streptococcus pyogenes (68, 69) with a calcium phosphate-based protocol adapted from ref. 70. pFH1t-UTG and FUCas9Cherry vectors express GFP and mCherry as reporter markers, respectively. Forty-eight hours posttransfection, cells were sorted for mCherry and GFP expression. Sorted cells were treated with 2 µg/mL doxycycline for 12 to 14 d as expression of guide RNA is under the control of Tet-On system. After 14 d, cells were stained with anti-BTN3 (BT3 103.2) and an APC-conjugated secondary antibody (Dianova). The stained cells were sorted for BTN3− (BTN3A-CRISPR) or BTN3low (BTN3A1-CRISPR) cells. Sorted cells were seeded in single-cell dilutions in 96-well plates and each single-cell–derived clone was tested for surface expression of BTN3. Those single-cell clones, which exhibited reduced or no BTN3 surface expression, were tested subsequently tested at DNA level. The above-mentioned CRISPR sequences target exon 3 (IgC) of BTN3As. To validate the loss of gene mutation in targeted genes, the genomic IgC exon was amplified and cloned via TOPO TA cloning, followed by sequencing of clones to validate the gene mutation. BTN3 genes were expressed by retroviral transduction (63) in 293T knockout cells using a modified phNGFR vector (phNGFR linker mCherry). This vector allows the transcription of a gene of interest in frame with an mCherry gene. Human BTN3A1 (GenBank: NM_007048.5) was cloned into phNGFR linker mCherry using the primers huBTN3-EcoRI-fwd and huBTN3-BglII-rev (SI Appendix, Table S1). The EcoRI and BamHI restriction sites of the vector were used. The primers vpBTN3-MfeI-fwd and vpBTN3-BglII-rev were applied in a Phusion PCR (Thermo Scientific) for the cloning of alpaca BTN3 [GenBank: MG029164 (33, 38)] into phNGFR using EcoRI and BamHI restriction sites. vpBTN3 H351R mutant was generated by fusion of overlap-PCR products amplified by vpBTN3-MfeI-fwd/vpBTN3_H351RmutRev and vpBTN3_H351RmutFw/vpBTN3-BglII-rev primers using phNGFR linker mCherry-vpBTN3 plasmid as template. The fused PCR products were cloned as above. Two chimeric BTN3 molecules were comprised of the extracellular domains of BTN3 (ED: BTN3-V and BTN3-C) of alpaca and the transmembrane and intracellular parts (ID) of human or vice versa (see sequence in SI Appendix, Fig. S1). The first molecule used human BTN3A1 ED and alpaca BTN3 ID (hu/vp BTN3) and was cloned in two steps. The primers huBTN3-EcoRI-fwd and hu/vpBTN3-rev were used for the amplification with Phusion Polymerase (Thermo Scientific) of human ED from a plasmid carrying the whole gene and the primers hu/vpBTN3-fwd and vpBTN3-BglII-rev for alpaca ID. The outer primers huBTN3-EcoRI-fwd and vpBTN3-BglII-rev were used in a second PCR to generate a complete hu/vpBTN3 gene, which was cloned into phNGFR mCherry using EcoRI and BamHI. The same strategy was applied for the second molecule using alpaca BTN3 ED and human BTN3A1 ID (vp/huBTN3) with the primers vpBTN3-MfeI-fwd and hu/vpBTN3-rev for alpaca ED as well as hu/vpBTN3-fwd and huBTN3-BglII-rev. Again, EcoRI and BamHI restriction sites of phNGFR mCherry were used.
Generation of TCR Transduced Reporter Lines.
TCR transductants were used in stimulation assays to assess TCR reactivity. γδ TCRs were retrovirally transduced in the rat αβ TCR-bearing 53/4 rat/mouse CD28 (r/mCD28) hybridoma (40). The r/m chimeric CD28 molecule leads to an increased mIL-2 release of responder cells (BW58 or 53/4 transductants) upon binding to counter ligands like CD80 on the APC line (59, 61) and is essential for PAg-dependent mIL-2 production of murine reporter cell lines expressing a human Vγ9Vδ2 TCR in coculture with Raji cells (62). The human Vγ9Vδ2 TCR (huTCR) clonotype (TCR MOP) used in this study has been described previously (40). Alpaca TCR vpTCR(Jδ2) sequences were generated by ligation of three fragments from full-length Vγ9 [pEGZ vpVγ9 (3)] or Vδ2 [pIH vpVδ2 cl.8 (3)] and partial CDR3+TRJ spanning sequences from TCR clonotype analysis (pCR4-TOPO) with the In-Fusion HD Cloning Kit (Takara Bio). Fragment 1 spanned most of TRV and fragment 3 most of TRC and both were amplified from full-length TCR sequences, whereas fragment 2 spanned the CDR3 and TRJ and was amplified from partial TCR sequences originating from clonotype analysis. The restriction sites EcoRI and BamHI of the vectors pEGZ (Vγ9) or pIH (Vδ2) were used. Primers for vpTCR(Jδ2) where: Vγ9-fragment1 (Vγ9ins1-EcoRI-fwd+Vγ9ins1-rev), Vγ9-fragment2 (Vγ9ins2-fwd+Vγ9ins2-rev), Vγ9-fragment3 (Vγ9ins3-fwd+Vγ9ins3-BamHI-rev), Vδ2-fragment1 (Vδ2-EcoRI-fwd+Vδ2ins1-rev), Vδ2-fragment2 (Vδ2ins2-fwd+Vδ2ins2-rev), Vδ2-fragment3 (Vδ2ins3-fwd+Vδ2ins3-BamHI-rev). Joining of all three fragments was performed using the outer primers Vδ2ins1-3-EcoRI-fwd and Vδ2ins1-3-BamHI-rev. The V(D)J transition of the TCR vpTCR(Jδ4) sequence was originally identified by single-cell PCR (scTCR4) as described above. The respective γδ TCR amplicons were generated using the same approach and the outer primers already applied for vpTCR(Jδ2) andVγ9ins1-EcoRI-fwd, Vγ9ins3-BamHI-rev as well as Vδ2ins1-EcoRI-fwd were used.
Isothermal Titration Calorimetry.
Proteins samples for ITC were purified over a Superdex 200 into 20 mM Hepes pH 7.4, 150 mM NaCl. Concentration of peak fractions was determined from A280 using the theoretical extinction coefficient, pooled, and diluted to 100 μM. IPP was obtained from Echelon Biosciences, while HMBPP and pyrophosphate were obtained from Sigma Aldrich. ITC data for Fig. 5 were collected using 100 μM alpaca B30.2 protein in the cell and 2 mM IPP or HMBPP in the syringe on a MicroCal iTC200 (GE Healthcare Life Sciences). The alpaca B30.2 protein and pyrophosphate negative control for SI Appendix, Fig. S12 experiments were carried out at concentrations of 30 μM and 600 μM, respectively. For SI Appendix, Fig. S13, alpaca B30.2 wild-type and mutant ITC samples were diluted to 30 μM due to relative instability of the mutants, while the PAg in the syringe was diluted to 300 μM to avoid premature saturation of B30.2 binding to HMBPP.
Additionally, the PAg in the syringe was diluted to 300 μM. Experiments were carried out at 25 °C using an initial injection of 0.4 μL followed by 19 2-μL injections. ITC binding fits were determined after reference subtraction of injections of the ligand into buffer-only from the experimental results. Fits were determined using the Microcal Origin software. The stoichiometry (N) of the IPP–alpaca B30.2 interaction was set to 1 during fitting due to weak affinity. Buffer controls use the same concentration of ligand as the protein–ligand experiments they are plotted with. While there is no protein present to compute a molar ratio for these controls, the volume and concentration of ligand injected at each time point is identical for controls and experiments, and so the same x axis is used for clarity.
Protein Expression and Purification.
The alpaca B30.2 domain was cloned into pET28a with an N terminus six-HIS tag followed by a thrombin protease site. The construct was expressed in BL21 strain Escherichia coli. Cells were grown to OD600 = 0.6 in LB at 37 °C and induced with 0.5 mL 1 M isfopropyl-β-d-thiogalactopyranoside (IPTG) per liter of culture for 4 h at room temperature. Protein was harvested and purified using Ni-NTA (Qiagen) IMAC chromatography in 20 mM Tris pH8.0, 400 mM NaCl, 20 mM Imidazole, 4 mM BME, and eluted with 20 mM Tris pH 8.0, 400 mM NaCl, 250 mM Imidazole, 4 mM BME, and desalted into 10 mM Hepes pH 7.2, 150 mM NaCl, 0.02% azide, 4 mM BME using an Econo-Pac 10DG column (Bio-Rad). Protein was cleaved overnight using thrombin protease at 4 °C before using for the ITC experiment.
Data Analysis and Statistics.
Sequence data were analyzed using Chromas Lite 2.1.1 for AB1 files, Clustal Omega for DNA and protein alignments and National Center for Biotechnology Information Basic Local Alignment Search Tool (BLAST) for alignment calculations and database analysis. Sequence alignments were modified with BioEdit and color-coded with the default settings of the program. Ensembl, GenBank, and IMGT databases were used as resources for sequence data and homology analysis. The presence of the alpaca BTN3 as singleton gene and lack of BTN3 paralogues, respectively, was confirmed by BLAST searches against recently published high coverage whole-genome shotgun sequences with 72.5-fold genome coverage (JEMW01000000, contig JEMW010322411.1) and 260-fold genome coverage (ABRR030000000 contig ABRR03077292.1) (71). The program SerialCloner 2.6.1 was applied for cloning and restriction site analysis and FlowJo 7.6.5 and 8.8.7 for analysis of flow cytometry data. Statistical analysis was carried out using Graph Pad Prism and for ANOVA post hoc tests, P values adjusted by GraphPad Prism calculation are shown.
Data Availability Statement.
All data are contained in the main text and SI Appendix. For primary data, please contact the corresponding author.
Supplementary Material
Acknowledgments
We thank Lea Schröter for help with experiments for revision; Niklas Beyersdorf for reviewing the manuscript and valuable input; and Marco Herold, Walter and Eliza Hall Institute of Medical Research, Melbourne, and Ingolf Berberich, Institute of Virology and Immunobiology, Wuerzburg for CrispR/Cas9 vectors and expression vectors and for help in establishing the CrispR/Cas9 technology in the laboratory of T.H. The work of A.S.F., M.M.K., and T.H. was supported by the Deutsche Forschungsgemeinschaft Grants DFG-He 2346-7/1 and 8/1 (part of Grant FOR 2799: Receiving and translating signals via the gamma-delta T Cell receptor) and Wilhelm-Sander-Stiftung Grant 2013.907.2.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. W.K.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909474117/-/DCSupplemental.
References
- 1.Vantourout P., Hayday A., Six-of-the-best: Unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 13, 88–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karunakaran M. M., Herrmann T., The Vγ9Vδ2 T cell antigen receptor and butyrophilin-3 A1: Models of interaction, the possibility of co-evolution, and the case of dendritic epidermal T cells. Front. Immunol. 5, 648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karunakaran M. M., Göbel T. W., Starick L., Walter L., Herrmann T., Vγ9 and Vδ2 T cell antigen receptor genes and butyrophilin 3 (BTN3) emerged with placental mammals and are concomitantly preserved in selected species like alpaca (Vicugna pacos). Immunogenetics 66, 243–254 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Borst J., et al. , Non-random expression of T cell receptor gamma and delta variable gene segments in functional T lymphocyte clones from human peripheral blood. Eur. J. Immunol. 19, 1559–1568 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Casorati G., De Libero G., Lanzavecchia A., Migone N., Molecular analysis of human gamma/delta+ clones from thymus and peripheral blood. J. Exp. Med. 170, 1521–1535 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabelitz D., He W., The multifunctionality of human Vγ9Vδ2 γδ T cells: Clonal plasticity or distinct subsets? Scand. J. Immunol. 76, 213–222 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Morita C. T., Jin C., Sarikonda G., Wang H., Nonpeptide antigens, presentation mechanisms, and immunological memory of human Vgamma2Vdelta2 T cells: Discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol. Rev. 215, 59–76 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Delfau M.-H., Hance A. J., Lecossier D., Vilmer E., Grandchamp B., Restricted diversity of V γ 9-JP rearrangements in unstimulated human γ/δ T lymphocytes. Eur. J. Immunol. 22, 2437–2443 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Pauza C. D., Cairo C., Evolution and function of the TCR Vgamma9 chain repertoire: It’s good to be public. Cell. Immunol. 296, 22–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita C. T., et al. , Direct presentation of nonpeptide prenyl pyrophosphate antigens to human gamma delta T cells. Immunity 3, 495–507 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Harly C., et al. , Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood 120, 2269–2279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., et al. , Butyrophilin 3A1 plays an essential role in prenyl pyrophosphate stimulation of human Vγ2Vδ2 T cells. J. Immunol. 191, 1029–1042 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vavassori S., et al. , Butyrophilin 3A1 binds phosphorylated antigens and stimulates human γδ T cells. Nat. Immunol. 14, 908–916 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Di Marco Barros R., et al. , Epithelia use butyrophilin-like molecules to shape organ-specific γδ T cell compartments. Cell 167, 203–218.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blazquez J. L., Benyamine A., Pasero C., Olive D., New insights into the regulation of γδ T cells by BTN3A and other BTN/BTNL in tumor immunity. Front. Immunol. 9, 1601 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbee S. D., et al. , Skint-1 is a highly specific, unique selecting component for epidermal T cells. Proc. Natl. Acad. Sci. U.S.A. 108, 3330–3335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyden L. M., et al. , Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat. Genet. 40, 656–662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J., Liu Y., Lau Y.-L., Tu W., γδ-T cells: An unpolished sword in human anti-infection immunity. Cell. Mol. Immunol. 10, 50–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harly C., Peigné C. M., Scotet E., Molecules and mechanisms implicated in the peculiar antigenic activation process of human Vγ9Vδ2 T cells. Front. Immunol. 5, 657 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rincon-Orozco B., et al. , Activation of V γ 9V δ 2 T cells by NKG2D. J. Immunol. 175, 2144–2151 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Chen Z. W., Multifunctional immune responses of HMBPP-specific Vγ2Vδ2 T cells in M. tuberculosis and other infections. Cell. Mol. Immunol. 10, 58–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoeres T., Smetak M., Pretscher D., Wilhelm M., Improving the efficiency of Vγ9Vδ2 T-cell immunotherapy in cancer. Front. Immunol. 9, 800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gober H. J., et al. , Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J. Exp. Med. 197, 163–168 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kistowska M., et al. , Dysregulation of the host mevalonate pathway during early bacterial infection activates human TCR gamma delta cells. Eur. J. Immunol. 38, 2200–2209 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Kunzmann V., et al. , Stimulation of gammadelta T cells by aminobisphosphonates and induction of antiplasma cell activity in multiple myeloma. Blood 96, 384–392 (2000). [PubMed] [Google Scholar]
- 26.Wilhelm M., et al. , Gammadelta T cells for immune therapy of patients with lymphoid malignancies. Blood 102, 200–206 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Fournié J. J., et al. , What lessons can be learned from γδ T cell-based cancer immunotherapy trials? Cell. Mol. Immunol. 10, 35–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher J. P. H., Heuijerjans J., Yan M., Gustafsson K., Anderson J., γδ T cells for cancer immunotherapy: A systematic review of clinical trials. OncoImmunology 3, e27572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberl M., et al. , Microbial isoprenoid biosynthesis and human gammadelta T cell activation. FEBS Lett. 544, 4–10 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Kazen A. R., Adams E. J., Evolution of the V, D, and J gene segments used in the primate gammadelta T-cell receptor reveals a dichotomy of conservation and diversity. Proc. Natl. Acad. Sci. U.S.A. 108, E332–E340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palakodeti A., et al. , The molecular basis for modulation of human Vγ9Vδ2 T cell responses by CD277/butyrophilin-3 (BTN3A)-specific antibodies. J. Biol. Chem. 287, 32780–32790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., et al. , Conservation of nonpeptide antigen recognition by rhesus monkey V gamma 2V delta 2 T cells. J. Immunol. 170, 3696–3706 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Fichtner A. S., Karunakaran M. M., Starick L., Truman R. W., Herrmann T., The armadillo (Dasypus novemcinctus): A witness but not a functional example for the emergence of the butyrophilin 3/Vγ9Vδ2 system in placental mammals. Front. Immunol. 9, 265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandstrom A., et al. , The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vγ9Vδ2 T cells. Immunity 40, 490–500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y., et al. , A structural change in butyrophilin upon phosphoantigen binding underlies phosphoantigen-mediated Vγ9Vδ2 T cell activation. Immunity 50, 1043–1053.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Willcox C. R., et al. , Butyrophilin-like 3 directly binds a human Vgamma4(+) T cell receptor using a modality distinct from clonally-restricted antigen. Immunity 51, 813–825.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis W. C., et al. , Flow cytometric analysis of an immunodeficiency disorder affecting juvenile llamas. Vet. Immunol. Immunopathol. 74, 103–120 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Karunakaran M. M., “The evolution of Vγ9Vδ2 T cells,” PhD dissertation, Julius-Maximilians-University Wuerzburg, Wuerzburg (2014). .
- 39.Wang H., Fang Z., Morita C. T., Vgamma2Vdelta2 T cell receptor recognition of prenyl pyrophosphates is dependent on all CDRs. J. Immunol. 184, 6209–6222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starick L., et al. , Butyrophilin 3A (BTN3A, CD277)-specific antibody 20.1 differentially activates Vγ9Vδ2 TCR clonotypes and interferes with phosphoantigen activation. Eur. J. Immunol. 47, 982–992 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Vantourout P., et al. , Heteromeric interactions regulate butyrophilin (BTN) and BTN-like molecules governing γδ T cell biology. Proc. Natl. Acad. Sci. U.S.A. 115, 1039–1044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhodes D. A., et al. , Activation of human γδ T cells by cytosolic interactions of BTN3A1 with soluble phosphoantigens and the cytoskeletal adaptor periplakin. J. Immunol. 194, 2390–2398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nerdal P. T., et al. , Butyrophilin 3A/CD277-dependent activation of human γδ T cells: Accessory cell capacity of distinct leukocyte populations. J. Immunol. 197, 3059–3068 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Compte E., Pontarotti P., Collette Y., Lopez M., Olive D., Frontline: Characterization of BT3 molecules belonging to the B7 family expressed on immune cells. Eur. J. Immunol. 34, 2089–2099 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Riaño F., et al. , Vγ9Vδ2 TCR-activation by phosphorylated antigens requires butyrophilin 3 A1 (BTN3A1) and additional genes on human chromosome 6. Eur. J. Immunol. 44, 2571–2576 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Afrache H., Pontarotti P., Abi-Rached L., Olive D., Evolutionary and polymorphism analyses reveal the central role of BTN3A2 in the concerted evolution of the BTN3 gene family. Immunogenetics 69, 379–390 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Nguyen K., et al. , The butyrophilin 3A1 intracellular domain undergoes a conformational change involving the juxtamembrane region. FASEB J. 31, 4697–4706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peigné C. M., et al. , The juxtamembrane domain of butyrophilin BTN3A1 controls phosphoantigen-mediated activation of human Vγ9Vδ2 T cells. J. Immunol. 198, 4228–4234 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Wang H., Nada M. H., Tanaka Y., Sakuraba S., Morita C. T., Critical roles for coiled-coil dimers of butyrophilin 3A1 in the sensing of prenyl pyrophosphates by human Vγ2Vδ2 T cells. J. Immunol. 203, 607–626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dustin M. L., Scotet E., Olive D., An X-ray vision for phosphoantigen recognition. Immunity 50, 1026–1028 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Nei M., Rooney A. P., Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39, 121–152 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams E. J., Gu S., Luoma A. M., Human gamma delta T cells: Evolution and ligand recognition. Cell. Immunol. 296, 31–40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghosh A. K., et al. , Highly potent HIV-1 protease inhibitors with novel tricyclic P2 ligands: Design, synthesis, and protein-ligand X-ray studies. J. Med. Chem. 56, 6792–6802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boehm T., et al. , Evolution of alternative adaptive immune systems in vertebrates. Annu. Rev. Immunol. 36, 19–42 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Zaidi I., et al. , γδ T cells are required for the induction of sterile immunity during irradiated sporozoite vaccinations. J. Immunol. 199, 3781–3788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Salihi K. A., Invited review: Camelids zoonotic diseases. J. Camelid Sci. 11, 1–20 (2018). [Google Scholar]
- 57.Glimcher L. H., McKean D. J., Choi E., Seidman J. G., Complex regulation of class II gene expression: Analysis with class II mutant cell lines. J. Immunol. 135, 3542–3550 (1985). [PubMed] [Google Scholar]
- 58.Holst J., et al. , Scalable signaling mediated by T cell antigen receptor-CD3 ITAMs ensures effective negative selection and prevents autoimmunity. Nat. Immunol. 9, 658–666 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Kreiss M., et al. , Contrasting contributions of complementarity-determining region 2 and hypervariable region 4 of rat BV8S2+ (Vbeta8.2) TCR to the recognition of myelin basic protein and different types of bacterial superantigens. Int. Immunol. 16, 655–663 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Lühder F., et al. , Topological requirements and signaling properties of T cell-activating, anti-CD28 antibody superagonists. J. Exp. Med. 197, 955–966 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pyz E., et al. , The complementarity determining region 2 of BV8S2 (V beta 8.2) contributes to antigen recognition by rat invariant NKT cell TCR. J. Immunol. 176, 7447–7455 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Li J.-Q., “Modulating the expression of enzymes of isoprenoid synthesis: Effects on Vgamma9Vdelta2 T cell activation and tumor cell growth,” PhD dissertation, Julius-Maximilians-University Wuerzburg, Wuerzburg (2010).
- 63.Soneoka Y., et al. , A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23, 628–633 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cossarizza A., et al. , Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur. J. Immunol. 49, 1457–1973 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ravens S., et al. , Human γδ T cells are quickly reconstituted after stem-cell transplantation and show adaptive clonal expansion in response to viral infection. Nat. Immunol. 18, 393–401 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Kashani E., et al. , A clonotypic Vγ4Jγ1/Vδ5Dδ2Jδ1 innate γδ T-cell population restricted to the CCR6+CD27− subset. Nat. Commun. 6, 6477 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aubrey B. J., et al. , An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 10, 1422–1432 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Valente L. J., et al. , Therapeutic response to non-genotoxic activation of p53 by Nutlin3a is driven by PUMA-mediated apoptosis in lymphoma cells. Cell Rep. 14, 1858–1866 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Sambrook J., Russell D. W., Calcium-phosphate-mediated transfection of eukaryotic cells with plasmid DNAs. CSH Protoc. 2006, pdb.prot3871 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Richardson M. F., et al. , Chromosome-level alpaca reference genome VicPac3.1 improves genomic insight into the biology of New World camelids. Front. Genet. 10, 586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the main text and SI Appendix. For primary data, please contact the corresponding author.