Significance
An explanation has been lacking for the suppressive action of antagonists of growth hormone-releasing hormone receptors (GHRH-Rs) on cancers that do not express GHRH-Rs, an established target of the antagonists. We demonstrate here that esophageal squamous cell carcinoma (ESCC), a representative cancer type that barely expresses GHRH-Rs, responds to GHRH-R antagonists. Hypoxia induces GHRH-R splice variant 1 (SV1) and activates a key glycolytic enzyme. Glycolytic metabolism and tumor progression are promoted by activation of SV1 and reversed by the GHRH-R antagonist MIA-602. A high expression of SV1 in ESCC patients predicts a poor prognosis. These findings document the importance of SV1 as a target of GHRH-R antagonists and underline the therapeutic potential of GHRH-R antagonists against SV1-expressing cancers.
Keywords: splicing isoform of GHRH-R, GHRH-R antagonist, PFKM, glycolysis, hypoxia
Abstract
The extrahypothalamic growth hormone-releasing hormone (GHRH) and its cognate receptors (GHRH-Rs) and splice variants are expressed in a variety of cancers. It has been shown that the pituitary type of GHRH-R (pGHRH-R) mediates the inhibition of tumor growth induced by GHRH-R antagonists. However, GHRH-R antagonists can also suppress some cancers that do not express pGHRH-R, yet the underlying mechanisms have not been determined. Here, using human esophageal squamous cell carcinoma (ESCC) as a model, we were able to reveal that SV1, a known splice variant of GHRH-R, is responsible for the inhibition induced by GHRH-R antagonist MIA-602. We demonstrated that GHRH-R splice variant 1 (SV1) is a hypoxia-driven promoter of tumor progression. Hypoxia-elevated SV1 activates a key glycolytic enzyme, muscle-type phosphofructokinase (PFKM), through the nuclear factor kappa B (NF-κB) pathway, which enhances glycolytic metabolism and promotes progression of ESCC. The malignant actions induced by the SV1–NF-κB–PFKM pathway could be reversed by MIA-602. Altogether, our studies demonstrate a mechanism by which GHRH-R antagonists target SV1. Our findings suggest that SV1 is a hypoxia-induced oncogenic promoter which can be an alternative target of GHRH-R antagonists.
Aberrant RNA splicing is a common characteristic of cancers (1–3). Products of alternative splicing may display functions distinct from their canonical full-length transcripts, tending to mediate constitutively active proto-oncogenes, regulating cancer stem cells, promoting metastasis, and developing resistance to therapy (4–6). Emerging evidence strongly suggests that hypoxia is a key driver of alternative splicing in cancers (7). However, splice variants that occur during hypoxia and their roles in oncogenesis are much less understood.
A stimulatory loop formed by tumor-derived growth hormone-releasing hormone (GHRH) and its receptors has been shown to promote the growth of many cancers, which can be blocked by antagonists of GHRH’s cognate receptor (GHRH-R) (8–10). Thus, the pituitary-type GHRH receptor (pGHRH-R), a canonical full-length transcript, has been detected in many human neoplastic cells and tissues, mediating the antiproliferative effects of GHRH-R antagonists (8, 9, 11–14). However, the underlying mechanisms of the antitumor activities of GHRH-R antagonists are far from being elucidated. For example, GHRH-R antagonists exhibit antitumor effects in the subgroup of cancers that do not harbor GHRH-R overexpression, such as ovarian cancer, pancreatic cancer, and melanoma (15–17), suggesting that there may be alternative targets responding to these antagonists. The splice variant type 1 (SV1) of GHRH-R was detected in human cancers in the laboratory of one of us (A.V.S.) (18) and was demonstrated to possess ligand-dependent as well as ligand-independent activity (19). SV1 was enriched in extrapituitary neoplastic tissues (20). However, the functional role of SV1 in human malignancies is largely unknown. Moreover, it remains to be clarified whether SV1 acts as an alternative target of GHRH-R antagonists in a subgroup of cancers that express barely detectable levels of GHRH-R but high levels of SV1.
In this study, we showed that SV1 was induced by hypoxia in esophageal squamous cell carcinoma (ESCC), which barely harbors GHRH-R overexpression. Using cellular and animal studies, we detected an unappreciated hypoxia-SV1-inflammation-metabolic signaling in ESCC, providing evidence for the functionality of GHRH-R antagonists in a pattern common in all cancers.
Results
GHRH-R Splice Variant SV1 Mediates the Inhibitory Effect of GHRH-R Antagonist MIA-602 in Human ESCC.
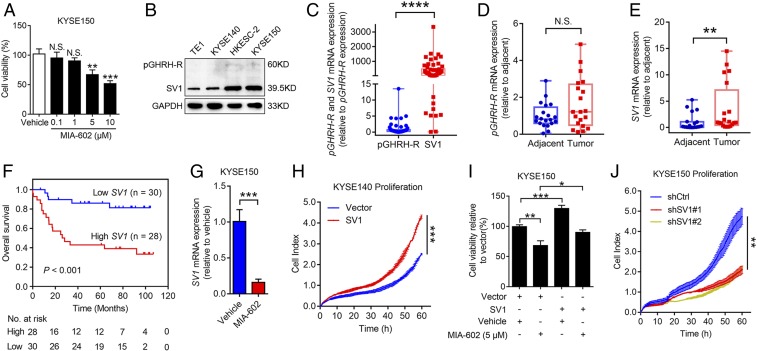
GHRH-R antagonists can inhibit the proliferation of a variety of human malignancies, some of which express low levels of GHRH-R (15, 21). It was claimed that the GHRH-R protein was barely detectable in human esophageal cancer (22). To investigate whether GHRH-R antagonists have inhibitory effects on human ESCC, ESCC cells KYSE150 and HKESC-2 were treated with different concentrations of MIA-602 for 48 h. MIA-602 began to exert significant inhibitory effects on cell viability at 5 μM in both cells (P < 0.01 for 5 μM, and P < 0.001 for 10 μM) (Fig. 1A and SI Appendix, Fig. S1A). Subsequently, a real-time cell analyzer (RTCA) was used to investigate the dynamic behavior of migration and invasion of KYSE150 cells exposed to MIA-602 (SI Appendix, Fig. S1 B and C). We found that cell migration and invasion were both significantly inhibited by MIA-602 treatment, compared to KYSE150 cells treated with the vehicle solution. To explain these results, we examined the expression of pGHRH-R, the known target of GHRH-R antagonists, in a panel of human ESCC cells, as well as two immortalized esophageal epithelial cells (NE2 and NE083) as normal controls. Nearly all these cells expressed very low pGHRH-R (SI Appendix, Fig. S1D), indicating that pGHRH-R is probably not the target of GHRH-R antagonists in ESCC. One of the splice variants of GHRH-R, SV1, was reported to be highly expressed in several types of cancer (23). Therefore, we determined the mRNA level for SV1 in these cells. SV1 had relatively high expression in ESCC cells, especially in HKESC-2 cells, whereas the normal cells, NE2 and NE083, expressed relatively low levels of SV1 (SI Appendix, Fig. S1D). Immunoblotting analysis further confirmed that the protein level for SV1 was higher than that of pGHRH-R in TE1, KYSE140, HKESC-2, and KYSE150 cells, with a more enriched pattern in HKESC-2 and KYSE150 cells (Fig. 1B).
Fig. 1.
MIA-602 inhibits ESCC cell progression through SV1. (A) Viability of KYSE150 cells treated with MIA-602 (0.1, 1, 5, or 10 μM) or vehicle solution for 48 h measured by cell-counting kit 8 (CCK-8) assay. (B) Protein levels of pGHRH-R and SV1 in four ESCC cells. GAPDH was used as an internal control. (C) RT-qPCR analyses of pGHRH-R and SV1 in 58 human ESCC specimens. (D and E) RT-qPCR analyses of pGHRH-R (D) and SV1 (E) in 20 human ESCC tissues and paired adjacent noncancerous tissues. (F) Kaplan–Meier analysis showing that OS was significantly better in patients with low expression of SV1 than in those with high expression. (G) KYSE150 cells treated with MIA-602 or vehicle; levels of SV1 were determined by RT-qPCR. (H) Proliferation of SV1-overexpressing KYSE140 cells monitored by the xCELLigence RTCA dual-plate (DP) system (ACEA Biosciences). Quantitative analysis of the cell index at 60 h is shown. (I) Viability of SV1-overexpressing KYSE150 cells treated with MIA-602 or vehicle measured by CCK-8 assay. (J) Proliferation of SV1-knockdown KYSE150 cells monitored by the xCELLigence RTCA DP system. Quantitative analysis of the cell index at 60 h is shown. Error bars indicate SEM. N.S., not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by one-way ANOVA with post hoc intergroup comparisons (A, I, and J) or student’s t test (C–E, G, and H); n = 3 in each group (A, G, and I).
We compared the levels of pGHRH-R and SV1 in 58 human primary ESCCs, and the results showed that SV1 was more abundant in tumors than pGHRH-R (P < 0.0001) (Fig. 1C). Moreover, a comparison between tumor and adjacent nontumor tissues revealed insignificant levels of pGHRH-R, while SV1 was significantly higher in tumor tissues (P < 0.01 for Fig. 1E) (Fig. 1 D and E). Receiver operator characteristic (ROC) analysis identified an optimal cutoff score of 0.012 for RT-qPCR, which categorized 48.28% (28 of 58) for overexpression of SV1 (SI Appendix, Fig. S1E). SV1 expression was positively correlated with the largest tumor dimension (P = 0.006), pathological nodal (pN) status (P = 0.004), and pathological stage of tumor nodal metastasis (pTNM) (P = 0.034), as indicated by correlation assay (SI Appendix, Table S1), supporting that SV1 is highly expressed in human ESCC and is closely associated with disease progression. Furthermore, Kaplan‒Meier survival analysis revealed a shorter overall survival (OS) for ESCC patients with increasing expression of SV1 (log-rank test, P < 0.001) (Fig. 1F). the multivariate Cox regression model showed that SV1 expression is an independent prognostic factor for patients with ESCC [hazard ratio (HR) = 4.269, 95% CI = 1.547–11.775, P = 0.005] (SI Appendix, Table S2). Survival and multivariate analyses confirmed the clinical relevance of SV1 as a prognostic marker for OS in patients with ESCC.
Notably, administration of MIA-602 decreased expression of SV1 in KYSE150 and HKESC-2 cells (P < 0.001) (Fig. 1G and SI Appendix, Fig. S1F). These data suggest that SV1 is a potential intrinsic oncogene and a target for GHRH-R antagonists in ESCC. We then stably overexpressed SV1 in TE1 and KYSE140 cells to assess the role of SV1 (P < 0.001) (SI Appendix, Fig. S1 G–J). Cell proliferation monitored by real-time cell analyzer (RTCA) showed that overexpression of SV1 resulted in significantly enhanced cell proliferation in both cells, compared with the vector control cells, respectively (P < 0.001 for Fig. 1H; P < 0.01 for SI Appendix, Fig. S1K) (Fig. 1H and SI Appendix, Fig. S1K). Moreover, the overexpression of SV1 successfully reversed the decrease in cell viability induced by MIA-602 in KYSE150 cells (Fig. 1I and SI Appendix, Fig. S1 L and M). To confirm whether SV1 mediates the inhibitory effect of MIA-602, low SV1 expressors (TE1 and KYSE140) were treated with different concentrations of MIA-602 and subjected to cell viability assay. We found that MIA-602 did not exert significant inhibitory effects until the concentration reached 10 μM in both cells (P < 0.05 for 10 μM in KYSE140 cells, and P < 0.01 for 10 μM in TE1 cells) (SI Appendix, Fig. S2 A and B), which was higher than the concentration of 5 μM MIA-602 required in high SV1 expressors (KYSE150 and HKESC-2) (Fig. 1A and SI Appendix, Fig. S1A). However, inhibition of viability was observed in KYSE140 cells overexpressing SV1 with as little as 1 μM MIA-602 (P < 0.01 for 1 and 2.5 μM, and P < 0.001 for 5 μM in KYSE140-SV1 cells) (SI Appendix, Fig. S2C). In addition, results from colony formation assay showed that a concentration of 2.5 μM MIA-602 could significantly reduce the number of colonies of KYSE150 cells but not that of KYSE140 cells (P < 0.05 for 1 μM in KYSE150 cells and 5 μM in KYSE140 cells; P < 0.01 for 2.5 and 5 μM in KYSE150 cells) (SI Appendix, Fig. S2D). Furthermore, stable KYSE150 cells expressing SV1 shRNA were established for detecting the role of SV1 with or without MIA-602 treatment (P < 0.001 for SI Appendix, Fig. S2E) (SI Appendix, Fig. S2 E and F). Cell proliferation monitored by RTCA showed that knockdown of SV1 decreased cell proliferation in KYSE150 cells (P < 0.01) (Fig. 1J). MIA-602 did not exert significant inhibitory effects even when the concentration reached 5 μM in SV1-knockdown KYSE150 cells (SI Appendix, Fig. S2G). Collectively, these results confirm that SV1 was the target of the GHRH-R antagonist MIA-602 and was able to mediate MIA-602’s inhibitory effects in human ESCC.
SV1 Induced by Hypoxia Contributes to Aberrant Glycolysis.
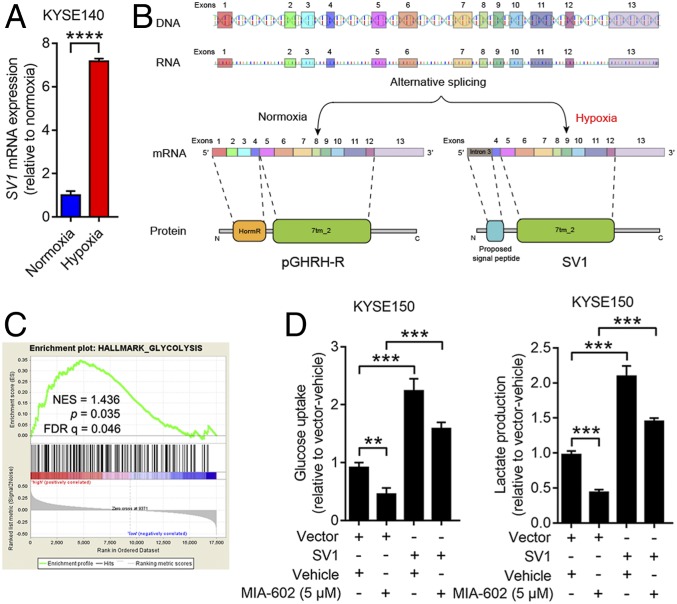
Recently, hypoxia within tumor tissues was indicated as a driver of alternative splicing that contributes to tumorigenic development (24). To test the hypothesis that hypoxia in ESCC leads to alternative splicing of pGHRH-R into SV1, we examined the transcript of SV1 on ESCC cells grown under normoxia and hypoxia. A significant increase in SV1 expression was seen in KYSE140 and TE1 cells grown under hypoxia (P < 0.0001 for both) (Fig. 2A and SI Appendix, Fig. S3A). This finding supports the speculation that hypoxia induces elevation of SV1 through driving the alternative splicing of GHRH-R (Fig. 2B). Hypoxic cancer cells are often forced into aberrantly increased glycolysis (25). We observed significantly higher lactate production and glucose uptake in ESCC cells under hypoxia, compared with cells grown under normoxia (SI Appendix, Fig. S3 B and C). Results from gene set enrichment analysis (GSEA) using microarray dataset GSE47404 showed that the levels of mRNA for SV1 significantly correlated with the glycolytic pathways in ESCC (P = 0.035) (Fig. 2C). These results suggest that SV1 was induced by hypoxia and resulted in promotion of glycolysis in ESCC. To validate whether GHRH-R antagonists could interfere with the glycolysis enhanced by SV1, we evaluated the glycolytic activities of ESCC cells after treatment with MIA-602. Exposure to MIA-602 significantly decreased lactate production and glucose uptake of KYSE150 cells, and similar results were obtained in HKESC-2 cells (P < 0.001 for all) (SI Appendix, Fig. S3 D and E). Importantly, the overexpression of SV1 further reversed the decreased glycolysis caused by treatment with MIA-602 (Fig. 2D). Thus, expression of SV1 in ESCC was induced by hypoxia and resulted in enhanced glycolysis, which could be inhibited by GHRH-R antagonists.
Fig. 2.
Hypoxia-induced SV1 enhances glycolysis. (A) Expression of SV1 measured by RT-qPCR in KYSE140 cells pretreated at normoxia or hypoxia for 24 h. (B) Schematic of full-length GHRH-R and SV1 structures (DNA, mRNA, and protein). (C) SV1 expression positively correlates with the glycolysis pathway according to a GSEA plot (GSE47404, n = 71). (D) Glucose uptake and lactate production measured in SV1-overexpressing cells treated with MIA-602 or vehicle. Error bars indicate SEM. **P < 0.01, ***P < 0.001, ****P < 0.0001 by student’s t test (A) or one-way ANOVA with post hoc intergroup comparisons (D); n = 3 in each group (A and D).
SV1 Regulates Glycolytic Metabolism Activity through NF-κB–PFKM Axis.
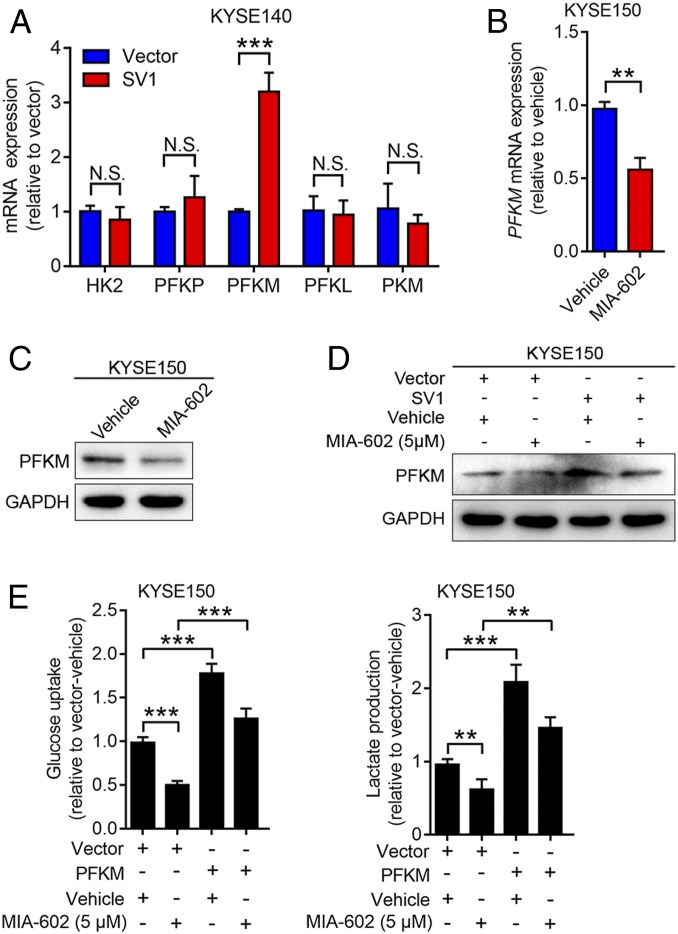
We further investigated the underlying mechanisms by which SV1 regulates glycolytic metabolism in ESCC cells. RT-qPCR was conducted to detect the expression of key enzymes regulating glycolysis in KYSE140 cells overexpressing SV1. Among the enzymes screened, muscle-type phosphofructokinase (PFKM) was the only one elevated by overexpression of SV1, and similar results were observed in TE1 cells stably overexpressing SV1 (P < 0.001 for both) (Fig. 3A and SI Appendix, Fig. S4A). Expression of PFKM in KYSE150 and HKESC-2 cells was obviously decreased by treatment with MIA-602, as revealed by RT-qPCR and immunoblotting analyses (P < 0.01 for Fig. 3B and SI Appendix, Fig. S4B) (Fig. 3 B and C and SI Appendix, Fig. S4 B and C), indicating that PFKM is a critical downstream molecule induced by SV1. Furthermore, the overexpression of SV1 reversed the expression of PFKM that was suppressed by treatment with MIA-602 (Fig. 3D and SI Appendix, Fig. S4D). TE1 and KYSE140 cells stably overexpressing PFKM were subjected to RTCA assays, and increased proliferation was observed in both cells (SI Appendix, Fig. S4 E–H). Moreover, the lactate production and glucose uptake that were reduced by MIA-602 could be reversed by PFKM overexpression (Fig. 3E). These findings collectively suggest that PFKM is a functional downstream target of SV1 to regulate glycolytic metabolism.
Fig. 3.
SV1 regulates glycolysis through PFKM. (A) mRNA levels of five key glycolytic enzymes (HK2, three isoforms of PFK, and PKM) in SV1-overexpressing KYSE140 cells determined by RT-qPCR. (B and C) Expression of PFKM in KYSE150 cells treated with MIA-602 for 48 h and analyzed by RT-qPCR (B) and immunoblotting (C). GAPDH was used as an internal control. (D) Expression of PFKM in SV1-overexpressing cells treated with MIA-602 or vehicle and analyzed by immunoblotting. GAPDH was used as an internal control. (E) Glucose uptake and lactate production measured in PFKM-overexpressing cells treated with MIA-602 or vehicle. Error bars indicate SEM. N.S., not significant; **P < 0.01, ***P < 0.001 by student’s t test (A and B) or one-way ANOVA with post hoc intergroup comparisons (E); n = 3 in each group (A, B, and E).
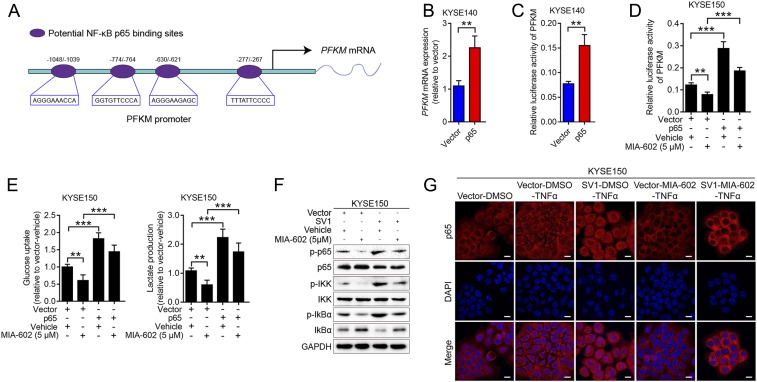
We previously demonstrated that nuclear factor kappa B (NF-κB) mediates the action of GHRH-R antagonists to inhibit the growth of gastric cancer (14). NF-κB signaling not only participates in the progression of ESCC but also stimulates glycolytic activity (26, 27). Given the facts that NF-κB is a well-known transcription factor and that we observed alteration of PFKM transcription in ESCC cells, we wondered if the regulation of PFKM transcription by SV1 was NF-κB dependent. Bioinformatic analysis revealed the presence of binding sites for NF-κB p65 in the PFKM promoter region, suggesting that NF-κB p65 might regulate the transcription of PFKM (Fig. 4A). Stable cells expressing p65 or p65 shRNA were established for detecting the involvement of p65 in glycolytic metabolism regulated by SV1-PFKM (SI Appendix, Fig. S5 A and B). PFKM transcription increased in p65-overexppressed cells, but decreased in p65-silenced cells (P < 0.01 for Fig. 4B; P < 0.001 for SI Appendix, Fig. S5C) (Fig. 4B and SI Appendix, Fig. S5C). Luciferase assay revealed that forced expression of p65 significantly increased the activity of the PFKM promoter, whereas knockdown of p65 decreased it (P < 0.01 for both) (Fig. 4C and SI Appendix, Fig. S5D). Next, we found that p65 could reverse the PFKM promoter activity, lactate production, and glucose uptake that were decreased by MIA-602 treatment (Fig. 4 D and E). Immunoblotting analysis showed that the NF-κB pathway was suppressed by MIA-602 treatment (SI Appendix, Fig. S5E). Moreover, activation of the canonical NF-κB pathway by stimulation with tumor necrosis factor alpha (TNF-α) resulted in increased nucleal translocation of p65 in ESCC cells, whereas MIA-602 treatment dramatically attenuated the translocation of p65 to the nucleus (SI Appendix, Fig. S5F), demonstrating a strong blocking effect of MIA-602 on the activation of NF-κB signaling. Further, overexpression of SV1 reversed the suppression of the NF-κB pathway that was conferred by MIA-602 treatment (Fig. 4F). The nuclear translocation of p65 was blocked by MIA-602, but reappeared after SV1 overexpression (Fig. 4G). Taken together, these results strongly support involvement of the NF-κB pathway in glycolytic metabolism regulated by SV1-PFKM.
Fig. 4.
Cancer cell glycolysis regulated by SV1-PFKM is NF-κB dependent. (A) Graphic representation of the four putative NF-κB p65 binding sites in the PFKM proximal promoter. (B) mRNA level of PFKM in p65-overexpressing cells determined by RT-qPCR. (C) The PFKM luciferase reporter was transfected into KYSE140-p65 cells and control vector cells, and the relative PFKM promoter activities were measured based on the luciferase activities. (D and E) Relative PFKM promoter activities (D), glucose uptake, and lactate production (E) measured in p65-overexpressing cells treated with MIA-602 or vehicle. (F) SV1-overexpressing cells treated with MIA-602 or vehicle before being harvested for immunoblot analyses of the labeled antigens. GAPDH was used as an internal control. (G) Subcellular localization of p65 (red) in KYSE150 cells as analyzed by immunofluorescence assay. Nuclei stained with DAPI (blue). Error bars indicate SEM. **P < 0.01, ***P < 0.001 by student’s t test (B and C) or one-way ANOVA with post hoc intergroup comparisons (D and E); n = 3 in each group (B–E). (Scale bars, 10 μm.)
MIA-602 Inhibits Tumor Growth by Targeting SV1–NF-κB–PFKM In Vivo.
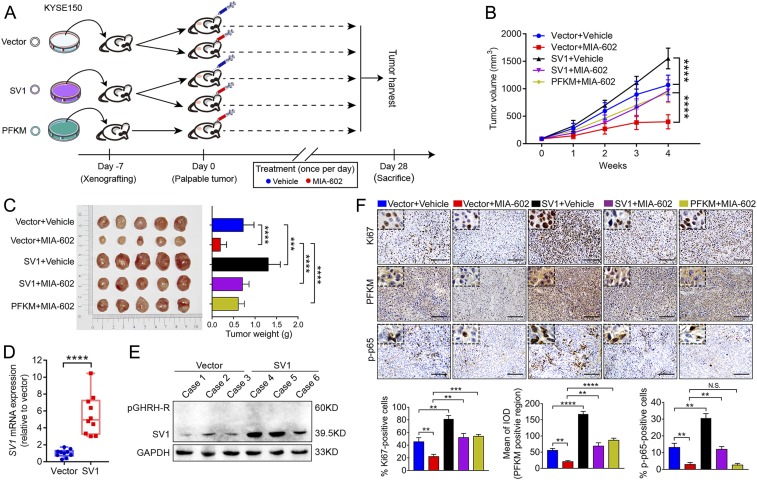
To demonstrate that GHRH-R antagonists are capable of inhibiting tumor growth by targeting the SV1-PFKM axis in vivo, we established xenograft tumor models by subcutaneous (s.c.) inoculation of KYSE150-SV1, KYSE150-PFKM, and KYSE150-Vector cells into nude mice (Fig. 5A). As demonstrated by the tumor growth curves, SV1-overexpressing tumor cells exhibited enhanced growth (P < 0.0001) (Fig. 5B). Administration of MIA-602 remarkably inhibited tumor growth in mice bearing KYSE150-Vector cells (P < 0.001) (Fig. 5B). Of note, both SV1 and PFKM overexpression partially counteracted the inhibitory effects on tumor growth induced by MIA-602 (Fig. 5B). Corresponding tumor sizes and weights were obtained on day 28 (Fig. 5C). Importantly, the effective up-regulation of the mRNA for SV1 and protein levels in KYSE150-SV1 cells was confirmed in harvested tumors (P < 0.0001 for Fig. 5D) (Fig. 5 D and E). Inhibition of tumor cell proliferation by MIA-602 treatment was also reflected by immunohistochemical staining for the proliferation marker Ki67 (Fig. 5F). The down-regulation of the PFKM and NF-κB pathways by MIA-602 treatment was also demonstrated by immunohistochemistry (Fig. 5F). Thus, MIA-602 inhibits tumor growth mediated by SV1–NF-κB–PFKM signaling in vivo.
Fig. 5.
MIA-602 suppresses SV1-mediated ESCC tumor growth in vivo. (A) Scheme indicating the timing of xenografting and longitudinal treatment. (B) Tumor growth curves of KYSE150-SV1 and KYSE150-PFKM cells treated with MIA-602 or vehicle. (C) Tumors harvested on day 28 (Left) and average weights of tumors (Right). (D and E) Expression of SV1 detected by RT-qPCR (D) and immunoblotting (E) in tumor tissues from mice bearing KYSE150-SV1 or KYSE150-Vector cells. GAPDH was used as an internal control. (F) Representative images of immunohistochemistry of Ki67, PFKM, and p-p65 in tumor sections derived from mice (Top). Plots of percentages or mean of integrated optical density (IOD) of five groups of cells expressing the indicated proteins (Bottom). Error bars indicate SEM. N.S., not significant; **P < 0.01, ***P < 0.001, ****P < 0.0001 by one-way ANOVA with post hoc intergroup comparisons; n = 10 in each group. (Scale bars, 50 μm.)
Discussion
In this study, we provided experimental and clinical evidence to demonstrate the significance of the GHRH-R splicing variant SV1 in the progression and prognosis of ESCC. Both in vitro and in vivo studies indicate that hypoxia-induced SV1 promotes ESCC through a previously unknown mechanism that activates the inflammation-metabolic signaling of NF-κB–PFKM. Our results document that GHRH-R antagonists exert inhibitory effects by targeting SV1 in a subgroup of cancers that do not harbor overexpression of GHRH-R.
The presence of pGHRH-R and its response to GHRH-R antagonists had been previously demonstrated in various human cancers, including breast, prostatic, and gastric cancers, and renal cell carcinoma (11, 13, 14, 28). However, there also exist some tumor types which do not express high levels of pGHRH-R but which respond to GHRH and GHRH-R antagonists (15–17), implying that there are alternative targets. The splice variant SV1 has the greatest structural similarity to pGHRH-R, is widely expressed by different primary human and experimental cancers, and is considered the most likely functional splice variant mediating the effects of GHRH analogs in tumors (9, 20). ESCC is one of the most common malignancies of the digestive tract, with a poor prognosis and a high mortality rate (29–32). By analyzing a large group of patients and cells, we revealed a very low level of mRNA for pGHRH-R but a highly enriched SV1 transcript in ESCC. Furthermore, the significance of SV1 in malignant progression and clinical outcomes had not been appreciated previously. By analysis of a patient cohort with follow-up of clinicopathological information, the overexpression of SV1 was identified as an independent prognostic predictor for patients with ESCC. Future studies are required to confirm these findings across the spectrum of multiple cohorts in multiple centers. These data predict the contribution of SV1 to progression of ESCC and emphasize SV1 as a potential therapeutic target in human cancers. Combining these results with the finding that MIA-602, a highly potent GHRH-R antagonist, exerts antineoplastic effects in ESCC cells, we can consider ESCC a representative model to demonstrate that SV1 mediates the therapeutic effects of GHRH-R antagonists in human tumors with low expression of pGHRH-R. The expression of pGHRH-R and SV1, as well as the effects of GHRH-R antagonists against squamous-cell carcinoma, were previously largely unknown. Our evidence indicates that SV1 is the main form of GHRH-R in squamous cell carcinomas that respond to GHRH-R antagonists (22, 33). Our studies also indicate that in squamous cell carcinoma SV1 can be used as a therapeutic target shared with pGHRH-R.
Compared with the canonical full-length GHRH-R, the first three exons in SV1 are replaced by a fragment of retained intron 3; the rest of the coding region of SV1 is identical with that of pGHRH-R. The protein product of SV1 differs from the full-length receptor in a small part of the N-terminal extracellular domain, which could serve as a proposed signal peptide (18). In this study, we found that increasing splicing of pGHRH-R into SV1 was induced by hypoxia in ESCC. Alternative splicing in tumor cells is one of the cellular adaptations that are largely promoted by the hypoxic microenvironment in solid tumors. Given that intron-containing mRNAs are the most frequent splicing products that result from hypoxia (34–36), it is reasonable to predict that the enhanced expression of SV1 in ESCC is induced by hypoxia. We also demonstrated that SV1 in ESCC cells possesses both ligand-dependent and ligand-independent functions. Cells transfected with SV1 expression vectors exhibited a strong induction of cell proliferation, migration, and invasion. Overexpression of SV1 is sufficient to abrogate suppression of the NF-κB pathway that was induced by MIA-602 treatment. This supports the view that SV1 acts as a tumor promoter intrinsically in the absence of the ligand GHRH. More importantly, our results show that SV1 modulates the inflammation-metabolic signaling of NF-κB–PFKM to sustain high proliferation rates of tumor cells, implying that isoforms of GHRH-R form a complex regulatory network in human malignancy. Furthermore, we provide both in vitro and in vivo data showing that SV1 promotes tumor growth and that this effect can be reversed by the GHRH-R antagonist MIA-602, supporting the ligand-dependent and ligand-independent functions of SV1 (19, 37, 38).
Widespread changes in gene expression, including alternative splicing of metabolic enzymes, have been implicated in the process of hypoxia-induced metabolic adaptation of cancer cells (34, 39, 40). In this study, we defined a previously undocumented molecular pathway by which hypoxia acts through a splice variant of a membrane hormone receptor to elevate the glycolytic enzyme PFKM, indicating that hypoxia-induced metabolic changes in cancer cells may involve a more complex regulation. Increasing evidence has shown that cancer cells exhibit a metabolic reprogramming toward aerobic glycolysis to ensure high levels of energy supply and biomass production to support tumor growth and progression (41, 42). Hypoxia-induced SV1 triggers a PFKM-mediated metabolic rewiring, as evidenced by enhanced glucose consumption and the simultaneously increased production of lactate. Increased glucose flux fuels the proliferation of cancer cells. Increased lactate secretion is associated with an acidic tumor microenvironment that contributes to enhanced invasiveness (43, 44). Our findings illustrate that SV1 promotes growth and metastasis of cancer through the induction of PFKM and aberrant glycolytic metabolism.
PFKM is one of the isozymes encoded by phosphofructokinase-1 (PFK-1), which catalyzes the second irreversible step in the glycolytic pathway by phosphorylating fructose-6-phosphate to form fructose-1, 6-bisphosphate (45, 46). It is the key regulatory enzyme mediating changes in glycolysis in cancer cells (45, 46). However, the importance of PFKM and its upstream regulators in tumorigenic progression remains largely unknown. We found that SV1 up-regulates PFKM in an NF-κB–dependent manner, supporting the view that SV1 is a key regulator and that NF-κB is a previously unrecognized transcription activator of PFKM. Glycolytic inhibitors, including pharmaceuticals that directly inhibit PFK-1, have been considered for cancer treatment. One major challenge of these inhibitors is their potential toxicity to certain normal tissues that also use glucose as their main energy source, such as brain, retinae, and testis (47). Interestingly, MIA-602 effectively decreased PFKM in both ESCC cells and ESCC-bearing mice. Given that the side effects of GHRH analogs are minimal (9, 48), we propose that GHRH-R antagonists might be more practically used as glycolytic inhibitors to treat cancers. However, this finding needs to be validated clinically.
In summary, these studies addressed a long-standing question, which is why subgroups of cancers with low expression of pGHRH-R respond to GHRH-R antagonists, establishing the role of SV1 as a hypoxia-driven spliced tumor promoter and as a mediator linking hypoxia and glycolysis in cancer. MIA-602 counteracts NF-κB–PFKM signaling suppressing aberrant glycolytic metabolism and tumor progression in vitro and in vivo by inhibiting SV1. SV1 is highlighted as a therapeutic target of GHRH-R antagonists in cancer. Demonstration of the importance of SV1 may shed light on how to select the appropriate patients with cancers which do not harbor GHRH-R overexpression for the use of GHRH-R antagonists.
Materials and Methods
Clinical Patients and Samples.
All specimens of primary ESCC, along with adjacent noncancerous tissues, were from patients who had undergone radical surgery without preoperative therapy at the Cancer Hospital of Shantou University Medical College (CHSUMC) during 2011–2012. All samples were histopathologically and clinically confirmed as ESCC. Fifty-eight surgical samples of ESCC were freshly collected by snap-freezing in liquid nitrogen in a tumor-banking protocol, and were used for RNA extraction and RT-qPCR analysis. All clinical research protocols of this study were reviewed and approved by the Ethics Committee of Shantou University Medical College. Written informed consent was obtained from patients in accordance with the Declaration of Helsinki.
Peptides and Chemicals.
GHRH-R antagonist MIA-602 was synthesized in the laboratory of A.V.S in Miami. For in vitro experiments, the peptides were dissolved in dimethyl sulfoxide (DMSO) to form a 5-mM solution, and further diluted with cell culture medium to the concentration indicated. The final concentration of DMSO in the culture medium was 0.1% (vol/vol). For the in vivo study, animals were treated daily by s.c. administration of 5 µg GHRH-R antagonist MIA-602 or vehicle solutions (0.1% DMSO in 5.5% mannitol; Sigma).
Tumor Xenografts.
A total of 5 × 105 KYSE150 cells was resuspended in 100 μL phosphate-buffered saline (PBS) and injected s.c. into the flanks of 6-wk-old female nude mice (Vital River Laboratory Animal Technology Co.). Animal experiments were reviewed and approved by the Ethics Committee of Shantou University Medical College.
Additional materials and methods are described in SI Appendix, Materials and Methods.
Data Availability Statement.
All data discussed in the paper are available in the manuscript or SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by a grant in part from the National Natural Science Foundation of China (81773087 and 81572876 to H.Z.), the Science and Technology Planning Project of Guangdong Province of China (2019A030317024 to H.Z.), and the Shantou University-Technion Research Program (43209504 to H.Z.). The work in Miami, Florida, was supported by the Medical Research Service of the Department of Veterans Affairs and the University of Miami Miller School of Medicine, Sylvester Comprehensive Cancer Center (to A.V.S.).
Footnotes
Competing interest statement: The Sponsor declares a conflict of interest. A.V.S. is a coinventor on the patent for growth hormone-releasing hormone analogs, assigned to the University of Miami, Miami, FL, and the Veterans Affairs Medical Center, Miami, FL. The other authors declare no conflict of interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913433117/-/DCSupplemental.
References
- 1.Oltean S., Bates D. O., Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Lin Y., et al. , Evaluation of salivary exosomal chimeric GOLM1-NAA35 RNA as a potential biomarker in esophageal carcinoma. Clin. Cancer Res. 25, 3035–3045 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Zhang H., et al. , Aberrant chimeric RNA GOLM1-MAK10 encoding a secreted fusion protein as a molecular signature for human esophageal squamous cell carcinoma. Oncotarget 4, 2135–2143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sibley C. R., Blazquez L., Ule J., Lessons from non-canonical splicing. Nat. Rev. Genet. 17, 407–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dvinge H., Kim E., Abdel-Wahab O., Bradley R. K., RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heddleston J. M., et al. , Hypoxia inducible factors in cancer stem cells. Br. J. Cancer 102, 789–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanopka A., Cell survival: Interplay between hypoxia and pre-mRNA splicing. Exp. Cell Res. 356, 187–191 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Barabutis N., Schally A. V., Growth hormone-releasing hormone: Extrapituitary effects in physiology and pathology. Cell Cycle 9, 4110–4116 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Schally A. V., Varga J. L., Engel J. B., Antagonists of growth-hormone-releasing hormone: An emerging new therapy for cancer. Nat. Clin. Pract. Endocrinol. Metab. 4, 33–43 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Villanova T., et al. , Antagonists of growth hormone-releasing hormone (GHRH) inhibit the growth of human malignant pleural mesothelioma. Proc. Natl. Acad. Sci. U.S.A. 116, 2226–2231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muñoz-Moreno L., Schally A. V., Prieto J. C., Carmena M. J., Bajo A. M., Growth hormone-releasing hormone receptor antagonists modify molecular machinery in the progression of prostate cancer. Prostate 78, 915–926 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Fahrenholtz C. D., et al. , Preclinical efficacy of growth hormone-releasing hormone antagonists for androgen-dependent and castration-resistant human prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 111, 1084–1089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez R., et al. , Antagonists of growth hormone-releasing hormone suppress in vivo tumor growth and gene expression in triple negative breast cancers. Oncotarget 3, 988–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan J., et al. , Growth hormone-releasing hormone receptor antagonists inhibit human gastric cancer through downregulation of PAK1-STAT3/NF-κB signaling. Proc. Natl. Acad. Sci. U.S.A. 113, 14745–14750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rekasi Z., et al. , Antagonists of growth hormone-releasing hormone and vasoactive intestinal peptide inhibit tumor proliferation by different mechanisms: Evidence from in vitro studies on human prostatic and pancreatic cancers. Endocrinology 141, 2120–2128 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Szalontay L., et al. , Novel GHRH antagonists suppress the growth of human malignant melanoma by restoring nuclear p27 function. Cell Cycle 13, 2790–2797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klukovits A., et al. , Novel antagonists of growth hormone-releasing hormone inhibit growth and vascularization of human experimental ovarian cancers. Cancer 118, 670–680 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Rekasi Z., Czompoly T., Schally A. V., Halmos G., Isolation and sequencing of cDNAs for splice variants of growth hormone-releasing hormone receptors from human cancers. Proc. Natl. Acad. Sci. U.S.A. 97, 10561–10566 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiaris H., et al. , Ligand-dependent and -independent effects of splice variant 1 of growth hormone-releasing hormone receptor. Proc. Natl. Acad. Sci. U.S.A. 100, 9512–9517 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havt A., et al. , The expression of the pituitary growth hormone-releasing hormone receptor and its splice variants in normal and neoplastic human tissues. Proc. Natl. Acad. Sci. U.S.A. 102, 17424–17429 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szepeshazi K., et al. , Antagonists of growth hormone-releasing hormone (GH-RH) inhibit in vivo proliferation of experimental pancreatic cancers and decrease IGF-II levels in tumours. Eur. J. Cancer 36, 128–136 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Hohla F., et al. , Differential expression of GHRH receptor and its splice variant 1 in human normal and malignant mucosa of the oesophagus and colon. Int. J. Oncol. 33, 137–143 (2008). [PubMed] [Google Scholar]
- 23.Bellyei S., et al. , GHRH antagonists reduce the invasive and metastatic potential of human cancer cell lines in vitro. Cancer Lett. 293, 31–40 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Bowler E., et al. , Hypoxia leads to significant changes in alternative splicing and elevated expression of CLK splice factor kinases in PC3 prostate cancer cells. BMC Cancer 18, 355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Outschoorn U. E., Peiris-Pagés M., Pestell R. G., Sotgia F., Lisanti M. P., Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 113 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Lin C., et al. , Nkx2-8 downregulation promotes angiogenesis and activates NF-κB in esophageal cancer. Cancer Res. 73, 3638–3648 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Kawauchi K., Araki K., Tobiume K., Tanaka N., p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 10, 611–618 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Halmos G., et al. , Human renal cell carcinoma expresses distinct binding sites for growth hormone-releasing hormone. Proc. Natl. Acad. Sci. U.S.A. 97, 10555–10560 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong H., et al. , Reciprocal androgen receptor/interleukin-6 crosstalk drives oesophageal carcinoma progression and contributes to patient prognosis. J. Pathol. 241, 448–462 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Feng Y., et al. , Metformin promotes autophagy and apoptosis in esophageal squamous cell carcinoma by downregulating Stat3 signaling. Cell Death Dis. 5, e1088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., et al. , Metformin induces human esophageal carcinoma cell pyroptosis by targeting the miR-497/PELP1 axis. Cancer Lett. 450, 22–31 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Dioufa N., et al. , Growth hormone-releasing hormone receptor splice variant 1 is frequently expressed in oral squamous cell carcinomas. Horm. Cancer 3, 172–180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brady L. K., et al. , Transcriptome analysis of hypoxic cancer cells uncovers intron retention in EIF2B5 as a mechanism to inhibit translation. PLoS Biol. 15, e2002623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han J., et al. , Hypoxia is a key driver of alternative splicing in human breast cancer cells. Sci. Rep. 7, 4108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dvinge H., Bradley R. K., Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 7, 45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiaris H., et al. , Expression of a splice variant of the receptor for GHRH in 3T3 fibroblasts activates cell proliferation responses to GHRH analogs. Proc. Natl. Acad. Sci. U.S.A. 99, 196–200 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barabutis N., et al. , Stimulation of proliferation of MCF-7 breast cancer cells by a transfected splice variant of growth hormone-releasing hormone receptor. Proc. Natl. Acad. Sci. U.S.A. 104, 5575–5579 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semenza G. L., HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 123, 3664–3671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christofk H. R., et al. , The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Koppenol W. H., Bounds P. L., Dang C. V., Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Preter G., et al. , Inhibition of the pentose phosphate pathway by dichloroacetate unravels a missing link between aerobic glycolysis and cancer cell proliferation. Oncotarget 7, 2910–2920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liberti M. V., Locasale J. W., Correction to: ‘The Warburg effect: How does it benefit cancer cells?’: [Trends in Biochemical Sciences, 41 (2016) 211]. Trends Biochem. Sci. 41, 287 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Zhang F., et al. , Phosphofructokinase-1 negatively regulates neurogenesis from neural stem cells. Neurosci. Bull. 32, 205–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi W., et al. , Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelicano H., Martin D. S., Xu R. H., Huang P., Glycolysis inhibition for anticancer treatment. Oncogene 25, 4633–4646 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Zarandi M., et al. , Synthesis and structure-activity studies on novel analogs of human growth hormone releasing hormone (GHRH) with enhanced inhibitory activities on tumor growth. Peptides 89, 60–70 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are available in the manuscript or SI Appendix.