Abstract
Autophagy refers to the lysosomal degradation of damaged or superfluous components and is essential for metabolic plasticity and tissue integrity. This evolutionarily conserved process is particularly vital to mammalian post-mitotic cells such as neurons, which face unique logistical challenges and must sustain homoeostasis over decades. Defective autophagy has pathophysiological importance, especially for human neurodegeneration. The present-day definition of autophagy broadly encompasses two distinct yet related phenomena: non-selective and selective autophagy. In this minireview, we focus on established and emerging concepts in the field, paying particular attention to the physiological significance of macroautophagy and the burgeoning world of selective autophagy pathways in the context of the vertebrate nervous system. By highlighting established basics and recent breakthroughs, we aim to provide a useful conceptual framework for neuroscientists interested in autophagy, in addition to autophagy enthusiasts with an eye on the nervous system.
Keywords: autophagy, mitochondria, neurodegeneration, organelles, neurons, metabolism
Introduction: neural complexity and quality control
Membranous organelles make intimate contacts with each other and serve as platforms for multiple molecular processes executed by signalling proteins. This crosstalk occurs with an astounding degree of intracellular dynamism against the backdrop of both phenotypic and environmental context. Amidst this cellular landscape, all components are subject to damage and dysfunction. Accordingly, eukaryotic cells have evolved sophisticated quality control mechanisms to sustain homoeostasis and function. For the human nervous system, sustaining homoeostasis over decades is particularly important. With its trillions of connections, mammalian neural architecture presents a unique physiological conundrum. Neurons are post-mitotic, have high metabolic demands and are often expansive in their morphological configuration and synaptic complexity. Each neural process and projection represents a distinct biochemical compartment, and the seamless interplay of numerous quality control processes is required to safeguard system integrity. To maintain organellar integrity within highly arboreous neurons that project over considerable distances, these quality control pathways are highly dependent upon both intracellular (anterograde, retrograde) and extracellular trafficking mechanisms (transcellular signalling).
One such quality control process is autophagy, a mechanism that evolved to sustain cells during instances of nutrient deprivation [1–3]. The term ‘autophagy’ derives from the Greek ‘to eat oneself’, and its use in language dates back to the 1800s [4]. Autophagy is essential for eukaryotic homoeostasis and has profound importance in the nervous system. The basic idea underpinning autophagy is that defective or superfluous subcellular components are sequestered and delivered to the lysosome by various routes for destruction. The autophagic turnover of intracellular components is thought to fuel metabolism.
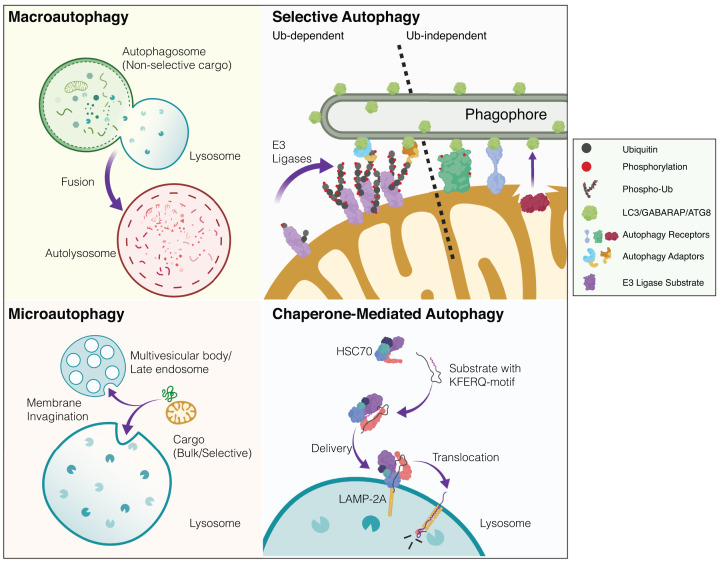
Several distinct forms of autophagy exist and these are categorised according to how cargo is recognised, processed and delivered to acidic endolysosomal compartments (Figure 1 provides a generalised graphical overview of autophagy modalities). Macroautophagy refers to the non-selective or ‘bulk’ sequestration of cytoplasmic constituents (organelles, protein aggregates, intracellular pathogens) into a transient double-membrane bound autophagosome. The autophagosome eventually fuses with an acidic compartment of the endolysosomal system (autolysosome or amphisome in the case of late endosome–autophagosome fusion), destroying its contents. This pathway contrasts with microautophagy, where cytoplasmic components enter the late endosome or lysosome via membrane invagination. Even more distinct than this, chaperone-mediated autophagy (CMA) degrades substrate proteins harbouring specific motifs (typically ‘Lys-Phe-Glu-Arg-Gln’ (KFERQ)-like motif) that are recognised by chaperones and translocated into lysosomes via specific membrane receptors [5] (Figure 1). In this brief review, we highlight some essential and emerging concepts in autophagy research, with a particular emphasis on the contribution of autophagy signalling to mammalian neural integrity.
Figure 1. Simplified overview of mammalian autophagy modalities.
There are several distinct modes of autophagy in eukaryotic cells. These subtypes are defined according to how cargo is recognised and delivered to the acidic or degradative cellular subcompartments (i.e. lysosomes or late endosomes). Macroautophagy: cytoplasmic constituents such as organelles and protein aggregates are non-selectively engulfed in a double-membrane bound autophagosome. This bulk sequestration of cellular matter results in degradation upon the formation of the autolysosome (autophagosome-lysosome fusion). The homoeostatic balance between degradation and biosynthesis is ensured by the process of autophagosome-lysosome reformation (not depicted). Microautophagy: in contrast with autophagosomal delivery, cargo destined for destruction enters the endolysosomal system via membrane invagination. This occurs in non-selective and selective manners with chaperones. Multi-vesicular bodies (MVBs) are also known to participate in microautophagy. Selective autophagy: distinct substrates (organelles, proteinaceous/membranous aggregates, pathogens) are recognised via priming by cargo-specific receptors. Selective autophagy of organelles (organellophagy) can occur in a ubiquitin-dependent or -independent fashion. The depicted example shows mitophagy where in response to stress, ubiquitin E3 ligases can trigger mitochondrial ubiquitylation, thereby recruiting specific adaptor proteins that engage the autophagy machinery. In ubiquitin-independent selective autophagy, specific receptors directly engage LC3/GABARAP/ATG8 proteins to drive the encapsulation and elimination of the organelle. Lipids such as cardiolipin and ceramide have also been reported as mitophagy receptors (not depicted). Although mitophagy is largely studied as an intracellular process, transcellular mitophagy (known as axonal transmitophagy in neurons) has been described in the CNS (not depicted). Mitochondrial subdomains can also be excised and delivered to the lysosome via mitochondrial-derived vesicles (MDVs, not shown). Chaperone-mediated autophagy (CMA): substrate proteins containing a distinct motif are selectively recognised by chaperones and delivered to lysosomes where they undergo translocation and destruction. Created with BioRender.
Autophagy essentials: a generalised overview
Macroautophagy is induced in response to many stimuli (e.g. starvation, hypoxia and infection) and the mechanistic regulation of this process is highly conserved across eukaryotes. These induction signals are often used to study macroautophagy in the laboratory, yet their interplay in modulating macroautophagy in vivo remains less clear.
The most upstream signalling component in mammalian autophagy is ULK1, a kinase complex (consisting of ULK1, FIP200, ATG13, ATG101) that initiates autophagosome biogenesis [6]. RNAi-mediated or small molecule inhibition of ULK1 is sufficient to suppress autophagy [7–11]. The ULK1-dependent phosphorylation of downstream substrates (class III PI3K VPS34) stimulates the biosynthesis of phosphatidylinositol-3-phosphate (PI3P) for phagophore nucleation [12,13]. Starvation-induced autophagy requires the VPS34-BECN1 PI-3 kinase and the ULK1 protein kinase for phagophore initiation [14]. Growth factor deprivation and a decreased energy state act as particularly potent induction stimuli for autophagy, with the latter stimulus activating the master metabolic regulator AMPK [15]. The mammalian target of rapamycin (mTOR) is a master nutrient sensor, and active mTORC1 associates with ULK1 [9,16,17]. Reciprocally, mTOR inhibition (either by nutrient deprivation or chemical inhibition) is as a powerful signal that activates macroautophagy [18], leading to its dissociated from, and enhanced kinase activity of ULK1. Independently of its essential role in autophagy, ULK1 has also been implicated in axonal branching [19,20].
The precise source of autophagic membranes has been a hot topic in the field for many years, with many cellular compartments predicted to contribute to precursor membranes. Because of its well-documented association with isolation membranes, subdomains of the endoplasmic reticulum (ER) are regarded to make a crucial contribution to autophagosome formation [21,22]. In addition, ER–Golgi hybrid intermediates, the Golgi complex and mitochondria have also been predicted to contribute to autophagosome biogenesis [23,24]. In recent years, increasing evidence has also mounted in favour of recycling endosomes as a critical source of autophagic membranes [25,26].
The efficiency of autophagosome membrane expansion, maturation and completion involves two ubiquitin conjugation systems: ATG12-ATG5-ATG16L1 and the Ubiquitin-like LC3 subfamily, respectively (also referred to as ATG8-like family). These conjugation systems stimulate the expansion of the autophagosome by promoting the recruitment of small ubiquitin-like ATG8 proteins to membrane-bound phosphatidylethanolamine (PE). These include the LC3 and GABARAP protein subfamilies, which are integral to autophagy. Cleaved LC3-I conjugates with PE, leading to LC3-II formation and ultimately autophagosome maturation [27]. Due to dynamic cycles of synthesis and degradation, LC3 levels fluctuate within cells. Because LC3-II is associated with autophagosomes and accumulates with chemical inhibition (e.g. Bafilomycin A1, chloroquine), it has been used extensively as a biochemical readout of autophagic flux, however there are caveats to this approach [28]. For more in-depth coverage of this highly complex process, the reader is directed to recent reviews [29–32].
Even when mammalian cells have ample access to nutrients, they still recycle damaged or superfluous organelles with exquisite specificity. Distinct from macroautophagy, this is known as ‘selective autophagy’, and is achieved through specialised receptors that confer specificity on the cellular component destined for elimination [33]. Recent in vivo research demonstrates that many selective autophagy pathways are likely to operate constitutively within tissues (e.g. mitophagy). However, for reasons of experimental tractability, these pathways have clasically been studied as induced ‘damage responses’ in proliferating cells under conditions of extreme stress, due to the high mechanistic resolution afforded by this approach [29,34]. Sophisticated approaches in mouse genetics and optical reporters now enable researchers to reconcile the in vivo significance of in vitro mechanisms. Selective autophagy has also been referred to as ‘organellophagy’, and constitutes a burgeoning area of the field [35], where the delineation of multiple selective autophagy pathways continues in ever greater detail.
Neural macroautophagy in mammalian health and survival
The continual development of facile assays to monitor macroautophagy underpins our present-day knowledge of this process in health and disease [36]. For many decades, researchers relied heavily upon electron microscopy (EM) to visualise morphically distinct autophagic structures in cells and tissues. Indeed, pioneering contributions to histochemistry and EM by Alex B. Novikoff [37] were critical in the early days of the autophagy field and enabled crucial insights by De Duve, Palade and others. Although EM remains the gold standard for high-resolution ultrastructural studies, many proponents recognise its limitations for tissue-based physiological studies. EM is highly labour-intensive, difficult to optimise for high-throughput analyses and requires specialist expertise for execution and accurate interpretation [38]. A pioneering study in 2004 by Noboru Mizushima and colleagues generated a fluorescent autophagy reporter mouse (LC3-GFP), enabling the systemic visualisation of autophagosomes in mammalian organ systems for the first time [39]. This important tool provided a snapshot of steady-state LC3-dependent macroautophagy (hereafter referred to as basal) levels across multiple tissues. The authors conducted extensive validation, showing the pronounced induction of macroautophagy in response to food restriction in mice, and a dramatic reduction in autophagy in Atg5-null mice [40]. The autophagic response to starvation differs between organ systems and tissues, indicating the complexity of this process in vivo. Originally, CNS neurons were reported to have minimal levels of basal macroautophagy, with reduced autophagy induction in response to starvation. This observation contrasted greatly with other organ systems (e.g. liver, heart, pancreas), where pronounced time-dependent changes in autophagic induction were observed [39]. However, later studies reported a dramatic induction of autophagy in cortical neurons, Purkinje cells and hypothalamic neurons by fasting [41,42].
Classically, macroautophagy serves to sustain metabolism and homoeostasis by ensuring an adequate pool of essential constituents through the catabolism of cellular components. In cultured cells and tissues, nutrient deprivation induces a pronounced macroautophagy response [43]. In terms of timeframe, this response occurs rapidly in cultured adherent cells in vitro but requires at least 24 h for a noticeable elevation above basal levels for some mammalian tissues. Although basal macroautophagy in the CNS is reported to occur at low levels and minimally induced upon starvation (compared to other tissues), it has profound importance in the nervous system [39]. This was elegantly demonstrated in two landmark studies that employed conditional genetic ablation of neuronal Atg5 and Atg7 (Atg5flox/flox; nestin-Cre and Atg7flox/flox; nestin-Cre) [44,45]. The selective loss of basal macroautophagy in neurons triggered profound phenotypes in these mice, characterised by severe locomotor and gait dysfunction, the intraneuronal accumulation of proteinaceous inclusions, coupled with widespread neuronal loss. In addition to this, mice with Purkinje-neuron-specific deletion of Atg7 (Atg7flox/flox; Pcp2-Cre), exhibited cell-autonomous dystrophy with terminal axon degeneration [46].
Taken together, this provided a solid physiological basis for basal macroautophagy as a critical modulator of mammalian neural integrity. Furthermore, it provided an essential proof-of-principle for autophagy as a critical mechanism to neutralise aggregate-prone proteins, which are a hallmark of many neurodegenerative diseases. Importantly, the disruption of many discrete steps in the autophagy pathway is associated with the molecular pathophysiology of several neurodegenerative diseases [15,47,48].
Kuma and colleagues [49] also showed the importance of autophagy in the early stages of neonatal life, often termed the ‘starvation phase’. Constitutive autophagy appears to proceed at a low level, with a dramatic, programmed elevation observed during the initial hours of postnatal development [49].
An elegant study by Yoshii and colleagues [50] has recently clarified the physiological significance of neuronal macroautophagy for systemic health and survival. Although a whole-body genetic deletion of Atg5 results in neonatal lethality, this phenotype can be dramatically rescued by the selective restoration of macroautophagy in neurons (Atg5−/−; Nse-Atg5). In this remarkable demonstration, these surviving animals have preserved neuronal autophagy, but are deficient in Atg5-dependent macroautophagy in all other extra-neural tissues. Although these animals survive because they can feed, they still exhibit phenotypic abnormalities with hypogonadism and anaemia [50]. Nonetheless, this work demonstrates the importance of neural autophagy for mammalian health and survival.
Although the loss of macroautophagy in the nervous system is clearly deleterious, it is important to recognise that autophagy inhibition represents an advantageous therapeutic strategy in certain contexts [51]. Recently, the systemic inhibition of autophagy had striking anti-tumorigenic effects in a pre-clinical model of KRAS-driven lung cancer [52]. However, these benefits were offset by the onset of severe neuropathology, leading to early death in these animals [53]. Conversely, agents that enhance autophagy are considered to have potential to attenuate age-associated cognitive decline. Related to this, autophagy has recently been suggested to promote memory formation by modulating hippocampal-dependent adaptive responses in mice [54]. Enhanced autophagy activation has recently been associated with beneficial cognitive effects in a mouse model of Fragile-X syndrome [55].
Autophagy-related factors also fulfil critical non-canonical (autophagy-independent) roles in the nervous system. For example, ULK1 signalling makes an important autophagy-independent contribution to axonal guidance in mammals [19,20]. Conditional ablation of both Ulk1/2 in mouse neurons causes selective impairments in axonal tract formation, leading to abnormal forebrain development in vivo [20]. Conditional Ulk1/2 -deficient mice also exhibit a progressive loss of hippocampal CA1 pyramidal neurons at 8 weeks of age [56]. Furthermore, loss of the key ULK1 interactors FIP200 and ATG101 in model organisms affects neural integrity in vivo. Conditional loss of FIP200 in the mouse nervous system leads to cerebellar neurodegeneration [57]. Loss of Drosophila Atg101 affects neural homoeostasis [58].
Macroautophagy in mammalian neuropathology
As described in the general overview and elaborated in greater depth in the associated references, macroautophagy is a complicated multistep process. Strikingly, disrupting almost any step in this pathway has disease significance, and dysfunctional autophagy is associated with a diverse range of inherited and idiopathic neurodegenerative disorders [59]. The discovery of ubiquitin/p62-positive intraneuronal aggregates in Atg7flox/flox; Nes-Cre mice, provided an important proof-of-concept demonstration that macroautophagy is a crucial neuroprotective mechanism in mammals [60]. Proteinaceous aggregates are a pathological hallmark of several neurodegenerative diseases (e.g. ‘proteinopathies’, e.g. tauopathies, synucleinopathies depending on the particular disease aetiology, in addition to Huntington’s disease, Amyotrophic Lateral Sclerosis (ALS) and others). Elegant mechanistic work in cultured cells combined with mouse genetics has dramatically expanded this domain – please consult Table 1 for a list of autophagy-associated mouse models with neurological phenotypes. For an excellent disease-specific discussion of evidence implicating autophagy and aggregate clearance in specific neurodegenerative disorders, please consult Menzies and colleagues, 2017 [15]. It is not surprising that autophagy now represents an attractive therapeutic target for neurodegenerative disease and beyond [61].
Table 1. Autophagy mouse models and known phenotypes.
| Gene | Yeast homologue | Model | Phenotype/findings | Reference |
|---|---|---|---|---|
| Atg3 | Atg3 | Atg3−/− | Neonatal lethality | [62] |
| Atg5 | Atg5 | Atg5−/− | Neonatal lethality | [46] |
| Atg5 | Atg5 | Neural specific Atg5−/− | Progressive deficits in motor neurons; accumulation of intraneuronal inclusion bodies | [45,63,64] |
| Atg5 | Atg5 | Atg5 deletion in POMC neurons | No reported phenotype | [65] |
| Atg7 | Atg7 | Atg7−/− | Neonatal lethality | [49] |
| Atg7 | Atg7 | Atg7 flox/flox | Liver tumours; neurodegeneration | [63,64,66] |
| Atg9a | Atg9 | Atg9a−/− | Neonatal lethality | [65] |
| Atg12 | Atg12 | Atg12-/- | Neonatal lethality | [67] |
| Atg12 | Atg12 | Atg12 deletion in POMC neurons | Diet-induced obesity | [67] |
| Atg13 | Atg13 | Atg13−/− | Embryonic lethality | [68] |
| Atg16L1 | Atg16 | Atg16L1 mutant mouse | Neonatal lethality | [44] |
| Becn1 | Atg6/Vps30 | Beclin1−/− | Neonatal lethality | [69–72] |
| Gabarap | Atg8 | Gabarap KO mouse | No reported phenotype | [73] |
| Lc3 | Atg8 | Lc3−/− | Normal development | [74] |
| Pik3c3/Vps34 | Vps34 | Pik3c3/Vps34 specific deletion in sensory neuron | Rapid neurodegeneration | [75] |
| Pik3c3/Vps34 | Vps34 | Pik3c3/Vps34−/− | Embryonic lethality | [76] |
| Ulk1 | Atg1 | Ulk1−/− | Delayed mitochondrial clearance in reticulocytes | [77] |
| Ulk2 | Atg1 | Ulk2−/− | No pheynotype | [78] |
| Ulk1/2 | Atg1 | Ulk1/2−/− | Neonatal lethality | [78] |
| Ulk1/2 | Atg1 | Ulk1/2-cDKO | Early embryonic and perinatal lethality/abnormal axon guidance | [20] |
| FIP200/Rb1cc1 | Atg17 | FIP200 f/f mouse | Cerebellar degeneration | [57] |
| Wipi4 | Atg18 | Wdr45flox/Y | Poor motor coordination | [79] |
| Specific deletion in CNS | Impaired learning and memory; extensive axon swelling with numerous axonal spheroids |
The recent discovery of human mutations in some core autophagy and autophagy-associated genes has cemented its vital role in neurodegeneration (Table 2). De novo mutations in the WDR45/ATG18 gene are associated with a distinct case of Neurodegeneration with Brain Iron Accumulation (NBIA). This gene encodes WIPI4 and couples PI3P synthesis to LC3 lipidation in autophagosome biogenesis [80]. The clinical features of NBIA include early-onset global developmental delay and progressive neurological deterioration (Parkinsonism, dystonia and dementia developing by early adulthood) [81].
Table 2. Disease-associated mutations in human core ATG genes.
| Gene | Yeast homologue | Disease | Reference |
|---|---|---|---|
| WDR45 | Atg18/Atg21 | NBIA | [81] |
| WDR45 | Atg18/Atg21 | SENDA (static encephalopathy of childhood with neurodegeneration in adulthood) | [82] |
| WDR45 | Atg18/Atg21 | BPAN (β-propeller protein-associated neurodegeneration) | [83,84] |
| ATG5 | Atg5 | Congenital ataxia, mental detardation, developmental delay | [85] |
Additionally, the manifestation of static encephalopathy of childhood with neurodegeneration in adulthood (SENDA) is associated with human mutations in WDR45. SENDA is a variant of NBIA reported by Kruer and colleagues in 2012 [82], with affected individuals exhibiting childhood intellectual impairments and severe dystonia-Parkinsonism in adulthood. Patient-derived cell lines exhibited lower autophagic activity combined with the accumulation of aberrant early autophagic structures [83]. SENDA is also known as β-propeller protein-associated neurodegeneration (BPAN), and a patient is reported to carry de novo heterozygous splice-site mutations of WDR45 [84]. Nes-Wdr45 fl/Y mice exhibit motor coordination and learning defects, accompanied by axon swelling [79].
Regarding the core ATG machinery, the first mutation in human ATG5 was recently reported. Two childhood siblings presented with cerebellar ataxia, hypoplasia and lack of coordination. These neurological findings were the result of a point mutation in human ATG5 (E122D), leading to a weak ATG5–ATG12 interaction [85]. As diagnostic resolution continues to advance in clinical genetics, it will be exciting to determine the exact prevalence of patients with autophagy-associated mutations and neurological symptoms.
The emerging importance of selective autophagy in mammalian neural integrity
Although we have a vast knowledge about the molecular regulation and physiological significance of macroautophagy, selective autophagy pathways have emerged as crucial pathways in their own right [27,33,34,86]. Selective autophagy refers to the targeted delivery of defective or superfluous organelles to the lysosome for elimination and typically operates in ubiquitin-dependent or -independent fashion. In ubiquitin-dependent turnover, cellular sensing of defective organelles/components activates specific ubiquitin ligase enzymes that decorate cargo with ubiquitin chains of varying topologies. This ‘organellar ubiquitylation’ can serve as a distinct ‘eat-me’ signal that engages specific autophagy adaptor proteins, resulting in the recruitment of the autophagy machinery and elimination of the organelle. The number of selective autophagy receptors has expanded dramatically in recent years (detailed in Table 3). For ubiquitin-dependent selective autophagy, deubiquitylases (DUBs) are potent regulators that oppose this process.
Table 3. Mammalian selective autophagy receptors.
| Autophagosomal substrate | Receptors |
|---|---|
| Mitochondria/mitophagy | NIX/BNIP3 [87], FUNDC1 [88], p62 [89], HDAC6 [90], OPTN1 [91], NDP52 [92], NIPSNAP [93], PHB2 [94], FKBP8 [95], Cardiolipin [96] |
| Peroxysomes/pexophagy | P62 [97,98], NBR1 [97], PEX3 [99], PEX2 [100], PEX5 [101], ACBD5(Atg37) [102] |
| Ribosomes/ribophagy | NUFIP1 (Nuclear fragile X-mental retardation-interacting protein 1) [103] |
| Lipid droplets/lipophagy | Macroautophagy/no receptors are identified [104,105] |
| Protein aggregates/aggrephagy | P62 [106], NBR1 [107], ALFY [107,108], OPTN [106], HDAC6 [109], BAG3 [110], NDP52 [111] |
Like macroautophagy, which occurs at basal levels and can also be induced by specific stimuli, selective autophagy pathways also occur at basal levels and can be induced in response to stress. The following section provides some historical context, and attempts to elaborate the most recent concepts in this field. We pay particular attention to mitochondrial turnover, given the significant advances made in this domain.
Mitophagy
Mitophagy in healthy tissues
The turnover of mitochondria (termed mitophagy) is arguably the most intensively studied pathway in the selective autophagy field. Mitochondria are dynamic signalling organelles that orchestrate myriad aspects of mammalian metabolism from energetics to epigenetics. Beyond ATP production and burning fuel, these pleiotropic powerhouses contribute to the biosynthesis of nucleotides, amino acids, haem, Fe–S clusters, sterols, as well as playing critical roles in organelle biogenesis and calcium homoeostasis [112]. Consequently, mitochondrial dysfunction is associated with a plethora of neurodegenerative disease states [34,113–115].
Despite the modern-day limelight it receives, it seems that mitochondrial turnover has captured the imagination of researchers for decades. At the time of writing this review, the first known reference to mitochondrial turnover dates to the pioneering work of Margaret Reed Lewis and Warren Lewis, in their 1915 cell biology masterpiece which examined mitochondrial dynamics using Janus green dye in tissue culture experiments [116]. They observed that under certain circumstances, mitochondria can ‘degenerate’ [116]. Interestingly, the term ‘mitochondrial degeneration’ appears again in a diabetes-associated case report from Edinburgh in 1928, but there is insufficient detail to provide a context for any reference to selective elimination [117]. Subsequently, important EM studies in the 1950s by Sam Clark Jr. [118] resolved mitochondria within endolysosomal structures in various mammalian tissues. The first direct paper on the selective elimination of mitochondria appears to come from Novikoff in 1962, in his original study of mitochondrial degeneration [119]. It is of interest that 6 years earlier, Novikoff also identified ‘dense bodies’, which linked degradative cellular compartments with mitochondria and other organelles [120]. The reader should note that in all of these aforementioned investigations, researchers were studying basal levels of mitochondrial turnover (often in healthy wild-type, unstressed animals), which precedes the present-day focus on stress-induced mitophagy in cell culture.
Akin to macroautophagy, it was challenging to observe mitophagy in cells and tissues until the advent of fluorescent reporter mice (these experimental challenges are outlined in [38]). Recent studies characterising the mito-Keima and mito-QC mitophagy reporter mouse models have been instrumental in illuminating mitochondrial turnover in vivo [121,122]. For an excellent review of reporter mice and physiological autophagy, please consult Kuma and colleagues [40]. The mitochondrial matrix localisation of mt-Keima and mitochondrial outer membrane localisation of mito-QC demonstrate an abundance of mitochondria in lysosomes (mitolysosomes) across a variety of organ systems) [121–125]. Importantly, both reporter models provide an unambiguous demonstration that mitophagy occurs at steady-state levels within mammalian tissues. In terms of mitochondrial turnover in the vertebrate nervous system, studies in mito-QC mice have illuminated the spatial nature of mitophagy within a variety of neural subtypes in vivo. These comparative investigations revealed a robust level of mitophagy in A9 dopaminergic neurons and cerebellar Purkinje neurons, with a striking spatial restriction of turnover to neuronal somata [122–124]. Additionally, microglia and cerebral vasculature also had noticeably high levels of mitochondrial turnover [122]. Recent data utilising these reporters also demonstrated the conservation of basal mitophagy from mammals to flies [126,127].
Mitophagy has also been shown to contribute to CNS development. Elegant work by Boya and colleagues demonstrated the importance of mitophagy in retinal neurodevelopment during the metabolic transition towards glycolysis [128]. Furthermore, high levels of basal mitophagy have also been observed in photoreceptor neurons [129].
An important concept that is pertinent to the complexity of the mammalian nervous system is that the process of mitophagy may not always be intracellular. Davis and colleagues [130] used a mitophagy reporter combined with EM analyses to demonstrate the extrusion of mitochondria from retinal ganglion axonal evulsions, and degraded by lysosomes within neighbouring astrocytes. Interestingly, this atypical mitophagy modality termed as ‘axonal transmitophagy’ was observed to occur under steady-state conditions [130]. Conversely, neurons have also been described to acquire mitochondria from glia following ischaemic injury [131]. It is tempting to speculate that such mechanisms may facilitate long range homeostasis in the vertebrate nervous system. Collectively, these studies highlight that much more work is required to decipher the differential mechanisms and significance of neural mitophagy in vivo.
Mitophagy in neuropathology
Mitochondrial and neural integrity are intimately linked. In particular, generations of researchers have sought to clarify the mysterious relationship between mitochondrial and motor dysfunction in the context of Parkinson’s disease (PD). The first known studies of mitochondrial integrity in PD-patient biospecimens emerged from two reports in the 1970s, from Japan and Sweden, respectively [132,133]. Although severely limited in both scope and resolution, these early papers postulate a conceptual link between mitochondrial function and Parkinsonism [133]. Relevant mechanistic links emerged from pioneering work by Langston and colleagues [134] who discovered that the Parkinsonian neurotoxin MPTP exhibits a mitochondrial mode of action. In the late 1980s, reduced mitochondrial activity was also observed in PD patient brain-specimens [135], although it remains contentious whether or not this is a generalised feature of Parkinsonism [136]. A direct genetic link between mitochondrial dysfunction and PD emerged in 2004 by Suomalainen and colleagues, who discovered L-DOPA responsive Parkinsonism in patients with defective mtDNA maintenance, conferred by mutations in POLG [137].
Because the cause(s) of idiopathic PD remain elusive, rare inherited forms of familial Parkinsonism have provided important molecular insights into potential mechanisms of pathophysiology. Many of the numerous loci that are causative for EOPD (early-onset PD) influence mitochondrial signalling [115]. Present-day interest in mitophagy stems from the vital observation that two of these PD-related proteins, PINK1 (encoded by PARK6) and Parkin (encoded by PARK2) can orchestrate the dramatic clearance of mitochondria in cultured proliferating cells under highly defined conditions. This pathway has been reviewed extensively in the literature (reviewed in [34,114,115,138]).
Briefly, PINK1 is a mitochondrial-associated ubiquitin kinase [139], and Parkin is a cytosolic RBR E3 ubiquitin ligase that exists in an inactive (autoinhibited) state. Upon loss of mitochondrial membrane potential, PINK1 becomes stabilised and activated on depolarised mitochondria, and Parkin is recruited to the damaged organelle. PINK1 then phosphorylates both Parkin and ubiquitin at their respective Ser65 residues. The binding of phospho-Ser65-ubiquitin to Parkin, primes it for full activation by PINK1 at ParkinSer65. These events drive a feed-forward activation loop of mitochondrial ubiquitylation, whereby damaged mitochondria become decorated in a coat of ubiquitin and selectively eliminated via autophagy (reviewed in [114,115,140]).
Aside from Parkin, other factors have been implicated in mitochondrial ubiquitylation, including ARIH1, FBX07, Gp78, MARCH5, MUL1, RNF5, RNF185, SIAH1 and SMURF1 [34,114]. Yun and colleagues discovered an exciting compensatory role for MUL1 (also known as MULAN and MAPL) [141,142] during the loss of PINK1/Parkin signalling in flies and mice [143]. Lastly, DUBs appear to be critical negative regulators of mitophagy in vitro, but also influence pexophagy [144]. Bingol and colleagues first reported that USP30 antagonises Parkin-mediated mitophagy [145]. The contribution of USP30 to mitophagy in mammalian tissues remains to be clarified, and the analysis of mitophagy in Usp30 mutant animals will surely prove interesting.
Mitophagy is not just a ubiquitin-dependent process and as such, many ubiquitin-independent mitophagy receptors have also been identified. These include FUNDC1, NIX/BNIP3L, NIPSNAP, PHB2, FKBP8 and Cardiolipin (Table 3) [34,114]. Aside from mitochondrial depolarisation, PINK1/Parkin-independent mitophagy can be robustly induced by iron chelation and hypoxia [114,115] For a recent and authoritative overview of these pathways, please consult Montava-Garriga and Ganley [146].
A provocative reassessment of mitophagy in vivo
It has been challenging to assess the contribution of mitophagy to adult neural integrity, because we do not yet know of any candidate genes that modulate the selective, steady-state autophagy of mitochondria in vivo, i.e. mitophagy-deficient mammals are yet to be described. Genetics studies have demonstrated that macroautophagy-deficient animals exhibit developmental lethality [45,46,62–64] (Table 1). Thus, it seems reasonable to assume that animals unable to neutralise mitochondrial damage via mitophagy would be equally compromised, or at least exhibit phenotypic severity from an early age. This is particularly relevant given the importance of programmed mitophagy in CNS development [125,128]. Furthermore, given the importance ascribed to mitochondrial quality control in sustaining tissue homoeostasis [112,147], is it likely that a mitophagy-deficient animal would even survive?
As previously discussed in this review, numerous historical studies have documented mitochondria within degradative cellular compartments, yet recent research on basal mitophagy has provoked some level of controversy in the field [34,140,148]. Why has such a contentious debate ensued? Over the past decade, many reviews and research papers have cited the PINK1-Parkin signalling pathway to be a ‘master regulator’ of mitophagy, and posit defective mitophagy to be the elusive cause of PD. Although this was an intriguing hypothesis and is still regarded by some as an attractive therapeutic target for PD, the regulation of mitophagy in vivo appears to be far more complicated than cell culture models would predict. Furthermore, there are notable caveats that have been well described with respect to PINK1/Parkin-dependent mitophagy [27,34,114,126]. For example, the induction of PINK1/Parkin-dependent mitophagy in proliferating cancer cells requires the use of toxic agents (e.g. protonophores such as CCCP) in concert with extremely high levels of Parkin expression. Recently, the activation of depolarisation-induced mitophagy was shown to be affected by the concentration of serum components in tissue culture media, specifically bovine serum albumin [149]. Until recently, there was a lack of suitable biochemical tools to detect endogenous PINK1 protein in mouse tissues under stringent conditions (i.e. verifying the specificity of antibody reagents with suitable KO controls and verification by mass spectrometry [124]). Despite this, it is noteworthy that the endogenous signalling pathway can be activated in cultured primary neurons, although neural mitophagy was unaffected in both mice and primary PD-patient cells lacking a functional PINK1/Parkin pathway [123].
Pink1 and Parkin mutant mice do not recapitulate the authentic hallmarks of human PD [150], and substantial levels of genetic and mitotoxic stress are often required to elicit measurable phenotypes in KO animals (although recent knock-in strategies have resulted in selective phenotypes [123]). Even though these interventions lead to motor dysfunction and neurodegeneration in some animals, they are not accompanied by alterations in nigrostriatal mitophagy [151,152]. Furthermore, several converging studies now show that basal mitophagy proceeds unhindered in tissues from both Pink1 KO animals and Parkin mutant mice with loss of E3 enzyme activity [123,129]. These observations of PINK1-Parkin independent mitochondrial turnover have been replicated in flies, mice, non-human primates and cell culture systems [126,153–155]. Intriguingly, although stress is required to recapitulate relevant PD phenotypes in adult Pink1/Parkin KO mice, this is not the case with rat and non-human primate models [156–158]. Thus, further work will be needed to clarify the significance of these differential phenotypes that manifest in a species-specific manner.
The manifestation of neuropathology in familial PD has informative distinctions from the idiopathic condition. Intracellular inclusions known as Lewy bodies were first described in 1919 by Konstantin Tretiakoff and are regarded as a classical hallmark of PD pathology. Indeed, recent ultrastructural studies of postmortem Lewy body composition from human PD patients underscore the importance of macroautophagy and organellar integrity in the nervous system [159]. Despite this, inclusion pathology is not a consistent feature of PINK1/Parkin-associated PD in mouse models or patients (some Lewy pathology has been documented in case reports from PINK1 and PARKIN patients, although this appears to be extremely rare) [160]. These differential aetiologies merit consideration, especially when considering defective mitophagy as the sole pathological cause of any complex disease.
Because basal mitophagy is not affected in animals lacking a functional PINK1-Parkin pathway, does this imply that the PINK1-Parkin signalling is completely irrelevant for mitophagy? We do not regard this assertion to be constructive or accurate, especially given the complexity of signalling in the context of the mammalian nervous system. Mutations in both PINK1 (PARK6) and Parkin (PARK2) lead to early-onset PD. Furthermore, the recent discovery of a new human PD mutation at Parkin S65 (S65N) demonstrates that loss of PINK1-dependent Parkin activation compromises striatal and motor function in mice and humans [123]. Under basal conditions, mitophagy was unaffected in mouse tissues and patient-derived cells with mutant ParkinSer65. The fact that mitophagy can proceed in the absence of functional PINK1-Parkin signalling, demonstrates that mammalian tissues require more than a single pathway to modulate mitochondrial turnover. In addition to its association with PD, mitophagy has also been implicated other neurodegenerative disease states. A recent study by Fang and colleagues [161] has implicated defective mitophagy in the pathophysiology of Alzheimer’s disease (AD).
Our view is that mitophagy is a context-dependent process, consisting of distinct basal and stress-induced pathways. This nuanced perspective is consistent with the growing consensus espoused in recent comprehensive reviews on this topic [34,121]. At present, the tissue-specific factors that modulate basal mitophagy in vivo remain to be defined. However, the most physiological regulators of steady-state mitophagy described to date appear to be NIX/BNIP3L [87], ULK1 and p62 [89]. Mutant Nix or Ulk animals exhibit an accumulation of abnormal mitochondria within reticulocytes in vivo (Table 3), and p62 is required for hepatic mitophagy [155].
PINK1/Parkin in vivo: if not mitophagy, then what?
Even if the loss of PINK1/Parkin does not influence steady-state mitophagy in vivo, the requirement of this pathway to sustain homoeostasis during profound levels of cellular stress is exceptionally intriguing. Thus, a key question remains: what is the mechanistic contribution of this pathway to neural integrity?
Rather than influencing steady-state mitochondrial turnover in vivo, it is conceivable that PINK1 and Parkin could orchestrate mitophagy/another form of mitochondrial quality control during highly defined conditions of extreme stress. In line with this, Pink1/Parkin KO mice crossed with mitochondrial mutator PolgD257A mice and subjected to exhaustive exercise, a STING-mediated inflammatory response accompanied the onset of neurodegenerative phenotypes [152]. Although there were no reported differences in basal mitophagy between unstressed Pink1 mutant animals, this intriguing experimental paradigm revealed defective cardiac mitophagy in aged mutant animals subjected to extreme stress, with circulating mtDNA driving a neuroinflammatory response [152]. Despite a reported mitophagy defect in the hearts of double-mutant animals (Pink1 or Parkin KO mice crossed to PolgD257A mutator mice, aged and subjected to exhaustive exercise) it is unclear whether mitophagy was affected in the PD-relevant nigrostriatal pathway. It is also interesting to note that mass spectrometry analysis revealed minimal differences in phospho-ubiquitin levels between WT and Pink1 KO animals at steady-state levels. These emerging links between mitochondrial and immune integrity are especially interesting. A 2014 study from the laboratory of Mike Murphy discovered that PolgD257A mice have elevated levels of circulating pro-inflammatory cytokines upon ageing (TNF-α, IL-1β) [162]. Yet, a comparative analysis of serum cytokine profiles between the uncrossed mutator mice in the Sliter and colleagues study suggests a very different immunological profile. Nonetheless, it will be intriguing to interrogate these findings and dissociate the mechanistic contribution of PINK1-Parkin signalling from the already complex background of mutator mice.
Recent Drosophila studies also support the notion that PINK1-Parkin signalling is dispensable for basal mitophagy, but may also play a role in stress-induced mitophagy [126,127]. Independent of mitophagy, other studies have delineated the mechanics of how matrix-localised mtDNA can activate cGAS-STING signalling [163], and have also linked cGAS-STING signalling to macroautophagy [164].
It is important to note that PINK1 and Parkin have also been implicated in the formation of ‘mitochondrial-derived vesicles’ (MDVs), a discrete form of quality control, whereby defective mitochondrial components or cargo-specific regions undergo a process of (1) budding (2) excision from mitochondria and (3) delivery to the endolysosomal system for elimination (reviewed in [165]). Although mitophagy and the MDV pathway ultimately have the same terminal destination (the lysosome), the cargo-specific nature of the MDV pathway represents a more targeted variant of quality control. It will be exciting to unearth the spatial regulation of MDVs in vivo, as has been done for mitophagy and macroautophagy. The PD-associated genes VPS35, PINK1 and PARKIN have been linked to the generation of particular MDV subsets.
These exciting links between mitochondrial integrity, immunity and PD are also consistent with pioneering collaborative research from the laboratories of Heidi McBride and Michel Desjardins, who discovered the phenomenon of mitochondrial antigen presentation (MitAP) in 2016 [166]. In the absence of PINK1-Parkin signalling, a pro-inflammatory stimulus (via lipopolysaccharide treatment) promoted the formation of MDVs and mitochondrial antigens on MHC-I molecules at the surface of antigen-presenting cells [161]. Although these findings suggested an autoimmune component to PD, it remained unclear how to reconcile this with both mitochondrial and nigrostriatal integrity, the latter of which are largely unperturbed in most PINK1/Parkin mutant mice.
Building on their previous discovery of MitAP and independent converging studies that demonstrate PINK1 is dispensable for basal mitophagy in vivo, a recent landmark study from Matheoud and colleagues [167] clarifies the contribution of both MitAP and PINK1-Parkin signalling to striatal integrity in vivo. By subjecting Pink1 KO mice to intestinal infection, they observed the initiation of MitAP and the induction of cytotoxic cellular subset: mitochondrial-specific CD8+ T cells [87]. Strikingly, this was accompanied by PD-relevant phenotypes (loss of striatal arborisation and L-DOPA responsive motor dysfunction). Although it remains unclear if these cytotoxic T cells directly drive the degeneration of dopaminergic neurons, these findings both extend our understanding of MitAP and the mitophagy-independent function of PINK1-Parkin signalling in the modulation of adaptive immunity. This paradigm is particularly noteworthy in pre-clinical terms, as mitochondrial stress was not required for the onset of neurological phenoytpes in Pink1 KO mice. The collective resolution of these experimental stress paradigms (infection, extreme stress) to the onset of human neurodegeneration constitutes an exciting area of future research.
It will also be crucial to expand upon these emergent mitophagy-independent functions of PINK1-Parkin signalling, and clarify their cell-specific roles in vivo [34,147]. This line of enquiry will be particularly important, as the manifestation of neuropathology in PINK1/Parkin PD patients is distinct from idiopathic PD cases. As other EOPD loci are known to influence cellular trafficking and autophagy (e.g. LRRK2), it will be exciting to decipher the interplay of these additional PD-related factors to mitochondrial quality control in vivo.
Could mitophagy therapeutics halt neurodegeneration?
Mitochondrial signalling has emerged as an attractive drug development area [168], and the search for small molecule compounds that could modulate mitophagy in tissues is an important domain of research. In a small-scale human trial, the metabolite Urolithin A (UA) was well tolerated and exerted beneficial effects on mitochondrial metabolism [169]. Although UA modulates mitophagy in Caenorhabditis elegans and influences mitochondrial function in rodent models [170], it remains to be determined how UA administration influences mitophagy signalling in human tissues at steady-state levels.
Because no disease-modifying therapies exist to halt PD progression, the PINK1-Parkin pathway has attracted considerable attention as a prospective translational target for PD [140,171]. Recent advances in understanding the atomic structure of PINK1 will surely provide a much-needed blueprint for these efforts [140,144,172]. Although targeting this particular pathway may indeed provide a meaningful benefit for a subset of PD patients, much pre-clinical work will be required to relate these effects to nigrostriatal mitophagy in vivo. The continual refinement of new tools to monitor selective autophagy in tissues will surely help to facilitate this [40,125].
Our perspective is that mitophagy-related drug discovery efforts should not hinge on a single signalling pathway. Thus, any anti-climactic or adverse outcomes from any PINK1/Parkin-associated therapeutic strategy should not be conflated with a therapeutic setback for mitophagy. Exciting new data by Matheoud and colleagues demonstrate the mitophagy-independent, physiologically relevant consequences of Pink1 ablation should merit serious consideration for those interested in targeting this pathway for PD.
Our evidence-based interpretation of the available data is that different physiological contexts necessitate different modalities and mechanisms of mitochondrial turnover [34,114,147]. Depending on when, where and how mitochondrial networks become compromised, our cells have devised more than a single mechanism to destroy imperfect powerhouses. This perspective does not conflict with previous findings but instead seeks to integrate our current understanding of mitophagy into a constructive framework that can drive meaningful translational benefit. It is widely accepted that proliferating cancer cells employ myriad molecular strategies to assert their pathological supremacy [173]. Are the elusive degenerative mechanisms of the mammalian nervous system likely to be any less complex?
Our concerns stem from the sobering state of drug discovery in neurodegenerative disease. A 2018 study reported a 99.6% failure rate for AD drug development with a further significant setback announced this year [174,175]. For neurodegeneration to be ‘the next oncology’ [176] and for PD to escape the same fate of AD, a different strategy will be required. We predict that rigorous efforts to understand the interplay of autophagy signalling pathways and their contribution to neural integrity in vivo will be vital in this area. Our perspective is consistent with established leaders calling attention to the multifaceted nature of neurodegenerative pathology [177].
Pexophagy
Pexophagy refers to the selective elimination of defective or damaged peroxisomes by autophagy and is receiving increased attention in the field. Peroxisomes are single-membrane bound organelles that fulfil vital metabolic functions in lipid, polyamine and amino acid catabolism, lipid biosynthesis, glyoxylate detoxification and ROS/NOS signalling [178]. Mitochondrial and peroxisomal biology are more intertwined than previously realised, illustrated by the striking discovery that mitochondria can generate newly born peroxisomes [179]. Defective peroxisomes are detrimental to nervous system function, illustrated by the many neurological phenotypes with disease mutations in peroxisome-associated genes. For example, the peroxisome biogenesis disorder (PBD) Zellweger syndrome (ZS) is a neurodevelopmental disease [180] characterised by the absence of peroxisomes, profound metabolic deficiencies, in addition to hypomyelination and sensineuronal degeneration. For a comprehensive review on peroxisomal function in health and disease, please consult Cho and colleagues [181].
Interestingly, the PBD-associated genes PEX2 [182] and PEX3 [183] encode mammalian pexophagy receptors. Mutant PEX2 is linked with Infertile Refsum disease (IRD) in humans [182]. Patients with IRD exhibit severe cognitive and behavioural impairments as well as subtle craniofacial abnormalities and retinitis pigmentosa.
Studies in cultured human cells have demonstrated the importance of PEX2 in modulating mammalian pexophagy [100]. In addition to pexophagy, PEX2 appears necessary for both starvation-induced and constitutive macroautophagy [100]. Genetic ablation of Pex2 in mice results in a dramatic loss of neuronal arborisation in cerebellar Purkinje neurons in vivo [184]. Studies have also linked PEX3 to peroxisome-ubiquitination-linked pexophagy, and NBR1 appears essential for PEX3-dependent pexophagy in cultured HeLa cells [99]. Homozygous mutations in PEX3 are also associated with severe cases of ZS [183]. The PEX5 peroxisome import receptor is reported to interact with mutated in ataxia telangictasia (ATM), a DNA-damage response-associated kinase, and activated ATM directs ULK1 to induce pexophagy [101]. ATM kinase plays an essential role in cellular homoeostasis, and mutations in ATM mutations cause neurodegeneration [185].
The Acyl-CoA binding domain containing protein5 (ACBD5) is a human orthologue of Atg35 and is also associated with pexophagy [186,102]. A pathogenic patient mutation in ACBD5 is associated with impaired fatty acid metabolism with retinal dystrophy and white matter disease [102].
Recently, the autophagy receptor NBR1 was discovered to be a pexophagy receptor [97]. Loss of endogenous NBR1 triggers an increase in catalase levels, and NBR1 overexpression targets peroxisomes to lysosomes, demonstrating a role for this protein in promoting constitutive pexophagy [97]. Interestingly, NBR1 is regarded to influence the formation of proteinaceous inclusions associated with neuropathology in PD, dementia and multiple system atrophy [187].
As previously mentioned, USP30 is a DUB that modulates Parkin-dependent mitochondrial clearance in cultured cells [188], yet its contribution to mitophagy in vivo remains to be determined. In a surprising twist, an elegant study from the Clague and Urbé laboratories recently demonstrated that USP30 associates with peroxisomes and modulates basal pexophagy [154,189].
Ribophagy
Research in yeast and plants has provided crucial insights into the selective autophagy of ribosomes, and the first demonstration of mammalian ribophagy was made recently in cultured cells using a Ribo-Keima flux reporter [190]. Like most autophagy pathways in cultured cells, basal ribophagic flux occurs at extremely low levels (<1% of ribosomes over a 24-h period). This increases when classical macroautophagy stimuli are applied, i.e. nutrient deprivation and mTOR inhibition promote VPS34-dependent ribophagic flux (∼10% of ribosomes over a 24-h period in human cells). Proteostatic stress using AS (arsenite) was also found to promote ATG5-dependent ribophagic flux.
Recently, an exciting study identified Nuclear fragile X-mental retardation-interacting protein 1 (NUFIP1) as a putative receptor for starvation-induced ribophagy [103]. NUFIP1 interacts with ribosomes and binds LC3B, promoting their autophagic delivery. Conversely, mammalian cells expressing mutant NUFIP1 (that cannot bind LC3B) exhibit impaired ribophagy.
The quality control of ribosomes is known to be maintained by a surveillance mechanism called ribosome-associated protein quality control (ROC) [191], and involves the E3 ubiquitin ligase Ltn1 Mutations in Ltn1 is reported to cause neurodegeneration in mice [191,192]. Given the spatial nature of translation in neurons [193], it will be certainly interesting to ascertain the role of ribophagy in sustaining neural integrity.
Aggrephagy
Aggrephagy refers to the selective elimination of proteinaceous aggregates, i.e. aberrantly folded ubiquitin-positive inclusions. Although neuronal protein aggregation occurs during healthy ageing, excessive accumulation is a hallmark of neurodegenerative pathology, including Lewy bodies in idiopathic PD, neurofibrillary τ tangles and amyloid plaques in AD, polyglutamine aggregates in Huntington’s disease [59,194] and ubiquitin/p62 positive inclusions in ALS. Several aggrephagy receptors have been identified (Table 3), and enhancing levels of aggrephagy has evolved as an attractive neuroprotective strategy with therapeutic promise. Interestingly, mutations in many aggrephagy receptors result in neurodegeneration: mutations in ALFY are associated with human microcephaly [59,194], OPTN1 for ALS and primary open angle glaucoma [195], in addition to p62 (SQSTM1) mutations which are associated with ALS, frontotemporal dementia (FTD) and distal myopathy [196,197].
Concluding remarks
Macroautophagy and selective autophagy are fundamental quality control mechanisms with demonstrable relevance for neural integrity. Their contribution to cell type-specific homoeostasis is only beginning to be elucidated, especially in the mammalian nervous system.
It is clear from the classical and contemporary data that a symphony of basal and stress-evoked pathways orchestrate cellular integrity. Further research will be vital to uncover how these pathways become activated in vivo and how they converge to sustain neural integrity during a lifetime of varying physiological contexts.
Acknowledgements
Due to space constraints, we were unable to provide a comprehensive citation of all the relevant primary literature. We apologise to those whom we have omitted. We thank members of the TGM lab for critical reading of the manuscript.
Abbreviations
- ACBD5
Acyl-CoA binding domain containing protein5
- AD
Alzheimer’s disease
- ALS
amyotrophic lateral sclerosis
- ATM
mutated in ataxia telangictasia
- DUB
deubiquitylase
- EM
electron microscopy
- EOPD
early-onset Parkinson’s disease
- ER
endoplasmic reticulum
- IRD
infertile Refsum disease
- MDV
mitochondrial-derived vesicle
- MitAP
mitochondrial antigen presentation
- mTOR
mammalian target of rapamycin
- NBIA
Neurodegeneration with Brain Iron Accumulation
- NUFIP1
nuclear fragile X-mental retardation-interacting protein 1
- PBD
peroxisome biogenesis disorder
- PD
Parkinson’s disease
- PE
phosphatidylethanolamine
- PI3P
phosphatidylinositol-3-phosphate
- SENDA
static encephalopathy of childhood with neurodegeneration in adulthood
- UA
Urolithin A
- ZS
Zellweger syndrome
- KO
Knockout (genetic ablation)
- ATG
Autophagy-related
- ULK1
Unc-51 like autophagy activating kinase 1 (Yeast Atg1)
- LC3
Microtubule-associated protein light chain 3
- CNS
Central Nervous System
Funding
This work was supported by the Academy of Finland; the Novo Nordisk Foundation; the Sigrid Jusélius Foundation; Sydäntutkimussäätiö and the University of Helsinki.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Mortimore G.E. and Pösö A.R. (1987) Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 7, 539–564 10.1146/annurev.nu.07.070187.002543 [DOI] [PubMed] [Google Scholar]
- 2.Meijer A.J. and Codogno P. (2004) Regulation and role of autophagy in mammalian cells. Int. J. Biochem. Cell Biol. 36, 2445–2462 10.1016/j.biocel.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 3.Xie Z. and Klionsky D.J. (2007) Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102EP–1109EP 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- 4.Ktistakis N.T. (2017) In praise of M. Anselmier who first used the term “autophagie” in 1859. Autophagy 13, 2015–2017 10.1080/15548627.2017.1367473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dice J.F. (1990) Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 15, 305–309 10.1016/0968-0004(90)90019-8 [DOI] [PubMed] [Google Scholar]
- 6.Itakura E. and Mizushima N. (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 10.4161/auto.6.6.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young A.R.J. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- 8.Chan E.Y.W., Kir S. and Tooze S.A. (2007) siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. [DOI] [PubMed] [Google Scholar]
- 9.Ganley I.G., Lam D.H., Wang J., Ding X., Chen S. and Jiang X. (2009) ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 10.1074/jbc.M900573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petherick K.J., Conway O.J.L., Mpamhanga C., Osborne S.A., Kamal A., Saxty B. et al. (2015) Erratum: Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290, 11376–11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan D.F., Chun M.G.H., Vamos M., Zou H., Rong J., Miller C.J. et al. (2015) Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 10.1016/j.molcel.2015.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell R.C., Tian Y., Yuan H., Park H.W., Chang Y.Y., Kim J. et al. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zachari M. and Ganley I.G. (2017) The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 61, 585–596 10.1042/EBC20170021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan E.Y.W., Kir S. and Tooze S.A. (2007) siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282, 25464–25474 10.1074/jbc.M703663200 [DOI] [PubMed] [Google Scholar]
- 15.Menzies F.M., Fleming A., Caricasole A., Bento C.F., Andrews S.P., Ashkenazi A. et al. (2017) Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034 10.1016/j.neuron.2017.01.022 [DOI] [PubMed] [Google Scholar]
- 16.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y. et al. (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 10.1091/mbc.e08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung C.H., Jun C.B., Ro S.-H., Kim Y.-M., Otto N.M., Cao J. et al. (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 10.1091/mbc.e08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livi G.P. (2019) Halcyon days of TOR: reflections on the multiple independent discovery of the yeast and mammalian TOR proteins. Gene 10.1016/j.gene.2018.12.046 [DOI] [PubMed] [Google Scholar]
- 19.Zhou X., Babu J.R., da Silva S., Shu Q., Graef I.A., Oliver T. et al. (2007) Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc. Natl. Acad. Sci. U.S.A. 104, 5842–5847 10.1073/pnas.0701402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang B., Iyengar R., Li-Harms X., Joo J.H., Wright C., Lavado A. et al. (2018) The autophagy-inducing kinases, ULK1 and ULK2, regulate axon guidance in the developing mouse forebrain via a noncanonical pathway. Autophagy 10.1080/15548627.2017.1386820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A. et al. (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T. and Yamamoto A. (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- 23.Young A.R.J., Chan E.Y.W., Hu X.W., Köchl R., Crawshaw S.G., High S. et al. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- 24.Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K. et al. (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puri C., Renna M., Bento C.F., Moreau K. and Rubinsztein D.C. (2013) Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154, 1285–1299 10.1016/j.cell.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb C.A., Yoshimori T. and Tooze S.A. (2013) The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N. (2018) A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20, 521–527 10.1038/s41556-018-0092-5 [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N. and Yoshimori T. (2007) How to interpret LC3 immunoblotting. Autophagy 542 [DOI] [PubMed] [Google Scholar]
- 29.Galluzzi L., Baehrecke E.H., Ballabio A., Boya P., Bravo-San Pedro J.M., Cecconi F. et al. (2017) Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine B. and Kroemer G. (2019) Biological functions of autophagy genes: a disease perspective. Cell 176, 11–42 10.1016/j.cell.2018.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto A. and Yue Z. (2014) Autophagy and its normal and pathogenic states in the brain. Annu. Rev. Neurosci. 37, 55–78 10.1146/annurev-neuro-071013-014149 [DOI] [PubMed] [Google Scholar]
- 32.Mizushima N., Yoshimori T. and Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 33.Rogov V., Dötsch V., Johansen T. and Kirkin V. (2014) Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167–178 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 34.Palikaras K., Lionaki E. and Tavernarakis N. (2018) Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013–1022 10.1038/s41556-018-0176-2 [DOI] [PubMed] [Google Scholar]
- 35.Okamoto K. (2014) Organellophagy: eliminating cellular building blocks via selective autophagy. J. Cell Biol. 205, 435–445 10.1083/jcb.201402054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshii S.R. and Mizushima N. (2017) Monitoring and measuring autophagy. Int. J. Mol. Sci. 18, 1865 10.3390/ijms18091865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novikoff A.B. and Shin W.Y. (1978) Endoplasmic reticulum and autophagy in rat hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 75, 5039–5042 10.1073/pnas.75.10.5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McWilliams T.G. and Ganley I.G. (2019) Investigating Mitophagy and mitochondrial morphology in vivo using mito-QC: a comprehensive guide. Methods Mol. Biol. 1880, 621–642 10.1007/978-1-4939-8873-0_41 [DOI] [PubMed] [Google Scholar]
- 39.Mizushima N., Yamamoto A., Matsui M., Yoshimori T. and Ohsumi Y. (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 10.1091/mbc.e03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuma A., Komatsu M. and Mizushima N. (2017) Autophagy-monitoring and autophagy-deficient mice. Autophagy 13, 1619–1628 10.1080/15548627.2017.1343770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alirezaei M., Kemball C.C., Flynn C.T., Wood M.R., Whitton J.L. and Kiosses W.B. (2010) Short-term fasting induces profound neuronal autophagy. Autophagy 6, 702–710 10.4161/auto.6.6.12376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaushik S., Rodriguez-Navarro J.A., Arias E., Kiffin R., Sahu S., Schwartz G.J. et al. (2011) Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 14, 173–183 10.1016/j.cmet.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo-Arozena A. et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.-G., Satoh T. et al. (2008) Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- 45.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R. et al. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- 46.Komatsu M., Wang Q.J., Holstein G.R., Friedrich V.L., Iwata J., Kominami E. et al. (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. U.S.A. 104, 14489–14494 10.1073/pnas.0701311104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levine B. and Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scrivo A., Bourdenx M., Pampliega O. and Cuervo A.M. (2018) Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol. 17, 802–815 10.1016/S1474-4422(18)30238-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T. et al. (2004) The role of autophagy during the early neonatal starvation period. Nature 432, 1032EP–1036EP 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- 50.Yoshii S.R., Kuma A., Akashi T., Hara T., Yamamoto A., Kurikawa Y. et al. (2016) Systemic analysis of Atg5-Null mice rescued from neonatal lethality by transgenic ATG5 expression in neurons. Dev. Cell 39, 116–130 10.1016/j.devcel.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 51.Kimmelman A.C. and White E. (2017) Autophagy and tumor metabolism. Cell Metab. 25, 1037–1043 10.1016/j.cmet.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo J.Y. and White E. (2013) Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy 9, 1636–1638 10.4161/auto.26123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karsli-Uzunbas G., Guo J.Y., Price S., Teng X., Laddha S.V., Khor S. et al. (2014) Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 10.1158/2159-8290.CD-14-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glatigny M., Moriceau S., Rivagorda M., Ramos-Brossier M., Nascimbeni A.C., Lante F. et al. (2019) Autophagy is required for memory formation and reverses age-related memory decline. Curr. Biol. 10.1016/j.cub.2018.12.021 [DOI] [PubMed] [Google Scholar]
- 55.Yan J., Porch M.W., Court-Vazquez B., Bennett M.V.L. and Zukin R.S. (2018) Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.1808247115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joo J.H., Wang B., Frankel E., Ge L., Xu L., Iyengar R. et al. (2016) The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol. Cell 10.1016/j.molcel.2016.05.030 [DOI] [PubMed] [Google Scholar]
- 57.Liang C.-C.C., Wang C., Peng X., Gan B. and Guan J.-L.L. (2010) Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 285, 3499–3509 10.1074/jbc.M109.072389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo T., Nan Z., Miao C., Jin X., Yang W., Wang Z. et al. (2019) The autophagy-related gene Atg101 in Drosophila regulates both neuron and midgut homeostasis. J. Biol. Chem. 10.1074/jbc.RA118.006069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menzies F.M., Fleming A. and Rubinsztein D.C. (2015) Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 16, 345–357 10.1038/nrn3961 [DOI] [PubMed] [Google Scholar]
- 60.Komatsu M., Waguri S., Koike M., Sou Y.-S., Ueno T., Hara T. et al. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- 61.Galluzzi L., Bravo-San Pedro J.M., Levine B., Green D.R. and Kroemer G. (2017) Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487–511 10.1038/nrd.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sou Y.-S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T. et al. (2008) The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell 19, 4762–4775 10.1091/mbc.e08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I. et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- 64.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S. et al. (2011) Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25, 795–800 10.1101/gad.2016211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malhotra R., Warne J.P., Salas E., Xu A.W. and Debnath J. (2015) Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy 11, 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I. et al. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H. et al. (2009) Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 106, 20842–20846 10.1073/pnas.0911267106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaizuka T. and Mizushima N. (2016) Atg13 is essential for autophagy and cardiac development in mice. Mol. Cell. Biol. 36, 585–595 10.1128/MCB.01005-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pickford F., Masliah E., Britschgi M., Lucin K., Narasimhan R., Jaeger P.A. et al. (2008) The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J. Clin. Invest. 118, 2190–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaeger P.A., Pickford F., Sun C.-H., Lucin K.M., Masliah E. and Wyss-Coray T. (2010) Regulation of amyloid precursor protein processing by the Beclin 1 complex. PLoS ONE 5, e11102 10.1371/journal.pone.0011102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yue Z., Jin S., Yang C., Levine A.J. and Heintz N. (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 100, 15077–15082 10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qu X. (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Sullivan G.A., Kneussel M., Elazar Z. and Betz H. (2005) GABARAP is not essential for GABA receptor targeting to the synapse. Eur. J. Neurosci. 22, 2644–2648 10.1111/j.1460-9568.2005.04448.x [DOI] [PubMed] [Google Scholar]
- 74.Cann G.M., Guignabert C., Ying L., Deshpande N., Bekker J.M., Wang L. et al. (2008) Developmental expression of LC3alpha and beta: absence of fibronectin or autophagy phenotype in LC3beta knockout mice. Dev. Dyn. 237, 187–195 10.1002/dvdy.21392 [DOI] [PubMed] [Google Scholar]
- 75.Zhou X., Wang L., Hasegawa H., Amin P., Han B.-X., Kaneko S. et al. (2010) Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc. Natl. Acad. Sci. U.S.A. 107, 9424–9429 10.1073/pnas.0914725107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X., Takatoh J. and Wang F. (2011) The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS ONE 6, e16358 10.1371/journal.pone.0016358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kundu M., Lindsten T., Yang C.-Y., Wu J., Zhao F., Zhang J. et al. (2008) Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112, 1493–1502 10.1182/blood-2008-02-137398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheong H., Lindsten T., Wu J., Lu C. and Thompson C.B. (2011) Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl. Acad. Sci. U.S.A. 108, 11121–11126 10.1073/pnas.1107969108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao Y.G., Sun L., Miao G., Ji C., Zhao H., Sun H. et al. (2015) The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy 11, 881–890 10.1080/15548627.2015.1047127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu Q., Yang P., Huang X., Hu W., Guo B., Wu F. et al. (2011) The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell 21, 343–357 10.1016/j.devcel.2011.06.024 [DOI] [PubMed] [Google Scholar]
- 81.Haack T.B., Hogarth P., Kruer M.C., Gregory A., Wieland T., Schwarzmayr T. et al. (2012) Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 91, 1144–1149 10.1016/j.ajhg.2012.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kruer M.C., Boddaert N., Schneider S.A., Houlden H., Bhatia K.P., Gregory A. et al. (2012) Neuroimaging features of neurodegeneration with brain iron accumulation. AJNR Am. J. Neuroradiol. 33, 407–414 10.3174/ajnr.A2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saitsu H., Nishimura T., Muramatsu K., Kodera H., Kumada S., Sugai K. et al. (2013) De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 45, 445–449.e1, 10.1038/ng.2562 [DOI] [PubMed] [Google Scholar]
- 84.Morikawa M., Takano K., Motobayashi M., Shiba N., Kosho T., Nakazawa Y. et al. (2017) Clinical features of a female with WDR45 mutation complicated by infantile spasms: a case report and literature review. Brain Dev. 39, 804–807 10.1016/j.braindev.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 85.Kim M., Sandford E., Gatica D., Qiu Y., Liu X., Zheng Y. et al. (2016) Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife 5, 213 10.7554/eLife.12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khaminets A., Behl C. and Dikic I. (2016) Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6–16 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- 87.Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A. et al. (2009) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P. et al. (2012) Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185 10.1038/ncb2422 [DOI] [PubMed] [Google Scholar]
- 89.Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J. et al. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- 90.Lee J.-Y., Nagano Y., Taylor J.P., Lim K.L. and Yao T.-P. (2010) Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 189, 671–679 10.1083/jcb.201001039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richter B., Sliter D.A., Herhaus L., Stolz A., Wang C., Beli P. et al. (2016) Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. U.S.A. 113, 4039–4044 10.1073/pnas.1523926113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L. et al. (2015) The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Princely Abudu Y., Pankiv S., Mathai B.J., Håkon Lystad A., Bindesbøll C., Brenne H.B. et al. (2019) NIPSNAP1 and NIPSNAP2 act as “Eat Me” signals for mitophagy. Dev. Cell 10.1016/j.devcel.2019.03.013 [DOI] [PubMed] [Google Scholar]
- 94.Wei Y., Chiang W.-C., Sumpter R., Mishra P. and Levine B. (2017) Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168, 224–238.e10 10.1016/j.cell.2016.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bhujabal Z., Birgisdottir Å.B., Sjøttem E., Brenne H.B., Øvervatn A., Habisov S. et al. (2017) FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 18, 947–961 10.15252/embr.201643147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chu C.T., Ji J., Dagda R.K., Jiang J.F., Tyurina Y.Y., Kapralov A.A. et al. (2013) Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 10.1038/ncb2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deosaran E., Larsen K.B., Hua R., Sargent G., Wang Y., Kim S. et al. (2013) NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126, 939–952 10.1242/jcs.114819 [DOI] [PubMed] [Google Scholar]
- 98.Kim P.K., Hailey D.W., Mullen R.T. and Lippincott-Schwartz J. (2008) Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U.S.A. 105, 20567–20574 10.1073/pnas.0810611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamashita S., Abe K., Tatemichi Y. and Fujiki Y. (2014) The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 10, 1549–1564 10.4161/auto.29329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sargent G., van Zutphen T., Shatseva T., Zhang L., Di Giovanni V., Bandsma R. et al. (2016) PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J. Cell Biol. 214, 677–690 10.1083/jcb.201511034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T. et al. (2015) ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 17, 1259–1269 10.1038/ncb3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferdinandusse S., Falkenberg K.D., Koster J., Mooyer P.A., Jones R., van Roermund C.W.T. et al. (2017) ACBD5 deficiency causes a defect in peroxisomal very long-chain fatty acid metabolism. J. Med. Genet. 54, 330–337 10.1136/jmedgenet-2016-104132 [DOI] [PubMed] [Google Scholar]
- 103.Wyant G.A., Abu-Remaileh M., Frenkel E.M., Laqtom N.N., Dharamdasani V., Lewis C.A. et al. (2018) NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758 10.1126/science.aar2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M. et al. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seok S., Fu T., Choi S.-E., Li Y., Zhu R., Kumar S. et al. (2014) Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 516, 108–111 10.1038/nature13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Korac J., Schaeffer V., Kovacevic I., Clement A.M., Jungblut B., Behl C. et al. (2013) Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 126, 580–592 10.1242/jcs.114926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Filimonenko M., Isakson P., Finley K.D., Anderson M., Jeong H., Melia T.J. et al. (2010) The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell 38, 265–279 10.1016/j.molcel.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clausen T.H., Lamark T., Isakson P., Finley K., Larsen K.B., Brech A. et al. (2010) p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330–344 10.4161/auto.6.3.11226 [DOI] [PubMed] [Google Scholar]
- 109.Lee J.-Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y.-S. et al. (2010) HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 29, 969–980 10.1038/emboj.2009.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gamerdinger M., Kaya A.M., Wolfrum U., Clement A.M. and Behl C. (2011) BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 12, 149–156 10.1038/embor.2010.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Newman A.C., Scholefield C.L., Kemp A.J., Newman M., McIver E.G., Kamal A. et al. (2012) TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS ONE 7, e50672 10.1371/journal.pone.0050672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nunnari J. and Suomalainen A. (2012) Mitochondria: in sickness and in health. Cell 148, 1145–1159 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Suomalainen A. and Battersby B.J. (2017) Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol. 491, 374. [DOI] [PubMed] [Google Scholar]
- 114.Rodger C.E., McWilliams T.G. and Ganley I.G. (2017) Mammalian mitophagy - from in vitro molecules to in vivo models. FEBS J. 33, 2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McWilliams T.G. and Muqit M.M. (2017) PINK1 and Parkin: emerging themes in mitochondrial homeostasis. Curr. Opin. Cell Biol. 45, 83–91 10.1016/j.ceb.2017.03.013 [DOI] [PubMed] [Google Scholar]
- 116.Lewis M.R. and Lewis W.H. (1915) Mitochondria (and other cytoplasmic structures) in tissue cultures. Am. J. Anat. 17, 339–401 10.1002/aja.1000170304 [DOI] [Google Scholar]
- 117.Susman W. (1928) Insulin and the diabetic pancreas. Edinb. Med. J. 35, 206 [Google Scholar]
- 118.Clark S.L. (1957) Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 3, 349–362 10.1083/jcb.3.3.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Novikoff A.B. and Essner E. (1962) Cytolysomes and mitochondrial degeneration. J. Cell Biol. 15, 140–146 10.1083/jcb.15.1.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Novikoff A.B., Beaufay H. and De Duve C. (1956) Electron microscopy of lysosomerich fractions from rat liver. J. Biophys. Biochem. Cytol. 2, 179–184 10.1083/jcb.2.4.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun N., Yun J., Liu J., Malide D., Liu C., Rovira I.I. et al. (2015) Measuring in vivo mitophagy. Mol. Cell. 60, 685–696 10.1016/j.molcel.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McWilliams T.G., Prescott A.R., Allen G.F.G., Tamjar J., Munson M.J., Thomson C. et al. (2016) mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333–345 10.1083/jcb.201603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McWilliams T.G., Barini E., Pohjolan-Pirhonen R., Brooks S.P., Singh F., Burel S. et al. (2018) Phosphorylation of Parkin at serine 65 is essential for its activation in vivo. Open Biol. 8, 10.1098/rsob.180108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McWilliams T.G., Prescott A.R., Montava-Garriga L., Ball G., Singh F., Barini E. et al. (2018) Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27, 439–449.e5 10.1016/j.cmet.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McWilliams T.G., Prescott A.R., Villarejo-Zori B., Ball G., Boya P. and Ganley I.G. (2019) A comparative map of macroautophagy and mitophagy in the vertebrate eye. Autophagy 15, 1296–1308 10.1080/15548627.2019.1580509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee J.J., Sanchez-Martinez A., Zarate A.M., Beninca C., Mayor U., Clague M.J. et al. (2018) Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J. Cell Biol. 27, jcb.201801044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim Y.Y., Um J.-H., Yoon J.-H., Kim H., Lee D.-Y., Lee Y.J. et al. (2019) Assessment of mitophagy in mt-Keima Drosophila revealed an essential role of the PINK1-Parkin pathway in mitophagy induction in vivo. FASEB J. 10.1096/fj.201900073R [DOI] [PubMed] [Google Scholar]
- 128.Esteban-Martínez L., Sierra-Filardi E., McGreal R.S., Salazar-Roa M., Mariño G., Seco E. et al. (2017) Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 36, 1688–1706 10.15252/embj.201695916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prescott A.R., Montava-Garriga L., Ball G., Singh F., Barini E., Muqit M.M.K. et al. (2018) Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27, 439–449.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Davis C.O., Kim K.-Y., Bushong E.A., Mills E.A., Boassa D., Shih T. et al. (2014) Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 111, 9633–9638 10.1073/pnas.1404651111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hayakawa K., Esposito E., Wang X., Terasaki Y., Liu Y., Xing C. et al. (2016) Transfer of mitochondria from astrocytes to neurons after stroke. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tomonaga M. and Tanabe H. (1973) Electron microscopic study of skeletal muscle changes in Parkinson diseases. Rinsho Shinkeigaku 13, 111–119 [PubMed] [Google Scholar]
- 133.Ahlqvist G., Landin S. and Wroblewski R. (1975) Ultrastructure of skeletal muscle in patients with Parkinson’s disease and upper motor lesions. Lab. Invest. [PubMed] [Google Scholar]
- 134.Langston J.W., Ballard P., Tetrud J.W. and Irwin I. (1983) Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 219, 979–980 10.1126/science.6823561 [DOI] [PubMed] [Google Scholar]
- 135.Schapira A.H., Gu M., Taanman J.W., Tabrizi S.J., Seaton T., Cleeter M. et al. (1998) Mitochondria in the etiology and pathogenesis of Parkinson’s disease. Ann. Neurol. 44, S89–98 10.1002/ana.410440714 [DOI] [PubMed] [Google Scholar]
- 136.Palin E.J.H., Paetau A. and Suomalainen A. (2013) Mesencephalic complex I deficiency does not correlate with parkinsonism in mitochondrial DNA maintenance disorders. Brain 10.1093/brain/awt160 [DOI] [PubMed] [Google Scholar]
- 137.Luoma P., Melberg A., Rinne J.O., Kaukonen J.A., Nupponen N.N., Chalmers R.M. et al. (2004) Parkinsonism, premature menopause, and mitochondrial DNA polymerase γ mutations: clinical and molecular genetic study. Lancet 10.1016/S0140-6736(04)16983-3 [DOI] [PubMed] [Google Scholar]
- 138.Whitworth A.J. and Pallanck L.J. (2017) PINK1/Parkin mitophagy and neurodegeneration—what do we really know in vivo? Curr. Opin. Genet. Dev. 44, 47–53 10.1016/j.gde.2017.01.016 [DOI] [PubMed] [Google Scholar]
- 139.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M.K., Harvey K., Gispert S. et al. (2004) Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- 140.Bayne A.N. and Trempe J.-F. (2019) Mechanisms of PINK1, ubiquitin and Parkin interactions in mitochondrial quality control and beyond. Cell. Mol. Life Sci. 10.1007/s00018-019-03203-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Neuspiel M., Schauss A.C., Braschi E., Zunino R., Rippstein P., Rachubinski R.A. et al. (2008) Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 18, 102–108 10.1016/j.cub.2007.12.038 [DOI] [PubMed] [Google Scholar]
- 142.Li W., Bengtson M.H., Ulbrich A., Matsuda A., Reddy V.A., Orth A. et al. (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yun J., Puri R., Yang H., Lizzio M.A., Wu C., Sheng Z.-H. et al. (2014) MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife 10.7554/eLife.01958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Komander D., Clague M.J. and Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- 145.Cornelissen T., Haddad D., Wauters F., Van Humbeeck C., Mandemakers W., Koentjoro B. et al. (2014) The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum. Mol. Genet. 23, 5227–5242 10.1093/hmg/ddu244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Montava-Garriga L. and Ganley I.G. (2019) Outstanding questions in mitophagy: what we do and do not know. J. Mol. Biol. 10.1016/j.jmb.2019.06.032 [DOI] [PubMed] [Google Scholar]
- 147.Jang J.Y., Blum A., Liu J. and Finkel T. (2018) The role of mitochondria in aging. J. Clin. Invest. 128, 3662–3670 10.1172/JCI120842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Baumann K. (2018) Mitochondria: the needless PINK1. Nat. Rev. Mol. Cell Biol. 10.1038/nrm.2018.4 [DOI] [PubMed] [Google Scholar]
- 149.Soutar M.P.M., Kempthorne L., Annuario E., Luft C., Wray S., Ketteler R. et al. (2019) FBS/BSA media concentration determines CCCP’s ability to depolarize mitochondria and activate PINK1-PRKN mitophagy. Autophagy 10.1080/15548627.2019.1603549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Perez F.A. and Palmiter R.D. (2005) Parkin-deficient mice are not a robust model of parkinsonism. Proc. Natl. Acad. Sci. U.S.A. 102, 2174–2179 10.1073/pnas.0409598102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pickrell A.M. and Youle R.J. (2015) The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sliter D.A., Martinez J., Hao L., Chen X., Sun N., Fischer T.D. et al. (2018) Parkin and PINK1 mitigate STING-induced inflammation. Nature 302, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Walsh T.G., van den Bosch M.T.J., Lewis K.E., Williams C.M. and Poole A.W. (2018) Loss of the mitochondrial kinase PINK1 does not alter platelet function. Sci. Rep. 8, 13843 10.1038/s41598-018-32716-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marcassa E., Kallinos A., Jardine J., Rusilowicz-Jones E.V, Martinez A., Kuehl S. et al. (2018) Dual role of USP30 in controlling basal pexophagy and mitophagy. EMBO Rep. 19, e45595 10.15252/embr.201745595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yamada T., Murata D., Adachi Y., Itoh K., Kameoka S., Igarashi A. et al. (2018) Mitochondrial stasis reveals p62-mediated ubiquitination in Parkin-independent mitophagy and mitigates nonalcoholic fatty liver disease. Cell Metab. 0, 588–604.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu J., Wang X., Lu Y., Duan C., Gao G., Lu L. et al. (2017) Pink1 interacts with α-synuclein and abrogates α-synuclein-induced neurotoxicity by activating autophagy. Cell Death Dis. 10.1038/cddis.2017.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Creed R.B. and Goldberg M.S. (2019) Analysis of α-synuclein pathology in PINK1 knockout rat brains. Front. Neurosci. 10.3389/fnins.2018.01034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kelm-Nelson C.A., Trevino M.A. and Ciucci M.R. (2018) Quantitative analysis of catecholamines in the Pink1−/− rat model of early-onset Parkinson’s disease. Neuroscience 10.1016/j.neuroscience.2018.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shahmoradian S.H., Lewis A.J., Genoud C., Hench J., Moors T.E., Navarro P.P. et al. (2019) Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 10.1038/s41593-019-0423-2 [DOI] [PubMed] [Google Scholar]
- 160.Truban D., Hou X., Caulfield T.R., Fiesel F.C. and Springer W. (2017) PINK1, Parkin, and mitochondrial quality control: what can we learn about Parkinson’s disease pathobiology? J. Parkinsons Dis. 10.3233/JPD-160989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fang E.F., Hou Y., Palikaras K., Adriaanse B.A., Kerr J.S., Yang B. et al. (2019) Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 25, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Logan A., Shabalina I.G., Prime T.A., Rogatti S., Kalinovich A.V., Hartley R.C. et al. (2014) In vivo levels of mitochondrial hydrogen peroxide increase with age in mtDNA mutator mice. Aging Cell 13, 765–768 10.1111/acel.12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Riley J.S., Quarato G., Cloix C., Lopez J., O’Prey J., Pearson M. et al. (2018) Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis. EMBO J. 10.15252/embj.201899238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nassour J., Radford R., Correia A., Fusté J.M., Schoell B., Jauch A. et al. (2019) Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 10.1038/s41586-019-0885-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sugiura A., McLelland G.-L., Fon E.A. and McBride H.M. (2014) A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 33, 2142–2156 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matheoud D., Sugiura A., Bellemare-Pelletier A., Laplante A., Rondeau C., Chemali M. et al. (2016) Parkinson’s disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell 166, 314–327 10.1016/j.cell.2016.05.039 [DOI] [PubMed] [Google Scholar]
- 167.Matheoud D., Cannon T., Voisin A., Penttinen A.-M., Ramet L., Fahmy A.M. et al. (2019) Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1-/- mice. Nature 571, 565–569 10.1038/s41586-019-1405-y [DOI] [PubMed] [Google Scholar]
- 168.Murphy M.P. and Hartley R.C. (2018) Mitochondria as a therapeutic target for common pathologies. Nat. Rev. Drug Discov. 10.1038/nrd.2018.174 [DOI] [PubMed] [Google Scholar]
- 169.Andreux P.A., Blanco-Bose W., Ryu D., Burdet F., Ibberson M., Aebischer P. et al. (2019) The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat. Metab. 10.1038/s42255-019-0073-4 [DOI] [PubMed] [Google Scholar]
- 170.Ryu D., Mouchiroud L., Andreux P.A., Katsyuba E., Moullan N., Nicolet-Dit-Félix A.A. et al. (2016) Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 22, 879–888 10.1038/nm.4132 [DOI] [PubMed] [Google Scholar]
- 171.Miller S. and Muqit M.M.K. (2019) Therapeutic approaches to enhance PINK1/Parkin mediated mitophagy for the treatment of Parkinson’s disease. Neurosci. Lett. 10.1016/j.neulet.2019.04.029 [DOI] [PubMed] [Google Scholar]
- 172.Kumar A., Tamjar J., Waddell A.D., Woodroof H.I., Raimi O.G., Shaw A.M. et al. (2017) Structure of PINK1 and mechanisms of Parkinson’s disease-associated mutations. Elife 6, 311 10.7554/eLife.29985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hanahan D. and Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 174.Selkoe D.J. (2019) Alzheimer disease and aducanumab: adjusting our approach. Nat. Rev. Neurol. 10.1038/s41582-019-0205-1 [DOI] [PubMed] [Google Scholar]
- 175.Cummings J., Reiber C. and Kumar P. (2018) The price of progress: funding and financing Alzheimer’s disease drug development. Alzheimers Dement. Transl. Res. Clin. Interv. 10.1016/j.trci.2018.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ehlers M.D. (2018) Neuroscience is the next oncology. Innov. Clin. Neurosci. 15, 15–16 [PMC free article] [PubMed] [Google Scholar]
- 177.Kara E., Marks J.D. and Aguzzi A. (2018) Toxic protein spread in neurodegeneration: reality versus fantasy. Trends Mol. Med. 10.1016/j.molmed.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 178.Wanders R.J.A., Waterham H.R. and Ferdinandusse S. (2015) Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Front. Cell Dev. Biol. 3, 83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sugiura A., Mattie S., Prudent J. and McBride H.M. (2017) Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542, 251–254 10.1038/nature21375 [DOI] [PubMed] [Google Scholar]
- 180.Zellweger H., Maertens P., Superneau D. and Wertelecki W. (1988) History of the cerebrohepatorenal syndrome of Zellweger and other peroxisomal disorders. South. Med. J. 81, 357–364 10.1097/00007611-198803000-00017 [DOI] [PubMed] [Google Scholar]
- 181.Cho D.-H., Kim Y.S., Jo D.S., Choe S.-K. and Jo E.-K. (2018) Pexophagy: molecular mechanisms and implications for health and diseases. Mol. Cells 41, 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shimozawa N., Imamura A., Zhang Z., Suzuki Y., Orii T., Tsukamoto T. et al. (1999) Defective PEX gene products correlate with the protein import, biochemical abnormalities, and phenotypic heterogeneity in peroxisome biogenesis disorders. J. Med. Genet. 36, 779–781 10.1136/jmg.36.10.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Muntau A.C., Mayerhofer P.U., Paton B.C., Kammerer S. and Roscher A.A. (2000) Defective peroxisome membrane synthesis due to mutations in human PEX3 causes Zellweger syndrome, complementation group G. Am. J. Hum. Genet. 67, 967–975 10.1086/303071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Faust P.L. (2003) Abnormal cerebellar histogenesis in PEX2 Zellweger mice reflects multiple neuronal defects induced by peroxisome deficiency. J. Comp. Neurol. 461, 394–413 10.1002/cne.10699 [DOI] [PubMed] [Google Scholar]
- 185.Choy K.R. and Watters D.J. (2018) Neurodegeneration in ataxia-telangiectasia: multiple roles of ATM kinase in cellular homeostasis. Dev. Dyn. 10.1002/dvdy.24522 [DOI] [PubMed] [Google Scholar]
- 186.Nazarko T.Y. (2014) Atg37 regulates the assembly of the pexophagic receptor protein complex. Autophagy 10, 1348–1349 10.4161/auto.29073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Odagiri S., Tanji K., Mori F., Kakita A., Takahashi H. and Wakabayashi K. (2012) Autophagic adapter protein NBR1 is localized in Lewy bodies and glial cytoplasmic inclusions and is involved in aggregate formation in α-synucleinopathy. Acta Neuropathol. 124, 173–186 10.1007/s00401-012-0975-7 [DOI] [PubMed] [Google Scholar]
- 188.Bingol B., Tea J.S., Phu L., Reichelt M., Bakalarski C.E., Song Q. et al. (2014) The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375 10.1038/nature13418 [DOI] [PubMed] [Google Scholar]
- 189.Ganley I.G. (2018) Organelle turnover: a USP30 safety catch restrains the trigger for mitophagy and pexophagy. Curr. Biol. 28, R842–R845 10.1016/j.cub.2018.06.067 [DOI] [PubMed] [Google Scholar]
- 190.An H. and Harper J.W. (2018) Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat. Cell Biol. 20, 135–143 10.1038/s41556-017-0007-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Joazeiro C. A.P. (2019) Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol. 10.1038/s41580-019-0118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chu J., Hong N.A., Masuda C.A., Jenkins B.V., Nelms K.A., Goodnow C.C. et al. (2009) A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cioni J.-M., Lin J.Q., Holtermann A.V, Koppers M., Jakobs M.A.H., Azizi A. et al. (2019) Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell 176, 56–72.e15 10.1016/j.cell.2018.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yamamoto A. and Simonsen A. (2011) The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol. Dis. 43, 17–28 10.1016/j.nbd.2010.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kadir R., Harel T., Markus B., Perez Y., Bakhrat A., Cohen I. et al. (2016) ALFY-controlled DVL3 autophagy regulates Wnt signaling, determining human brain size. PLoS Genet. 12, e1005919 10.1371/journal.pgen.1005919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wong Y.C. and Holzbaur E.L.F. (2014) Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U.S.A. 111, E4439–48 10.1073/pnas.1405752111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Li F., Xie X., Wang Y., Liu J., Cheng X., Guo Y. et al. (2016) Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins. Nat. Commun. 7, 12708 10.1038/ncomms12708 [DOI] [PMC free article] [PubMed] [Google Scholar]