Significance
Understanding the molecular mechanisms that maintain pancreas homeostasis in response to inflammatory stress is an urgent area of study due to the strong associations between pancreatitis and cancer and diabetes. Through the use of physiologically relevant mouse models, we identify an essential role for Men1 in the normal tissue response to both inflammatory and oncogenic stress. Loss of Men1 induces a proinflammatory gene expression program, associated with deregulation of Jund transcriptional activity. These results provide insights into a regulatory axis that maintains pancreas homeostasis in vivo.
Keywords: acute pancreatitis, Men1, Jund, MLL
Abstract
A more comprehensive understanding of the molecular mechanisms underlying pancreatic diseases, including pancreatitis and cancer, is essential to improve clinical management. MEN1 has established roles in epigenetic regulation and tumor suppression in the endocrine pancreas; however, intriguing recent data suggest MEN1 may also function in the exocrine pancreas. Using physiologically relevant genetic mouse models, we provide direct evidence that Men1 is essential for exocrine pancreas homeostasis in response to inflammation and oncogenic stress. Men1 loss causes increased injury and impaired regeneration following acute caerulein-induced pancreatitis, leading to more severe damage, loss of the normal acinar compartment, and increased cytokeratin 19-positive metaplasias and immune cell infiltration. We further demonstrate the Men1 protein is stabilized in response to insult, and loss of Men1 is associated with the overexpression of proinflammatory Jund target genes, suggesting that loss of Men1-mediated repression of Jund activity is, at least in part, responsible for the impaired response. Finally, we demonstrate that Men1 loss significantly accelerates mutant Kras-dependent oncogenesis. Combined, this work establishes Men1 as an important mediator of pancreas homeostasis in vivo.
Pancreas diseases represent a significant medical burden. The incidence of acute pancreatitis is increasing and is attributed to growing rates of obesity and gallstones, and is a significant cause of hospitalizations (1, 2). Chronic pancreatitis is associated with pancreatic ductal adenocarcinoma, which is one of the most deadly human cancers with a 5-y survival rate of only 9% (3). A more comprehensive view of the molecular mechanisms that maintain normal pancreas homeostasis is fundamental to understanding dysfunction and disease pathogenesis.
MEN1 (multiple endocrine neoplasia 1) is a well-established tumor suppressor in the endocrine pancreas. Rare germline mutations, generally nonsense or frameshift alterations, define the autosomal-dominant cancer predisposition syndrome, multiple endocrine neoplasia type 1 (MEN1) (4, 5). This syndrome is characterized by the presence of multiple tumors in the endocrine pancreas, parathyroid glands, and anterior pituitary, with additional tumor types occurring in some patients. Consistent with these germline observations, somatic MEN1 mutations have also been identified in sporadic human tumors, including pancreatic neuroendocrine tumors (PanNETs) and parathyroid tumors (6–8). Mouse models have further demonstrated that Men1 loss is sufficient to promote tumorigenesis. While homozygous deletion of Men1 results in embryo lethality at embryonic day (E)11.5 to 12.5, germline heterozygous mice faithfully mimic the human syndrome (9). Mice develop hyperplasias, and insulin-producing islet cell tumors as well as parathyroid adenomas as early as 9 mo of age, and tumors involving the thyroid, adrenal cortex, and pituitary by 16 mo of age. Conditional homozygous deletion of Men1 from the β-cells (RIP-Cre) (10), α-cells (Glu-Cre) (11, 12) or generally from all epithelial cells in the pancreas (Pdx1-Cre) (13) collectively results in islet cell hyperplasia and PanNET development, with the majority of tumors being functional insulinomas.
Mechanistically, MEN1 is part of a large macromolecular Set-domain histone methyltransferase complex, containing MLL2, ASH2L, RBBP5, and WDR5 (14). This complex mediates the methylation of histone 3 on lysine 4, and directly activates the expression of important cyclin-dependent kinase inhibitors CDKN1B (p27KIP1) and CDKN2C (p18INK4c) (15). Combined knockout of Cdkn1b and Cdkn2c in mice indeed results in a tumor spectrum similar to MEN1 syndrome with tumors developing in the pituitary, adrenal, thyroid, parathyroid, and pancreas, suggesting that loss of regulation of these genes is functionally contributing to tumorigenesis upon loss of Men1 (16). The other well-established transcriptional targets of MEN1/MLL are the clustered homeobox genes, Hoxa9, Hoxc6, and Hoxc8 (14, 17, 18), which are functionally important in the hematopoietic system. In this context MEN1 is oncogenic and is an essential cofactor in leukemias with translocations that produce MLL fusion proteins (19). The MEN1:MLL interaction is currently being pursued as a therapeutic target in these cancers. In addition to this epigenetic function, MEN1 interacts with other transcription factors to modulate their activity, although the functional significance of these interactions is not well characterized in vivo. Of note, MEN1 specifically associates with and inhibits the transcriptional activity of JUND, a member of the AP1 family (20). This association is specific to JUND and not other AP1 family members, including JUN, JUNB, and FOS.
While MEN1 clearly plays an essential tumor suppressor role in the endocrine pancreas, there are intriguing data suggesting that MEN1 may also play a functional role in the exocrine pancreas. A recent case report identified a concurrent PanNET and a pancreatic ductal adenocarcinoma (PDAC) in one patient with MEN1 syndrome (21). Moreover, decreased expression and promoter methylation of MEN1 have been reported in PDAC (22). We were therefore interested to directly evaluate the role of Men1 in the exocrine pancreas.
Using a physiologically relevant mouse model to genetically ablate Men1 expression throughout the pancreas, we demonstrate that Men1 plays an essential role in the normal homeostatic response to acute pancreatitis. We further demonstrate the Men1 protein is stabilized in response to stress and the altered response is not associated with the known MLL-dependent functions of Men1, but rather to its role as a negative regulator of the Jund transcription factor. Finally, we demonstrate that in addition to pancreatitis, Men1 is also important for the physiological response to oncogenic stress, as Men1 loss cooperates with mutant Kras to accelerate exocrine pancreas tumorigenesis.
Results
Men1 Deficiency Impairs Tissue Recovery following Acute Pancreatitis.
The pancreas is a remarkably regenerative organ. An intact response to and recovery from stress or insult are essential for normal tissue homeostasis of the pancreas, and prolonged injury or damage have been linked to diabetes and tumorigenesis (3, 23–25). To evaluate the function of Men1 in this context, we used the previously established conditional model of Men1 deficiency driven by the Pdx1-Cre transgene (10, 13) (Men1fl/flPdx1-CreTg, abbreviated as MC). Consistent with previous reports, there were no remarkable changes to the exocrine pancreas (SI Appendix, Fig. S1), despite robust reduction in Men1 protein levels (Fig. 1A). We next challenged the pancreas with the established model of caerulein-induced acute inflammation (25, 26). Briefly, caerulein is a cholecystokinin analog that stimulates the local release of digestive enzymes and results in edema, infiltration of immune cells and an almost complete loss of identifiable acinar cells. The organ recovers quickly and acinar cell morphology and mass are completely restored within 1 wk following insult, demonstrating the robustness of this regenerative system.
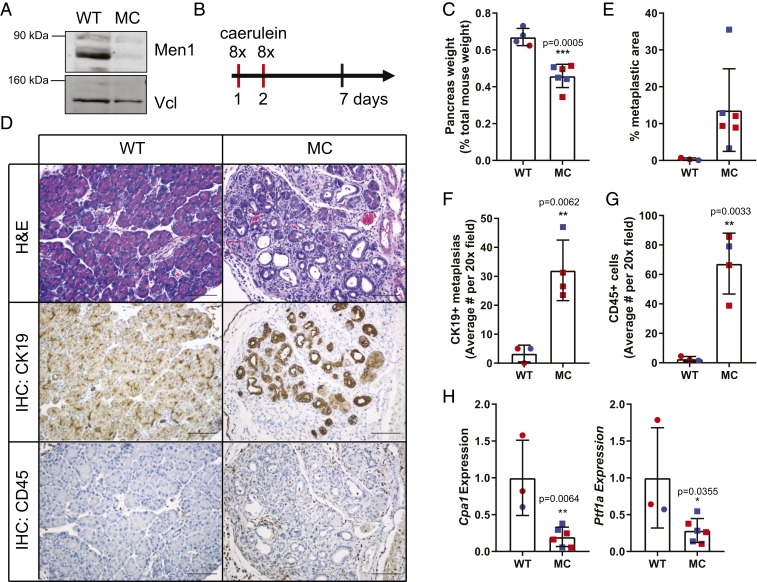
Fig. 1.
Men1 is essential for the normal pancreas response to acute pancreatitis. (A) Western blot analysis comparing wild-type (WT) to Men1-deficient (MC) pancreas. Men1 expression is compared to vinculin (Vcl) as a loading control. (B) Schematic representation of experimental induction of acute pancreatitis. (C) Pancreas weight 5 d following acute pancreatitis. (D) Pancreas histology 5 d following acute inflammation. Immunohistochemical staining for cytokertatin 19 (CK19) and CD45. Images are at 20×. (Scale bar, 100 μm.) (E) Quantification of percent metaplastic area as a percent of total pancreas area. (F and G) Quantification of immunohistochemical analysis for CK19 and CD45. (H) qRT-PCR analysis of acinar gene expression, Cpa1 and Ptf1a. Data are all presented as mean ± SD from individual mice, with female mice indicated in red and males in blue, with a Student’s t test for all statistical analysis.
We treated mice with eight hourly injections of caerulein, administered on 2 consecutive days, based on established protocols (27, 28) (Fig. 1B). Five days following the final treatment, mice were euthanized to evaluate pancreas histology and recovery from acute caerulien-induced pancreatitis (29). The pancreas weight of Men1-deficient mice was significantly reduced compared to wild-type (WT) controls (Fig. 1C), suggesting increased cell death in response to caerulein-induced damage. Histologically, the Men1-deficient pancreas demonstrated an impaired regenerative response to caerulein-induced injury, with limited normal acinar architecture at this time point, replaced by cytokeratin 19-positive ductal structures (metaplastic lesions, Fig. 1 D–F), and accompanied by significantly increased inflammatory infiltrate, highlighted by CD45 immunohistochemistry (Fig. 1 D and G). The percent of total pancreatic area containing metaplastic lesions was robustly increased in Men1-deficient pancreas, with the WT controls exhibiting only few areas of metaplasia (Fig. 1E). Moreover, there was also a significant decrease in the mRNA expression of the acinar markers Cpa1 and Ptf1a (Fig. 1H). We also evaluated pancreas histology 4 wk following the final treatment, and observe persistent damage in the MC pancreas, with a significant proportion of persistent metaplastic area (SI Appendix, Fig. S2). Combined, these data demonstrate that Men1 is essential for the normal pancreas response to caerulein-induced acute pancreatitis. Notably, consistent results were obtained from both female and male mice (Fig. 1).
Given the robust effects of Men1 loss on pancreas recovery following acute pancreatitis 5 d following treatment, we next investigated the timing of the tissue response. Wild-type and MC mice were treated with the same acute dosing of caerulein over 2 d, and subsequently euthanized at 24- and 72-h time points after the final injection. Representative histological images are presented in SI Appendix, Fig. S1A. In the wild-type pancreas, the 24-h time point revealed the expected histological changes, with immune cell infiltration and ductal transdifferentiation of the acinar compartment. By 72 h, the pancreas regained normal histology. More pronounced acinar to ductal metaplasia was evident in the MC pancreas at 24 h, and tissue injury persisted at the 72-h time point. Serum amylase and lipase levels can be used as confirmatory markers of pancreatitis, so we also evaluated levels in mice euthanized 17 to 18 h following the final caerulein treatment. There is a significant increase in serum amylase levels in caerulein-treated MC mice compared to the phosphate buffered saline (PBS)-treated controls (SI Appendix, Fig. S1B) validating the results of our histological and molecular analyses. In wild-type mice, at this time point, there is no significant difference in serum amylase between the two treatment groups (SI Appendix, Fig. S1B). Lipase levels were also increased upon caerulein treatment but the data were not statistically significant (SI Appendix, Fig. S1C).
Men1 Protein Is Stabilized following Caerulein Treatment.
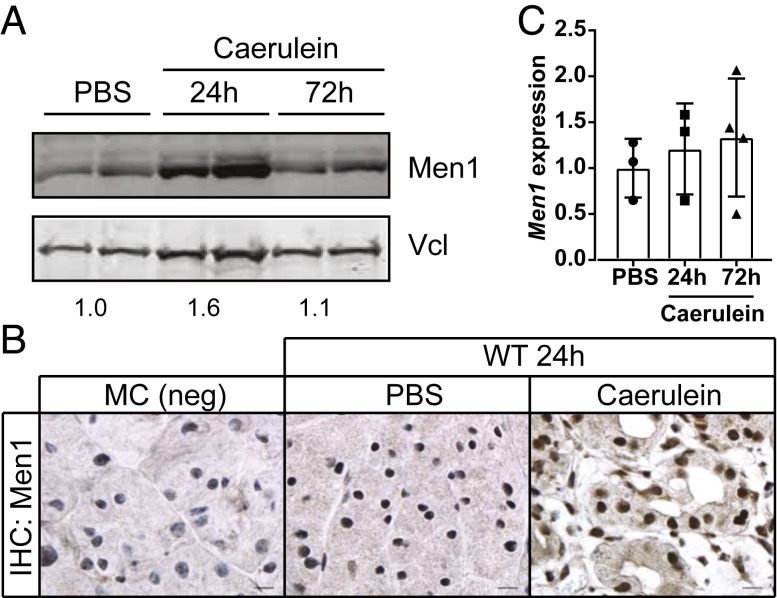
We next evaluated the effect of caerulein treatment and pancreatitis on the expression of Men1 in wild-type mice. Western blot analysis revealed an increase in Men1 protein 24 h following caerulein treatment compared to controls (Fig. 2A). Men1 protein subsequently returned to near baseline at 72 h, consistent with tissue recovery at this time point. Immunohistochemical analysis also revealed an increase in nuclear and cytoplasmic Men1 staining 24 h following caerulein treatment (Fig. 2B). Men1-deficient pancreas was used to validate the specificity of the staining. We found no significant difference in Men1 mRNA (Fig. 2C), suggesting an increase in Men1 protein synthesis or protein stability rather than gene transcription.
Fig. 2.
Men1 protein is stabilized in response to acute pancreatitis. (A) Western blot analysis of Men1 protein expression comparing PBS-treated control to caerulein-treated pancreas, 24 and 72 h following the final treatment. Protein expression is compared to vinculin (Vcl). Quantification of average Men1/Vlc signal is indicated below bands. (B) Immunohistochemical analysis for Men1 protein expression. Untreated MC pancreas is used as a negative control (Left). Staining was conducted on tissue samples 24 h following the final treatment with PBS control or caerulein. Images are at 100× magnification. (Scale bar, 10 μm.) (C) qRT-PCR analysis of relative Men1 mRNA 24 and 72 h following caerulein treatment. Data represent mean ± SD from individual mice.
Men1/MLL Target Genes Do Not Mediate the Pancreas Response to Acute Inflammation.
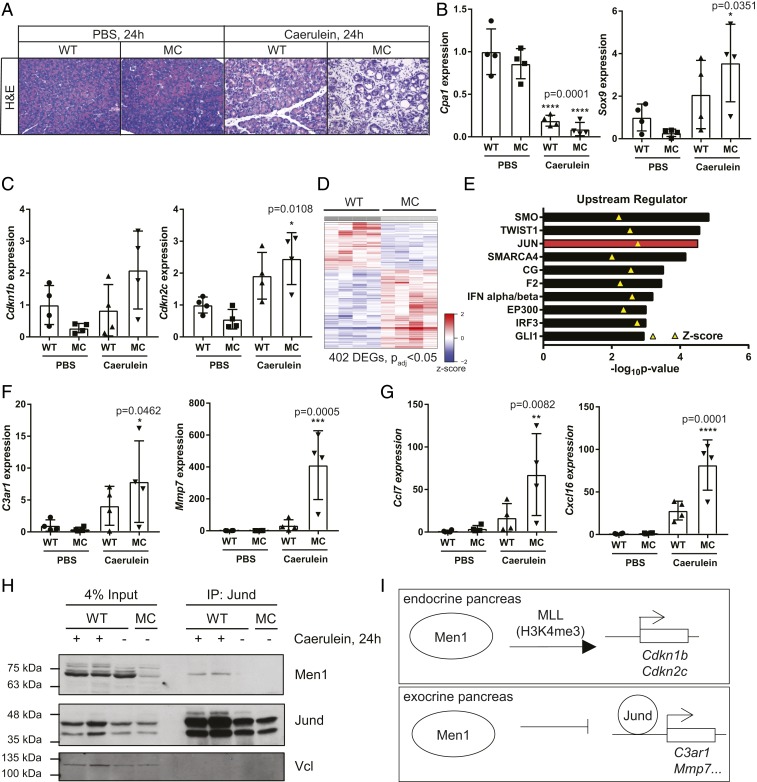
The major function that has been ascribed to Men1 as an endocrine tumor suppressor is as an epigenetic regulator of histone methylation through the MLL complex. Specifically MEN1 is important in the transcriptional activation of the cell cycle inhibitors CDKN1B (p27KIP1) and CDKN2C (p18INK4c). To investigate this function in the response to acute inflammation, we treated WT and MC mice with caerulein or PBS control and harvested pancreas 24 h following the final treatment (Fig. 3A). To assay the molecular response at this time point, we first evaluated the mRNA expression of the acinar gene Cpa1, and ductal gene Sox9. Caerulein treatment led to a significant down-regulation of Cpa1 in both genotypes compared to PBS-treated WT controls (****P = 0.0001, ANOVA with Dunnett’s multiple comparisons test), consistent with a loss of acinar cell state (Fig. 3B). At this time point we also observed a significant increase in Sox9 expression, but only in the MC pancreas, indicative of ductal metaplasia (Fig. 3B). We next evaluated the mRNA expression of Men1/MLL target genes Cdkn1b or Cdkn2c. If epigenetic transcriptional activator function is important in this context, we anticipated that the expression of these targets would be lost in the Men1-deficinent pancreas following caerulein treatment. Neither of these genes is down-regulated in this context (Fig. 3C). Cdkn2c actually shows a significant increase in expression compared to PBS-treated controls. Notably, in PBS-treated mice both Cdkn1b and Cdkn2c showed reduced, but not statistically significant, expression in MC mice compared to wild-type controls. Expression of Hoxc6 and Hoxc8, additional Men1/MLL target genes, was assayed but not detected. These data indicate that the established target genes of the Men1/MLL histone methyltransferase complex are not important in mediating the pancreas response to acute inflammation.
Fig. 3.
Molecular response to acute pancreatitis 24 h following treatment. (A) Representative images of pancreas histology 24 h following the final treatment. Images are at 20×. (Scale bar, 100 μm.) (B) qRT-PCR analysis of relative expression of Cpa1 (acinar) and Sox9 (ductal) gene. (C) qRT-PCR analysis of relative expression of Men1/MLL target genes, Cdkn1b and Cdkn2c. (D) Heatmap depicting differentially expressed genes in MC pancreas compared to WT control following chronic pancreatitis. (E) Ingenuity Upstream Regulator Analysis of RNA-sequencing data from chronic caerulein-inducted pancreatitis. The top 10 most significant predicted activators are presented. Z-score indicated with yellow triangle. (F) qRT-PCR analysis of genes identified as JUN targets in the analysis in D 24 h following acute caerulein-induced pancreatitis. (G) qRT-PCR validation of independent Jund target genes, Ccl7 and Cxcl16, 24 h following acute caerulein-induced pancreatitis. (H) Coimmunoprecipitation of Men1 with Jund. (I) Model of important Men1 functions in the endocrine and exocrine pancreas, respectively. All data are presented as mean ± SD from individual mice, relative to PBS-treated WT controls. Statistical analysis of gene expression was conducted with ANOVA with Dunnett’s multiple comparisons test to compare experimental samples to PBS-treated WT controls.
RNA-Sequence Analysis Identifies Jun as an Upstream Regulator of Differentially Expressed Genes in a Model of Chronic Inflammation.
We took advantage of a different study in the laboratory to explore the transcriptional changes associated with Men1 loss in the context of chronic pancreatitis. Caerulein was again used to induce inflammation, but mice were treated with four injections per day, 3 days a week for 3 wk to model chronic injury (30). Mice were euthanized 5 d following the final treatment and the pancreas of chronically treated MC mice showed a similar phenotype to the acute treatment, with an exacerbated injury and failure to recover compared to wild type (SI Appendix, Fig. S3A). RNA-sequence (RNA-seq) and differential gene expression analysis identified 402 differentially expressed genes (padj < 0.05) in the Men1-deficient pancreas compared to wild-type controls: 252 (62.7%) genes were up-regulated and 150 (37.3%) genes were down-regulated (Fig. 3D and Dataset S1). We next used Ingenuity Pathway Analysis (31) to conduct an upstream regulator analysis of the differentially expressed genes (Fig. 3E). Notable in the predicted activators was JUN, with one of the most significant P values (2.95E-05) and among the highest activation Z-scores (2.764). Moreover, previous data have identified MEN1 as a specific inhibitor of JUND transcriptional activity, in addition to its role as part of the MLL histone methyltransferase complex (20). Our analysis identified 10 genes with a measurement direction consistent with activation of JUN (SI Appendix, Fig. S3B). Of interest, several of these are inflammation-associated genes. Overexpression of Jun transcription factors also mediates a proinflammatory response and impaired recovery following pancreatitis in the context of Nr5a2 heterozygosity (32).
We next validated the expression of two of these genes in our acute pancreatitis assay at the 24-h time point. Both C3ar1 and Mmp7 were significantly induced only in MC pancreas compared to PBS-treated WT controls (Fig. 3F). We extended this analysis to two additional direct Jund targets that had previously been validated by chromatin immunoprecipitation (ChIP) and associated with an altered inflammatory response in the pancreas (32). Both Ccl7 and Cxcl16 also demonstrate significant induction only in the MC pancreas compared to control (Fig. 3G). Notably, expression of these genes is not induced by Men1 loss alone in PBS-treated controls, suggesting that caerulein treatment is required for their induction.
Previous data suggest Men1 directly binds and inhibits the transcriptional activity of Jund (20). As expected, we did not see robust changes in total Jund protein expression 24 h following caerulein treatment in MC pancreas compared to wild-type controls (SI Appendix, Fig. S4). We did, however, note a marked difference in the ratio of the two Jund bands in caerulean-treated pancreas compared to PBS-treated controls (SI Appendix, Fig. S4, marked by arrows). Control pancreas show a roughly equal distribution of the two Jund bands, whereas, the slower migrating band is the predominant species in caerulein-treated mice regardless of genotype. These data suggest that Jund is posttranslationally regulated, and likely activated, following caerulein treatment in vivo. We next evaluated the association between Men1 and Jund in wild-type mice 24 h following caerulein treatment. Men1 coimmunoprecipitates with Jund (Fig. 3H), demonstrating that these two proteins interact in the context of pancreatitis. Combined, these data indicate that Men1 loss promotes the expression of proinflammatory Jund target genes following caerulein-induced pancreatitis (Fig. 3I).
Men1 Loss Accelerates Mutant Kras Driven Oncogenesis.
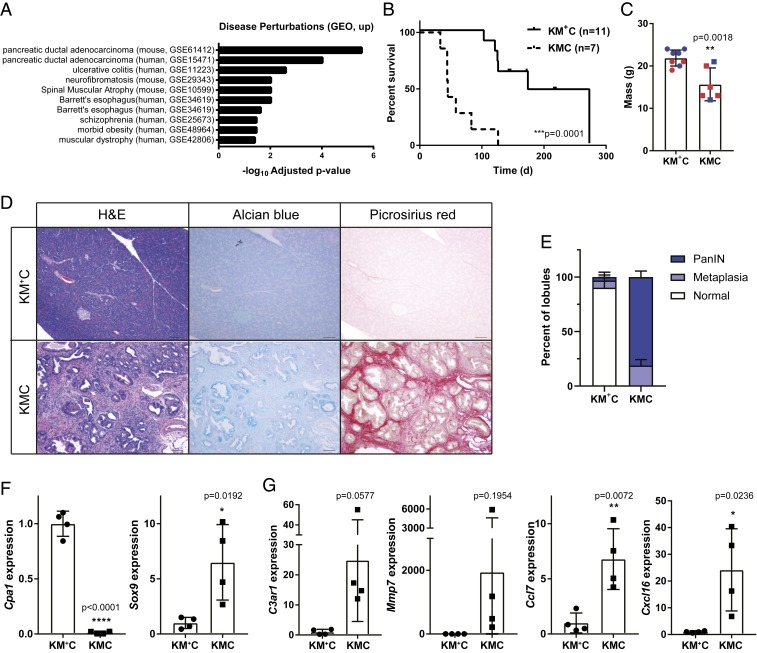
We next evaluated the differentially expressed genes from our RNA-seq data using Enrichr, an integrative web-based software application to rank enrichment terms (33, 34). From Enrichr, the disease perturbations from Gene Expression Omnibus (GEO) analysis revealed significant enrichment of genes up-regulated in two PDAC datasets: GSE61412, which compared gene expression in invasive mouse PDACs from Ink4a/Arffl/flKrasLSL-G12D/+Pdx1-CreTg mice compared to Cre-negative control pancreas (35), and GSE15471 which evaluated gene expression between matched tumor and normal samples from human PDAC patients (36). Thus, the gene expression changes caused by Men1 deficiency in our model of chronic pancreatitis show significant overlap with the gene expression changes in pancreatic ductal adenocarcinoma compared to normal tissue, in both mouse and human datasets (Fig. 4A). These associations, combined with data from other mouse models that clearly demonstrate that genetic perturbations that impair the homeostatic response to pancreatitis also cooperate with mutant Kras, the most frequently mutated gene in human PDAC, to accelerate disease progression (14, 37–39), suggest that loss of Men1 might also cooperate and promote neoplastic progression in the exocrine pancreas.
Fig. 4.
Men1 loss accelerates mutant Kras-driven oncogenesis. (A) Enricher analysis of disease perturbations (GEO, up). (B) Kaplan–Meier survival analysis. ***P = 0.0001, Log-rank (Mantel–Cox) test. (C) Weight of moribund KMC mice compared to age-matched controls. Male mice are indicated in blue and female in red. (D) Representative images of 6- to 7-wk-old pancreas samples stained with H&E, alcian blue, and picrosirius red. Images taken at 10× magnification. (Scale bar, 100 μm.) (E) Quantification of lesions present in pancreas sections represented as a percentage of total lobules, with mean ± SD from four independent mice presented. (F) qRT-PCR analysis of Cpa1 (acinar) and Sox9 (ductal) gene expression. (G) qRT-PCR analysis of Jund target genes. Weight and expression data are all presented as mean ± SD from individual mice with a Student’s t test for statistical analysis.
To directly test this hypothesis, we crossed the Men1fl allele to KrasLSL-G12D/+Pdx1-CreTg mice (KC), and established a small cohort of mice with homozygous Men1 loss (KrasLSL-G12D/+Men1fl/flPdx1-CreTg, KMC) and control mice heterozygous for the Men1fl allele (KrasLSL-G12D/+Men1fl/+Pdx1-CreTg, KM+C). Mice were born at the expected Mendelian ratio; however, the majority of mice with homozygous Men1 deletion became moribund within 2 mo of age and exhibited significantly reduced survival (P = 0.0001), with a median survival of only 45 d compared to 174 d for heterozygous controls (Fig. 4B). Moribund animals had hunched posture and were significantly smaller than littermate controls of the same age (Fig. 4C). Palpation of the sacroiliac bones suggested the mice were under conditioned or emaciated (40). The KM+C control mice reached humane endpoints for a number of reasons which were not directly related to pancreas pathology, including papillomas and rectal prolapse (SI Appendix, Table S1). On necropsy, the KMC pancreas was firm and notably distinct from KM+C mice euthanized at the same time point. We next compared pancreas histology. In all five KMC mice evaluated, the entire exocrine pancreas was replaced with ductal proliferation, with little or no acini remaining (SI Appendix, Fig. S5 A and B), suggesting that morbidity is likely a result of the general loss of exocrine pancreas function. The majority of the pancreatic lesions were PanIN-1s and PanIN-2s, with dilated ducts partially or completely lined by mucinous epithelium and containing goblet cells, highlighted by Alcian blue staining for mucins (Fig. 4D). The cells showed enlarged hyperchromatic, pencilate nuclei with focal pseudostratification. In a few dilated ducts, there was evidence of further disease progression to PanIN-3, lined by large pleomorphic cells with vesicular nuclei, prominent nucleoli, and exhibiting loss of polarity. PanIN lesions were associated with abundant collagen deposition assayed through picrosirius red staining (Fig. 4D). The pancreas from age-matched KM+C mice was generally histologically normal in all four mice evaluated, with the exception of a single mouse that exhibited chronic pancreatitis in one region of the pancreas (Fig. 4D and SI Appendix, Fig. S5C). We quantified the extent of disease in all mice by determining the percent of pancreas lobules that where normal, or contained metaplasias, or PanINs. The pancreas from KM+C mice contained an average of 90.2% normal lobules, focal areas of metaplasia in 6.5% of lobules, and isolated PanINs in 3.2% (Fig. 4E). Pancreas sections from KMC mice, however, had no normal lobules: 18.8% contained only metaplastic lesions and 81.2% contained PanINs (Fig. 4E).
We further evaluated the molecular response to Men1 loss in the pancreas of KMC mice to age-matched KM+C controls. As anticipated from tissue histology, there was a significant decrease in the expression of the acinar cell gene Cpa1, and a significant increase in the expression of the ductal gene Sox9 (Fig. 4F). This was accompanied by robust or significant increases in the expression of C3ar1, Mmp7, Ccl7, and Cxcl16, the same proinflammatory Jund target genes we evaluated in response to acute caerulein-induced pancreatitis (Fig. 4G). Combined these data demonstrate that Men1 loss, and the subsequent deregulation of Jund, can cooperate with oncogenic Kras to accelerate tumorigenesis.
Discussion
Through the use of genetic mouse models, we have identified a role for Men1 in the regulation of the inflammatory response of the pancreas in vivo. Men1 deficiency in the pancreas leads to robust defects in the homeostatic response following caerulein-induced pancreatitis and cooperates with mutant Kras to accelerate oncogenesis. Understanding the molecular mechanisms that regulate and maintain pancreas homeostasis is an urgent area of research due to the strong associations between pancreatitis and cancer. While recurrent MEN1 mutations have not been identified in human PDAC, there are many other mechanisms that can alter a pathway or gene. One study examined MEN1 protein expression by immunofluorescence in normal exocrine tissue compared to PDAC and identified reduced MEN1 expression in tumor tissue compared to controls (22). Moreover, using methylation-specific PCR they identified methylation of multiple CpGs in the MEN1 gene promoter in some cases, suggesting that altered epigenetic regulation may contribute to the reduced MEN1 protein expression in PDAC. Data from the Human Protein Atlas (www.proteinatlas.org) also support these observations (41). Twelve PDAC samples were stained with an antibody against MEN1, and in 8/12 (66.7%) samples, MEN1 protein was not detected, while staining was classified as low in 3/12 (25%) and medium in 1/12 (8.3%). Notably, none of the samples were classified as having high MEN1 staining. Future work to understand the mechanisms that regulate MEN1 protein levels and function in the pancreas and in PDAC will be instructive. Taken together, our results in combination with published and publicly available data strongly suggest that MEN1 may have tumor suppressor activity in the exocrine pancreas.
We also explored the mechanisms through which Men1 maintains pancreas homeostasis in response to caerulean-induced stress. Studies in mouse models have suggested that the primary tumor suppressor function for Men1 in the endocrine pancreas is through epigenetic regulation of gene expression as part of the MLL histone methyltransferase complex. Specifically, combined loss of the cell cycle inhibitors Cdkn1b and Cdkn2c promotes endocrine tumor development in the endocrine pancreas (16). Men1 loss, however, did not lead to reduced expression of these genes in response to caerulein treatment. Transcriptome analysis in a chronic model of caerulein-induced pancreatitis identified JUN as a candidate upstream regulator of the differentially expressed genes in the Men1-deficient pancreas. Previous studies have demonstrated that Men1 is also a direct negative regulator of Jund transcriptional activity (20). Indeed, Jund target genes are up-regulated in our assays, suggesting that increased Jun activity is contributing to the altered pancreas response. A recent study directly tested the functional impact of Jun family proteins in a similar model of impaired pancreas regeneration. Mice heterozygous for the orphan nuclear receptor Nr5a2 basally express elevated levels of proinflammatory genes. This thereby sensitizes the pancreas to damage, impairs regeneration, and promotes PDAC development in cooperation with mutant Kras (32, 39). Mechanistically, this was linked to up-regulation of AP1 family members Jun, Junb, and Jund in Nr5a2+/− pancreas, which increased promoter binding of Jun family proteins and induced the expression of inflammatory genes. Genetically, pancreas deletion of Jun rescued the phenotype of Nr5a2+/− mice and restored the normal tissue response to and recovery from caerulein-induced pancreatitis. While this study genetically manipulated Jun, AP1 family members share many common targets and these data support the proposed mechanisms of unrestricted Jund activity contributing to the phenotypes in our Men1-deficient model.
Our data also have potential clinical implications. We demonstrate that the homozygous loss of Men1 increases the severity of and impairs the recovery from an experimental model of pancreatitis. While rare, MEN1 syndrome patients inherit one mutant MEN1 allele and have an over 80% lifetime risk of developing pancreatic neuroendocrine tumors. As such, these patients undergo active surveillance for tumors. Once identified, it is challenging to predict which pancreas lesions are likely to progress and metastasize from imaging alone. Tissue sampling through fine needle aspirate or core biopsy provides the opportunity to obtain cells and tissue to evaluate features that might correlate with diseases status or progression, such as proliferation (Ki67). Approximately 1 to 3% of all patients receiving pancreas biopsies by endoscopic ultrasound and fine needle aspiration develop pancreatitis, and our data suggest that this number may be increased in the MEN1 patient population. As recommendations for screening continue to be modified, the potential risk of pancreatitis as identified through this work should be considered. Secondly, while MEN1 is an established tumor suppressor in endocrine tumors, recent work in MLL translocated leukemia suggests that MEN1 may also have tumor-promoting functions (19) and inhibitors of MEN1 have demonstrated therapeutic efficacy in preclinical models of this disease (42). MLL and JUND share the same MEN1 binding pocket (43); therefore, as potential small molecules of the MLL:MEN1 interaction advance for clinical use, our data suggest that it may be important to consider increased susceptibility to pancreatitis in the clinical evaluation.
Combined, our data identify Men1 as an essential mediator of the normal pancreas response to pancreatitis, and encourage future studies to understand the potential of Men1 to function as a tumor suppressor in pancreatic ductal adenocarcinomas.
Materials and Methods
Mice.
All mouse experiments were performed in compliance with the National Institutes of Health guidelines for animal research, and approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee. Mice were housed in individually ventilated cages, on individually ventilated cage racks, with up to five animals per cage after weaning. Direct cage bedding consisted of corn cob, Enviro-dri, Enrich-n-nest, and nestlets. The facility uses a 14-h light (7:00 AM to 9:00 PM) and 10-h dark (9:00 PM to 7:00 AM) cycle. Mice were fed a standard diet sterilized by irradiation (PicoLab Rodent Diet 5053, Purina) and were provided reverse osmosis chlorinated or acidified water ad libitum.
Men1 conditional knockout (stock no. 005109), Pdx1-CreTg (stock no. 014647), and KRasLSL-G12D (stock no. 008179) mice were previously described and obtained from The Jackson Laboratory (10, 44). Mice were maintained on a mixed background (C57BL/6, FVB, 129), Cre-negative littermates were used as functionally wild-type controls, and both male and female mice were used in our analyses. Genotyping primer sequences are provided in SI Appendix, Table S2.
Caerulein-Induced Acute Pancreatitis.
Acute pancreatitis was induced based on established protocols. Caerulein (MedChemExpress) was reconstituted in sterile PBS. Adult mice (6 to 10 wk old) were injected (individual dose: 75 μg/kg), given as eight hourly i.p. injections on 2 consecutive days during the light cycle. Mice were s.c. administered sustained release buprenorphine as an analgesic prior to the first caerulein injection.
Histology and Immunohistochemistry.
Tissues harvested from mice were fixed in 10% neutral buffered formalin and paraffin embedded. Four-micrometer sections were stained with hematoxylin and eosin (H&E) and examined by light microscopy. Tissue processing, paraffin embedding, sectioning, and H&E staining were performed by the MD Anderson Department of Veterinary Medicine and Surgery Histology Laboratory. Immunohistochemistry was performed using standard methods with citrate buffer for antigen retrieval and stained with antibodies against cytokeratin 19 (ab133496, Abcam), CD45 (550539, BD Pharmingen), and Men1 (ab3605, Abcam). Visualization was performed using ABC and DAB kits (Vector Laboratories) and slides were counterstained with hematoxylin.
Image Quantification.
Acinar to ductal metaplasias and PanIN lesions were quantified from whole pancreas sections scanned on an Aperio Scan Scope XT (Leica Biosystems) and manually annotated using Aperio ImageScope software (Leica Biosystems). For acute inflammation experiments, the percent of total pancreas area occupied by metaplasia was calculated for each sample. Cytokeratin 19-positive metaplasias and CD45-positive cells were manually quantified from four random and nonoverlapping 20× fields and averaged for each mouse. For quantification of metaplasias and PanIN lesions in the Kras cohort, individual pancreas lobules were counted and evaluated as normal, containing metaplasia, or containing PanIN. The number of lobules in KM+C pancreas was estimated based on the number of individual ductules present. Lobules were demarcated by surrounding collagen in KMC mice.
Western Blotting.
Tissue samples were flash frozen, pulverized, and lysed in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 6.5 mM Tris⋅HCl pH = 6.8, 25% glycerol, 10% β-mercaptoethanol). Protein extracts were separated by SDS/polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membranes, and probed with antibodies against Men1 (ab3605, Abcam), Jund (ab181615, Abcam), and Vinculin (V1931, Sigma). Proteins were visualized using Li-COR secondary antibodies and scanned on a Li-COR Odyssey Imager. Signal was quantified using ImageJ software (NIH).
Coimmunoprecipitation.
Flash frozen and pulverized pancreas was homogenized and then agitated for 2 h at 4 °C in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 1% Nonidet P-40 with protease inhibitor and phosphatase inhibitor). The samples were then centrifuged at 12,000 rpm for 20 min. The supernatant was taken for coimmunoprecipitation. One milligram of total protein lysate was incubated with 3 μg Jund antibody (ab181615, Abcam) overnight at 4 °C. The lysate was further incubated with 15 μL wet protein A-agarose beads (SC-2001, Santa Cruz Biotechnology) for 4 h at 4 °C. The beads were collected and washed three times with lysis buffer and protein was eluted with 1× SDS buffer. JunD and Men1 were detected by Western blot. The antibody for JunD (ab181615, Abcam) is diluted 1 in 1,000. The antibody for Men1 (A300-105A, Bethyl) is diluted 1 in 10,000. Light chain-specific secondary antibody (211-032-171, Jackson ImmunoReseach) was used and proteins were detected by chemiluminescence. Loading of inputs was assayed by immunoblotting for Vcl (V1931, Sigma), using a Li-COR secondary antibody and scanned on a Li-COR Odyssey Imager.
RNA Isolation and Quantitative PCR.
Flash frozen tissue was pulverized and homogenized in TRIzol RNA isolation reagent (Invitrogen). RNA was then purified using an RNeasy Mini Kit (Qiagen). RNA was then used as a template for first strand synthesis using the iScript Reverse Transcription Supermix (Bio-Rad). Quantitative RT-PCR was carried out using 2× SYBR Green qPCR master mix on a CFX384 Realtime System (Bio-Rad). Primer sequences are provided in SI Appendix, Table S2.
Differential Gene Expression Analysis from RNA-Seq Data.
STAR (version 2.6.0b) was used to align reads to the GRCm38.p6 reference genome (45). Samtools (version 1.8) was used to sort, convert between formats, and calculate mapping statistics (46). FastQC (0.11.5) was used to check for FASTQ read qualities. Gene annotation was carried out using GENCODE M19 (Ensembl 94) annotation, which was downloaded from the GENCODE project (47). Aligned reads were summarized at the gene level using HTseq-count (48). Differential gene expression was determined using R (3.5.1) and Bioconductor package DESeq2 (49). Read count was first prefiltered to remove extremely low expressed genes. DESeq2 was then used to carry out read count filtering, normalization, dispersion estimation, and identification of differential expression. DESeq2 modeled the counts using a negative binomial distribution, followed by the Walk test. The final adjusted P value was obtained using the Benjamini–Hochberg method.
Statistics.
All data are presented as mean ± SD. All statistical analyses were performed using GraphPad Prism 6 software, and P value of <0.05 was considered statistically significant. Comparisons between two groups were made with a Student’s t test, and comparisons among multiple groups were made with ANOVA with Dunnett’s multiple comparisons test to compare experimental samples to control treated wild-type controls. Comparison of survival curves was done using a Log-rank (Mantel–Cox) test.
Data Availability Statement.
RNA-seq data were deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession no. GSE145175.
Supplementary Material
Acknowledgments
We would like to thank Drs. Florencia McAllister and Anirban Maitra and members of the G.L. laboratory for helpful discussions. This research was supported by grants from the Neuroendocrine Tumor Research Foundation and NIH (NIH/National Cancer Institute R21 CA208463) to G.L. and a fellowship from the Canadian Institutes of Health Research (to A.R.W.). A.R.W. is the recipient of a Dodie P. Hawn Award in Genetics.
Footnotes
The authors declare no competing interest.
Data deposition: RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/ (accession no. GSE145175).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920017117/-/DCSupplemental.
References
- 1.Forsmark ChE., Vege S. S., Wilcox C. M., Acute pancreatitis. N. Engl. J. Med. 376, 598–599 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lee P. J., Papachristou G. I., New insights into acute pancreatitis. Nat. Rev. Gastroenterol. Hepatol. 16, 479–496 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hausmann S., Kong B., Michalski C., Erkan M., Friess H., The role of inflammation in pancreatic cancer. Adv. Exp. Med. Biol. 816, 129–151 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Giusti F., Marini F., Brandi M. L., “Multiple endocrine neoplasia type 1” in GeneReviews, Adam M. P., et al., Eds. (University of Washington, Seattle, WA, 1993). [Google Scholar]
- 5.Thakker R. V., Multiple endocrine neoplasia type 1 (MEN1) and type 4 (MEN4). Mol. Cell. Endocrinol. 386, 2–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiao Y., et al. , DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scarpa A., et al. ; Australian Pancreatic Cancer Genome Initiative , Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 543, 65–71 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Cromer M. K., et al. , Identification of somatic mutations in parathyroid tumors using whole-exome sequencing. J. Clin. Endocrinol. Metab. 97, E1774–E1781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crabtree J. S., et al. , A mouse model of multiple endocrine neoplasia, type 1, develops multiple endocrine tumors. Proc. Natl. Acad. Sci. U.S.A. 98, 1118–1123 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crabtree J. S., et al. , Of mice and MEN1: Insulinomas in a conditional mouse knockout. Mol. Cell. Biol. 23, 6075–6085 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J., et al. , Alpha cell-specific Men1 ablation triggers the transdifferentiation of glucagon-expressing cells and insulinoma development. Gastroenterology 138, 1954–1965 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Shen H. C., et al. , Multiple endocrine neoplasia type 1 deletion in pancreatic alpha-cells leads to development of insulinomas in mice. Endocrinology 151, 4024–4030 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen H. C., et al. , Recapitulation of pancreatic neuroendocrine tumors in human multiple endocrine neoplasia type I syndrome via Pdx1-directed inactivation of Men1. Cancer Res. 69, 1858–1866 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes C. M., et al. , Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell 13, 587–597 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Milne T. A., et al. , Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 102, 749–754 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franklin D. S., Godfrey V. L., O’Brien D. A., Deng C., Xiong Y., Functional collaboration between different cyclin-dependent kinase inhibitors suppresses tumor growth with distinct tissue specificity. Mol. Cell. Biol. 20, 6147–6158 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama A., et al. , Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24, 5639–5649 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne T. A., et al. , MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama A., et al. , The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123, 207–218 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Agarwal S. K., et al. , Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell 96, 143–152 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Karpathakis A., et al. , Pancreatic adenocarcinoma in a patient with multiple endocrine neoplasia 1 syndrome. Pancreas 42, 725–726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavallari I., et al. , Decreased expression and promoter methylation of the menin tumor suppressor in pancreatic ductal adenocarcinoma. Genes Chromosomes Cancer 48, 383–396 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Kleeff J., et al. , Chronic pancreatitis. Nat. Rev. Dis. Primers 3, 17060 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Carrière C., Young A. L., Gunn J. R., Longnecker D. S., Korc M., Acute pancreatitis markedly accelerates pancreatic cancer progression in mice expressing oncogenic Kras. Biochem. Biophys. Res. Commun. 382, 561–565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerra C., et al. , Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291–302 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Hyun J. J., Lee H. S., Experimental models of pancreatitis. Clin. Endosc. 47, 212–216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp J. L., et al. , Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris J. P. 4th, Cano D. A., Sekine S., Wang S. C., Hebrok M., Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 120, 508–520 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 30.McAllister F., et al. , Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell 25, 621–637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krämer A., Green J., Pollard J. Jr., Tugendreich S., Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cobo I., et al. , Transcriptional regulation by NR5A2 links differentiation and inflammation in the pancreas. Nature 554, 533–537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen E. Y., et al. , Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuleshov M. V., et al. , Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillaumond F., et al. , Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 112, 2473–2478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badea L., Herlea V., Dima S. O., Dumitrascu T., Popescu I., Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 55, 2016–2027 (2008). [PubMed] [Google Scholar]
- 37.Mallen-St Clair J., et al. , EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev. 26, 439–444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krah N. M., et al. , The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. eLife 4, e07125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flandez M., et al. , Nr5a2 heterozygosity sensitises to, and cooperates with, inflammation in KRas(G12V)-driven pancreatic tumourigenesis. Gut 63, 647–655 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Burkholder T., Foltz C., Karlsson E., Linton C. G., Smith J. M., Health evaluation of experimental laboratory mice. Curr. Protoc. Mouse Biol. 2, 145–165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlen M., et al. , A pathology atlas of the human cancer transcriptome. Science 357, eaan2507 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Borkin D., et al. , Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 27, 589–602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J., et al. , The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature 482, 542–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hingorani S. R., et al. , Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrow J., et al. , GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data were deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession no. GSE145175.