Abstract
Bacterial natural products display astounding structural diversity, which, in turn, endows them with a remarkable range of biological activities that are of significant value to modern society. Such structural features are generated by biosynthetic enzymes that construct core scaffolds or perform peripheral modifications, and can thus define natural product families, introduce pharmacophores and permit metabolic diversification. Modern genomics approaches have greatly enhanced our ability to access and characterize natural product pathways via sequence-similarity-based bioinformatics discovery strategies. However, many biosynthetic enzymes catalyse exceptional, unprecedented transformations that continue to defy functional prediction and remain hidden from us in bacterial (meta)genomic sequence data. In this Review, we highlight exciting examples of unusual enzymology that have been uncovered recently in the context of natural product biosynthesis. These suggest that much of the natural product diversity, including entire substance classes, awaits discovery. New approaches to lift the veil on the cryptic chemistries of the natural product universe are also discussed.
Bacterial natural products (NPs) are specialized metabolites that encompass an extraordinary breadth of different biological activities, many of which are of considerable value to society. NPs or NP-inspired compounds represent ~65% of all small-molecule approved drugs1 used in medicine to treat infectious diseases, cancers or as immunosuppressants2, and they are also applied extensively in agriculture3. While NPs have been an extremely productive source of new leads for the chemicals that are integral to modern life, rapid increases in resistance to antibiotics, cancer chemotherapies and pesticides pose significant threats to medicine and agriculture4,5.
The limited success of combinatorial libraries and high-throughput screening efforts to generate new drug candidates between 1995 and 2005 has led to renewed interest in academia in mining microorganisms for new bioactive compounds6. The range of bioactivities encountered in NPs is a direct consequence of the astonishingly diverse but distinct area of chemical space they occupy. Relative to synthetic compounds, NPs typically comprise complex ring systems, more stereogenic centres, more carbon, hydrogen and oxygen atoms and higher densities of functional groups (FIG. 1). The biosynthetic enzymes responsible for introducing such structural complexity have undergone many rounds of natural selection for the production of metabolites with chemical features that facilitate specific and effective interactions with important biological targets. Therefore, such enzymes represent particularly attractive synthetic tools for the development of novel therapeutics. As biocatalysts, they offer various advantages over conventional chemical catalysts and expand the synthetic chemist’s toolkit to include transformations for which total synthesis routes are currently limited (for example, selective C–H bond activation)7,8. Functional characterization of such enzymes is, therefore, a major area of research in the NP biosynthesis field. The search for new enzyme-catalysed chemistry promises access to ligands that modulate novel macromolecular targets9, critical to replacing the therapeutics and agrochemicals that have become ineffective. This Review explores the world of bacterial NP biosynthesis as a rich reservoir of untapped biochemical and structural novelty.
Fig. 1. Bacterial natural product chemical diversity.
Natural product (NP) examples described in this Review are illustrated. Compounds are coloured according to the section within this Review in which they are discussed. Orange, polyketide synthase/nonribosomal peptide synthetase-derived NPs; purple, terpenes; cyan, ribosomally synthesized and post-translationally modified peptides (RiPPs); magenta, NPs with non-signature biosynthetic origins. Note that closthioamide (28) and 6-thioguanine (29) are coloured magenta but are not RiPPs. 1, Obafluorin; 2, kutzneride 1; 3, curacin A; 4, nocardicin A; 5, pyrroindomycin A; 6, spinosyn A; 7, TMC-86A; 8, ikarugamycin; 9, saframycin A; 10, rhizoxin; 11, pederin; 12, leinamycin; 13, metatricycloene; 14, oocydin B; 15, albicidin; 16, saxitoxin; 17, dynemicin A; 18, clostrubin; 19, 12-epi-hapalindole U; 20, sodorifen; 21, longestin; 22, teleocidin B; 23, pentalenolactone; 24, bottromycin; 25, klebsazolicin; 26, saalfelduracin; 27, thioviridamide; 28, closthioamide; 29, 6-thioguanine; 30, nosiheptide; 31, streptide; 32, polytheonamide B; 33, microvionin; 34, ammosamide; 35, crocagin A; 36, andrimid; 37, belactosin C; 38, bicyclomycin; 39, indolmycin; 40, roseoflavin.
The biosynthetic genes encoding the pathways to NPs are usually co-located on the chromosome in biosynthetic gene clusters (BGCs). This paradigm facilitates their identification and characterization, and advances in next-generation DNA sequencing have created a massive data resource to mine for BGCs encoding novel NP structures and enzymology, a process greatly aided by a wealth of evolving bioinformatics tools10–15. However, such computational approaches are largely dependent on predefined rulesets that target known signature NP biosynthetic genes (BOX 1), thereby limiting the identification of pathways that employ uncharacterized enzymatic mechanisms. Novel NP biosynthetic enzymes have traditionally been identified by linking isolated NPs with unusual structures to their cognate BGCs using a retro-biosynthetic logic: well-characterized gene functions are allocated to specific biosynthetic steps and candidate genes for unusual chemical features are identified by a process of elimination from those that remain functionally unassigned.
Box 1. Signature natural product biosynthetic enzymology.
Natural products (NPs) are traditionally classified according to the type(s) of enzymes employed for their biosynthesis, and these signature enzymes direct biosynthetic gene cluster identification by genome-mining bioinformatics tools.
Polyketides are assembled from small acyl-coenzyme A building blocks by polyketide synthases (PKSs)18,224,225. PKSs can be classified into various types based on their architecture and functionality: type I refers to multi-enzyme complexes composed of linearly arranged and covalently attached catalytic domains, type II systems consist of discrete and monofunctional enzymes, and chalcone-synthase-like enzymes are designated as type III. In bacteria, polyketide NPs are commonly assembled by type I PKSs that are organized into modules, with each module being responsible for a single extension of the growing polyketide chain by one malonyl unit. Minimally, chain extension requires the activity of three characteristic domains present in each module: acyltransferase (AT) domains load malonyl units onto acyl carrier protein (ACP) domains, which tether substrates and polyketide intermediates, and ketosynthase domains, which catalyse Claisen-type condensation reactions between the growing polyketide chain and an extender unit. Trans-AT PKSs77 lack AT domains, instead employing free-standing proteins that act in trans. Further structural and stereochemical diversity can be introduced into the growing polyketide scaffold by additional accessory domains that alter the substituents and oxidation states of ACP-tethered intermediates. Thioesterase domains are typically found as the terminal domain in PKS systems, in which they catalyse polyketide release by hydrolysis or macrocyclization.
Nature has evolved multiple mechanisms of constructing peptidic NPs, many of which are derived from the post-translational modification (PTm) of ribosomally synthesized peptides120. These peptides typically comprise a core peptide fused to a leader or follower peptide (or both). The leader or follower serves as the recognition element for various PTM enzymes that install new structural features. Subsequent proteolytic cleavage yields the mature core peptide, sometimes with concomitant N-to-C cyclization and/or export out of the cell. Independently of the ribosome, nonribosomal peptide synthetases (NRPSs) are also responsible for the biosynthesis of numerous bioactive peptide NPs19. They exhibit modular architecture analogous to type I PKSs, with each NRPS module comprising three core domains responsible for extending the growing peptide chain by a single amino acid or carboxylic acid extender unit. Adenylation domains activate substrates, peptidyl carrier protein domains tether substrates and peptide intermediates, and condensation domains catalyse amide bond formation between them. Structural and stereochemical diversity is typically achieved via two mechanisms: the use of a wide range of noncanonical amino acid substrates and peptide tailoring reactions catalysed by additional NRPS accessory domains and/or trans-acting tailoring enzymes19.
Terpenes are typically derived from the prenyltransferase-catalysed extension of a dimethylallyl phosphate starter unit by one or more isopentenyl diphosphate extender units. The resulting polyisoprenoid intermediates are subsequently transformed by terpene cyclases, the signature enzymes for this class. Terpene cyclases catalyse diphosphate release from their linear substrates, promoting a wealth of cationic cyclization and rearrangement cascades that result in an astonishingly diverse range of monocyclic and polycyclic hydrocarbon scaffolds97,100.
When biosynthetic pathways are not active under a particular condition or their cognate NP cannot be detected (that is, they are ‘silent’), identifying genes or BGCs encoding novel chemistry can be extremely difficult, as they are either orphan or cryptic in nature. These two terms are frequently used to describe the status of BGCs with respect to the NPs they encode16 but can also be conversely applied to isolated NPs that have not been linked to their cognate biosynthetic loci. For the purposes of this Review, we have adapted both terms to reflect the status of discrete genes involved in NP biosynthesis. Biosynthetic genes defined as either orphan or cryptic can be thought of as being ‘hidden in plain sight’ — their function cannot be elucidated from in silico analysis alone. Such chemistry is not only ‘hidden’ in the genomes of poorly studied bacteria such as those that are as-yet uncultivated but also in taxa that have been studied over many decades, suggesting that a wealth of enzymes for unprecedented chemistries await discovery.
In this Review, we highlight recently characterized examples of exciting new chemical transformations and NP families resulting from previously ‘hidden’ enzymes. All NPs discussed are illustrated in FIG. 1 and are numbered throughout the text. Within selected NP classes, where possible, we proceed from orphan or cryptic enzymes that introduce unusual, discrete chemical moieties into NP scaffolds to those that perform transformations that define entirely new NP families. In addition, we summarize studies demonstrating that further novelty remains to be uncovered in the NP universe. We conclude by considering new approaches for the identification of novel chemistry from in silico data alone and the future prospects of the field.
Thiotemplated NP biosynthesis
Nature has evolved two core logics for the biosynthesis of NPs17, either via the action of individual enzymes acting on free substrates or by modular enzymes that assemble small, covalently bound building blocks to longer product chains. In the latter case, thioesters are used to activate monomer acyl groups for subsequent nucleophile capture, and all substrates, intermediates and products remain covalently tethered to the enzyme during biosynthesis. Two major NP classes are assembled following such logic: polyketides, which are constructed from acyl-coenzyme A (CoA) monomers by polyketide synthases (PKSs), and nonribosomal peptides (NRPs), which are constructed from amino and carboxylic acids by NRP synthetases (NRPSs)18,19 (BOX 1). Modules minimally comprise three functional components that are necessary for catalysing extension of a specific tethered intermediate by one additional monomer unit and typically act in series to catalyse discrete, consecutive biosynthetic steps. This phenomenon, epitomized by the 6-deoxyerythronolide B synthase PKSs encoded in the erythromycin BGC20, is known as the collinearity rule and often allows the prediction of the resulting NP structure based solely on PKS or NRPS sequence data. However, exceptions that defy bioinformatic prediction and deviate from the canonical biosynthetic logic continue to be discovered, hinting at a wealth of as-yet-uncovered chemical diversity.
Noncanonical substrate biosynthesis
The use of alternative building blocks is an important mechanism for generating NP chemical diversity, particularly for NRPSs, which incorporate a broad range of nonproteinogenic amino and carboxylic acids19,21 that are often the products of rare or unprecedented transformations.
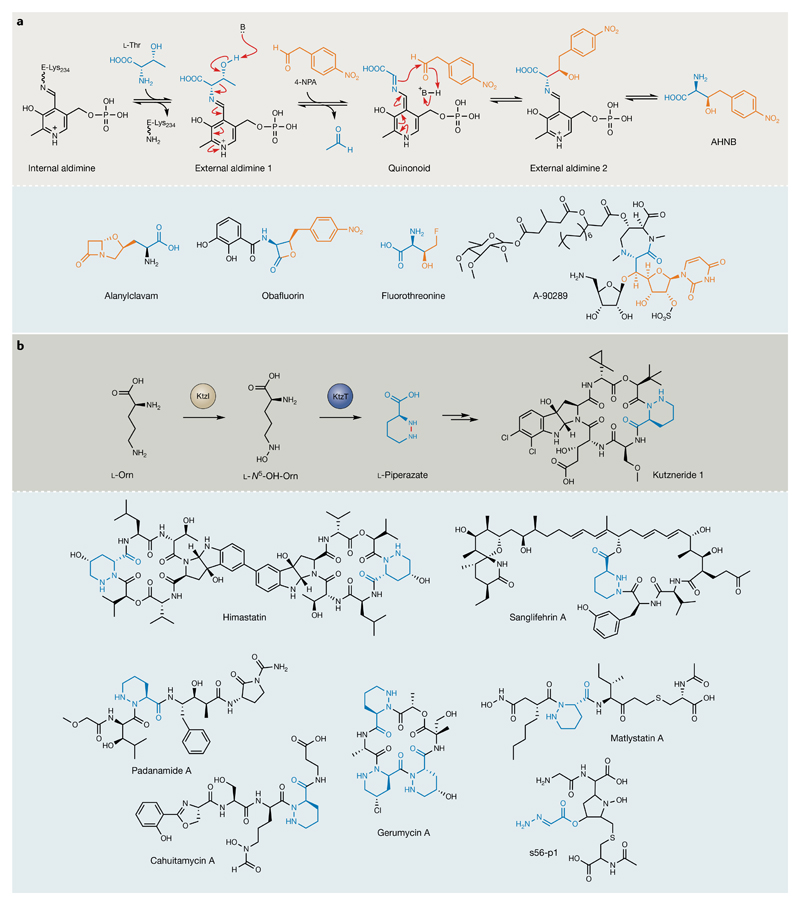
The β-lactone antibiotic obafluorin (1) produced by Pseudomonas fluorescens, for example, is assembled from 2,3-dihydroxybenzoic acid and an unusual (2S,3R)-2-amino-3-hydroxy-4-(4-nitrophenyl) butanoate (AHNB) unit22,23. Although previously predicted to be generated via a thiamine-diphosphate-dependent mechanism24, AHNB is, in fact, the product of a rare, pyridoxal-phosphate (PLP)-dependent, l-Thr transaldolase (l-TTA), ObaG, that catalyses sequential retro-aldol cleavage and crossed aldol reactions (FIG. 2a). Initially annotated as a putative serine hydroxymethyl-transferase, phylogenetic analysis revealed that ObaG forms a discrete evolutionary lineage22. The clade also contains LipK and its homologues and fluorothreonine transaldolase (FTase), the only two other characterized l-TTAs, responsible for the biosynthesis of the 5′-C-glycyluridine moiety in lipopeptidyl nucleoside antibiotics and fluorothreonine, respectively25,26 (FIG. 2a). This analysis also identified a putative l-TTA involved in the biosynthesis of the 5S-clavam antibiotic alanylclavam27 (OrfA), which was previously annotated as a glycine hydroxymethyltransferase, perhaps explaining why the biosynthetic step catalysed by this enzyme and its homologues has remained enigmatic in a number of clavam biosynthetic pathways28.
Fig. 2. Remarkable transformations during nonribosomal peptide synthetase substrate biosynthesis.
a | Proposed mechanism for the l-Thr transaldolase (l-TTA), ObaG, during obafluorin biosynthesis22. Grey panel: pyridoxal-phosphate binds l-Thr to form an external aldimine that undergoes retro-aldol cleavage to yield a glycine enolate. An aldol-type reaction with 4-nitrophenyacetaldehyde (4-NPA) yields (2S,3R)-2-amino-3-hydroxy-4-(4-nitrophenyl)butanoate (AHNB). The new bond formed in this transformation is indicated in red. Blue panel: other natural products that incorporate l-TTA-derived, β-OH-α-amino acid substrates (proposed for alanylclavam)25–27. In each case, aldehyde and l-Thr-substrate-derived moieties are highlighted in orange and blue, respectively. b | The KtzT-catalysed haem-dependent cyclization of l-N5-OH-Orn to form l-piperazate31 (grey panel). KtzI is an l-Orn N-hydroxylase. Blue panel: other natural products that are purported to employ a KtzT homologue or a mechanism involving N-hydroxylation to generate an activated intermediate susceptible to intramolecular attack by an amino group in the formation of N–N bonds during their biosynthesis32–38. The relevant corresponding moieties are highlighted in blue.
Another example is the kutznerides (2), antifungal hexapeptidolactones produced by Kutzneria sp. 744 that consist of an unusual t-butyl glycolate monomer and five different nonproteinogenic amino acids29. Among these is l-piperazate, a six-membered, cyclic amino acid with an N–N bond, a feature found in various bioactive NPs30, yet no enzyme had been characterized that catalyses N–N bond formation. Recently however, the haem-dependent enzyme KtzT was shown to be responsible, catalysing the N–N bond-forming cyclization of l-N5-OH-Orn to generate l-piperazate31 (FIG. 2b). Homologues of ktzT have been identified in the BGCs of a number piperazate-containing NPs, including the cahuitamycins32, gerumycins33, matlystatins34, pandanamides35, sanglifehrins36 and himastatin37. An analogous mechanism involving an intermediate that is activated by N-hydroxylation before intramolecular attack by an amino group has also been reported recently for s56-p1 (REF.38).
Divergent cis-acting domains
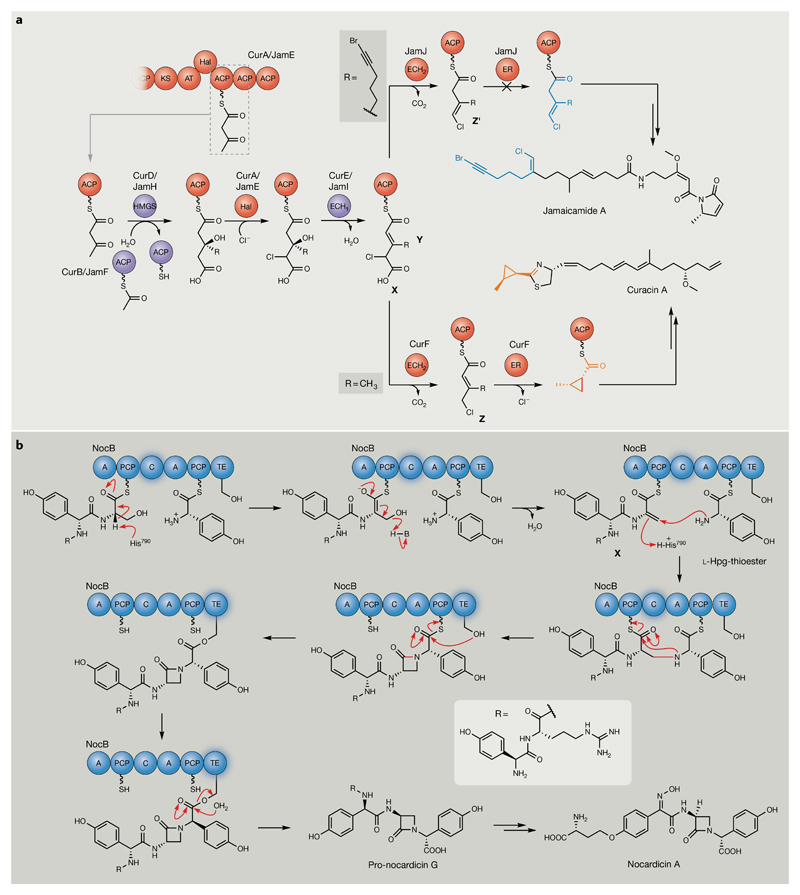
Once substrates are tethered as thioesters on PKS and NRPS carrier protein domains, further diversification is achieved during rounds of extension by additional accessory domains. While the canonical functions of these domains have been well characterized18,19, many have diverged to perform novel transformations that are extremely challenging to predict bioinformatically. Curacin A (3) is a potent anti-proliferative agent39 produced by the cyanobacterium Moorea producens that has provided unique insights into ‘hidden’ noncanonical PKS enzymology40–43. Curacin A contains a cyclopropyl ring, a cis-double bond and a terminal alkene, structural features that would not be readily predicted by analysing the BGC responsible for its production40. Remarkably, early enzymes of the curacin BGC share high homology with those of jamaicamide A (FIG. 3a), a sodium channel inhibitor that is also of cyanobacterial origin but comprises a different set of unusual moieties, including a vinyl chloride group and an alkynyl bromide44. During biosynthesis, an Fe2+/α-ketoglutarate-dependent halogenase (HAL) domain in CurA/JamE produces an acyl carrier protein (ACP)-bound γ-chloro-3-hydroxy-3-methylglutaryl intermediate (X) that is subsequently dehydrated by a discrete enoyl-CoA hydratase (ECH1), CurB/JamF, to afford an α,β-enoyl-γ-chloroglutaryl-ACP intermediate (Y)45–47 (FIG. 3a). The pathways diverge with subsequent decarboxylation activity of the CurF and JamJ ECH2 domains, which share 59% sequence similarity, yet result in the formation of α,β-enoyl-γ-chloro-ACP (Z) and β,γ-enoyl-γ-chloro-ACP (Z′) products, respectively. In the curacin pathway, this process sets the stage for subsequent modification by the CurF enoyl reductase (ER) domain via an atypical cyclopropanation reaction involving hydride addition from NADPH and substitution of chlorine. The homologous ER domain in JamJ has no apparent biosynthetic function. The origins of this exceptional cyclopropanase activity were localized to divergence in the amino acid composition of a 15-mer loop region found in the catalytic centre of the CurF ER domain, which otherwise appears to be a canonical PKS ER domain47. An Arg residue present within this loop is proposed to stabilize the chloride ion that is formed during ring formation, consistent with substitution experiments.
Fig. 3. Noncanonical polyketide synthase/nonribosomal peptide synthetase reactions involving tethered thioester intermediates.
a | Enoyl reductase (ER)-catalysed cyclopropanation during curacin biosynthesis. The curacin and jamaicamide biosynthetic pathways diverge at the β-branching step, which is initiated by decarboxylase (ECH2)-catalysed decarboxylation, resulting in α–β double-bond and β–γ double-bond formation in respective curacin and jamaicamide enoyl-γ-chloro-acyl carrier protein (ACP) intermediates45,46. Subsequent activity of an unusual cis-acting ER domain in CurF catalyses cyclopropane ring formation (highlighted in orange)47. The homologous ER domain in JamJ is unreactive towards its respective ECH2 product (unmodified ECH2 thioester product, highlighted in blue). The new bond formed to close the cyclopropane ring is highlighted in red. b | On-line tailoring reactions catalysed by divergent condensation (C) and thioesterase (TE) domains during nocardicin biosynthesis. The terminal C domain in NocB catalyses initial elimination of water, followed by cyclization to yield the β-lactam pharmacophore48. The resulting thioester intermediate is subsequently transferred to the NocB TE domain, which performs an unprecedented epimerization of the l-(p-hydroxyphenyl)glycine (l-Hpg) moiety, before canonical hydrolysis to release nocardicin G50. Nonribosomal peptide synthetase domains catalysing the specific transformations illustrated are highlighted and new bonds formed to generate the β-lactam ring by the C domain are shown in red. Polyketide synthase domains and nonribosomal peptide synthetase domains are highlighted in red and blue, respectively. Trans-acting enzymes are highlighted in purple. A, adenylation; AT, acyltransferase; ECH1, dehydratase; Hal, halogenase; HMGS, 3-hydroxy-3-methylglutaryl CoA synthase; KS, ketosynthase; PCP, peptidyl carrier protein.
In addition to accessory domains, core catalytic domains have also evolved to perform new functions. Among domains homologous to NRPS condensation (C) domains, for example, are the well-characterized heterocyclization (Cy) and epimerization (E) domains19; however, further homologues have also diverged to catalyse unprecedented reactions. This is exemplified by the NocB C domain from Nocardia uniformis, which is responsible for synthesis of the integrated β-lactam pharmacophore present in the antibiotic nocardicin A48 (4). This reaction is proposed to proceed via an initial E1cB (elimination unimolecular conjugate base) elimination of water from a seryl residue to generate a peptidyl carrier protein (PCP)-linked dehydroalanyl tetrapeptide thioester (X)49, mediated by a single additional His residue located directly on the N-terminal of the C domain H*HHxxxDG active-site signature (FIG. 3b). Subsequent β-addition of l-(p-hydroxyphenyl)glycine (l-Hpg) results in C–N bond formation. The NocB thioesterase (TE) domain has also evolved an additional tailoring function, catalysing peptide epimerization before the cleavage of pro-nocardicin G50 (FIG. 3b). This domain is highly selective for monocyclic β-lactam substrates and fails to hydrolyse linear peptide test substrates, underlining an important gatekeeping function during biosynthesis. The examples described here illustrate how subtle changes in primary amino acid can substantially alter domain functionality in thiotemplate-based enzymes, highlighting the propensity for novel enzymatic transformations to be incorrectly functionally annotated.
Unusual trans-acting tailoring enzymes
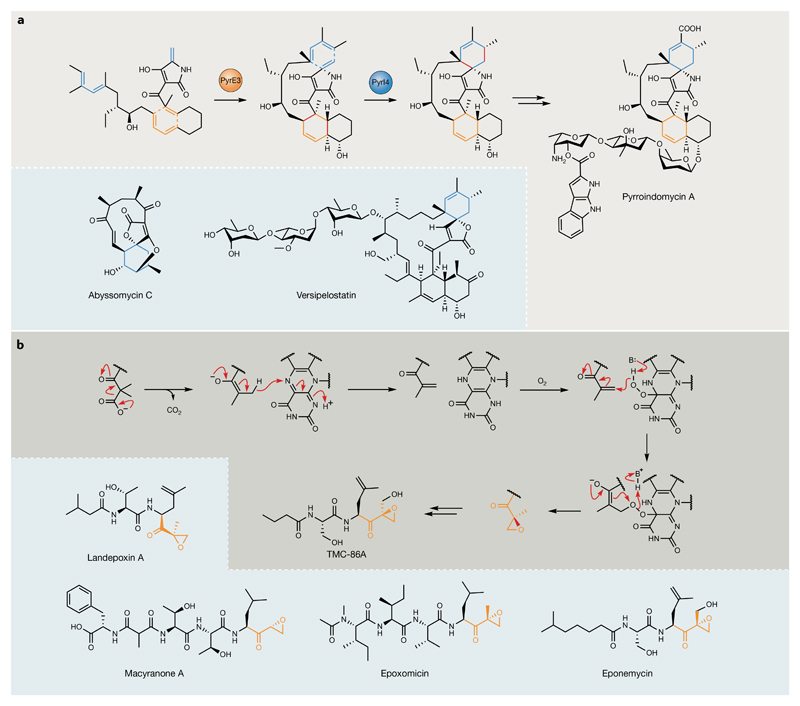
In addition to modifications by functionally divergent domains, polyketides and NRPs are often subject to further tailoring events catalysed by discrete enzymes acting either in trans during assembly or following scaffold release chain termination19,51. The Diels–Alder reaction is a [4+2] cycloaddition that is of enormous synthetic value in the preparation of substituted, transannular, six-membered carbocycles. More than 400 natural compounds have been suggested to be biosynthesized by formal Diels–Alder reactions52, resulting in decades of speculation as to the existence of natural enzymes capable of performing this highly prized transformation. Very recently, several putative ‘Diels–Alderases’ have been characterized in PKS/NRPS pathways that originate from diverse evolutionary origins53–58. The spirotetramate pyrroindomycin (5) antibiotics are a notable example because two consecutive [4+2] transformations occur during their maturation (FIG. 4a). These cyclizations are catalysed by the dedicated cyclases PyrE3 and PyrI4 (REF.56), which act in tandem to convert two pairs of 1,3-diene and alkene groups in a linear intermediate (X) into the two cyclohexene rings of the dialkyldecalin system and tetramate spiro-conjugate moieties, respectively. PyrE3 and PyrI4 were initially identified on the basis that they were functionally unassigned and shared homology with genes in the pathways of structurally related spirotetronate polyketides. PyrE3 belongs to the flavoenzyme superfamily, whereas PyrI4 resembles proteins of unknown function, again emphasizing the diverse, ‘hidden’ origins of Diels–Alderase candidates in nature. PyrI4 homologues from abyssomicin57 and versipelostatin55 biosynthesis have also been recently characterized as spirotetramate-forming cyclases (FIG. 4a). SpnF, an enzyme purported to catalyse a [4+2] cycloaddition during the biosynthesis of the insecticidal macrolide spinosyn A (6) by Saccharopolyspora spinosa, possesses an S-adenosyl methionine (SAM)-dependent methyltransferase fold, representing yet another enzymatic origin for this desirable transformation53,54.
Fig. 4. Unusual post-polyketide synthase/nonribosomal peptide synthetase enzymology.
a | Enzymatic [4+2] cycloadditions during pyrroindomycin biosynthesis. Grey panel: Diels–Alder-type cyclizations catalysed by PyrE3 (orange) and PyrI4 (blue)56. New bonds formed are highlighted in red. Blue panel: six-membered rings (highlighted in blue) installed by PyrI4 homologues during abyssomycin57 and versipelostatin55 biosynthesis. b | Grey panel: TmcF catalyses a remarkable decarboxylation–dehydrogenation–oxygenation transformation to generate the epoxyketone moiety present in TMC-86A62. The new bond formed to close the epoxide ring is highlighted in red. Blue panel: select examples of natural product proteasome inhibitors that are proposed to employ TmcF homologues during their biosynthesis to install α/β-epoxyketone warhead moieties59–62. Epoxyketone moieties introduced by TmcF and its homologues are highlighted in orange.
Post-assembly-line tailoring reactions are perhaps a less common source of chemical diversity in NRPS pathways but also hide exciting enigmatic chemistries. Just one recent example is a family of widespread flavin-dependent enzymes characterized in the biosynthesis of α,β-epoxyketone proteasome inhibitors (for example, TMC-86A, 7)59–62. These enzymes catalyse a remarkable decarboxylation–dehydrogenation–monooxygenation cascade that converts α-dimethyl-β-keto intermediates into the corresponding α,α-methyl-α,β-epoxyketones (FIG. 4b), thereby installing the warhead critical to proteasome-inhibitory activity62.
NP family-defining PKS and NRPS transformations
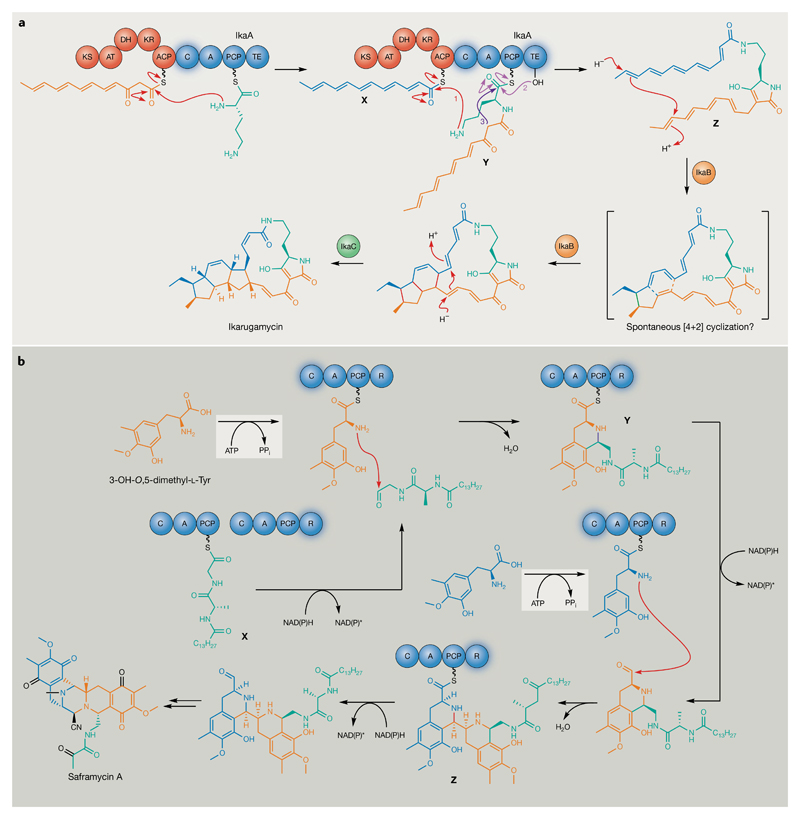
Beyond catalysing discrete tailoring reactions during scaffold assembly, thiotemplate-based enzymes can also perform unique transformations that define completely novel NP families. The polycyclic tetramate macrolactams represent a prime example and are an emerging new family of PKS–NRPS hybrid products from phylogenetically diverse bacteria that exhibit a range of potent and selective inhibitory activities63–71. This family comprises a structurally characteristic tetramic acid moiety embedded in a macrolactam that is fused to a stereochemically complex polycyclic carbacycle66. Astonishingly, such structural complexity is the product of only three enzymes, as demonstrated recently for ikarugamycin (8) biosynthesis72–74. An iterative bimodular PKS–NRPS hybrid, IkaA, is responsible for performing 13 individual bond-forming reactions, assembling two distinct hexaketidic polyene precursors that are attached to the N-termini of l-Orn, before catalysing a subsequent cyclization to afford the tetramic acid moiety with concomitant intermediate release (FIG. 5a). The as-indacene moiety is subsequently introduced by the oxidoreductase IkaB and alcohol dehydrogenase IkaC, which together selectively define all the stereocentres in the ikarugamycin 5,6,5 tricycle (FIG. 5a). IkaABC are, thus, responsible for the construction of 15 C–C and two C–N bonds and create eight stereogenic centres.
Fig. 5. Polyketide synthase-catalysed and nonribosomal peptide synthetase-catalysed transformations that define novel natural product families.
a | Members of the polycyclic tetramate macrolactam family of polyketides are produced by the activities of only three enzymes, as illustrated for ikarugamycin72–74. New bonds formed during successive cyclizations are highlighted in red. Steps 1–3 illustrate amide bond formation between thioester intermediates X and Y by the IkaA C domain (step 1), transfer of the resulting thioester intermediate to the IkaA thioesterase domain active site Ser residue (step 2) and thioesterase-catalysed intramolecular attack to yield the pyrroline moiety with concomitant release of intermediate Z (step 3). It is not clear whether formation of the six-membered ring is spontaneous or enzyme-catalysed. The timing and nature of the isomerization event that results in the introduction of a cis double-bond in the final ikarugamycin pathway product is also not clear; however, the IkaC homologue OX4 is proposed to be responsible for cis double-bond introduction during the biosynthesis of heat-stable antifungal factor, a polycyclic tetramate macrolactam produced by Lysobacter enzymogenes226–228. b | A single nonribosomal peptide synthetase module catalyses iterative Pictet–Spengler-type cyclizations and reductions of structurally different intermediates to assemble the characteristic scaffold of the tetrahydroisoquinoline antibiotics76. Structurally different SfmC reductase (R) domain peptidyl thioester intermediates are indicated by X–Z. Adenylations of 3-OH-O,5-dimethyl-l-Tyr residues before condensation (C)-domain-catalysed cyclization by the SfmC adenylation (A) domain are indicated in grey. New bonds formed by the C domain are highlighted in purple. Individual substrates are highlighted in different colours so that their fate can be tracked through the mechanism illustrated. ACP, acyl carrier protein; AT, acyltransferase; DH, dehydratase; KR, ketoreductase; KS, ketosynthase; PCP, peptidyl carrier protein; TE, thioesterase.
An equally remarkable example is the tetrahydroisoquinoline antibiotics produced by various soil and symbiotic marine bacteria75. During the biosynthesis of saframycin A (9), a monomodular NRPS, SfmC, catalyses a remarkable, seven-step transformation to assemble the complex saframycin scaffold76 (FIG. 5b). SfmC comprises C, adenylation (A), PCP and reductase (R) domains, which act iteratively to assemble the characteristic pentacyclic tetrahydroisoquinoline core of this compound family from different dipeptidyl substrates. The well-characterized R domain usually generates aldehyde intermediates by reductive chain release; however, the SfmC R domain is exceptional in that it reduces three structurally different peptidyl thioesters (X–Z). The resulting intermediates are substrates for the SfmC C domain that catalyses iterative Pictet–Spengler cyclizations on consecutive reduced intermediates, completing this astonishing NRPS-catalysed transformation. Ikarugamycin and saframycin A illustrate the challenges associated with determining product structures for even seemingly canonical, thiotemplate-based enzymes, particularly when such systems act iteratively and utilize multiple, diverse substrates. However, such examples show that unprecedented transformations and entirely new, widespread NP families await discovery, even in systems that look relatively unexceptional based on primary sequence data.
Trans-acyltransferase PKS biosynthesis
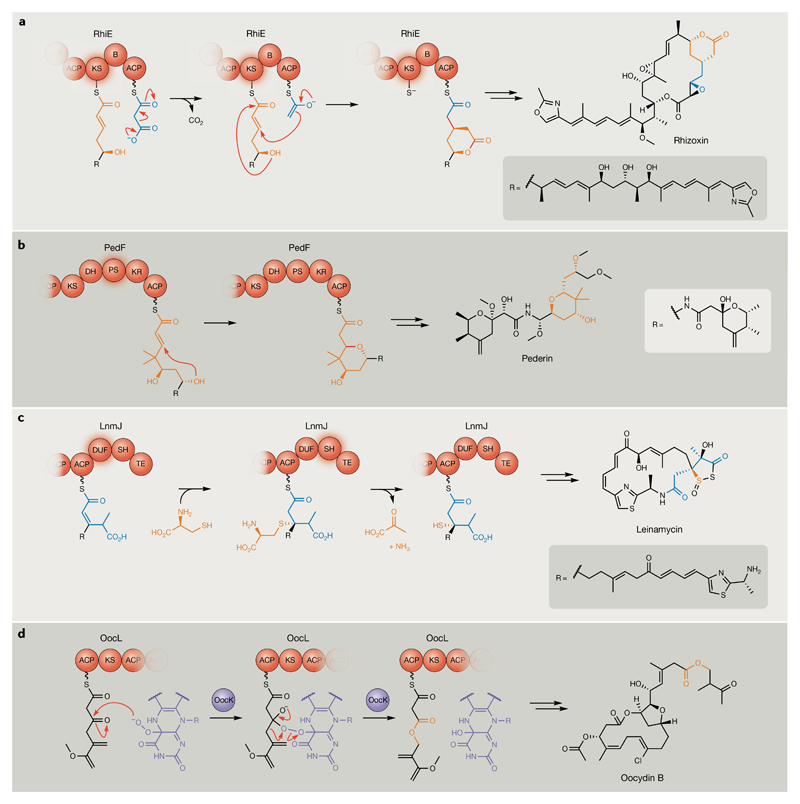
The canonical biosynthetic model for complex polyketide biosynthesis has been expanded to include a second type of multi-modular polyketide system, termed the trans-acyltransferase (AT) PKSs77. Despite exhibiting multi-modular architecture, trans-AT PKSs lack AT domains, instead employing free-standing proteins acting in trans. Trans-AT PKSs evolved in a mosaic-like fashion, independently of so-called cis-AT PKSs, and exhibit enormous architectural diversity, including features such as unusual domain orders, unique domains, non-elongating modules, intermodular domain activity and split modules. Such architectures have historically made trans-AT BGCs difficult to identify, providing an additional interpretation to ‘hidden in plain sight’; the bacillaene BGC remained enigmatic for >10 years owing to its confusing architecture suggesting a nonfunctional pathway78, and such BGCs continue to evade detection by current bioinformatics tools79. Trans-AT PKSs are replete with ‘hidden’ transformations that introduce even further chemical diversity into the final products. Recently published examples include a novel type of branching module involved during rhizoxin (10) biosynthesis that installs a δ-lactone by a Michael-type addition80,81, multiple examples of pyran synthases (domains and discrete enzymes) from diverse evolutionary origins that construct five-membered and six-membered cyclic ethers via oxa-Michael conjugate additions (for example, pederin, 11)82–84 and a domain of unknown function (DUF)/cysteine lyase (SH) didomain that generates a sulfur heterocycle during leinamycin (12) biosynthesis85 (FIG. 6a–c).
Fig. 6. Newly characterized on-line trans-acyltransferase polyketide synthase transformations.
a | Ketoreductase (KS)-catalysed vinolygous β-branching and lactonization during rhizoxin biosynthesis80,81. New bonds formed during this transformation are highlighted in red. b | Pyran synthase (PS)-domain-catalysed cyclic ether formation during pederin biosynthesis82. The new bond formed to close the pyran ring is highlighted in red. c | Sulfur insertion catalysed by the sequential activities of cis-acting domain of unknown function (DUF) and rare cysteine lyase domains during leinamycin biosynthesis85. d | Oxygen insertion into the growing oocydin polyketide backbone catalysed by the trans-acting Baeyer–Villiger monooxygenase, OocK90. OocK and its flavin cofactor are coloured purple. Further processing of the OocK-modified moiety is proposed to be due to additional enzymatic activities in the oocydin producer. Trans-acyltransferase polyketide synthase domains catalysing the specific transformations illustrated and the resulting chemical moieties generated are highlighted in orange. ACP, acyl carrier protein; B, branching domain; DH, dehydratase; KR, ketoreductase; SH, cysteine lyase; TE, thioesterase.
Evidence of further unusual chemistry in the PKS/NRPS realm
With the precipitous drop in genetic sequencing costs in recent years, we now have access to many thousands of bacterial (meta)genomes, which are beginning to reveal the extent of chemical novelty that remains to be explored within genomic dark matter. Sivonen et al. conducted a broad analysis of 2,699 genomes covering the three domains of life to assess the frequency and distribution of PKS and NRPS pathways86. They identified 3,339 BGCs (89% in bacterial genomes), the majority of which have unknown final products. Remarkably, 10% of bacterial clusters also lacked modular architecture, hinting at the existence of overlooked NP families and raising the possibility that further cryptic PKS and NRPS systems might exist that evade detection by current bioinformatics tools. Examples like metatricycloene (13)87, which is the product of an iterative type II PKS, hint at the biosynthetic novelty that might be hidden in such non-modular pathways. In a more recent study of BGCs in the genomes of understudied soil bacteria88, 59% of predicted biosynthetic proteins had no ≥50%-length homologue in the Minimum Information about a Biosynthetic Gene cluster (MIBiG) repository10, further highlighting the potential for chemical novelty.
For trans-AT PKS pathways, the search for novel chemistry is aided by the insight that evolutionarily related ketosynthase (KS) domains usually accept intermediates bearing similar moieties around the thioester α-to-γ region89. Phylogenetic analysis can thus be utilized to identify distinct ‘orphan’ KS domain clades for which the chemical structure of the substrate intermediate thioester region is unknown, thereby revealing candidates for possible novel enzymology in preceding modules. This approach is exemplified by the recent identification of a KS domain proposed to accept an ester function as a common progenitor moiety, and led to the biochemical characterization of an unusual Baeyer–Villiger-type monooxygenase that catalyses a rare oxygen insertion during chain elongation during oocydin (14) biosynthesis by Serratia plymuthica90 (FIG. 6d). Many KS clades remain uncharacterized from orphan trans-AT BGCs and, although some will likely have evolved in parallel to those with known substrate types, many others might recognize new chemical moieties and lead to further expansions of the basic functional repertoire of PKSs.
Ongoing studies of thiotemplated pathways continue to reveal new, uncharacterized biosynthetic enzymes and domain types (for example, Alb04, albicidin, 15 (REF.91)), novel module compositions and domain organizations (for example, SxtA, saxitoxin, 16 (REF.92)) and single enzymes for multiple discrete scaffolds (for example, DynE8, dynemicin A, 17 (REFS93,94)). In addition, the first known polyketide from a strictly anaerobic bacterium, Clostridium beijerinckii, was also recently described95,96. Clostrubin (18) is a pentacyclic polyphenol that comprises a benzo[a]tetraphene ring topology that is unprecedented among NPs and, therefore, represents a novel polyketide family with an as-yet-unknown BGC. The studies described here serve to illustrate that, even among the best-studied NP classes to date, there remains much biosynthetic diversity to be discovered.
Terpenes
In contrast to thiotemplate-based assembly lines, many other NP classes are constructed by discrete enzymes that catalyse independent biosynthetic steps. With ~60,000 described compounds from 400 distinct structural families97, terpenes are the most chemically diverse NP class, yet they are assembled from simple C5 isoprenoid building blocks (BOX 1). Chemical diversity is introduced through two key mechanisms: first, cyclization of polyisoprenoid intermediates by terpene cyclases (TCs) to form monocyclic or polycyclic terpene products, and second, downstream diversification by tailoring enzymes. The assignment of cyclization topologies to TCs remains beyond the capability of modern computational methods and the identification and characterization of the enzymes themselves can also be challenging98, creating the possibility of cryptic TCs that evade bioinformatic detection. However, among those that can currently be identified, recent biochemical investigations continue to reveal a wealth of new TC-catalysed transformations hidden in bacterial genomes99,100.
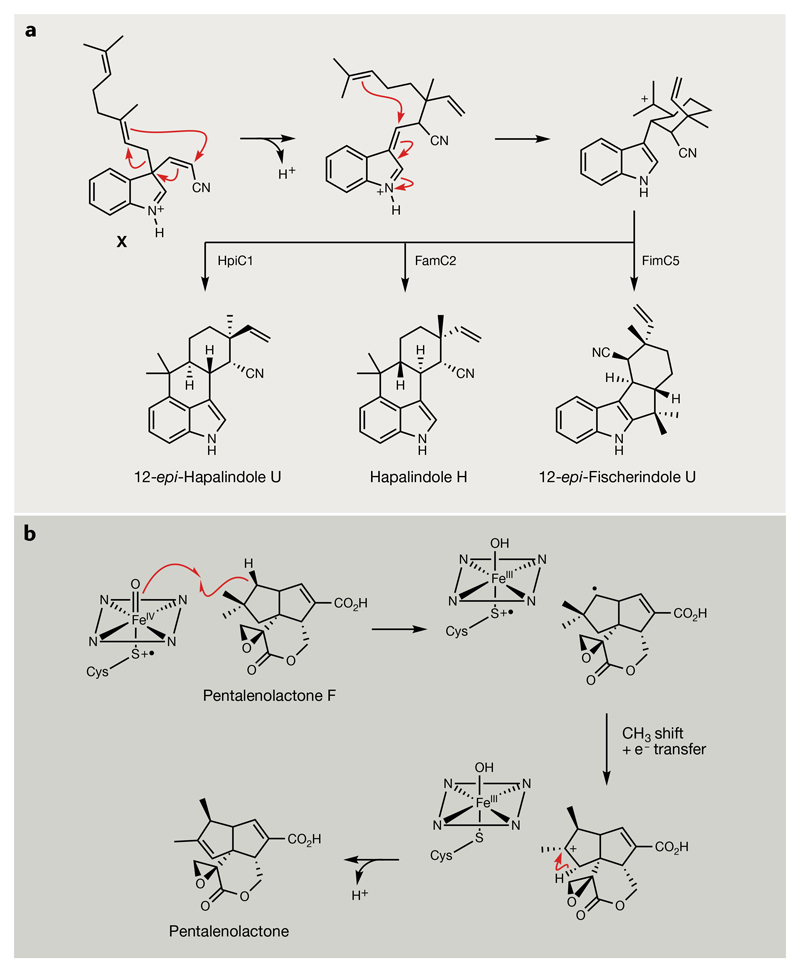
In addition to canonical TCs and their extraordinary array of cyclization patterns, unrelated enzymes can further expand the chemical diversity of terpenoids. Several noteworthy examples occur in the context of hybrid pathways that cyclize prenylated precursors101, as exemplified by the hapalindole alkaloids (19) produced by members of the Stigonematales cyanobacterial order. These alkaloids are derived from 3-geranyl-3-isocyanylvinyl indolenine, which is the product of an aromatic prenyltransferase that couples cis-3-isocyanylvinyl indole with geranyl diphosphate102–105. The resulting bicyclic intermediate (X) is the substrate for a new class of calcium-dependent ‘Stig’ cyclases that perform a remarkable series of transformations to yield the tricyclic and tetracyclic scaffolds of their respective NPs, including a rare Cope-like rearrangement, 6-exo-trig cyclization and an electrophilic aromatic substitution (FIG. 7a). This cascade provides a highly economical route to unique bioactive chemical structures with exquisite stereospecificity and regiospecificity.
Fig. 7. Unusual transformations from terpene biosynthesis.
a | Cyclization of the common 3-geranyl-3-isocyanylvinyl indolenine intermediate (X) catalysed by Stig cyclases. The remarkable transformation catalysed involves a rare Cope-like rearrangement, a 6-exo-trig cyclization and an electrophilic aromatic substitution102–105. Different Stig cyclases possess specific stereoselectivity and regioselectivity, as illustrated by the different products generated by HpiC1, FamC2 and FimC5. b | Proposed mechanism of oxidative rearrangement in the conversion of pentalenolactone F into pentalenolactone by the cytochrome P450, PntF117,229. The mechanism of transient neopentyl cation intermediate formation remains to be experimentally verified.
In order to overcome the challenges associated with identifying canonical TCs bioinformatically, profile-based hidden Markov models have been successfully applied to identify 262 TCs in bacterial genomes106. Not only are terpenoid BGCs widely distributed across bacteria but many TC families have been identified which comprise no characterized members, an observation also made during a recent phylogenetic analysis of 2,728 bacterial TC sequences107. Many of the TCs identified in these studies appear to be transcriptionally silent but their expression in an engineered Streptomyces heterologous host has enabled the identification of a number of unprecedented terpene NPs108, and novel TC families will surely continue to be discovered regularly. Indeed, it was recently shown that the organic volatile sodorifen (20) produced by S. plymuthica 4Rx13 is the product of a TC109,110, a remarkable finding given that every C atom in the bicyclic ring structure of the compound is substituted with either a methyl or methylene group, which previously obscured the biosynthetic origins of this molecule111. Unusual methylations of C5 building blocks have also been reported for the biosynthesis of a number of other structurally unique bacterial terpenes, with longestin112 (21) and the teleocidins113 (22) representing additional characterized examples.
Following cyclization, terpenoid NP scaffolds are commonly subject to further tailoring reactions, including acetylation, glycosylation and methylation. Oxidative tailoring steps are predominantly performed by members of the cytochrome P450 (CYP) superfamily, which are among the most versatile biocatalysts in nature114. CYPs have undergone significant divergent evolution to catalyse an extraordinarily broad range of transformations, the majority of which cannot be predicted bioinformatically115,116. One example is PntM, which catalyses the complex rearrangement of pentalenolactone F to pentalenolactone (23), a sesquiterpene antibiotic produced by >30 different Streptomyces species. The PntM-catalysed transformation proceeds via an oxidatively generated neopentyl cation intermediate, following substrate hydrogen abstraction and subsequent electron transfer (FIG. 7b), which is unprecedented for known CYP-catalysed reactions117. This carbocation then undergoes stereospecific migration of a methyl group to complete this unusual rearrangement.
RiPP pathways
The increasing accessibility of bacterial genomes and improvements in software algorithms for the identification of ribosomally synthesized and post-translationally modified peptide (RiPP) pathways12,15,118,119 have led to a rapid expansion in recent years in the number of previously cryptic, yet widespread, families120, all of which employ a common biosynthetic logic (BOX 1). However, many RiPP NPs carry extensive, NRP-like modifications that can obscure their ribosomal origins120–124 and small, unannotated, precursor peptide substrates remain difficult to identify. Consequently, the RiPP class might represent one of the largest untapped reservoirs of NP diversity120. Beyond variations in core peptide length and amino acid composition, a plethora of post-translational modification (PTM) enzymes can introduce chemical diversity into peptides. Such PTMs can confer considerable advantages over unmodified linear peptides, including improved target affinity and resistance to proteolytic degradation. This section focuses on two particular RiPP-associated enzyme families that have garnered a great deal of recent attention in the literature for their remarkable and synthetically challenging modifications.
YcaO proteins
While the majority of PTMs occur on amino acid side chains bearing nucleophilic functional groups, YcaO proteins represent a rare example of PTM enzymes that modify the peptide backbone125, catalysing reactions involving ATP-dependent amide activation126. The role of YcaO domain proteins in azole and azoline ring formation in a number of different RiPP families has been known for some time and the cyclodehydration reaction they catalyse (FIG. 8Aa) has been biochemically and structurally characterized in a number of RiPP family biosynthetic pathways126. Recently, however, several new peptide backbone modifications have been reported for YcaO enzymes.
Fig. 8. Post-translational modifications of ribosomally synthesized and post-translationally modified peptide natural products.
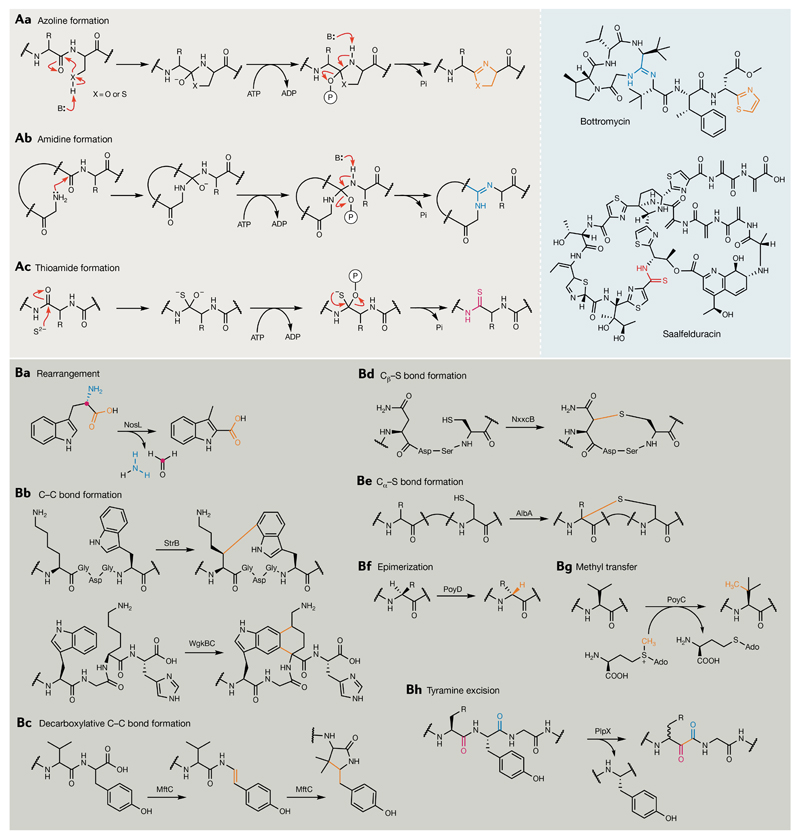
A | The three characterized ATP-dependent, amide backbone modifications currently known to be catalysed by members of the YcaO superfamily. Grey panel: azole and azoline formation (part Aa, orange), amidine formation (part Ab, blue) and thioamide formation (part Ac, magenta). Blue panel: examples of ribosomally synthesized and post-translationally modified peptides (RiPPs) comprising one or more of the above YcaO-catalysed modifications are also illustrated and each modification has been highlighted with its respective colour. B | Summary of post-translational modifications catalysed by members of the radical S-adenosyl methionine (rSAM) superfamily in the context of RiPP biosynthesis. Colours are used to highlight new bonds formed and/or the fate of particular atoms following (rSAM) enzyme activity.
The streptomycete antibiotic bottromycin (24) represents the first member of a RiPP family with characteristic macrocyclic amidine and thiazole functionalities, the latter likely the product of a YcaO cyclodehydratase. Intriguingly, the bottromycin BGC encodes two YcaO proteins, which suggests that the additional copy may perform macroamidine formation. Indeed, an untargeted metabolomics approach allowed the characterization of novel pathway intermediates and the order of reactions to be elucidated127, ultimately identifying the YcaO proteins BtmE and BtmF as being responsible for thiazole and macroamidine formation, respectively. The functions of both enzymes have also recently been confirmed biochemically128,129. The proposed macrocyclization catalysed by BtmF is analogous to cyclodehydration, with the terminal amide of the linear bottromycin precursor peptide serving as the nucleophile for attack of a downstream amide carbonyl (FIG. 8Ab). Remarkably, a recent example of a bifunctional YcaO protein, KlpD, was characterized in the biosynthesis of the ribosome inhibitor klebsazolicin (25) by Klebsiella pneumoniae130. The enzyme catalyses both azoline and macroamide formation, using a range of different nucleophiles to install three thiazole heterocycles, one oxazole cycle and a six-membered amidine ring131.
One final peptide backbone modification ascribed to YcaO family proteins during RiPP biosynthesis is the installation of thioamide bonds. During recent efforts to expand the thiopeptide family of RiPPs, Mitchell and co-workers successfully isolated and characterized saalfelduracin (26) from Amycolatopsis saalfeldensis, a structurally unique thiopeptide bearing a rare thioamide moiety132. Heterologous expression experiments confirmed the role of a YcaO–TfuA-like protein pair in thioamidation (FIG. 8Ac), thereby decrypting the previously unknown origins of the thioamide moieties present in other peptides (for example, thiopeptin and Sch 18640)132 and consistent with the involvement of YcaO–TfuA-like protein pairs in the biosynthesis of thioviridamide (27)-like RiPPs133–137. Thioamide bonds are also present in a number of other non-RiPP NPs, such as closthioamide (28), a remarkable symmetrical peptide comprising six thioamide moieties138,139, and 6-thioguanine (29), a guanosine nucleotide mimic with potent DNA-targeting and RNA-targeting cytotoxicity140,141. In these examples however, thioamide formation is catalysed by members of a novel family of alpha hydrolase (AANH)-like proteins and not YcaO proteins139,141.
Although only characterized very recently, YcaO proteins catalyse diverse, exciting, new modifications of peptide backbones. In a recent bioinformatics survey of YcaO proteins, sequence similarity network (SSN) analyses revealed the existence of many potential, uncharacterized, isofunctional YcaO protein subfamilies129, a number of which were found to occur in a broad range of different BGC contexts, hinting at a treasure trove of novel transformations that remain to be uncovered.
Radical SAM enzymes
The radical SAM (rSAM) enzymes are a rapidly expanding superfamily that currently comprises >250,000 members142,143. They are known to catalyse a diverse array of chemically challenging transformations, many of which involve the selective activation of inert C–H bonds. Each transformation is initiated by a common mechanism of radical generation via an organometallic Ω intermediate144, in which a reduced [4Fe–4S] cluster cofactor homolytically cleaves the SAM 5′–C–S bond to generate Met and a highly reactive 5′-deoxyadenosyl radical (dAdo·). This radical usually abstracts a substrate hydrogen, setting the stage for an incredibly diverse range of transformations that convert resulting substrate radicals into products. rSAM enzymes are ubiquitous in nature and are involved in many fundamental biochemical processes145 but have recently been recognized to also catalyse an astonishing range of PTMs in RiPP biosynthetic pathways146 (FIG. 8B), many of which define entire RiPP subfamilies. Here, we focus on select examples of novel chemistries that are radical in more than one sense of the word.
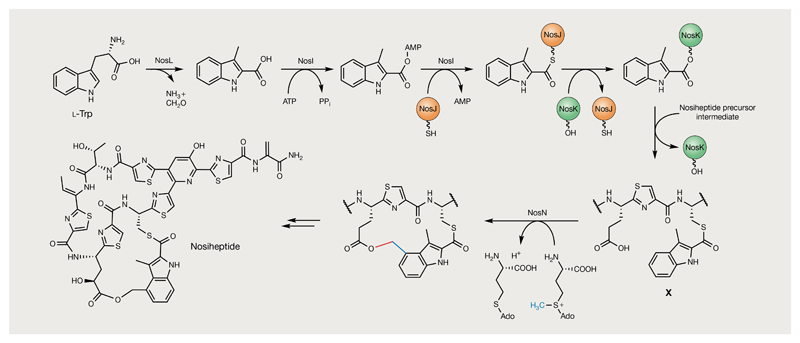
Nosiheptide (30) is a streptomycete thiopeptide antibiotic that comprises an unusual indolic side chain system, the biosynthesis of which involves a particularly remarkable succession of enzyme-catalysed transformations (FIG. 9). The indolic acid moiety is derived from an l-Trp that undergoes a complex rearrangement to 3-methyl-2-indolic acid (MIA), involving an intriguing migration of the l-Trp carboxylate to the C2 position of the indole ring with concomitant elimination of formaldehyde and ammonia, catalysed by the rSAM enzyme NosL147 (FIG. 8Ba). Unusually, NosL initiates this intricate transformation by abstracting an H-atom from the α-amino group, rather than a carbon centre148–151. The subsequent formation of the indolic side-ring system from MIA involves the action of a novel, discrete, indoyl thiolation protein that was previously unannotated (NosJ) and a unique α/β-hydrolase fold protein (NosK) that transfers the indoyl to a linear pentathiazolyl peptide intermediate Cys residue that remains selectively unmodified152–154 (FIG. 9). In order to close the ring, rSAM chemistry is again employed by NosN, a unique class C rSAM methyltransferase that catalyses not only C1 transfer to Cys-bound MIA at the C4 position but also subsequent ester bond formation between the resulting methylene moiety and Glu6 in the pentathiazolyl intermediate155. Thiopeptide BGCs have been known for almost a decade and represent one of the most extensively studied RiPP classes120; however, examples such as nosiheptide illustrate how many different novel functionalities can be harboured in just a single pathway, an enthralling prospect for future genome-mining endeavours.
Fig. 9. Formation of the side-ring system during the biosynthesis of nosiheptide.
3-Methyl-2-indolic acid (MIA) is formed by the rearrangement of l-Trp catalysed by NosL and is subsequently introduced into the nosiheptide side-ring system by NosIJK147–154. These enzymes transfer the MIA moiety to an unmodified Cys residue in a linear pentathiazolyl nosiheptide intermediate. NosN is proposed to methylate MIA at the C4 position to generate a key methylene radical intermediate that is subsequently linked via an ester bond to Glu6 in the pentathiazolyl intermediate (X)155. The S-adenosyl methionine (SAM)-derived methyl group introduced by NosN is highlighted in blue and the bond formed with Glu6 to close the nosiheptide side-ring system is highlighted in red.
In addition to complex rearrangements of RiPP scaffolds, rSAM enzymes can also catalyse completely novel mechanisms of C–C bond formation at positions distinct from those achieved by nucleophilic mechanisms156. StrB from Streptococcus thermophilus performs an intriguing crosslinking reaction during the biosynthesis of the quorum-sensing-regulated signalling peptide streptide (31)157, the founding member of a new family of RiPP NPs from streptococci158–161. StrB crosslinks non-activated C atoms in a Lys and a Trp side chain in the StrA core peptide (FIG. 8Bb) via an electrophilic aromatic substitution mechanism162 to generate a cyclic peptide product. Structural data for the StrB homologue SuiB have provided additional insight into the key role of the substrate leader sequence in positioning the core peptide in the enzyme active site for cyclization158. The StrB mechanism of C–C bond formation is distinct from that employed by MftC. The latter involves two separate substrate hydrogen abstractions and proceeds via an initial oxidative decarboxylation step to install a Val–Tyr (tyramine) crosslink in the mycobacterial signalling metabolite mycofactocin163,164 (FIG. 8Bc). SSN analysis of streptide-like BGCs present in other Streptococcus species identified a number of discrete clusters that can be defined by conserved motifs present within precursor peptides160. Characterization of rSAM PTM enzymes associated with these precursors has unveiled even further transformations, including a remarkable cyclization reaction, in which two new C–C bonds between Lys and Trp are formed160, and β-thioether bond formation161 (catalysed by WgkBC and NxxcB, respectively; FIG. 8Bb,d). Previously, rSAM-installed thioether bonds were only known to occur at α carbons in RiPP NPs165–167 (FIG. 8Be). Diverse rSAM-mediated modifications are thus clearly hidden within even highly similar BGC contexts and further exploration of the streptococcal RiPP network will likely continue to yield unusual radical chemistry.
As a final example of rSAM-based chemistry previously ‘hidden’ in the dark matter of microbial genomes, our laboratory’s own work on uncultivated microbiota of marine sponges168,169 has revealed them to be a treasure trove of novel NP enzymology. The polytheonamides (32) are highly cytotoxic, pore-forming, β-helical peptides biosynthesized by a ‘Candidatus Entotheonella’ factor symbiont of the sponge Theonella swinhoei and are among the largest and most complex NPs characterized to date123,170. Owing to their complexity, the polytheonamides were initially thought to be NRPS products, but identification of the biosynthetic genes revealed that they originate from a ribosomal pathway123. The peptide substrates in these pathways comprise a nitrile hydratase leader peptide (NHLP)-like domain, the characteristic feature of members of the poorly studied proteusin RiPP family. rSAM-based modifications are central to polytheonamide biosynthesis, which involves an astonishing cascade of ~50 PTMs that are distributed across a core peptide spanning 49 residues171. The rSAM epimerase PoyD encoded in the polytheonamide BGC is responsible for the introduction of 18 d-amino acid residues in the peptide backbone171–173 (FIG. 8Bf), resulting in an alternating l-amino acid and d-amino acid organization that allows polytheonamide to adopt its characteristic β-helical structure and is necessary for peptide solubility173,174. PoyB/C are cobalamin-dependent rSAM enzymes that, together, are predicted to catalyse 17 C-methylations on non-activated sp3 carbon centres in the PoyA core171 (FIG. 8Bg). PoyC has further been shown to catalyse up to three methylations of an N-terminal Thr residue171, concomitant with PoyF-catalysed dehydration of Thr, which, together, are necessary for formation of the unusual polytheonamide N-terminal t-butyl group that improves membrane insertion and bioactivity of these pore-forming peptides.
The Structure Function Linkage Database175 contains 113,775 rSAM protein sequences, of which >50% comprise functional domains that are yet to be assigned to a specific family. This diversity indicates a wealth of as-yet-undescribed enzymatic transformations within this rapidly growing superfamily. Indeed, in a phylogenetic profiling study conducted in 2011 (REF.176), a number of new candidate families of predicted rSAM maturation enzymes were identified, several of which remain to be functionally characterized, including a family predicted to utilize selenocysteine-containing substrates. rSAM NP PTM enzymes continue to be discovered and characterized with high frequency, and genome-mining efforts in our laboratory to uncover further proteusin-like BGCs in cyanobacterial species recently led to the discovery of a completely novel rSAM enzyme and RiPP family, the spliceotides177. These rSAM enzymes catalyse an unprecedented splicing reaction that results in the net loss of one tyramine equivalent from the peptide backbone and the introduction of an α-keto-β-amino moiety (PlpX; FIG. 8Bh). With so many uncharacterized members within this enormous superfamily, newly discovered radical transformations will surely continue to redefine what we believe to be possible chemically in nature.
Untapped chemistry in the RiPP universe
How many more RiPP families remain to be discovered? This number is very difficult to estimate but new RiPP families continue to be regularly reported in the literature, suggesting that we are only beginning to appreciate the full extent of RiPP chemical diversity. In many cases, the discovery of novel RiPPs expand the chemical diversity known for previously characterized classes, as exemplified by nosiheptide. An even more recent example is microvionin (33) from Microbacterium arborescens, the first member of a new lanthipeptide subfamily that is N-terminally lipidated with an unusual guanidino fatty acid178. The proteusin family currently only comprises the polytheonamides as representatives, yet many sequenced cyanobacterial genomes contain proteusin-like BGCs, suggesting a large, diverse and virtually uncharacterized family of NPs173. The spliceotides (putative BGCs present in 10% of cyanobacterial genomes)177 similarly represent a large family for which no end-NP is currently known, indicating vast areas of uncharted chemical space that remain to be explored in the NP universe.
Completely novel RiPP classes also continue to be discovered regularly. The pyrroloquinoline alkaloids are intriguing examples179, the biosynthetic genetic determinants of which remained cryptic for many years. The BGC for ammosamide (34) in Streptomyces sp. CNR-698 features open reading frames typically associated with lantibiotic biosynthesis, a remarkable discovery given the relatively simple structure of the final product, which is composed of a single, modified Trp residue believed to be derived from the precursor peptide180. The biochemical role of many of the other genes in the BGC remain enigmatic but include a predicted F420-dependent oxidase involved in primary amide biosynthesis, a transformation not previously known for such enzymes. The pyrroloquinoline alkaloids and other newly discovered RiPP families (for example, crocagins181, 35) illustrate the challenge of identifying precursor peptides from primary sequence data, particularly when only a limited number of residues contribute to the final product, but represent an enticing prospect in terms of new RiPP families and chemistries that await discovery in microbial (meta)genomic ‘dark matter’.
Pathways lacking signature biosynthetic genes
Nature has evolved a plethora of mechanisms for the assembly of small bioactive compounds, the BGCs of which can often remain enigmatic owing to expectations that newly discovered metabolites belong to previously characterized NP types. NRPSs, for example, are traditionally considered to be nature’s go-to machinery for catalysing amide bond formation independently of the ribosome during NP biosynthesis, but in recent years, many pathways have been characterized that employ alternative strategies for assembling amino and carboxylic acid substrates182,183. Examples include a transglutaminase homologue that catalyses N-acyl-β-peptide link formation during andrimid (36) biosynthesis in various γ-proteobacterial species184, the use of ATP-grasp enzymes such as that involved in the assembly of the β-lactone belactosin (37)185 and cyclodipeptide synthases that utilize activated aminoacyl-tRNAs to biosynthesize diketopiperazine NPs, such as bicyclomycin (38)186–188.
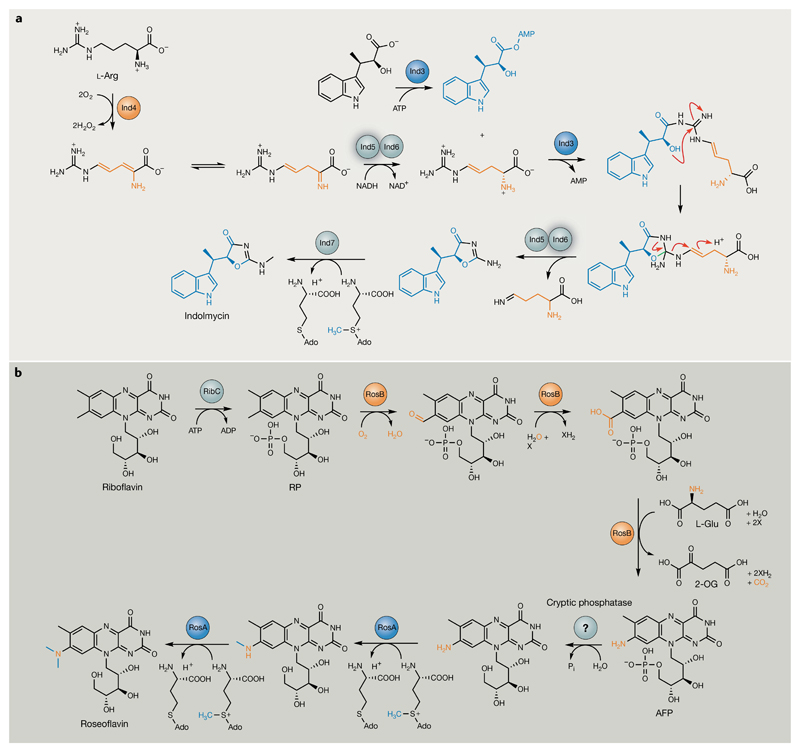
Indolmycin (39) is a potent antibacterial compound produced by various bacteria that is derived from the condensation of two nonproteinogenic amino acids, indolmycenic acid and d-4,5-dehydroarginine. However, this reaction is also not catalysed by an NRPS but by an unusual phenyl acetate CoA ligase homologue, Ind3, which additionally catalyses formation of the characteristic oxazolinone ring present in the final product189 (FIG. 10a). The pathway also comprises an unusual PLP-dependent aminotransferase, Ind4, which performs an unexpected, four-electron oxidation of l-Arg involving a non-activated C–C bond190. This O2-dependent activity is rare for PLP-dependent enzymes, which must typically protect carbanionic reaction intermediates from undesirable electrophilic attack. The indolmycin BGC remained elusive for >50 years and was finally identified not by a retro-biosynthetic rationale but by targeting the putative self-resistance gene for the antibiotic189. Resistance-guided genome mining is becoming an increasingly popular strategy of tying compounds to their cognate BGCs191–193 and, moreover, can reveal the biological significance of unusual chemistries, which is central to their downstream application.
Fig. 10. Unusual enzymology from natural product pathways that lack signature biosynthetic genes.
a | Biosynthesis of indolmycin from l-Arg and indolmycenic acid. Following Ind4 oxidation of l-Arg, the resulting unstable imine product is selectively reduced by the d-specific, NADH-dependent reductase Ind5, preventing off-pathway reactions, such as deamination, from occurring190. Ind3 catalyses an ATP-dependent condensation of d-4,5-dehydroarginine with indolmycenic acid, resulting in the formation of the indolmycin oxazoline ring (new bond highlighted in red)189. Ind6 (embedded in a complex with Ind5) performs an unusual gatekeeping role to ensure release of the correct leaving group, again to prevent the generation of off-pathway products. Ind5 and Ind6 are highlighted for their respective transformations. The N-methyltransferase Ind7 completes biosynthesis. b | Roseoflavin biosynthesis from riboflavin196. The ATP-dependent flavokinase RibC first catalyses riboflavin-5′-phosphate (RP) formation. RosB (orange) performs the subsequent three transformations to generate 8-demethyl-8-amino-riboflavin-5′-phosphate (AFP) in the presence of thiamine and l-Glu, with 2-oxoglutarate (2-OG) produced as a by-product195,196. The RibC-installed phosphate group is subsequently removed by a cryptic phosphatase, before sequential N-methylations catalysed by the S-adenosyl methionine (SAM)-dependent dimethyltransferase RosA194 (blue). X represents a cryptic hydrogen acceptor. Modifications catalysed by RosA and RosB have been highlighted in their respective colours.
In many cases, NPs are derived from the tailoring and modification of primary metabolites without any signature enzyme required for a dedicated assembly step and can thus be extremely challenging to identify. This is exemplified by the biosynthetic pathway to the antimetabolite roseoflavin (40) in Streptomyces species, which, surprisingly, only requires the activity of three enzymes to convert riboflavin-5′-phosphate (RP) into roseoflavin (FIG. 10b): a SAM-dependent dimethyltransferase (RosA), a flavodoxin-type protein (RosB) and a cryptic phosphatase194–196. RosA was identified in cell-free extract biochemical assays and performs the two terminal methylations of the 8-demethyl-8-amino-riboflavin (AF) aromatic amino group to form roseoflavin194. The remaining biosynthetic genes remained elusive for some time however, because they do not cluster with rosA in the S. davawensis genome. Consequently, rosB was only discovered following extensive cosmid library screening and systematic deletion analysis196. Although RosB contains a putative RP binding site, no function could be ascribed to the enzyme. Remarkably, biochemical analysis of recombinant RosB revealed that it is the first member of a novel enzyme class that catalyses an incredible multistep reaction involving an oxidative cascade and decarboxylation and transamination steps that convert RP and l-Glu into 8-demethyl-8-amino-RP (AFP)195,196 (FIG. 10b). Clearly, even with a reference NP structure, novel enzymology can still remain hidden from us in the known genomes of producers. This is made especially challenging when biosynthetic genes involved in a single pathway are not clustered and/or if single uncharacterized enzymes perform multiple biosynthetic steps. This realization presents a great challenge in terms of uncovering such cryptic pathways but further highlights the exciting possibility that a far greater wealth of NP chemistry remains to be uncovered in bacterial genomes than expected.
Unlocking nature’s diversity
How much novel NP enzymology awaits discovery? This question of unknown unknowns is currently almost impossible to answer, yet examples that were previously ‘hidden in plain sight’ continue to be reported in the literature with high frequency, implying that a great wealth of uncharted chemistry remains in the bacterial NP world, consistent with recent predictions197,198. The use of classical and reverse genetics techniques to map known molecules to their cognate BGCs has already resulted in the discovery of many new biochemical transformations and will continue to be an important means of unlocking nature’s diversity. However, the post-genomic era has created a deluge of sequence data that has revealed significant unexplored potential encoded in the ‘dark matter’ of bacterial (meta)genomes lying dormant in ‘silent’ BGCs, which has led to a transitioning to genes-to-molecules-based approaches to search for novel NPs.
The bottleneck of transcriptional silence continues to drive innovation and a plethora of different approaches exist to activate silent pathways199–201. Advances in analytical techniques (mass spectrometry, NMR imaging and data analysis software)202–205 allow us to better detect and characterize compounds produced in minute quantities, a major bottleneck in the field, lifting the veil on ‘silent’ NPs to further increase the number of unusual molecule leads, in addition to identifying critical pathway intermediates that may aid in deciphering cryptic pathways. The ability to isolate and characterize unstable, cryptic NPs such as colibactin206 is also improving with advances in our understanding of NP biosynthesis and its regulation, in addition to the development of reactivity-based screening protocols that target specific functional groups207,208. The integration of proteomic and transcriptomic data provides us with further information about the differential expression of BGCs209, in addition to offering a potential means by which non-clustered cryptic biosynthetic pathways can be identified and characterized. Developments in cultivation techniques210 and cultivation-independent meta-omics approaches211,212 are also providing us with greater access to poorly studied taxa that possess distinct NP profiles that might harbour novel chemistry.
One important consideration is that there is no guarantee that silent BGCs will encode novel chemistry, raising the question of how to prioritize systems for further study given that we certainly cannot experimentally characterize all NP biosynthetic pathways. While current bioinformatics tools and databases such as MIBiG10 facilitate dereplication and allow us to identify NP BGCs that encode novel end-products11–15, a significant limitation is the paucity of reliable functional annotation143, which precludes the identification of candidate enzymes and domains that catalyse novel transformations during NP biosynthesis. As illustrated in this Review, subtle changes in active-site residue composition can significantly alter the chemistry and catalysis of an enzyme, and the extension of curated annotations to other database entries based on sequence homology alone can, therefore, lead to incorrect functional predictions that leave novel enzymology effectively ‘hidden’. How, then, can we mine specifically for NP biosynthetic pathways that comprise enzymes catalysing unprecedented transformations based on in silico data alone? Will we ever be able to decrypt all NP BGCs to achieve a ‘nowhere-to-hide’ scenario for NP enzymatic novelty?
Decrypting biosynthetic ‘dark matter’
In order to mitigate the possibility of overlooking novel enzymology owing to unspecific or incorrect annotation, several approaches have been implemented to identify enzymes that have evolutionarily diverged to perform alternate functions in the context of NP biosynthesis and even offer opportunities to access cryptic pathways — significant steps towards a ‘nowhere-to-hide’ scenario.
Genomic enzymology strategies143 explore protein functional diversity in the context of entire enzyme superfamilies, allowing recognition of sequence and structure attributes that are conserved for specific functions. SSNs213 provide a holistic illustration of the pairwise relationships that result from all-by-all sequence comparisons of particular enzyme superfamilies and can be adjusted to permit the identification of isofunctional protein families. If these comprise no characterized members, then it can be indicative of a novel function. The integration of information regarding genomic neighbours can further begin to place members of uncharacterized protein families into functional contexts and could be a valuable tool for the future identification of novel BGC types214,215.
The ClusterFinder algorithm also employs a more global view to detecting NP pathways and is concerned with identifying genomic regions that are enriched in Pfam domains that occur frequently in known NP BGCs, rather than signature biosynthetic genes216. Cryptic clusters can be identified in this way because biosynthetic pathways that produce entirely different products employ members of many of the same enzyme superfamilies115,116,146,217–220. This approach was successfully applied in a systematic effort to identify and characterize BGCs encoded across 1,154 diverse bacterial genomes and led to the identification of a previously unrecognized family of arylpolyene-encoding BGCs that are distributed across a wide range of bacterial phyla216.
EvoMining221 represents a final approach, which applies an evolutionary logic based on the notion that enzymes involved in NP biosynthesis represent paralogues of primary metabolic enzymes that have undergone duplication and divergence to identify novel enzymology. Phylogenetic analysis permits the identification of functionally divergent enzymes that have been repurposed in the context of NP biosynthesis, encoded both in BGCs that employ known biosynthetic mechanisms and also in cryptic pathways that currently evade genome-mining tool detection. Using this approach, a completely novel biosynthetic pathway for arseno-organic metabolites was shown to be present in S. coelicolor and S. lividans221, indicating that chemical novelty remains to be discovered, even in the genomes of model NP producers.
Outlook
Post-genomic methods have created a new era of NP research that offers unprecedented opportunities for pharmacological and biochemical discoveries. A wealth of previously unexpected chemistry is evidenced by the examples in this Review and by recent estimations of the large numbers of structurally unique NPs that remain to be uncovered197,198. The continuous evolution of innovative bioinformatics algorithms and tools to analyse the ever-increasing volumes of sequence and spectral data we generate will be critical for the success of these efforts11 and will benefit greatly from global, community-based endeavours to effectively catalogue and curate these data and to experimentally link them to biochemical functions10,222,223. Besides investigating chemical and enzyme functions, it will be important to understand the role of NPs in bacterial ecology. Such insights will also help to reveal novel cellular targets, ecosystems that are hot spots of metabolic diversity and resources of orthogonal chemistry that create new opportunities for downstream applications in pharmacology and biotechnology.
Acknowledgements
T.A.S. is financially supported by a European Molecular Biology Organization (EMBO) Long-Term Fellowship (ALTF 344-2018). The authors are additionally grateful for funding from the European Research Council (ERC; ERC Advanced Project SynPlex), the Swiss National Science Foundation (SNF; 31003A_146992/1 and NRP 72 ‘Antimicrobial resistance’, 407240_167051), the Helmut Horten Foundation and the Promedica Foundation. The authors would also like to thank S. Leopold-Messer for comments on the figures.
Footnotes
Jörn Piel: 0000-0002-2282-8154
Author contributions
T.A.S. researched the literature and wrote the article, in addition to producing all of the figures. J.P. contributed to discussion, writing and reviewing/editing of the manuscript before submission.
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Chemistry thanks T. Simpson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Orphan
Biosynthetic genes that can be detected bioinformatically in a natural product biosynthetic gene cluster context but are functionally unassigned or have been assigned an incorrect or nonspecific function.
Cryptic
Functionally unassigned genes not previously recognized as belonging to natural product biosynthesis.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Demain AL. Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol. 2014;41:185–201. doi: 10.1007/s10295-013-1325-z. [DOI] [PubMed] [Google Scholar]
- 3.Cantrell CL, Dayan FE, Duke SO. Natural products as sources for new pesticides. J Nat Prod. 2012;75:1231–1242. doi: 10.1021/np300024u. [DOI] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oerke E-C, Dehne H-W. Safeguarding production-losses in major crops and the role of crop protection. Crop Prot. 2004;23:275–285. [Google Scholar]
- 6.Katz L, Baltz RH. Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 7.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 8.Sheldon RA, Woodley JM. Role of biocatalysis in sustainable chemistry. Chem Rev. 2018;118:801–838. doi: 10.1021/acs.chemrev.7b00203. [DOI] [PubMed] [Google Scholar]
- 9.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 10.Medema MH, et al. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat Chem Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skinnider MA, et al. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc Natl Acad Sci USA. 2016;113:E6343–E6351. doi: 10.1073/pnas.1609014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blin K, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skinnider MA, Merwin NJ, Johnston CW, Magarvey NA. PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017;45:W49–W54. doi: 10.1093/nar/gkx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tietz JI, et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol. 2017;13:470–478. doi: 10.1038/nchembio.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross H, et al. The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem Biol. 2007;14:53–63. doi: 10.1016/j.chembiol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Bernhardt P, O’Connor SE. Opportunities for enzyme engineering in natural product biosynthesis. Curr Opin Chem Biol. 2009;13:35–42. doi: 10.1016/j.cbpa.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 19.Süssmuth RD, Mainz A. Nonribosomal peptide synthesis-principles and prospects. Angew Chem Int Ed. 2017;56:3770–3821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- 20.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 21.Walsh CT, O’Brien RV, Khosla C. Nonproteinogenic amino acid building blocks for nonribosomal peptide and hybrid polyketide scaffolds. Angew Chem Int Ed. 2013;52:7098–7124. doi: 10.1002/anie.201208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott TA, Heine D, Qin Z, Wilkinson B. An L-threonine transaldolase is required for L-threo-β-hydroxy-α-amino acid assembly during obafluorin biosynthesis. Nat Commun. 2017;8 doi: 10.1038/ncomms15935. 15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaffer JE, Reck MR, Prasad NK, Wencewicz TA. β-Lactone formation during product release from a nonribosomal peptide synthetase. Nat Chem Biol. 2017;13:737–744. doi: 10.1038/nchembio.2374. [DOI] [PubMed] [Google Scholar]
- 24.Herbert RB, Knaggs AR. Biosynthesis of the antibiotic obafluorin from r-aminophenylalanine and glycine (glyoxylate) J Chem Soc Perkin Trans 1. 1992 doi: 10.1039/P19920000109. [DOI] [Google Scholar]
- 25.Deng H, Cross SM, McGlinchey RP, Hamilton JT, O’Hagan D. In vitro reconstituted biotransformation of 4-fluorothreonine from fluoride ion: application of the fluorinase. Chem Biol. 2008;15:1268–1276. doi: 10.1016/j.chembiol.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Barnard-Britson S, et al. Amalgamation of nucleosides and amino acids in antibiotic biosynthesis: discovery of an L-threonine:uridine-5′-aldehyde transaldolase. J Am Chem Soc. 2012;134:18514–18517. doi: 10.1021/ja308185q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zelyas NJ, Cai H, Kwong T, Jensen SE. Alanylclavam biosynthetic genes are clustered together with one group of clavulanic acid biosynthetic genes in Streptomyces clavuligerus. J Bacteriol. 2008;190:7957–7965. doi: 10.1128/JB.00698-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen SE. Biosynthesis of clavam metabolites. J Ind Microbiol Biotechnol. 2012;39:1407–1419. doi: 10.1007/s10295-012-1191-0. [DOI] [PubMed] [Google Scholar]
- 29.Fujimori DG, et al. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc Natl Acad Sci USA. 2007;104:16498–16503. doi: 10.1073/pnas.0708242104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blair LM, Sperry J. Natural products containing a nitrogen-nitrogen bond. J Nat Prod. 2013;76:794–812. doi: 10.1021/np400124n. [DOI] [PubMed] [Google Scholar]
- 31.Du YL, He HY, Higgins MA, Ryan KS. A heme-dependent enzyme forms the nitrogen-nitrogen bond in piperazate. Nat Chem Biol. 2017;13:836–838. doi: 10.1038/nchembio.2411. [DOI] [PubMed] [Google Scholar]
- 32.Park SR, et al. Discovery of cahuitamycins as biofilm inhibitors derived from a convergent biosynthetic pathway. Nat Commun. 2016;7 doi: 10.1038/ncomms10710. 10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sit CS, et al. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc Natl Acad Sci USA. 2015;112:13150–13154. doi: 10.1073/pnas.1515348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leipoldt F, et al. Warhead biosynthesis and the origin of structural diversity in hydroxamate metalloproteinase inhibitors. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01975-6. 1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du YL, Dalisay DS, Andersen RJ, Ryan KS. N-Carbamoylation of 2,4-diaminobutyrate reroutes the outcome in padanamide biosynthesis. Chem Biol. 2013;20:1002–1011. doi: 10.1016/j.chembiol.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 36.Qu X, et al. Cloning, sequencing and characterization of the biosynthetic gene cluster of sanglifehrin A, a potent cyclophilin inhibitor. Mol Biosyst. 2011;7:852–861. doi: 10.1039/c0mb00234h. [DOI] [PubMed] [Google Scholar]
- 37.Ma J, et al. Biosynthesis of himastatin: assembly line and characterization of three cytochrome P450 enzymes involved in the post-tailoring oxidative steps. Angew Chem Int Ed. 2011;50:7797–7802. doi: 10.1002/anie.201102305. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda K, et al. Discovery of unprecedented hydrazine-forming machinery in bacteria. J Am Chem Soc. 2018;140:9083–9086. doi: 10.1021/jacs.8b05354. [DOI] [PubMed] [Google Scholar]
- 39.Verdier-Pinard P, et al. Structure-activity analysis of the interaction of curacin A, the potent colchicine site antimitotic agent, with tubulin and effects of analogs on the growth of MCF-7 breast cancer cells. Mol Pharmacol. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 40.Chang Z, et al. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 41.Gu L, et al. Polyketide decarboxylative chain termination preceded by O-sulfonation in curacin a biosynthesis. J Am Chem Soc. 2009;131:16033–16035. doi: 10.1021/ja9071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gehret JJ, et al. Terminal alkene formation by the thioesterase of curacin A biosynthesis: structure of a decarboxylating thioesterase. J Biol Chem. 2011;286:14445–14454. doi: 10.1074/jbc.M110.214635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiers WD, Dodge GJ, Sherman DH, Smith JL, Aldrich CC. Vinylogous dehydration by a polyketide dehydratase domain in curacin biosynthesis. J Am Chem Soc. 2016;138:16024–16036. doi: 10.1021/jacs.6b09748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edwards DJ, et al. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 45.Gu L, et al. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khare D, et al. Conformational switch triggered by α-ketoglutarate in a halogenase of curacin A biosynthesis. Proc Natl Acad Sci USA. 2010;107:14099–14104. doi: 10.1073/pnas.1006738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khare D, et al. Structural basis for cyclopropanation by a unique enoyl-acyl carrier protein reductase. Structure. 2015;23:2213–2223. doi: 10.1016/j.str.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaudelli NM, Long DH, Townsend CA. β-Lactam formation by a non-ribosomal peptide synthetase during antibiotic biosynthesis. Nature. 2015;520:383–387. doi: 10.1038/nature14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long DH, Townsend CA. Mechanism of integrated β-lactam formation by a nonribosomal peptide synthetase during antibiotic synthesis. Biochemistry. 2018;57:3353–3358. doi: 10.1021/acs.biochem.8b00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaudelli NM, Townsend CA. Epimerization and substrate gating by a TE domain in β-lactam antibiotic biosynthesis. Nat Chem Biol. 2014;10:251–258. doi: 10.1038/nchembio.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keatinge-Clay AT. The uncommon enzymology of cis-acyltransferase assembly lines. Chem Rev. 2017;117:5334–5366. doi: 10.1021/acs.chemrev.6b00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minami A, Oikawa H. Recent advances of Diels-Alderases involved in natural product biosynthesis. J Antibiot. 2016;69:500–506. doi: 10.1038/ja.2016.67. [DOI] [PubMed] [Google Scholar]
- 53.Kim HJ, Ruszczycky MW, Choi SH, Liu YN, Liu HW. Enzyme-catalysed [4 + 2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fage CD, et al. The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition. Nat Chem Biol. 2015;11:256–258. doi: 10.1038/nchembio.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashimoto T, et al. Biosynthesis of versipelostatin: identification of an enzyme-catalyzed [4 + 2]-cycloaddition required for macrocyclization of spirotetronate-containing polyketides. J Am Chem Soc. 2015;137:572–575. doi: 10.1021/ja510711x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian Z, et al. An enzymatic [4 + 2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat Chem Biol. 2015;11:259–265. doi: 10.1038/nchembio.1769. [DOI] [PubMed] [Google Scholar]
- 57.Byrne MJ, et al. The catalytic mechanism of a natural Diels-Alderase revealed in molecular detail. J Am Chem Soc. 2016;138:6095–6098. doi: 10.1021/jacs.6b00232. [DOI] [PubMed] [Google Scholar]
- 58.Jeon BS, Wang SA, Ruszczycky MW, Liu HW. Natural [4 + 2]-cyclases. Chem Rev. 2017;117:5367–5388. doi: 10.1021/acs.chemrev.6b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schorn M, et al. Genetic basis for the biosynthesis of the pharmaceutically important class of epoxyketone proteasome inhibitors. ACS Chem Biol. 2014;9:301–309. doi: 10.1021/cb400699p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Owen JG, et al. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proc Natl Acad Sci USA. 2015;112:4221–4226. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keller L, et al. Macyranones: structure, biosynthesis, and binding mode of an unprecedented epoxyketone that targets the 20S proteasome. J Am Chem Soc. 2015;137:8121–8130. doi: 10.1021/jacs.5b03833. [DOI] [PubMed] [Google Scholar]
- 62.Zabala D, et al. A flavin-dependent decarboxylase-dehydrogenase-monooxygenase assembles the warhead of α,β-epoxyketone proteasome inhibitors. J Am Chem Soc. 2016;138:4342–4345. doi: 10.1021/jacs.6b01619. [DOI] [PubMed] [Google Scholar]
- 63.Jomon K, Kuroda Y, Ajisaka M, Sakai H. A new antibiotic, ikarugamycin. J Antibiot. 1972;25:271–280. doi: 10.7164/antibiotics.25.271. [DOI] [PubMed] [Google Scholar]
- 64.Hasumi K, Shinohara C, Naganuma S, Endo A. Inhibition of the uptake of oxidized low-density lipoprotein in macrophage J774 by the antibiotic ikarugamycin. Eur J Biochem. 1992;205:841–846. doi: 10.1111/j.1432-1033.1992.tb16848.x. [DOI] [PubMed] [Google Scholar]
- 65.Luo T, Fredericksen BL, Hasumi K, Endo A, Garcia JV. Human immunodeficiency virus type 1 Nef-induced CD4 cell surface downregulation is inhibited by ikarugamycin. J Virol. 2001;75:2488–2492. doi: 10.1128/JVI.75.5.2488-2492.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blodgett JA, et al. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc Natl Acad Sci USA. 2010;107:11692–11697. doi: 10.1073/pnas.1001513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Popescu R, et al. Ikarugamycin induces DNA damage, intracellular calcium increase, p38 MAP kinase activation and apoptosis in HL-60 human promyelocytic leukemia cells. Mutat Res. 2011;709–710:60–66. doi: 10.1016/j.mrfmmm.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Luo Y, et al. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4 doi: 10.1038/ncomms3894. 2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding Y, et al. Alteramide B is a microtubule antagonist of inhibiting Candida albicans. Biochim Biophys Acta. 2016;1860:2097–2106. doi: 10.1016/j.bbagen.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding Y, et al. HSAF-induced antifungal effects in Candida albicans through ROS-mediated apoptosis. RSC Adv. 2016;6:30895–30904. doi: 10.1039/C5RA26092B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malcomson B, et al. Connectivity mapping (ssCMap) to predict A20-inducing drugs and their antiinflammatory action in cystic fibrosis. Proc Natl Acad Sci USA. 2016;113:E3725–E3734. doi: 10.1073/pnas.1520289113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Antosch J, Schaefers F, Gulder TA. Heterologous reconstitution of ikarugamycin biosynthesis in E. coli. Angew Chem Int Ed. 2014;53:3011–3014. doi: 10.1002/anie.201310641. [DOI] [PubMed] [Google Scholar]
- 73.Zhang G, et al. Mechanistic insights into polycycle formation by reductive cyclization in ikarugamycin biosynthesis. Angew Chem Int Ed. 2014;53:4840–4844. doi: 10.1002/anie.201402078. [DOI] [PubMed] [Google Scholar]
- 74.Greunke C, Glöckle A, Antosch J, Gulder TA. Biocatalytic total synthesis of ikarugamycin. Angew Chem Int Ed. 2017;56:4351–4355. doi: 10.1002/anie.201611063. [DOI] [PubMed] [Google Scholar]
- 75.Scott JD, Williams RM. Chemistry and biology of the tetrahydroisoquinoline antitumor antibiotics. Chem Rev. 2002;102:1669–1730. doi: 10.1021/cr010212u. [DOI] [PubMed] [Google Scholar]
- 76.Koketsu K, Watanabe K, Suda H, Oguri H, Oikawa H. Reconstruction of the saframycin core scaffold defines dual Pictet-Spengler mechanisms. Nat Chem Biol. 2010;6:408–410. doi: 10.1038/nchembio.365. [DOI] [PubMed] [Google Scholar]
- 77.Helfrich EJ, Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2016;33:231–316. doi: 10.1039/c5np00125k. [DOI] [PubMed] [Google Scholar]
- 78.Butcher RA, et al. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc Natl Acad Sci USA. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ueoka R, Bortfeld-Miller M, Morinaka BI, Vorholt JA, Piel J. Toblerols: cyclopropanol-containing polyketide modulators of antibiosis in Methylobacteria. Angew Chem Int Ed. 2018;57:977–981. doi: 10.1002/anie.201709056. [DOI] [PubMed] [Google Scholar]
- 80.Heine D, Bretschneider T, Sundaram S, Hertweck C. Enzymatic polyketide chain branching to give substituted lactone, lactam, and glutarimide heterocycles. Angew Chem Int Ed. 2014;53:11645–11649. doi: 10.1002/anie.201407282. [DOI] [PubMed] [Google Scholar]
- 81.Sundaram S, Heine D, Hertweck C. Polyketide synthase chimeras reveal key role of ketosynthase domain in chain branching. Nat Chem Biol. 2015;11:949–951. doi: 10.1038/nchembio.1932. [DOI] [PubMed] [Google Scholar]
- 82.Pöplau P, Frank S, Morinaka BI, Piel J. An enzymatic domain for the formation of cyclic ethers in complex polyketides. Angew Chem Int Ed. 2013;52:13215–13218. doi: 10.1002/anie.201307406. [DOI] [PubMed] [Google Scholar]
- 83.Luhavaya H, et al. Enzymology of pyran ring a formation in salinomycin biosynthesis. Angew Chem Int Ed. 2015;127:13826–13829. doi: 10.1002/ange.201507090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sung KH, et al. Insights into the dual activity of a bifunctional dehydratase-cyclase domain. Angew Chem Int Ed. 2018;57:343–347. doi: 10.1002/anie.201707774. [DOI] [PubMed] [Google Scholar]
- 85.Ma M, Lohman JR, Liu T, Shen B. C-S bond cleavage by a polyketide synthase domain. Proc Natl Acad Sci USA. 2015;112:10359–10364. doi: 10.1073/pnas.1508437112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci USA. 2014;111:9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iqbal HA, Low-Beinart L, Obiajulu JU, Brady SF. Natural product discovery through improved functional metagenomics in Streptomyces. J Am Chem Soc. 2016;138:9341–9344. doi: 10.1021/jacs.6b02921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crits-Christoph A, Diamond S, Butterfield CN, Thomas BC, Banfield JF. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature. 2018;558:440–444. doi: 10.1038/s41586-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 89.Nguyen T, et al. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 90.Meoded RA, et al. A polyketide synthase component for oxygen insertion into polyketide backbones. Angew Chem Int Ed. 2018;57:11644–11648. doi: 10.1002/anie.201805363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cociancich S, et al. The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat Chem Biol. 2015;11:195–197. doi: 10.1038/nchembio.1734. [DOI] [PubMed] [Google Scholar]
- 92.Chun SW, Hinze ME, Skiba MA, Narayan ARH. Chemistry of a unique polyketide-like synthase. J Am Chem Soc. 2018;140:2430–2433. doi: 10.1021/jacs.7b13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cohen DR, Townsend CA. A dual role for a polyketide synthase in dynemicin enediyne and anthraquinone biosynthesis. Nat Chem. 2018;10:231–236. doi: 10.1038/nchem.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cohen DR, Townsend CA. Characterization of an anthracene intermediate in dynemicin biosynthesis. Angew Chem Int Ed. 2018;57:5650–5654. doi: 10.1002/anie.201802036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pidot S, Ishida K, Cyrulies M, Hertweck C. Discovery of clostrubin, an exceptional polyphenolic polyketide antibiotic from a strictly anaerobic bacterium. Angew Chem Int Ed. 2014;53:7856–7859. doi: 10.1002/anie.201402632. [DOI] [PubMed] [Google Scholar]
- 96.Behnken S, Hertweck C. Cryptic polyketide synthase genes in non-pathogenic Clostridium spp. PLoS One. 2012;7:e29609. doi: 10.1371/journal.pone.0029609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vattekkatte A, Garms S, Brandt W, Boland W. Enhanced structural diversity in terpenoid biosynthesis: enzymes, substrates and cofactors. Org Biomol Chem. 2018;16:348–362. doi: 10.1039/c7ob02040f. [DOI] [PubMed] [Google Scholar]
- 98.Yamada Y, et al. Terpene synthases are widely distributed in bacteria. Proc Natl Acad Sci USA. 2015;112:857–862. doi: 10.1073/pnas.1422108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baunach M, Franke J, Hertweck C. Terpenoid biosynthesis off the beaten track: unconventional cyclases and their impact on biomimetic synthesis. Angew Chem Int Ed. 2015;54:2604–2626. doi: 10.1002/anie.201407883. [DOI] [PubMed] [Google Scholar]
- 100.Dickschat JS. Bacterial terpene cyclases. Nat Prod Rep. 2016;33:87–110. doi: 10.1039/c5np00102a. [DOI] [PubMed] [Google Scholar]
- 101.Heide L. Prenyl transfer to aromatic substrates: genetics and enzymology. Curr Opin Chem Biol. 2009;13:171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 102.Li S, et al. Hapalindole/ambiguine biogenesis is mediated by a Cope rearrangement, C-C bond-forming cascade. J Am Chem Soc. 2015;137:15366–15369. doi: 10.1021/jacs.5b10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li S, et al. Decoding cyclase-dependent assembly of hapalindole and fischerindole alkaloids. Nat Chem Biol. 2017;13:467–469. doi: 10.1038/nchembio.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu Q, Liu X. Discovery of a calcium-dependent enzymatic cascade for the selective assembly of hapalindole-type alkaloids: on the biosynthetic origin of hapalindole U. Angew Chem Int Ed. 2017;56:9062–9066. doi: 10.1002/anie.201703932. [DOI] [PubMed] [Google Scholar]
- 105.Newmister SA, et al. Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis. Nat Chem Biol. 2018;14:345–351. doi: 10.1038/s41589-018-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA. 2008;105:7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rinkel J, Lauterbach L, Rabe P, Dickschat JS. Two diterpene synthases for spiroalbatene and cembrene A from Allokutzneria albata. Angew Chem Int Ed. 2018;57:3238–3241. doi: 10.1002/anie.201800385. [DOI] [PubMed] [Google Scholar]
- 108.Yamada Y, et al. Novel terpenes generated by heterologous expression of bacterial terpene synthase genes in an engineered Streptomyces host. J Antibiot. 2015;68:385–394. doi: 10.1038/ja.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Domik D, et al. A terpene synthase is involved in the synthesis of the volatile organic compound sodorifen of Serratia plymuthica 4Rx13. Front Microbiol. 2016;7:737. doi: 10.3389/fmicb.2016.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.von Reuss S, et al. Sodorifen biosynthesis in the rhizobacterium Serratia plymuthica involves methylation and cyclization of MEP-derived farnesyl pyrophosphate by a SAM-dependent C-methyltransferase. J Am Chem Soc. 2018;140:11855–11862. doi: 10.1021/jacs.8b08510. [DOI] [PubMed] [Google Scholar]
- 111.Weise T, et al. VOC emission of various Serratia species and isolates and genome analysis of Serratia plymuthica 4Rx13. FEMS Microbiol Lett. 2014;352:45–53. doi: 10.1111/1574-6968.12359. [DOI] [PubMed] [Google Scholar]
- 112.Ozaki T, et al. Enzymatic formation of a skipped methyl-substituted octaprenyl side chain of longestin (KS-505a): involvement of homo-IPP as a common extender unit. Angew Chem Int Ed. 2018;57:6629–6632. doi: 10.1002/anie.201802116. [DOI] [PubMed] [Google Scholar]
- 113.Awakawa T, et al. A methyltransferase initiates terpene cyclization in teleocidin B biosynthesis. J Am Chem Soc. 2014;136:9910–9913. doi: 10.1021/ja505224r. [DOI] [PubMed] [Google Scholar]
- 114.Nelson DR. Cytochrome P450 diversity in the tree of life. Biochim Biophys Acta Proteins Proteom. 2018;1866:141–154. doi: 10.1016/j.bbapap.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang X, Li S. Expansion of chemical space for natural products by uncommon P450 reactions. Nat Prod Rep. 2017;34:1061–1089. doi: 10.1039/c7np00028f. [DOI] [PubMed] [Google Scholar]
- 116.Greule A, Stok JE, De Voss JJ, Cryle MJ. Unrivalled diversity: the many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat Prod Rep. 2018;35:757–791. doi: 10.1039/c7np00063d. [DOI] [PubMed] [Google Scholar]
- 117.Duan L, Jogl G, Cane DE. The cytochrome P450-catalyzed oxidative rearrangement in the final step of pentalenolactone biosynthesis: substrate structure determines mechanism. J Am Chem Soc. 2016;138:12678–12689. doi: 10.1021/jacs.6b08610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Agrawal P, Khater S, Gupta M, Sain N, Mohanty D. RiPPMiner: a bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 2017;45:W80–W88. doi: 10.1093/nar/gkx408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van Heel AJ, et al. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Banerjee S, Hansen JN. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J Biol Chem. 1988;263:9508–9514. [PubMed] [Google Scholar]
- 122.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 123.Freeman MF, et al. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science. 2012;338:387–390. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 124.Walsh CT. Blurring the lines between ribosomal and nonribosomal peptide scaffolds. ACS Chem Biol. 2014;9:1653–1661. doi: 10.1021/cb5003587. [DOI] [PubMed] [Google Scholar]
- 125.Müller MM. Post-translational modifications of protein backbones: unique functions, mechanisms, and challenges. Biochemistry. 2018;57:177–185. doi: 10.1021/acs.biochem.7b00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Burkhart BJ, Schwalen CJ, Mann G, Naismith JH, Mitchell DA. YcaO-dependent posttranslational amide activation: biosynthesis, structure, and function. Chem Rev. 2017;117:5389–5456. doi: 10.1021/acs.chemrev.6b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Crone WJ, et al. Dissecting bottromycin biosynthesis using comparative untargeted metabolomics. Angew Chem Int Ed. 2016;55:9639–9643. doi: 10.1002/anie.201604304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Franz L, Adam S, Santos-Aberturas J, Truman AW, Koehnke J. Macroamidine formation in bottromycins is catalyzed by a divergent YcaO enzyme. J Am Chem Soc. 2017;139:18158–18161. doi: 10.1021/jacs.7b09898. [DOI] [PubMed] [Google Scholar]
- 129.Schwalen CJ, et al. In vitro biosynthetic studies of bottromycin expand the enzymatic capabilities of the YcaO superfamily. J Am Chem Soc. 2017;139:18154–18157. doi: 10.1021/jacs.7b09899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Metelev M, et al. Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel. Nat Chem Biol. 2017;13:1129–1136. doi: 10.1038/nchembio.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Travin DY, et al. Biosynthesis of translation inhibitor klebsazolicin proceeds through heterocyclization and N-terminal amidine formation catalyzed by a single YcaO enzyme. J Am Chem Soc. 2018;140:5625–5633. doi: 10.1021/jacs.8b02277. [DOI] [PubMed] [Google Scholar]
- 132.Schwalen CJ, Hudson GA, Kille B, Mitchell DA. Bioinformatic expansion and discovery of thiopeptide antibiotics. J Am Chem Soc. 2018;140:9494–9501. doi: 10.1021/jacs.8b03896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Frattaruolo L, Lacret R, Cappello AR, Truman AW. A genomics-based approach identifies a thioviridamide-like compound with selective anticancer activity. ACS Chem Biol. 2017;12:2815–2822. doi: 10.1021/acschembio.7b00677. [DOI] [PubMed] [Google Scholar]
- 134.Izawa M, Kawasaki T, Hayakawa Y. Cloning and heterologous expression of the thioviridamide biosynthesis gene cluster from Streptomyces olivoviridis. Appl Environ Microbiol. 2013;79:7110–7113. doi: 10.1128/AEM.01978-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Izumikawa M, et al. Novel thioviridamide derivative - JBIR-140: heterologous expression of the gene cluster for thioviridamide biosynthesis. J Antibiot. 2015;68:533–536. doi: 10.1038/ja.2015.20. [DOI] [PubMed] [Google Scholar]
- 136.Kawahara T, et al. Neothioviridamide, a polythioamide compound produced by heterologous expression of a Streptomyces sp. cryptic RiPP biosynthetic gene cluster. J Nat Prod. 2018;81:264–269. doi: 10.1021/acs.jnatprod.7b00607. [DOI] [PubMed] [Google Scholar]
- 137.Kjaerulff L, et al. Thioholgamides: thioamide-containing cytotoxic RiPP natural products. ACS Chem Biol. 2017;12:2837–2841. doi: 10.1021/acschembio.7b00676. [DOI] [PubMed] [Google Scholar]
- 138.Lincke T, Behnken S, Ishida K, Roth M, Hertweck C. Closthioamide: an unprecedented polythioamide antibiotic from the strictly anaerobic bacterium Clostridium cellulolyticum. Angew Chem Int Ed. 2010;49:2011–2013. doi: 10.1002/anie.200906114. [DOI] [PubMed] [Google Scholar]
- 139.Dunbar KL, et al. Genome editing reveals novel thiotemplated assembly of polythioamide antibiotics in anaerobic bacteria. Angew Chem Int Ed. 2018;57:14080–14084. doi: 10.1002/anie.201807970. [DOI] [PubMed] [Google Scholar]
- 140.Coyne S, et al. Biosynthesis of the antimetabolite 6-thioguanine in Erwinia amylovora plays a key role in fire blight pathogenesis. Angew Chem Int Ed. 2013;52:10564–10568. doi: 10.1002/anie.201305595. [DOI] [PubMed] [Google Scholar]
- 141.Litomska A, et al. Enzymatic thioamide formation in a bacterial antimetabolite pathway. Angew Chem Int Ed. 2018;57:11574–11578. doi: 10.1002/anie.201804158. [DOI] [PubMed] [Google Scholar]
- 142.Broderick JB, Duffus BR, Duschene KS, Shepard EM. Radical S-adenosylmethionine enzymes. Chem Rev. 2014;114:4229–4317. doi: 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gerlt JA. Genomic enzymology: web tools for leveraging protein family sequence-function space and genome context to discover novel functions. Biochemistry. 2017;56:4293–4308. doi: 10.1021/acs.biochem.7b00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Byer AS, et al. Paradigm shift for radical S-adenosyl-l-methionine reactions: the organometallic intermediate Ω is central to catalysis. J Am Chem Soc. 2018;140:8634–8638. doi: 10.1021/jacs.8b04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mehta AP, et al. Radical S-adenosylmethionine (SAM) enzymes in cofactor biosynthesis: a treasure trove of complex organic radical rearrangement reactions. J Biol Chem. 2015;290:3980–3986. doi: 10.1074/jbc.R114.623793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mahanta N, Hudson GA, Mitchell DA. Radical S-adenosylmethionine enzymes involved in RiPP biosynthesis. Biochemistry. 2017;56:5229–5244. doi: 10.1021/acs.biochem.7b00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang Q, et al. Radical-mediated enzymatic carbon chain fragmentation-recombination. Nat Chem Biol. 2011;7:154–160. doi: 10.1038/nchembio.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nicolet Y, Zeppieri L, Amara P, Fontecilla-Camps JC. Crystal structure of tryptophan lyase (NosL): evidence for radical formation at the amino group of tryptophan. Angew Chem Int Ed. 2014;53:11840–11844. doi: 10.1002/anie.201407320. [DOI] [PubMed] [Google Scholar]
- 149.Ji X, Li Y, Ding W, Zhang Q. Substrate-tuned catalysis of the radical S-adenosyl-l-methionine enzyme NosL involved in nosiheptide biosynthesis. Angew Chem Int Ed. 2015;54:9021–9024. doi: 10.1002/anie.201503976. [DOI] [PubMed] [Google Scholar]
- 150.Sicoli G, et al. Fine-tuning of a radical-based reaction by radical S-adenosyl-l-methionine tryptophan lyase. Science. 2016;351:1320–1323. doi: 10.1126/science.aad8995. [DOI] [PubMed] [Google Scholar]
- 151.Bhandari DM, Fedoseyenko D, Begley TP. Mechanistic studies on the radical SAM enzyme tryptophan lyase (NosL) Meth Enzymol. 2018;606:155–178. doi: 10.1016/bs.mie.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 152.Badding ED, et al. Rerouting the pathway for the biosynthesis of the side ring system of nosiheptide: the roles of NosI, NosJ, and NosK. J Am Chem Soc. 2017;139:5896–5905. doi: 10.1021/jacs.7b01497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ding W, et al. Biosynthesis of the nosiheptide indole side ring centers on a cryptic carrier protein NosJ. Nat Commun. 2017;8:437. doi: 10.1038/s41467-017-00439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Qiu Y, et al. Thiolation protein-based transfer of indolyl to a ribosomally synthesized polythiazolyl peptide intermediate during the biosynthesis of the side-ring system of nosiheptide. J Am Chem Soc. 2017;139:18186–18189. doi: 10.1021/jacs.7b11367. [DOI] [PubMed] [Google Scholar]
- 155.LaMattina JW, et al. NosN, a radical S-adenosylmethionine methylase, catalyzes both C1 transfer and formation of the ester linkage of the side-ring system during the biosynthesis of nosiheptide. J Am Chem Soc. 2017;139:17438–17445. doi: 10.1021/jacs.7b08492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yokoyama K, Lilla EA. C-C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products. Nat Prod Rep. 2018;35:660–694. doi: 10.1039/c8np00006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schramma KR, Bushin LB, Seyedsayamdost MR. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat Chem. 2015;7:431–437. doi: 10.1038/nchem.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Davis KM, et al. Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition. Proc Natl Acad Sci USA. 2017;114:10420–10425. doi: 10.1073/pnas.1703663114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schramma KR, Seyedsayamdost MR. Lysine-tryptophan-crosslinked peptides produced by radical SAM enzymes in pathogenic streptococci. ACS Chem. Biol. 2017;12:922–927. doi: 10.1021/acschembio.6b01069. [DOI] [PubMed] [Google Scholar]
- 160.Bushin LB, Clark KA, Pelczer I, Seyedsayamdost MR. Charting an unexplored streptococcal biosynthetic landscape reveals a unique peptide cyclization motif. J Am Chem Soc. 2018;140:17674–17684. doi: 10.1021/jacs.8b10266. [DOI] [PubMed] [Google Scholar]
- 161.Caruso A, Bushin LB, Clark KA, Martinie RJ, Seyedsayamdost MR. Radical approach to enzymatic b-thioether bond formation. J Am Chem Soc. 2018;141:990–997. doi: 10.1021/jacs.8b11060. [DOI] [PubMed] [Google Scholar]
- 162.Schramma KR, Forneris CC, Caruso A, Seyedsayamdost MR. Mechanistic investigations of lysine-tryptophan cross-link formation catalyzed by streptococcal radical S-adenosylmethionine enzymes. Biochemistry. 2018;57:461–468. doi: 10.1021/acs.biochem.7b01147. [DOI] [PubMed] [Google Scholar]
- 163.Bruender NA, Bandarian V. The radical S-adenosyl-l-methionine enzyme MftC catalyzes an oxidative decarboxylation of the C-terminus of the MftA peptide. Biochemistry. 2016;55:2813–2816. doi: 10.1021/acs.biochem.6b00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Khaliullin B, Ayikpoe R, Tuttle M, Latham JA. Mechanistic elucidation of the mycofactocin-biosynthetic radical S-adenosylmethionine protein, MftC. J Biol Chem. 2017;292:13022–13033. doi: 10.1074/jbc.M117.795682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Flühe L, et al. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat Chem Biol. 2012;8:350–357. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- 166.Flühe L, et al. Two [4Fe-4S] clusters containing radical SAM enzyme SkfB catalyze thioether bond formation during the maturation of the sporulation killing factor. J Am Chem Soc. 2013;135:959–962. doi: 10.1021/ja310542g. [DOI] [PubMed] [Google Scholar]
- 167.Wieckowski BM, et al. The PqqD homologous domain of the radical SAM enzyme ThnB is required for thioether bond formation during thurincin H maturation. FEBS Lett. 2015;589:1802–1806. doi: 10.1016/j.febslet.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 168.Wilson MC, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 169.Mori T, et al. Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts. Proc Natl Acad Sci USA. 2018;115:1718–1723. doi: 10.1073/pnas.1715496115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hamada T, Matsunaga S, Yano G, Fusetani N. Polytheonamides A and B, highly cytotoxic, linear polypeptides with unprecedented structural features, from the marine sponge. Theonella swinhoei. J Am Chem Soc. 2005;127:110–118. doi: 10.1021/ja045749e. [DOI] [PubMed] [Google Scholar]
- 171.Freeman MF, Helf MJ, Bhushan A, Morinaka BI, Piel J. Seven enzymes create extraordinary molecular complexity in an uncultivated bacterium. Nat Chem. 2017;9:387–395. doi: 10.1038/nchem.2666. [DOI] [PubMed] [Google Scholar]
- 172.Morinaka BI, et al. Radical S-adenosyl methionine epimerases: regioselective introduction of diverse D-amino acid patterns into peptide natural products. Angew Chem Int Ed. 2014;53:8503–8507. doi: 10.1002/anie.201400478. [DOI] [PubMed] [Google Scholar]
- 173.Morinaka BI, Verest M, Freeman MF, Gugger M, Piel J. An orthogonal D2O-based induction system that provides insights into d-amino acid pattern formation by radical S-adenosylmethionine peptide epimerases. Angew Chem Int Ed. 2017;56:762–766. doi: 10.1002/anie.201609469. [DOI] [PubMed] [Google Scholar]
- 174.Renevey A, Riniker S. The importance of N-methylations for the stability of the b6.3-helical conformation of polytheonamide B. Eur Biophys J. 2017;46:363–374. doi: 10.1007/s00249-016-1179-1. [DOI] [PubMed] [Google Scholar]
- 175.Akiva E, et al. The structure-function linkage database. Nucleic Acids Res. 2014;42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Haft DH, Basu MK. Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J Bacteriol. 2011;193:2745–2755. doi: 10.1128/JB.00040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Morinaka BI, et al. Natural noncanonical protein splicing yields products with diverse β-amino acid residues. Science. 2018;359:779–782. doi: 10.1126/science.aao0157. [DOI] [PubMed] [Google Scholar]
- 178.Wiebach V, et al. The anti-staphylococcal lipolanthines are ribosomally synthesized lipopeptides. Nat Chem Biol. 2018;14:652–654. doi: 10.1038/s41589-018-0068-6. [DOI] [PubMed] [Google Scholar]
- 179.Antunes EM, Copp BR, Davies-Coleman MT, Samaai T. Pyrroloiminoquinone and related metabolites from marine sponges. Nat Prod Rep. 2005;22:62–72. doi: 10.1039/b407299p. [DOI] [PubMed] [Google Scholar]
- 180.Jordan PA, Moore BS. Biosynthetic pathway connects cryptic ribosomally synthesized posttranslationally modified peptide genes with pyrroloquinoline alkaloids. Cell Chem Biol. 2016;23:1504–1514. doi: 10.1016/j.chembiol.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Viehrig K, et al. Structure and biosynthesis of crocagins: polycyclic posttranslationally modified ribosomal peptides from Chondromyces crocatus. Angew Chem Int Ed. 2017;56:7407–7410. doi: 10.1002/anie.201612640. [DOI] [PubMed] [Google Scholar]
- 182.Ogasawara Y, Dairi T. Biosynthesis of oligopeptides using ATP-grasp enzymes. Chemistry. 2017;23:10714–10724. doi: 10.1002/chem.201700674. [DOI] [PubMed] [Google Scholar]
- 183.Ulrich EC, van der Donk WA. Cameo appearances of aminoacyl-tRNA in natural product biosynthesis. Curr Opin Chem Biol. 2016;35:29–36. doi: 10.1016/j.cbpa.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fortin PD, Walsh CT, Magarvey NA. A transglutaminase homologue as a condensation catalyst in antibiotic assembly lines. Nature. 2007;448:824–827. doi: 10.1038/nature06068. [DOI] [PubMed] [Google Scholar]
- 185.Wolf F, et al. Biosynthesis of the β-lactone proteasome inhibitors belactosin and cystargolide. Angew Chem Int Ed. 2017;56:6665–6668. doi: 10.1002/anie.201612076. [DOI] [PubMed] [Google Scholar]
- 186.Meng S, et al. A six-oxidase cascade for tandem C-H bond activation revealed by reconstitution of bicyclomycin biosynthesis. Angew Chem Int Ed. 2018;57:719–723. doi: 10.1002/anie.201710529. [DOI] [PubMed] [Google Scholar]
- 187.Patteson JB, Cai W, Johnson RA, Santa Maria KC, Li B. Identification of the biosynthetic pathway for the antibiotic bicyclomycin. Biochemistry. 2018;57:61–65. doi: 10.1021/acs.biochem.7b00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vior NM, et al. Discovery and biosynthesis of the antibiotic bicyclomycin in distantly related bacterial classes. Appl Environ Microbiol. 2018;84:e02828–17. doi: 10.1128/AEM.02828-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Du YL, Alkhalaf LM, Ryan KS. In vitro reconstitution of indolmycin biosynthesis reveals the molecular basis of oxazolinone assembly. Proc Natl Acad Sci USA. 2015;112:2717–2722. doi: 10.1073/pnas.1419964112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Du YL, et al. A pyridoxal phosphate-dependent enzyme that oxidizes an unactivated carbon-carbon bond. Nat Chem Biol. 2016;12:194–199. doi: 10.1038/nchembio.2009. [DOI] [PubMed] [Google Scholar]
- 191.Thaker MN, et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol. 2013;31:922–927. doi: 10.1038/nbt.2685. [DOI] [PubMed] [Google Scholar]
- 192.Alanjary M, et al. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 2017;45:W42–W48. doi: 10.1093/nar/gkx360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Panter F, Krug D, Baumann S, Müller R. Self-resistance guided genome mining uncovers new topoisomerase inhibitors from myxobacteria. Chem Sci. 2018;9:4898–4908. doi: 10.1039/c8sc01325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jankowitsch F, et al. A novel N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase (RosA) catalyzing the two terminal steps of roseoflavin biosynthesis in Streptomyces davawensis. J Biol Chem. 2011;286:38275–38285. doi: 10.1074/jbc.M111.292300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jhulki I, Chanani PK, Abdelwahed SH, Begley TP. A remarkable oxidative cascade that replaces the riboflavin C8 methyl with an amino group during roseoflavin biosynthesis. J Am Chem Soc. 2016;138:8324–8327. doi: 10.1021/jacs.6b02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schwarz J, Konjik V, Jankowitsch F, Sandhoff R, Mack M. Identification of the key enzyme of roseoflavin biosynthesis. Angew Chem Int Ed. 2016;55:6103–6106. doi: 10.1002/anie.201600581. [DOI] [PubMed] [Google Scholar]
- 197.Doroghazi JR, et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG. Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci USA. 2017;114:5601–5606. doi: 10.1073/pnas.1614680114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 200.Wang B, Guo F, Dong SH, Zhao H. Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat Chem Biol. 2018;15:111–114. doi: 10.1038/s41589-018-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xu F, et al. A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat Chem Biol. 2019;15:161–168. doi: 10.1038/s41589-018-0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Mass spectrometry of natural products: current, emerging and future technologies. Nat Prod Rep. 2014;31:718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Krug D, Müller R. Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat Prod Rep. 2014;31:768–783. doi: 10.1039/c3np70127a. [DOI] [PubMed] [Google Scholar]
- 204.Gruene T, et al. Rapid structure determination of microcrystalline molecular compounds using electron diffraction. Angew Chem Int Ed. 2018;57:16313–16317. doi: 10.1002/anie.201811318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jones CG, et al. The cryoEM method microED as a powerful tool for small molecule structure determination. ACS Cent Sci. 2018;4:1587–1592. doi: 10.1021/acscentsci.8b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Balskus EP. Colibactin: understanding an elusive gut bacterial genotoxin. Nat Prod Rep. 2015;32:1534–1540. doi: 10.1039/c5np00091b. [DOI] [PubMed] [Google Scholar]
- 207.Castro-Falcón G, Hahn D, Reimer D, Hughes CC. Thiol probes to detect electrophilic natural products based on their mechanism of action. ACS Chem Biol. 2016;11:2328–2336. doi: 10.1021/acschembio.5b00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Maxson T, et al. Targeting reactive carbonyls for identifying natural products and their biosynthetic origins. J Am Chem Soc. 2016;138:15157–15166. doi: 10.1021/jacs.6b06848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Machado H, Tuttle RN, Jensen PR. Omics-based natural product discovery and the lexicon of genome mining. Curr Opin Microbiol. 2017;39:136–142. doi: 10.1016/j.mib.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Berdy B, Spoering AL, Ling LL, Epstein SS. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat Protoc. 2017;12:2232–2242. doi: 10.1038/nprot.2017.074. [DOI] [PubMed] [Google Scholar]
- 211.Wilson MC, Piel J. Metagenomic approaches for exploiting uncultivated bacteria as a resource for novel biosynthetic enzymology. Chem Biol. 2013;20:636–647. doi: 10.1016/j.chembiol.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 212.Charlop-Powers Z, Milshteyn A, Brady SF. Metagenomic small molecule discovery methods. Curr Opin Microbiol. 2014;19:70–75. doi: 10.1016/j.mib.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Copp JN, Akiva E, Babbitt PC, Tokuriki N. Revealing unexplored sequence-function space using sequence similarity networks. Biochemistry. 2018;57:4651–4662. doi: 10.1021/acs.biochem.8b00473. [DOI] [PubMed] [Google Scholar]
- 214.Zhao S, et al. Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks. eLife. 2014;3:e03275. doi: 10.7554/eLife.03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Calhoun S, et al. Prediction of enzymatic pathways by integrative pathway mapping. eLife. 2018;7:e31097. doi: 10.7554/eLife.31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Cimermancic P, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Perry C, de Los Santos ELC, Alkhalaf LM, Challis GL. Rieske non-heme iron-dependent oxygenases catalyse diverse reactions in natural product biosynthesis. Nat Prod Rep. 2018;35:622–632. doi: 10.1039/c8np00004b. [DOI] [PubMed] [Google Scholar]
- 218.Gao SS, Naowarojna N, Cheng R, Liu X, Liu P. Recent examples of a-ketoglutarate-dependent mononuclear non-haem iron enzymes in natural product biosyntheses. Nat Prod Rep. 2018;35:792–837. doi: 10.1039/c7np00067g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Du YL, Ryan KS. Pyridoxal phosphate-dependent reactions in the biosynthesis of natural products. Nat Prod Rep. 2018;36:430–457. doi: 10.1039/c8np00049b. [DOI] [PubMed] [Google Scholar]
- 220.Tolmie C, Smit MS, Opperman DJ. Native roles of Baeyer-Villiger monooxygenases in the microbial metabolism of natural compounds. Nat Prod Rep. 2018;36:326–353. doi: 10.1039/c8np00054a. [DOI] [PubMed] [Google Scholar]
- 221.Cruz-Morales P, et al. Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model Streptomycetes. Genome Biol Evol. 2016;8:1906–1916. doi: 10.1093/gbe/evw125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gerlt JA, et al. The enzyme function initiative. Biochemistry. 2011;50:9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 225.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 226.Yu F, et al. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lou L, et al. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc. 2011;133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Xu L, Wu P, Wright SJ, Du L, Wei X. Bioactive polycyclic tetramate macrolactams from Lysobacter enzymogenes and their absolute configurations by theoretical ECD calculations. J Nat Prod. 2015;78:1841–1847. doi: 10.1021/acs.jnatprod.5b00099. [DOI] [PubMed] [Google Scholar]
- 229.Wang X, Shi J, Liu Y. Oxidative rearrangement mechanism of pentalenolactone F catalyzed by cytochrome P450 CYP161C2 (PntM) Inorg Chem. 2018;57:8933–8941. doi: 10.1021/acs.inorgchem.8b00860. [DOI] [PubMed] [Google Scholar]