Introductory
T. gondii has a complex life cycle typified by an asexual development taking place in vertebrate, and a sexual reproduction which occurs exclusively in felids and thereby is less studied. The developmental transitions rely on changes in gene expression patterns, and recent studies have assigned roles for chromatin shapers, including histone modifications, in establishing specific epigenetic programs for each given stage. Here, we identified T. gondii microrchidia (MORC) protein as an upstream transcriptional repressor of sexual commitment. MORC, in partnership with Apetala (AP2) transcription factors, was shown to recruit the histone deacetylase HDAC3, thereby impeding the chromatin accessibility of the genes predestined to be exclusively expressed in sexual stages. We found that MORC-depleted cells underwent marked transcriptional changes, resulting in the expression of a specific repertoire of genes, and revealing a shift from asexual proliferation to sexual differentiation. MORC acts as a master regulator that directs the hierarchical expression of secondary AP2 factors, with these latter potentially contributing to the unidirectionality of the life cycle. Thus, MORC plays a cardinal role in the T. gondii life cycle, and its conditional depletion offers a way to study the parasite’s sexual development in vitro, and is proposed as an alternative to the requirement of cat infections.
Keywords: Toxoplasma gondii, gene expression, sexual commitment, epigenetic, chromatin remodeling, microrchidia protein
Introduction
Toxoplasma gondii is a prevalent single-celled eukaryotic parasite causing toxoplasmosis, a food-borne zoonotic disease, with life-threatening potentials in cases of immune-suppressed individuals and of unborn fetuses. T. gondii has a dynamic life cycle that alternates between multiple host species. The asexual development takes place in a wide range of mammals and birds, identified as the intermediate hosts, while the sexual reproduction occurs exclusively in felids, being the definitive hosts1.
Detailed transcriptome and proteome studies conducted throughout the life cycle have uncovered that large numbers of mRNAs/proteins were exclusively expressed in one or the other of the developmental stages. Toxoplasma has evolved a remarkably dynamic as well as precise genetic reprogramming, yet the mechanisms linking between gene expression and stage transitions remain poorly understood, mainly because of the limited access in vivo to the sexual stages and also because of current methodological limitations in maintaining these stages in cell culture.
A complex gathering several AP2 transcription factors (TFs), which emerged in Apicomplexa phylum as key regulators of life cycle transitions2,3, along with the histone deacetylase 3 (HDAC3) was suspected to operate as a chromatin-engaged molecular motor that would drive stage-specific genes expression in T. gondii4,5. Interestingly, HDAC3 co-purified with a microrchidia (MORC) homolog in T. gondii4, which thus far hasn’t been further studied. MORC proteins in plants and animals act on chromatin compaction6–12, and in this study we establish the T. gondii MORC to be a master regulator of the developmental trajectories throughout the parasite life cycle, including changes occurring over the course of sexual commitment.
Results
MORC, a highly conserved ATPase protein, forms complexes gathering HDAC3 and distinct AP2-related transcription factors
The MORC family is typified by the aggregation of a gyrase, Hsp90, histidine kinase, and MutL (GHKL) and S5 domains, together forming a catalytically active ATPase module13,14 (Fig. 1a). MORC is highly conserved across eukaryotes, excepting fungi, yet is present in apicomplexan parasites, suggesting a relatively early origin in nucleated cells15 (Fig. 1b and SI Fig. 1). From an evolutionary point of view, the apicomplexan version of the GHKL ATPase domain are actually closer to the one of animals than of plants (Fig. 1b). In this respect, Apicomplexans have a unique version of the MORC ATPase15 that is rimmed by Kelch-repeat β-propellers, and a CW-type zinc finger domain, the latter domain being reported as a histone PTM reader16 that is present in the metazoan MORC proteins as well (Fig. 1a, c).
Fig. 1. MORC, a highly conserved ATPase protein, interacts with HDAC3 in T. gondii.
a, MORC withholds 3 predicted structured domains: KELCH motifs, a conserved HATPase catalytic domain and a C-terminal CW type zinc finger domain, all displayed using cartoon secondary structures in blue, red and orange respectively. Structural predictions were modeled using the Phyre2 server and are represented by an enclosed surface cartoon diagram using UCSF Chimera. b, Schematic circular phylogram showing the relationships between MORC proteins (detailed in SI Fig. 1). The MORC domains have arisen early in eukaryotic evolution before the divergence of plants and animals. In this context, apicomplexan MORC family shares its speciation event with metazoans but not with land plants. Apicomplexan parasites have only one gene copy coding for MORC, while other species have expanded family. c, Representative domain architectures of T. gondii MORC domains and of related proteins are shown. MORC is encoded by the gene TGME49_305340. d, Nuclear location of MORC (green) and HDAC3 (red) in HFF cells infected with parasites expressing an HA-Flag (HF)-tagged copy of MORC. Cells were co-stained with Hoechst DNA-specific dye. Scale bar, 5 μm. e, MORC forms a partnership with HDAC3. Size-exclusion chromatography of MORC-containing complexes after FLAG-affinity selection. The fractions were analyzed on silver-stained SDS–PAGE gels (top) and processed with Western blots to detect MORC–HF (anti-HA) and HDAC3 (bottom). f, Mass spectrometry-based proteomic analysis of Flag elution identified MORC and its partners. The identities of the proteins are indicated on the right. g-h, the purification was repeated using HFF cells infected with a type II (Pru) strain expressing a HF-tagged copy of MORC. Flag-affinity eluates were analyzed by western blot to detect MORC–HA–FLAG and HDAC3 and by mass spectrometry-based proteomic to identify shared complexes between the type I (RH) and type II (Pru) strains. d-h All data are representative of three independent biological experiments.
MORC was originally shown to co-purify with HDAC3 in tachyzoite4. Immunofluorescence analysis of intracellular parasites revealed that HDAC3 and MORC diffusely spread throughout the nucleus in a punctate pattern and they frequently overlapped (Fig. 1d). The highly strong and specific association between MORC and HDAC3 was further confirmed by reverse immunoprecipitation and mass-spectrometry-based proteomic analysis, using knock-in parasite lines expressing a tagged version of MORC (Fig.1e-h and SI Table 1). Together, these data pointed out a cooperative action between multiple TFs (co-elution of a set of 10 AP2-related TFs, ED Fig. 1a) embedded within a high molecular-weight chromatin complex (~500-800 kDa, Fig.1e) that couples lysine deacetylation with an ATPase-mediated chromatin remodeling machinery6–7, 12.
MORC and HDAC3 co-localize extensively at the genome
To further explore how MORC and HDAC3 partner in vivo, we examined their genome-wide distributions using chromatin immunoprecipitation followed by deep sequencing (ChIPseq; GSE136060). In parallel, we generated high-resolution profiles of six permissive and repressive histone post-translational modifications (PTMs) marks in the cystogenic type II (Pru) strain (this study and 17). Overall, the inspection of these individual ChIPseq tracks revealed a strong overlap between the binding sites of the HDAC3 and MORC cistromes (Fig. 2a and SI Fig. 2a-b), with about 90% of the peaks found located at intergenic regions (Fig. 2b). Zooming into a more detailed gene level, revealed that MORC and HDAC3 consistently overlapped with hypoacetylated chromatin, which likely outcomes from the deacetylation activity of HDAC3 (SI Fig. 2c-d). At transcribed genes, MORC and HDAC3 seem to be acting as barriers to demarcate active chromatin regions typified by distinct acetylation levels, or to polarize promoter regions, thereby likely forming blueprints for the acquisition and maintenance of pre-determined gene expression profiles throughout the tachyzoite cell cycle (SI Fig. 2c-d). At many other regions, MORC and HDAC3 were co-enriched nearby silenced genes embedded in chromatin that was marked by both H3K9me3 (a repressive mark) and H3K14ac (an activation mark), a dual PTM signature reported recently as a poised chromatin state characteristic of developmentally regulated genes in T. gondii17 (SI Fig. 2e). In this respect, MORC acts as a chromatin repressor that insulates neighboring genes by potentially blocking transcriptional activators influences.
Fig. 2. HDAC3’s recruitment to the chromatin is mediated by MORC.
a, Scatterplot comparing the enrichment levels between MORC and HDAC3. ChIP-seq data from HFF cells infected with Pru MORC-HF (left) and Pru MORC-mAID-HA_KD (right) left untreated (UT) were analyzed. The x- and y-axis show the average tag count of the enrichment (n=8322 genes). Pairwise correlations were calculated using Pearson correlation coefficient (r). b, Distribution of MORC-, HDAC3- and PTM-occupied regions relatively to the T. gondii reference genome annotation. c, MORC-mAID-HA protein expression levels upon 7 hours of adding IAA, displayed by IFA on cells infected with RH MORC-mAID-HA. Fixed and permeabilized parasites were probed with MORC-mAID-HA (red) and GAP45 (green). d, Time course analysis of the expression levels of MORC-mAID-HA. Samples were taken at the indicated time periods after addition of IAA and probed with antibodies to HA and HDAC3. IAA-treated (3 days) parasites were also washed, incubated with fresh media in the absence of IAA (2 days) and subjected to a western analysis. Same experiment was repeated three times and a representative blot is displayed. e, Coexpression of MORC-mAID-HA (green) and HDAC3 (red) in RH MORC KD left untreated (UT) or treated with IAA for 16 hours. On the right, nuclear areas marked by HDAC3 and MORC at a higher magnification. f, Genome-wide MORC and HDAC3 occupancy profiles in Pru MORC KD left UT and treated with IAA for 30 hours. The average signals profiles of each protein were plotted across a -2 kb to +6 kb region with respect to T. gondii genes ATG. The y-axis shows the average tag count of the enrichment. g, IGB view of MORC and HDAC3 enrichment across T. gondii Chromosome X before and after MORC degradation, assessing the extent of the effect of MORC on the HDAC3 chromatin occupancy. The y-axis depicts read density. Experiment in c and e were conducted three times and representative images are displayed. Scale bar, 10 μm.
HDAC3 binds to chromatin in a MORC-dependent manner
We then examined whether MORC is required for the recruitment of HDAC3 to the chromatin, by investigating the genome-wide occupancy of the deacetylase, in the absence of MORC. Because of the essential requirement of MORC in T. gondii (fitness score = -4.69)18, we employed the Auxin-Inducible Degron (AID) system19, that we improved (ED Fig. 2a and SI Table 2), to acutely and reversibly deplete the MORC protein below detectable levels. The Indole-3-Acetic Acid (IAA) treatment of the edited parasites triggered a highly specific and near-complete clearance of the pool of MORC-mAID-HA protein upon 2 and 16 hours in RH and Pru strains, respectively (Fig. 2c-d and ED Fig. 2b-c).
Consequent to MORC’s degradation, HDAC3 protein levels remained relatively stable, at least at early times following the depletion (Fig. 2d) with no apparent change in its nuclear location being detected (Fig. 2e). To further investigate the functional interplay between MORC and HDAC3, we then performed ChIPseq in the context of MORC’s conditional knockdown. As expected, the IAA addition was accompanied by a near-complete loss of MORC chromatin enrichment when compared to untreated cells (Fig. 2f-g). Concomitant with the loss of MORC, we detected a severe reduction in HDAC3’s binding, with a minimal HDAC3 retention at few loci (Fig. 2f-g). The entirety of these latter was initially MORC enriched, therefore this observation could be attributed to the different antibodies withholding distinct immunoprecipitating abilities, thereby stressing the HDAC’s targeting dependency on its interaction with MORC. Remarkably, the loss of HDAC3 on chromatin produced a remarkable and selected increase in histone acetylation nearby, herein underlining the deacetylation function of the enzyme (ED Fig. 2d).
MORC’s protein depletion phenocopies HDAC3’s inhibition in inducing targeted genes expression
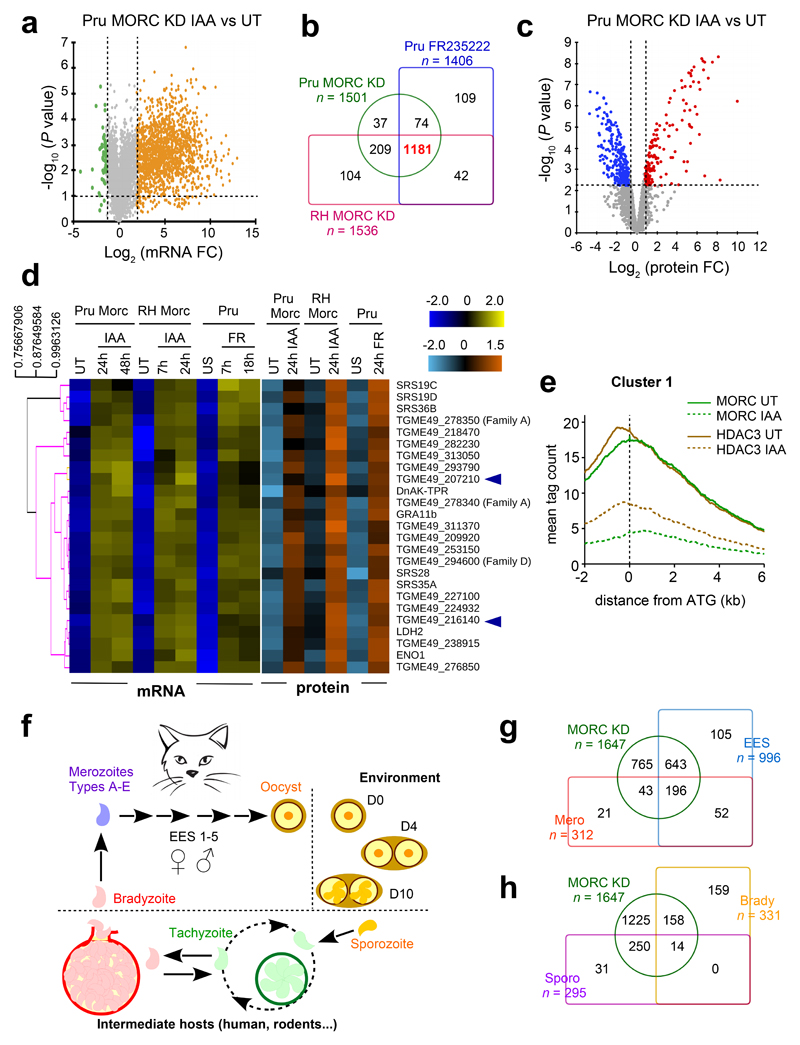
To define the effects of MORC’s depletion on transcription, we conducted detailed time-course measurements of mRNA levels, by RNA-sequencing (RNAseq), following MORC’s depletion in both RH and Pru strains (SI Table 3 and GSE136123). In parallel, we investigated the transcriptional alterations caused by the exposure of the parasites to a sub-lethal concentration of FR235222, a specific inhibitor of HDAC3’s activity5 (Fig. 3a and ED Fig. 3a). The most evident effect of either MORC’s depletion or HDAC3’s inhibition, was that a sizeable fraction of mRNAs (corresponding to ~1181 genes; Fig. 3b) accumulated gradually overtime, and this regardless of the strain type. 1647 genes, hereinafter collectively referred to as cluster 1, were specifically silenced by MORC (Fig. 3b and SI Table 4). We then investigated whether the robust expression of T. gondii genes upon MORC’s depletion was also reflected by the corresponding proteins abundance, by monitoring the proteome changes upon the addition of either IAA or FR235222 (Fig. 3c and ED Fig. 3b). For several genes, the changes in mRNA levels strongly correlated with changes in protein levels (Fig. 3d and SI Table 5). We corroborated these data by generating several knock-in parasite lines to show that MORC’s depletion or HDAC3’s inhibition was able to elicit the expression of epitope tagged target proteins (ED Fig. 3c and in the following figures). Importantly, the combined ChIPseq and RNAseq analysis revealed, at the genome-wide level, that the majority of genes in cluster 1 were direct targets of MORC and its partner HDAC3 (Fig. 3e). Effectively, the IAA addition was followed by a near-complete loss of MORC’s enrichment and a concomitant reduction of HDAC3’s occupancy in the vicinity of the 5’ ends of the cluster 1 genes (Fig. 3e); nevertheless it is worth mentioning that a significant fraction of the genes of cluster 1 (n=498, cluster 1b) was found to be displaying a lack of occupancy of MORC as well as of HDAC3 nearby (SI Fig. 3 and Supplementary discussion). Taken together, our results provide strong evidence that MORC, in partnership with HDAC3, plays a crucial and broad role in silencing the transcription of a precise set of genes in tachyzoite.
Fig. 3. MORC’s protein depletion phenocopies HDAC3’s inhibition by inducing the expression of sexual stage-specific genes.
a, Volcano plot displaying gene expression variations between the contexts of untreated (UT) versus IAA-treated (24 hours) Pru MORC KD parasites (n=8914, SI Table 3). The orange and green dots indicate the number of significantly up- and down-regulated genes, respectively, using adjusted p < 0.01 (Bonferroni-corrected) and ± 2-fold change as the cut-off threshold corresponding to each comparison. b, Venn diagram comparing upregulated mRNAs upon MORC depletion in RH and Pru strains as well as in the context of HDAC3 inhibition by FR235222. c, Volcano plot illustrating changes in protein expression between Pru MORC KD left UT and treated with IAA for 24 hours (n=1673 proteins, SI Table 5). Overexpressed proteins upon MORC depletion are indicated in red and under-expressed ones in blue. Benjamini-Hochberg FDR = 1.02 % (p-value ≤ 0.00631). d, Heatmap showing hierarchical clustering analysis (Pearson correlation) of the selected cluster 1 genes and their corresponding proteins differentially regulated upon MORC depletion through different strain/induction combinations. Parasites were left unstimulated (US) or treated with FR235222 to inhibit HDAC3. The color scale bar indicates log2 fold changes. e, Genome-wide MORC and HDAC3 occupancy profiles at cluster 1 genes in Pru MORC KD left UT and treated with IAA for 30 hours. The average signal profiles of each protein were plotted over a -2 kb to +6 kb region with respect to T. gondii genes ATG. The y-axis shows the average tag count of the enrichment. f, Scheme of T. gondii life cycle presenting the different stages of the lytic asexual cycle in bottom and of the sexual stages on the top left, with the differentiation events occurring in the corresponding hosts and environments. g-h, Venn diagrams illustrating the overlap between the MORC-regulated cluster 1 genes (n = 1647) and the RNAs detected in the different life cycle stages (data source: ToxoDB).
MORC and HDAC3 silence merozoite-primed sexual commitment
We have provided strong evidence that the cluster 1 genes were not initially expressed in tachyzoite, revealing a repressive role for MORC at this stage, and allowing us to infer that they might be expressed at other times during the life cycle (Fig. 3f). Interestingly, comparative RNA-sequencing analysis, using existing datasets generated, from early and mature bradyzoites20, merozoites21, from longitudinal studies on enteroepithelial stages (EES)22 or from three developmental stages of oocysts23, revealed that MORC’s depletion pre-dominantly induced the expression of sexually-specific expressed genes (~55% of cluster 1, including merozoite genes) and, to a lesser extent, sporozoite- (15%) and bradyzoite-specific genes (~10%) (Fig. 3g-h and SI Table 4). The enteroepithelial cycle starts two days following the ingestion of tissue cysts by the cat, during which the merozoites initiate gametes formation1 (Fig. 3f). Although the tachyzoite and the merozoite are both fast-replicating as well as being metabolically similar, yet they can be readily distinguished by their peculiar transcriptional signatures, typified by 312 transcripts that are exclusively expressed in merozoites, while 453 genes are strictly expressed in the tachyzoite stage21,24. Remarkably, ~85% of the merozoite-specific transcripts (n = 268/312) were induced upon MORC’s depletion (Fig. 4a-b and SI Table 4). This correlation is also observed in the abundance profiles of the proteins of the same cluster, including genuine merozoite markers like GRA11a and GRA11b25 and a large set of merozoite-restricted surface proteins (SRS), predicted to promote gamete development and fertilization21 (Fig. 4b). To better document this phenotype, we performed a targeted knock-in, within the MORC-mAID-HA strain of a reporter gene, TGME49_243940, of which the expression was predefined as being restricted to the merozoite stage (Fig. 4b). The IAA treatment of these edited parasites led first to the near-complete clearance of MORC as expected, and second to the concomitant accumulation of the TGME49_243940-mCherry protein (Fig. 4c). Similarly, the systematic CRISPR-mediated gene disruption of MORC’s partners resulted in the severe induction of the TGME49_243940-HA protein (ED Fig. 4a), for which the trigger was also achieved following the chemical inhibition of HDAC3 (ED Fig. 4b). As foreseen, in the MORC-depleted cells context, a substantial reduction in the chromatin occupancies of both HDAC3 and MORC was observed nearby the predefined 268 merozoite genes (Fig. 4d-e). In this regard, the loss of HDAC3’s binding resulted often in an increased levels of histone acetylation at the promoter regions, thereby inducing the transcriptional activation of merozoite-specific genes including TGME49_243940 (Fig. 4e). This acetylation pattern was mostly observed at the genes directly regulated by MORC. Together, these data position MORC as a central and early checkpoint of sexual commitment.
Fig. 4. MORC’s degradation, the same as HDAC3’s inhibition induce the expression of merozoite-restricted transcripts.
a, Venn diagram illustrating the overlap between the MORC-regulated genes in RH and in Pru strains, and the 312 genes whose expression were reported to be exclusive to merozoite. b, Heatmap showing mRNA hierarchical clustering analysis (Pearson correlation) of selected merozoite-specific genes and their corresponding proteins differentially regulated upon MORC depletion through different strain/induction combinations. The color scale bar indicates log2 fold changes. c, IFA of HFF infected with parasites harboring an mCherry endogenously tagged merozoite specific reporter gene (TGME49_243940) within the lineage RH MORC-mAID-HA. Fixed and permeabilized parasites were probed with HA (green) and mCherry (red). Scale bar, 10 μm. d, Genome-wide occupancy profiles of MORC and HDAC3 at merozoite-specific genes (n=268) in Pru MORC KD left UT and treated with IAA for 30 hours. e-f, Density profiles are displayed over representative regions of Chromosomes VI and (e) and II (f). Chromosomal positions are indicated on x-axis. The ChIP-seq profiles were obtained using antibodies directed against various histone marks, HDAC3 and HA (MORC detection) and chromatin sampled from a Pru MORC KD strain left untreated (UT) and or treated with IAA for 24 hours. RNA-seq data through different strain/induction combinations are shown in black. The y-axis depicts read density from ChIP-seq and RPKM (Reads Per Kilobase of transcript per Million mapped reads) values for RNA-seq data. The merozoite- and axonemal-specific genes TGME49_243940 (e) and TgPF16 (TGME49_297820; f) are shown in dark blue, respectively. The experiment was repeated independently twice with similar results. g, IFA of HFF infected with parasites harboring an endogenously FLAG-tagged axonemal-specific reporter gene (TgPF16) within the lineage RH MORC-mAID-HA. Fixed and permeabilized parasites were co-stained with FLAG (red) and Hoechst DNA-specific dye (blue). Scale bar, 5 μm. Experiments in c and g were conducted more than three times and representative images are displayed.
MORC and HDAC3 maintain a great number of sexual stages genes in a sustained repressive chromatin state
Following their release from the schizonts, merozoites would initiate gamete formation (Fig. 3f). Recent comparative transcriptomic study of sampled EES harvested from kittens, defined five stages with distinct transcriptional profiles (EES1-5), and ranging from the early merogony to the late sexual stages which include gametes and oocysts22. Remarkably, 51% of the genes that were found to be significantly upregulated in the MORC-depleted cells, are defined as expressed strictly during the sexual cycle, which is well above the EES specific proportion of the entire genome (estimated to ~12%)22 (Fig. 3g and SI Table 4). Cluster 1 is greatly enriched in a subset of genes coding for micro- and macrogamete-specific proteins holding putative important roles regarding fertilization and oocyst wall formation (ED Fig. 4c-d and ED Fig. 5a-b).
The male gamete of T. gondii is characterized by the presence of two flagella; which resemble, in many aspects, that of eukaryotic sperm22. The majority, if not all of the proteins behind microgametes main features, such as those involved in the axonemal cytoskeleton (one basal body protein, three Radial Spoke Protein, eleven dyneins) and in the intraflagellar transport (six IFT proteins, nine kinesins), were labeled as being regulated targets of MORC (ED Fig. 6). TgPF16, a sperm-associated antigen26–28, displayed a direct MORC/HDAC3-mediated gene silencing (Fig. 4f). To examine the localization of TgPF16, we transiently expressed TgPF16 fused to a C-terminal Flag within the MORC-mAID-HA parasite line. Upon IAA addition, TgPF16 was induced and found associated to growing cytoplasmic axonemes, and occasionally, the protein was detected along the emerging flagella of a presumably male gamete, with a patchy distribution resembling the described PbPF16 labeling in Plasmodium28 (Fig. 4g). MORC also regulates the Hapless 2 (HAP2) gene which expression was shown to be restricted to male gametes in different organisms, ranging from protists to flowering plants and insects29,30 (ED Fig. 5c). HAP2 is required in both Plasmodium and Toxoplasma for the gamete’s fusion and the subsequent fertilization22,31,32. Even though the sexual development occurs in several flavors, yet it seems that the alternation between the haploid and diploid states, the progression from one differentiated stage to the other, as well as the maintenance of a given stage epigenetic signature, are strictly controlled by MORC and HDAC3.
MORC and HDAC3 silence oocyst-specific gene expression
We next explored whether MORC’s depletion stimulated, with a magnitude as strong as the aforementioned observations, the expression of oocyst-specifically expressed genes, and that was by comparing the data we produced to the reported transcriptomes stemming from three time points, representing immature (at day 0), maturing (at day 4) and mature (at day 10) stages of the oocyst development, respectively23 (Fig. 3f). Remarkably, ~89% (n= 264/295) of the genes defined as being strictly expressed in the oocysts, were found to be induced in the MORC-depleted cells, which is well above the oocyst-specific proportion of the entire genome (corresponding to roughly 4%)23 (Fig. 3h, SI Fig. 4, and SI Table 4). In this respect, the acute degradation of MORC was sufficient to induce almost all the proteins that are usually present in fully sporulated oocysts (Fig. 5a), a result we have substantiated by detecting, in the MORC-depleted cells, a G1/19 mAb-reactive sporozoite-specific protein33 (Fig. 5b). This was also corroborated by our proteomic analysis which identified, with high confidence, SporoSAG34, the hallmark surface antigen in sporozoites (Fig. 5a and SI Table 5). The SporoSAG locus displays the canonical bivalent chromatin state “poised” for transcription, along with a local co-occupancy of MORC and HDAC3 (ED Fig. 7a). At this locus and at all loci sharing the specificity for this same stage, we observed a concomitant loss of MORC and of its partner HDAC3 from the chromatin upon MORC’s depletion (Fig. 5c and ED Fig. 7). Notably, the induction of SporoSAG’s expression reached an unmatched level (mRNA levels > 8,000 RPKM) following either the sustained degradation of MORC or the inhibition of HDAC3 (ED Fig. 7a). A larger group of proteins following similar expression patterns were uncovered (Detailed in Supplementary discussion and SI Fig. 4 and ED Fig. 7), emphasizing a critical role of MORC and HDAC3 in the development of oocysts, a key stage for the spread of T. gondii.
Fig. 5. MORC regulates developmental transitions at multiple checkpoints.
a, Heatmap showing mRNA hierarchical clustering analysis (Pearson correlation) of selected oocyst/sporozoite-specific genes and their corresponding proteins differentially regulated upon MORC depletion through different strain/induction combinations. The abundance of their transcripts in the various stages of development, namely tachyzoite, bradyzoite/cyst, merozoite, EES and oocyst was displayed. The color scale bar indicates log2 fold changes. b, Expression of a sporozoite-specific protein upon MORC depletion in RH strain was measured by IFA. Parasites were probed with G1/19 mAb-reactive sporozoite-specific protein (in red) and GAP45 (green). c, Genome-wide MORC and HDAC3 occupancy profiles at sporozoite-specific genes (n=264) in Pru MORC KD left UT and treated with IAA for 30 hours. d, Expression levels of bradyzoite- and cyst-specific markers upon MORC depletion in RH strain were measured by IFA. Fixed and permeabilized parasites were probed with CC2, BCLA or BAG1 (green) along with Dolichos biflorus Agglutinin (DBA) lectin (red). e, Time course analysis of expression levels of bradyzoite (BCLA and BAG1)- and merozoite (TGME49_243940)-specific markers after MORC-mAID-HA depletion. Samples were taken at the indicated time periods after addition of IAA and probed with antibodies against HA, BCLA, BAG1 and mCherry. The experiment was repeated three times and a representative blot is presented. f-g, IFA of HFF infected with MORC-HA-mAID RH parasites expressing a targeted knock-in merozoite specific gene (TGME49_243940). Immunofluorescence analysis of individual vacuoles upon MORC depletion, with a co-staining using a bradyzoite marker (BCLA, green) or a merozoite marker (red, TGME49_243940-mCherry). h, the differential expression of the two markers was assessed by counting the number of vacuoles (n= 100) labeling one or the other markers or both, over a time course. Experiments in b, d, f and g were conducted more than three times and representative images are displayed. Scale bars, 10 μm for b and g and 5 μm for d and f.
MORC depletion triggers a tachyzoite-to-bradyzoite switch, a mandatory step towards sexual development
Once in a cat’s enterocyte, the bradyzoite would convert into merozoite, priming distinctly the initiation in vivo of the sexual cycle (Fig. 3f). Despite the evidence we brought of MORC acting as a repressor at strategic points along the life cycle continuum, it still remained to be determined whether the commitment into merozoite, that is triggered in vitro by MORC’s depletion, would also require the compulsory passage through the bradyzoite stage1. In this respect, we identified a significant, yet a limited number of bradyzoite-specific genes (n=172, Fig. 3h) which expression was induced in the MORC-depleted cells (ED Fig. 8a). These genes are coding for surface antigens (BAG1, BSR4/p36, BRP1), metabolic enzymes (LDH2, ENO1) and cyst-wall associated proteins (CST1, CC2, BCLA) (Fig. 5d-e, ED Fig. 8 and SI Fig. 5a-c) and they seem to be regulated by MORC in a similar manner to the precedingly described genes. In this regard, they display, at the vicinity of their promoters, the co-binding of MORC and HDAC3, which disappeared with the loss of MORC, resulting in their transcriptional induction (ED Fig. 8b and SI Fig. 5a-c). Additionally, we have created transgenic parasite strains expressing tagged versions of bradyzoite-specific enzymes ENO1 and LDH2, and showed that their expression was silenced in a HDAC3-dependent manner (SI Fig. 5a-b). The cyst wall is highly glycosylated and stains easily with the Dolichos biflorus Agglutinin (DBA) lectin. We observed a clear DBA accumulation at the MORC-depleted vacuoles, likely as a consequence of the CST1-SRS44 induction, a gene known to code for a cyst wall protein responsible for DBA’s reactivity35 (Fig. 5d). Furthermore, the induction of BCLA, a hallmark of latent tissue cyst (Dard et al. unpublished), was compared to the induction of the merozoite marker TGME49_243940. Intriguingly, TGME49_243940 protein level increased at the very beginning of MORC’s degradation, peaked at 48 hours and declined sharply afterwards, whereas BCLA was only detectable far after the complete clearance of MORC, and its expression level was sustained overtime (Fig. 5e). Immunofluorescence analysis of individual vacuoles, revealed a non-homogeneous labeling following MORC’s degradation, displaying a great number of vacuoles expressing either BCLA or TGME49_243940-mCherry, in an almost mutually exclusive manner (Fig. 5f-h). This observation is consistent with the initial findings which stated that roughly half (=159/331; Fig. 3h) of bona fide bradyzoite genes were still kept repressed in the MORC-depleted cells. One would speculate that this partial induction of bradyzoite-specific genes could be due to some repressor acting downstream of the MORC pathway. In this regard, MORC-depleted cells expressed AP2IX-9, a transcriptional repressor initially suspected to restrict Toxoplasma’s commitment towards developing mature bradyzoite tissue cysts36 (Fig. 6a). These data provide a solid baseline for a model in which MORC’s degradation induces AP2IX-9 to prevent the merozoite from converting back into a bradyzoite, hence elucidating the molecular mechanisms behind the maintenance of the unidirectionality of the sexual stage development (Fig. 3f).
Fig. 6. MORC guides developmental trajectories by employing downstream regulating pathways.
a, Heatmap showing mRNA hierarchical clustering analysis (Pearson correlation) of AP2 transcription factors shown to be regulated by MORC depletion. The abundance of their transcripts in the various stages of development, namely tachyzoite, bradyzoite/cyst, merozoite, enteroepithelial stages (EES) and oocyst stages is displayed. The color scale bar indicates log2 fold changes. The AP2 factors were color coded correspondingly with the color of the life stage during which they are the most expressed. b, IFA of HFF infected with parasites harboring an endogenously FLAG tagged merozoite-specific AP2IV-3 within the lineage RH MORC-mAID-HA. Fixed and permeabilized parasites were probed with FLAG (red) and Hoechst DNA-specific dye (blue). Scale bar, 10 μm. The experiment was conducted more than three times and representative images are displayed. c, Heatmap showing mRNA hierarchical clustering analysis (Pearson correlation) of selected MORC-regulated genes through different strain/induction combinations. The presented genes belong to the set predefined as exclusively expressed in tachyzoites and repressed in merozoites. The color scale bar indicates log2 fold changes. d, Genome-wide H4K31me1 enrichment profiles at cluster 1 genes in Pru MORC KD left UT and treated with IAA for 30 hours. e, Average nucleosome occupancy plotted based on the relative distance to the nearest annotated ATG, using Danpos250 to define the most likely position of nucleosomes. Nucleosomes occupancy at all T. gondii genes (on top) or on the MORC-regulated cluster 1 genes (in bottom) in the context of MORC depletion are shown. f, Density profiles are displayed at the merozoite-specific TGME49_238915 gene. MORC’s and HDAC3’s occupancies, the H4K31 methylation enrichment, the corresponding transcript level (black), are all displayed before and after MORC’s depletion. Similarly, the surrounding nucleosome occupancy and distribution (in purple) are displayed based on the MNase-seq data that were conducted before and after MORC degradation. Chromosomal positions are indicated on x-axis. The experiment was repeated independently twice with similar results. g, A proposed model describing the mechanism underlying the MORC complex-mediated modulations of the T. gondii life cycle.
MORC depletion induces a downstream network of secondary transcriptional regulators to guide developmental trajectories
Besides balancing bradyzoite gene expression, MORC’s depletion was shown to concomitantly restrain sets of genes defined as being expressed exclusively in tachyzoite and not in merozoite (ED Fig. 9 and Fig. 6c)21. In view of the small number of genes found to be repressed following the absence of MORC (n=126 genes, Fig. 3a and SI Table 3), one could argue in favor of this transcriptional output resulting from secondary transcription factors. In fact, four AP2 factors (indicated in blue, Fig. 6a) displayed the same restricted mRNA patterns, both in the context of MORC’s depletion and in the merozoite stage, thereby they might be acting as transcriptional activators in tachyzoite. In our current model, the MORC controlled developmental trajectories are likely directed downstream by a hierarchical expression of other regulators, which in turn, would dictate the setting of a distinct pre-determined transcriptional program for a given stage (Fig. 6g). For instance, three MORC-regulated AP2 factors were strictly expressed in merozoite (indicated in green, Fig. 6a). A representative of these potential regulators, AP2IV-3, which is clearly induced in the MORC-depleted cell (Fig. 6b and ED Fig. 10a), shares a homologous DNA binding domain with AP2-G (ED Fig. 10b-c), the master transcriptional regulator of gametocytogenesis in Plasmodium falciparum37, thus underlining a possible convergence of function within the phylum. The serial expression, upon MORC depletion, of as many as 17 AP2, including ones which expression is restricted to micro/macrogametes or sporozoites (Fig. 6a) suggests a sequential transcriptional modulation mediated by AP2 TFs throughout the life cycle (Fig. 6g). Even though the functions of AP2s as activators or repressors remain to be determined, yet they still represent tenable candidates for apprehending the priming for a permissive or for a transcriptionally silent state of chromatin, respectively. In this regard, chromatin opening was synergistically enhanced by the deposition of H4K31me1 on gene bodies upon MORC depletion (Fig. 6d, f and SI Fig. 6a-d), a mark that was associated with the progression of the elongating RNA polymerase II in T. gondii17. Nucleosomes positioning also plays a critical role in gene transcriptional regulation38. In fact, a genome-wide nucleosome mapping allowed us to explore the genomic properties of the chromatin upon MORC’s degradation. Overall, nucleosomes distribution was not affected by MORC’s depletion, however their occupancy, especially in 5′ upstream gene regions (SI Fig. 7a-f), increased drastically within the cluster 1 gene group (Fig. 6e), and this correlated positively with an increase in the mRNA levels of the downstream genes (Fig. 6f). Importantly, the MNase-protected footprints gave rise to similar increase patterns regarding the pan-acetylated histone H4 ChIP-control (SI Fig. 7g). This is somehow counterintuitive, as transcription factors would usually bind nucleosome-depleted regions as well as the higher-expressing genes would tend to have greater degrees of nucleosomes depletion at their promoter regions. Yet, few exceptions were reported, such as the ones concerning pioneering factors which preferentially bind regions of high nucleosome occupancy38–40. This apparent increase of occupancy is further deliberated in the Supplementary Discussion. These results considered, we provide evidence that MORC is a critical determinant of nucleosomes positioning, yet whether this effect stems from its intrinsic ATP-dependent remodeling activity, as reported in human and plant cells6,10,41, remains to be determined.
Discussion
In this study, we provide compelling evidence that MORC is a crucial regulator on multiple levels throughout the T. gondii life cycle. In this respect, MORC’s acute degradation allowed us to map, in great detail, the gene expression changes occurring during the sexual development, revealing a role for MORC and for HDAC3 in gene silencing. We came out with a model in which MORC establishes multiple partnerships with HDAC3 along with primary AP2 TFs (ED Fig. 1a) in order to control genetic regulatory circuits, with these latter consisting of clusters of secondary AP2 TFs (Fig. 6a) which, in turn, would influence the gene expression of the particular stage they belong to (Fig. 6g). Therefore, the diversity of the MORC-AP2 TFs complexes might be holding a layer of regulation of the differential cell fate switches. How MORC is implicated in the cell fate decision remains enigmatic. This could be adjusted by the external cues from the different host cells environments that the parasite faces throughout its life cycle, leading potentially to a more adapted genetical reprogramming of the different stage specific signatures (see Supplementary Discussion).
The study of the sexual stages of T. gondii harbors technical difficulties, and the demand for alternatives to cat models is increasing due to the rising ethical challenges42. Within the same vein of stem cell research, this study paves the way for an in vitro epigenetic reprogramming of the T. gondii sexual development, using MORC as a cornerstone.
Methods
Parasites and human cell culture
Human primary fibroblasts (HFFs, ATCC® CCL-171™) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen) supplemented with 10% heat inactivated Fetal Bovine Serum (FBS) (Invitrogen), 10 mM (4-(2-hydroxyethyl)-1-piperazine ethanesulphonic acid) (HEPES) buffer pH 7.2, 2 mM L-glutamine and 50 μg/ml of penicillin and streptomycin (Invitrogen). Cells were incubated at 37°C in 5% CO2. The Toxoplasma strains used in this study and listed in SI Table 2 were maintained in vitro by serial passage on monolayers of HFFs. The cultures were free of mycoplasma, as determined by qualitative PCR.
Reagents
The following primary antibodies were used in the immunofluorescence, immunoblotting and/or ChIP assays: rabbit home-made anti-BCLA, rabbit anti-TgHDAC3 (RRID: AB_2713903), rabbit anti-H4K31ac (RRID: AB_2811024), rabbit anti-H4K31me1 (RRID: AB_2811025), rabbit anti-TgGAP45 (gift from Pr. Dominique Soldati, University of Geneva), rat anti-CC2 (gift from W. Bohne and U. Gross), mouse anti-TgBAG1, mouse anti-G1/19 (gift from Pr. Gereon Schares, Friedrich-Loeffler-Institut), mouse anti-HA tag (Roche, RRID: AB_2314622), rabbit anti-HA Tag (Cell Signaling Technology, RRID: AB_1549585), rabbit anti-mCherry (Cell Signaling Technology, RRID: AB_2799246), rabbit anti-FLAG (Cell Signaling Technology, RRID: AB_2798687), rabbit anti-H3K14ac (Diagenode, RRID:AB_2713906), H3K9me3 (Millipore, RRID:AB_916348), H3K4me3 (Diagenode, RRID:AB_2616052), rabbit Anti-acetyl-Histone H4, pan (Lys 5,8,12) (Millipore, RRID:AB_310270). Immunofluorescence secondary antibodies were coupled with Alexa Fluor 488 or Alexa Fluor 594 (Thermo Fisher Scientific). Secondary antibodies used in Western blotting were conjugated to alkaline phosphatase (Promega) or horseradish peroxidase.
Auxin induced degradation
Depletion of MORC-AID-HA was achieved with 3-Indoleacetic acid (IAA, Sigma-Aldrich # 45533) as described by Brown KM et al., 201719. A stock of 500 mM IAA dissolved in 100% EtOH at 1:1,000 was used to deplete mAID-tagged proteins at a final concentration of 500 μM. Mock treatment consisted of an equivalent volume of 100% EtOH at a final concentration of 0.0789%, wt/vol. To monitor the degradation of AID-tagged proteins, parasites grown in HFF monolayers were treated with auxin, or ethanol alone, for different time intervals at 37 °C. Following treatment, parasites were harvested and analyzed by Immunofluorescence or Western blotting. The MORC complete clearance was successful upon 2 hours and 16 hours of IAA treatment in the type I (RH) and type II (Pru), respectively. For the RNA-sequencing analysis, IAA induction in type I took place for both 7 and 24 hours before the RNA extraction, and in type II the timings were fixed to 24 and 48 hours upon treatment. For the proteome-wide analysis, the treatment took place for 24h in RH strain and 36h for PRU strain. For the ChIP-seq analysis, the treatment took place for 24h in RH strain and for 32h for the PRU strain.
HDAC3 inhibition using FR235222
For the RNA-seq essays, FR235222 was used at a final concentration of 25 nM for 7h and 18h; for the IFA of reporter genes, it was used at 100 nM for 18 to 24h, and for the proteome the concentration was fixed at 25 nM and proteins were extracted 24 hours upon treatment.
Immunofluorescence microscopy
T. gondii infecting HFF cells grown on coverslips were fixed in 3% formaldehyde for 20 min at room temperature, permeabilized with 0.1% (v/v) Triton X-100 for 15 min and blocked in Phosphate buffered saline (PBS) containing 3% (w/v) BSA. The cells were then incubated for 1 hour with primary antibodies followed by the addition of secondary antibodies conjugated to Alexa Fluor 488 or 594 (Molecular Probes). Nuclei were stained for 10 min at room temperature with Hoechst 33258 at 2 μg/ mL in PBS. After four washes in PBS, coverslips were mounted on a glass slide with Mowiol mounting medium, and images were acquired with a fluorescence ZEISS ApoTome.2 microscope and images were processed by ZEN software (Carl Zeiss, Inc.).
Western blot
Immunoblot analysis of protein was performed as described in Bougdour et al., 201343. Briefly, ~107 cells were lysed in 50 μl lysis buffer (10 mM Tris-HCl, pH6.8, 0.5 % SDS [v/v], 10% glycerol [v/v], 1 mM EDTA and protease inhibitors cocktail) and sonicated. Proteins were separated by SDS-PAGE, transferred to a polyvinylidene fluoride membrane (Immobilon-P; EMD Millipore) by liquid transfer, and Western blots were probed using appropriate primary antibodies followed by alkaline phosphatase or horseradish peroxidase-conjugated goat secondary antibodies. Signals were detected using NBT-BCIP (Amresco) or enhanced chemiluminescence system (Thermo Scientific).
Plasmid construction
The plasmids and primers for gene of interest (GOI) used in this work are listed in SI Table 2. To construct the vector pLIC-GOI-HAFlag, the coding sequence of GOI was amplified using primers LIC-GOI-Fwd and LIC-GOI-Rev using T. gondii genomic DNA as template. The resulting PCR product was cloned into the pLIC-HF-dhfr or pLIC-mCherry-dhfr vectors using the Ligation Independent Cloning (LIC) cloning method43. The plasmid pTOXO_Cas9-CRISPR was described in Sangaré et al., 201644. Twenty mers-oligonucleotides corresponding to specific GOI were cloned using Golden Gate strategy. Briefly, primers GOI-gRNA-Fwd and GOI-gRNA-Rev containing the sgRNA targeting GOI genomic sequence were phosphorylated, annealed and ligated into the pTOXO_Cas9-CRISPR plasmid linearized with BsaI, leading to pTOXO_Cas9-CRISPR::sgGOI. Just two transgenic components are needed to implement the auxin-inducible degron (AID) system, a plant auxin receptor called transport inhibitor response 1 (TIR1) and a POI tagged with an AID. We engineered a type I RHΔku80 and a type II lines of T. gondii to stably express Tir1 from Oryza sativa tagged with Ty and controlled by a promoter selected for a moderate expression of the chimeric protein. The plasmid pModProm1-TiR1-TY1-3DHFR (DNA sequence in SI Table 2) was DNA synthetized and then cloned in pUC57 simple by Genscript. The chimeric construct was inserted within the UPRT locus. We also created a pLIC vector containing a codon-optimized for T. gondii expression DNA block with the mAID from Arabidopsis thaliana auxin-responsive protein IAA17E66-S133, as defined in Brown KM, 201719, in frame with a HA tag (SI Table 2).
Toxoplasma gondii transfection
T. gondii strains were electroporated with vectors in cytomix buffer (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/ KH2PO4 pH 7.6, 25 mM HEPES pH7.6, 2 mM EGTA, 5 mM MgCl2) using a BTX ECM 630 machine (Harvard Apparatus). Electroporation was performed in a 2 mm cuvette at 1.100V, 25 Ω and 25 μF. When needed, the antibiotic (concentration) used for drug selection was chloramphenicol (20 μM), mycophenolic acid (25 μg/ml) with xanthine (50 μg/ml), pyrimethamine (3 μM), or 5-fluorodeoxyuracil (10 μM). Stable transgenic parasites were selected with the appropriate antibiotic, single-cloned in 96 well plates by limiting dilution and verified by immunofluorescence assay or genomic analysis.
Chromatographic purification of MORC-containing complex
T. gondii extracts from RH∆ku80 or Pru∆ku80 cells stably expressing HAFlag-tagged MORC, were incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 1 hour at 4°C. Beads were washed with 10-column volumes of BC500 buffer (20 mM Tris-HCl, pH 8.0, 500 mM KCl, 20% glycerol, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, and protease inhibitors). Bound polypeptides were eluted stepwise with 250 μg/ml FLAG peptide (Sigma Aldrich) diluted in BC100 buffer. For size-exclusion chromatography, protein eluates were loaded onto a Superose 6 HR 10/30 column equilibrated with BC500. Flow rate was fixed at 0.35 ml/min, and 0.5-ml fractions were collected.
Mass spectrometry-based Interactome analyses
Protein bands were excised from colloidal blue stained gels (Thermo Fisher Scientific) before in-gel digestion using modified trypsin (Promega, sequencing grade) as previously described45. Resulting peptides were analyzed by online nanoliquid chromatography (UltiMate 3000 and UltiMate 3000 RSLCnano, Thermo Scientific, for respectively RH and Pru strains) coupled to tandem MS (LTQ-Orbitrap Velos Pro and Q-Exactive Plus, Thermo Scientific, for respectively RH and Pru strains). Peptides were sampled on a 300 μm x 5 mm PepMap C18 precolumn and separated on a 75 μm x 250 mm C18 PepMap column (Thermo Scientific) using 25-min gradients. MS and MS/MS data were acquired using Xcalibur (Thermo Scientific). Peptides and proteins were identified using Mascot (version 2.6) through concomitant searches against the Toxoplasma gondii database (ME49 taxonomy, version 30 downloaded from ToxoDB46, the Uniprot database (Homo sapiens taxonomy, March 2019 version), classical contaminant database and the corresponding reversed databases. Trypsin was chosen as the enzyme and two missed cleavages were allowed. Precursor and fragment mass error tolerances were set at respectively 10 ppm and 0.6 Da. Peptide modifications allowed during the search were: Carbamidomethyl (C, fixed), Acetyl (Protein N-term, variable) and Oxidation (M, variable). The Proline software (http://proline.profiproteomics.fr) was used to filter the results: conservation of rank 1 PSMs, PSM homology threshold ≤ 0.01, PSM score ≥ 25, and minimum of 1 specific peptide per identified protein group. Proline was then used to perform a compilation and grouping of the protein groups identified in the different samples. Proteins from the contaminant database were discarded from the final list of identified proteins. MS1-based label free quantification of the protein groups was performed using Proline to infer intensity-based absolute quantification (iBAQ) values that were used to rank identified Toxoplasma proteins in the interactomes. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016846.
Mass spectrometry-based Proteome-wide analyses
HFF cells were grown to confluence and infected with type I (RHΔku80 or RHΔku80 MORC KD) or type II (PruΔku80 or PruΔku80 MORC KD) strains before lysis in 8M urea and 50mM HEPES. Extracted proteins were reduced using 20mM of dithiothreitol for 1h at 37°C before alkylation with 55mM of iodoacetamide for 45 min at room temperature in the dark. The samples were then diluted using ammonium bicarbonate to obtain a urea concentration of 4M. Proteins were digested with LysC (Promega) at a ratio of 1:200 during 4h at 37°C. The samples were diluted again using ammonium bicarbonate to obtain a urea concentration of 1M. Proteins were then digested with Trypsin (Promega) at a ratio of 1:200 overnight at 37°C. Resulting peptides were purified by C18 reverse phase chromatography (Sep-Pak C18, Waters) before drying down before fractionation by tip-based strong cation exchange (3M Empore). For this, peptides were dissolved in 5% acetonitrile, 1% TFA and eluted in 4 fractions (F1: 100 mM ammonium acetate, 20% ACN, 0.5 % formic acid; F2: 175 mM ammonium acetate, 20% ACN, 0.5 % formic acid; F3: 375 mM ammonium acetate, 20% ACN, 0.5 % formic acid; F4: 80% acetonitrile, 5% ammonium hydroxide) before desalting using C18 reverse phase chromatography (Ultra-Micro SpinColumns, Harvard Apparatus). Technical triplicates were performed. NanoLC-MS/MS analyses were performed using an Ultimate 3000 RSLCnano coupled to a Q-Exactive Plus (Thermo Scientific). Peptides were sampled on a 300 μm × 5 mm PepMap C18 precolumn and separated on a 75 μm × 250 mm PepMap C18 column (Thermo Scientific) using a 120-min gradient. MS and MS/MS data were acquired using the Xcalibur software (Thermo Scientific). RAW files were processed using MaxQuant48 version 1.6.2.10. Spectra were searched against the Toxoplasma gondii database (ME49 taxonomy, version 30 downloaded from ToxoDB46, the Uniprot database (Homo sapiens taxonomy, April 2019 version), the frequently observed contaminants database embedded in MaxQuant47, and the corresponding reverse databases. Trypsin was chosen as the enzyme and two missed cleavages were allowed. Precursor and fragment mass error tolerances were set at their default values. Peptide modifications allowed during the search were: Carbamidomethyl (C, fixed), Acetyl (Protein N-term, variable) and Oxidation (M, variable). Minimum number of peptides and razor + unique peptides were set to 1. Maximum false discovery rates were set to 0.01 at PSM and protein levels. The match between runs option was activated. Statistical analyses were performed using ProStaR48. Peptides and proteins identified in the reverse and contaminant databases or matching to human sequences were discarded. Only proteins quantified in at least 3 replicates of one condition were conserved. After log2 transformation, protein intensities were normalized using the summed intensities of a pool of proteins for which transcripts were found invariable in RNAseq dataset (0.91 < fold change < 1.1). Missing values were then imputed (slsa method for Partially Observed Values and DetQuantile with quantile set to 2.5 and factor set to 1 for Missing Entirely in the Condition). Statistical testing was conducted using limma. Differentially abundant proteins were sorted out using the following cut-offs: log2(fold change) ≥ 0.8 or ≤ -0.8 and p-values allowing to reach an FDR ~1% according to the Benjamini-Hochberg estimator. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016845.
Chromatin Immunoprecipitation and Next Generation Sequencing in Toxoplasma gondii
HFF cells were grown to confluence and infected with type II (Pru∆ku80 or Pru∆ku80 MORC KD) strains. Harvested intracellular parasites were crosslinked with formaldehyde (final concentration 1%) for 8 min at room temperature and the crosslinking was stopped by addition of glycine (final concentration 0.125M) for 5 min at room temperature. Crosslinked chromatin was lysed in ice-cold lysis buffer (50 mM HEPES KOH pH7.5, 14 0mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.125% triton X-100, protease inhibitor cocktail) and sheared in shearing buffer (1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 10 mM Tris pH 8.0, protease inhibitor cocktail) by sonication using a Diagenode Biorupter. Samples were sonicated, for 16 cycles (30 seconds ON and 30 seconds OFF), to 200-500 base-pair average size. Immunoprecipitation was carried out using sheared chromatin, 5% BSA, protease inhibitor cocktail, 10% triton X-100, 10% deoxycholate, DiaMag Protein A-coated magnetic beads (Diagenode) and antibodies targeting histone PTM or protein of interest. A rabbit IgG antiserum was used as a control mock. After overnight incubation at 4°C on rotating wheel, chromatin-antibody complexes were washed and eluted from beads by using iDeal ChIP-seq kit for Histones (Diagenode) according to the manufacturer’s protocol. Samples were de-crosslinked by heating for 4 hours at 65°C. DNA was purified by using IPure kit (Diagenode) and quantified by using Qubit Assays (Thermo Fisher Scientific) according to the manufacturer's protocol. For ChIP-seq, purified DNA was used to prepare libraries and then sequenced by Arraystar (USA).
Library Preparation, Sequencing and Data analysis (Arraystar)
ChIP-Sequencing library preparation was performed according to Illumina’s protocol Preparing Samples for ChIP Sequencing of DNA. Library Preparation: 10 ng DNA of each sample was converted to phosphorylated blunt-ended with T4 DNA polymerase, Klenow polymerase and T4 polymerase (NEB); An ‘A’ base was added to the 3' end of the blunt phosphorylated DNA fragments using the polymerase activity of Klenow (exo minus) polymerase (NEB); Illumina's genomic adapters were ligated to the A tailed DNA fragments; PCR amplification was performed to enrich ligated fragments using Phusion High Fidelity PCR Master Mix with HF Buffer (Finnzymes Oy). The enriched product of ~200-700 bp was cut out from gel and purified. Sequencing: The library was denatured with 0.1M NaOH to generate single-stranded DNA molecules, and loaded onto channels of the flow cell at 8pM concentration, amplified in situ using TruSeq Rapid SR cluster kit (#GD-402-4001, Illumina). Sequencing was carried out by running 100 cycles on Illumina HiSeq 4000 according to the manufacturer’s instructions. Data Analysis: After the sequencing platform generated the sequencing images, the stages of image analysis and base calling were performed using Off-Line Basecaller software (OLB V1.8). After passing Solexa CHASTITY quality filter, the clean reads were aligned to T. gondii reference genome (Tgo) using BOWTIE (V2.1.0). Aligned reads were used for peak calling of the ChIP regions using MACS V1.4.0. Statistically significant ChIP-enriched regions (peaks) were identified by comparison of two samples, using a p-value threshold of 10-5. Then the peaks in each sample were annotated by the overlapped gene using the newest T. gondii database. The EXCEL/BED format file containing the ChIP-enriched regions was generated for each sample. Data visualization: The mapped 100 bp reads represent enriched DNA fragments by ChIP experiment. Any region of interest in the raw ChIP-seq signal profile can be directly visualized in genome browser. We use 10-bp resolution intervals (10-bp bins) to partition the stacked reads region, and count the number of reads in each bin. All the 10 bp resolution ChIP-seq profiles of each sample are saved as UCSC wig format files, which can be visualized in T. gondii Genome Browser (http://protists.ensembl.org/Toxoplasma_gondii/Info/Index). All these raw and processed files can be found at Gene Expression Omnibus (GEO) Series GSE136060.
RNA-seq and sequence alignment
Total RNAs were extracted and purified using TRIzol (Invitrogen, Carlsbad, CA, USA) and RNeasy Plus Mini Kit (Qiagen). RNA quantity and quality were measured by NanoDrop 2000 (Thermo Scientific). RNA integrity was assessed by standard non-denaturing 1.2% TBE agarose gel electrophoresis. The ribosomal large subunits from Toxoplasma and the host cells was used to verify that the ratio between Toxoplasma RNA versus host RNA was equivalent between the different biological samples, thus indicating that the samples had equivalent infection rates. For each condition, total RNAs from two independent biological replicates were pooled to prepare cDNA libraries which were then sequenced using Illumina technology in a single replicate dataset. RNA-sequencing was performed by GENEWIZ (South Plainfield, NJ, USA). Briefly, the RNA quality was checked with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, California, USA) and Illumina TruSEQ RNA library prep and sequencing reagents were used following the manufacturer's recommendations (Illumina, San Diego, CA, USA). The samples were paired-end multiplex sequenced (2 × 125 bp) on the Illumina Hiseq 2500 platform and generated at least 70 million reads for each sample. The RNA-Seq reads (FASTQ) were processed and analyzed using the Lasergene Genomics Suite version 14 (DNASTAR, Madison, WI, USA) using default parameters. The paired-end reads were uploaded onto the SeqMan NGen (version 14, DNASTAR. Madison, WI, USA) platform for reference-based assembly using either the Mus musculus genome package (GRCm38.p3) or the Toxoplasma Type II ME49 strain (ToxoDB-24, ME49 genome) as reference template. The ArrayStar module (version 14, DNASTAR. Madison, WI, USA) was used for normalization, differential gene expression and statistical analysis of uniquely mapped paired-end reads using the default parameters. The expression data quantification and normalization were calculated using the RPKM (Reads Per Kilobase of transcript per Million mapped reads) normalization method. All these raw and processed files can be found at GEO Series GSE136123.
MNase Digestion
15 min formaldehyde cross-linking was performed and the optimal MNase digestion time was determined. Nuclei were mildly sonicated to free cross-linked nucleosomes from nuclear membranes. Soluble chromatin was collected and de-cross-linked directly (MNase-Seq) or used for chromatin immunoprecipitation. Chromatin was then resuspended in 1 mL pre-warmed Buffer M containing 320 mM of sucrose and 1 mM CaCl2, after which 1U MNase (Sigma) was added. The Mnase incubation time was set to 15min at 37°C and the reaction was stopped by adding EDTA (10 ul of 0.5M/ 1 ml of extract). DNA was extracted from nucleosomes by eliminating using Proteinase K (PK) for 1hour at 37C, this was followed by a standard phenol-chloroform-isoamyl alcohol DNA extraction and the DNA was then NaCl/ethanol precipitated with the addition of a final glycogen precipitation step. DNA was then purified by using IPure kit (Diagenode) and quantified by using Qubit Assays (Thermo Fisher Scientific) according to the manufacturer's protocol. Purified DNA was used to prepare libraries and then sequenced by Arraystar (USA). Positioned nucleosomes were called using the DANPOS software (Version 2.2.1) as described previously in Chen K et al. 201349.
Phylogenetic analysis of HATPase domain-containing MORC proteins
The HATPase sequences were identified by a blastp analysis using the Swissprot database. The sampled HATPase domains were aligned using ClustalW software. Neighbor-joining tree was computed using the ClustalX software.
Software and Statistical analyses
Volcano plots, scatter plots, and histograms were generated with Prism 7. Sample sizes were not predetermined and chosen according to previous literature. All experiments were performed in biological replicates to allow for statistical analyses. No method of randomization was used. All experiments were performed in independent biological replicates as stated for each experiment in the manuscript. All corresponding treatment and control samples from ChIP-seq and RNA-seq were processed at the same time to minimize technical variation. Investigators were not blinded during the experiments. Experiments were performed in biological replicates and provided consistent statistically relevant results.
Extended Data
Extended Data Fig. 1. Domain architectures and expression levels during the life cycle of MORC and partners.
a, Representative domain architectures of T. gondii MORC and partners identified by mass spectrometry-based proteomic are shown. Domains were predicted by SMART and PFAM: ELM2 (Egl-27 and MTA1 homology 2), PHD (plant homeodomain), and AP2 (APETALA2) b, Heatmap representation of MORC and partners gene expression in different life cycle stages (source: ToxoDB). Gene expression values were mean log2 transformed and median centered for clustering. Transcriptomic data from tachyzoite, merozoite, longitudinal studies on enteroepithelial stages (EES1 to EES5), immature (day 0), maturing (day 4) and mature (day 10) stages of oocyst development and cysts from chronically infected mice were used. c, Mapping of domains and identified phosphorylation and ubiquitination sites detected by mass-spectrometry (source: ToxoDB).
Extended Data Fig. 2. mAID-based MORC inducible KD system successfully ablated its expression on the protein post-translational level in both type I (RH) and type II (Pru) strains.
a, Auxin-inducible degradation system for controlling protein stability in T. gondii. We first engineered RH and Pru strains ectopically expressing the plant auxin receptor called transport inhibitor response 1 (TIR1). We chose the UPRT locus to integrate TIR1 under the control of a promoter allowing a mild expression of the protein tagged to Ty. We utilized a mini-AID (mAID) tagging LIC system for conditional MORC depletion. The resulting cell lines are referred to as MORC-KD hereafter. Conditional depletion of MORC-mAID-HA is reliant on auxin (IAA), TIR1, and the proteasome. b, Depletion of MORC-mAID-HA, upon addition of IAA for 16 hours, was measured by IFA in cells infected with Pru MORC KD. Fixed and permeabilized parasites were probed with MORC-mAID-HA (red) and GAP45 (green). MORC-depleted cells displayed strong inhibition of proliferation when compared to untreated cells. Yet, this growth defect phenotype was completely reversed upon IAA washout, indicating that MORC depletion while impeding the cell cycle progression does not kill the parasites. Graph on the right, in situ quantification of nuclear MORC-mAID-HA using IFA. The horizontal bars represent the mean ± s.d. of the nuclear MORC intensity from three independent experiments (n = 50 nuclei per dot). The p-values were calculated using one-way ANOVA. Scale bar, 10 μm. c, in situ quantification (related to Fig. 2c) of nuclear MORC-mAID-HA using IFA in RH MORC KD as described above. d, Smoothed and background-subtracted tag density profiles are displayed over representative regions of Chr. V (top) and X (bottom). The ChIP-seq profiles were obtained with antibodies directed against pan-acetylated histone H4, HDAC3 and HA (MORC detection) from chromatin sampled from a Pru MORC KD strain left untreated (UT) and or treated with IAA for 24 hours. The experiment was repeated independently twice with similar results.
Extended Data Fig. 3. MORC protein depletion, and HDAC3 inhibition induces gene expression.
a, Volcano plots showing gene expression differences identified from comparison of UT versus FR235222 in Pru MORC KD (left graph) and of UT versus IAA (24 hours) in RH MORC KD (right graph) (n=8914 genes, SI Table 3). The orange and green dots indicate the number of significantly up- and down-regulated genes, respectively, using adjusted p < 0.01 (Bonferroni-corrected) and ± 2-fold change as the cut-off threshold corresponding to each comparison. X-axis showing log2 fold change, Y-axis showing -log10(p value). b, Volcano plots illustrating changes in protein expression between UT versus FR235222 in Pru MORC KD (left graph, n=2566 proteins, Benjamini-Hochberg FDR = 1 %, p-value ≤ 0.00794) and of UT versus IAA (24 hours) in RH MORC KD (right graph, n=2139 proteins, Benjamini-Hochberg FDR = 1.01%, p-value ≤ 0.00501). Overexpressed proteins upon MORC depletion are indicated in red and under-expressed ones in blue. c, Pru strains were engineered to endogenously epitope tag with HA two MORC-regulated genes, TGME49_207210 and TGME49_216140. Expression was monitored following HDAC3 chemical inactivation by FR235222 and HA staining. Scale bars, 10 μm. Experiments were conducted more than three times and representative images are displayed.
Extended Data Fig. 4. MORC alongside HDAC3 represses the expression of sexual stages-specific genes.
a, A type II (ME49) strain was engineered to endogenously expressed a HA tag version of the merozoite gene TGME49_238915. Genetic inactivation of HDAC3, MORC and AP2-XII-1 promote TGME49_243940 protein expression (in red). The efficiency of genetic disruption in Cas9-expressing parasites was monitored by cas9-GFP expression (in green). Scale bar, 5 μm. b, Expression of TGME49_243940 was monitored following HDAC3 chemical inactivation by FR235222 and HA staining (in red). Scale bar, 10 μm. c, Heatmap showing hierarchical clustering analysis of selected MORC-regulated genes from cluster 1 through different strain/induction combinations, including the abundance of their transcripts in the various stages of development, namely tachyzoite, bradyzoite/cyst, merozoite, enteroepithelial stages (EES) and oocyst stages. The color scale bar indicates log2 fold changes. d, Expression of TGME49_316130 was monitored following HDAC3 chemical inactivation by FR235222 and HA staining (in red). Scale bar, 10 μm. Experiments in a, b and d were conducted more than three times and representative images are displayed.
Extended Data Fig. 5. MORC KD derepresses proteins involved in merozoite and fertilization.
a, Smoothed and background-subtracted tag density profiles are displayed over Chr. VI. The ChIP-seq profiles were obtained with antibodies directed against various histone marks, HDAC3 and HA (MORC detection) from chromatin sampled from a Pru MORC KD strain left untreated (UT) and or treated with IAA for 24 hours. RNA-seq data through different strain/induction combinations are shown in black. The y-axis depicts read density from ChIP-seq and RPKM values for RNA-seq data. The merozoite-specific gene TGME49_238915 is shown in dark blue. b, Pru strain was engineered to endogenously expressed HA epitope-tagged TGME49_238915. Expression was monitored following HDAC3 chemical inactivation by FR235222 and HA staining. Scale bar, 10 μm. Experiment was conducted more than three times and representative images are displayed. c, Density profiles are displayed on Chromosome V at the TgHAP2 (TGME49_285940) locus. d, Density profiles are displayed on Chromosome X around a merozoite-specific cluster of tandemly repeated genes (SRS48 family). (a, c-d) The experiment was repeated independently twice with similar results. Chromosomal positions are indicated on x-axis.
Extended Data Fig. 6. MORC regulates microgamete related genes including those coding for flagella components.
Heatmap representation of RNA-seq data portraying the number of genes involved in microgamete biology, alongside their levels of expression upon MORC depletion/HDAC3 inhibition through different strain/induction combinations. The abundance of their transcripts in the various stages of development, namely tachyzoite, bradyzoite/cyst, merozoite, enteroepithelial stages (EES) and oocyst stages was displayed. The color scale bar indicates log2 fold changes. The genes were divided in sets grouping together genes involved in axonemal cytoskeleton, and genes that harbor potential domains involved in intraflagellar transportation.
Extended Data Fig. 7. MORC depletion induces the expression of genes involved in oocyst wall formation and coding for sporozoite-specific markers.
a, Density profiles are displayed on Chromosome VIIb at the SporoSAG/SRS28 (TGME49_258550) locus, encoding for the hallmark surface antigen in sporozoite. The ChIP-seq profiles were obtained with antibodies directed against various histone PTMs, HDAC3 and HA (MORC detection) from chromatin sampled from a Pru MORC KD strain left UT and or treated with IAA for 24 hours. RNA-seq data through different strain/induction combinations are shown in black. The y-axis depicts read density from ChIP-seq and RPKM values for RNA-seq data. b, Density profiles are displayed on Chromosome XI at the TgHowp1 (TGME49_316890) locus. TgHOWP1 is a representative example of proteins expressed in the early stages of wall formation as revealed by the accumulation of its transcripts in both late developmental EES5 and in immature (D0) oocyst (ToxoDB data). c, Density profiles are displayed on Chromosome VIIb at the TGME49_262110 locus. Expression of TGME49_262110 was monitored following HDAC3 chemical inactivation by FR235222 and HA staining (in green). Experiments was conducted more than three times and representative images are displayed. Scale bar, 5 μm. (a-c) The experiment was repeated independently twice with similar results. Chromosomal positions are indicated on x-axis.
Extended Data Fig. 8. MORC depletion derepresses partly bradyzoite-specific genes.
a, Heatmap showing hierarchical clustering analysis of selected bradyzoite-specific and MORC-regulated genes from cluster 1 through different strain/induction combinations. The abundance of their transcripts in the various stages of development, namely tachyzoite, bradyzoite/cyst, merozoite, enteroepithelial stages (EES) and oocyst stages was displayed. The color scale bar indicates log2 fold changes. b, Density profiles are displayed on Chromosome VIIb at the Bag1 (TGME49_259020) locus, encoding for the hallmark surface antigen in bradyzoite. The ChIP-seq profiles were obtained with antibodies directed against various histone PTMs, HDAC3 and HA (MORC detection) from chromatin sampled from a Pru MORC KD strain left UT and or treated with IAA for 24 hours. RNA-seq data through different strain/induction combinations are shown in black. The y-axis depicts read density from ChIP-seq and RPKM values for RNA-seq data. The experiment was repeated independently twice with similar results. Chromosomal positions are indicated on x-axis.
Extended Data Fig. 9. MORC depletion causes indirect repression of tachyzoites specific genes.
a-b, Density profiles are displayed at the tachyzoite-specific ROP16 (a) and RON2 (b) genes. The ChIP-seq profiles were obtained with antibodies directed against various histone marks, HDAC3 and HA (MORC detection) from chromatin sampled from a Pru MORC KD strain left UT and or treated with IAA for 24 hours. RNA-seq data through different strain/induction combinations are shown in black. The y-axis depicts read density from ChIP-seq and RPKM values for RNA-seq data. The experiment was repeated independently twice with similar results. Chromosomal positions are indicated on x-axis.
Extended Data Fig. 10. MORC regulates the expression of AP2IV-3, a Plasmodium falciparum AP2-G homologous protein.
a, Density profiles are displayed at the merozoite-specific AP2IV-3 gene. The ChIP-seq profiles were obtained with antibodies directed against various histone marks, HDAC3 and HA (MORC detection) from chromatin sampled from a Pru MORC KD strain left UT and or treated with IAA for 24 hours. RNA-seq data through different strain/induction combinations are shown in black. The y-axis depicts read density from ChIP-seq and RPKM values for RNA-seq data. The experiment was repeated independently twice with similar results. Chromosomal positions are indicated on x-axis. b, Representative domain architectures of T. gondii AP2IV-3 and P. falciparum AP2-G, displaying the approximate AP2 domain position within the protein sequence. c, Protein alignment of the AP2IV-3 and AP2-G respective AP2 domains, with the amino acid homology shown in red.
Supplementary Material
Acknowledgments
We are grateful to the developers of the ToxoDB.org Genome Resource. ToxoDB and EuPathDB are part of the National Institutes of Health/National Institutes of Allergy and Infectious Diseases (NIH/NIAID)-funded Bioinformatics Resource Center. We thank Dr. Guillaume Communie for his help computing Danpos software. This work was supported by the Laboratoire d'Excellence (LabEx) ParaFrap [ANR-11-LABX-0024], the Agence Nationale pour la Recherche [Project HostQuest, ANR-18-CE15-0023 and ProFI grant, ANR-10-INSB-08-01], the European Research Council [ERC Consolidator Grant N°614880 Hosting TOXO to M.A.H] and Fondation pour la Recherche Médicale [FRM FDT201904008364 to Farhat DC].
Footnotes
ORCiD: 0000-0002-2547-8233
Data availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request. The RNAseq demultiplexed FASTQ files and gene-wise quantifications have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Serie accession number GSE136123. The ChIPseq data have been deposited to the GEO Datasets under accession number GSE136060. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD016846 (MORC interactome) and PXD016845 (proteome-wide analyses).
Author Contributions
M.-A.H. supervised the research. M.-A.H. and D.C.F. generated the genetic tools, performed and analyzed all the genome-wide studies (RNA-Seq, ChIP-Seq, Mnase-assay), conducted most of biochemical experiments and the immunofluorescence assays. C.D., D.C. and F.S. generated biochemical reagents. L.B. and P.-J.D.-B. performed mass spectrometric analysis. Y.C. guided the mass spectrometry experiments. C.S. performed structural modeling and analyzed the Mnase assay. A.B. computed and analyzed the RNA sequencing data. P.O. and M.B. analyzed the ChIP-seq data. M.-A.H. and D.C.F. wrote the paper.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Dubey JP, Lindsay DS, Speer CA. Structures of Toxoplasma gondii Tachyzoites, Bradyzoites, and Sporozoites and Biology and Development of Tissue Cysts. Clin Microbiol Rev. 1998;11:267–299. doi: 10.1128/cmr.11.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaji S. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Research. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim K. The Epigenome, Cell Cycle, and Development in Toxoplasma. 2018:21. doi: 10.1146/annurev-micro-090817-062741. [DOI] [PubMed] [Google Scholar]
- 4.Saksouk N, et al. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bougdour A, et al. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J Exp Med. 2009;206:953–966. doi: 10.1084/jem.20082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moissiard G, et al. MORC Family ATPases Required for Heterochromatin Condensation and Gene Silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorković ZJ. MORC proteins and epigenetic regulation. Plant Signaling & Behavior. 2012;7:1561–1565. doi: 10.4161/psb.22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris CJ, et al. Arabidopsis AtMORC4 and AtMORC7 Form Nuclear Bodies and Repress a Large Number of Protein-Coding Genes. PLOS Genetics. 2016;12:e1005998. doi: 10.1371/journal.pgen.1005998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiser NE, et al. MORC-1 Integrates Nuclear RNAi and Transgenerational Chromatin Architecture to Promote Germline Immortality. Developmental Cell. 2017;41:408–423.e7. doi: 10.1016/j.devcel.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastor WA, et al. MORC1 represses transposable elements in the mouse male germline. Nature Communications. 2014;5 doi: 10.1038/ncomms6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tchasovnikarova IA, et al. Hyperactivation of HUSH complex function by Charcot–Marie–Tooth disease mutation in MORC2. Nature Genetics. 2017;49:1035–1044. doi: 10.1038/ng.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, et al. The Gene-Silencing Protein MORC-1 Topologically Entraps DNA and Forms Multimeric Assemblies to Cause DNA Compaction. Molecular Cell. 2019;75:700–710.e6. doi: 10.1016/j.molcel.2019.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue N. New gene family defined by MORC, a nuclear protein required for mouse spermatogenesis. Human Molecular Genetics. 1999;8:1201–1207. doi: 10.1093/hmg/8.7.1201. [DOI] [PubMed] [Google Scholar]
- 14.Iyer LM, Abhiman S, Aravind L. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biology Direct. 2008;3:8. doi: 10.1186/1745-6150-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. International Journal for Parasitology. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Andrews FH, et al. Multivalent Chromatin Engagement and Inter-domain Crosstalk Regulate MORC3 ATPase. Cell Reports. 2016;16:3195–3207. doi: 10.1016/j.celrep.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sindikubwabo F, et al. Modifications at K31 on the lateral surface of histone H4 contribute to genome structure and expression in apicomplexan parasites. eLife. 2017;6 doi: 10.7554/eLife.29391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidik SM, et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell. 2016;166:1423–1435.e12. doi: 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown KM, Long S, Sibley LD. Plasma Membrane Association by N-Acylation Governs PKG Function in Toxoplasma gondii. mBio. 2017;8:e00375–17. doi: 10.1128/mBio.00375-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pittman KJ, Aliota MT, Knoll LJ. Dual transcriptional profiling of mice and Toxoplasma gondii during acute and chronic infection. BMC Genomics. 2014;15:806. doi: 10.1186/1471-2164-15-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hehl AB, et al. Asexual expansion of Toxoplasma gondii merozoites is distinct from tachyzoites and entails expression of non-overlapping gene families to attach, invade, and replicate within feline enterocytes. BMC Genomics. 2015;16:66. doi: 10.1186/s12864-015-1225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramakrishnan C, et al. An experimental genetically attenuated live vaccine to prevent transmission of Toxoplasma gondii by cats. Sci Rep. 2019;9 doi: 10.1038/s41598-018-37671-8. 1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritz HM, et al. Transcriptomic analysis of toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PLoS ONE. 2012;7:e29998. doi: 10.1371/journal.pone.0029998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behnke MS, Zhang TP, Dubey JP, Sibley LD. Toxoplasma gondii merozoite gene expression analysis with comparison to the life cycle discloses a unique expression state during enteric development. BMC Genomics. 2014;15:350. doi: 10.1186/1471-2164-15-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramakrishnan C, Walker RA, Eichenberger RM, Hehl AB, Smith NC. The merozoite-specific protein, TgGRA11B, identified as a component of the Toxoplasma gondii parasitophorous vacuole in a tachyzoite expression model. International Journal for Parasitology. 2017 doi: 10.1016/j.ijpara.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Smith EF. PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. The Journal of Cell Biology. 1996;132:359–370. doi: 10.1083/jcb.132.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sapiro R, et al. Male Infertility, Impaired Sperm Motility, and Hydrocephalus in Mice Deficient in Sperm-Associated Antigen 6. Molecular and Cellular Biology. 2002;22:6298–6305. doi: 10.1128/MCB.22.17.6298-6305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straschil U, et al. The Armadillo Repeat Protein PF16 Is Essential for Flagellar Structure and Function in Plasmodium Male Gametes. PLoS ONE. 2010;5:e12901. doi: 10.1371/journal.pone.0012901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark T. HAP2/GCS1: Mounting evidence of our true biological EVE? PLoS Biol. 2018;16:e3000007. doi: 10.1371/journal.pbio.3000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori T, Hirai M, Kuroiwa T, Miyagishima S. The Functional Domain of GCS1-Based Gamete Fusion Resides in the Amino Terminus in Plant and Parasite Species. PLoS ONE. 2010;5:e15957. doi: 10.1371/journal.pone.0015957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes & Development. 2008;22:1051–1068. doi: 10.1101/gad.1656508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angrisano F, et al. Targeting the Conserved Fusion Loop of HAP2 Inhibits the Transmission of Plasmodium berghei and falciparum. Cell Reports. 2017;21:2868–2878. doi: 10.1016/j.celrep.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gondim LFP, et al. Characterization of an IgG monoclonal antibody targeted to both tissue cyst and sporocyst walls of Toxoplasma gondii. Experimental Parasitology. 2016;163:46–56. doi: 10.1016/j.exppara.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Radke JR, et al. Identification of a sporozoite-specific member of the Toxoplasma SAG superfamily via genetic complementation: T. gondii sporozoite developmental antigens. Molecular Microbiology. 2004;52:93–105. doi: 10.1111/j.1365-2958.2003.03967.x. [DOI] [PubMed] [Google Scholar]
- 35.Tomita T, et al. The Toxoplasma gondii Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence. PLoS Pathogens. 2013;9:e1003823. doi: 10.1371/journal.ppat.1003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radke JB, et al. ApiAP2 transcription factor restricts development of the Toxoplasma tissue cyst. Proceedings of the National Academy of Sciences. 2013;110:6871–6876. doi: 10.1073/pnas.1300059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poran A, et al. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature. 2017;551:95–99. doi: 10.1038/nature24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radman-Livaja M, Rando OJ. Nucleosome positioning: How is it established, and why does it matter? Developmental Biology. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teif VB, et al. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- 41.Li D-Q, et al. MORC2 Signaling Integrates Phosphorylation-Dependent, ATPase-Coupled Chromatin Remodeling during the DNA Damage Response. Cell Reports. 2012;2:1657–1669. doi: 10.1016/j.celrep.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wadman M. Closure of U.S. Toxoplasma lab draws ire. doi: 10.1126/science.364.6436.109. 2. [DOI] [PubMed] [Google Scholar]
- 43.Bougdour A, et al. Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell Host Microbe. 2013;13:489–500. doi: 10.1016/j.chom.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Sangaré LO. Unconventional endosome-like compartment and retromer complex in Toxoplasma gondii govern parasite integrity and host infection. Nature Communications. 2016;7 doi: 10.1038/ncomms11191. 11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salvetti A, et al. Nuclear Functions of Nucleolin through Global Proteomics and Interactomic Approaches. J Proteome Res. 2016;15:1659–1669. doi: 10.1021/acs.jproteome.6b00126. [DOI] [PubMed] [Google Scholar]
- 46.Gajria B, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Research. 2007;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 48.Wieczorek S, et al. DAPAR & ProStaR: software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics. 2017;33:135–136. doi: 10.1093/bioinformatics/btw580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen K, et al. DANPOS: Dynamic analysis of nucleosome position and occupancy by sequencing. Genome Research. 2013;23:341–351. doi: 10.1101/gr.142067.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.