Significance
Among all influenza pandemics, the 1918 influenza is considered to be the worst pandemic in human history. NS1 is a multifunctional virulence factor of influenza viruses and interacts with many host proteins. Here we present the mechanism underlying the molecular recognition of the p85β subunit of human phosphoinositide 3-kinase by the NS1 of the 1918 strain. We find that the structure of 1918 NS1 is highly dynamic, whereas NS1 of a seasonal influenza strain is mostly static. Moreover, the two NS1 proteins bind to p85β with drastically different binding affinities and kinetics. Our findings provide a mechanistic insight into strain-dependent behaviors of NS1 proteins, which remains elusive despite its importance in understanding the virulence of influenza viruses.
Keywords: conformational dynamics, influenza virus, nonstructural protein 1
Abstract
The 1918 influenza A virus (IAV) caused the most severe flu pandemic in recorded human history. Nonstructural protein 1 (NS1) is an important virulence factor of the 1918 IAV. NS1 antagonizes host defense mechanisms through interactions with multiple host factors. One pathway by which NS1 increases virulence is through the activation of phosphoinositide 3-kinase (PI3K) by binding to its p85β subunit. Here we present the mechanism underlying the molecular recognition of the p85β subunit by 1918 NS1. Using X-ray crystallography, we determine the structure of 1918 NS1 complexed with p85β of human PI3K. We find that the 1918 NS1 effector domain (1918 NS1ED) undergoes a conformational change to bind p85β. Using NMR relaxation dispersion and molecular dynamics simulation, we identify that free 1918 NS1ED exists in a dynamic equilibrium between p85β-binding–competent and –incompetent conformations in the submillisecond timescale. Moreover, we discover that NS1ED proteins of 1918 (H1N1) and Udorn (H3N2) strains exhibit drastically different conformational dynamics and binding kinetics to p85β. These results provide evidence of strain-dependent conformational dynamics of NS1. Using kinetic modeling based on the experimental data, we demonstrate that 1918 NS1ED can result in the faster hijacking of p85β compared to Ud NS1ED, although the former has a lower affinity to p85β than the latter. Our results suggest that the difference in binding kinetics may impact the competition with cellular antiviral responses for the activation of PI3K. We anticipate that our findings will increase the understanding of the strain-dependent behaviors of influenza NS1 proteins.
Influenza A virus (IAV) is responsible for the majority of seasonal flu cases resulting in more than 30,000 deaths every year in the United States alone (1). Moreover, occasional emergence of pandemic IAV incapacitates vaccines and infects millions of people worldwide. Four major flu pandemics have occurred in the past 100 y, most recently in 2009 (2). The deadliest one, which is termed “Spanish flu,” occurred in 1918 and resulted in more than 50 million deaths worldwide (3). Although there has been considerable progress in understanding the origin and pathogenicity of the 1918 IAV (4), molecular bases of its high virulence remain unclear. It is therefore of particular interest to elucidate mechanisms underlying molecular recognition of host factors by viral proteins of 1918 IAV.
Nonstructural protein 1 (NS1) of IAV has attracted considerable attention because of its role as a multifunctional virulence factor during the viral infection cycle (5–8). Thus, it is generally considered a potential drug target for the treatment of IAV infections (9–11). The primary functions of NS1 are to antagonize host innate immune responses (12), such as the expression of type I interferon (IFN) (13–15), and to increase viral replication (7, 16, 17). NS1 consists of an RNA binding domain (RBD) and an effector domain (ED), followed by an intrinsically disordered C-terminal tail (CTT) (Fig. 1A and refs. 18 and 19). The RBD binds viral double-stranded RNA and inhibits IFN-induced oligonucleotide A synthetase (20, 21). ED and CTT play key roles in interfering with host antiviral immune processes by binding to a number of host proteins (8, 15, 22–26).
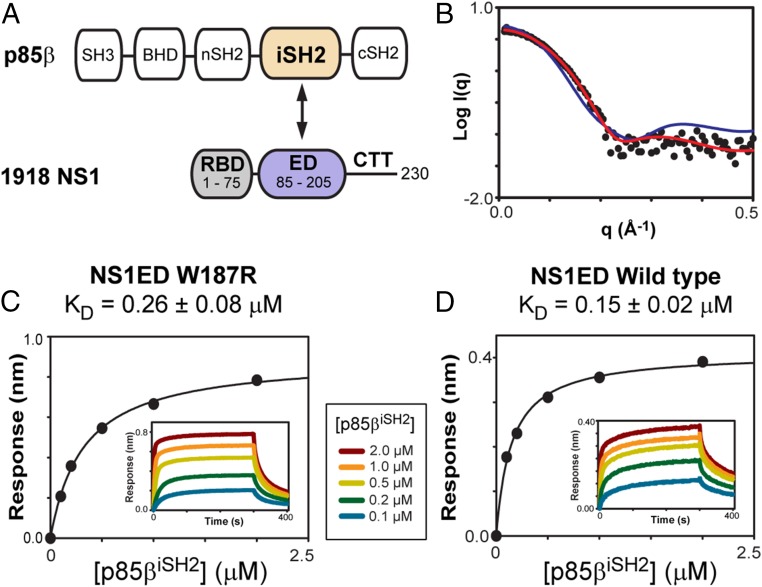
Fig. 1.
Interaction between 1918 NS1 and p85β. (A) Domain organizations of p85β and NS1. Interacting domains between the proteins are highlighted with colors. (B) Small-angle X-ray scattering data of free 1918 NS1ED: experimental (closed circles), monomeric NMR structure-derived (red line), and dimeric crystal structure-derived (blue line) data. BLI-derived binding isotherms of 1918 NS1ED (C) W187R and (D) wild type with p85βiSH2. (Insets) Representative binding sensorgrams. See also SI Appendix, Fig. S1.
One important function of NS1 during the infection cycle is to activate the phosphoinositide 3-kinase (PI3K) signaling pathway (8, 16, 22, 23, 27, 28). Class IA PI3K is a heterodimeric enzyme composed of a regulatory p85β subunit and a catalytic p110 subunit (29). The binding of NS1ED to the iSH2 domain in the p85β (p85βiSH2) activates the p110 catalytic subunit (23, 27). NS1-mediated activation of the PI3K pathway results in increased IAV virulence by delaying cellular apoptosis (16) and/or by changing the cellular distribution of PI3K (30–32).
The multifunctional activity of NS1 depends on interactions with a variety of host proteins. Thus, elucidating the binding mechanism between NS1 and host proteins provides important insights into influenza virulence at the molecular level. So far, however, only a few structures of NS1 complexed with host factors have been determined (15, 23, 24). Moreover, the structure of 1918 NS1 in complex with host proteins remained unknown. As a result, molecular recognition mechanisms of host proteins by 1918 NS1 remain poorly understood.
Here, to gain mechanistic insights into the molecular recognition between 1918 NS1 and human p85βiSH2, we determine the crystal structure of the complex. Using NMR spectroscopy and molecular dynamics (MD) simulation, we reveal that the free 1918 NS1ED undergoes a dynamic conformational transition between p85β-binding–incompetent and –competent states in the sub-millisecond (sub-ms) timescale. Moreover, we find that NS1 proteins of different influenza strains undergo varying degrees of conformational dynamics in the p85-binding region. Strain-dependent differences in the function of NS1 have been reported (31–35), although their underlying mechanisms remain elusive. Thus, for a detailed analysis, we compare the structure and conformational dynamics of 1918 NS1ED with those of NS1ED from a seasonal influenza strain (A/Udorn [Ud]/72 H3N2). Intriguingly, we find that the two proteins exhibit drastic differences not only in their major conformations and the sub-ms dynamics but also in binding affinities and kinetics to p85β. Our results indicate that 1918 NS1ED can occupy p85β faster than Ud NS1ED. These findings suggest that the faster binding kinetics of 1918 NS1ED may play an important role in the competition with host innate immune responses during the infection cycle.
Results and Discussion
Binding of 1918 NS1ED and p85βiSH2.
Isolated NS1ED tends to form a homodimer (36–38), while interactions with some host proteins, including p85β, are mediated by its monomeric form (23, 39). To investigate the molecular basis of p85β recognition by the 1918 NS1 monomer, we incorporated an W187R substitution, which was shown to prevent NS1ED homodimerization and precipitation (37). However, a recent crystal structure of free 1918 NS1ED W187A indicated a dimeric form (10), although both W187A and W187R mutations were shown to form a monomer for NS1 of different influenza viruses (37).
To ascertain the monomeric state of free 1918 NS1ED W187R, we performed small-angle X-ray scattering (SAXS). The experimental SAXS profile matched well with the one calculated based on the monomeric NMR structure (Fig. 1B), with experimental and structure-derived Rg values of 15 Å and 14 Å, respectively. We also confirmed that the W187R substitution does not affect the binding to p85βiSH2 by comparing the in vitro binding affinities (KD) of 1918 NS1ED wild type and W187R mutant (Fig. 1 C and D and SI Appendix, Fig. S1). This result is consistent with the solution NMR structure of 1918 NS1ED W187R (40), indicating that the protein exists as a monomer.
Structure of the Complex Between 1918 NS1 and p85β.
We determined crystal structures of 1918 NS1ED W187A and W187R in complex with p85βiSH2 at 2.95-Å and 2.75-Å resolution, respectively (Fig. 2A and SI Appendix, Table S1). The two structures are virtually identical to each other except for the mutated residue (residue 187) (Fig. 2A and SI Appendix, Fig. S2), demonstrating that the mutation does not affect the interaction with p85β. Both structures showed two complex molecules in an asymmetric unit (SI Appendix, Fig. S2), which was attributed to a crystallographic dimer of the complex: Previous studies identified that NS1ED forms a 1:1 complex with p85β (23, 39). Overall, our structures are similar to that of the complex between the NS1ED of influenza strain Puerto Rico (PR8)/8/34 (H1N1) and bovine p85βiSH2 (23) (see SI Appendix, Fig. S3 for the detailed structural comparison).
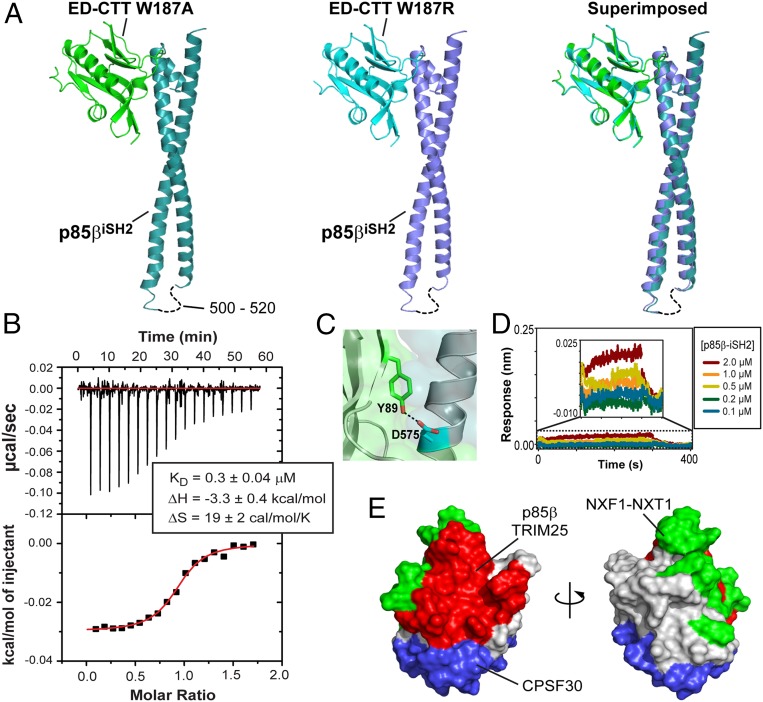
Fig. 2.
Binding mode between 1918 NS1ED and p85βiSH2. (A) Crystal structures of the complex between 1918 NS1ED and p85βiSH2: (Left) 1918 NS1ED W187A, (Middle) W187R, and (Right) superimposed. (B) ITC traces and binding isotherm of the titration of 1918 NS1ED W187R into p85βiSH2. Solid line represents the fit to the binding isotherm with a 1:1 binding model. Fit values are the average of two repeats. (C) Hydrogen bond between Y89 and D575. (D) BLI binding sensorgram of NS1ED Y89F and p85βiSH2. (Inset) Expanded view of the sensorgram. The BLI data for the NS1ED wild type is shown in Fig. 1D. (E) Surface representation of 1918 NS1ED with binding interfaces for p85β (red), TRIM 25 (red), CPSF 30 (blue), and NXF1-NXT1 (green). Overlapped regions were not depicted separately for clarity.
The complex interface is largely hydrophobic, with hydrophobic and hydrophilic occluded surface areas of ∼1,120 Å2 and 530 Å2, respectively (see SI Appendix, Fig. S2 for all interface residues). This composition of the interface is manifested in the thermodynamic origins of the binding free energy. The isothermal titration calorimetry (ITC) result indicated a favorable enthalpy, reflecting specific interactions at the interface, and even more favorable entropy, consistent with the burial of the large apolar surface area upon formation of the complex (41, 42) (Fig. 2B).
The structures also indicated that Y89 in 1918 NS1ED forms a hydrogen bond to D575 of p85βiSH2 (Fig. 2C). Y89 is highly conserved in human IAVs (SI Appendix, Fig. S4), and previous studies showed that the hydrogen bond is important for the activation of PI3K by NS1 from some influenza strains (23, 27, 28). Using biolayer interferometry (BLI), we found that the Y89F mutation in 1918 NS1 abolishes the binding to p85β (Fig. 2D). Our structure indicates that the large effect on the binding energetics might be due to the burial of the hydrogen bond in the hydrophobic binding interface (43).
NS1 Makes the Most of Its Surface Area for Interactions with Diverse Host Proteins.
Structural studies conducted by our laboratory and by other research groups have so far determined four complex structures of NS1 and host proteins (15, 23, 24, 44). All of the interactions were mediated exclusively by NS1ED, highlighting the importance of understanding the molecular recognition mechanism of NS1ED. Notably, these structures demonstrate that the majority of the NS1ED surface is exploited to interact with a wide range of host factors (Fig. 2E). While many studies have shown that viral proteins use intrinsically disordered regions to interact with multiple host proteins (45), it is remarkable that a well-ordered domain like NS1ED makes the most of its surface area for disparate binding interfaces.
The overlap of binding surfaces suggests that mutational effects of NS1 might be difficult to unravel at the cellular level solely based on a specific protein–protein interaction. Another implication is a competitive binding of NS1 with multiple host factors. For example, the binding sites for NXF1–NTF2 (nuclear RNA export factor 1–nuclear transport factor 2) and CPSF30 (cleavage and polyadenylation specificity factor) partially overlap (Fig. 2E), implying their mutually exclusive binding to NS1. Indeed, it was indicated that the interaction of NS1 with NXF1–NTF2 is likely to occur earlier than interaction with CPSF30 during infection (44). Thus, we expect that more structures of NS1 complexed with host factors will help understand the differential function of NS1 during infection.
Free 1918 NS1ED Undergoes Conformational Transition to Bind p85βiSH2.
Recently, structures of free 1918 NS1ED W187A and W187R mutants were reported (10, 40). This allowed us to compare the structures of 1918 NS1ED between the free and p85β-bound states. Intriguingly, we found that 1918 NS1ED adopts substantially distinct conformations between the two states (Fig. 3 A and B). Both W187A and W187R mutants showed highly similar conformational difference between the free and bound forms, indicating that the difference is not associated with a specific mutation (SI Appendix, Fig. S5).
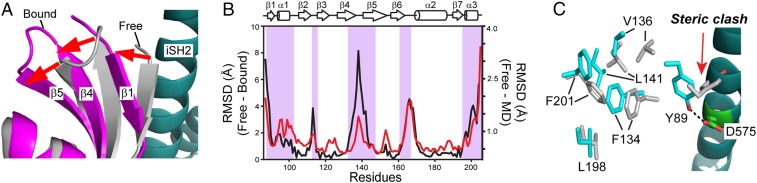
Fig. 3.
Free 1918 NS1ED has a p85β-BI conformation. (A) Conformational change of 1918 NS1ED upon binding p85β. Crystal structures of free (gray) and p85β-bound (magenta) NS1ED W187A, respectively. (B) rmsd plot between the free and p85β-bound NS1ED (black line) and average rmsd (red line) during 100-ns MD simulation. Shaded regions correspond to the p85β-binding interface. (C) Hydrophobic residues in free (gray) and p85β-bound (cyan) states of NS1ED W187A.
The rmsd of 1918 NS1ED between the free and p85β-bound states showed that several regions undergo a considerable (rmsd >1 Å) conformational change upon binding to p85β (Fig. 3B). Notably, β1–β4–β5 strands as a whole are twisted away from the p85β-binding interface to avoid steric clash (Fig. 3A). We found that the conformational change is accompanied by a large-scale rearrangement of hydrophobic residues. In the free form, a set of hydrophobic residues (F134, V136, and L141) forms a hydrophobic cluster with L198 and F201 in the C-terminal α-helix (Fig. 3C). In the bound form, however, these residues have drastically different conformations, resulting in the overall shift of the β1–β4–β5 strands. These residues are highly conserved across IAV strains (SI Appendix, Fig. S5). Moreover, this conformational change is essential for the hydrogen bond between Y89NS1 and D575p85β. The side chain of Y89NS1 in the free form points in the opposite direction to that in the bound form, and if there is no conformational change Y89NS1 sterically clashes with p85β (Fig. 3C). Hereinafter, we refer to the structures of the free and p85β-bound 1918 NS1ED as “binding-incompetent” (BI) and “binding-competent” (BC) forms, respectively.
Dynamic Conformational Sampling by Free 1918 NS1ED.
Our previous study indicated that free 1918 NS1ED contains structurally plastic regions (40). Intriguingly, we found that the same regions undergo conformational change upon binding to p85βiSH2 (SI Appendix, Fig. S6), suggesting that the transition between the BI and BC conformations occurs in the free state. To test the hypothesis, we performed 100-ns MD simulation using the NMR structure (Protein Data Bank [PDB] ID code 6NU0) as an initial model. Indeed, the rmsd profile of the simulation was highly similar to that between the free (i.e., BI) and p85β-bound (i.e., BC) forms (Fig. 3B), indicating that free 1918 NS1ED undergoes dynamic conformational sampling between the BI and BC forms. We verified the simulation result by examining NMR relaxation parameters. Conformational exchange in the microsecond-to-millisecond timescale increases the product of NMR R1 and R2 rate constants, R1*R2 (46). We found that all of the residues with significantly elevated R1*R2 values (>mean + SEM) are located in regions where a large conformational change occurs upon binding to p85β (Fig. 4A). These results are also consistent with the finding that four resonance peaks (residues 139, 141, 143, and 144) in β4–β5 loop region were missing in the 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum because of the dynamic motion in the intermediate NMR timescale (Fig. 4A).
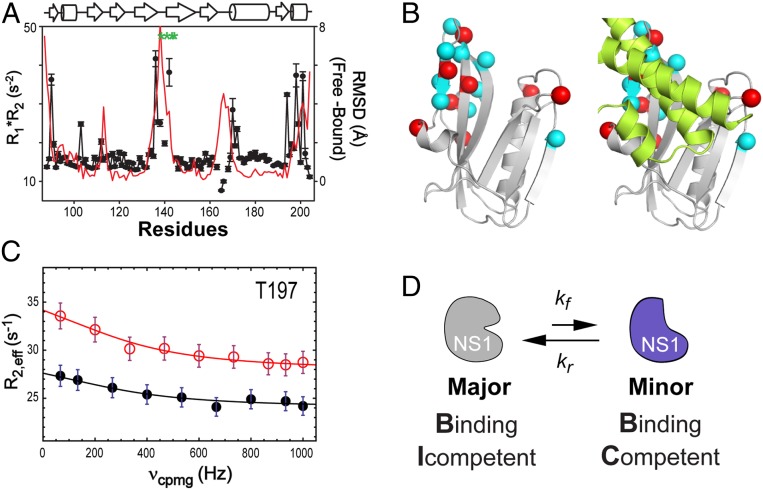
Fig. 4.
Conformational dynamics of free 1918 NS1ED. (A) NMR R1*R2 (closed circles) of 1918 NS1ED W187R and rmsd (red line) between the free and p85β-bound forms. Green asterisks correspond to the missing residues in a HSQC spectrum. (B) Residues that exhibited detectable Rex are shown as spheres (Left). Residues in group A are shown as red spheres. (Right) The dynamic residues are located at the p85βiSH2 (lemon)-binding site. (C) A representative 15N CPMG-RD profile (T197) of free NS1-ED recorded at 800 MHz (red open circles) and 600 MHz (black closed circles). Solid lines correspond to the global fit of the data. (D) Schematic showing a conformational exchange of free 1918 NS1ED between the BI and BC conformations.
Using NMR 15N R2 Carr–Purcell–Meiboom–Gill relaxation dispersion (CPMG-RD) (47–49), we further characterized the conformational dynamics of free 1918 NS1ED W187R. A total of 17 15N resonances exhibited exchange contribution (Rex) in the experiment. All of the residues are located in regions that undergo conformational change upon binding to p85βiSH2 (Fig. 4B). Eight of them exhibited Rex larger than 2.5 s−1 (Fig. 4C), which is adequate for quantitative data fitting. Thus, we limited our CPMG-RD analysis to these residues (group A). The remaining nine resonances (group B) were used for confirmatory purposes only; the RD data were fitted with variables precalculated using group-A resonances.
To calculate the exchange rate constant kex (= kf + kr) between two conformational states of free 1918 NS1ED (Fig. 4D), the NMR CPMG-RD data were fitted using the Carver and Richards equation (ref. 50 and Fig. 4C). The fitting results of all analyzed residues are shown in SI Appendix, Fig. S7. Global fitting including all group-A resonances yielded kex = 2,459 ± 205 s−1. The population of the minor species (pminor) was estimated to be ∼8% of the entire population: The χ2 surface along the pminor showed a broad basin in the range between 5 and 10% (SI Appendix, Fig. S8). Based on the kex and pminor values, we estimated that the forward and reverse rate constants (kf and kr) between the major and minor species are 99 to 246 s−1 and 2,188 to 2,360 s−1, respectively. This result showed that major species transiently samples the minor conformation with a lifetime less than 500 μs. To test whether the residues (group B) excluded from quantitative analysis undergo the same conformational exchange, we fitted the dispersion data of the residues in group B using the precalculated exchange parameters (kex and pminor) as fixed variables. The fitting curves showed reasonable agreement with data (SI Appendix, Fig. S7B), albeit qualitative, indicating a concerted conformational exchange process.
Both NMR and crystal structures of free 1918 NS1ED are in the BI conformation (10, 40), indicating that the major species in the dynamic equilibrium corresponds to the BI form. In contrast, direct structural characterization of the minor species is not feasible because of its low population and short lifetime. Instead, multiple lines of data indicated that the minor species has the BC conformation. First, our MD simulation showed that the conformational fluctuation of the free protein is highly similar to that between the free (BI) and p85β-bound (BC) conformations (Fig. 3B). Second, all of the residues that showed sub-ms dynamics are located in the regions that undergo conformational change upon binding to p85βiSH2 (Fig. 4 A and B). Third, CPMG-RD provided conformational information of the minor species. If the minor species corresponds to the BC conformation, its conformation must be similar to the p85β-bound form. In this case, the chemical shift difference obtained by fitting to the CPMG-RD profile (∆ωCPMG = ωmajor,BI − ωminor,BC) correlates with the measured chemical shift difference between the free and bound states (∆δ = δfree − δbound). We indirectly estimated δbound using a computer program SHIFTX2 (51) and crystal structure of 1918 NS1ED bound to p85βiSH2. This is because direct measurement of δbound was not possible due to the extremely low solubility of the 1918 NS1ED:p85βiSH2 complex. A qualitative comparison of ∆ωCPMG of the group-A residues with ∆δSHIFTX showed a reasonable agreement (SI Appendix, Fig. S9). Taken together, these results indicate that free 1918 NS1ED undergoes conformational sampling between the BI and BC conformations in the sub-ms timescale.
Conformational Dynamics of NS1 Proteins from Different Influenza Viruses.
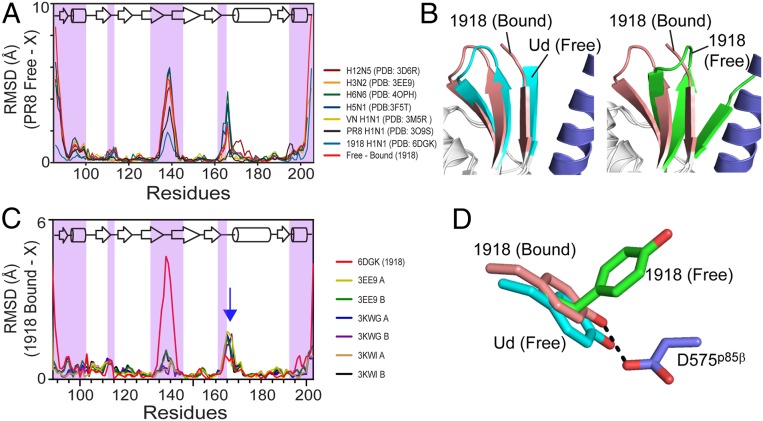
Our structural analysis indicated that the BI–BC dynamics of 1918 NS1ED is essential for binding to p85β. To ascertain whether NS1 proteins of different influenza strains also undergo similar conformational dynamics, we compared the backbone rmsd of seven free NS1ED structures from five influenza strains with that of the PR8 strain (PDB ID code 2GX9). Surprisingly, these structures exhibited a high conformational variability in the p85β-binding region; however, their overall rmsd patterns were similar to the conformational change of 1918 NS1ED upon binding to p85βiSH2 (red line in Fig. 5A). These diverse structures captured under different crystallization conditions collectively showed intrinsic conformational plasticity of free NS1ED proteins. The comparison suggests that NS1 proteins of different influenza viruses undergo varying degrees of conformational dynamics in the p85β-binding interface.
Fig. 5.
Strain-dependent conformational diversity of NS1 proteins. (A) rmsd plots of NS1 proteins from diverse influenza viruses with respect to the structure of PR8 (H1N1, PDB ID code 2GX9). p85β-binding regions are shaded with magenta. X represents individual PDB coordinates. (B) Superimposed structure of p85β-bound 1918 NS1ED and (Left) free Ud NS1ED and (Right) free 1918 NS1ED. (C) rmsd plots of free Ud NS1ED proteins with respect to p85β-bound 1918 NS1ED. Red line: rmsd between free and bound 1918 NS1ED. Blue arrow: residues 164 to 171. (D) Conformations of Y89 in p85β-bound (brown), free (green) 1918 NS1ED, and free Ud NS1ED (cyan).
To further test the hypothesis in detail, we compared structures and conformational dynamics of 1918 NS1ED (H1N1) to those of Ud NS1ED (H3N2). Intriguingly, the structure of free Ud NS1ED was similar to the BC conformation of 1918 NS1 (Fig. 5B). To avoid any bias in the comparison, we compared all crystal structures of free Ud NS1ED available in the PDB (Fig. 5C). We found that all of the conformations of free Ud NS1ED were consistently close to the BC conformation and dissimilar to the BI conformation (red line in Fig. 5C), regardless of their crystallization conditions, ED dimeric state, and mutations at residue 187. Moreover, the side-chain position of Y89 in free Ud NS1 was similar to that in the BC state of 1918 NS1ED (Fig. 5D). These results indicate that Ud NS1ED, as opposed to 1918 NS1, populates BC-like conformation in its free state and may not need a large conformational change to bind p85β.
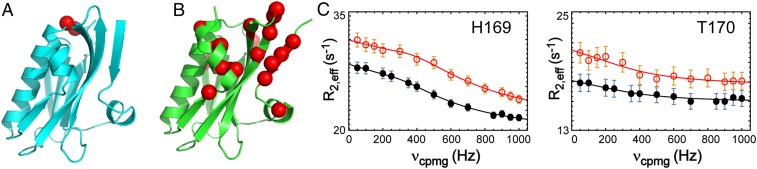
To further examine the intrinsic conformational dynamics of free Ud NS1ED, we conducted 15N CPMG-RD analysis. Indeed, free Ud NS1ED did not exhibit Rex, except for only two residues, H169 and T170 (Fig. 6A). This is in stark contrast to the 1918 NS1ED in which 17 residues undergo sub-ms dynamics (Fig. 6B). A global fitting of the CPMG-RD data of the two residues yielded kex = 2,200 ± 170 s−1 and pminor = 1% (Fig. 6C). For comparison, we provided the CPMG-RD profiles of the corresponding 17 residues in Ud NS1ED (SI Appendix, Fig. S10). Notably, there was no overlap between the dynamic residues of the two proteins. For example, Y89, a key interface residue, did not exhibit Rex in Ud NS1ED, which is consistent with the crystal structures (Fig. 5D). Nevertheless, it remains to be further determined whether the conformational change is required for Ud NS1ED to bind p85βiSH2. The two dynamic residues in Ud NS1ED are not in the direct binding interface; however, they are located nearby the p85β-binding interface. The two residues are in the β6–α2 loop (residues 164 to 171), in which residues 161 to 165 form direct contacts with p85β. In this light, it is worth mentioning that residues 164 to 171 showed an elevated rmsd in all Ud NS1 structures (blue arrow in Fig. 5C). Although the p85β-bound structure of Ud NS1 will be required to assess the role of the subtle conformational change in binding, our results suggest that the degree of the conformational change and spatial distribution of dynamic residues are remarkably different between the two strains.
Fig. 6.
Strain-dependent structure and dynamics of NS1ED. Structures of (A) free Ud and (B) free 1918 NS1ED proteins. Residues that exhibited Rex are shown in red spheres. (C) The 15N CPMG-RD profiles of two dynamic residues in Ud NS1ED: 800 MHz (red open circles) and 600 MHz (black closed circles). Solid lines correspond to the global fit of the data.
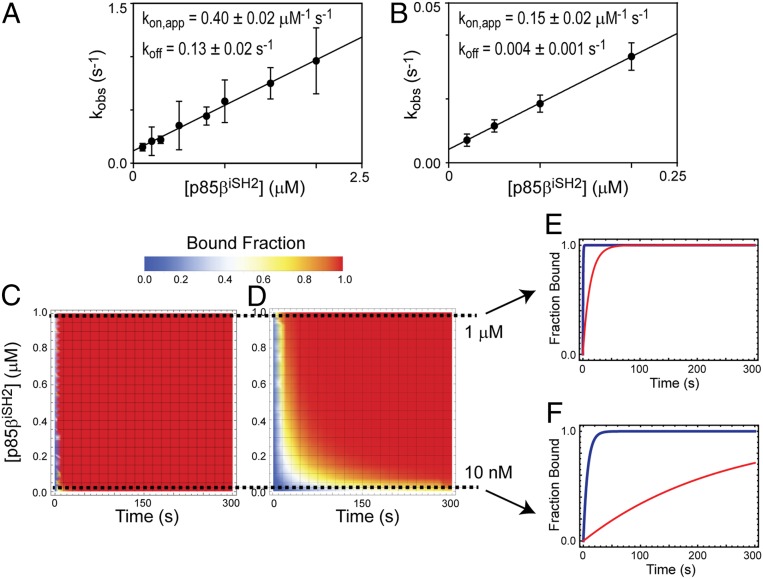
To ascertain the effect of the structural and dynamic differences on the interaction with p85β, we compared binding affinities and kinetics of 1918 and Ud NS1s for p85β. The KD value of Ud NS1ED and p85β was 50 nM (SI Appendix, Fig. S11), which is six times lower than that of 1918 NS1ED. More surprisingly, the two NS1s exhibited noticeably different binding kinetics to p85β (Fig. 7 A and B). Especially, Ud NS1ED dissociates from p85β ∼32 times more slowly than 1918 NS1ED. Although the structure of Ud NS1ED complexed with p85β is not available, residues at the p85β-binding interface are well-conserved between the two strains: Only 2 out of 20 interface residues have conservative substitutions between the two proteins (L95I and M98L) and the rest are identical. To test whether the two mutations affected binding, we grafted the residues of Ud NS1ED to the 1918 protein, that is, L95I/M98L, in the 1918 background. As expected, the grafting did not noticeably change the binding affinity and kinetics (SI Appendix, Fig. S12). Thus, the sequence difference outside the p85β-binding surface between the two proteins induced the remarkable difference in the conformational dynamics and consequently the binding properties.
Fig. 7.
Binding kinetics of 1918 and Ud NS1s to p85β. BLI-derived kobs values for binding of A 1918 and (B) Ud NS1ED to p85βiSH2. The solid line corresponds to the linear fit to the data. Error bars reflect the SD of three replicate measurements. The slope and y intercept of the linear regression curve correspond to kon and koff, respectively. Numerical simulations of increase in p85β-bound fraction of C 1918 and (D) Ud NS1ED proteins as functions of time and p85β concentration. Simulated time-dependent changes in p85-bound fractions for 1918 (blue) and Ud (red) NS1ED proteins at two different concentrations of p85β: (E) 1 µM and (F) 10 nM.
To further understand how the conformational dynamics of NS1ED affected the observed binding kinetics, we analyzed the binding kinetics in combination with NMR-derived dynamics data (SI Appendix, Fig. S13 and Supplementary Discussion). The result of the integrated analysis further supported our hypothesis in which the minor state of free 1918 NS1ED has a conformation competent for binding to p85β. Moreover, the result demonstrated that the conformational dynamics step significantly modulated the observed binding rate constant. It is worth noting the difference in the koff values between 1918 and Ud NS1s (Fig. 7 A and B), which might be due to the differences in the structure and/or dynamics of their p85β-bound state. However, both NS1 proteins had extremely low solubility when bound to p85β, limiting the study of conformational dynamics in complex states.
What is the functional implication of our results? PI3K is a signaling hub implicated not only in proviral responses but also in antiviral responses. For example, IAVs and other viruses, such as HIV, activate PI3K to induce proviral responses (22, 52, 53), whereas RIG-I (retinoic acid-inducible gene I) also activates PI3K upon recognizing viral RNA, resulting in the onset of antiviral responses (54, 55). Thus, we speculate that the rapid hijacking of p85β (i.e., PI3K) might be an important advantage for IAVs in competing with the host antiviral innate immune system.
We examined whether the NS1 from two different IAV strains, 1918 and Ud, demonstrate different rates of increase in p85β-bound fraction. For this, we conducted a simple numerical simulation of saturation kinetics based on the measured kon and koff values and simple pseudo-first-order approximation ([NS1ED] < [p85β]): This might mimic the early phase of NS1 expression during the infection cycle. Because the cellular concentration of p85β is unknown, we simulated the saturation kinetics between 1 nM and 1,000 nM of p85β (Fig. 7 C and D). Intriguingly, despite the higher affinity of Ud NS1, its small kon and koff values increased the time required to saturate p85β, compared to 1918 NS1. In contrast, larger kon and koff values of 1918 NS1 enables the protein to reach to the saturation more rapidly. Fig. 7 E and F show the saturation kinetics at two concentrations of p85β, 10 nM and 1,000 nM. Despite the high similarity in their overall structures, NS1 proteins of diverse influenza strains showed strain-dependent functional differences (23, 31, 32, 56, 57). However, its underlying mechanism remains elusive. Our results suggest that the conformational dynamics and binding kinetics of NS1 proteins need to be explored to fully understand the strain-dependent behavior of IAVs.
Materials and Methods
Protein Sample Preparation.
Genes encoding 1918 and Ud NS1 and p85βiSH2 proteins were prepared by gene-synthesis service from Genscript. All NS1 proteins (wild type, W187A, W187R, and Y89F mutants) were expressed in BL21 (DE3) Escherichia coli cells with a His6 and SUMO tags and purified by Ni2+ NTA column and gel-filtration chromatography. The p85βiSH2 (residues 435 to 599) domain was expressed in BL21 (DE3) E. coli and purified in the same as NS1 proteins. Purity of protein samples was confirmed using sodium dodecyl sulfate polyacrylamide gel electrophoresis; all samples were >95% pure.
BLI Experiments.
The binding of p85βiSH2 to surface-immobilized NS1 constructs was measured at 25 °C using an Octet RED biolayer interferometer (Pall ForteBio). His6-tagged NS1 proteins were immobilized on Ni-NTA biosensor tips. The buffer was 20 mM sodium phosphate (pH 7.0), 100 mM NaCl, 1% bovine serum albumin, and 0.4 M trehalose. Association and dissociation phases were measured for 300 s and 100 s, respectively. All measurements were performed at least three times (SI Appendix, Fig. S1). The steady-state KD values were determined by averaging five final signals in the association phase and fitting them using a 1:1 binding model as follows:
| [1] |
where ∆R and ∆Rmax are the change and the maximum signal change, respectively. Pt is the total protein concentration and Lt is the total analyte concentration at each titration point.
Overall Binding Kinetics.
To measure binding kinetics of 1918 NS1ED and p85βiSH2, the association phase was fitted with a double-exponential function. The fast-phase rate constants correspond to kobs while the slow-phase corresponds to nonspecific binding of p85βiSH2 to BLI sensor tips. To measure binding kinetics of Ud NS1ED and p85βiSH2, the association phase was fitted with a single-exponential function. The kobs value changed linearly with the concentration of p85βiSH2. kon and koff values were obtained by linear fitting to the plot of kobs vs. [p85βiSH2]. Numerical simulations of saturation kinetics were conducted by calculating , where kobs was calculated using measured kon and koff values and the relationship kobs = kon*[p85βiSH2] + koff. The two-dimensional density plot of saturation kinetics as a function of [p85βiSH2] was obtained by Mathematica (Wolfram Research Inc.).
NMR 15N Relaxation Measurements.
The 15N CPMG-RD experiments were recorded at two static magnetic fields (600 and 800 MHz) with CPMG frequencies (νcpmg), ranging from 50 to 1,000 Hz, as pseudo-three-dimensional experiments with a constant-time CPMG evolution period of 30 ms and 40 ms for 1918 and Ud NS1ED, respectively. The uncertainty of R2,eff was estimated by comparing the peak intensities of duplicated experiments. The CPMG-RD profile was fitted using the Carver–Richards equation (50). To slow down protein precipitation during NMR RD measurement, we added 0.4 M trehalose in the sample. We confirmed that the observed R2,eff is not induced by trehalose, by comparing the ∆R2,eff (the difference of R2,eff collected at two CPMG frequencies, 50 Hz and 1,000 Hz) to the R1*R2 values acquired without trehalose (SI Appendix, Fig. S14). Moreover, chemical shifts of backbone amide resonances were not affected by trehalose (SI Appendix, Fig. S14). We confirmed that the observed dynamics is not induced by dimerization of NS1ED by comparing peak intensities of the dynamic residues as a function of protein concentration (SI Appendix, Fig. S15). Measurements for 15N R1 and R2 rate constants were performed as described elsewhere (41). All NMR data were collected at 298 K on Bruker Avance III 600-MHz and 800-MHz spectrometers equipped with a cryogenic probe at the Biomolecular NMR facility (Texas A&M University). Backbone resonance assignments for 1918 and Ud NS1ED proteins were obtained from the Biological Magnetic Resonance Bank (BMRB; http://www.bmrb.wisc.edu/); BMRB accession numbers for 1918 and Ud NS1ED are 12032 and 16376, respectively.
Chemical Shifts Prediction Using SHIFTX2.
Prediction of chemical shifts from structural models was performed using the SHIFTX2 (51). The chemical shift for the BC form was predicted using the NS1 structure in the complex with p85βiSH2 (PDB ID code 6OX7) after removing p85βiSH2. The chemical shifts for free state was derived from 1H-15N HSQC spectrum of free 1918 NS1ED.
Crystallography.
The 1918 NS1 W187A (residues 86 to 230):p85βiSH2 complex was crystallized at 277 K by hanging-drop vapor diffusion in 20 mM Tris (pH 7.1), 80 mM NaCl, and 0.7 M trehalose. A 100 μM of NS1-ED was mixed with a 100 μM p85β-iSH2 to prepare the complex. The 1918 NS1 W187R (residues 86 to 230):p85βiSH2 complex was crystallized at 277 K by hanging-drop vapor diffusion in 20 mM sodium phosphate (pH 7.0) and 80 mM NaCl. The crystals were flash-frozen in liquid nitrogen in the reservoir solution containing 25% (vol/vol) glycerol. X-ray diffraction datasets were collected at 120 K using an R-AXIS IV++ image plate detector mounted on a Rigaku MicroMax 007HF X-ray generator. The data were processed using iMosflm in the CCP4 package (58). The structure was determined using previously reported structure of PR8 NS1-ED complexed with bovine iSH2 (PDB ID code 3L4Q) as a search model using the Phenix package (59). The structures were remodeled and refined with Coot (60) and the Phenix package. Crystal structures of 1918 NS1-ED W187A and W187R in complex with p85β-iSH2 are deposited in the PDB (ID codes 6U28 and 6OX7).
SAXS.
The X-ray scattering measurement was conducted at the Life Science X-ray Scattering (LiX) beamline (61) of the National Synchrotron Light Source II, Brookhaven National Laboratory. Online size-exclusion chromatography SAXS was used to eliminate aggregation in the sample and ensure the quality of the scattering data. Data were collected at room temperature, using a Shimadzu high-performance liquid chromatography (HPLC) system and a flow cell developed in house for X-ray scattering measurements. One hundred fifty microliters of a protein sample at 100 μM was injected into a Superdex Increase 200 10/300 GL column (GE Healthcare) at a flow rate of 0.45 mL/min. The eluent from the column was split 3:1 into the X-ray flow cell and the ultraviolet absorbance and refractive index detectors on the HPLC system. Scattering images were collected continuously at 1-s exposure per frame. Python scripts developed at the beamline were used for data processing as previously described (62), including merging of scattering data from multiple detectors for each X-ray exposure and buffer scattering subtraction on the selected sample frames. The program Scatter was used for Guinier analysis and estimation of Rg. The theoretical SAXS profile and Rg for NS1-ED structure was calculated using MultiFoXS (63).
MD Simulation.
For simulation, we used CHARMM (64) version c42a1 with param 36 all-atom force field (65). The NMR structure of free 1918 NS1ED in the BI form (PDB ID code 6NU0) was used as an initial structure. The system was solvated with TIP3P water molecules in a cubic box with a side length of 66 Å. The system was then electrically neutralized with Cl− and Na+ ions at ∼50 mM. The system was subjected to a four-stage energy minimization. At each stage, it underwent 100 steps of the steepest descent method followed by 300 steps of the adapted basis Newton–Raphson method. During energy minimization, a gradually decreasing harmonic constraint was applied to the backbone heavy atoms in the protein. Spring constants of the harmonic constraints were 5 kcal/mol·Å2 (stage 1), 1 kcal/mol·Å2 (stage 2), 0.1 kcal/mol·Å2 (stage 3), and 0 (no constraint at stage 4). The system was then heated from 0 K to 300 K for 100 ps and equilibrated for 200 ps under 1-atm pressure, with harmonic constraints applied to backbone heavy atoms in the protein. Spring constants of the harmonic constraint were 1 kcal/mol·Å2 (heating) and 0.5 kcal/mol·Å2 (equilibration). The system then underwent a preparatory run for 1 ns. During the preparatory run, only the Cα atoms were harmonically restrained with a spring constant of 0.25 kcal/mol·Å2. The production run was carried out under constant volume and at 300 K (NVT) without any constraint applied to the protein. The SHAKE algorithm was used to fix the length of covalent bonds involving hydrogen atoms. The integration step size was 2 fs. The cutoff distance for taking account of nonbonded interaction was 12 Å. The particle-mesh Ewald summation method was used to account for long-range electrostatic interactions. The Domain Decomposition (DOMDEC) module of CHARMM was used for efficient parallelization (66). The system contains a total of 27,268 atoms. Coordinates were saved every 5 ps. To calculate rmsd, all of the trajectories were first superimposed relative to the reference structure using backbone heavy atoms. At each time step, the rmsd value for a selected set of atoms was calculated as the root-mean-square of the distances between the atoms at current trajectory and the atoms at the reference structure.
ITC Measurements.
Samples were dialyzed into 20 mM sodium phosphate (pH 7.0) and 80 mM NaCl overnight. Data were acquired in duplicate at 25 °C using a Microcal VP-ITC instrument; 100 μM NS1ED was in the syringe and 10 μM p85βiSH2 was in the cell. The Ka, ∆H, and ∆S were directly obtained from fitting the data to a 1:1 binding model using Origin software (Microcal).
Sequence Conservation.
All human IAV sequences (4,023 nonduplicated sequences) were obtained from the NCBI influenza Research Database (https://www.fludb.org/brc/home.spg?decorator=influenza). The sequences were aligned using MUSCLE (Multiple Sequence Comparison by Log-Expectation) algorithm and the sequence logo was generated using WebLogo (67).
Data Availability.
Atomic coordinates of the crystal structures were deposited in the PDB (ID codes 6U28 and 6OX7). NMR resonances of 1918 and Ud NS1ED are available from the BMRB. All data discussed in the paper will be made available to readers. All DNA plasmids for expressing 1918 and Ud NS1ED will be available upon request.
Supplementary Material
Acknowledgments
We thank Profs. Josh Wand and Tatyana Igumenova for their critical manuscript reading. Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under grant R01GM127723. The LiX beamline is part of the Life Science Biomedical Technology Research resource, cofunded by the NIGMS under grant P41 GM111244 and by the US Department of Energy Office of Biological and Environmental Research under grant KP1605010, with additional support from NIH grant S10 OD012331. The operation of NSLS-II is supported by US Department of Energy, Office of Basic Energy Sciences, under contract DE-SC0012704. Simulations were performed on machines at the Texas A&M High-Performance Research Computing Facility.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.E.K. is a guest editor invited by the Editorial Board.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, https://www.wwpdb.org (PDB ID codes 6U28 and 6OX7).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920582117/-/DCSupplemental.
References
- 1.Thompson W. W., et al. , Estimating influenza-associated deaths in the United States. Am. J. Public Health 99 (suppl. 2), S225–S230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tscherne D. M., García-Sastre A., Virulence determinants of pandemic influenza viruses. J. Clin. Invest. 121, 6–13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kash J. C., et al. , Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443, 578–581 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tumpey T. M., et al. , Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310, 77–80 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Krug R. M., Garcia-Sastre A., “The NS1 protein: A master regulator of host and viral functions” in Textbook of Influenza, Webster R. G., Monto A. S., Braciale T. J., Lamb R. A., Eds. (Wiley-Blackwell, ed. 2, 2013), pp. 114–132. [Google Scholar]
- 6.Hale B. G., Randall R. E., Ortín J., Jackson D., The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89, 2359–2376 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Klemm C., Boergeling Y., Ludwig S., Ehrhardt C., Immunomodulatory nonstructural proteins of influenza A viruses. Trends Microbiol. 26, 624–636 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Heikkinen L. S., et al. , Avian and 1918 Spanish influenza a virus NS1 proteins bind to Crk/CrkL Src homology 3 domains to activate host cell signaling. J. Biol. Chem. 283, 5719–5727 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Engel D. A., The influenza virus NS1 protein as a therapeutic target. Antiviral Res. 99, 409–416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinpeter A. B., Jureka A. S., Falahat S. M., Green T. J., Petit C. M., Structural analyses reveal the mechanism of inhibition of influenza virus NS1 by two antiviral compounds. J. Biol. Chem. 293, 14659–14668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu D., et al. , Novel influenza virus NS1 antagonists block replication and restore innate immune function. J. Virol. 83, 1881–1891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krug R. M., Functions of the influenza A virus NS1 protein in antiviral defense. Curr. Opin. Virol. 12, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Sastre A., et al. , Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252, 324–330 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Rajsbaum R., et al. , Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 8, e1003059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koliopoulos M. G., et al. , Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition. Nat. Commun. 9, 1820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrhardt C., et al. , Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81, 3058–3067 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X., et al. , An NS-segment exonic splicing enhancer regulates influenza A virus replication in mammalian cells. Nat. Commun. 8, 14751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., et al. , Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4, 896–899 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Bornholdt Z. A., Prasad B. V., X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature 456, 985–988 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min J. Y., Krug R. M., The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 7100–7105 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin C., et al. , Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J. Biol. Chem. 282, 20584–20592 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Ehrhardt C., et al. , Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 8, 1336–1348 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Hale B. G., et al. , Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc. Natl. Acad. Sci. U.S.A. 107, 1954–1959 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das K., et al. , Structural basis for suppression of a host antiviral response by influenza A virus. Proc. Natl. Acad. Sci. U.S.A. 105, 13093–13098 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Q., et al. , The molecular mechanisms underlying the hijack of host proteins by the 1918 Spanish influenza virus. ACS Chem. Biol. 12, 1199–1203 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Obenauer J. C., et al. , Large-scale sequence analysis of avian influenza isolates. Science 311, 1576–1580 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Hale B. G., Jackson D., Chen Y. H., Lamb R. A., Randall R. E., Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. U.S.A. 103, 14194–14199 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ylösmäki L., Schmotz C., Ylösmäki E., Saksela K., Reorganization of the host cell Crk(L)-PI3 kinase signaling complex by the influenza A virus NS1 protein. Virology 484, 146–152 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Liu P., Cheng H., Roberts T. M., Zhao J. J., Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 8, 627–644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallacher M., et al. , Cation currents in human airway epithelial cells induced by infection with influenza A virus. J. Physiol. 587, 3159–3173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayllon J., Hale B. G., García-Sastre A., Strain-specific contribution of NS1-activated phosphoinositide 3-kinase signaling to influenza A virus replication and virulence. J. Virol. 86, 5366–5370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayllon J., García-Sastre A., Hale B. G., Influenza A viruses and PI3K: Are there time, place and manner restrictions? Virulence 3, 411–414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jureka A. S., Kleinpeter A. B., Cornilescu G., Cornilescu C. C., Petit C. M., Structural basis for a novel interaction between the NS1 protein derived from the 1918 influenza virus and RIG-I. Structure 23, 2001–2010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark A. M., Nogales A., Martinez-Sobrido L., Topham D. J., DeDiego M. L., Functional evolution of influenza virus NS1 protein in currently circulating human 2009 pandemic H1N1 viruses. J. Virol. 91, e00721-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kainov D. E., et al. , Differential effects of NS1 proteins of human pandemic H1N1/2009, avian highly pathogenic H5N1, and low pathogenic H5N2 influenza A viruses on cellular pre-mRNA polyadenylation and mRNA translation. J. Biol. Chem. 286, 7239–7247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayllon J., Russell R. J., García-Sastre A., Hale B. G., Contribution of NS1 effector domain dimerization to influenza A virus replication and virulence. J. Virol. 86, 13095–13098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aramini J. M., et al. , Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: An interface with multiple functions. J. Biol. Chem. 286, 26050–26060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerry P. S., et al. , A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS One 6, e17946 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrillo B., et al. , The influenza A virus protein NS1 displays structural polymorphism. J. Virol. 88, 4113–4122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen Q., Cho J. H., The structure and conformational plasticity of the nonstructural protein 1 of the 1918 influenza A virus. Biochem. Biophys. Res. Commun. 518, 178–182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatt V. S., Zeng D., Krieger I., Sacchettini J. C., Cho J. H., Binding mechanism of the N-terminal SH3 domain of CrkII and proline-rich motifs in cAbl. Biophys. J. 110, 2630–2641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perozzo R., Folkers G., Scapozza L., Thermodynamics of protein-ligand interactions: History, presence, and future aspects. J. Recept. Signal Transduct. Res. 24, 1–52 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Kortemme T., Baker D., A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang K., et al. , Structural basis for influenza virus NS1 protein block of mRNA nuclear export. Nat. Microbiol. 4, 1671–1679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davey N. E., Travé G., Gibson T. J., How viruses hijack cell regulation. Trends Biochem. Sci. 36, 159–169 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Kneller J. M., Lu M., Bracken C., An effective method for the discrimination of motional anisotropy and chemical exchange. J. Am. Chem. Soc. 124, 1852–1853 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Loria J. P., Rance M., Palmer A. G., A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999). [Google Scholar]
- 48.Carr H. Y., Purcell E. M., Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 94, 630–638 (1954). [Google Scholar]
- 49.Meiboom S., Gill D., Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 29, 688–691 (1958). [Google Scholar]
- 50.Carver J. P., Richards R. E., A general state-site solution for the chemical exchange produced dependence of T2 upon the Carr-Purcell pulse separation. J. Magn. Reson. 6, 89–105 (1972). [Google Scholar]
- 51.Han B., Liu Y., Ginzinger S. W., Wishart D. S., SHIFTX2: Significantly improved protein chemical shift prediction. J. Biomol. NMR 50, 43–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell G. R., et al. , Induction of autophagy by PI3K/MTOR and PI3K/MTOR/BRD4 inhibitors suppresses HIV-1 replication. J. Biol. Chem. 293, 5808–5820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diehl N., Schaal H., Make yourself at home: Viral hijacking of the PI3K/Akt signaling pathway. Viruses 5, 3192–3212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hrincius E. R., et al. , Phosphatidylinositol-3-kinase (PI3K) is activated by influenza virus vRNA via the pathogen pattern receptor Rig-I to promote efficient type I interferon production. Cell. Microbiol. 13, 1907–1919 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Yeon S. H., Song M. J., Kang H. R., Lee J. Y., Phosphatidylinositol-3-kinase and Akt are required for RIG-I-mediated anti-viral signalling through cross-talk with IPS-1. Immunology 144, 312–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayman A., et al. , Variation in the ability of human influenza A viruses to induce and inhibit the IFN-beta pathway. Virology 347, 52–64 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Lopes A. M., Domingues P., Zell R., Hale B. G., Structure-guided functional annotation of the influenza A virus NS1 protein reveals dynamic evolution of the p85β-binding site during circulation in humans. J. Virol. 91, e01081-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collaborative Computational Project, Number 4 , The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 59.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DiFabio J., et al. , The life science x-ray scattering beamline at NSLS-II. AIP Conf. Proc. 1741, 030049 (2016). [Google Scholar]
- 62.Yang L., Using an in-vacuum CCD detector for simultaneous small- and wide-angle scattering at beamline X9. J. Synchrotron Radiat. 20, 211–218 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Schneidman-Duhovny D., Hammel M., Tainer J. A., Sali A., FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44, W424–W429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brooks B. R., et al. , CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hart K., et al. , Optimization of the CHARMM additive force field for DNA: Improved treatment of the BI/BII conformational equilibrium. J. Chem. Theory Comput. 8, 348–362 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hynninen A. P., Crowley M. F., New faster CHARMM molecular dynamics engine. J. Comput. Chem. 35, 406–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of the crystal structures were deposited in the PDB (ID codes 6U28 and 6OX7). NMR resonances of 1918 and Ud NS1ED are available from the BMRB. All data discussed in the paper will be made available to readers. All DNA plasmids for expressing 1918 and Ud NS1ED will be available upon request.