Significance
Efficient ATP synthesis requires the coordinated functions of proteins and RNAs produced from both the nuclear and mitochondrial genomes. However, the importance of coevolution between the genomes in maintaining these interactions is highly debated. Here we assess the role of coevolution within populations by comparing the nuclear and mitochondrial genotypes of high- and low-fitness hybrids between genetically divergent populations of a marine copepod. High-fitness hybrids demonstrated elevated mitochondrial adenosine triphosphate synthesis and large biases for nuclear alleles from the same population as their mitochondrial genome. These results suggest that selection strongly favors coevolved mitochondrial and nuclear genes in natural populations. Disruption of mitonuclear compatibility, as may occur during secondary contact between populations, results in substantial reductions in hybrid fitness.
Keywords: copepod, mitonuclear, coevolution, intergenomic, incompatibilities
Abstract
Oxidative phosphorylation, the primary source of cellular energy in eukaryotes, requires gene products encoded in both the nuclear and mitochondrial genomes. As a result, functional integration between the genomes is essential for efficient adenosine triphosphate (ATP) generation. Although within populations this integration is presumably maintained by coevolution, the importance of mitonuclear coevolution in key biological processes such as speciation and mitochondrial disease has been questioned. In this study, we crossed populations of the intertidal copepod Tigriopus californicus to disrupt putatively coevolved mitonuclear genotypes in reciprocal F2 hybrids. We utilized interindividual variation in developmental rate among these hybrids as a proxy for fitness to assess the strength of selection imposed on the nuclear genome by alternate mitochondrial genotypes. Developmental rate varied among hybrid individuals, and in vitro ATP synthesis rates of mitochondria isolated from high-fitness hybrids were approximately two-fold greater than those of mitochondria isolated from low-fitness individuals. We then used Pool-seq to compare nuclear allele frequencies for high- or low-fitness hybrids. Significant biases for maternal alleles were detected on 5 (of 12) chromosomes in high-fitness individuals of both reciprocal crosses, whereas maternal biases were largely absent in low-fitness individuals. Therefore, the most fit hybrids were those with nuclear alleles that matched their mitochondrial genotype on these chromosomes, suggesting that mitonuclear effects underlie individual-level variation in developmental rate and that intergenomic compatibility is critical for high fitness. We conclude that mitonuclear interactions can have profound impacts on both physiological performance and the evolutionary trajectory of the nuclear genome.
Oxidative phosphorylation in the mitochondria is central to the functioning of essentially all eukaryotic cells and thus is critical for the majority of complex life (1–4). Over evolutionary time most mitochondrial genes have translocated to the nucleus, but a small number that are necessary for adenosine triphosphate (ATP) generation are still encoded within metazoan mitochondria: typically 13 protein-coding, 2 ribosomal RNA, and 22 transfer RNA (tRNA) genes in bilaterian animals (5). These genes require functional interactions with nuclear-encoded proteins, and thus mitochondrial performance relies upon integration between the nuclear and mitochondrial genomes (1–3). Consequently, there is predicted to be strong selection for mitonuclear compatibility between interacting genes (i.e., coevolution) in isolated populations and species (6, 7).
If strong selection leads to coevolved mitonuclear interactions within populations, then one would predict that these interactions might be disrupted by hybridization when isolated populations experience secondary contact (8). Indeed, mismatches between mitochondrial-encoded and nuclear-encoded alleles in hybrids can have profound negative phenotypic consequences for many traits (8), and this “hybrid breakdown” has been demonstrated across many eukaryotic taxa, ranging from diseases in humans (9) to life-history effects in invertebrates (10–12). These mitonuclear examples of Bateson–Dobzhansky–Muller incompatibilities (13–15) may have important implications for key biological processes, including development of postzygotic isolation between species (16–18) and potential health consequences of mitochondrial replacement therapies in humans (19). However, the ubiquity and relevance of these implications have been questioned (7, 20). Therefore, determining the extent to which mitonuclear interactions influence evolution of the nuclear genome and the degree to which intergenomic incompatibilities result in negative fitness consequences is critical for understanding the role of mitochondrial DNA in shaping the physiological performance and evolution of eukaryotes.
In the current study, we address these issues with interpopulation hybrids of San Diego, California (SD) and Santa Cruz, California (SC) Tigriopus californicus. This species of copepod is found in supralittoral tidepools along the west coast of North America from Baja California, Mexico, to Alaska, United States, with extremely low gene flow between isolated populations on different rocky outcrops (21). This isolation has led to high levels of genetic divergence among populations (21–26), and F2 hybrids from laboratory crosses between T. californicus populations typically display breakdown of mitochondrial ATP synthesis and several fitness-related life-history traits, including fecundity and developmental rate (10, 11, 27–29). The loss of performance in hybrids is recovered by backcrosses to the maternal, but not the paternal, parental population (11, 30), which, since mitochondrial DNA is maternally inherited (8), clearly implicates a role for mitonuclear interactions in hybrid breakdown in this species.
Here, we reasoned that, if there is strong selection for mitonuclear compatibility throughout ontogeny (6), then there should be clear physiological and genetic associations with variation in fitness-related traits among F2 hybrids. Specifically, we hypothesized that high-fitness hybrids have improved mitochondrial performance compared to low-fitness hybrids and that this improved performance is associated with biases for maternal nuclear alleles that match the mitochondrial genotype in both SD♀×SC♂ and SC♀×SD♂ high-fitness hybrids (i.e., biases for different parental alleles in each cross). We utilized interindividual differences in developmental rate and ATP synthesis rate among hybrids in combination with Pool-seq to test these hypotheses and to assess the potential strength of selection for compatible mitochondrial and nuclear genomes in eukaryotes.
Results
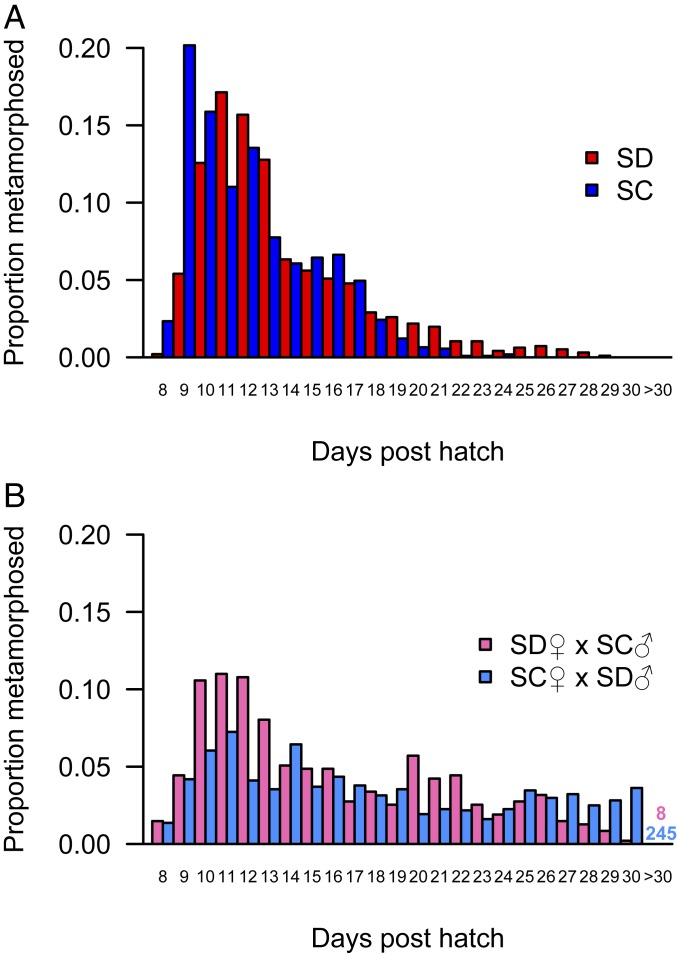
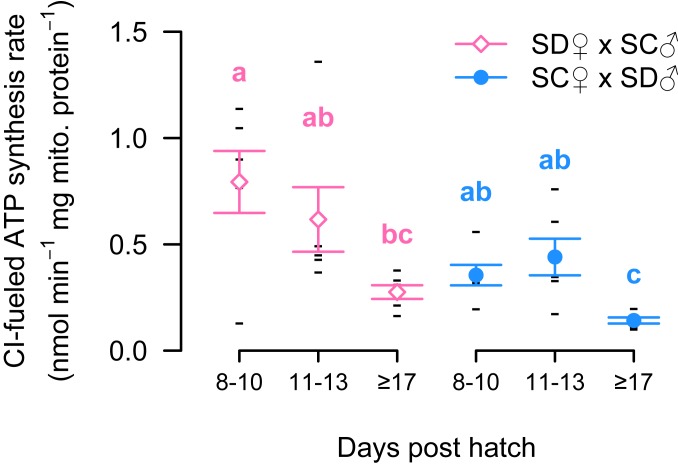
Developmental rates were similar in both parental populations of T. californicus with metamorphosis to the copepodid I stage occurring 8 to 22 days post hatch (dph) for ∼98% of nauplii (maximum dph of 29 and 24 for SD and SC, respectively; Fig. 1A). In contrast, the distributions of developmental times among F2 hybrids from both reciprocal crosses demonstrated substantial shifts toward greater dph to metamorphosis compared to the parental populations, which is consistent with hybrid breakdown (Fig. 1B). In both crosses, metamorphosis was observed 8 to 30 dph with 8 of 473 SD♀×SC♂ nauplii and 245 of 1,242 SC♀×SD♂ nauplii still present on day 30, which were scored as >30 dph. Preliminary data for pure SD and SC nauplii suggest that the majority of offspring underwent metamorphosis 9 to 16 dph, and as a result F2 hybrids were split into 8 to 10, 11 to 13 and ≥17 dph groups to assess maximal mitochondrial ATP synthesis rates. Complex I-fueled ATP synthesis rates were significantly affected by both cross (F1,30 = 11.32; P = 2.1 × 10−3) and developmental group (F2,30 = 13.44; P = 6.8 × 10−5) with no interaction between factors (F2,30 = 0.44; P = 0.65), and post hoc tests indicated that faster developing (8 to 10 dph) copepods had higher ATP synthesis rates than more slowly developing (≥17 dph) copepods in both crosses (P ≤ 0.04; Fig. 2). ATP synthesis rates in copepods with intermediate developmental rates (11 to 13 dph) were similar to those of faster developing hybrids in the SC♀×SD♂ cross (P = 0.99) and intermediate between faster and slower developing hybrids in the SD♀×SC♂ cross (P ≥ 0.13; Fig. 2).
Fig. 1.
Developmental time to metamorphosis for T. californicus nauplii as proportions of all individuals. (A) SD (red; n = 963) and SC (blue; n = 1,071). (B) SD♀×SC♂ (pink; n = 473) and SC♀×SD♂ (light blue; n = 1,242) F2 hybrids. The pink and light blue numbers in B display the number of nauplii remaining at 30 dph for each cross.
Fig. 2.
Maximal complex I (CI)-fueled ATP synthesis rates for adult F2 hybrids that metamorphosed 8 to 10, 11 to 13, and ≥17 dph for both reciprocal crosses (mean ± SEM for SD♀×SC♂: empty pink diamonds and for SC♀xSD♂: filled light blue circles; individual data points: black dashes; n = 6 per group). Shared lowercase letters indicate groups that do not differ significantly.
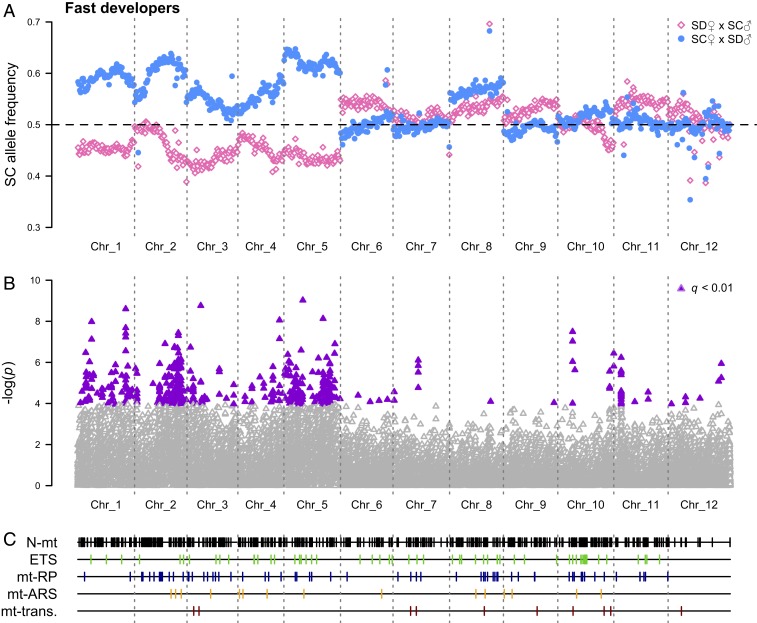
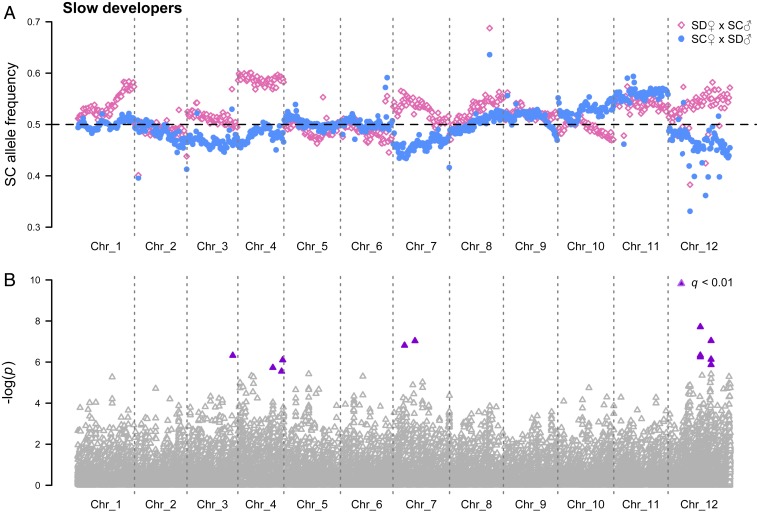
F2 hybrids from a second set of reciprocal crosses were divided into those that metamorphosed 8 to 12 or >22 dph (fast or slow developers, respectively), and Pool-seq was used to test if nuclear allele frequencies responded to differences in mitochondrial genotype between the crosses (i.e., SD versus SC). Comparisons between reciprocal fast-developing hybrids demonstrated significant differences in nuclear allele frequencies across large regions of chromosomes 1 to 5 (Fig. 3 and Dataset S1), and these deviations were consistent with substantial biases favoring maternal (i.e., coevolved) alleles in both crosses. In contrast, significant allele frequency deviations between the crosses were essentially absent in slow developers with few differences in individual single-nucleotide polymorphisms (SNPs) (Fig. 4). Tests based on individual SNPs have relatively low power to detect allele frequency variations in F2 hybrids. Thus, we performed an additional exploratory analysis using Kolmogorov–Smirnov (KS) tests, which have greater statistical power but increase the possibility of false positives (see SI Appendix for details). In fast developers, these tests confirmed excesses of coevolved alleles on chromosomes 1 to 5 (P ≤ 5.8 × 10−4), but also found biases for paternal (i.e., mismatched) alleles on chromosomes 6 and 9 (P = 1.6 × 10−4 for both), and in slow developers, mismatched alleles were in excess on chromosomes 1, 3, 4, and 7 (P ≤ 7.4 × 10−4).
Fig. 3.
SC allele frequencies for 759 250-kb chromosomal windows in fast developing F2 hybrids. (A) SD♀×SC♂: empty pink diamonds and SC♀xSD♂: filled light-blue circles. (B) Statistical results for individual loci with ≥80× coverage (significant differences: filled purple triangles). (C) The locations of 599 nuclear-encoded mitochondrial genes (all N-mt genes: black; classes of NO-mt genes—electron transport system [ETS]: green; mitochondrial ribosomal proteins [mt-RP]: dark blue; mitochondrial aminoacyl tRNA synthetases [mt-ARS]: orange; mitochondrial transcription and DNA replication [mt-trans.]: dark red).
Fig. 4.
SC allele frequencies for 759 250-kb chromosomal windows in slow developing F2 hybrids. (A) SD♀×SC♂: empty pink diamonds and SC♀×SD♂: filled light-blue circles). (B) Statistical results for individual loci with ≥80× coverage (significant differences: filled purple triangles).
As a secondary examination of potential mitonuclear effects on F2 hybrid allele frequencies and fitness, we compared fast and slow developers within each reciprocal cross. When SNPs were tested independently, there was limited support for variation between fast and slow developers, most likely due to relatively small differences between these pools (SI Appendix, Fig. S1). In general, the results of exploratory analyses with KS tests between fast and slow developers suggest similar patterns of variation as the intercross comparisons between fast developers (SI Appendix, Fig. S1). Fast developers had higher coevolved allele frequencies than slow developers on chromosomes 1, 3, 4, and 5 in SD♀×SC♂ hybrids and on chromosomes 1, 2, 3, 4, 5, 7, and 8 in SC♀×SD♂ hybrids. Only one chromosome demonstrated the opposite pattern with elevated mismatched allele frequencies in fast developers compared to slow developers on chromosome 11 in the SC♀×SD♂ cross. Taken together, our results suggest that mitonuclear interactions are major genetic factors contributing to interindividual variation in developmental rate among F2 hybrids and that in the majority of cases at least partial maintenance of coevolved mitonuclear genotypes is critical for enhanced performance in this fitness-related trait.
Discussion
Mitochondrial DNA contains relatively few genes, but because of the functional products encoded by these genes and their interactions with nuclear gene products, differences in mitochondrial genotype have been predicted to exert strong selection pressures on the nuclear genome throughout ontogeny (6). Our study demonstrates variations in developmental rate, ATP synthesis rate, and nuclear allele frequencies among F2 hybrids that are consistent with strong selection favoring compatible mitonuclear interactions within even a single generation. Mitochondrial genotype had an overall effect on variation in ATP synthesis rate between the reciprocal crosses, but in both crosses fast developers had higher synthesis rates than slow developers. In the high-fitness (i.e., fast-developing) hybrids, there were also substantial deviations from expected neutral nuclear allele frequencies of 0.5 that favored alleles from the same population as the mitochondrial genome. Effects of mitonuclear coevolution were not evenly spread across the nuclear genome, but involved at least 5 of the 12 chromosomes with clear deviations favoring coevolved alleles on chromosomes 1 to 5. Relative to previous studies in T. californicus hybrids (31–33), this clear pattern toward partial recovery of coevolved mitonuclear genotypes is most likely a consequence of selecting individuals based on variation in a fitness-related trait that has been correlated with mitochondrial performance in this species (30).
Although the average chromosome-wide allele frequency deviations favoring coevolved nuclear alleles in the current study may appear modest (ranging from 0.032 to 0.120 with some chromosomal regions reaching ∼0.147; Fig. 3), the magnitudes of these deviations need to be interpreted relative to general expectations for F2 hybrids. In T. calfornicus, there is little evidence for selection against heterozygous F2 hybrids (31, 33), and F1 hybrids between SD and SC (heterozygous across all fixed SNPs) generally show enhanced fitness compared to parentals (11). Therefore, it is likely that the major allele frequency deviations in our study are consequences of negative effects associated with one of the two possible homozygous genotypes. As a result, given Mendelian segregation ratios of 1:2:1 in F2 hybrids, the most extreme biases for maternal alleles observed here are likely indicative of up to 77 to 91% deficits of homozygous paternal genotypes in fast developers on some regions of these chromosomes. These calculations exclude any error associated with the allele frequencies estimated for our DNA pools; however, even if these deficits represent moderate overestimates, they are sufficiently large that our data clearly demonstrate strong selection favoring mitonuclear compatibility. Additionally, we observed little evidence for allele frequency variation consistent with nuclear-only effects. If these potential effects are examined as in Lima et al. (32), only frequency variations on chromosome 8 in fast developers and on chromosome 11 in slow developers may be indicative of modest effects of nuclear genetic variation alone. Yet, deviations favoring coevolved alleles in fast developers were rarely symmetrical between the reciprocal crosses. This may simply reflect sampling or technical variation associated with our experiment, and the effects of mitonuclear incompatibilities are not necessarily of equal magnitudes in reciprocal crosses (6). An alternative possibility is that relatively weak nuclear-only effects on chromosomes 1 to 5 also shape allele frequency variation in our study. Regardless, our results support a key role for mitonuclear incompatibilities in loss of fitness in these hybrids.
Previous studies have demonstrated at least three candidate mechanisms involved in coevolution in T. californicus: electron transport system complex activities (11, 34–37), mitochondrial transcription (38), and mitonuclear ribosomal interactions (39). Yet, these candidate gene studies do not directly reveal the number or relative importance of mitonuclear incompatibilities contributing to hybrid breakdown in this species. In comparison, our Pool-seq approach provides an unbiased examination of the genomic architecture of breakdown of developmental rate in hybrids between SD and SC. The increased frequencies of maternal alleles across multiple genomic regions in our most fit hybrids clearly indicate a polygenic basis for mitonuclear coevolution, which may be attributable to the high level of divergence between the mitochondrial genomes of these populations (21.7%) (22). However, due to the central role of the mitochondrion in metabolism, even minor disruption of mitonuclear interactions may have major fitness effects (6, 8). For example, mutations in a single nuclear-encoded mitochondrial tRNA synthetase and one mitochondrial tRNA lead to mitochondrial dysfunction in Drosophila hybrids (12). The allele frequency variation in our slowly developing hybrids is likely consistent with large effects of relatively few interactions in T. californicus as well. Despite an approximately two-fold reduction in both developmental rate and ATP synthesis rate (Figs. 1 and 2), strong deviations favoring paternal alleles in slow developers were largely absent in our study (with the exception of chromosome 4 in SD♀×SC♂; Fig. 4). Therefore, our data indicate that mismatched genotypes across most sites of mitonuclear interactions on chromosomes 1 to 5 are not necessary to observe these substantial negative fitness effects. Instead, it is likely that different subsets of these potential mismatches among F2 hybrids are sufficient to cause similar decreases in developmental rate. This is in stark contrast to the situation in high-fitness hybrids in which large biases favoring maternal alleles were observed across chromosomes 1 to 5, suggesting that highly compatible mitonuclear genotypes are necessary for high fitness.
Of the 1,000 to 1,500 nuclear-encoded mitochondrial (N-mt) genes (nuclear genes encoding products that are imported into the mitochondria) in metazoans, at least 180 are expected to have intimate functional interactions with either mitochondrial DNA or mitochondrial-encoded gene products (NO-mt genes) (40). Barreto et al. (22) identified 599 putative N-mt genes, including 139 NO-mt genes, in the T. californicus genome (Fig. 3C). Although our data begin to resolve which of these candidates may play the largest roles in intergenomic coevolution, there was little resolution of allele frequency deviations beyond the level of chromosomes in our hybrids. This is likely a consequence of only a single opportunity for interpopulation recombination in F2 hybrids (41) or the involvement of multiple loci on the same chromosome (42). Although N-mt genes were not more common on the chromosomes with biases for coevolved alleles than on other chromosomes in our study, chromosomes demonstrating allelic biases tended to have relatively higher ratios of NO-mt genes to other N-mt genes (SI Appendix, Fig. S2), which is consistent with a disproportionate role for NO-mt genes in mitonuclear interactions.
Taken together, our data demonstrate strong selection against disruption of coevolved genes following hybridization and, conversely, strong selection for intergenomic compatibility within populations and species. These effects of mitonuclear interactions were sufficiently strong in T. californicus that selection for rapid development within a single generation identified key sites of mitonuclear interactions across the genome. Mitonuclear coevolution in this species may be exceptionally strong (27), but our results also suggest that even small numbers of mitonuclear incompatibilities may result in substantial losses of fitness. Thus, the findings of the current study demonstrate the possibility of large effects of intergenomic incompatibilities in eukaryotes and suggest that these incompatibilities have the potential to contribute to reproductive isolation between populations across many taxa (16–18).
Materials and Methods
Adult copepods were collected from intertidal splash pools near San Diego, California (SD: 32° 45′ N, 117° 15′ W) and Santa Cruz, California (SC: 36° 56′ N, 122° 02′ W) and were split into 200-mL laboratory cultures in glass beakers containing filtered seawater at 20 °C, 36 parts per thousand salinity, and 12 h light:12 h dark. Culturing procedures generally followed the methods of Tsuboko-Ishii and Burton (43), but virgin females of each population were obtained by separating precopulatory breeding pairs (44, 45). Separated males and females were used to make reciprocal interpopulation crosses: 40 matings for ATP synthesis assays and 120 matings for Pool-seq for each reciprocal. Mature (red) egg sacs were dissected from gravid parental and F1 females, and developmental time to metamorphosis in offspring (i.e., from hatching to copepodid stage I) was scored individually as in Harada et al. (46).
For ATP synthesis assays, F2 hybrids from each reciprocal cross were divided into those that metamorphosed 8 to 10, 11 to 13, and ≥17 dph and were allowed to reach adulthood. Assays for six pools of six adults were conducted for each group following the protocols of Harada et al. (46), and variation in synthesis rates among groups was assessed by two-way ANOVA with cross and developmental group as factors followed by Tukey post hoc tests in R v3.4.0 (The R Foundation, Vienna; α = 0.05). For Pool-seq, F2 hybrids from each reciprocal cross were grouped into those that metamorphosed 8 to 12 dph (“fast developers”) or >22 dph (“slow developers”). A group of 180 adults was pooled for each group, and genomic DNA was isolated by phenol-chloroform extraction (47). Whole-genome 150-bp paired-end sequencing was performed at Novogene (Sacramento, CA) on a NovaSeq. 6000 (Illumina, San Diego, CA). Sequencing reads were trimmed and filtered as in Lima et al. (32) and mapped to the SD T. californicus reference genome v2.1 and an updated reference genome for the SC population (22) with BWA MEM v0.7.12 (48). Allele frequencies at fixed SNPs between the parental populations were determined with PoPoolation2 (49) as described elsewhere (32, 41). As large blocks of parental chromosomes are inherited together in F2 hybrids due to only one generation of interpopulation recombination (41), allele frequencies for 1,910,010 SNPs with ≥50× coverage were then averaged for 250-kb windows along each chromosome (759 windows total). Frequency differences between pools were assessed by calculation of Z-statistics as in Huang et al. (50) for individual SNPs with ≥80× coverage (42,502 SNPs; α = 0.01). As these tests have low power to detect small allele frequency deviations, such as those expected in most cases in F2 hybrids (32, 41), we also performed additional exploratory analyses at the chromosomal-level using KS tests similar to previously published approaches (32, 41). The numbers of SNPs, windows, SNPs per window (μ ± σ), coverages, summary allele frequencies, and KS test P values for each chromosome are presented in SI Appendix, Tables S1 and S2. Additional details for all methods used in the current study are also provided in SI Appendix, Supplemental Methods.
Data Availability.
The raw sequencing reads and associated sample metadata generated in this report have been deposited in the National Center for Biotechnology Information Sequence Read Archive and BioProject databases (PRJNA606908), and all other datasets have been deposited in the Dryad Digital Repository (DOI: 10.5061/dryad.x69p8czf4) (51).
Supplementary Material
Acknowledgments
The current study was funded by National Science Foundation Grants DEB1556466 and IOS1754347 (to R.S.B.). We thank Drs. Felipe Barreto and Thiago Lima for advice regarding the sequencing methods and analyses and Laura Furtado, Antonia Bock, and Rebecca Pak for assistance with copepod culturing.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The raw sequencing reads and associated sample metadata generated in this report have been deposited in the National Center for Biotechnology Information Sequence Read Archive and BioProject databases (PRJNA606908), and all other datasets have been deposited in the Dryad Digital Repository (DOI: 10.5061/dryad.x69p8czf4).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910141117/-/DCSupplemental.
References
- 1.Hill G. E., Mitonuclear ecology. Mol. Biol. Evol. 32, 1917–1927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane N., Power, Sex, Suicide: Mitochondria and the Meaning of Life (Oxford University Press, 2005). [Google Scholar]
- 3.Rand D. M., Haney R. A., Fry A. J., Cytonuclear coevolution: The genomics of cooperation. Trends Ecol. Evol. 19, 645–653 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Wallace D. C., Colloquium paper: Bioenergetics, the origins of complexity, and the ascent of man. Proc. Natl. Acad. Sci. U.S.A. 107 (suppl. 2), 8947–8953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin L., Blumberg A., Barshad G., Mishmar D., Mito-nuclear co-evolution: The positive and negative sides of functional ancient mutations. Front. Genet. 5, 448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill G. E., et al. , Assessing the fitness consequences of mitonuclear interactions in natural populations. Biol. Rev. Camb. Philos. Soc. 94, 1089–1104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sloan D. B., Havird J. C., Sharbrough J., The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol. Ecol. 26, 2212–2236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton R. S., Barreto F. S., A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol. Ecol. 21, 4942–4957 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Wallace D. C., Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 51, 440–450 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Burton R. S., Hybrid breakdown in developmental time in the copepod Tigriopus californicus. Evolution 44, 1814–1822 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Ellison C. K., Burton R. S., Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution 62, 631–638 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Meiklejohn C. D., et al. , An incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 9, e1003238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateson W., “Heredity and variation in modern lights” in Darwin and Modern Science, Seward A. C., Ed. (Cambridge University Press, 1909), pp. 85–101. [Google Scholar]
- 14.Dobzhansky T. H., Genetics and the Origin of Species (Columbia University Press, 1937). [Google Scholar]
- 15.Muller H. J., Isolating mechanisms, evolution, and temperature. Biol. Symp. 6, 71–125 (1942). [Google Scholar]
- 16.Gershoni M., Templeton A. R., Mishmar D., Mitochondrial bioenergetics as a major motive force of speciation. BioEssays 31, 642–650 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Hill G. E., Mitonuclear coevolution as the genesis of speciation and the mitochondrial DNA barcode gap. Ecol. Evol. 6, 5831–5842 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill G. E., Reconciling the mitonuclear compatibility species concept with rampant mitochondrial introgression. Integr. Comp. Biol. 59, 912–924 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Reinhardt K., Dowling D. K., Morrow E. H., Medicine. Mitochondrial replacement, evolution, and the clinic. Science 341, 1345–1346 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Eyre-Walker A., Mitochondrial replacement therapy: Are mito-nuclear interactions likely to be a problem? Genetics 205, 1365–1372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burton R. S., Genetic evidence for long term persistence of marine invertebrate populations in an ephemeral environment. Evolution 51, 993–998 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Barreto F. S., et al. , Genomic signatures of mitonuclear coevolution across populations of Tigriopus californicus. Nat. Ecol. Evol. 2, 1250–1257 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Burton R. S., Lee B. N., Nuclear and mitochondrial gene genealogies and allozyme polymorphism across a major phylogeographic break in the copepod Tigriopus californicus. Proc. Natl. Acad. Sci. U.S.A. 91, 5197–5201 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edmands S., Phylogeography of the intertidal copepod Tigriopus californicus reveals substantially reduced population differentiation at northern latitudes. Mol. Ecol. 10, 1743–1750 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Pereira R. J., Barreto F. S., Pierce N. T., Carneiro M., Burton R. S., Transcriptome-wide patterns of divergence during allopatric evolution. Mol. Ecol. 25, 1478–1493 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Peterson D. L., et al. , Reproductive and phylogenetic divergence of tidepool copepod populations across a narrow geographical boundary in Baja California. J. Biogeogr. 40, 1664–1675 (2013). [Google Scholar]
- 27.Burton R. S., Ellison C. K., Harrison J. S., The sorry state of F2 hybrids: Consequences of rapid mitochondrial DNA evolution in allopatric populations. Am. Nat. 168 (suppl. 6), S14–S24 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Edmands S., Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution 53, 1757–1768 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Ganz H. H., Burton R. S., Genetic differentiation and reproductive incompatibility among Baja California populations of the copepod Tigriopus californicus. Mar. Biol. 123, 821–827 (1995). [Google Scholar]
- 30.Ellison C. K., Burton R. S., Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution 60, 1382–1391 (2006). [PubMed] [Google Scholar]
- 31.Foley B. R., Rose C. G., Rundle D. E., Leong W., Edmands S., Postzygotic isolation involves strong mitochondrial and sex-specific effects in Tigriopus californicus, a species lacking heteromorphic sex chromosomes. Heredity 111, 391–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lima T. G., Burton R. S., Willett C. S., Genomic scans reveal multiple mito-nuclear incompatibilities in population crosses of the copepod Tigriopus californicus. Evolution 73, 609–620 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Pritchard V. L., et al. , Interpopulation hybridization results in widespread viability selection across the genome in Tigriopus californicus. BMC Genet. 12, 54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison J. S., Burton R. S., Tracing hybrid incompatibilities to single amino acid substitutions. Mol. Biol. Evol. 23, 559–564 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Rawson P. D., Burton R. S., Functional coadaptation between cytochrome c and cytochrome c oxidase within allopatric populations of a marine copepod. Proc. Natl. Acad. Sci. U.S.A. 99, 12955–12958 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willett C. S., Burton R. S., Viability of cytochrome c genotypes depends on cytoplasmic backgrounds in Tigriopus californicus. Evolution 55, 1592–1599 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Willett C. S., Burton R. S., Environmental influences on epistatic interactions: Viabilities of cytochrome c genotypes in interpopulation crosses. Evolution 57, 2286–2292 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Ellison C. K., Burton R. S., Genotype-dependent variation of mitochondrial transcriptional profiles in interpopulation hybrids. Proc. Natl. Acad. Sci. U.S.A. 105, 15831–15836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barreto F. S., Burton R. S., Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol. Biol. Evol. 30, 310–314 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Burton R. S., Pereira R. J., Barreto F. S., Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst. 44, 281–302 (2013). [Google Scholar]
- 41.Lima T. G., Willett C. S., Using Pool-seq to search for genomic regions affected by hybrid inviability in the copepod T. californicus. J. Hered. 109, 469–476 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Willett C. S., Lima T. G., Kovaleva I., Hatfield L., Chromosome-wide impacts on the expression of incompatibilities in hybrids of Tigriopus californicus. G3 (Bethesda) 6, 1739–1749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuboko-Ishii S., Burton R. S., Individual culturing of Tigriopus copepods and quantitative analysis of their mate-guarding behavior. J. Vis. Exp. 139, e58378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burton R. S., Mating system of the intertidal copepod Tigriopus californicus. Mar. Biol. 86, 247–252 (1985). [Google Scholar]
- 45.Burton R. S., Feldman M. W., Swisher S. G., Linkage relationships among five enzyme-coding gene loci in the copepod Tigriopus californicus: A genetic confirmation of achiasmiatic meiosis. Biochem. Genet. 19, 1237–1245 (1981). [DOI] [PubMed] [Google Scholar]
- 46.Harada A. E., Healy T. M., Burton R. S., Variation in thermal tolerance and its relationship to mitochondrial function across populations of Tigriopus californicus. Front. Physiol. 10, 213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J., Russell D. W., Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harb. Protoc. 2006, pdb-rot4455 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kofler R., Pandey R. V., Schlötterer C., PoPoolation2: Identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27, 3435–3436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang W., et al. , Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc. Natl. Acad. Sci. U.S.A. 109, 15553–15559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Healy T., Burton R., Data from “Strong selective effects of mitochondrial DNA on the nuclear genome.” Dryad Digital Repository. 10.5061/dryad.x69p8czf4. Deposited 26 February 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing reads and associated sample metadata generated in this report have been deposited in the National Center for Biotechnology Information Sequence Read Archive and BioProject databases (PRJNA606908), and all other datasets have been deposited in the Dryad Digital Repository (DOI: 10.5061/dryad.x69p8czf4) (51).