ABSTRACT
Bacterioplankton play a pivotal role in marine ecosystems. However, their temporal dynamics and underlying control mechanisms are poorly understood in tropical regions such as the Red Sea. Here, we assessed the impact of bottom-up (resource availability) and top-down (viruses and heterotrophic nanoflagellates) controls on bacterioplankton abundances by weekly sampling a coastal central Red Sea site in 2017. We monitored microbial abundances by flow cytometry together with a set of environmental variables including temperature, salinity, dissolved organic and inorganic nutrients and chlorophyll a. We distinguished five groups of heterotrophic bacteria depending on their physiological properties relative nucleic acid content, membrane integrity and cell-specific respiratory activity, two groups of Synechococcus cyanobacteria and three groups of viruses. Viruses controlled heterotrophic bacteria for most of the year, as supported by a negative correlation between their respective abundances and a positive one between bacterial mortality rates and mean viral abundances. On the contrary, heterotrophic nanoflagellates abundance covaried with that of heterotrophic bacteria. Heterotrophic nanoflagellates showed preference for larger bacteria from both the high and low nucleic acid content groups. Our results demonstrate that top-down control is fundamental in keeping heterotrophic bacterioplankton abundances low (< 5 × 10 5 cells mL−1) in Red Sea coastal waters.
Keywords: heterotrophic bacteria, Synechococcus, heterotrophic nanoflagellate, viruses, top-down control, Red Sea
The weekly variations of bacterioplankton standing stocks in the central Red Sea coastal water are apparently controlled by top-down regulators (viruses and heterotrophic nanoflagellates).
INTRODUCTION
Planktonic prokaryotes represent the largest living biomass of aquatic ecosystems (Whitman, Coleman and Wiebe 1998) and are therefore key components of marine food webs. Roughly half of the ocean's primary production is processed by heterotrophic bacteria and archaea (Ducklow 1999; Pomeroy et al. 2007), making them the main agents in the transformation and remineralization of organic matter (Azam et al. 1983; Ogawa et al. 2001; Benner and Amon 2015). Marine picocyanobacteria, mainly pertaining to the genera Synechococcus and Prochlorococcus (Scanlan 2012; Flombaum et al. 2013), are in turn significant contributors to ocean primary production (Li 1994; Arrigo 2005), forming the base of the food web in oligotrophic environments (Hagström et al. 1988; Fuhrman, Cram and Needham 2015; Armengol et al. 2019).
The variation in the stocks of bacterioplankton is ultimately regulated by the availability of nutrients (bottom-up control) and the mortality caused by protistan grazing and viral lysis (top-down control) (Gasol, Pedrós-Alió and Vaqué 2002; Tanaka and Rassoulzadegan 2004; Weinbauer 2004). Bottom-up control of autotrophic and heterotrophic bacteria has been extensively demonstrated (Ducklow and Carlson 1992; Church 2008; Marañón et al. 2012), frequently using chlorophyll a concentration as a proxy of the availability of organic and inorganic nutrients (Bouvier, Del Giorgio and Gasol 2007; Moràn, Ducklow and Erickson 2011; Lyngsgaard et al. 2017). However, weak relationships between nutrients and bacterial biomass or activity (e.g. Sommer 2000; Bettarel et al. 2002; Gomes et al. 2015) or the absence of them (Gasol, Pedrós-Alió and Vaqué 2002; Longnecker et al. 2010) have been also claimed as evidence of effective top-down control by viruses and protistan grazers. With surface abundances of ∼107 viral particles mL−1 (Weinbauer 2004; Suttle 2007; Lara et al. 2017), viral infection is one of the most significant factors leading to bacterioplankton loss (Hurst 2000), accelerating the transformation of particulate organic matter (POM) from the lysed cells to dissolved organic matter (DOM) (Dell'Anno, Corinaldesi and Danovaro 2015). Among protistan grazers, heterotrophic nanoflagellates (HNF), typically present at concentrations of 103 cells mL−1 in the upper layers of the ocean (Sherr and Sherr 2002), are the most important predators of bacteria (Pernice et al. 2015; Worden et al. 2015), providing a link from low to high trophic levels through the microbial loop (Azam et al. 1983; Sherr and Sherr 2002).
Comparative analyses of the relationships between the abundance of bacteria and their major mortality agents (Miki and Jacquet 2008; Bunse and Pinhassi 2017) have revealed all possible options: (i) viral lysis is responsible for the largest fraction losses of heterotrophic bacteria and cyanobacteria in oligotrophic systems (Boras et al. 2009; Bouvy et al. 2011; Vaqué et al. 2017); (ii) protistan grazers are the dominant agents of bacterial mortality (Choi, Hwang and Cho 2003; Ng and Liu 2016; Livanou et al. 2019) and (iii) both protistan grazing and viral lysis cause similar loss rates (Fuhrman and Noble 1995; Hwang and Cho 2002). The relative contribution of HNF and viruses to bacterial mortality has also been shown to vary with seasons (Bouvy et al. 2015; Fuhrman, Cram and Needham 2015; Bunse and Pinhassi 2017) and depth (Herndl et al. 2008; Nagata et al. 2010; Lara et al. 2017). Moreover, although bacterioplankton are the preferred food source of HNF (Sanders, Caron and Berninger 1992; Boenigk and Arndt 2002), they can also feed on viruses (Bettarel et al. 2005; Miki and Yamamura 2005).
This complex interplay between bottom-up and top-down controls is even more poorly understood when it comes to temporal variations along the annual cycle (Šolić et al. 2009; Fuhrman, Cram and Needham 2015). Typically, the seasonality of heterotrophic bacterioplankton controls has been studied much more extensively from the bottom-up (e.g. Ducklow 1999; Christaki et al. 2001; Degerman et al. 2013; Huete-Stauffer et al. 2015) than from the top-down perspective (e.g. Solid and Krstulovic 1994; Šestanović et al. 2004; Tsai, Gong and Hung 2013; Tsai, Gong and Shiau 2015). Most of the studies addressing both controls simultaneously were conducted in temperate and polar regions (e.g. Maranger et al. 2015; Bowman et al. 2017; Morán et al. 2018). In the Mediterranean, Šolić et al. (2009) concluded that bacterioplankton stocks were strongly regulated by bottom-up control during colder months and top-down regulated during the warmer season. In the vast area comprised between the tropics, temperature seems to play a minor role in regulating bacterioplankton abundances and activity as a consequence of strong nutrient limitation or high mortality rate due to HNF and viruses (Morán et al. 2017; Field et al. 1998). However, only a few studies have documented the relative importance of substrate availability and mortality factors in regulating bacterial standing stocks (i.e. Dufour and Torréton 1996; Calbet, Landry and Nunnery 2001; Bock et al. 2018) in tropical waters.
The Red Sea is one of these undersampled tropical regions, comprising the hottest and saltiest deep waters of the planet (Ngugi et al. 2012; Berumen et al. 2019). Bacterioplankton studies are scarce in this marine basin, especially those focused on the role of top-down control. Sommer (2000), Sommer et al. (2002), Berninger and Wickham (2005), Wickham, Claessens and Post (2015) and Weisse (1989) assessed bacterial mortality due to protistan grazing, while Muhling et al. (2005) and Dekel-Bird et al. (2015) estimated the impact of viral lysis on cyanobacteria at a few locations, especially the Gulf of Aqaba. However, none of these studies included the full annual variability. Investigations including both bottom-up and top-down bacterioplankton drivers are not frequent in tropical waters (e.g. Rejas, Muylaert and De Meester 2005; Gregoracci et al. 2012; Bock et al. 2018) and in the Red Sea we are only aware of the work of Wickham, Claessens and Post (2015) and Berninger and Wickham (2005). The goal of this study is to shed more light on the specific roles of resource availability and mortality due to viral lysis and protistan grazing on the standing stocks of coastal Red Sea planktonic bacteria by assessing them jointly on a weekly basis, a high and unusual sampling frequency for this type of studies regardless of the location. A recent publication suggests that heterotrophic bacterial standing stocks in shallow waters of the central Red Sea were top-down controlled by protistan gazers while their growth rates and efficiencies in DOM transformation were bottom-up controlled (Silva et al. 2019). In order to confirm this hypothesis and include viruses in the top-down control, we have evaluated the relative effect of both types of control on the standing stocks and physiological properties (i.e. cell and genome size, membrane integrity and respiratory activity) of planktonic bacteria with a high temporal resolution. By weekly monitoring surface waters in a shallow environment of the central Red Sea, we aimed at testing that: (i) top-down control has a stronger impact than bottom-up in regulating bacterioplankton standing stocks and (ii) both viral lysis and protistan grazing are important sources of bacterioplankton mortality.
MATERIAL AND METHODS
Experimental site and sample collection
Surface water was collected in 9 L acid-washed polycarbonate carboys on a weekly basis from 7 February 2017 until 29 January 2018 from the harbor of King Abdullah University of Science and Technology (KAUST), located north of Thuwal, Saudi Arabia (22° 18.412' N 39° 6.172' E).
Physicochemical variables and inorganic nutrients
Temperature and salinity were measured immediately prior to sampling with a YSI probe (Pro Plus). Inorganic nutrients were analyzed in 15 mL samples, previously filtered through pre-combusted (470°C for 5 h) glass fiber filters of 0.7 µm nominal pore size (Whatman GF/F) and stored at −20°C. A segmented flow analyzer was used to measure nitrate (NO3−), nitrite (NO2−) and phosphate (PO43−) concentrations, following the methods mentioned in Hansen and Koroleff (1999). Standards were prepared with nutrient-free seawater. Dissolved inorganic nitrogen (DIN) was estimated as the sum of nitrate and nitrite concentrations (DIN = [NO3−] + [NO2−]).
Chlorophyll a concentration was used as a proxy of phytoplankton biomass. Seawater was sequentially filtered through three 47 mm Millipore polycarbonate filters of decreasing pore-size (20, 2 and 0.2 µm). Chlorophyll a was extracted by sonicating the filters with 90% acetone. The fluorescence of each sample was measured by a fluorometer (Turner design, Trilogy) calibrated with a chlorophyll standard (Anacystis nidulans, Sigma-Aldrich).
Dissolved organic carbon (DOC) and total dissolved nitrogen (TDN) were determined from 40 mL samples filtered through pre-combusted (470°C for 5 h) glass fiber filters of 0.7 µm nominal pore size (Whatman GF/F), immediately acidified to pH 1–2 by adding 200 µL of H3PO4 (85%) and stored at 4 C. High-temperature catalytic oxidation (HTCO) on a Shimadzu TOC-l with a total nitrogen unit was used for the analysis. Reference materials of deep-sea carbon (42–45 μmol C L−1 and 31–33 μmol N L−1) and low carbon water (1–2 μmol C L−1) provided by D.A. Hansell (Univ. of Miami) were used to monitor the ultimate accuracy of DOC and TDN measurements. Dissolved organic nitrogen (DON) was calculated as TDN—DIN.
C. High-temperature catalytic oxidation (HTCO) on a Shimadzu TOC-l with a total nitrogen unit was used for the analysis. Reference materials of deep-sea carbon (42–45 μmol C L−1 and 31–33 μmol N L−1) and low carbon water (1–2 μmol C L−1) provided by D.A. Hansell (Univ. of Miami) were used to monitor the ultimate accuracy of DOC and TDN measurements. Dissolved organic nitrogen (DON) was calculated as TDN—DIN.
Flow cytometric analyses
All analyses were done with a FACSCanto II flow cytometer (BD Biosciences) following the protocols described in Gasol and Morán (2015). About 1 µm diameter latex fluorescent beads (Molecular Probes) were used as internal standards for the different fluorescence and light scatter signals. At least daily during the analyses we measured the actual flow rate (µL min−1) by weighting 1 mL of Milli-Q water before and after running the sample for 5 minutes (Gasol and Morán 2015). The software FCS Express 5 (DenNovo Software) was used for all post-acquisition analyses.
Two major groups of autotrophic bacteria (Synechococcus and Prochlorococcus) could be distinguished based on the relative values of the right angle side scatter (SSC) and the autofluorescence signals of their natural pigments (red for chlorophyll a and orange for phycoerythrin). Cyanobacteria abundances and cellular properties were analyzed fresh in 600 µL of seawater samples after adding 10 µL of beads solution at a concentration of 105 beads mL−1.
The abundance of heterotrophic bacteria was determined with different methods that allowed us to distinguish between groups of different relative nucleic acid content, membrane physical state and respiratory activity, which has been collectively termed as ‘single-cell physiological structure’ (Giorgio and Gasol 2008; Moràn, Ducklow and Erickson 2011). High nucleic acid (HNA) and low nucleic acid (LNA) bacteria were distinguished according to their relative SSC and green fluorescence signals after staining with a nucleic acid fluorochrome. Seawater samples (1800 µL) were fixed with 180 µL of glutaraldehyde plus paraformaldehyde (final concentrations of 1% and 0.5%, respectively). The samples were incubated in the dark for 10 minutes and stored at −80°C until analysis. 400 µL of the thawed sample were then stained with 4 µL of SYBR Green I (Molecular Probes) at 1000X concentration, incubated in the dark for 10 minutes at room temperature and ran in the flow cytometer after adding 10 µL of beads solution (106 mL−1). The cell sizes (µm3) of the HNA and LNA populations were calculated by converting the relative SSC values to cell diameter and then to biovolume assuming a spherical shape as described in detail in Calvo-Díaz and Morán (2006).
Live and Dead cells were identified based on the state of their membrane, intact for Live cells and damaged for Dead cells, using the nucleic acid double staining method as originally described by Grégori et al. (2003). SYBR Green I penetrates the cell membrane of both Live and Dead bacteria, while propidium iodide (PI) stains only bacteria that have damaged cell membranes. 400 µL of fresh seawater were simultaneously stained with 4 µL of SYBR Green I (Molecular Probes) and 4 µL of PI (Molecular Probes), incubated in the dark for 10 minutes at room temperature and ran in the flow cytometer after adding 10 µL of beads solution (106 mL−1).
The stain 5-cyano-2,3-ditolyl tetrazolium chloride (CTC), which competes with molecular oxygen in the electron transport chain, reducing the CTC to precipitates into an insoluble red fluorescent formazan salt, which allows us to identify and enumerate actively respiring cells (CTC+). 250 µL of fresh seawater were stained with 28 µL CTC and incubated in the dark for 90 minutes at room temperature before being ran in the flow cytometer after the addition of 10 µL of beads solution (105 mL−1).
Heterotrophic nanoflagellates abundance
HNF were detected by flow cytometry in green and red fluorescence and relative SSC cytograms following Christaki et al. (2011a). At each sampling date, duplicates of 4400 µL of seawater were fixed with glutaraldehyde (1.5% final concentration), incubated for 10 minutes in the dark at room temperature, deep frozen in liquid nitrogen and stored at −80°C until analysis. 1000 µL of the thawed samples were stained with SYBR Green I at 1000X concentration, incubated for 10 minutes in the dark at room temperature and ran in the flow cytometer after adding 10 µL of beads solution (106 mL−1).
Viral abundance
Viruses populations were identified based on their relative nucleic acid content. Three groups of viruses were detected by flow cytometry according to their relative SSC and green fluorescence signals following the protocol optimized by Brussaard (2004). Duplicate samples of 1500 µL of seawater were fixed with glutaraldehyde (2% final concentration) previously filtered through 0.2 µm Millipore polycarbonate filters. After incubating them for 15 minutes in the dark, samples were deep frozen in liquid nitrogen and stored at −80°C until analysis. 25 µL of the thawed samples were diluted in 475 µL of 1X Tris-EDTA buffer at pH 8, which had been previously autoclaved and filtered through a 0.2 µm Millipore polycarbonate filter. Samples were stained with 5 µL of SYBR Green I (Molecular Probes) that was previously diluted to 100X in Milli-Q water. Samples were incubated in a 80°C water bath for 10 minutes, then left to cool for 5 minutes in the dark at room temperature and ran in the flow cytometer after adding 10 µL of beads solution (107 mL−1). A control was performed by running 500 µL of 0.2 µm Millipore polycarbonate filtered and autoclaved Tris-EDTA buffer and 5 µL of SYBR Green I (Molecular Probes). The control apparent abundance values were subtracted from each analyzed sample.
Incubation experiments
In a series of experiments conducted monthly between December 2015 and March 2017, we performed short-term incubations of KAUST Harbor surface seawater. Triplicates 2000 mL samples, which had been pre-filtered through GF/C (1.2 µm nominal pore-size) in order to remove HNF and other larger planktonic organisms, were incubated for 6 days. Details of the experimental setup and calculations can be found in Silva et al. (2019). These experiments were initially intended to calculate the specific growth rates and carrying capacities (maximum bacterial abundance) of heterotrophic bacteria in natural conditions. However, in all the experiments we were also able to detect a decrease in their abundances after reaching the carrying capacities (usually at 2–3 days), which can only be attributed to viral lysis since HNF had been removed. Similarly to specific growth rates, mortality rates were calculated as the absolute slope of the natural logarithm of abundance vs. time for each decaying phase.
Data analysis
RStudio software was used to perform linear regressions, standard and paired t-tests, Pearson correlations, as well as analysis of variance (ANOVA) and post-hoc Tukey HSD tests for assessing temporal differences between seasons. All figures presented in this study were made by JMP Pro 14.1.0.
RESULTS
Physicochemical characteristics and bottom-up control
Weekly variations of temperature, salinity and nutrient concentrations are shown in Fig. 1. Temperature and salinity (Fig. 1A) behaved very similarly during most of the year. Temperature reached maximum values during summer (34.4°C in August) whereas the lowest values were observed in January (21.9°C). The highest salinity was also observed during summer (40.1 in September), with lower values (37.9–39.0) found from November through March and sporadic drops scattered all over the year. Inorganic nutrient concentrations were variable along the year. Nitrate concentration ranged widely from 1.24 to 44.61 µmol L−1, with generally lower values found in winter-early spring (Fig. 1B). Phosphate concentration was generally low, showing values consistently higher than 0.10 µmol L−1 only from February through May with little variation (ca. 0.05 µmol L−1) for the rest of the year (Fig. 1C). DOC concentration ranged from 81.3 to 152.2 µmol L−1 (Fig. 1D) and it gradually increased from spring until late summer, peaking in October. DON concentration (1.4–18.7 µmol L−1) also increased from spring to summer but it kept at relatively high values (ca. 10 µmol L−1) until the end of the year (Fig. 1E). DOC and DON concentrations were weakly correlated resulting in variable C: N ratios (5.0–25.7) and DOC was also positively correlated with temperature (Table 1).
Figure 1.
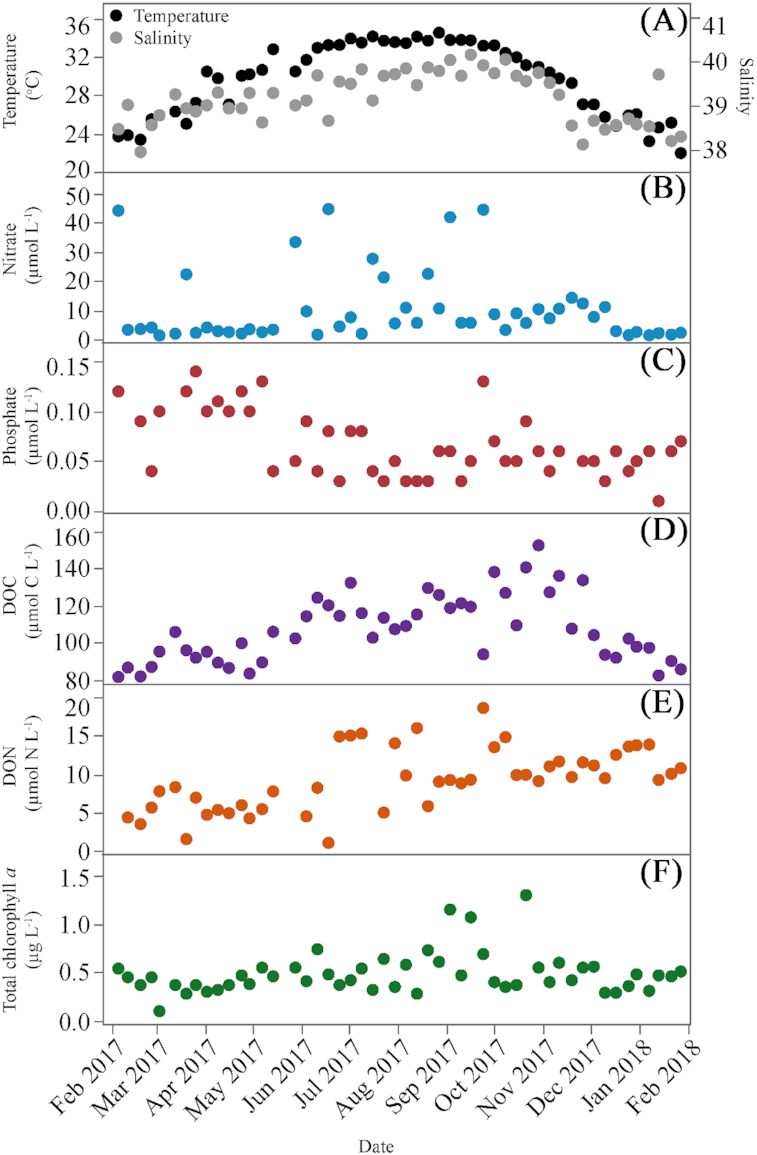
Weekly variability of environmental variables at KAUST Harbor during the sampled period. (A) temperature and salinity, (B) nitrate concentration, (C) Phosphate concentration, (D) DOC concentration, (F) DON concentration and (F) Total chlorophyll a concentration.
Table 1.
Correlation matrix (Pearson r) between environmental variables, T (temperature), S (salinity), DOC, DON, NO−3 (nitrate), PO3−4 (phosphate), Total Chl (total chlorophyll a), log10 transformed HNA, LNA, Live, CTC+ bacteria, LF Syn (Low fluorescence Synechococcus), HF Syn (High fluorescence Synechococcus), V1, V2, V3 and HNF abundances. Only significant values (*P < 0.05, **P < 0.01 and ***P < 0.001) are included here, n = 44–51.
| T | S | DOC | DON | NO−3 | PO3−4 | Total Chl | HNA | LNA | Live | CTC+ | LF Syn | HF Syn | V1 | V2 | V3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | 0.76*** | |||||||||||||||
| DOC | 0.63*** | 0.53*** | ||||||||||||||
| DON | – | – | 0.29* | |||||||||||||
| NO−3 | – | – | – | – | ||||||||||||
| PO3−4 | – | – | −0.33* | −0.31* | – | |||||||||||
| Total Chl | 0.47** | 0.40** | 0.41** | – | 0.30* | – | ||||||||||
| HNA | −0.36** | −0.39** | −0.33** | −0.49*** | −0.31*- | – | – | |||||||||
| LNA | – | – | – | – | – | – | – | 0.28* | ||||||||
| Live | −0.34** | −0.33** | – | – | −0.47*** | – | – | 0.56*** | 0.45*** | |||||||
| CTC+ | – | 0.28* | – | – | – | – | – | – | 0.30* | – | ||||||
| LF Syn | 0.51*** | 0.30* | 0.34** | – | – | – | – | 0.37** | – | – | ||||||
| HF Syn | −0.82*** | −0.73*** | −0.46*** | −0.31* | – | – | – | 0.40** | – | 0.31* | – | – | ||||
| V1 | 0.74*** | 0.53*** | 0.40** | – | – | 0.27* | −0.27* | – | −.030* | – | – | 0.72*** | ||||
| V2 | 0.44*** | – | – | – | – | – | – | – | – | – | – | 0.30* | 0.41** | 0.41** | ||
| V3 | 0.29* | – | – | – | −0.27* | – | – | – | – | – | – | 0.40** | −0.31* | – | 0.58*** | |
| HNF | −0.53*** | −0.55*** | – | – | – | – | 0.30* | – | – | – | – | 0.63*** | −0.62*** | – | – |
Total chlorophyll a concentration ranged from 0.10 to 1.30 µg L−1 (Fig. 1F), with maxima (>0.70 µg L−1) intermittently observed mostly in late summer and early fall. The contribution of the different chlorophyll a size-classes showed a noticeable seasonal pattern (Fig. S1, Supporting Information). Microplankton chlorophyll a contribution showed significantly higher values in summer (35%) than in winter and fall (ANOVA and Tukey HSD, P = 0.0008, df = 50). Nanopicoplankton chlorophyll a, with a mean annual contribution of 29%, did not show any significant differences among seasons. The contribution of picoplankton to total chlorophyll a was higher than the other two fractions during the whole year (mean 47%), with a maximum contribution in January (72%). Picophytoplankton summer contribution was significantly lower than in winter and fall (ANOVA and Tukey HSD, P = 0.0001, df = 50). Total chlorophyll a showed positive correlations with temperature, salinity and DOC concentration (Table 1).
Autotrophic and heterotrophic bacteria
Picophytoplankton were dominated by cyanobacteria (mostly Synechococcus) at the study site. Total Synechococcus abundance ranged from 1.08 to 11.18 × 104 cells mL−1 (Fig. 2A), with consistently low values (<5 × 104 cells mL−1) found in early winter and maxima in fall. Two groups of Synechococcus cells were distinguished based on their chlorophyll a and phycoerythrin fluorescence signals. The low fluorescence (LF) group was always the most abundant, ranging from 50% to 100% of total cells, while the high fluorescence (HF) one was present only at the beginning (January to March) and the end (October to December) of the year, with a mean contribution of 33% from December through February (Fig. 3A).
Figure 2.
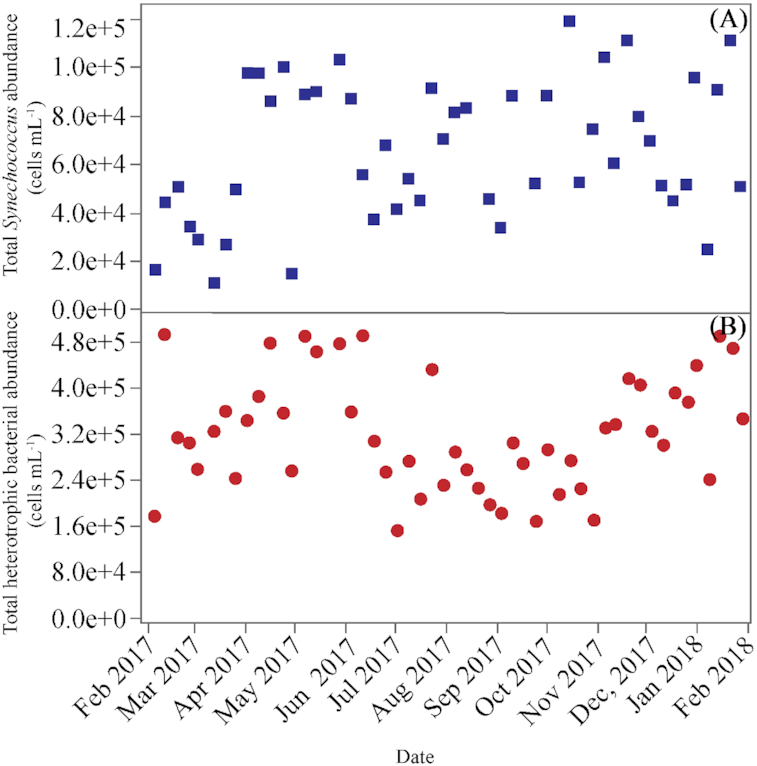
Weekly variability of total abundance of (A), Synechococcus (LF Synechococcus + HL Synnechococcus) and (B), heterotrophic bacteria (HNA+LNA) at KAUST Harbor during the sampled period.
Figure 3.
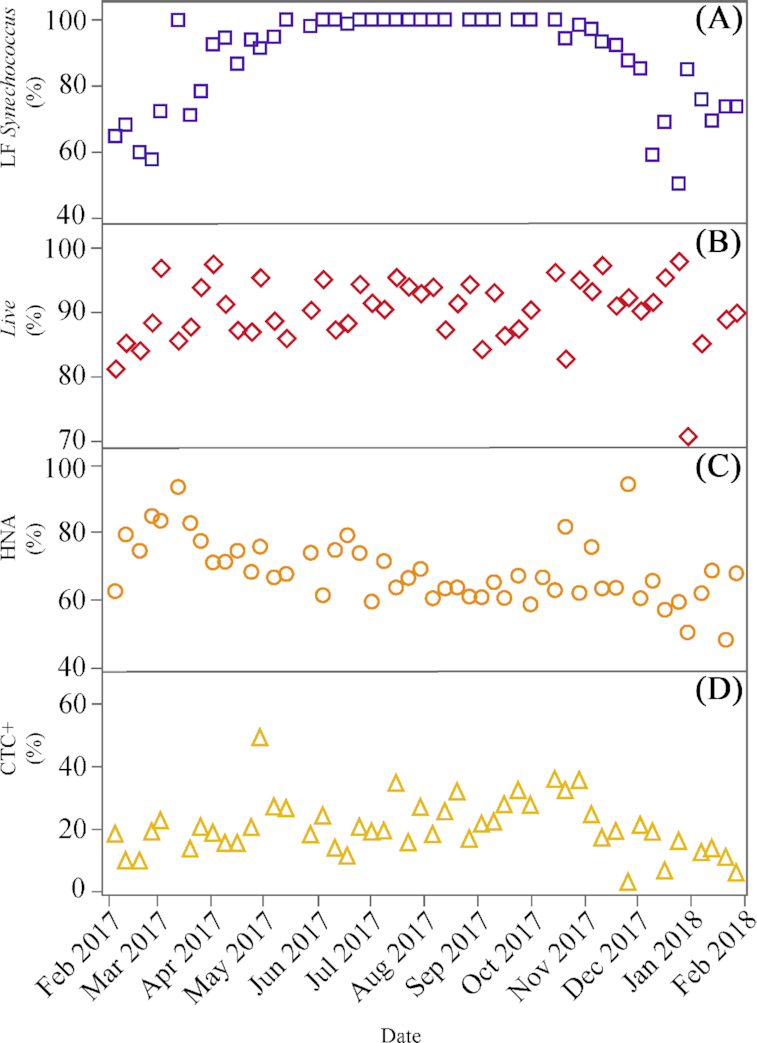
Weekly contributions (%) to total cyanobacteria and heterotrophic bacterial abundance of (A), low fluorescence (LF) Synechococcus (relative to the sum of LF and HF groups), (B), Live bacteria (relative to the sum of Live and Dead bacteria), (C), HNA bacteria (relative to the sum of HNA and LNA bacteria) and (D), CTC+ cells (relative to the sum of HNA and LNA bacteria) at KAUST Harbor.
The total abundance of heterotrophic bacteria (i.e. the sum of LNA and HNA cells) ranged from 1.55 to 4.97 × 105 cells mL−1, and showed a seasonal pattern with two relative peaks in late spring and winter and significantly lower values in summer (ANOVA and Tukey HSD, P = 0.0003, df = 48) (Fig. 2B). The sum of Live and Dead bacteria showed as expected a very similar seasonal pattern, with values ranging from 1.69 to 6.17 × 105 cells mL−1. Although LNA+HNA cell abundances were on average 3% lower than Live + Dead ones (Live + Dead abundance = 69 286 + 0.84 x LNA+HNA abundance, r2= 0.55, p<0.0001, n = 47), there was no systematic underestimation (paired t-test, P = 0.09, n = 47). Total heterotrophic bacterial abundance was negatively correlated with DOC concentration (r = −0.33, P = 0.01, n = 49). The percentage of Live bacteria was always higher than 80% except a low value of 70% found in January 2018. The highest values (95%) were reached occasionally without any clear seasonal pattern (Fig. 3B). HNA cells prevailed over their LNA counterparts, although their contribution to total numbers (48–94%) tended to decrease along the sampled period (Fig. 3C). Actively respiring bacteria (CTC+) contributed between 5 to 48% to total abundance and displayed a weak seasonality with maximum values generally observed between May and October (Fig. 3D).
Heterotrophic nanoflagellates and viruses
The abundance of heterotrophic nanoflagellates ranged from 86 to 1627 cells mL−1, and showed a conspicuous bimodal distribution (Fig. 4A) with two relative maxima observed in April and January and minima in summer (July and August). Summer HNF abundances were significantly lower than the other seasons (ANOVA, Tukey HSD, P = 0.0001, df = 48).
Figure 4.
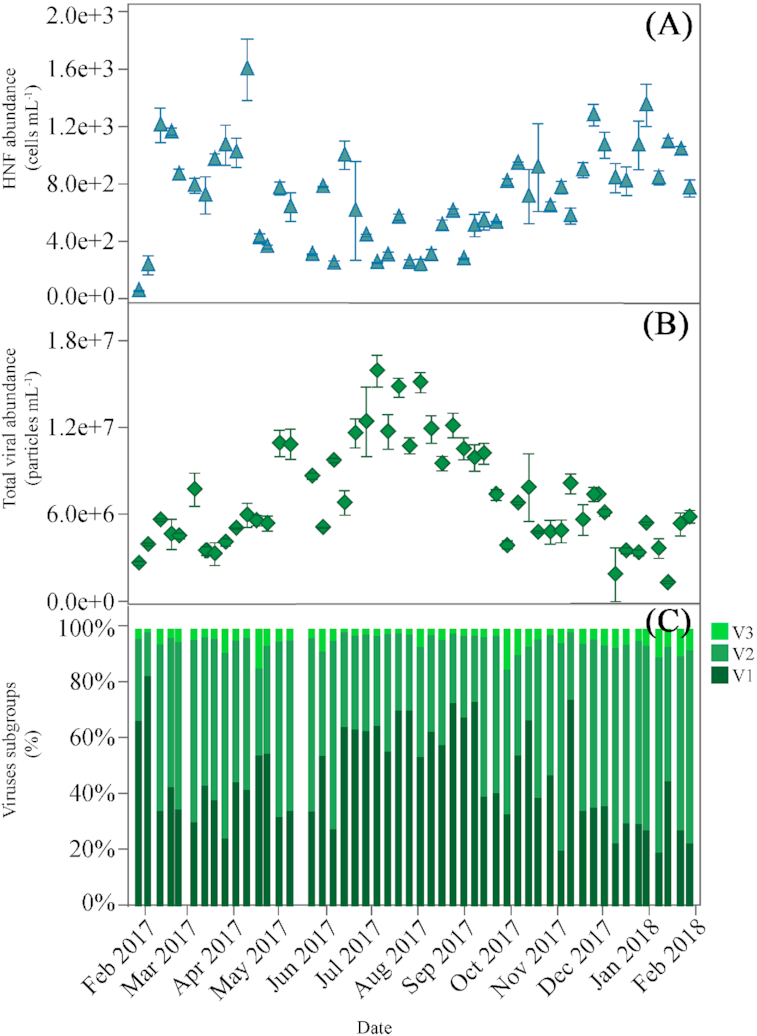
Weekly variability of the abundance of (A), HNF (mean ± SE cells mL−1), (B), Total viruses (V1+V2+V3) (mean ± SE particles mL−1) and (C), Contribution to total numbers (%) of each of the three subgroups of viruses (V1, V2 and V3) according to their relative nucleic acid content (V1: low, V2: medium and V3: high) at KAUST Harbor station.
Three subgroups of free viral particles were consistently distinguished in our samples based on their relative green fluorescence as a proxy for nucleic acid content. Two to three flow cytometric populations of differing nucleic acid content are frequently found in aquatic ecosystems (e.g. Johannessen et al. 2017; Tsai, Gong and Liu 2018). Following the usual coding (Brussaard 2004), V1 corresponds to the lowest nucleic acid content, V2 to the medium and V3 to the highest. Total viral abundance (V1+V2+V3) ranged from 1.30 to 15.91 × 106 particles mL−1 and displayed a strong seasonal pattern (Fig. 4B), with minima in winter and maxima (>107 particles ml−1) from June through September. Summer abundances were significantly higher than the other seasons (ANOVA, Tukey HSD, P = 0.0001, df = 49). Temperature and DOC concentration were positively correlated with total viral abundance (r = 0.72, P = 0.0001 and r = 0.40, P = 0.003, respectively, n = 50).
The percent contribution of each viral subgroup is shown in Fig. 4C. V1 was generally the most abundant group, ranging from 19% to 83% of total counts. Its seasonality was similar to that of total viral abundance, with maximum contributions over 40% found from July to September. V2 group contribution ranged from 15% to 74% with maximum values in spring and fall and minimum in summer. V3 was the least abundant group, ranging from 1.2% to 14% of the total. The abundance of V1viruses showed a positive correlation with DOC (Table 1) and negative with total bacterial abundance (r = −0.36, P = 0.01, n = 49).
The summer maximum in total viral abundance coincided with the minimum in HNF abundance and vice-versa. As a consequence, the total abundances of viruses and HNF in our data set were negatively correlated (r = −0.56, P = 0.0001, n = 48, Fig. 7).
Figure 7.
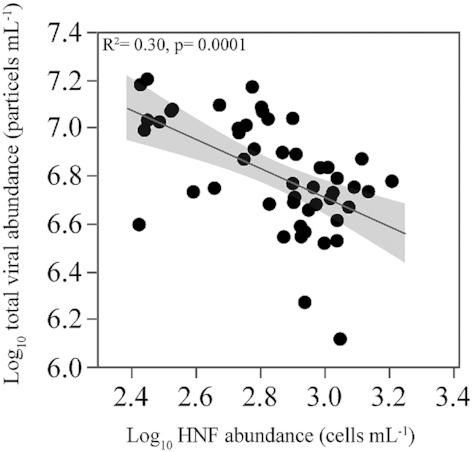
Relationship between the abundance of heterotrophic nanoflagellates and total viruses for the entire data set. Values are log10 transformed. Fitted line represents the ordinary least squares linear regressions model.
Top-down control of bacterioplankton communities
HNF were positively correlated with total bacterial abundance (r = 0.41, P = 0.003, n = 47, increasing to r = 0.52, P = 0.0002 if we exclude one outlier, Fig. 5A). The corresponding ratio of bacteria to HNF ranged from 223 to 2087 (data not shown), with maxima observed in spring and summer, but without any clear temporal pattern. To better test the relationship between HNF and heterotrophic bacterial abundance we used the qualitative model proposed by Gasol (1994), aimed at providing a framework of the main factors controlling HNF abundance. The maximum attainable abundance (MAA) line in this model describes the variability of predator and prey abundances on the assumption that HNF only predate on heterotrophic bacteria, while the mean realized abundance (MRA) line determines whether bottom-up or top-down controls are acting on the abundance of HNF (Gasol 1994). Depending on the position of the points relative to the MAA and the MRA lines, we can infer which control mechanisms are affecting HNF abundance. Points on the MAA line indicate a tight bottom-up control of HNF by their prey while points below the MRA line indicate top-down control of HNF (e.g. by ciliates or other larger heterotrophs). All our data fell below the MAA line and most of the points appeared between the MAA and MRA lines, with some of them (ca. 1/5) below the latter (Fig. 5A). HNF also seemed to have an effect on the cell size of heterotrophic bacteria, as the increase in HNF abundance was paralleled by weak though significant decreases in the size of both HNA and LNA cells (r = −0.40, P = 0.004, n = 47 and r = −0.33, P = 0.02, n = 48, respectively, Fig. 5B).
Figure 5.
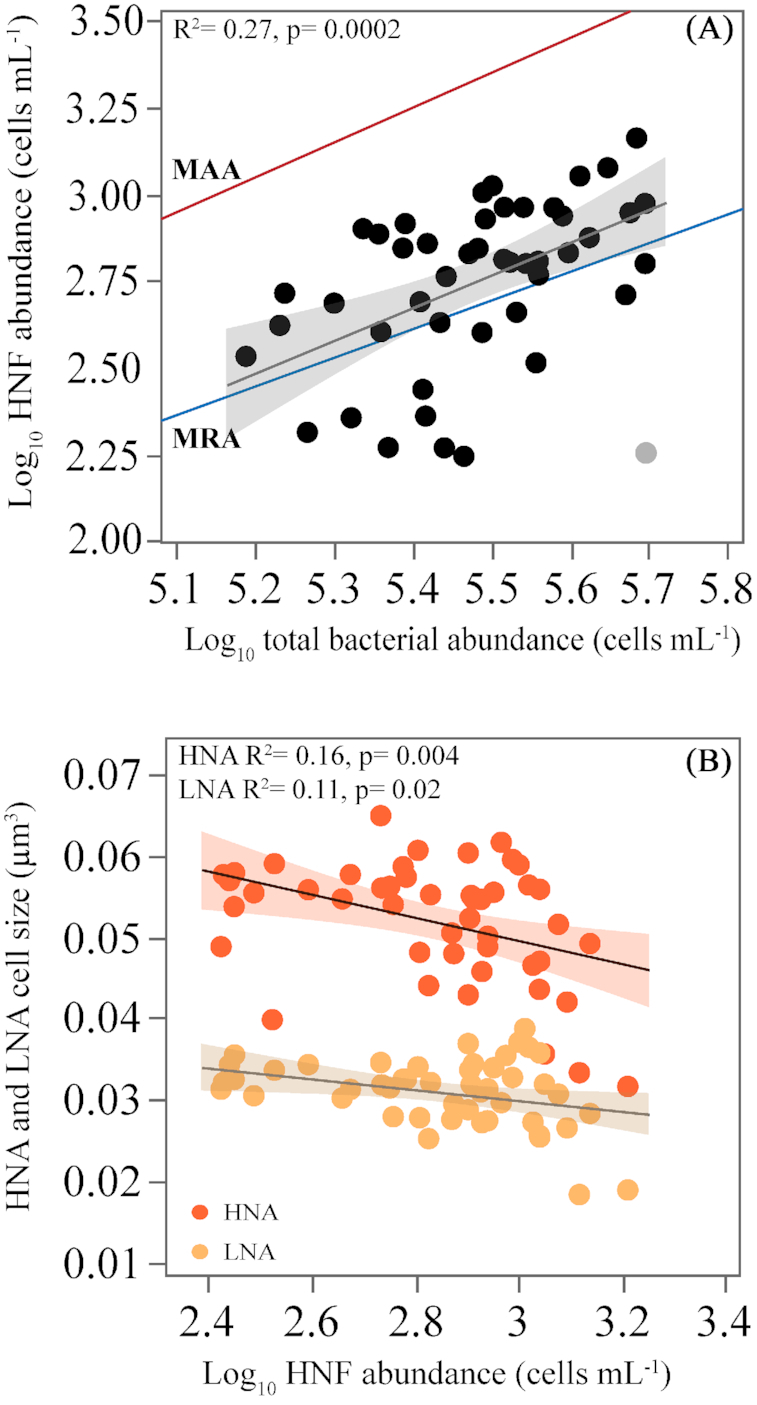
(A), Relationship between the abundances of heterotrophic nanoflagellates and heterotrophic bacteria, compared with the empirical model by Gasol (1994). MAA is the maximum attainable abundance and MRA is mean realized abundance (see the text for details), (light color symbol is excluded from the regression) and (B), Relationship between the cell size of LNA and HNA and HNF abundance. All abundances are log10 transformed. Fitted lines represent the ordinary least squares linear regressions model.
Apparent virus-mediated heterotrophic bacterial mortality was noticeable for most of the year, except for the first two months in which the abundance of viruses was rather stable (mean 4.43 × 106 particles mL−1). However, viral abundance started to increase in parallel to the decrease in bacterial abundance from April to July (Fig. 2B), followed by a decrease in viral abundance from August onwards until the last collected sample (Fig. 4B). We observed a negative relationship between log-transformed viral and bacterial abundances for the period from April 2017 to January 2018 (r = −0.33, P = 0.03, n = 41). As a consequence of opposite changes in viral and heterotrophic bacterial abundances, the virus to bacterium ratio or VBR (6–68, data not shown) displayed a very clear seasonal signal, with significantly higher values during summer relative to the other seasons (ANOVA, Tukey HSD, P = 0.0001, df = 48). We also found a positive relationship between VBR and DOC concentration ( r = 0.43, P = 0.001, n = 51).
The carrying capacities of total heterotrophic bacteria in the incubation experiments of pre-filtered water (see Material and Methods) conducted in 2016 ranged from 3.61 to 10.18 × 105 cells mL−1 (Silva et al. 2019), much higher than the actual abundances observed in that year or in our study (uniformly < 5 × 105 cells mL−1, Fig. 2B). Apparent viral mortality rates in 2016 ranged from 0.19 to 0.85 d−1, showing a clear seasonality with maximum values observed in January and August (Fig. 6A). Viral-induced mortality positively correlated with bacterial carrying capacities (r = 0.78, P = 0.01, n = 9). Interestingly, if we compare these mortality rates with the monthly mean viral abundances found here, a strong positive relationship emerged comparing the datasets from March to December (i.e. excluding the period in which bacteria and viruses were not negatively correlated, cf. Figs. 2B and 4B), with viral abundance explaining as much as 64% of the variance in estimated mortality rates (r = 079, P = 0.01, n = 9, Fig. 6B).
Figure 6.
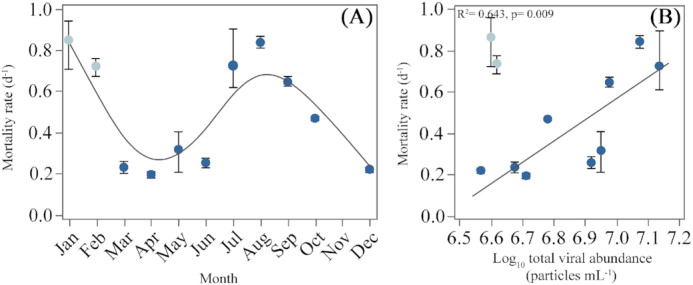
(A), Estimated (mean ± SE) total heterotrophic bacteria (HNA+LNA) mortality rates caused by viruses obtained from 2016 experiments detailed in Silva et al. (2019) and (B), Relationship between the mortality rates of (A) and the log10 transformed monthly averages of total viral abundance (V1+V2+V3) of this study (see Fig. 4 A for weekly data), (light color symbols are excluded from the regression).
DISCUSSION
Here, we used weekly monitoring over 12 months to assess the joint effect of bottom-up and top-down controls on coastal bacterioplankton in the Red Sea after a recent study on heterotrophic bacterial growth rates at the same study site in the previous year suggested their stocks were strongly controlled by heterotrophic nanoflagellates predation (Silva et al. 2019). Both autotrophic and heterotrophic bacterial abundances were similarly low in these two studies.
Temperature and salinity covaried tightly in 2017 (Fig. 1A), showing values similar to previous reports in the central and northern parts of the Red Sea (Ngugi et al. 2012; Kürten et al. 2016; Calleja, Al-Otaibi and Morán 2019), including KAUST Harbor (Silva et al. 2019). The fact that these authors presented monthly rather than weekly values might be the cause for the lack of relationship between temperature and salinity in their study. Short-lived, sudden changes in salinity are not rare in the study site (Fig. 1A). Due to its location in a semi-enclosed bay, KAUST Harbor experienced occasionally very high concentrations of nitrate (i.e. > 10 µmol L−1, Fig. 1B). These nitrate concentrations were higher than in other surveys conducted further north (Badran 2001; Berninger and Wickham 2005; Devassy et al. 2017), but could be linked to dust deposition events, since they were strongly correlated with silicate concentrations (r = 0.96, P < 0001, n = 51). Moreover, phosphate concentrations were not very different from values reported in the Gulf of Aqaba (Klinker et al. 1978; Wickham, Claessens and Post 2015). Even without the contribution of ammonium (not available for 2017), the inorganic nutrients N:P ratio was always higher than 16, except on one occasion (12.9 in March), indicating that P rather than N was generally limiting in our study site (Downing 1997; Mackey et al. 2007; Severiano et al. 2012). Consequently, total chlorophyll a concentration was rather low (<0.6 µg L−1) year-round except for a few values found in late summer and fall (Fig. 1F). Our chlorophyll a concentration was nevertheless slightly higher than the values usually recorded (1.15–1.36 µg L−1) in the Gulf of Aqaba (Berninger and Wickham 2005). Similar to other oligotrophic waters (e.g. Chen et al. 2015; López‐Urrutia and Morán 2015; Salhi et al. 2018) including the Red Sea (Gradinger, Weisse and Pillen 2009; Shibl et al. 2016; Kheireddine et al. 2017; Kheireddine et al. 2018), chlorophyll a in KAUST Harbor was dominated year-round by the smallest size fraction, although picophytoplankton contribution exceeded 50% only occasionally (Fig. S1, Supporting Information).
When addressing the role of top-down controls on heterotrophic bacteria, we should not forget their autotrophic counterparts (cyanobacteria), only slightly larger in equivalent spherical diameter, especially Prochlorococcus (Marie, Simon and Vaulot 2005; Morán et al. 2015). Similarly to shallow environments elsewhere (e.g. Partensky, Blanchot and Vaulot 1999; Pan et al. 2005; Lin et al. 2010; Dhib et al. 2017) Synechococcus was the only cyanobacteria genus consistently present at our sampling site (Ansari et al., submitted). Although other authors occasionally found Prochlorococcus in a coastal tropical estuary in the North Indian ocean (Mitbavkar et al. 2012), at KAUST Harbor we detected Prochlorococcus only 6 times (data not shown). On average, total Synechococcus abundance was 6.55 × 104 cells mL−1, equivalent to 20% of total heterotrophic bacterial abundance. Synechococcus abundance was similar to previous reports from the northern Red Sea (Muhling et al. 2005; Post et al. 2011; Wickham, Claessens and Post 2015), but lower than in other tropical coastal waters (e.g. Agawin et al. 2003; Ahlgren et al. 2014; Ribeiro et al. 2016). Two groups of Synechococcus with high and low contents of phycoerythrin (PE) chromophores, composed of both phycourobilin (PUM) and phycoerythrobin (PEB) (Katano and Nakano 2006), were also detected in other oligotrophic systems (Mitbavkar et al. 2012; Xia et al. 2017; Taucher et al. 2018), including the Red Sea (Veldhuis and Kraay 1993). Although there was not a clear seasonal pattern in Synechococcus total numbers (Fig. 2A), we did observe a clear seasonality in the contribution of the two groups identified by flow cytometry, with the low fluorescence group (LF) as the single one present from spring to fall (Fig. 3A). Some studies have indicated that Synechococcus of differing (PE) chromophores contents respond differently to changing environmental conditions (Palenik 2001) through a distinct growth capability (Hirose et al. 2008).
Heterotrophic bacteria begun to be monitored weekly at KAUST Harbor in July 2015 (Ansari et al., submitted). The total abundances obtained with the methods we used in this study (i.e. the sum of HNA and LNA, and of Live and Dead cells) were significantly correlated and clustered around the 1:1 line (data not shown), indicating the suitability of both methods for estimating total abundance (Vives-Rego, Lebaron and Nebe-von Caron 2000; Morán and Calvo-Díaz 2009). Total abundance peaked in spring as in the previous year, although 2017 showed slightly higher abundances (Silva et al. 2019). The fact that the abundances at KAUST Harbor were generally lower than in other oligotrophic waters (e.g. values > 106 cells mL−1 were reported in Longnecker, Sherr and Sherr 2006; Šantić et al. 2012; Mojica, Carlson and Behrenfeld 2019), the few other reports available for the Red Sea (Weisse 1989; Grossart and Simon 2002; Ansari et al. 2015; Calbet et al. 2015; Kürten et al. 2015) could be related to low chlorophyll a values but most likely to strong top-down control (see Silva et al. 2019 and below). HNA bacteria consistently dominated in our ecosystem (Fig. 3C), contrary to other tropical sites (Andrade et al. 2003; Gregoracci et al. 2012; Segovia et al. 2018). The earlier hypothesis that LNA bacteria were mostly dead or inactive cells (Servais et al. 1999; Berman et al. 2001; Lebaron et al. 2001) is not supported by our data, since Dead cells as assessed by the nucleic acid double staining method (Grégori et al. 2001; Giorgio and Gasol 2008) were only a minor contribution of total numbers and the abundances of Dead and LNA bacteria were not correlated. Furthermore, Silva et al. (2019) observed occasionally notably high growth rates of the LNA group (0.33–1.08 d−1) in incubation experiments from the same sampling site. The very high contribution of Live bacteria year-round is in agreement with studies conducted elsewhere (Lasternas and Agustí 2014; Gomes et al. 2015; Huete-Stauffer et al. 2015), suggesting that Dead cells are not easily found in most natural environments. More notably, we also observed a relatively high contribution of actively respiring bacteria at KAUST Harbor (Fig. 3D), with CTC+ cells averaging 22% of the total bacterial counts, within the range found in other coastal waters (Smith 1998; Longnecker, Sherr and Sherr 2005). Notably lower contributions (<2%) were found in richer waters of the NE Atlantic (Franco-Vidal and Morán 2011; Morán and Calvo-Díaz 2009). Site-specific differences seem important for the contribution of CTC+ cells since Gasol and Arístegui (2007) found values from 20 to 40%, similar to ours, in the oligotrophic waters of Gran Canaria Island (Gasol and Arístegui 2007).
DOC concentration positively correlated with total chlorophyll a concentration (Table 1), suggesting that at our site a significant DOC portion could be freshly derived from phytoplankton (Avril 2002; Raimbault, Garcia and Cerutti 2008). Heterotrophic bacteria have been traditionally linked to chlorophyll a in studies focusing on bottom-up control (e.g. Bird and Kalff 1984; Pace and Cole 1994: Vázquez-Domínguez et al. 2008; Gomes et al. 2015), since higher chlorophyll a content has been demonstrated to support higher bacterial abundance over large trophic gradients (Gasol and Duarte 2000). Covariance between DOC concentrations and heterotrophic bacterial standing stocks and activity have also been frequently reported (Alonso-Sáez et al. 2008; Fouilland and Mostajir 2010; Lonborg et al. 2011). The lack of positive correlations between DOC and any of the different single-cell bacterial groups assessed here (Table 1) may rather suggest that bulk DOC concentration was not a good indicator of the concentration of labile compounds. The significant, negative correlation between HNA bacterial abundance and nutrient concentrations (especially DON, Table ;1) might also be interpreted as that this group increased their numbers at the expense of depleting organic and inorganic nutrients, indeed indirectly supporting the bottom-up control. Although our sampling frequency aimed at capturing short-term dynamics, the specific growth rates of heterotrophic bacteria at KAUST Harbor (Silva et al. 2019) were too high even for weekly snapshots. These authors have demonstrated a fast uptake of DOM and bottom-up control of activity was indeed suggested by the correlation between bacterial growth efficiency and DON concentration (Silva et al. 2019). The positive correlation between DOC concentration and total viral abundance and VBR, previously documented by Suttle (2007) and Danovaro et al. (2011) , may in turn indicate that viral lysis was indeed contributing to increase DOC concentration (Stets and Cotner 2008; Auguet et al. 2005; Middelboe and Lyck 2002). In summary, we expected DOC, DON or chlorophyll a concentrations to show significant, positive covariations with bacterioplankton populations if their abundances were strongly bottom-up controlled. The lack of them would rather point to a greater top-down control on an annual basis, as recently found at the study site when comparing the carrying capacities in the presence and absence of protistan grazers (Silva et al. 2019). Since evidence of bottom-up regulation of heterotrophic bacterial growth rates and efficiencies was indeed shown by Silva et al. (2019), our results thus agree with the contention that bottom-up processes regulate the productivity of heterotrophic bacteria while top-down control regulate their biomass (Pace and Cole 1996; Gasol, Pedrós-Alió and Vaqué 2002).
To disentangle the role of viruses and heterotrophic nanoflagellates as controls of aquatic bacterial communities, it is important to address the dynamics of the three groups together (Cram, Parada and Fuhrman 2016; Vaqué et al. 2017). The variability in total viral abundance observed in this study fits well with observations conducted in other oligotrophic surface waters such as the Mediterranean (Siokou-Frangou et al. 2010; Christaki et al. 2011b) and other subtropical (Tsai, Gong and Shiau 2015; Gainer et al. 2017) and tropical (Winter et al. 2008; Lara et al. 2017) regions. Maximum abundances in summer were also frequently detected elsewhere (Parsons et al. 2012; Brum et al. 2016; Johannessen et al. 2017). As previously observed in many studies (e.g. Larsen et al. 2004; Brussaard et al. 2008b; Mojica et al. 2016; Johannessen et al. 2017), we managed to consistently distinguish three subgroups of viruses based on their nucleic acid content (Fig. 4C). It is interesting to note that, in spite of its lower fluorescence signal and presumably genome size (Suttle 2007; Personnic et al. 2009; Zhong et al. 2014), the V1 subgroup, which was dominant in summer and early fall reaching up to 73% of total numbers, was however overtaken by the V2 subgroup during the rest of the year (average 54% from late fall to spring). Although we are aware of the hidden, presumably high diversity found within flow cytometry-defined groups (Brussaard et al. 2008a), low fluorescence viruses were linked to HNA bacteria and V2 to phytoplankton in studies carried out in the Arctic Ocean (Li and Dickie 2001; Payet and Suttle 2008). Similar relationships were found between the V2 and V3 subgroups and the abundance of LF and HF Synechococcus cyanobacteria (Table 1). The infection rate is influenced by host morphology (Jasna et al. 2018), and since Synechococcus are larger than heterotrophic bacteria, our results suggest that larger viruses would also lyse larger host cells in accordance with Danovaro et al. (2011). Moreover, viral diversity has been linked to flow cytometric subgroups by Martínez, Swan and Wilson (2014). These authors showed that V1 viruses were linked to bacteriophages, generally myoviruses and podoviruses, while V2 viruses were dominated by phytoplankton viruses, particularly Phycodnaviridae and Myoviridae, and V3 viruses were mostly giant viruses related to the Mimiviridae and Phycodnaviridae families.
The standing stocks of heterotrophic bacteria were apparently controlled by viruses for most of the sampled period, as reflected in the negative relationship found between their respective total abundances. The summer decrease in total bacterial abundance (Fig. 2B) could thus be seen as a result of viral attack, followed by a resumed, slight increase in bacterial abundance when viruses were not able to find enough bacteria to infect (Fig. 4B). Positive relationships between viral and bacterial abundances are widespread across aquatic environments (e.g. Alonso et al. 2001; Weinbauer 2004; De Corte et al. 2012; Tsai, Gong and Hung 2013; Wigington et al. 2016), but our relationship was markedly the opposite. Negative correlations have however been observed at one NW Mediterranean site (Trabelsi and Rassoulzadegan 2011) and in the Bermuda Atlantic Time Series (BATS, Parsons et al. 2012). At BATS viruses reached their maximum abundance during summer and they were negatively correlated especially to the abundance of the SAR11 clade, which dominated heterotrophic bacteria year-round, and were subjected to higher infection levels according to Zhao et al. (2013) and Alonso-Sáez, Morán and Clokie (2018). This situation can be explained by the ‘Kill the winner’ hypothesis (Fuhrman and Suttle 1993; Thingstad et al. 2014; Storesund et al. 2015), in which viruses are attacking the most abundant host population. Recently, other authors have claimed that the small cells belonging to the SAR11 clade are resistant (Zhao et al. 2013) or less prone to viral infection (Hewson and Fuhrman 2007). At the study site, thanks to the relatively fine temporal resolution of our sampling, we were able to document a direct and sustained predator-prey dynamic between viruses and their heterotrophic bacterial hosts.
The virus to bacterium ratio (VBR) has been frequently used to estimate the strength of the relationship between viruses and their potential host communities (Maranger and Bird 1995; Wigington et al. 2016). In our dataset, VBR increased from ca. 10 during most of the year to ca. 50 during summer. This 5-fold change could be explained by a strong seasonal shift to higher bacterial mortality rates during summer (Li and Dickie 2001; Berdjeb et al. 2011). Parada et al. (2008) reported that the increase in water temperature can affect viral production by enhancing their attachment to host cells. As reported in various aquatic ecosystems (Yates, Gerba and Kelley 1985; De Corte et al. 2012), we also found a strong positive relationship between temperature and total viral abundance, explaining 52% of its variance.
Bacterioplankton mortality rate in the absence of protistan grazers has been frequently used to estimate the impact of phages on the loss of their host cells (Suttle 1994; Tsai, Gong and Hung 2013; Tsai, Gong and Chao 2016). In addition to our own weekly observations, we show here additional evidence of bacterial top-down regulation by viruses using the experimental measurements conducted at the study site in the previous 16 months (Silva et al. 2019). Once protistan grazers were excluded, we would have expected that bacterial abundance remained stable or slightly decreased after reaching their carrying capacity. Rather, marked decays of up to 0.84 d−1 were consistently observed (Fig. 6A). Moreover, we found that bacterial mortality rate was higher in the absence of grazers than when the entire microbial community was present, suggesting that the higher complexity of their interactions also affected viral infection rates (Wommack and Colwell 2000; Danovaro et al. 2011). Assuming that it is the host density that triggers viral activity (Wiggins and Alexander 1985; Parsons et al. 2012), the correlations between viral-induced mortality and both mean viral abundance and bacterial carrying capacities in samples taken in two consecutive year, strongly indicate that viruses have greater effects once bacteria reach high abundances, thus explaining the opposite annual patterns in the abundances reported here.
Together with viruses, HNF are the major agents of top-down control of bacteria in aquatic ecosystems (Choi, Hwang and Cho 2003; Bouvy et al. 2011; Kopylov et al. 2016). Our HNF abundance was similar to previous reports in other oligotrophic environments (Christaki et al. 2001; Tanaka and Rassoulzadegan 2002; Tsai et al. 2015) including the Red Sea Gulfs of Aden (Weisse 1989) and Gulf of Aqaba (Berninger and Wickham 2005; Nakajima et al. 2017), with seasonality opposite to ours (El-Serehy, Al-Rasheid and Shafik 2012). The variability of the bacteria to HNF ratio also fell within the range observed in both coastal (Sugai et al. 2016) and open ocean systems (Christaki et al. 2011a). The positive relationship between bacterial abundance and HNF on an annual basis has been extensively documented (e.g. Weisse and MacIsaac 2000; Calbet 2001; Tsai et al. 2015; Baltar et al. 2016), including the Red Sea (Berninger and Wickham 2005; El-Serehy et al. 2013) However, the increase in HNF was not proportional to the increase in bacteria (Fig. 5A), which would be expected if heterotrophic nanoflagellates were only dependent on bacteria as a source of food (Pernthaler 2005), as represented by the MAA line in Gasol's (1994) model. Our data fell below this line, meaning that for a given abundance of heterotrophic bacteria there could potentially be more HNF than currently observed. Since the linear regression model explained only 27% of the variance in HNF abundance, our results suggest that either heterotrophic bacteria were not the only source of food for HNF or that there was also a top-down control on heterotrophic nanoflagellates themselves by larger protists such as ciliates or rotifers (Güde 1988; Gasol and Vaqué 1993; Gasol 1994; Segovia et al. 2016).
Interestingly, at the same study site, Silva et al. (2019) observed in 2016 that the carrying capacities of heterotrophic bacteria in the absence of HNF reached twice (in August and November) one million cells per mL, representing 67% more than the average value (3.18 × 105 cells mL−1) observed during our study period, in strong support of the hypothesis that both top-down controls impact heterotrophic bacterial abundances along the annual cycle. Contrary to viruses, we failed to detect direct Lotka-Volterra oscillations between the abundance of heterotrophic bacteria and HNF as in other studies (e.g. Azam et al. 1983; Weisse 1989; Baltar et al. 2016). The overall effect of HNF was still visible in bacterial cell size: both LNA and HNA cells tended to be smaller when the abundance of HNF increased (Fig. 5B), in agreement with the HNF selective preference for larger bacterial prey (Vaqué, Casamayor and Gasol 2001; Comte et al. 2006; Baltar et al. 2016). Although LNA cells were on average 58% smaller than their HNA counterparts, the results indicate that HNF showed prey-size preference also on LNA cells (Thomas et al. 2011). Together with their moderate growth rates (Silva et al. 2019), our results do not support that the smaller size and nutritious quality of LNA bacteria protect them from protistan grazing in the coastal Red Sea (Jürgens and Güde 1994).
To sum up, after one year of weekly observations, the standing stocks of heterotrophic bacteria at this coastal Red Sea site seem to be more subject to top-down than to bottom-up control. Although microbial food web dynamics need even shorter temporal scale assessments, our weekly sampling showed clear opposite variations of viruses and HNF abundances, resulting in a significant negative relationship between both bacterial top-down control (Fig. 7) as recently shown by Vaqué et al. (2019) in the Arctic. Also similar to NE Pacific coastal waters (Pasulka, Samo and Landry 2015), these results suggest that viral lysis and protistan grazing were indeed dominating Red Sea coastal bacterioplankton losses in different periods of the year. An alternative explanation would be the ‘kill the killer of the winner’ process (Miki and Yamamura 2005), in which protists feed on the viruses that infect the dominant heterotrophic bacteria, so that our findings would evidence a direct interaction between grazers and viruses (Miki and Jacquet 2008). Bettarel, Sime‐Ngando and Amblard ( 2003), and Jacquet et al. (2010) provided evidence for this direct HNF control on viral abundances although the interactions between viruses and protistan grazers are still poorly understood.
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank our colleagues Abbrar Labban and Najwa Al-Otaibi for their help through the course of this research.
FUNDING
This work was supported by King Abdullah University of Science and Technology (KAUST) baseline funding to XAGM.
Conflict of interest. None declared.
REFERENCES
- Agawin NSR, Duarte CM, Agustí S et al.. Abundance, biomass and growth rates of Synechococcus sp. in a tropical coastal ecosystem (Philippines, South China Sea). Estuar Coast Shelf Sci. 2003;56:493–502. [Google Scholar]
- Ahlgren NA, Noble A, Patton AP et al.. The unique trace metal and mixed layer conditions of the Costa Rica upwelling dome support a distinct and dense community of Synechococcus. Limnol Oceanogr. 2014;59:2166–84. [Google Scholar]
- Alonso-Sáez L, Morán XAG, Clokie MR. Low activity of lytic pelagiphages in coastal marine waters. ISME J. 2018;12:2100–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Sáez L, Vázquez-Domínguez E, Cardelús C et al.. Factors Controlling the Year-Round Variability in Carbon Flux Through Bacteria in a Coastal Marine System. Ecosystems. 2008;11:397–409. [Google Scholar]
- Alonso MC, Jimenez-Gomez F, Rodriguez J et al.. Distribution of virus-like Particles in an OligotrophicMarine Environment (Alboran Sea, Western Mediterranean). Microb Ecol. 2001;42:407–15. [DOI] [PubMed] [Google Scholar]
- Andrade L, Gonzalez AM, Araujo FV et al.. Flow cytometry assessment of bacterioplankton in tropical marine environments. J Microbiol Methods. 2003;55:841–50. [DOI] [PubMed] [Google Scholar]
- Ansari MI, Harb M, Jones B et al.. Molecular-based approaches to characterize coastal microbial community and their potential relation to the trophic state of Red Sea. Sci Rep. 2015;5:9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol L, Calbet A, Franchy G et al.. Planktonic food web structure and trophic transfer efficiency along a productivity gradient in the tropical and subtropical Atlantic Ocean. Sci Rep. 2019;9:2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2005;437:349. [DOI] [PubMed] [Google Scholar]
- Auguet JC, Montanie H, Delmas D et al.. Dynamic of Virioplankton Abundance and Its Environmental Control in the Charente Estuary (France). Microbial Ecology. 2005;50:337–49. [DOI] [PubMed] [Google Scholar]
- Avril B. DOC dynamics in the northwestern Mediterranean Sea (DYFAMED site). Deep Sea Res Part II Top Stud Oceanogr. 2002;49:2163–82. [Google Scholar]
- Azam F, Fenchel T, Field JG et al.. The Ecological Role of Water-Column Microbes in the Sea. Mar Ecol Prog Ser. 1983;10:257–63. [Google Scholar]
- Badran MI. Dissolved oxygen, Chlorophyll a and nutrients: Seasonal cycles in waters of the gulf of Aquaba, Red Sea. Aquat Ecosyst Health Manag. 2001;4:139–50. [Google Scholar]
- Baltar F, Palovaara J, Unrein F et al.. Marine bacterial community structure resilience to changes in protist predation under phytoplankton bloom conditions. ISME J. 2016;10:568–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner R, Amon RMW. The Size-Reactivity Continuum of Major Bioelements in the Ocean. Annu Rev Mar Sci. 2015;7:185–205. [DOI] [PubMed] [Google Scholar]
- Berdjeb L, Pollet T, Domaizon I et al.. Effect of grazers and viruses on bacterial community structure and production in two contrasting trophic lakes. BMC Microbiol. 2011;11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman T, Kaplan B, Chava S et al.. Metabolically active bacteria in Lake Kinneret. Aquat Microb Ecol. 2001;23:213–24. [Google Scholar]
- Berninger U-G, Wickham SA. Response of the microbial food web to manipulation of nutrients and grazers in the oligotrophic Gulf of Aqaba and northern Red Sea. Mar Biol. 2005;147:1017–32. [Google Scholar]
- Berumen ML, Voolstra CR, Daffonchio D et al.. The Red Sea: Environmental Gradients Shape a Natural Laboratory in a Nascent Ocean. In: Voolstra CR, Berumen ML (eds). Coral Reefs of the Red Sea. Cham: Springer International Publishing, 2019, 1–10. [Google Scholar]
- Bettarel Y, Dolan JR, Hornak K et al.. Strong, weak, and missing links in a microbial community of the N.W. Mediterranean Sea. FEMS Microbiol Ecol. 2002;42:451–62. [DOI] [PubMed] [Google Scholar]
- Bettarel Y, Sime-Ngando T, Bouvy M et al.. Low consumption of virus-sized particles by heterotrophic nanoflagellates in two lakes of the French Massif Central. Aquat Microb Ecol. 2005;39:205–9. [Google Scholar]
- Bettarel Y, Sime‐ngando T, Amblard C. 8 Grazing by heterotrophic nanoflagellates on virus- and bacteria-sized particles in two lakes of different trophy. J Phycol. 2003;39:3–4. [Google Scholar]
- Bird DF, Kalff J. Empirical Relationships between Bacterial Abundance and Chlorophyll Concentration in Fresh and Marine Waters. Can J Fish Aquat Sci. 1984;41:1015–23. [Google Scholar]
- Bock N, Wambeke FV, Dion M et al.. Microbial community structure in the western tropical South Pacific. Biogeosciences. 2018;15:3909–25. [Google Scholar]
- Boenigk J, Arndt H. Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Antonie Van Leeuwenhoek. 2002;81:465–80. [DOI] [PubMed] [Google Scholar]
- Boras JA, Sala MM, Vázquez-Domínguez E et al.. Annual changes of bacterial mortality due to viruses and protists in an oligotrophic coastal environment (NW Mediterranean). Environ Microbiol. 2009;11:1181–93. [DOI] [PubMed] [Google Scholar]
- Bouvier T, Del Giorgio PA, Gasol JM. A comparative study of the cytometric characteristics of high and low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ Microbiol. 2007;9:2050–66. [DOI] [PubMed] [Google Scholar]
- Bouvy M, Bettarel Y, Bouvier C et al.. Trophic interactions between viruses, bacteria and nanoflagellates under various nutrient conditions and simulated climate change. Environ Microbiol. 2011;13:1842–57. [DOI] [PubMed] [Google Scholar]
- Bouvy M, Got P, Bettarel Y et al.. Importance of predation and viral lysis for bacterial mortality in a tropical western Indian coral-reef ecosystem (Toliara, Madagascar). Mar Freshw Res. 2015;66:1009–17. [Google Scholar]
- Bowman JS, Amaral-Zettler LA, Rich JJ et al.. Bacterial community segmentation facilitates the prediction of ecosystem function along the coast of the western Antarctic Peninsula. ISME J. 2017;11:1460–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brum JR, Hurwitz BL, Schofield O et al.. Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 2016;10:437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard CPD, Timmermans KR, Uitz J et al.. Virioplankton dynamics and virally induced phytoplankton lysis versus microzooplankton grazing southeast of the Kerguelen (Southern Ocean). Deep Sea Res Part II Top Stud Oceanogr. 2008a;55:752–65. [Google Scholar]
- Brussaard CPD, Wilhelm SW, Thingstad F et al.. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2008b;2:575–8. [DOI] [PubMed] [Google Scholar]
- Brussaard CPD. Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol. 2004;70:1506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunse C, Pinhassi J. Marine bacterioplankton seasonal succession dynamics. Trends Microbiol. 2017;25:494–505. [DOI] [PubMed] [Google Scholar]
- Calbet A, Agersted MD, Kaartvedt S et al.. Heterogeneous distribution of plankton within the mixed layer and its implications for bloom formation in tropical seas. Sci Rep. 2015;5:11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calbet A, Landry MR, Nunnery S. Bacteria-flagellate interactions in the microbial food web of the oligotrophic subtropical North Pacific. Aquat Microb Ecol. 2001;23:283–92. [Google Scholar]
- Calbet A. Bacteria-flagellate interactions in the microbial food web of the oligotrophic subtropical North Pacific. Aquat Microb Ecol. 2001;23:283–92. [Google Scholar]
- Calleja ML, Al-Otaibi N, Morán XAG. Dissolved organic carbon contribution to oxygen respiration in the central Red Sea. Sci Rep. 2019;9:4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Díaz A, Morán XAG. Seasonal dynamics of picoplankton in shelf waters of the southern Bay of Biscay. Aquat Microb Ecol. 2006;42:159–74. [Google Scholar]
- Chen M, Liu H, Song S et al.. Size-fractionated mesozooplankton biomass and grazing impact on phytoplankton in northern South China Sea during four seasons. Deep Sea Res Part II Top Stud Oceanogr. 2015;117:108–18. [Google Scholar]
- Choi DH, Hwang CY, Cho BC. Comparison of virus- and bacterivory-induced bacterial mortality in the eutrophic Masan Bay, Korea. Aquat Microb Ecol. 2003;30:117–25. [Google Scholar]
- Christaki U, Courties C, Massana R et al.. Optimized routine flow cytometric enumeration of heterotrophic flagellates using SYBR Green I. Limnol Oceanogr Methods. 2011a;9:329–39. [Google Scholar]
- Christaki U, Giannakourou A, Van Wambeke F et al.. Nanoflagellate predation on auto- and heterotrophic picoplankton in the oligotrophic Mediterranean Sea. J Plankton Res. 2001;23:1297–310. [Google Scholar]
- Christaki U, Wambeke FV, Lefevre D et al.. Microbial food webs and metabolic state across oligotrophic waters of the Mediterranean Sea during summer. Biogeosciences. 2011b;8:1839–52. [Google Scholar]
- Church MJ. Resource Control of Bacterial Dynamics in the Sea. Microbial Ecology of the Oceans, New Jersey, USA: John Wiley & Sons, Ltd, 2008, 335–82. [Google Scholar]
- Comte J, Jacquet S, Viboud S et al.. Microbial community structure and dynamics in the largest natural french lake (Lake Bourget). Microb Ecol. 2006;52:72–89. [DOI] [PubMed] [Google Scholar]
- Cram JA, Parada AE, Fuhrman JA. Dilution reveals how viral lysis and grazing shape microbial communities: Viral lysis and grazing shape microbial communities. Limnol Oceanogr. 2016;61:889–905. [Google Scholar]
- Danovaro R, Corinaldesi C, Dell'Anno A et al.. Marine viruses and global climate change. FEMS Microbiol Rev. 2011;35:993–1034. [DOI] [PubMed] [Google Scholar]
- De Corte D, Sintes E, Yokokawa T et al.. Links between viruses and prokaryotes throughout the water column along a North Atlantic latitudinal transect. ISME J. 2012;6:1566–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degerman R, Dinasquet J, Riemann L et al.. Effect of resource availability on bacterial community responses to increased temperature. Aquat Microb Ecol. 2013;68:131–42. [Google Scholar]
- Dekel-Bird NP, Sabehi G, Mosevitzky B et al.. Host-dependent differences in abundance, composition and host range of cyanophages from the Red Sea. Environ Microbiol. 2015;17:1286–99. [DOI] [PubMed] [Google Scholar]
- Dell'Anno A, Corinaldesi C, Danovaro R. Virus decomposition provides an important contribution to benthic deep-sea ecosystem functioning. Proc Natl Acad Sci USA. 2015;112:E2014–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devassy RP, El-Sherbiny MM, Al-Sofyani AM et al.. Spatial variation in the phytoplankton standing stock and diversity in relation to the prevailing environmental conditions along the Saudi Arabian coast of the northern Red Sea. Mar Biodivers. 2017;47:995–1008. [Google Scholar]
- Dhib A, Denis M, Ziadi B et al.. Assessing ultraphytoplankton and heterotrophic prokaryote composition by flow cytometry in a Mediterranean lagoon. Environ Sci Pollut Res. 2017;24:13710–21. [DOI] [PubMed] [Google Scholar]
- Downing JA. Marine nitrogen: Phosphorus stoichiometry and the global N:P cycle. Biogeochemistry. 1997;37:237–52. [Google Scholar]
- Ducklow HW, Carlson CA. Oceanic bacterial production. In: Marshall KC. (ed). Advances in Microbial Ecology. Boston, MA: Springer US, 1992, 113–81. [Google Scholar]
- Ducklow HW. The bacterial component of the oceanic euphotic zone. FEMS Microbiol Ecol. 1999;30:1–10. [Google Scholar]
- Dufour PH, Torréton J-P. Bottom-up and top-down control of bacterioplankton from eutrophic to oligotrophic sites in the tropical northeastern Atlantic Ocean. Deep Sea Res Part Oceanogr Res Pap. 1996;43:1305–20. [Google Scholar]
- El-Serehy HA, Al-Rasheid KA, Al-Quraishi S et al.. Abundance and trophodynamics of surface microbial loop populations in the northern Red Sea. Aquat Ecosyst Health Manag. 2013;16:51–61. [Google Scholar]
- El-Serehy HA, Al-Rasheid KA, Shafik H. Microbial loop populations: their abundances and trophodynamics in the gulf of Aqaba, Red Sea. Turk J Fish Aquat Sci. 2012;12:565–73. [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT et al.. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science. 1998;218:237–40. [DOI] [PubMed] [Google Scholar]
- Flombaum P, Gallegos JL, Gordillo RA et al.. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci. 2013;110:9824–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouilland E, Mostajir B. Revisited phytoplanktonic carbon dependency of heterotrophic bacteria in freshwaters, transitional, coastal and oceanic waters. FEMS Microbiol Ecol. 2010;73:419–29. [DOI] [PubMed] [Google Scholar]
- Franco-Vida L, Morán XAG. Relationships Between Coastal Bacterioplankton Growth Rates and Biomass Production: Comparison of Leucine and Thymidine Uptake with Single-Cell Physiological Characteristics. Microbial ecology. 2011;61:328–41. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Cram JA, Needham DM. Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol. 2015;13:133–46. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Noble RT. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr. 1995;40:1236–42. [Google Scholar]
- Fuhrman JA, Suttle CA. Viruses in Marine Planktonic Systems. Oceanography. 1993;6:51–63. [Google Scholar]
- Gainer PJ, Pound HL, Larkin AA et al.. Contrasting seasonal drivers of virus abundance and production in the North Pacific Ocean. PLoS One. 2017;12:e0184371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasol JM, Arístegui J. Cytometric evidence reconciling the toxicity and usefulness of CTC as a marker of bacterial activity. Aquat Microb Ecol. 2007;46:71–83. [Google Scholar]
- Gasol JM, Duarte CM. Comparative analyses in aquatic microbial ecology: how far do they go? FEMS Microbiol Ecol. 2000;31:99–106. [DOI] [PubMed] [Google Scholar]
- Gasol JM, Morán XAG. Flow cytometric determination of microbial abundances and its use to obtain indices of community structure and relative activity. Hydrocarbon and Lipid Microbiology Protocols. Berlin, Heidelberg: Springer, 2015, 159–87. [Google Scholar]
- Gasol JM, Pedrós-Alió C, Vaqué D. Regulation of bacterial assemblages in oligotrophic plankton systems: results from experimental and empirical approaches. Antonie Van Leeuwenhoek. 2002;81:435–52. [DOI] [PubMed] [Google Scholar]
- Gasol JM, Vaqué D. Lack of coupling between heterotrophic nanoflagellates and bacteria: A general phenomenon across aquatic systems? Limnol Oceanogr. 1993;38:657–65. [Google Scholar]
- Gasol JM. A framework for the assessment of top-down vs bottom-up control of heterotrophic nanoflagellate abundance. Mar Ecol Prog Ser. 1994;113:291–300. [Google Scholar]
- del Giorgio PA, Gasol JM. Physiological structure and single-cell activity in marine bacterioplankton. Microbial Ecology of the Oceans, New York, USA: John Wiley & Sons, Ltd, 2008, 243–98. [Google Scholar]
- Gomes A, Gasol JM, Estrada M et al.. Heterotrophic bacterial responses to the winter–spring phytoplankton bloom in open waters of the NW Mediterranean. Deep Sea Res Part Oceanogr Res Pap. 2015;96:59–68. [Google Scholar]
- Gradinger R, Weisse T, Pillen T. Significance of Picocyanobacteria in the Red Sea and the Gulf of Aden. Bot Mar. 2009;35:245–50. [Google Scholar]
- Gregoracci GB, Nascimento JR, Cabral AS et al.. Structuring of Bacterioplankton Diversity in a Large Tropical Bay. PLoS One. 2012;7:e31408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossart H-P, Simon M. Bacterioplankton dynamics in the Gulf of Aqaba and the northern Red Sea in early spring. Mar Ecol Prog Ser. 2002;239:263–76. [Google Scholar]
- Grégori G, Citterio S, Ghiani A et al.. Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl Env Microbiol. 2001;67:4662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grégori G, Denis M, Seorbati S et al.. Resolution of Viable and Membrane-Compromised Free Bacteria in Aquatic Environments by Flow Cytometry. Curr Protoc Cytom. 2003;23:11–15. [DOI] [PubMed] [Google Scholar]
- Güde H. Direct and indirect influences of crustacean zooplankton on bacterioplankton of Lake Constance. Hydrobiologia. 1988;159:63–73. [Google Scholar]
- Hagström Å, Azam F, Andersson A et al.. Microbial loop in an oligotrophic pelagic marine ecosystem: possible roles of cyanobacteria and nanoflagellates in the organic fluxes. Mar Ecol Prog Ser. 1988;49:171–8. [Google Scholar]
- Hansen HP, Koroleff F. Determination of nutrients. In: Grasshoff K, Kremling K, Ehrhardt M (eds). Methods of Seawater Analysis. Weinheim, Germany: Wiley-VCH Verlag GmbH, 1999, 159–228. [Google Scholar]
- Herndl GJ, Agogué H, Baltar F et al.. Regulation of aquatic microbial processes: the ‘microbial loop’ of the sunlit surface waters and the dark ocean dissected. Aquat Microb Ecol. 2008;53:59–68. [Google Scholar]
- Hewson I, Fuhrman JA. Characterization of Lysogens in Bacterioplankton Assemblages of the Southern California Borderland. Microb Ecol. 2007;53:631–8. [DOI] [PubMed] [Google Scholar]
- Hirose M, Katano T, Hayami Y et al.. Changes in the abundance and composition of picophytoplankton in relation to the occurrence of a Kyucho and a bottom intrusion in the Bungo Channel, Japan. Estuar Coast Shelf Sci. 2008;76:293–303. [Google Scholar]
- Huete-Stauffer TM, Arandia-Gorostidi N, Díaz-Pérez L et al.. Temperature dependences of growth rates and carrying capacities of marine bacteria depart from metabolic theoretical predictions. FEMS Microbiol Ecol. 2015;91:fiv111. [DOI] [PubMed] [Google Scholar]
- Hurst CJ. Viral Ecology. Academic Press, 2000. [Google Scholar]
- Hwang CY, Cho BC. Virus-infected bacteria in oligotrophic open waters of the East Sea, Korea. Aquat Microb Ecol. 2002;30:1–9. [Google Scholar]
- Jacquet S, Miki T, Noble R et al.. Viruses in aquatic ecosystems: important advancements of the last 20 years and prospects for the future in the field of microbial oceanography and limnology. Adv Oceanogr Limnol. 2010;1:97–141. [Google Scholar]
- Jasna V, Ram ASP, Parvathi A et al.. Differential impact of lytic viruses on prokaryotic morphopopulations in a tropical estuarine system (Cochin estuary, India). PLoS One. 2018;13:e0194020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen TV, Larsen A, Bratbak G et al.. Seasonal Dynamics of Haptophytes and dsDNA Algal Viruses Suggest Complex Virus-Host Relationship. Viruses. 2017;9:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens K, Güde H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser. 1994;112:169–88. [Google Scholar]
- Katano T, Nakano SI. Growth rates of Synechococcus types with different phycoerythrin composition estimated by dual-laser flow cytometry in relationship to the light environment in the Uwa Sea. Journal of Sea Research. 2006;55:182–190. [Google Scholar]
- Kheireddine M, Ouhssain M, Claustre H et al.. Assessing pigment-based phytoplankton community distributions in the Red Sea. Front Mar Sci. 2017;4 DOI:10.3389/fmars.2017.00132. [Google Scholar]
- Kheireddine M, Ouhssain M, Organelli E et al.. Light absorption by suspended particles in the red sea: effect of phytoplankton community size structure and pigment composition. J Geophys Res Oceans. 2018;123:902–21. [Google Scholar]
- Klinker J, Reiss Z, Kropach C et al.. Nutrients and biomass distribution in the Gulf of Aqaba (Elat), Red Sea. Mar Biol. 1978;45:53–64. [Google Scholar]
- Kopylov AI, Sazhin AF, Zabotkina EA et al.. Viruses, bacteria, and heterotrophic nanoflagellates in Laptev Sea plankton. Oceanology. 2016;56:789–98. [Google Scholar]
- Kürten B, Al-Aidaroos AM, Kürten S et al.. Carbon and nitrogen stable isotope ratios of pelagic zooplankton elucidate ecohydrographic features in the oligotrophic Red Sea. Prog Oceanogr. 2016;140:69–90. [Google Scholar]
- Kürten B, Khomayis HS, Devassy R et al.. Ecohydrographic constraints on biodiversity and distribution of phytoplankton and zooplankton in coral reefs of the Red Sea, Saudi Arabia. Mar Ecol. 2015;36:1195–214. [Google Scholar]
- Lara E, Vaqué D, Sà EL et al.. Unveiling the role and life strategies of viruses from the surface to the dark ocean. Sci Adv. 2017;3:e1602565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A, Flaten GAF, Sandaa R-A et al.. Spring phytoplankton bloom dynamics in Norwegian coastal waters: Microbial community succession and diversity. Limnol Oceanogr. 2004;49:180–90. [Google Scholar]
- Lasternas S, Agustí S. The percentage of living bacterial cells related to organic carbon release from senescent oceanic phytoplankton. Biogeosciences. 2014;11:6377–87. [Google Scholar]
- Lebaron P, Servais P, Agogué H et al.. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl Env Microbiol. 2001;67:1775–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Zhu A, Xu Z et al.. Dynamics of photosynthetic picoplankton in a subtropical estuary and adjacent shelf waters. J Mar Biol Assoc UK. 2010;90:1319–29. [Google Scholar]
- Livanou E, Lagaria A, Santi I et al.. Pigmented and heterotrophic nanoflagellates: Abundance and grazing on prokaryotic picoplankton in the ultra-oligotrophic Eastern Mediterranean Sea. Deep Sea Research Part II: Topical Studies in Oceanography. 2019;164:100–11. [Google Scholar]
- Li WK. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: Measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–75. [Google Scholar]
- Li WKW, Dickie PM. Monitoring phytoplankton, bacterioplankton, and virioplankton in a coastal inlet (Bedford Basin) by flow cytometry. Cytometry. 2001;44:236–46. [DOI] [PubMed] [Google Scholar]
- Lonborg C, Martínez-García S, Teira E et al.. Bacterial carbon demand and growth efficiency in a coastal upwelling system. Aquat Microb Ecol. 2011;63:183–91. [Google Scholar]
- Longnecker K, Sherr BF, Sherr EB. Activity and phylogenetic diversity of bacterial cells with high and low nucleic acid content and electron transport system activity in an upwelling ecosystem. Appl Environ Microbiol. 2005;71:7737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker K, Sherr BF, Sherr EB. Variation in cell-specific rates of leucine and thymidine incorporation by marine bacteria with high and with low nucleic acid content off the Oregon coast. Aquat Microb Ecol. 2006;43:113–25. [Google Scholar]
- Longnecker K, Wilson MJ, Sherr EB et al.. Effect of top-down control on cell-specific activity and diversity of active marine bacterioplankton. Aquat Microb Ecol. 2010;58:153–65. [Google Scholar]
- Lyngsgaard MM, Markager S, Richardson K et al.. How Well Does Chlorophyll Explain the Seasonal Variation in Phytoplankton Activity? Estuaries Coasts. 2017;40:1263–75. [Google Scholar]
- López‐Urrutia Á, Morán XAG. Temperature affects the size-structure of phytoplankton communities in the ocean. Limnol Oceanogr. 2015;60:733–8. [Google Scholar]
- Mackey KRM, Labiosa RG, Calhoun M et al.. Phosphorus availability, phytoplankton community dynamics, and taxon-specific phosphorus status in the Gulf of Aqaba, Red Sea. Limnol Oceanogr. 2007;52:873–85. [Google Scholar]
- Maranger R, Bird D. Viral abundance in aquatic systems:a comparison between marine and fresh waters. Mar Ecol Prog Ser. 1995;121:217–26. [Google Scholar]
- Maranger R, Vaqué D, Nguyen D et al.. Pan-Arctic patterns of planktonic heterotrophic microbial abundance and processes: Controlling factors and potential impacts of warming. Prog Oceanogr. 2015;139:221–32. [Google Scholar]
- Marañón E, Cermeño P, Latasa M et al.. Temperature, resources, and phytoplankton size structure in the ocean. Limnol Oceanogr. 2012;57:1266–78. [Google Scholar]
- Marie D, , Simon N, Vaulot D. Phytoplankton cell counting by flow cytometry. Algal Culturing Techniques, San Diego, USA: Elsevier, 2005, 253–67. [Google Scholar]
- Martínez JM, Swan BK, Wilson WH. Marine viruses, a genetic reservoir revealed by targeted viromics. The ISME journal. 2014;8:1079–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelboe M, Lyck PG. Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquat Microb Ecol. 2002;27:187–94. [Google Scholar]
- Miki T, Jacquet S. Complex interactions in the microbial world: underexplored key links between viruses, bacteria and protozoan grazers in aquatic environments. Aquat Microb Ecol. 2008;51:195–208. [Google Scholar]
- Miki T, Yamamura N. Intraguild predation reduces bacterial species richness and loosens the viral loop in aquatic systems: ‘kill the killer of the winner’ hypothesis. Aquat Microb Ecol. 2005;40:1–12. [Google Scholar]
- Mitbavkar S, Rajaneesh KM, Anil AC et al.. Picophytoplankton community in a tropical estuary: Detection of Prochlorococcus-like populations. Estuar Coast Shelf Sci. 2012;107:159–64. [Google Scholar]
- Mojica KDA, Carlson CA, Behrenfeld MJ. Regulation of low and high nucleic acid fluorescent heterotrophic prokaryote subpopulations and links to viral-induced mortality within natural prokaryote-virus communities. Microb Ecol. 2019;79:213–30. [DOI] [PubMed] [Google Scholar]
- Mojica KDA, Huisman J, Wilhelm SW et al.. Latitudinal variation in virus-induced mortality of phytoplankton across the North Atlantic Ocean. ISME J. 2016;10:500–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moràn XAG, Ducklow HW, Erickson M. Single‐cell physiological structure and growth rates of heterotrophic bacteria in a temperate estuary (Waquoit Bay, Massachusetts). Limnol Oceanogr. 2011;56:37–48. [Google Scholar]
- Morán XAG, Alonso-Sáez L, Nogueira E et al.. More, smaller bacteria in response to ocean's warming? Proc R Soc B Biol Sci. 2015;282:20150371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán XAG, Calvo‐Díaz A, Arandia‐Gorostidi N et al.. Temperature sensitivities of microbial plankton net growth rates are seasonally coherent and linked to nutrient availability. Environ Microbiol. 2018;20:3798–810. [DOI] [PubMed] [Google Scholar]
- Morán XAG, Calvo‐Díaz A. Single-cell vs. bulk activity properties of coastal bacterioplankton over an annual cycle in a temperate ecosystem: Single-cell activity of coastal bacterioplankton. FEMS Microbiol Ecol. 2009;67:43–56. [DOI] [PubMed] [Google Scholar]
- Morán XAG, Gasol JM, Pernice MC et al.. Temperature regulation of marine heterotrophic prokaryotes increases latitudinally as a breach between bottom-up and top-down controls. Glob Chang Biol. 2017;23:3956–64. [DOI] [PubMed] [Google Scholar]
- Muhling M, Fuller NJ, Millard A et al.. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ Microbiol. 2005;7:499–508. [DOI] [PubMed] [Google Scholar]
- Nagata T, Tamburini C, Arístegui J et al.. Emerging concepts on microbial processes in the bathypelagic ocean – ecology, biogeochemistry, and genomics. Deep Sea Res Part II Top Stud Oceanogr. 2010;57:1519–36. [Google Scholar]
- Nakajima R, Tanaka Y, Guillemette R et al.. Effects of coral-derived organic matter on the growth of bacterioplankton and heterotrophic nanoflagellates. Coral Reefs. 2017;36:1171–9. [Google Scholar]
- Ngugi DK, Antunes A, Brune A et al.. Biogeography of pelagic bacterioplankton across an antagonistic temperature-salinity gradient in the Red Sea. Mol Ecol. 2012;21:388–405. [DOI] [PubMed] [Google Scholar]
- Ng WHA, Liu H. Diel periodicity of grazing by heterotrophic nanoflagellates influenced by prey cell properties and intrinsic grazing rhythm. J Plankton Res. 2016;38:636–51. [Google Scholar]
- Ogawa H, Amagai Y, Koike I et al.. Production of Refractory Dissolved Organic Matter by Bacteria. Science. 2001;292:917–20. [DOI] [PubMed] [Google Scholar]
- Pace ML, Cole JJ. Comparative and experimental approaches to top-down and bottom-up regulation of bacteria. Microb Ecol. 1994;28:181–93. [DOI] [PubMed] [Google Scholar]
- Pace ML, Cole JJ. Regulation of bacteria by resources and predation tested in whole-lake experiments. Limnol Oceanogr. 1996;41:1448–60. [Google Scholar]
- Palenik B. Chromatic Adaptation in Marine Synechococcus Strains. Appl Environ Microbiol. 2001;67:991–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan LA, Zhang LH, Zhang J et al.. On-board flow cytometric observation of picoplankton community structure in the East China Sea during the fall of different years. FEMS Microbiol Ecol. 2005;52:243–53. [DOI] [PubMed] [Google Scholar]
- Parada V, Baudoux A-C, Sintes E et al.. Dynamics and diversity of newly produced virioplankton in the North Sea. ISME J. 2008;2:924–36. [DOI] [PubMed] [Google Scholar]
- Parsons RJ, Breitbart M, Lomas MW et al.. Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J. 2012;6:273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partensky F, Blanchot J, Vaulot D. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters : a review. Bulletin-Institut Oceanographique Monaco-Numero Special-. 1999;19:457–476. [Google Scholar]
- Pasulka AL, Samo TJ, Landry MR. Grazer and viral impacts on microbial growth and mortality in the southern California Current Ecosystem. J Plankton Res. 2015;37:320–36. [Google Scholar]
- Payet JP, Suttle CA. Physical and biological correlates of virus dynamics in the southern Beaufort Sea and Amundsen Gulf. J Mar Syst. 2008;74:933–45. [Google Scholar]
- Pernice MC, Forn I, Gomes A et al.. Global abundance of planktonic heterotrophic protists in the deep ocean. ISME J. 2015;9:782–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537. [DOI] [PubMed] [Google Scholar]
- Personnic S, Domaizon I, Dorigo U et al.. Seasonal and spatial variability of virio-, bacterio-, and picophytoplanktonic abundances in three peri-alpine lakes. Hydrobiologia. 2009;627:99–116. [Google Scholar]
- Pomeroy LR, leB. Williams PJ, Azam F et al.. The microbial loop. Oceanography. 2007;20:28–33. [Google Scholar]
- Post AF, Penno S, Zandbank K et al.. Long term seasonal dynamics of Synechococcus Population Structure In The Gulf Of Aqaba, Northern Red Sea. Front Microbiol. 2011;2:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimbault P, Garcia N, Cerutti F. Distribution of inorganic and organic nutrients in the South Pacific Ocean - evidence for long-term accumulation of organic matter in nitrogen-depleted waters. Biogeosciences. 2008;5:281–98. [Google Scholar]
- Rejas D, Muylaert K, De Meester L. Trophic interactions within the microbial food web in a tropical floodplain lake (Laguna Bufeos,Bolivia). Rev Biol Trop. 2005;53:85–96. [PubMed] [Google Scholar]
- Ribeiro CG, dos Santos AL, Marie D et al.. Pico and nanoplankton abundance and carbon stocks along the Brazilian Bight. PeerJ. 2016;4:e2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhi N, Zmerli Triki H, Molinero JC et al.. Seasonal variability of picophytoplankton under contrasting environments in northern Tunisian coasts, southwestern Mediterranean Sea. Mar Pollut Bull. 2018;129:866–74. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Caron DA, Berninger U-G. Relationships between bacteria and heterotrophic nanoplankton in marine and fresh waters: an inter-ecosystem comparison. Mar Ecol Prog Ser. 1992;86:1–14. [Google Scholar]
- Šantić D, Krstulović N, Šolić M et al.. HNA and LNA bacteria in relation to the activity of heterotrophic bacteria. Acta Adriat Int J Mar Sci. 2012;53:25–39. [Google Scholar]
- Scanlan DJ. Marine Picocyanobacteria. In: Whitton BA. (ed). Ecology of Cyanobacteria II: Their Diversity in Space and Time. Dordrecht: Springer Netherlands, 2012, 503–33. [Google Scholar]
- Segovia BT, Domingues CD, Meira BR et al.. Coupling between heterotrophic nanoflagellates and bacteria in fresh waters: Does latitude make a difference? Front Microbiol. 2016;7:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia BT, Meira BR, Lansac-Toha FM et al.. Growth and cytometric diversity of bacterial assemblages under different top–down control regimes by using a size-fractionation approach. J Plankton Res. 2018;40:129–41. [Google Scholar]
- Servais P, Courties C, Lebaron P et al.. Coupling Bacterial Activity Measurements with Cell Sorting by Flow Cytometry. Microb Ecol. 1999;38:180–9. [DOI] [PubMed] [Google Scholar]
- Šestanović S, Šolić M, Krstulović N et al.. Seasonal and vertical distribution of planktonic bacteria and heterotrophic nanoflagellates in the middle Adriatic Sea. Helgol Mar Res. 2004;58:83. [Google Scholar]
- Severiano J dos S, Moura A do N, Magalhães EM de M et al.. Study about Top-Down and Bottom-Up Controls in Regulating the Phytoplankton Biomass in a Eutrophic Reservoir in Northeastern Brazil. J Water Resour Prot. 2012;04:616. [Google Scholar]
- Sherr EB, Sherr BF. Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek. 2002;81:293–308. [DOI] [PubMed] [Google Scholar]
- Shibl AA, Haroon MF, Ngugi DK et al.. Distribution of Prochlorococcus Ecotypes in the Red Sea Basin Based on Analyses of rpoC1 Sequences. Front Mar Sci. 2016;3, DOI:10.3389/fmars.2016.00104. [Google Scholar]
- Silva L, Calleja ML, Huete-Stauffer TM et al.. Low Abundances but High Growth Rates of Coastal Heterotrophic Bacteria in the Red Sea. Front Microbiol. 2019;9, DOI:10.3389/fmicb.2018.03244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siokou-Frangou I, Christaki U, Mazzocchi MG et al.. Plankton in the open Mediterranean Sea: a review. Biogeosciences. 2010;7:1543–86. [Google Scholar]
- Smith EM. Coherence of microbial respiration rate and cell-specific bacterial activity in a coastal planktonic community. Aquat Microb Ecol. 1998;16:27–35. [Google Scholar]
- Šolić M, Krstulovic N, Vilibic I et al.. Variability in the bottom-up and top-down controls of bacteria on trophic and temporal scales in the middle Adriatic Sea. Aquat Microb Ecol. 2009;58:15–29. [Google Scholar]
- Solid M, Krstulovic N. Role of predation in controlling bacterial and heterotrophic nanoflagellate standing stocks in the coastal Adriatic Sea: seasonal patterns. MARINE ECOLOGY-PROGRESS SERIES. 1994;114:219–35. [Google Scholar]
- Sommer U, Berninger UG, Böttger-Schnack R et al.. Grazing during early spring in the Gulf of Aqaba and the northern Red Sea. Mar Ecol Prog Ser. 2002;239:251–61. [Google Scholar]
- Sommer U. Scarcity of medium-sized phytoplankton in the northern Red Sea explained by strong bottom-up and weak top-down control. Mar Ecol Prog Ser. 2000;197:19–25. [Google Scholar]
- Stets EG, Cotner JB. The influence of dissolved organic carbon on bacterial phosphorus uptake and bacteria-phytoplankton dynamics in two Minnesota lakes. Limnol Oceanogr. 2008;53:137–47. [Google Scholar]
- Storesund JE, Erga SR, Ray JL et al.. Top-down and bottom-up control on bacterial diversity in a western Norwegian deep-silled fjord. FEMS Microbiol Ecol. 2015;91, DOI:10.1093/femsec/fiv076. [DOI] [PubMed] [Google Scholar]
- Sugai Y, Tsuchiya K, Kuwahara VS et al.. Bacterial growth rate and the relative abundance of bacteria to heterotrophic nanoflagellates in the euphotic and disphotic layers in temperate coastal waters of Sagami Bay, Japan. J Oceanogr. 2016;72:577–87. [Google Scholar]
- Suttle CA. Marine viruses — major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. [DOI] [PubMed] [Google Scholar]
- Suttle CA. The significance of viruses to mortality in aquatic microbial communities. Microb Ecol. 1994;28:237–43. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Rassoulzadegan F. Full-depth profile (0–2000 m) of bacteria, heterotrophic nanoflagellates and ciliates in the NW Mediterranean Sea: Vertical partitioning of microbial trophic structures. Deep Sea Res Part II Top Stud Oceanogr. 2002;49:2093–107. [Google Scholar]
- Tanaka T, Rassoulzadegan F. Vertical and seasonal variations of bacterial abundance and production in the mesopelagic layer of the NW Mediterranean Sea: bottom-up and top-down controls. Deep Sea Res Part Oceanogr Res Pap. 2004;51:531–44. [Google Scholar]
- Taucher J, Arístegui J, Bach LT et al.. Response of subtropical phytoplankton communities to ocean acidification under oligotrophic conditions and during nutrient fertilization. Front Mar Sci. 2018;5, DOI:10.3389/fmars.2018.00330. [Google Scholar]
- Thingstad TF, Våge S, Storesund JE et al.. A theoretical analysis of how strain-specific viruses can control microbial species diversity. Proc Natl Acad Sci. 2014;111:7813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Berdjeb L, Sime-Ngando T et al.. Viral abundance, production, decay rates and life strategies (lysogeny versus lysis) in Lake Bourget (France). Environ Microbiol. 2011;13:616–30. [DOI] [PubMed] [Google Scholar]
- Trabelsi A, Rassoulzadegan F. Effect of bacterial community dynamics on DOC seasonal changes in the north-western Mediterranean Sea. J Plankton Res. 2011;33:1249–62. [Google Scholar]
- Tsai AY, Gong G-C, Chao CF. Contribution of Viral Lysis and Nanoflagellate Grazing to Bacterial Mortality at Surface Waters and Deeper Depths in the Coastal Ecosystem of Subtropical Western Pacific. Estuaries Coasts. 2016;39:1357–66. [Google Scholar]
- Tsai AY, Gong G-C, Huang YW et al.. Estimates of bacterioplankton and Synechococcus spp. mortality from nanoflagellate grazing and viral lysis in the subtropical Danshui River estuary. Estuar Coast Shelf Sci. 2015;153:54–61. [Google Scholar]
- Tsai AY, Gong G-C, Hung J. Seasonal variations of virus- and nanoflagellate-mediated mortality of heterotrophic bacteria in the coastal ecosystem of subtropical western Pacific. Impact Short-Term Warm Seas Var Bact Growth Grazing Viral Lysis Coast Waters Taiwan. 2013;10:3055–65. [Google Scholar]
- Tsai AY, Gong G-C, Lui H. Seasonal variations in virioplankton and picoplankton in semi-enclosed and open coastal waters. Atoms. Ocean. Sci. 2018;29:465–72. [Google Scholar]
- Tsai AY, Gong G-C, Shiau W. Viral lysis and nanoflagellate grazing on prokaryotes: effects of short-term warming in a coastal subtropical marine system. Hydrobiologia. 2015;751:43–54. [Google Scholar]
- Vaqué D, Boras JA, Torrent-Llagostera F et al.. Viruses and Protists Induced-mortality of Prokaryotes around the Antarctic Peninsula during the Austral Summer. Front Microbiol. 2017;8:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaqué D, Casamayor E, Gasol J. Dynamics of whole community bacterial production and grazing losses in seawater incubations as related to the changes in the proportions of bacteria with different DNA content. Aquat Microb Ecol. 2001;25:163–77. [Google Scholar]
- Vaqué D, Lara E, Arrieta JM et al.. Warming and CO2 enhance arctic heterotrophic microbial activity. Front Microbiol. 2019;10:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuis MJW, Kraay GW. Cell abundance and fluorescence of picoplankton in relation to growth irradiance and nitrogen availability in the red sea. Neth J Sea Res. 1993;31:135–45. [Google Scholar]
- Vives-Rego J, Lebaron P, Nebe-von Caron G. Current and future applications of flow cytometry in aquatic microbiology. FEMS Microbiol Rev. 2000;24:429–48. [DOI] [PubMed] [Google Scholar]
- Vázquez-Domínguez E, Duarte CM, Agustí S et al.. Microbial plankton abundance and heterotrophic activity across the Central Atlantic Ocean. Prog Oceanogr. 2008;79:83–94. [Google Scholar]
- Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–81. [DOI] [PubMed] [Google Scholar]
- Weisse T, MacIsaac E. Significance and fate of bacterial production in oligotrophic lakes in British Columbia. Can J Fish Aquat Sci. 2000;57:96–105. [Google Scholar]
- Weisse T. The microbial loop in the Red Sea: dynamics of pelagic bacteria and heterotrophic nanoflagellates. Mar Ecol Prog Ser. 1989;55:241–50. [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham SA, Claessens M, Post AF. Ciliates, microbes and nutrients: interactions in the seasonally mixed Gulf of Aqaba. J Plankton Res. 2015;37:258–71. [Google Scholar]
- Wiggins BA, Alexander M. Minimum bacterial density for bacteriophage replication: Implications for significance of bacteriophages in natural ecosystems. Appl Env Microbiol. 1985;49:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigington CH, Sonderegger D, Brussaard CPD et al.. Re-examination of the relationship between marine virus and microbial cell abundances. Nat Microbiol. 2016;1:15024. [DOI] [PubMed] [Google Scholar]
- Winter C, Moeseneder MM, Herndl GJ et al.. Relationship of geographic distance, depth, temperature, and viruses with prokaryotic communities in the Eastern Tropical Atlantic Ocean. Microb Ecol. 2008;56:383–9. [DOI] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: Viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Follows MJ, Giovannoni SJ et al.. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science. 2015;347:1257594. [DOI] [PubMed] [Google Scholar]
- Xia X, Guo W, Tan S et al.. Synechococcus assemblages across the salinity gradient in a salt wedge estuary. Front Microbiol. 2017;8:1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates MV, Gerba CP, Kelley LM. Virus persistence in groundwater. Appl Environ Microbiol. 1985;49:778–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Temperton B, Thrash JC et al.. Abundant SAR11 viruses in the ocean. Nature. 2013;494:357–60. [DOI] [PubMed] [Google Scholar]
- Zhong X, Pradeep Ram AS, Colombet J et al.. Variations in Abundance, Genome Size, Morphology, and Functional Role of the Virioplankton in Lakes Annecy and Bourget over a 1-Year Period. Microb Ecol. 2014;67:66–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


