Abstract
Artificial intelligence (AI) continues to garner substantial interest in medical imaging. The potential applications are vast and include the entirety of the medical imaging life cycle from image creation to diagnosis to outcome prediction. The chief obstacles to development and clinical implementation of AI algorithms include availability of sufficiently large, curated, and representative training data that includes expert labeling (eg, annotations). Current supervised AI methods require a curation process for data to optimally train, validate, and test algorithms. Currently, most research groups and industry have limited data access based on small sample sizes from small geographic areas. In addition, the preparation of data is a costly and time-intensive process, the results of which are algorithms with limited utility and poor generalization. In this article, the authors describe fundamental steps for preparing medical imaging data in AI algorithm development, explain current limitations to data curation, and explore new approaches to address the problem of data availability.
© RSNA, 2020
Learning Objectives:
After reading the article and taking the test, the reader will be able to:
■ List the different steps needed to prepare medical imaging data for development of machine learning models
■ Discuss the new approaches that may help address data availability to machine learning research in the future
■ Identify properly de-identified medical data according to the U.S. Health Insurance Portability and Accountability Act (HIPAA) and European General Data Protection Regulation (GDPR) standards
Accreditation and Designation Statement
The RSNA is accredited by the Accreditation Council for Continuing Medical Education (ACCME) to provide continuing medical education for physicians. The RSNA designates this journal-based SA-CME activity for a maximum of 1.0 AMA PRA Category 1 Credit™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
Disclosure Statement
The ACCME requires that the RSNA, as an accredited provider of CME, obtain signed disclosure statements from the authors, editors, and reviewers for this activity. For this journal-based CME activity, author disclosures are listed at the end of this article.
Summary
Supervised artificial intelligence (AI) methods for evaluation of medical images require a curation process for data to optimally train, validate, and test algorithms. The chief obstacles to development and clinical implementation of AI algorithms include availability of sufficiently large, curated, and representative training data that includes expert labeling (eg, annotations).
Essentials
■ Image data availability is an important hurdle for implementation of artificial intelligence (AI) in the clinical setting.
■ AI researchers need to be aware of the data source and potential biases, which may affect generalizability of AI algorithms.
■ New approaches such as federated learning, interactive reporting, and synoptic reporting may help to address data availability in the future; however, curating and annotating data, as well as computational requirements, are substantial barriers.
Introduction
Artificial intelligence (AI), as a field defined broadly by the engineering of computerized systems able to perform tasks that normally require human intelligence, has substantial potential in the medical imaging field (1). Machine learning and deep learning algorithms have been developed to improve workflows in radiology or to assist the radiologist by automating tasks such as lesion detection or medical imaging quantification. Workflow improvements include prioritizing worklists for radiologists (2,3), triaging screening mammograms (4), reducing or eliminating gadolinium-based contrast media for MRI (5,6), and reducing the radiation dose of CT imaging by advancing image noise reduction (7–9). Automatic lesion detection by using machine learning has been applied to many imaging modalities and includes detection of pneumothorax (10,11), intracranial hemorrhage (12), Alzheimer disease (13), and urinary stones (14). Automatic quantification of medical images includes assessing skeletal maturity on pediatric hand radiographs (15), coronary calcium scoring on CT images (16), prostate classification at MRI (17), breast density at mammography (18), and ventricle segmentation at cardiac MRI (19,20). Yet substantial implementation and regulatory challenges have made application of AI models in clinical practice difficult and limited the potential of these advancements. Nearly all limitations can be attributed to one substantial problem: lack of available image data for training and testing of AI algorithms.
Currently, most research groups and companies have limited access to medical images, while the small sample sizes and lack of diverse geographic areas hinder the generalizability and accuracy of developed solutions (21). Although small data sets may be sufficient for training of AI algorithms in the research setting, large data sets with high-quality images and annotations are still essential for supervised training, validation, and testing of commercial AI algorithms. This is especially true in the clinical setting and is well outlined by Park and Han (22).
Most health care systems are not adequately equipped to share large amounts of medical images. Even when development is possible, medical data are often stored in disparate silos, which is not optimal for medical AI development that can be broadly used in clinical practice(s). Furthermore, simply achieving access to large quantities of image data is insufficient to allay these shortcomings. Adequate curation, analysis, labeling, and clinical application are critical to achieving high-impact clinically meaningful AI algorithms. We describe a process of labeling, curating, and sharing medical image data for AI algorithm development, followed by an in-depth discussion of alternative strategies to achieve responsible data sharing and applications in AI algorithm development for optimal clinical impact. To date, to our knowledge, this is the first work that gives an overview of the process of medical imaging data preparation for machine learning.
Conflicts of Interest
Data and information were controlled by authors who are not industry employees. Two authors (W.A.K. and C.H.) are industry employees of Segmed (Palo Alto, Calif), a company that delivers machine learning training data for medical imaging. Four authors (M.J.W., J.W., H.H., and M.P.L.) are advisors and stockholders of the same company. However, for this project none of the authors received financial or research support from the industry and the current project itself also was not funded.
Data Preparation Overview
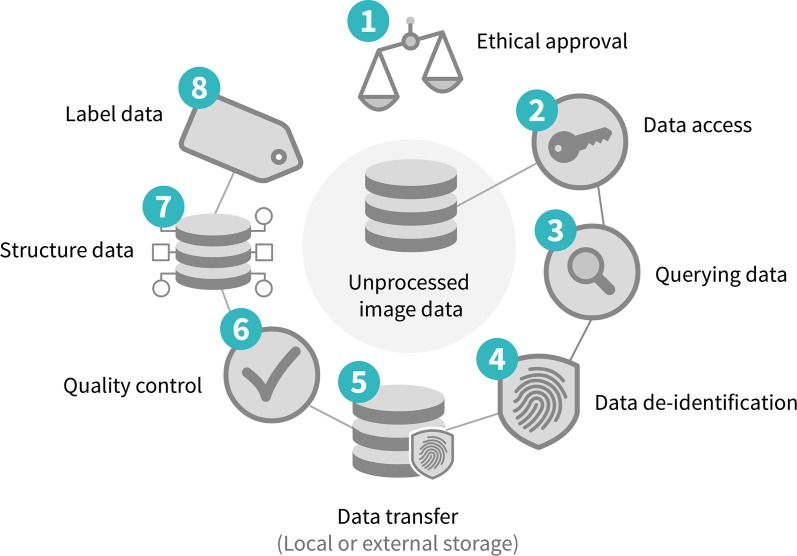
Before medical images can be used for the development of an AI algorithm, certain steps need to be taken. Typically, approval from the local ethical committee is required before medical data may be used for development of a research or a commercial AI algorithm. An institutional review board needs to evaluate the risks and benefits of the study to the patients. In many cases existing data are used, which requires a retrospective study. Because the patients in this type of study do not need to undergo any additional procedures, explicit informed consent is generally waived. With clinical trials, each primary investigator may need to provide approval to share data on their participants. In case of a prospective study, where study data are gathered prospectively, informed consent is necessary. After ethical approval, relevant data needs to be accessed, queried, properly de-identified, and securely stored. Any protected health information needs to be removed both from the Digital Imaging and Communications in Medicine (DICOM) metadata, as well as from the images (23). If the data are intended for open-source research efforts, then additional human inspection of each image is standard because some images contain free-form annotations that have been scanned and cannot be removed reliably with automated methods. The quality and amount of the images vary with the target task and domain. The next step is to structure the data in homogenized and machine-readable formats (24). The last step is to link the images to ground-truth information, which can be one or more labels, segmentations, or electronic phenotype (eg, biopsy or laboratory results). The entire process to prepare medical images for AI development is summarized in Figure 1.
Figure 1:
Diagram shows process of medical image data handling.
Accessing and Querying Data
Developers of AI algorithms are typically not located within a hospital and therefore often do not have direct access to medical imaging data through the picture archiving and communication system (PACS), especially when AI researchers are developing commercial algorithms. Access to PACS environments is limited to accredited professionals such as physicians, technologists, PACS managers, and clinical scientists. Making data accessible to AI developers is challenging and requires multiple steps, including de-identification of data (described later). The ideal approach is collaboration between clinicians and AI developers, either in-house or through collaborative research agreements.
Once data are accessible to AI developers, different strategies are available to search for medical images and clinical data. Custom search queries may, for example, consist of strings, international classification of disease codes, and current procedural terminology codes. Data can be systematically searched and extracted from hospital PACS and electronic medical records by using PACS or radiology information system search engines. For example, many PACS vendors allow user access to metadata such as annotations, creator, series and image number, and unique target lesion names and relations. These data can be exported in some PACS and further managed by other systems such as electronic medical records, cancer databases, and oncologist or other provider databases (25). Alternatively, software packages are available to simplify the process of data querying (26–28).
De-Identification
Although written informed consent from patients is not always necessary, according to the U.S. Health Insurance Portability and Accountability Act, or HIPAA, and the European General Data Protection Regulation, both retrospectively and prospectively gathered data require proper de-identification. Sensitive information includes but is not limited to name, medical record number, and date of birth. A complete list of the 18 HIPAA identifiers is shown in Table 1. Identifiable information is commonly present in the DICOM metadata (header) and multiple tools are available to automatically remove this information (29). DICOM de-identification profiles are defined for a range of applications and used as the basis for de-identification workflows implemented in the Radiological Society of North America Clinical Trial Processor and the Cancer Imaging Archive (30). Besides the DICOM metadata, protected health information may also be embedded in images, which is often the case with US examinations or radiographs that are scanned into a health care system. Removal of embedded information requires more advanced de-identification methods such as optical character recognition (31) and human review for handwriting on scanned images not always recognized by automated methods. Care must also be taken not to inadvertently mix data sets, because doing so increases the individual risk of reidentification through cross-linking of nonrelated data points (32). Finally, medical data can be anonymized with k-anonymity, which transforms an original data set containing protected health information to prevent potential intruders from determining the patient’s identity (33). For posting radiology data in open-source research efforts, the DICOM metadata is often removed completely or converted to another format such as Neuroimaging Informatics Technology Initiative, or NIFTI, which retains only voxel size and patient position. Totally removing the DICOM metadata for open-source research efforts prevents privacy issues but reduces the value of data, because metadata is important for AI algorithm development.
Table 1:
Protected Health Information Identifiers according to the Health Insurance Portability and Accountability Act
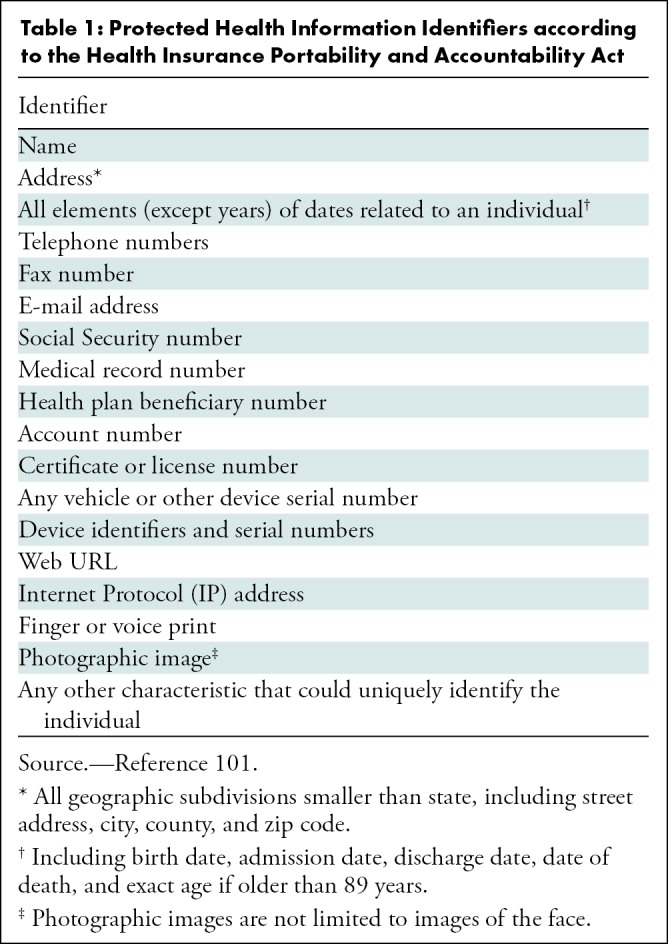
Important protected health information that can be potentially overlooked, yet can act as “identity signatures,” include the HIPAA items full-face photos and comparable images, as well as biometric identifiers (ie, retinal scan and fingerprints). For example, head and neck CT data can qualify as comparable images. With widespread volumetric acquisition and ease of three-dimensional reformatting, the soft-tissue kernels or filters allow facial reconstruction that can identify the patient. Until there is a secure digital encryption method to alter identification without compromising clinical information, those making data publicly available need to take potential biometric signatures into consideration.
Data Storage
Data are commonly transferred to either a local data storage (single-center study) or an external data storage (multicenter study or commercial AI development). Data are usually stored at an on-premise server; however, with current cloud-based developments, data are increasingly stored in the cloud. Advantages of on-premise data storage include data safety and availability, but the potential of sharing data with other institutions is limited. Cloud-based data storage, on the other hand, is becoming more secure, improves the possibilities of sharing data, and provides data backup. Disadvantages of cloud-based storage include costs and the need for a fast internet connection.
Resampling Medical Images
Image perception of medical image data are relatively complex compared with nonmedical image perception tasks. Most convolutional neural networks for classification of images are trained and tested on two-dimensional images with fewer than 300 × 300 pixels (34). Medical images, however, exceed these dimensions; the in-plane spatial resolution is generally higher than 300 × 300 pixels, and many medical image studies are three-dimensional instead of two-dimensional. Training convolutional neural networks with images larger than 300 × 300 pixels is possible; however, computers with strong computational power are necessary. This problem is most relevant in high-resolution applications; examples include CT of the inner ear or full-field digital mammography. Solutions include downsampling of the image resolution or patch-based evaluation of only image parts with relevant information (eg, focus on the aortic region in an algorithm developed for aortic dissection segmentation). However, patch-based methods frequently have high computational demands and are time consuming to train. Model training can also be simplified by classifying labels to healthy (scale 0) and diseased at different levels; for example, from less severely diseased (scale 2) to more severely diseased (scale 4) (35).
Besides DICOM files, which contain metadata and image slices, other file types are also available. AI development with raw MRI or CT data (before images are reconstructed) is gaining interest and has a potentially valuable role. Advantages include an increased amount of information captured in raw data, and disadvantages include the large storage space needed and difficult interpretation of raw data without reconstructed images.
Choosing Appropriate Label and Ground Truth Definition
Current AI algorithms for medical image classification tasks are generally based on a supervised learning approach. This means that before an AI algorithm can be trained and tested, the ground truth needs to be defined and linked to the image. The term ground truth typically refers to information acquired from direct observation (such as biopsy or laboratory results). Image labels are annotations performed by medical experts such as radiologists. These annotations can be considered ground truth if imaging is the reference standard (eg, pneumothorax). Choosing the appropriate label for a given imaging AI application requires a balance between finding the best discriminating categories (ie, normal vs emergent) and clinically relevant granularity (ie, subtype of liver lesion) depending on the desired task. With the exception of AI methods that enhance image quality, medical images in isolation are generally not suitable for developing diagnostic AI models unless associated with a diagnosis through the free-text radiology report (which require additional labeling strategies discussed below), expert consensus, segmentation, or an applied ground truth label such as electronic phenotyping (1).
Although extracting structured labels from the radiology report text by using natural language processing may be ultimately the most scalable approach, researchers need to be cautious of the error rates both in the natural language processing techniques and the original text reports. In large quantities, it is known that AI algorithms can be trained on relatively low-quality data, but knowing the true ground truth for a given task to correlate with the imaging findings is the ideal (Fig 2). Medical imaging alone is considered ground truth for certain diagnoses, including intracranial hemorrhage, fractures, renal stone, and aortic dissection. However, the majority of findings is not definitive on the basis of imaging examinations alone and requires further follow-up, pathologic diagnosis, or clinical outcomes to achieve ground truth (ie, lung cancer, liver mass, pneumonia, etc). For example, an opacity on a chest radiograph has an extensive differential. It is difficult to know the ground truth without obtaining surgical, pathologic, genomic, or clinical outcome data (Fig 3). There are also situations in which one modality may support a diagnosis but require definitive confirmation by using another modality. For example, a head CT might have findings supporting a diagnosis of stroke, but an MRI could definitively confirm. Depending on the ultimate task or purpose of the AI algorithm, ground truth definition may require confirmatory clinical labeling beyond the radiology opinion or report such as a pathologic or surgical report, clinical outcome, or both. Accumulation of this clinical information for a large number of patients can be resource intensive and is often referred to as electronic phenotyping (36). Querying of nonimaging data such as clinical outcomes and patient demographics can often not be performed through PACS, but requires extraction of information from electronic medical records.
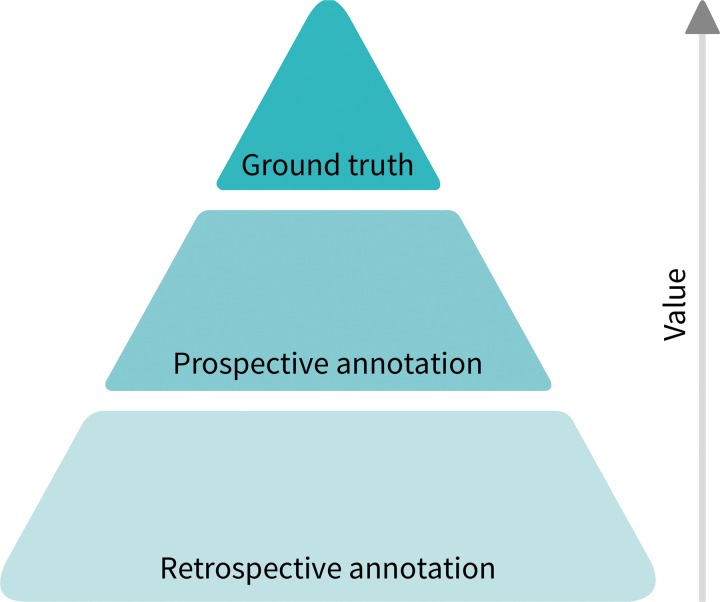
Figure 2:
Diagram shows value hierarchy of imaging annotation. Most useful but least abundant is ground truth (pathologic, genomic, or clinical outcome data). Prospective annotation is incredibly valuable due to availability of contemporaneous information (clinical and/or laboratory data). By comparison, retrospective annotations are least valuable.
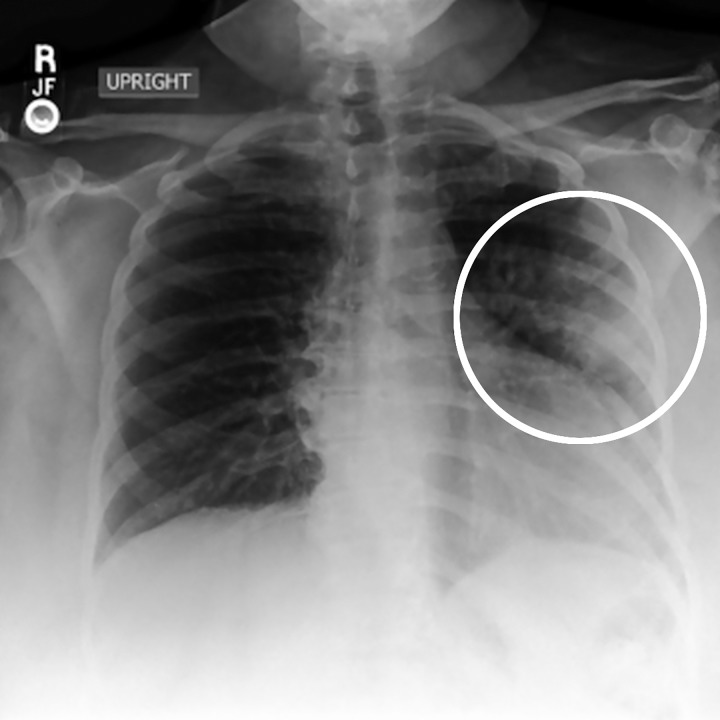
Figure 3:
Image in posterior-anterior direction shows nonspecific abnormality on chest radiograph. Application of most accurate label for nonspecific finding such as opacity in left lung (circle) is challenging in absence of other clinical and laboratory data.
In general, imaging data can be labeled in a variety of ways including structured label(s), image annotations, image segmentations, and/or electronic phenotypes (1,37). More often, application of the imaging diagnosis based on expert interpretation or a consensus of experts based on a reinterpretation of the images, or free-text report is used (10). Another approach to labeling is through the use of segmentations including, for example, outlining lung nodules at CT of the chest (Fig 4).
Figure 4a:
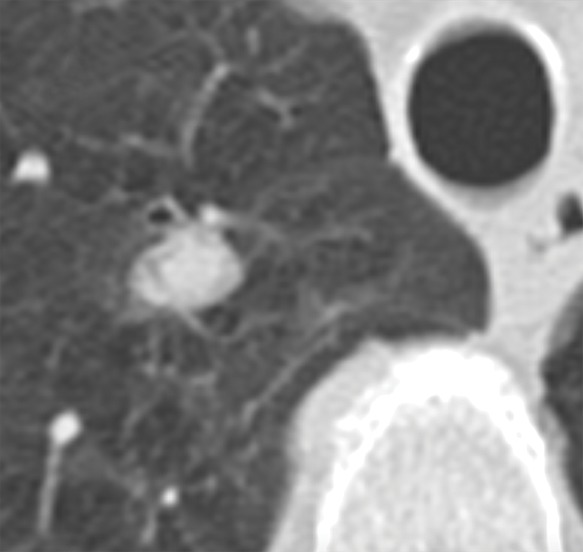
Axial images show medical image segmentations performed by experts. (a) CT examination of patient with lung nodule. (b) Nodule is independently and blindly segmented by three medical experts with free open-source software package (Horos, version 3.3.5; Nimble d/b/a Purview, Annapolis, Md). (c) Magnified image of segmentations. There are differences between segmentations; however, these differences are small and not clinically relevant.
Figure 4b:
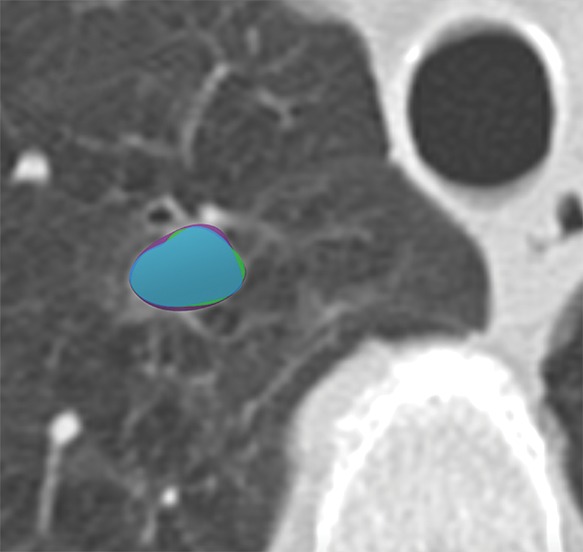
Axial images show medical image segmentations performed by experts. (a) CT examination of patient with lung nodule. (b) Nodule is independently and blindly segmented by three medical experts with free open-source software package (Horos, version 3.3.5; Nimble d/b/a Purview, Annapolis, Md). (c) Magnified image of segmentations. There are differences between segmentations; however, these differences are small and not clinically relevant.
Figure 4c:
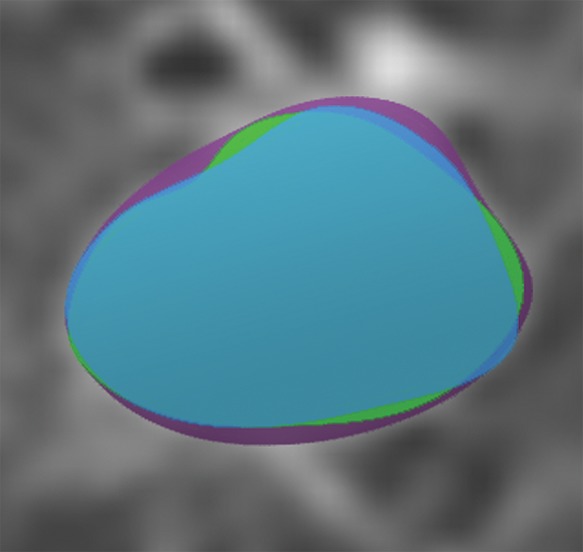
Axial images show medical image segmentations performed by experts. (a) CT examination of patient with lung nodule. (b) Nodule is independently and blindly segmented by three medical experts with free open-source software package (Horos, version 3.3.5; Nimble d/b/a Purview, Annapolis, Md). (c) Magnified image of segmentations. There are differences between segmentations; however, these differences are small and not clinically relevant.
Ground Truth or Label Quality
Accurate ground truth definition or image labels for a large number of radiology examinations are required to build accurate medical imaging AI models (38,39). There are guidelines for reporting diagnostic imaging aiming toward structured reporting, which would immensely reduce the effort needed to extract useful imaging labels. At present, however, the overwhelming majority of reports remain composed of free text (40). Novel semantic reporting systems that aim to index and codify free-text reports in real time are being developed, but are currently not widely available for large populations. As a result, most centers attempting to use retrospective data are faced with large volumes of imaging studies and narrative reports that require substantial effort to label. Currently, there are many approaches to perform retrospective labeling, ranging from simple manual labeling by radiologists to automated approaches that can extract structured information from the radiology report and/or electronic medical record (41).
Outside of medical applications, manual labeling is a commonly used approach in acquiring labeled imaging data for AI applications. Large corporations often hire nonexperts to hand review and label large amounts of data needed to support automated services such as ranking web search results, providing recommendations, or displaying relevant ads (42,43). This approach can be effective in medical imaging as well, but is impractical in most cases when used on large populations because it is extremely time consuming (and costly) to use medical experts, particularly for advanced modalities such as CT, PET, or MRI. When a relatively small number of images are needed for AI development, medical expert labeling and segmentation may be feasible. Segmentation performance can be evaluated by using either the Dice coefficient or the more advanced simultaneous truth and performance level estimation, or STAPLE, algorithm (44). The STAPLE algorithm compares segmentations and computes a probabilistic estimate of the true segmentation. For narrowly focused applications such as colonic polyp classification and kidney segmentation, crowdsourcing of labels by nonexperts may be feasible (45,46). Heim et al (47) compared segmentations of the liver performed by nonexperts, engineers with domain knowledge, medical students, and radiologists. Despite the finding that the crowd needed more time, accuracy was similar between these groups. Crowdsourcing challenges include inaccuracy with anatomic variations and pathologies, quality control, and ethical issues such as sharing medical images with the crowd. Crowd-sourced labeling is mostly performed with web-based tools, which are freely available (48,49).
One solution is to extract information from the report of imaging findings through rule-based natural language processing (50,51) or recurrent neural networks (52,53). One of the most useful natural language processing methods is called topic modeling, which summarizes a data set with a large amount of text to obtain gross insight over the data set. This approach characterizes document content based on key terms and estimates topics contained within documents. For example, documents associated with “brain MRI” would comprise key terms such as axial, contrast, MRI, sagittal, brain, enhancement, et cetera. Another class of architectures, recurrent neural networks, are neural network–based models that can be trained on a small sample of reports and rapidly achieve performance levels of the state-of-the-art more traditional natural language processing tools (54,55). Recurrent neural networks represent an important improvement to language modeling because a dependency of a word in narrative language can occur long distances apart, such as “No evidence for acute or subacute infarction.” In this example, the “No” is far from the target “infarction.” It can be confusing for traditional natural language processing tools but picked up with recurrent neural networks (54). As a result, strategies for extracting structured labels from unstructured text have emerged that have shown a great deal of promise for limited applications to apply structured labels in large populations and generate large labeled data sets of imaging studies (2,56).
Because radiology reports are most often unstructured and not created specifically for the development of AI algorithms, the extracted information contains noise (ie, has a relatively low quality). Neural networks can still be relatively robust when trained with noisy labels (57). However, one should be careful when using noisy labels for the development of clinically applicable algorithms because every labeling error could be translated to a decrease in algorithm accuracy. It is estimated that 2%–20% of radiology reports contain demonstrable errors (58).
Lastly, there is a trend toward interactive reporting where the radiologist report contains hypertext directly connected to image annotations (59). Such annotations have been used effectively for labeling of open-source data sets (60). Measurements can be performed in advance of radiologists by radiology preprocessors that improve annotation quality while saving radiologists time (61). Preliminary work on prospective labeling is showing that two-diameter measurements and ovals are better than one-diameter measurements and much better than arrows (62). In addition to structured reporting, to the level of synoptic reporting, this should contribute significantly to increased prospective expert-labeled data. Interactive reporting is becoming more common where radiologists routinely label images in three dimensions and connect directly to hypertext descriptions in their report. This may be a potential solution to the local labeling issue with research just beginning. Nevertheless, substantial collaborative efforts may ultimately be needed to arrive at widely adopted reporting and standardization of labeling of imaging studies such that interoperability of data sets and subsequent models is possible.
Data Sets
Development of AI algorithms by using supervised learning requires large and heterogeneous training, validation, and testing data sets.
Data Set Types
Similar to conventional regression modeling, AI models are trained by inputting medical images linked to ground truth outcome variables (eg, pneumothorax). Generally, the training imaging data set is larger than the validation and testing data sets in ratios of 80:10:10 or 70:15:15. To ensure generalizability of the AI algorithm, bias of the training data set should be limited. If an AI algorithm is trained with images from a European institution and the algorithm is used in an Asian population, then performance may be affected by population or disease prevalence bias. Similarly, if all the imaging training data were acquired by using one kind of imaging machine, it may not work as well on machines from other manufacturers, known as vendor or single-source bias. It is thus advised to use images from multiple diverse sources, or at least images representing the target population or health system in which the algorithm is to be deployed. After the algorithm is trained, a validation data set is needed to fine-tune the algorithm hyperparameters and to check for overfitting. Note that validation in AI algorithm development has a different meaning than in conventional statistical modeling. Here, validation means tuning of the algorithm until the final performance of the model is evaluated with a testing data set. Multiple internal validation methods are available; however, independent validation in an external data set is preferred over internal validation to properly evaluate generalizability (63). Even if an electronic phenotype is available (eg, biopsy results of a lung nodule), annotations are needed for training and validation data sets to inform the algorithm of the location of the specific lung nodule to allow the algorithm to better understand the images. The testing data set functions as the reference standard and is used to evaluate the performance of the algorithm. In multiple conditions, imaging is the reference standard (eg, pneumothorax), where high-quality annotations are needed for the testing imaging data set because this data set functions as the reference standard. The quality and veracity of the testing data set is arguably more important than that of the training set because this data set is used for performance testing and regulatory approval.
Data Set Size
To ensure generalizability, large training data sets are often essential. For specific targeted applications or populations, relatively small data sets (hundreds of cases) may be sufficient. Large sample sizes are especially required in populations with substantial heterogeneity or when differences between imaging phenotypes are subtle (35). The algorithm performance for computer vision tasks increases logarithmically with increased training data volume (64,65). Therefore, a proper sample size is needed. The main questions for the power calculation include the following: (a) which cases need to be included in the sample to allow for generalizability in a larger population, and (b) how many cases are needed to show an effect (66). The sample size calculation for test data sets should use traditional power calculation methods to estimate the sample size. In general, the development of generalizable AI algorithms in medical imaging requires statistically powered data sets in the order of hundreds of thousands or millions, which is problematic for many researchers and developers.
One partial solution for this problem may be semisupervised learning. Fully annotated data sets are needed for supervised learning, whereas semisupervised learning uses a combination of annotated and unannotated images to train an algorithm (67,68). Semisupervised learning may allow for a limited number of annotated cases; however, large data sets of unannotated images are still needed.
Another potential future solution to increase data sample size may be the generation of synthetic data through generative adversarial networks (69). Generative adversarial networks have the potential to synthesize unlimited numbers of high-quality realistic images that can be added to training data sets for development of detection and classification algorithms. First results in synthesized radiographs and mammograms are promising. However, limited evidence is available, especially when abnormalities are present on images (69,70).
Data Sources
Most academically developed AI algorithms in medical imaging have been trained, validated, and tested with local data from a single institution (1). Whereas multi-institutional data from different geographic areas would include a wide variety of imaging machines, ethnicities, and pathologies, single-institutional data are commonly used due to lack of access to multi-institutional data. Many medical centers lack motivation and resources to share data with other institutions or companies that develop AI algorithms due to regulatory and privacy issues, although medical image data can be shared without violating General Data Protection Regulation or HIPAA regulations with proper de-identification methods and secure data handling. Currently, medical image data are stored in isolated decentralized silos, limiting the development of generalizable unbiased AI algorithms, which could theoretically be solved by having centralized data storage systems. When data are being made available to AI developers, appropriate data management is essential. Wilkinson et al (71) describe the FAIR (findability, accessibility, interoperability, and reusability) principle for good data management.
Open-Source Data Sets
An increasing number of data sets has been open sourced to address the problem of data access in medical research. Data sets are available in a wide range of domains from neuroimaging (72–77), breast imaging (36,78), chest radiographs (41,79), knee MRI (80), body CT (60), and others; a list of well-known open-source data sets is given in Table 2. Whereas open-source data sets stimulate the development of novel AI algorithms in the medical imaging field, there are important limitations. First, there is a wide variety of number and quality of images and availability of metadata and clinical information. Second, some open-source data sets are (partly) acquired by using outdated machines, contain low-quality images, lack expert labeling or data curation, or have a sample size that is too small to reach high-quality algorithms that can be used clinically. Moreover, many open-source data sets are restricted to noncommercial (research only) use (79). This is a major limitation for researchers wishing to develop marketable algorithms, as commercial adoption is a common avenue for clinical deployment.
Table 2:
Large Open-Source Medical Imaging Data Sets
Bias
One of the most important limitations of training AI algorithms based on data from a single institution or from multiple institutions in a small geographic area is sampling bias. If an AI algorithm trained this way is applied to a different geographic area, then results of the algorithm may be unreliable due to differences between the sample population and target population (81). Other sources of bias include differences in age, proportions of race and sex, use of imaging machines (vendors, types, acquisition protocols), and prevalence of diseases. There may even be biases that researchers are unaware of, such as variations in local practice. For many medical applications there is a substantial variability between experts who evaluate images, which is true in clinical practice because it is inherent when labels and segmentations are manually created. This variability may result in biased labels and segmentations that may be mitigated by having multiple experts evaluating the same case (82,83). However, substantial costs and time delays often limit image assessment by multiple experts. Another important reason for using training data from a widespread geographic region is the availability of AI algorithms to patients in developing countries. Most AI algorithms are developed by research groups and companies who have access to training data sets with images from patients in developed countries (84).
Data Format
The two key types of formats relevant to AI application development are image data formats and image annotation formats. Nearly all PACS store medical images in DICOM format, which is the international standard for image objects. However, groups who collect images may convert them from DICOM to other formats such as portable networks graphics, or PNG, tagged image file format, or TIFF, or NIFTI for ease of distribution. However, one should keep in mind that important DICOM metadata are removed with these conversions. Image converting programs are sufficiently prevalent and accessible that there is usually no problem accessing and using image data acquired from multiple institutions in AI application development.
Unlike with image data, image annotations are not stored in a single common format. A major limitation of current commercial imaging systems that acquire image annotations (eg, for tracking cancer lesions [85–91]) is that they generally do not store annotations in a format that permits reuse for AI development. Image annotations are commonly stored in PACS and other systems as DICOM presentation state objects (92), which often vary among vendors and from which it is difficult to extract regions of interest, and usually these objects do not contain image labels. Even if they use DICOM structured reporting, or DICOM-SR (93), which provides different use case–specific templates for storing explicit details of image annotations, similar kinds of annotation data across systems may be stored by using different types of DICOM SR templates that thwart interoperability and reuse of annotations for AI development when acquiring them from different sites or even from different commercial systems within a single site. An important image annotation format for saving regions of interest is the DICOM segmentation map format (92,94), which is part of the DICOM standard. For nongraphic annotations, namely image labels such as radiologic findings or diagnoses, the annotation and image markup, or AIM, format (95–97) was developed and recently incorporated into the DICOM-SR standard (92). A few vendors of AI products and PACS have begun supporting this standard, as well as the open-source ePAD web-based image viewing and annotation tool (48). Adopting these standards for storing these image annotation data will enable multicenter sharing, aggregating, and repurposing of image data for studying new quantitative imaging biomarkers.
Federated Learning
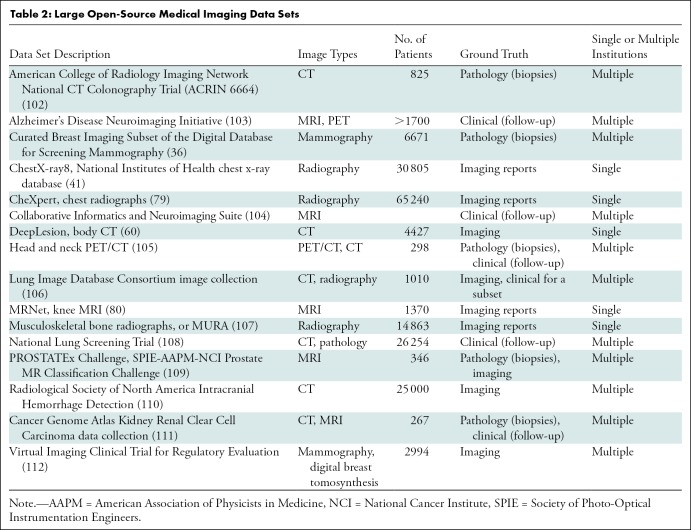
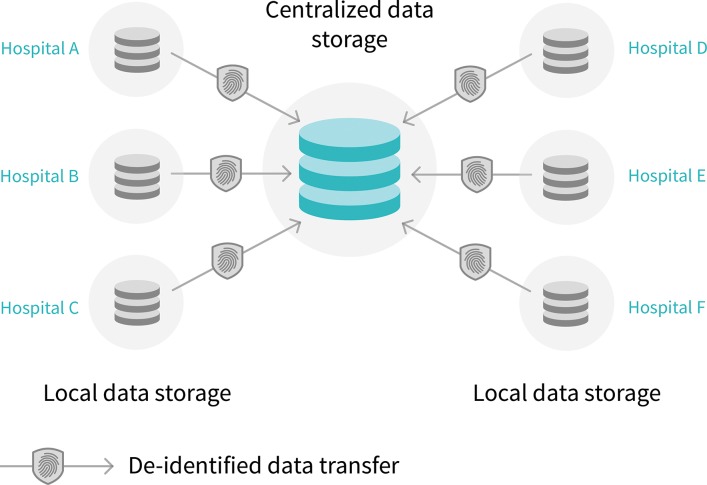
In 2017, Google (Mountain View, Calif) introduced federated learning, a potential solution for the availability of data for AI algorithm development (98). With current practice, de-identified data are transferred from the hospital (or silo) to a central storage system, whereas with federated learning the data stay in the hospital while the algorithm can be trained locally at multiple locations (Fig 5). Moreover, the algorithm itself takes up substantially less storage compared with image data. Therefore, distribution of algorithm training across institutions may be a viable solution.
Figure 5a:
Diagram shows centralized versus federated learning. (a) Current artificial intelligence (AI) model development is through centralized model, in which de-identified data are transferred to centralized data storage system where AI algorithm can be developed. (b) In the future, federated learning may be used, in which data stays in each hospital. With federated learning, instead of transferring data outside each hospital, data stays in hospitals and AI model is sent to and trained in hospitals.
Figure 5b:
Diagram shows centralized versus federated learning. (a) Current artificial intelligence (AI) model development is through centralized model, in which de-identified data are transferred to centralized data storage system where AI algorithm can be developed. (b) In the future, federated learning may be used, in which data stays in each hospital. With federated learning, instead of transferring data outside each hospital, data stays in hospitals and AI model is sent to and trained in hospitals.
Different approaches have been proposed, including parallel and nonparallel training methods. Parallel training methods were developed to speed up algorithm training by splitting the data set in separate samples. Different models are trained on each split of the data and finally the gradients are transferred to a central model (99,100). With nonparallel training, a sequential or cyclical method is used where the model is updated with data from each institution. Federated learning has not yet been evaluated extensively in the medical imaging field. Chang et al (35) simulated image classification with nonparallel federated learning across four institutions and found better performance than with models based on single institutions and similar performance as centrally hosted patient data.
Despite the potential benefits of this technique, there are important problems that need to be solved before federated learning can be applied. First, scalability is limited because image annotation and labeling needs to be performed to an agreed standard per site, because data cannot leave an institution. Second, substantial computation resources may need to be replicated and placed within each facility for federated learning. Third, preprocessing and organizing the data for ingestion by the algorithm (an estimated 80% or more of the effort) is challenging in the federated approach, because the visibility of data to the algorithm developers is impeded. Fourth, variations in terminology across sites requires mapping to a common controlled terminology. Fifth, only gradient information is shared with algorithm developers, which is a step toward protecting protected health information data leaving the hospital because raw image data does not have to be shared. However, sensitive information may still be present in the gradient information. Lastly, there will be heterogeneity across different institutions in terms of patient populations, data volume, data format, et cetera. However, although federated learning is an attractive model, it is not yet applicable in the clinical setting due to its very early phase. Federated solutions to data structuring, labeling, and computing—as well as agreed cross-site standardization of all data formats—will need to be developed for this approach to achieve large-scale adoption.
Data Label Relationship to Future Implementation
It may never be possible to constrain medical imaging to a finite number of labels deterministically. While most of the AI research and solutions in medical imaging today are still carried out solving specific isolated tasks and based on curated data labeling, this is an approach at odds with the desired future state of a continuous learning environment enabling the autonomous incremental adaption to an ever more complex medical system. Practically, this will require the infrastructure to update the prediction model to take into account different data distributions or new information. Data curation and labeling strategies will therefore adapt to new AI techniques continuously learning from streaming (even multimodal) data, which will challenge any static approach to data labeling and training.
Conclusion
Image data availability is an important hurdle for implementation of artificial intelligence (AI) in the clinical setting. AI researchers need to be aware of the data source and potential biases, which may affect generalizability of AI algorithms. New approaches such as federated learning, interactive reporting, and synoptic reporting may help to address data availability in the future. However, curating and annotating data, as well as computational requirements, are substantial barriers.
Supported in part by the Intramural Research Program of the National Institutes of Health (NIH) Clinical Center, the National Library of Medicine of the NIH (R01LM012966), Stanford Child Health Research Institute (Stanford NIH-National Center for Advancing Translational Sciences Clinical and Translational Science Awards [UL1 TR001085]), and the National Cancer Institute of the NIH (U01CA142555, 1U01CA190214, 1U01CA187947, 1U01CA242879).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The mention of commercial products herein does not imply endorsement by the National Institutes of Health or the Department of Health and Human Services.
Disclosures of Conflicts of Interest: M.J.W. Activities related to the present article: is cofounder and shareholder of Segmed. Activities not related to the present article: is a consultant for Arterys; has grants/grants pending with American Heart Association, Philips Healthcare, and Stanford University. Other relationships: disclosed no relevant relationships. W.A.K. Activities related to the present article: is cofounder, shareholder, and chief technology officer of Segmed. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. C.H. Activities related to the present article: is cofounder, shareholder, and chief executive officer of Segmed. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. J.W. Activities related to the present article: is cofounder and shareholder of Segmed. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. D.F. Activities related to the present article: is shareholder of Segmed. Activities not related to the present article: receives research support from Siemens Healthineers and GE Healthcare; received payment for lectures including service on speakers bureaus from Siemens Healthineers; has ownership interest in iSchemaView. Other relationships: disclosed no relevant relationships. H.H. Activities related to the present article: is advisory board member of Segmed; is consultant for Smart Reporting. Activities not related to the present article: holds stock/stock options in Segmed. Other relationships: disclosed no relevant relationships. L.R.F. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is board member of Journal of Digital Imaging; has two government patents; has research agreement with Carestream Health/Philips; receives author royalties from Springer. Other relationships: disclosed no relevant relationships. R.M.S. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: receives royalties from iCAD, Philips, Ping An, and ScanMed; receives research support from Ping An and NVIDIA. Other relationships: disclosed no relevant relationships. D.L.R. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: has grants/grants pending with National Institutes of Health (NIH). Other relationships: disclosed no relevant relationships. M.P.L. Activities related to the present article: is shareholder and advisory board member for Segmed. Activities not related to the present article: is a consultant for Bunker Hill and Nines AI; received research grant from the National Library of Medicine of the NIH. Other relationships: disclosed no relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- DICOM
- Digital Imaging and Communications in Medicine
- PACS
- picture archiving and communication system
References
- 1.Langlotz CP, Allen B, Erickson BJ, et al. A Roadmap for Foundational Research on Artificial Intelligence in Medical Imaging: From the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology 2019;291(3):781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunnmon JA, Yi D, Langlotz CP, Ré C, Rubin DL, Lungren MP. Assessment of Convolutional Neural Networks for Automated Classification of Chest Radiographs. Radiology 2019;290(2):537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prevedello LM, Erdal BS, Ryu JL, et al. Automated Critical Test Findings Identification and Online Notification System Using Artificial Intelligence in Imaging. Radiology 2017;285(3):923–931. [DOI] [PubMed] [Google Scholar]
- 4.Yala A, Schuster T, Miles R, Barzilay R, Lehman C. A Deep Learning Model to Triage Screening Mammograms: A Simulation Study. Radiology 2019;293(1):38–46. [DOI] [PubMed] [Google Scholar]
- 5.Gong E, Pauly JM, Wintermark M, Zaharchuk G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reson Imaging 2018;48(2):330–340. [DOI] [PubMed] [Google Scholar]
- 6.Zhang N, Yang G, Gao Z, et al. Deep Learning for Diagnosis of Chronic Myocardial Infarction on Nonenhanced Cardiac Cine MRI. Radiology 2019;291(3):606–617. [DOI] [PubMed] [Google Scholar]
- 7.Willemink MJ, Noël PB. The evolution of image reconstruction for CT-from filtered back projection to artificial intelligence. Eur Radiol 2019;29(5):2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative Adversarial Networks for Noise Reduction in Low-Dose CT. IEEE Trans Med Imaging 2017;36(12):2536–2545. [DOI] [PubMed] [Google Scholar]
- 9.Yang Q, Yan P, Zhang Y, et al. Low-Dose CT Image Denoising Using a Generative Adversarial Network With Wasserstein Distance and Perceptual Loss. IEEE Trans Med Imaging 2018;37(6):1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang EJ, Park S, Jin KN, et al. Development and Validation of a Deep Learning-Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs. JAMA Netw Open 2019;2(3):e191095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang EJ, Nam JG, Lim WH, et al. Deep Learning for Chest Radiograph Diagnosis in the Emergency Department. Radiology 2019;293(3):573–580. [DOI] [PubMed] [Google Scholar]
- 12.Arbabshirani MR, Fornwalt BK, Mongelluzzo GJ, et al. Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digit Med 2018;1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding Y, Sohn JH, Kawczynski MG, et al. A Deep Learning Model to Predict a Diagnosis of Alzheimer Disease by Using 18F-FDG PET of the Brain. Radiology 2019;290(2):456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parakh A, Lee H, Lee JH, et al. Urinary Stone Detection on CT Images Using Deep Convolutional Neural Networks: Evaluation of Model Performance and Generalization. Radiol Artif Intell 2019;1(4):e180066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a Deep-Learning Neural Network Model in Assessing Skeletal Maturity on Pediatric Hand Radiographs. Radiology 2018;287(1):313–322. [DOI] [PubMed] [Google Scholar]
- 16.de Vos BD, Wolterink JM, Leiner T, de Jong PA, Lessmann N, Isgum I. Direct Automatic Coronary Calcium Scoring in Cardiac and Chest CT. IEEE Trans Med Imaging 2019;38(9):2127–2138. [DOI] [PubMed] [Google Scholar]
- 17.Schelb P, Kohl S, Radtke JP, et al. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology 2019;293(3):607–617. [DOI] [PubMed] [Google Scholar]
- 18.Lehman CD, Yala A, Schuster T, et al. Mammographic Breast Density Assessment Using Deep Learning: Clinical Implementation. Radiology 2019;290(1):52–58. [DOI] [PubMed] [Google Scholar]
- 19.Ngo TA, Lu Z, Carneiro G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal 2017;35:159–171. [DOI] [PubMed] [Google Scholar]
- 20.Tao Q, Yan W, Wang Y, et al. Deep Learning-based Method for Fully Automatic Quantification of Left Ventricle Function from Cine MR Images: A Multivendor, Multicenter Study. Radiology 2019;290(1):81–88. [DOI] [PubMed] [Google Scholar]
- 21.Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E. Convolutional Neural Networks for Radiologic Images: A Radiologist’s Guide. Radiology 2019;290(3):590–606. [DOI] [PubMed] [Google Scholar]
- 22.Park SH, Han K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology 2018;286(3):800–809. [DOI] [PubMed] [Google Scholar]
- 23.MIRC. MIRC CTP. RSNA. https://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor. Published 2019. Accessed October 2019.
- 24.Harvey H, Glocker B. A Standardised Approach for Preparing Imaging Data for Machine Learning Tasks in Radiology. In: Ranschaert E, Morozov S, Algra P, eds. Artificial Intelligence in Medical Imaging. Cham, Switzerland: Springer, 2019. [Google Scholar]
- 25.Goyal N, Apolo AB, Berman ED, et al. ENABLE (Exportable Notation and Bookmark List Engine): an Interface to Manage Tumor Measurement Data from PACS to Cancer Databases. J Digit Imaging 2017;30(3):275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuance . mPower Clinical Analytics for medical imaging. https://www.nuance.com/healthcare/diagnostics-solutions/radiology-performance-analytics/mpower-clinical-analytics.html. Published 2019. Accessed October 2019. [Google Scholar]
- 27.Stanford-Research-Informatics-Center. STAnford Research Repository (STARR) . http://med.stanford.edu/starr-tools.html. Published 2019. Accessed October 2019.
- 28.Illuminate. InSight. https://goilluminate.com/solution/insight/. Published 2019. Accessed October 2019.
- 29.Aryanto KY, Oudkerk M, van Ooijen PM. Free DICOM de-identification tools in clinical research: functioning and safety of patient privacy. Eur Radiol 2015;25(12):3685–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore SM, Maffitt DR, Smith KE, et al. De-identification of Medical Images with Retention of Scientific Research Value. RadioGraphics 2015;35(3):727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Google-Healthcare . De-identifying DICOM data. https://cloud.google.com/healthcare/docs/how-tos/dicom-deidentify. Published 2019. Accessed October 2019.
- 32.Price WN, 2nd, Cohen IG. Privacy in the age of medical big data. Nat Med 2019;25(1):37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El Emam K, Dankar FK. Protecting privacy using k-anonymity. J Am Med Inform Assoc 2008;15(5):627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey H, Heindl A, Khara G, et al. Deep Learning in Breast Cancer Screening. In: Ranschaert E, Morozov S, Algra P, eds. Artificial Intelligence in Medical Imaging. Cham, Switzerland: Springer, 2019. [Google Scholar]
- 35.Chang K, Balachandar N, Lam C, et al. Distributed deep learning networks among institutions for medical imaging. J Am Med Inform Assoc 2018;25(8):945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee RS, Gimenez F, Hoogi A, Miyake KK, Gorovoy M, Rubin DL. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci Data 2017;4:170177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013;20(e1):e147–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brodley CE, Friedl MA. Identifying mislabeled training data. J Artif Intell Res 1999;11(1):131–167. [Google Scholar]
- 39.Sheng VS, Provost F, Ipeirotis PG. Get another label? improving data quality and data mining using multiple, noisy labelers. Proceedings of the 14th ACM SIGKDD international conference on Knowledge discovery and data mining, Las Vegas, August 24–27, 2008. New York, NY: Association for Computing Machinery, 2008; 614–622. [Google Scholar]
- 40.European Society of Radiology (ESR). ESR paper on structured reporting in radiology. Insights Imaging 2018;9(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Peng Y, Lu L, et al. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, July 21–26, 2017. Piscataway, NJ: IEEE, 2017. [Google Scholar]
- 42.Yoshii K, Goto M, Komatani K, et al. An efficient hybrid music recommender system using an incrementally-trainable probabilistic generative model. IEEE Trans Audio Speech Lang Process 2008;16(2):435–447. [Google Scholar]
- 43.Conway D, White JM. Machine Learning for Email: Spam Filtering and Priority Inbox. Sebastopol, Calif: O’Reilly Media, 2011. [Google Scholar]
- 44.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 2004;23(7):903–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen TB, Wang S, Anugu V, et al. Distributed human intelligence for colonic polyp classification in computer-aided detection for CT colonography. Radiology 2012;262(3):824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehta P, Sandfort V, Gheysens D, et al. Segmenting The Kidney On CT Scans Via Crowdsourcing. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, April 8–11, 2019. Piscataway, NJ: IEEE, 2019; 829–832. [Google Scholar]
- 47.Heim E, Roß T, Seitel A, et al. Large-scale medical image annotation with crowd-powered algorithms. J Med Imaging (Bellingham) 2018;5(3):034002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubin DL, Ugur Akdogan M, Altindag C, Alkim E. ePAD: An Image Annotation and Analysis Platform for Quantitative Imaging. Tomography 2019;5(1):170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urban T, Ziegler E, Lewis R, et al. LesionTracker: Extensible Open-Source Zero-Footprint Web Viewer for Cancer Imaging Research and Clinical Trials. Cancer Res 2017;77(21):e119–e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lakhani P, Kim W, Langlotz CP. Automated detection of critical results in radiology reports. J Digit Imaging 2012;25(1):30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakhani P, Kim W, Langlotz CP. Automated extraction of critical test values and communications from unstructured radiology reports: an analysis of 9.3 million reports from 1990 to 2011. Radiology 2012;265(3):809–818. [DOI] [PubMed] [Google Scholar]
- 52.Lipton ZC, Berkowitz J, Elkan C. A Critical Review of Recurrent Neural Networks for Sequence Learning. arXiv 2015;1506.00019 [preprint]. https://arxiv.org/abs/1506.00019. Posted 2015. Accessed January 31, 2020. [Google Scholar]
- 53.Sutskever I, Martens J, Hinton G. Generating text with recurrent neural networks. Proceedings of the 28th International Conference on International Conference on Machine Learning. Bellevue, Wash: Omnipress, 2011; 1017–1024. [Google Scholar]
- 54.Banerjee I, Ling Y, Chen MC, et al. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif Intell Med 2019;97:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc 2017;24(2):361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen MC, Ball RL, Yang L, et al. Deep Learning to Classify Radiology Free-Text Reports. Radiology 2018;286(3):845–852. [DOI] [PubMed] [Google Scholar]
- 57.Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc 2016;23(6):1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brady A, Laoide RO, McCarthy P, McDermott R. Discrepancy and error in radiology: concepts, causes and consequences. Ulster Med J 2012;81(1):3–9. [PMC free article] [PubMed] [Google Scholar]
- 59.Folio LR, Machado LB, Dwyer AJ. Multimedia-enhanced Radiology Reports: Concept, Components, and Challenges. RadioGraphics 2018;38(2):462–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan K, Wang X, Lu L, Summers RM. DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J Med Imaging (Bellingham) 2018;5(3):036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Do HM, Spear LG, Nikpanah M, et al. Augmented Radiologist Workflow Improves Report Value and Saves Time: A Potential Model for Implementation of Artificial Intelligence. Acad Radiol 2020;27(1):96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Do HM, Farhadi F, Xu Z, et al. AI Radiomics in a Monogenic Autoimmune Disease: Deep Learning of Routine Radiologist Annotations Correlated with Pathologically Verified Lung Findings. Oak Brook, Ill: Radiological Society of North America, 2019. [Google Scholar]
- 63.Summers RM, Handwerker LR, Pickhardt PJ, et al. Performance of a previously validated CT colonography computer-aided detection system in a new patient population. AJR Am J Roentgenol 2008;191(1):168–174. [DOI] [PubMed] [Google Scholar]
- 64.Halevy A, Norvig P, Pereira F. The Unreasonable Effectiveness of Data. IEEE Intell Syst 2009;24(2):8–12. [Google Scholar]
- 65.Sun C, Shrivastava A, Singh S, et al. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era. 2017 IEEE International Conference on Computer Vision (ICCV), Venice, October 22–29, 2017. Piscataway, NJ: IEEE, 2017; 843–852. [Google Scholar]
- 66.Eng J. Sample size estimation: how many individuals should be studied? Radiology 2003;227(2):309–313. [DOI] [PubMed] [Google Scholar]
- 67.Kingma DP, Rezende DJ, Mohamed S, et al. Semi-Supervised Learning with Deep Generative Models. arXiv 2014:1406.5298 [preprint]. https://arxiv.org/abs/1406.5298. Posted 2014. Accessed January 31, 2020.
- 68.Schlegl T, Seeböck P, Waldstein SM, et al. Unsupervised Anomaly Detection with Generative Adversarial Networks to Guide Marker Discovery. arXiv 2017:1703.05921 [preprint]. https://arxiv.org/abs/1703.05921. Posted 2017. Accessed Accessed January 31, 2020. [DOI] [PubMed] [Google Scholar]
- 69.Korkinof D, Rijken T, O'Neill M, et al. High-Resolution Mammogram Synthesis using Progressive Generative Adversarial Networks. arXiv 2018:1807.03401 [preprint]. https://arxiv.org/abs/1807.03401. Posted 2018. Accessed Accessed January 31, 2020. [Google Scholar]
- 70.Salehinejad H, Colak E, Dowdell T, Barfett J, Valaee S. Synthesizing Chest X-Ray Pathology for Training Deep Convolutional Neural Networks. IEEE Trans Med Imaging 2019;38(5):1197–1206. [DOI] [PubMed] [Google Scholar]
- 71.Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3(1):160018 [Published addendum appears in Sci Data 2019;6(1):6.] 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4(1):170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Di Martino A, Yan CG, Li Q, et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 2014;19(6):659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jack CR, Jr, Bernstein MA, Fox NC, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008;27(4):685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mennes M, Biswal BB, Castellanos FX, Milham MP. Making data sharing work: the FCP/INDI experience. Neuroimage 2013;82:683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Menze BH, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Essen DC, Smith SM, Barch DM, et al. The WU-Minn Human Connectome Project: an overview. Neuroimage 2013;80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xi P, Shu C, Goubran R. Abnormality Detection in Mammography using Deep Convolutional Neural Networks. arXiv 2018:1803.01906 [preprint]. https://arxiv.org/abs/1803.01906. Posted 2018. Accessed January 31, 2020. [Google Scholar]
- 79.Irvin J, Rajpurkar P, Ko M, et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. arXiv 2019;1901.07031 [preprint]. https://arxiv.org/abs/1901.07031. Posted 2019. Accessed January 31, 2020. [Google Scholar]
- 80.Bien N, Rajpurkar P, Ball RL, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med 2018;15(11):e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toll DB, Janssen KJ, Vergouwe Y, Moons KG. Validation, updating and impact of clinical prediction rules: a review. J Clin Epidemiol 2008;61(11):1085–1094. [DOI] [PubMed] [Google Scholar]
- 82.Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016;316(22):2402–2410. [DOI] [PubMed] [Google Scholar]
- 83.Rajpurkar P, Irvin J, Ball RL, et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 2018;15(11):e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health 2019;1(6):PE271–E297. [DOI] [PubMed] [Google Scholar]
- 85.Siemens-Healthineers. syngo.via for Oncology. https://www.siemens-healthineers.com/medical-imaging-it/syngoviaspecialtopics/syngo-via-for-oncology. Published 2014. Accessed October 2019.
- 86.Mint-Medical. Mint Lesion. https://mint-medical.com/products-solutions/. Published 2014. Accessed October 2019.
- 87.Synaptive-Medical. ClearCanvas Workstation. http://clearcanvas.ca Published 2019. Accessed October 2019.
- 88.Ray L. Detailed and Precise Measurement with Lesion Management. Carestream Radiology. http://www.carestream.com/blog/2013/09/30/detailed-and-precise-measurement-with-lesion-management/. Published 2013. Accessed October 2019. [Google Scholar]
- 89.MIM-Software. MIMviewerand PET Edge. http://www.mimsoftware.com/products/radnuc. Published 2014. Accessed October 2019.
- 90.Three-Palm-Software. LesionOne – Lesion Tracking Application Framework. https://threepalmsoft.com/products/lesionone/. Published 2019. Accessed October 2019. [Google Scholar]
- 91.Folio LR, Sandouk A, Huang J, Solomon JM, Apolo AB. Consistency and efficiency of CT analysis of metastatic disease: semiautomated lesion management application within a PACS. AJR Am J Roentgenol 2013;201(3):618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.DICOM-Standards-Committee . DICOM PS3.3 2019e - Information Object Definitions. NEMA. http://dicom.nema.org/medical/dicom/current/output/chtml/part03/PS3.3.html. Published 2019. Accessed November 2019. [Google Scholar]
- 93.Clunie DA. DICOM structured reporting and cancer clinical trials results. Cancer Inform 2007;4:33–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fedorov A, Clunie D, Ulrich E, et al. DICOM for quantitative imaging biomarker development: a standards based approach to sharing clinical data and structured PET/CT analysis results in head and neck cancer research. PeerJ 2016;4:e2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Channin DS, Mongkolwat P, Kleper V, et al. The Annotation and Image Markup (AIM) Project; Version 2.0 Update. Society for Imaging Informatics in Medicine Annual Scientific Meeting, Minneapolis, 2010. LOCATION: PUBLISHER, 2010. [Google Scholar]
- 96.Rubin DL, Mongkolwat P, Kleper V, et al. Medical Imaging on the Semantic Web: Annotation and Image Markup. AAAI Spring Symposium Series, Semantic Scientific Knowledge Integration, Stanford, 2008. LOCATION: PUBLISHER, 2008. [Google Scholar]
- 97.caBIG-In-vivo-Imaging-Workspace. Annotation and Image Markup (AIM). https://wiki.nci.nih.gov/display/AIM/Annotation+and+Image+Markup+-+AIM. Accessed October 2019.
- 98.McMahan B, Ramage D. Federated Learning: Collaborative Machine Learning without Centralized Training Data. Google AI Blog. https://ai.googleblog.com/2017/04/federated-learning-collaborative.html. Published 2017. Accessed October 2019. [Google Scholar]
- 99.Dean J, Corrado GS, Monga R, et al. Large Scale Distributed Deep Networks. Adv Neural Inf Process Syst 2012;25:1–11. https://papers.nips.cc/paper/4687-large-scale-distributed-deep-networks. [Google Scholar]
- 100.Su H, Chen H. Experiments on Parallel Training of Deep Neural Network using Model Averaging. arXiv 2015:1507.01239 [preprint]. https://arxiv.org/abs/1507.01239. Posted 2015. Accessed January 31, 2020. [Google Scholar]
- 101.U.S. Department of Labor , Employee Benefits Security Administration. The Health Insurance Portability and Accountability Act (HIPAA). Washington, DC: U.S. Department of Labor, 2004. [Google Scholar]
- 102.Johnson CD, Chen MH, Toledano AY, et al. Accuracy of CT colonography for detection of large adenomas and cancers. N Engl J Med 2008;359(12):1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mueller SG, Weiner MW, Thal LJ, et al. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimers Dement 2005;1(1):55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Landis D, Courtney W, Dieringer C, et al. COINS Data Exchange: An open platform for compiling, curating, and disseminating neuroimaging data. Neuroimage 2016;124(Pt B):1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vallières M, Kay-Rivest E, Perrin LJ, et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep 2017;7(1):10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Armato SG, 3rd, McLennan G, Bidaut L, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011;38(2):915–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rajpurkar P, Irvin J, Bagul A, et al. MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs. arXiv 2018;1712.06957 [preprint]. https://arxiv.org/abs/1712.06957. Posted 2018. Accessed January 31, 2020. [Google Scholar]
- 108.National Lung Screening Trial Research Team , Aberle DR, Berg CD, et al. The National Lung Screening Trial: overview and study design. Radiology 2011;258(1):243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging 2014;33(5):1083–1092. [DOI] [PubMed] [Google Scholar]
- 110.Radiological Society of North America . RSNA Intracranial Hemorrhage Detection - Identify acute intracranial hemorrhage and its subtypes. Kaggle. https://www.kaggle.com/c/rsna-intracranial-hemorrhage-detection/overview. Published 2019. Accessed October 2019. [Google Scholar]
- 111.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Badano A, Graff CG, Badal A, et al. Evaluation of Digital Breast Tomosynthesis as Replacement of Full-Field Digital Mammography Using an In Silico Imaging Trial. JAMA Netw Open 2018;1(7):e185474. [DOI] [PMC free article] [PubMed] [Google Scholar]