Abstract
Although mutations in the Lamin A/C gene (LMNA) cause a variety of devastating diseases, the pathological mechanism is often unknown. Lamin A/C proteins play a crucial role in forming a meshwork under the nuclear membrane, providing the nucleus with mechanical integrity and interacting with other proteins for gene regulation. Most LMNA mutations result in heart diseases, including some types that primarily have heart disease as the main pathology. In this study, we used cells from patients with different LMNA mutations that primarily lead to heart disease. Indeed, it is a mystery why a mutation to the protein in every nucleus of the body manifests as a disease of primarily the heart in these patients. Here, we aimed to investigate if strains mimicking those within the myocardial environment are sufficient to cause differences in cells with and without the LMNA mutation. To test this, a stretcher device was used to induce cyclic strain upon cells, and viability/proliferation, cytoskeleton and extracellular matrix organization, and nuclear morphology were quantified. The properties of cells with Hutchinson-Gilford progeria syndrome (HGPS) were found to be significantly different from all other cell lines and were mostly in line with previous findings. However, the properties of cells from patients who primarily had heart diseases were not drastically different when compared to individuals without the LMNA mutation. Our results indicated that cyclic strain alone was insufficient to cause any significant differences that could explain the mechanisms that lead to heart diseases in these patients with LMNA mutations.
Keywords: lamin A/C, laminopathies, cyclic strain, heart mechanics, heart disease
Introduction
Genetic mutations that affect cellular functions and properties potentially develop into detrimental diseases in the body. One gene that is known to lead to pathologies, when mutated, is the Lamin A/C gene (LMNA) [1,2]. Lamins A and C, A-type lamins encoded by LMNA, are type V intermediate filament proteins present in all nucleated somatic cells in the body [3]. These proteins form a supportive meshwork, known as the nuclear lamina, which provides mechanical and structural support for the nuclear envelope [4]. In addition, A-type lamins interact with nucleoplasm and chromatin proteins allowing them to take part in both gene regulation and mechanical signaling [5–7]. Mutations to LMNA have been linked to a wide range of diseases, referred to as laminopathies, affecting multiple tissues and organs systems within the body [1,2]. Though each mutation variant may affect different parts of the body, most laminopathies are known to be associated with some form of heart disease [1,2]. Indeed, there are also LMNA mutations which primarily lead to heart diseases with no other detrimental pathologies [8–10]. Given that A-type lamins are found in every nucleated cell throughout the body, it is unclear why the heart is specifically vulnerable to the LMNA mutation.
Mutations in A-type lamins are known to make cells vulnerable to mechanical perturbations due to the decrease in stiffness and stability of the nuclear lamina [11–14]. For example, an impaired nuclear lamina can result in a loss of cell viability and functionality, especially in mechanically stressed tissues [12]. Indeed, when static strain was applied to cells with the mutation, increased deformability of the nuclear envelope and a higher percentage of dysmorphic nuclei were observed [12,13,15]. Although many parts of the body are exposed to strains, only the heart is subjected to repeated ∼1 Hz contractions over a lifetime. Thus, cardiomyocytes are uniquely exposed to nonstop cyclic strain which has not yet been investigated for its effects on cells with the LMNA mutation. If the consequences are similar to those observed in static strain, cardiomyocyte functionality and heart activity may also be negatively affected.
To understand the mechanisms leading to heart diseases in laminopathies, we investigated if strains mimicking those in myocardial environment alone are sufficient to demarcate cells with and without the LMNA mutation. To study this, skin biopsies were taken from patients with LMNA mutations who primarily exhibited heart disease [8–10]. In addition, these were compared to cells from healthy individuals with no mutations and from a patient with Hutchinson-Gilford progeria syndrome. Fibroblasts were specifically used as they were observed in a previous study to have functional pathologies in vitro [16] and are not normally subjected to cyclic strain. By subjecting these nonstretching cells to cyclic strain and observing the consequences on the nuclei, we can better clarify the role of dynamic mechanical strain in causing heart diseases. Thus, a monolayer of cells was uniaxially and cyclically stretched with magnitudes similar to the heart to investigate the effects on cell viability/proliferation, cytoskeleton and extracellular matrix organization, proportion of dysmorphic nuclei, and nuclear shape [17]. These experiments allowed us to address how the myocardial environment affects cells with the LMNA mutation and determine if the observed results were sufficient to explain pathways that lead to heart diseases in laminopathies.
Methods
Skin Fibroblast Acquisition.
Informed consent was obtained and approved by UC Irvine Institutional Review Board (IRB# 2014–1253) for this study. Skin fibroblasts were collected from three families with different LMNA mutations: Family A having three individuals with the heterozygous LMNA splice-site mutation (c.357-2A>G) [8], Family B having three individuals with the heterozygous LMNA nonsense mutation (c.736 C > T, pQ246X) in exon 4 [9], and Family C having three individuals with the heterozygous LMNA missense mutation (c.1003C>T, pR335W) in exon 6 [10]. In addition, for each family, fibroblasts were collected from three age and gender matched individuals without the mutation to serve as related negative controls. Unrelated negative control fibroblasts, referred to as donor cells, were purchased from Lonza (catalog# CC-2511) and Coriell (catalog# ND31845, AG14284). HGPS fibroblasts with a heterozygous LMNA G608G point mutation were obtained from Coriell Institute for Medical Research (Camden, NJ: catalog #AG11513) to serve as a positive control [18]. The individuals with LMNA mutations from the families were referred to as Patients. Negative Controls included individuals without mutations from the three families and the donors. For nomenclature, patient (P), control (C), or donor (D) are followed by the family designator (A, B, or C) and the preassigned number of the individual (1, 2, 3, or 4).
Cell Culture.
All cell lines were expanded to passage 16 for these experiments. Cells were cultured in media consisting of minimum essential media (MEM, ThermoFisher, Grand Island, NY), 10% Fetal Bovine Serum (FBS, ThermoFisher, Grand Island, NY), and 1% Hyclone Antibiotics Antimycotic solution (AB, GE Life Sciences, Utah). Cells were passaged at 80–100% confluency, using 0.05% Trypsin (Fisher Scientific, Hanover Park, IL).
Stretcher Experiments.
MechanoCulture FX-2 (CellScale, CDN), stretcher device, was used for all experiments. The actuator was programmed to execute manufacturer specified 15% uniaxial cyclic stretch at 1 Hz for 24 h or 20% uniaxial cyclic stretch at 1 Hz for 72-h depending on the experiment.
The wells were first washed with the phosphate buffered saline (PBS, ThermoFisher, Grand Island, NY). A 0.05 mg/mL fibronectin solution was then added to each well (Fisher Scientific, Hanover Park, IL) followed by a 2-h incubation for the adhesion of an isotropic fibronectin monolayer to the silicone well bottoms. After being washed with PBS to remove excess fibronectin, a 300 μL solution of 2.0 × 105 cells and culturing media was then seeded into each well and allowed to incubate for 24 h. After incubation, media was changed to a maintenance media consisting of MEM, 2% FBS, and 1% AB to maintain confluency. The stretcher device was then initiated with the desired stretching protocol and placed inside an incubator with 5% CO2 at 37 °C for the duration of the experiment. For the 72-h experiments, maintenance media was changed every two days.
Fixing and Immunofluorescent Staining.
The cells were fixed with a solution of 4% paraformaldehyde (VWR, Radnow, PA) and 0.05% Triton X-100 (Sigma-Aldrich, Saint Louis, MO). Once fixed, the cultures were stained for nuclei (4,6-Diamidino-2-Phenylindole Dihydrochloride, DAPI, ThermoFisher, Grand Island, NY), actin (Alexa Fluor 488 Phalloidin, ThermoFisher, Grand Island, NY), and fibronectin (polyclonal rabbit antihuman fibronectin, Sigma-Aldrich, Saint Louis, MO). Secondary staining was done for fibronectin using goat antirabbit IgG secondary antibodies (Alexa Fluor 750, ThermoFisher, Grand Island, NY). Afterward, the wells were punched out with a commercially purchased metal square hole puncher as high-resolution images could not be obtained through the membrane. The punched-out wells were flipped, mounted onto glass microscope slides with ProLong Gold Antifade Mountant (ThermoFisher, Grand Island, NY), and sealed with clear nail polish around the edges.
Imaging and Data Acquisition.
The samples were imaged with an IX-83 inverted motorized microscope (Olympus America, Center Valley, PA). Images were taken using an UPLFLN 40× oil immersion objective (Olympus America, Center Valley, PA) and a digital CCD camera ORCA-R2 C10600-10B (Hamamatsu Photonics, Shizuoka Prefecture, Japan). Ten fields of views randomly selected for each sample and imaged at 40× magnification (6.22 μm/pixel).
matlab Analysis.
A custom written matlab code was used to classify nuclei as normal or dysmorphic and measure nuclear properties [16]. Aspect ratio and area were also simultaneously calculated by the code during the process. Additionally, another in-house set of matlab codes was used to quantify the orientation order parameter (OOP) of actin and fibronectin [19].
Statistical Analysis.
Unless otherwise specified, statistical analysis was done using analysis of variance (ANOVA) one-way testing with Tukey's method. For nonparametric variables (i.e., nuclei defectiveness), the Kruskal Wallis tests was used followed by Dunn's multiple comparison test. Significance was defined as having a p-value of less than 0.05. Sample sizes for density, OOP, and percentage of defective nuclei were the number of individual wells for each group. Sample sizes for area and aspect ratio were the total number of individual nuclei for each group. A secondary statistical analysis was also done for area and aspect ratio where sample sizes were the individual patients. A summary of the sample sizes of each statistically tested group and the respective statistical tests for each quantification can be found in Tables 1 and 2.
Table 1.
Summary of sample sizes of statistically tested groups
| I. Metric | II. Experimental groups | III. Condition | IV. Compiled number of nuclei | V. Number of coverslips | VI. Number of individuals | VII. Test and sample size used |
|---|---|---|---|---|---|---|
| Density | Patients | Static | 14,629 | 95 | 9 | ANOVA coverslips (column V) |
| Stretch | 15,065 | 102 | ||||
| Negative Controls | Static | 17,492 | 112 | 10 | ||
| Stretch | 16,450 | 118 | ||||
| Positive Controls | Static | 5789 | 46 | 1 | ||
| Stretch | 5751 | 46 | ||||
| Actin OOP | Patients | Static | N/A | 94 | 9 | ANOVA coverslips (column V) |
| Stretch | 101 | |||||
| Negative Controls | Static | N/A | 112 | 10 | ||
| Stretch | 117 | |||||
| Positive Controls | Static | N/A | 47 | 1 | ||
| Stretch | 47 | |||||
| Fibronectin OOP | Patients | Static | N/A | 81 | 9 | ANOVA coverslips (column V) |
| Stretch | 88 | |||||
| Negative Controls | Static | N/A | 102 | 10 | ||
| Stretch | 107 | |||||
| Positive Controls | Static | N/A | 41 | 1 | ||
| Stretch | 41 | |||||
| % Defective nuclei | Patients | Static | 5823 | 94 | 9 | Kruskal–Wallis and Dunn's multiple comparison coverslips (column V) |
| Stretch | 5832 | 102 | ||||
| Negative Controls | Static | 7687 | 112 | 10 | ||
| Stretch | 7595 | 117 | ||||
| Positive Controls | Static | 3059 | 46 | 1 | ||
| Stretch | 2921 | 46 | ||||
| Aspect ratio and area | Patients A | Static | 738 | 33 | 3 | ANOVA nuclei (column IV) and individuals (column VI) |
| Stretch | 1012 | 40 | ||||
| Patients B | Static | 2445 | 30 | 3 | ||
| Stretch | 2480 | 30 | ||||
| Patients C | Static | 2640 | 31 | 3 | ||
| Stretch | 2340 | 32 | ||||
| Negative Controls | Static | 7687 | 112 | 10 | ||
| Stretch | 7595 | 117 | ||||
| Positive Controls | Static | 3059 | 46 | 1 | ||
| Stretch | 2921 | 46 |
Table 2.
Summary of sample sizes of statistically tested groups for 72-h experiments
| I. Metric | II. Experimental groups | III. Condition | IV. Compiled number of nuclei | V. Number of coverslips | VI. Test and sample size used |
|---|---|---|---|---|---|
| Density | Control A1 | Static | 2513 | 14 | ANOVA coverslips (column V) |
| Stretch | 2390 | 18 | |||
| Control A1 (72-h) | Static | 2602 | 16 | ||
| Stretch | 1713 | 13 | |||
| Patient A1 | Static | 2108 | 12 | ||
| Stretch | 2922 | 18 | |||
| Patient A1 | Static | 2293 | 13 | ||
| (72-h) | Stretch | 1839 | 15 | ||
| Actin OOP | Control A1 | Static | N/A | 14 | ANOVA coverslips (column V) |
| Stretch | 18 | ||||
| Control A1 (72-h) | Static | N/A | 16 | ||
| Stretch | 13 | ||||
| Patient A1 | Static | N/A | 12 | ||
| Stretch | 18 | ||||
| Patient A1 | Static | N/A | 13 | ||
| (72-h) | Stretch | 15 | |||
| Fibronectin OOP | Control A1 | Static | N/A | 6 | ANOVA coverslips (column V) |
| Stretch | 10 | ||||
| Control A1 (72-h) | Static | N/A | 16 | ||
| Stretch | 13 | ||||
| Patient A1 | Static | N/A | 6 | ||
| Stretch | 12 | ||||
| Patient A1 | Static | N/A | 13 | ||
| (72-h) | Stretch | 15 | |||
| % Defective nuclei | Control A1 | Static | 267 | 14 | Kruskal–Wallis and Dunn's multiple comparison coverslips (column V) |
| Stretch | 358 | 18 | |||
| Control A1 (72-h) | Static | 1554 | 16 | ||
| Stretch | 1080 | 13 | |||
| Patient A1 | Static | 292 | 12 | ||
| Stretch | 519 | 18 | |||
| Patient A1 | Static | 1368 | 13 | ||
| (72-h) | Stretch | 1223 | 15 | ||
| Aspect ratio and area | Control A1 | Static | 267 | 14 | ANOVA nuclei (column IV) |
| Stretch | 358 | 18 | |||
| Control A1 | Static | 1554 | 16 | ||
| (72-h) | Stretch | 1080 | 13 | ||
| Patient A1 | Static | 292 | 12 | ||
| Stretch | 519 | 18 | |||
| Patient A1 | Static | 1368 | 13 | ||
| (72-h) | Stretch | 1223 | 15 |
Results
Dynamic mechanical stimulation is one of the obvious differences between heart and other tissues in the body. To determine if cyclic strain alone is sufficient to cause issues, fibroblasts from cell lines with a LMNA mutation were exposed to simplified myocardial-like strains, 15% strain at 1 Hz for 24 h. Cells with known LMNA mutations leading primarily to heart disease (Patients), cells without LMNA mutations (Negative Controls), and HGPS cells (Positive Control) were cyclically stretched and analyzed for cell viability and cytoskeleton morphology.
Quantifying Cell Viability/Proliferation and Matrix Organization.
To determine if viability/proliferation is affected by cyclic stretching, nuclei were stained after both static and stretch protocols (Fig. 1(a), (i) and (ii)). As the cells were seeded at 2.0 × 105 cells/well for all conditions, the final density quantified from the nuclei stain is a measure of the combined viability and proliferation potential. Compared to both Patients and Negative Controls, the Positive Control generally exhibited a lower cell density (Fig. 1(a), (iii)). However, cyclically stretching the cells did not induce a change in viability/proliferation in either Patient or Positive Control groups. Interestingly, Patients cells also did not have a compromised cell viability/proliferation compared to the Negative Controls (Fig. 1(a), (iii)). To quantify the effects of the LMNA mutation on the cytoskeleton and the extracellular matrix, OOP was used as a measurement of organization for actin (Fig. 1(b)) and fibronectin (Fig. 1(c)) [19]. Without stretch, the tissues and underlying extracellular matrix remained isotropic (low OOP) with no differences observed in actin or fibronectin organization for all groups (Figs. 1(b), (iii) and 1(c), (iii), purple bars (print: black)). Conversely, cyclic stretching induced organization (high OOP) in both the tissues and the underlying fibronectin (Figs. 1(b), (iii) and 1(c), (iii), yellow bars (print: gray)). Interestingly, in the Positive Control, actin and fibronectin were less organized poststretch when compared to Negative Controls (Figs. 1(b), (iii) and 1(c), (iii)). Yet, there were no differences observed in either actin or fibronectin organization between Patients and Negative Controls (Figs. 1(b), (iii) and 1(c), (iii)).
Fig. 1.
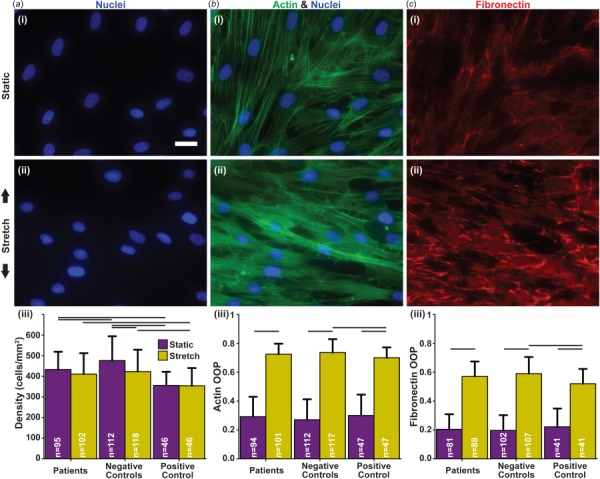
Consequences of cyclic strain on cell viability/proliferation and organization. (a) (i–ii) Example images of nuclei stained with DAPI (blue (print: gray)); (iii) cell density estimated by nuclei count. (b) (i–ii) example images of actin stained with Phalloidin (green (print: gray)); (iii) quantification of actin organization. (c) (i–ii) Example images of samples stained for fibronectin (red (print: gray)); (iii) quantification of extracellular matrix organization. (a–c) Scale bar: 25 μm; (i) stains in the static condition; (ii) stains in the stretch condition with black arrows indicating the direction of stretch; (iii) connecting lines represent significances within conditions and between corresponding groups (p < 0.05); sample sizes listed within the bars. Summary of statistical analysis and sample sizes found in Table 1.
Subtle Effects of Cyclic Strain on Nuclear Morphology.
To further assess if cells with LMNA mutations are vulnerable to cyclic strain, nuclear morphology was examined. Nuclei were automatically detected, categorized, and measured for morphological properties using custom software [16]. To determine if cells with the LMNA mutation are more vulnerable to deformities by cyclic strain, the percentage of defective nuclei was calculated in both static and stretch conditions (Fig. 2(a)). The nuclei of Positive Control cells were found to be generally more defective than Patients and Negative Controls for both conditions (Fig. 2(b)). However, no differences were found between Patients and Negative Controls in any of the conditions (Fig. 2(b)). Interestingly, exposing cells to cyclic stretch did not increase the percentage of defective nuclei for any of the groups (Fig. 2(b)).
Fig. 2.
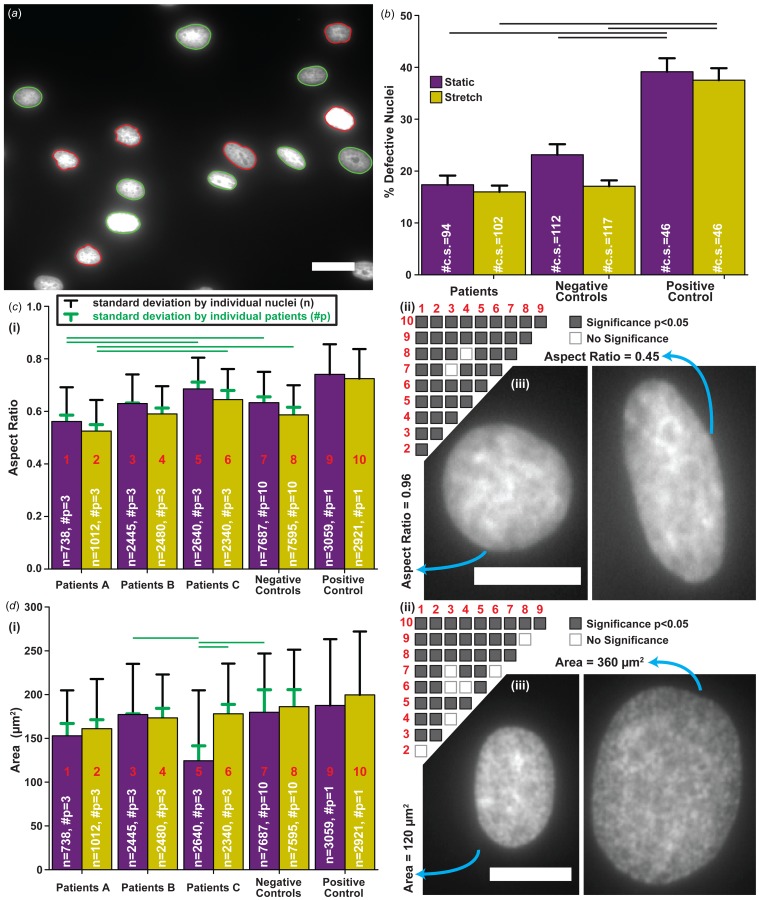
Consequences of cyclic strain on nuclear morphology. (a) Automatic nuclei detection, classification, and measurement of nuclear properties; nuclei are designated as defective (red (print: dark gray)) and normal (green (print: light gray)) [16]. (b) Percent of defective nuclei; sample size is the number of individual coverslips (#c.s.); error bars represent the standard error of the mean; connecting black lines represent significances between corresponding groups (p < 0.05). ((c) and (d)) (i) Summary of aspect ratio (c) and area (d); sample sizes: total nuclei number (n) and number of patients (#p); error bars represent standard deviation calculated for nuclei by pooling all coverslips and patients (black) or individual patients (green (print: light gray)); green lines (print: light gray) represent significances analyzed by individual patients (p < 0.05); (ii) significance matrix of (i) for individual nuclei, i.e., black error bars, with red (print: dark gray) numbers corresponding to the respective labeled group in plot; (iii) example nuclei with a high aspect ratio or small area (left) and a low aspect ratio or large area (right). All scale bars: 25 μm. Summary of statistical analysis and sample sizes found in Table 1. Data presented for separate individuals is plotted in Fig. S1 available in the Supplemental Materials on the ASME Digital Collection.
For a more detailed analysis, the aspect ratio (Fig. 2(c)) and area (Fig. 2(d)) of the nuclei were measured. Aspect ratio, a measure of eccentricity, is one for perfectly circular nuclei (Fig. 2(c), (iii), left) and less than one for elongated nuclei (Fig. 2(c), (iii), right). Each variation of the LMNA mutation responded differently to cyclic strain (Fig. 2(c), (i) and (ii)). The nuclei of the Patients A group were more elongated compared to the Negative Controls in both static and stretch conditions (Fig. 2(c), (i) and (ii)). In contrast, Patients C nuclei showed less elongation in general while Patient B nuclei showed no significant differences when compared to the Negative Controls (Fig. 2(c), (i) and (ii)). As expected, Positive Control nuclei were the least elongated among all cell lines for both static and stretch conditions (Fig. 2(c), (i) and (ii)) [16]. For nuclear area, cells with different LMNA mutations also exhibited varying responses (Fig. 2(d), (i) and (ii)): Nuclei ranged from small (Fig. 2(d), (iii), left) to large (Fig. 2(d), (iii), right) areas for both static and stretch conditions. Though when summarized, Patients A and B groups did not show a change in nuclear area poststretch while the Patients C group significantly increased in response to stretching (Fig. 2(d), (i) and (ii)). In addition, the Patients B group showed no differences when compared to the Negative Controls in the static and stretch conditions, unlike both Patients A and C groups (Fig. 2(d), (i) and (ii)). Similar to aspect ratio, Positive Control had the greatest nuclear area poststretch compared to all Patient groups and Negative Controls (Fig. 2(d), (i) and (ii)). When both area and aspect were compared by pooling individual patients instead of all patients and coverslips, many significances resulting from large sample sizes were removed. The Patients A group still had significantly more elongated nuclei when compared to the Patients C group and Negative Controls (Fig. 2(c), (i), green lines). For area, only the patients C group maintained a significant increase in nuclear area poststretch (Fig. 2(d), (i), green lines). Unlike all of the other cell-lines, upon exposure to cyclic stretching, the Patients C group exhibited a combination of nuclear area and aspect ratio changes indicating that either the nuclear volume increases or the height of the nuclei decreases in the Patients C group (see Fig. S3 available in the Supplemental Materials on the ASME Digital Collection).
Consequences of Exposure to Extensive Cyclic Strain.
The choice to expose fibroblasts to 24 h of strain was dictated by experimental convenience, but it can be argued that one day or normal 15% strain is an insufficient amount of time and/or stretch to cause LMNA mutation driven changes. We therefore cultured matched patient and negative control cell lines for 72-h while exposing the cells to a higher strain. As a result of applying cyclic strain for 72 h, Patient A1 cells (PA1), unlike Control A1 cells (CA1), showed decreased viability/proliferation (Fig. 3(b), yellow bars (print: gray)). Yet, neither CA1 nor PA1 showed increased amounts of defective nuclei as a result of longer exposure times to cyclic strain (Fig. 3(c)). To further examine the effects of prolonged cyclic stretching, organization of tissues and underlying extracellular matrix were examined along with nuclear morphology (Figs. 3(d)–3(g)). Indeed, as observed in the 24 h experiment, tissues remained isotropic with no differences in actin or fibronectin organization for all groups without stretching (Figs. 3(d) and 3(e), purple bars (print: black)). Correspondingly, the presence of the 72-h cyclic strain increased organization in actin and fibronectin for both CA1 and PA1 as it did for the 24 h experiment (Figs. 3(d) and 3(e), yellow bars). For nuclear morphology, only subtle differences were observed among both individuals under all exposure conditions (Figs. 3(f) and 3(g)). In addition, unlike CA1, exposure to cyclic strain did not induce an increase in nuclear area for PA1 from static to stretch conditions (Fig. 3(g), matching patterns of purple bars to yellow bars (print: black to gray)).
Fig. 3.
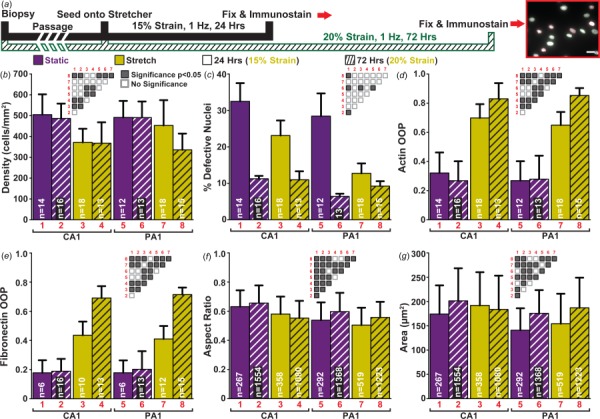
Exposure to extensive cyclic strain and its effects: (a) summary of the three different cyclic stretching regimes/experiments (insert scale bar: 25 μm); (b) cell density estimated by nuclei count; (c) percent of defective nuclei; error bars represent the standard error of the mean; (d) quantification of actin organization; (e) quantification of extracellular matrix organization; (f) summary of aspect ratio for all nuclei in each experiment; (g) summary of nuclear area for all nuclei in each experiment. (b)–(g) Static data shown in purple (print: black) and stretched data in yellow (print: gray). Patterns: solid bars for 24 h of 15% cyclic strain and hatch bars for 72-h of 20% cyclic strain; significance matrices of corresponding bars labeled in red (print: gray) (gray boxes p < 0.05, white boxes p > 0.05); sample sizes denoted in white within bars. ((b), (d)–(g)) Error bars represent the standard deviation. Summary of statistical analysis and sample sizes found in Table 2.
Discussion
In this work, we examined if simplified myocardial-like strains are sufficient to induce a difference between cells with and without LMNA mutations. To achieve this, human fibroblast lines were assessed for cytoskeleton and extracellular matrix organization, nuclear morphology, and viability after being exposed to uniaxial cyclic strain.
In analyzing the results, it is useful to note several differences between the dynamic mechanical environment inside the myocardium and on the planar silicon membranes. In the myocardium, with cells oriented in varying circumferential and longitudinal directions, at peak systole, a range of measured circumferential, radial, and longitudinal strains have been reported as −23% to −10%, 30 to 47%, −23 to 0%, respectively [20–29]. In contrast, uniaxial strains, similar to in vitro models [30–32] and a significant simplification of the myocardial environment, were used to generat data for Figs. 1–3. Additionally, there are known differences between cardiomyocytes and fibroblasts in their responses to strain; cardiomyocytes organize parallel to the direction of strain while fibroblasts organize orthogonally (see Fig. S2 is available in the Supplemental Materials on the ASME Digital Collection) [33–35]. Consequently, after the cells had organized (within 2-h), stretching exposed fibroblasts to strains orthogonal to their cytoskeleton, which is different from the strains experienced by cells in the myocardium. Given that cyclic strain is suspected to be an important factor in LMNA mutation-driven pathologies in the heart, these differences of how fibroblasts are exposed to strains should be considered when interpreting the results of these experiments.
The number of dysmorphic nuclei and viability/proliferation was first evaluated to see how strains similar to those within the myocardium affect cells with and without LMNA mutations. For our experiments, as the initial density was the same for all cell lines, the final density after each culturing period was used as a representative measure of viability/proliferation. If the results had shown significant differences between any of the experimental groups involving the Patients and Negative Controls, it would have been interesting to distinguish between viability and proliferation with either a live/dead viability assay or fluorescence-activated cell sorting to assess the number of apoptotic cells. However, as there were no significant differences in these experiments of the groups, the measurement viability/proliferation is sufficient. Furthermore, the number of dysmorphic nuclei was defined as the number of irregularly shaped nuclei instead of nuclei with blebbing since the former more frequently leads to nuclear membrane rupturing and consequentially DNA damage [36]. From the previous literature, both Lamin A/C knockout (Lmna−/−) and lamin C-only expression (LmnaLCO/LCO) mouse embryonic fibroblasts (MEFs) were found to have significantly higher amounts of irregular-shaped nuclei compared to those with normal LMNA expression [12,13,36]. Similarly, our experiments using human fibroblasts with HGPS, one of the more severe variation of LMNA mutations, had significantly higher amounts of dysmorphic nuclei compared to Negative Controls (Fig. 2(b)). Patients, known to have only one mutated LMNA allele [8–10], might be comparable to heterozygous Lamin A/C knockout (Lmna+/ −) MEFs [13]; both exhibited no significant increase in the number of dysmorphic nuclei when compared to their negative controls (Fig. 2(b) and see Ref. [13], respectively). For viability, Lammerding et al. quantified the number of apoptotic cells for MEFs with different LMNA mutations. Both Lmna−/− and LmnaLCO/LCO MEFs had similar viabilities compared to MEFs with normal LMNA expression, but when subjected to biaxial cyclic stretch, only the former had decreased viability [13]. In our experiments, HGPS had significantly lower cell viability compared to the Negative Controls in static conditions, but this was not exacerbated when cells were subjected to uniaxial cyclic stretching (Fig. 1(a), (iii)). For both Patients and Lmna+/ − MEFs, viability was not compromised or affected by cyclic stretching (Fig. 1(a), (iii) and see Ref. [13], respectively). The minor discrepancies seen in cell viability between our results and previous literature might be caused by not only the varying LMNA mutations in Patients but also the differences between uniaxial and biaxial cyclic stretching. Although there is a disparity between external stretching, our experiments, versus the strains applied to the nucleus within the cardiomyocytes, overall our results were mostly in line with what was expected from LMNA knockout mouse experiments [12,13,36].
To further examine LMNA mutations and their relation to heart diseases, nuclear morphology, cytoskeleton organization, and extracellular matrix organization were compared among the groups after exposure to cyclic stretching. As expected from the previous literature [33–35], uniaxial cyclic stretching induced actin and fibronectin organization for all cell lines (Figs. 1(b), (iii) and 1(c),(iii)). Predictably, since HGPS patients have severe skin abnormalities, HGPS cells had less organization poststretch when compared to Negative Controls (Figs. 1(b), (iii) and 1(c), (iii)). Interestingly, there were no differences in organization among Patients and Negative Controls for either static or stretch conditions (Figs. 1(b), (iii) and 1(c), (iii)). When area and eccentricity were examined in the previous literature, similar trends were observed where HGPS cells and Lmna−/− MEFs were rounder with nuclear areas comparable to their respective negative controls [12,37,38]. Indeed, for our experiments HGPS cells were also observed to be rounder and have similar sized nuclei when compared to Negative Controls (Figs. 2(c), (i)–(ii) and 2(d), (i)–(ii)). For Patient groups, the inconsistencies seen among them may suggest that different mutations may have their own subtle and diverse effects on the mechanical properties of the nucleus as seen in Fig. S3 which is available in the Supplemental Materials on the ASME Digital Collection [8–10].
The direct mechanisms of how the LMNA mutation affects cells and their nuclei are still largely unknown. However, it is possible that the mutation can have an effect on the cytoskeleton, and in turn, cellular morphology, which has been observed to influence nuclear shape and positioning [39,40]. While the tissue characterization performed in our experiments does not provide direct measurements of cell morphology (Fig. 2(b)), the lack of changes in actin organization in the three families compared to the negative controls suggests that cell morphology is not likely to be the cause of the subtle differences found between the nuclear shapes in the three families (Figs. 2(c), (i)–(ii) and 2(d), (i)–(ii) and see Fig. S3 available in the Supplemental Materials on the ASME Digital Collection). However, the possibility remains that there are subtle cell shape differences that could affect cell-lines with a mutation, which could be further investigated in the future with single cell experiments or by specifically staining for cell membrane proteins.
For each patient, it was also determined in a previous study that the amount of dysmorphic nuclei is negatively correlated to the age at which heart disease symptoms are first presented [16]. Thus, it was uncertain if subjecting these cell lines to cyclic stretching for just one day was adequate to induce disease-causing differences that take years to develop within these patients. Therefore, additional strain experiments were conducted on a patient cell line (PA1) with an early presentation age and the corresponding related control (CA1) to examine the effects of longer exposure times. When the percentage of defective nuclei was quantified, no significant changes were observed after prolonged exposure to cyclic strain for either CA1 or PA1 (Fig. 3(c)). However, PA1 did exhibit a loss in viability/proliferation after 72-h of higher magnitude cyclic stretching when compared to the 24 h condition, but it only decreased to levels similar to CA1 (Fig. 3(b), hatch bars). While applying higher strains may induce a greater response, it is beyond the physiological strain ranges, 10–20% [30–32], and thus the scope of this manuscript. Altogether, increasing exposure time to cyclic stretching and the strain to peak intensities within the physiological range did not deviate our findings greatly from the shorter experiments and results of previous publications.
Regardless of exposure time, cyclic strain was found to be insufficient to consistently affect the number of defective nuclei, nuclear morphology, viability, and cytoskeleton and extracellular matrix organization. Unlike HGPS individuals, cells from patients with primarily heart pathologies just exhibited subtle differences in nuclear morphology, but they were not drastically different compared to cell lines with normal LMNA genotype. Thus, simple cyclic strains in fibroblasts within the physiological ranges of strain were insufficient to promote differences that would explain pathways leading to the main pathology of heart disease in these patients with LMNA mutations.
Acknowledgment
The authors would like to thank to the LMNA gene mutation families that participated in the study, Linda McCarthy for her help in culturing the cell lines, and Hamza Atcha for his help in verifying the strains of the silicone membranes. We would also like to thank Professor Samuel Safran, Professor Eran Bouchbinder, Ohad Cohen, and Dan Deviri for discussions related to the work.
Contributor Information
Richard D. H. Tran, Cardiovascular Modeling Laboratory, The Edwards Lifesciences Center for Advanced Cardiovascular Technology, Department of Biomedical Engineering, University of California, 2131 Engineering Hall Irvine, Irvine, CA 92697-2700 e-mail: rdtran1@uci.edu
Mark Siemens, Cardiovascular Modeling Laboratory, The Edwards Lifesciences Center for Advanced Cardiovascular Technology, Department of Biomedical Engineering, University of California, 2131 Engineering Hall Irvine, Irvine, CA 92697-2700 e-mail: msiemens@uci.edu .
Cecilia H. H. Nguyen, Division of Genetics and Genomics, Department of Pediatrics, School of Medicine, University of California, 2042 Hewitt Hall Irvine, Irvine, CA 92697-3940 e-mail: cecilihn@uci.edu
Alexander R. Ochs, Cardiovascular Modeling Laboratory, The Edwards Lifesciences Center for Advanced Cardiovascular Technology, Department of Biomedical Engineering, University of California, 2131 Engineering Hall Irvine, Irvine, CA 92697-2700 e-mail: ochsa@uci.edu
Michael V. Zaragoza, Department of Pediatrics, Division of Genetics & Genomics, 2042 Hewitt Hall Irvine, Irvine, CA 92697-3940; Department of Biological Chemistry, University of California, School of Medicine, 2042 Hewitt Hall Irvine, Irvine, CA 92697-3940 e-mail: mzaragoz@uci.edu
Anna Grosberg, Cardiovascular Modeling Laboratory, The Edwards Lifesciences Center for Advanced Cardiovascular Technology, Center for Complex Biological Systems, Department of Biomedical Engineering, University of California, 2418 Engineering Hall Irvine, Irvine, CA 92697-2700;; Department of Chemical and Biomolecular Engineering, University of California, 2418 Engineering Hall Irvine, Irvine, CA 92697-2700 e-mail: grosberg@uci.edu
Funding Data
National Institutes of Health (Grant No. NIH 1 R01 HL129008-01 (Grosberg, Zaragoza), Funder ID: 10.13039/100000002).
References
- [1]. Brayson, D. , and Shanahan, C. M. , 2017, “Current Insights Into LMNA Cardiomyopathies: Existing Models and Missing LINCs,” Nucleus, 8(1), pp. 17–33. 10.1080/19491034.2016.1260798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Parnaik, V. K. , 2008, “Role of Nuclear Lamins in Nuclear Organization, Cellular Signaling, and Inherited Diseases,” Int. Rev. Cell Mol. Biol., 266, pp. 157–206. 10.1016/S1937-6448(07)66004-3 [DOI] [PubMed] [Google Scholar]
- [3]. Gruenbaum, Y. , and Medalia, O. , 2015, “Lamins: The Structure and Protein Complexes,” Curr. Opin. Cell Biol., 32, pp. 7–12. 10.1016/j.ceb.2014.09.009 [DOI] [PubMed] [Google Scholar]
- [4]. Shimi, T. , Kittisopikul, M. , Tran, J. , Goldman, A. E. , Adam, S. A. , Zheng, Y. , Jaqaman, K. , and Goldman, R. D. , 2015, “Structural Organization of Nuclear Lamins A, C, B1, and B2 Revealed by Superresolution Microscopy,” Mol. Biol. Cell, 26(22), pp. 4075–4086. 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Andres, V. , and Gonzalez, J. M. , 2009, “Role of A-Type Lamins in Signaling, Transcription, and Chromatin Organization,” J. Cell Biol., 187(7), pp. 945–957. 10.1083/jcb.200904124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Gonzalez, J. M. , Navarro-Puche, A. , Casar, B. , Crespo, P. , and Andres, V. , 2008, “Fast Regulation of AP-1 Activity Through Interaction of Lamin a/C, ERK1/2, and c-Fos at the Nuclear Envelope,” J. Cell Biol., 183(4), pp. 653–666. 10.1083/jcb.200805049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Zhong, N. , Radu, G. , Ju, W. , and Brown, W. T. , 2005, “Novel Progerin-Interactive Partner Proteins hnRNP E1, EGF, Mel 18, and UBC9 Interact With Lamin A/C,” Biochem. Biophys. Res. Commun., 338(2), pp. 855–861. 10.1016/j.bbrc.2005.10.020 [DOI] [PubMed] [Google Scholar]
- [8]. Zaragoza, M. V. , Fung, L. , Jensen, E. , Oh, F. , Cung, K. , McCarthy, L. A. , Tran, C. K. , Hoang, V. , Hakim, S. A. , and Grosberg, A. , 2016, “Exome Sequencing Identifies a Novel LMNA Splice-Site Mutation and Multigenic Heterozygosity of Potential Modifiers in a Family With Sick Sinus Syndrome, Dilated Cardiomyopathy, and Sudden Cardiac Death,” PLoS One, 11(5), p. e0155421. 10.1371/journal.pone.0155421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Zaragoza, M. V. , Hakim, S. A. , Hoang, V. , and Elliott, A. M. , 2017, “Heart-Hand Syndrome IV: A Second Family With LMNA-Related Cardiomyopathy and Brachydactyly,” Clin. Genet., 91(3), pp. 499–500. 10.1111/cge.12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Zaragoza, M. , Nguyen, C. , Widyastuti, H. , McCarthy, L. , and Grosberg, A. , 2017, “Dupuytren's and Ledderhose Diseases in a Family With LMNA-Related Cardiomyopathy and a Novel Variant in the ASTE1 Gene,” Cells, 6(4), p. 40. 10.3390/cells6040040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Broers, J. L. V. , Peeters, E. A. G. , Kuijpers, H. J. H. , Endert, J. , Bouten, C. V. C. , Oomens, C. W. J. , Baaijens, F. P. T. , and Ramaekers, F. C. S. , 2004, “Decreased Mechanical Stiffness in LMNA-/- Cells is Caused by Defective Nucleo-Cytoskeletal Integrity: Implications for the Development of Laminopathies,” Hum. Mol. Genet., 13(21), pp. 2567–2580. 10.1093/hmg/ddh295 [DOI] [PubMed] [Google Scholar]
- [12]. Lammerding, J. , Schulze, P. C. , Takahashi, T. , Kozlov, S. , Sullivan, T. , Kamm, R. D. , Stewart, C. L. , and Lee, R. T. , 2004, “Lamin A/C Deficiency Causes Defective Nuclear Mechanics and Mechanotransduction,” J. Clin. Invest., 113(3), pp. 370–378. 10.1172/JCI200419670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Lammerding, J. , Fong, L. G. , Ji, J. Y. , Reue, K. , Stewart, C. L. , Young, S. G. , and Lee, R. T. , 2006, “Lamins a and C but Not Lamin B1 Regulate Nuclear Mechanics,” J. Biol. Chem., 281(35), pp. 25768–25780. 10.1074/jbc.M513511200 [DOI] [PubMed] [Google Scholar]
- [14]. Sullivan, T. , Escalante-Alcalde, D. , Bhatt, H. , Anver, M. , Bhat, N. , Nagashima, K. , Stewart, C. L. , and Burke, B. , 1999, “Loss of A-Type Lamin Expression Compromises Nuclear Envelope Integrity Leading to Muscular Dystrophy,” J. Cell Biol., 147(5), pp. 913–919. 10.1083/jcb.147.5.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Zwerger, M. , Jaalouk, D. E. , Lombardi, M. L. , Isermann, P. , Mauermann, M. , Dialynas, G. , Herrmann, H. , Wallrath, L. L. , and Lammerding, J. , 2013, “Myopathic Lamin Mutations Impair Nuclear Stability in Cells and Tissue and Disrupt Nucleo-Cytoskeletal Coupling,” Hum. Mol. Genet., 22(12), pp. 2335–2349. 10.1093/hmg/ddt079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Core, J. Q. , Mehrabi, M. , Robinson, Z. R. , Ochs, A. R. , McCarthy, L. A. , Zaragoza, M. V. , and Grosberg, A. , 2017, “Age of Heart Disease Presentation and Dysmorphic Nuclei in Patients With LMNA Mutations,” PLoS One, 12(11), p. e0188256. 10.1371/journal.pone.0188256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Stoppel, W. L. , Kaplan, D. L. , and Black, L. D. , 2016, “Electrical and Mechanical Stimulation of Cardiac Cells and Tissue Constructs,” Adv. Drug Deliv. Rev., 96, pp. 135–155. 10.1016/j.addr.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Eriksson, M. , Brown, W. T. , Gordon, L. B. , Glynn, M. W. , Singer, J. , Scott, L. , Erdos, M. R. , Robbins, C. M. , Moses, T. Y. , Berglund, P. , Dutra, A. , Pak, E. , Durkin, S. , Csoka, A. B. , Boehnke, M. , Glover, T. W. , and Collins, F. S. , 2003, “Recurrent de Novo Point Mutations in Lamin a Cause Hutchinson-Gilford Progeria Syndrome,” Nature, 423(6937), pp. 293–298. 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Grosberg, A. , Alford, P. W. , McCain, M. L. , and Parker, K. K. , 2011, “Ensembles of Engineered Cardiac Tissues for Physiological and Pharmacological Study: Heart on a Chip,” Lab Chip, 11(24), pp. 4165–4173. 10.1039/c1lc20557a [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Hurlburt, H. M. , Aurigemma, G. P. , Hill, J. C. , Narayanan, A. , Gaasch, W. H. , Vinch, C. S. , Meyer, T. E. , and Tighe, D. A. , 2007, “Direct Ultrasound Measurement of Longitudinal, Circumferential, and Radial Strain Using 2-Dimensional Strain Imaging in Normal Adults,” Echocardiography, 24(7), pp. 723–731. 10.1111/j.1540-8175.2007.00460.x [DOI] [PubMed] [Google Scholar]
- [21]. Ng, A. C. T. , Delgado, V. , Bertini, M. , van der Meer, R. W. , Rijzewijk, L. J. , Shanks, M. , Nucifora, G. , Smit, J. W. A. , Diamant, M. , Romijn, J. A. , de Roos, A. , Leung, D. Y. , Lamb, H. J. , and Bax, J. J. , 2009, “Findings From Left Ventricular Strain and Strain Rate Imaging in Asymptomatic Patients With Type 2 Diabetes Mellitus,” Am. J. Cardiol., 104(10), pp. 1398–1401. 10.1016/j.amjcard.2009.06.063 [DOI] [PubMed] [Google Scholar]
- [22]. Gjesdal, O. , Hopp, E. , Vartdal, T. , Lunde, K. , Helle-Valle, T. , Aakhus, S. , Smith, H.-J. , Ihlen, H. , and Edvardsen, T. , 2007, “Global Longitudinal Strain Measured by Two-Dimensional Speckle Tracking Echocardiography is Closely Related to Myocardial Infarct Size in Chronic Ischaemic Heart Disease,” Clin. Sci., 113(6), pp. 287–296. 10.1042/CS20070066 [DOI] [PubMed] [Google Scholar]
- [23]. Kouzu, H. , Yuda, S. , Muranaka, A. , Doi, T. , Yamamoto, H. , Shimoshige, S. , Hase, M. , Hashimoto, A. , Saitoh, S. , Tsuchihashi, K. , Miura, T. , Watanabe, N. , and Shimamoto, K. , 2011, “Left Ventricular Hypertrophy Causes Different Changes in Longitudinal, Radial, and Circumferential Mechanics in Patients With Hypertension: A Two-Dimensional Speckle Tracking Study,” J. Am. Soc. Echocardiography, 24(2), pp. 192–199. 10.1016/j.echo.2010.10.020 [DOI] [PubMed] [Google Scholar]
- [24]. Serri, K. , Reant, P. , Lafitte, M. , Berhouet, M. , Le Bouffos, V. , Roudaut, R. , and Lafitte, S. , 2006, “Global and Regional Myocardial Function Quantification by Two-Dimensional Strain: Application in Hypertrophic Cardiomyopathy,” J. Am. Coll. Cardiol., 47(6), pp. 1175–1181. 10.1016/j.jacc.2005.10.061 [DOI] [PubMed] [Google Scholar]
- [25]. Wang, J. , Khoury, D. S. , Yue, Y. , Torre-Amione, G. , and Nagueh, S. F. , 2007, “Preserved Left Ventricular Twist and Circumferential Deformation, but Depressed Longitudinal and Radial Deformation in Patients With Diastolic Heart Failure,” Eur. Heart J., 29(10), pp. 1283–1289. 10.1093/eurheartj/ehn141 [DOI] [PubMed] [Google Scholar]
- [26]. Waldman, L. K. , Fung, Y. C. , and Covell, J. W. , 1985, “Transmural Myocardial Deformation in the Canine Left-Ventricle—Normal Invivo 3-Dimensional Finite Strains,” Circ. Res., 57(1), pp. 152–163. 10.1161/01.RES.57.1.152 [DOI] [PubMed] [Google Scholar]
- [27]. Yingchoncharoen, T. , Agarwal, S. , Popovic, Z. B. , and Marwick, T. H. , 2013, “Normal Ranges of Left Ventricular Strain: A Meta-Analysis,” J. Am. Soc. Echocardiogr., 26(2), pp. 185–191. 10.1016/j.echo.2012.10.008 [DOI] [PubMed] [Google Scholar]
- [28]. Kasner, M. , Gaub, R. , Sinning, D. , Westermann, D. , Steendijk, P. , Hoffmann, W. , Schultheiss, H.-P. , and Tschöpe, C. , 2010, “Global Strain Rate Imaging for the Estimation of Diastolic Function in HFNEF Compared With Pressure-Volume Loop Analysis,” Eur. J. Echocardiography, 11(9), pp. 743–751. 10.1093/ejechocard/jeq060 [DOI] [PubMed] [Google Scholar]
- [29]. Leitman, M. , Lysyansky, P. , Sidenko, S. , Shir, V. , Peleg, E. , Binenbaum, M. , Kaluski, E. , Krakover, R. , and Vered, Z. , 2004, “Two-Dimensional Strain—A Novel Software for Real-Time Quantitative Echocardiographic Assessment of Myocardial Function,” J. Am. Soc. Echocardiography, 17(10), pp. 1021–1029. 10.1016/j.echo.2004.06.019 [DOI] [PubMed] [Google Scholar]
- [30]. Nguyen, M.-D. , Tinney, J. P. , Ye, F. , Elnakib, A. A. , Yuan, F. , El-Baz, A. , Sethu, P. , Keller, B. B. , and Giridharan, G. A. , 2015, “Effects of Physiologic Mechanical Stimulation on Embryonic Chick Cardiomyocytes Using a Microfluidic Cardiac Cell Culture Model,” Anal. Chem., 87(4), pp. 2107–2113. 10.1021/ac503716z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Ghafar-Zadeh, E. , Waldeisen, J. R. , and Lee, L. P. , 2011, “Engineered Approaches to the Stem Cell Microenvironment for Cardiac Tissue Regeneration,” Lab Chip, 11(18), pp. 3031–3048. 10.1039/c1lc20284g [DOI] [PubMed] [Google Scholar]
- [32]. Marsano, A. , Conficconi, C. , Lemme, M. , Occhetta, P. , Gaudiello, E. , Votta, E. , Cerino, G. , Redaelli, A. , and Rasponi, M. , 2016, “Beating Heart on a Chip: A Novel Microfluidic Platform to Generate Functional 3D Cardiac Microtissues,” Lab Chip, 16(3), pp. 599–610. 10.1039/C5LC01356A [DOI] [PubMed] [Google Scholar]
- [33]. Matsuda, T. , Takahashi, K. , Nariai, T. , Ito, T. , Takatani, T. , Fujio, Y. , and Azuma, J. , 2004, “N-Cadherin-Mediated Cell Adhesion Determines the Plasticity for Cell Alignment in Response to Mechanical Stretch in Cultured Cardiomyocytes,” Biochem. Biophys. Res. Commun., 326(1), pp. 228–232. 10.1016/j.bbrc.2004.11.019 [DOI] [PubMed] [Google Scholar]
- [34]. Salameh, A. , Wustmann, A. , Karl, S. , Blanke, K. , Apel, D. , Rojas-Gomez, D. , Franke, H. , Mohr, F. W. , Janousek, J. , and Dhein, S. , 2010, “Cyclic Mechanical Stretch Induces Cardiomyocyte Orientation and Polarization of the Gap Junction Protein connexin43,” Circ. Res., 106(10), pp. 1592–1602. 10.1161/CIRCRESAHA.109.214429 [DOI] [PubMed] [Google Scholar]
- [35]. Greiner, A. M. , Chen, H. , Spatz, J. P. , and Kemkemer, R. , 2013, “Cyclic Tensile Strain Controls Cell Shape and Directs Actin Stress Fiber Formation and Focal Adhesion Alignment in Spreading Cells,” PLoS One, 8(10), p. e77328. 10.1371/journal.pone.0077328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Chen, N. Y. , Kim, P. , Weston, T. A. , Edillo, L. , Tu, Y. , Fong, L. G. , and Young, S. G. , 2018, “Fibroblasts Lacking Nuclear Lamins Do Not Have Nuclear Blebs or Protrusions but Nevertheless Have Frequent Nuclear Membrane Ruptures,” Proc. Natl. Acad. Sci. U. S. A., 115(40), pp. 10100–10105. 10.1073/pnas.1812622115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Choi, S. , Wang, W. , Ribeiro, A. J. S. , Kalinowski, A. , Gregg, S. Q. , Opresko, P. L. , Niedernhofer, L. J. , Rohde, G. K. , and Dahl, K. N. , 2011, “Computational Image Analysis of Nuclear Morphology Associated With Various Nuclear-Specific Aging Disorders,” Nucleus, 2(6), pp. 570–579. 10.4161/nucl.2.6.17798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Booth-Gauthier, E. A. , Du, V. , Ghibaudo, M. , Rape, A. D. , Dahl, K. N. , and Ladoux, B. , 2013, “Hutchinson-Gilford Progeria Syndrome Alters Nuclear Shape and Reduces Cell Motility in Three Dimensional Model Substrates,” Integr. Biol., 5(3), pp. 569–577. 10.1039/c3ib20231c [DOI] [PubMed] [Google Scholar]
- [39]. Lele, T. P. , Dickinson, R. B. , and Gundersen, G. G. , 2018, “Mechanical Principles of Nuclear Shaping and Positioning,” J. Cell Biol., 217(10), pp. 3330–3342. 10.1083/jcb.201804052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Lee, H. , Adams, W. J. , Alford, P. W. , McCain, M. L. , Feinberg, A. W. , Sheehy, S. P. , Goss, J. A. , and Parker, K. K. , 2015, “Cytoskeletal Prestress Regulates Nuclear Shape and Stiffness in Cardiac Myocytes,” Exp. Biol. Med. (Maywood), 240(11), pp. 1543–1554. 10.1177/1535370215583799 [DOI] [PMC free article] [PubMed] [Google Scholar]


