Abstract
The adult Hydra polyp continually renews all of its cells using three separate stem cell populations, but the genetic pathways enabling this homeostatic tissue maintenance are not well understood. We sequenced 24,985 Hydra single-cell transcriptomes and identified the molecular signatures of a broad spectrum of cell states, from stem cells to terminally differentiated cells. We constructed differentiation trajectories for each cell lineage and identified gene modules and putative regulators expressed along these trajectories, thus creating a comprehensive molecular map of all developmental lineages in the adult animal. In addition, we built a gene expression map of the Hydra nervous system. All together, we have generated a resource for addressing questions regarding the evolution of metazoan developmental processes and nervous system function.
Introduction
Hydrozoans have been at the center of fundamental discoveries in developmental biology, including animal regeneration and the first observation of stem cells (1, 2). Among hydrozoans, the cell populations and lineage relationships are best characterized in the freshwater polyp Hydra (Fig. 1A–D)(3–7). Homeostatic somatic maintenance of the adult Hydra polyp depends on the activity of every differentiation pathway, resulting in all cells being replaced approximately every 20 days (8). Hydra has three cell lineages (endodermal epithelial, ectodermal epithelial, and interstitial), and each is supported by its own stem cell population (Fig. 1A–D) (9). All epithelial cells in the body column are mitotic unipotent stem cells, resulting in continual displacement of cells toward the extremities. Epithelial stem cells differentiate to build the foot at the aboral end and the hypostome and tentacles at the oral end (Fig. 1A,C); differentiated cells are eventually shed from the extremities (10). Multipotent interstitial stem cells (ISCs) give rise to the three somatic cell types of the interstitial lineage — nematocytes, neurons, and gland cells (Fig. 1D) — and can also replace germline stem cells (GSCs) if they are experimentally depleted (6, 11, 12) (Fig. 1D). The cnidarian specific stinging cells, the nematocytes, are single-use cells; neurons and gland cells are closely associated with epithelial cells and thus are continually displaced and lost (13). Interstitial cells are maintained by three mechanisms: 1) Mitotic divisions of ISCs, progenitors, and gland cells (12), 2) ISC differentiation into neurons, nematocytes, and gland cells (5, 7), and 3) Neurons and gland cells change their expression and function with position (14, 15). Thus, cell identity in Hydra depends on coordinating stem cell differentiation and gene expression programs in a manner dependent on cell location. Understanding the molecular mechanisms underlying cellular differentiation and patterning in Hydra would be greatly facilitated by the creation of a spatial and temporal map of gene expression.
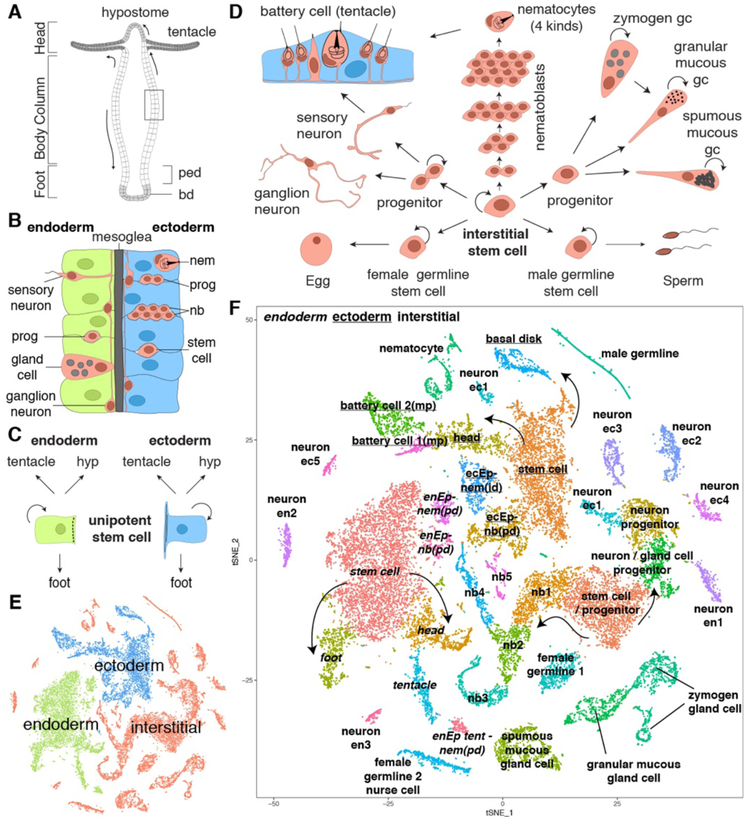
Figure 1. Hydra tissue composition and single cell RNA sequencing of 24,985 Hydra cells.
A) The Hydra body is a hollow tube with an adhesive foot at the aboral end (bd: basal disk, ped: peduncle) and a head with a mouth and a ring of tentacles at the oral end. The mouth opening is at the tip of a cone shaped protrusion — the hypostome. B) Enlargement of box in A. The body column consists of two epithelial layers (endoderm and ectoderm) separated by an extracellular matrix — the mesoglea. Cells of the interstitial cell lineage (red) reside in the interstitial spaces between epithelial cells, except gland cells which are integrated into the endodermal epithelium. Ectodermal cells can enclose nerve cells or nematocytes forming biological doublets. C) Epithelial cells of the body column are mitotic, have stem cell properties, and give rise to terminally differentiated cells of the hypostome (hyp), tentacles, and foot. D) Schematic of the interstitial stem cell lineage. The lineage is supported by a multipotent interstitial stem cell (ISC) that gives rise to neurons, gland cells, and nematocytes; ISCs are also capable of replenishing germline stem cells if they are lost. E) t-SNE representation of clustered cells colored by cell lineage. F) t-SNE representation of clustered cells annotated with cell state. Figures A-D adapted from (50). ec: ectodermal, en: endodermal, Ep: epithelial cell, gc: gland cell, id: integration doublet, mp: multiplet, nb: nematoblast, nem: differentiated nematocyte, pd: suspected phagocytosis doublet. id, mp, and pd are categories of biological doublets. Arrows indicate suggested transitions from stem cell populations to differentiated cells.
We used single-cell RNA sequencing (scRNA-seq) to complement this extensive knowledge of Hydra developmental processes. We collected ~25,000 Hydra single-cell transcriptomes covering a wide range of differentiation states and built differentiation trajectories for each lineage. These trajectories allowed us to identify putative regulatory modules that drive cell state specification, find evidence for a shared progenitor state in the gland cell and neural differentiation pathways, and explore gene expression changes along the oral-aboral axis. Finally, we generated a molecular map of the nervous system with spatial resolution, which provides opportunities to study mechanisms of neural network plasticity and nervous system evolution. We have made the single-cell data available at the Broad Institute’s Single Cell Portal. We anticipate that providing a comprehensive molecular map as a resource to the developmental biology and neuroscience communities will rapidly advance the ability of researchers to make discoveries using Hydra. Cnidarians such as Hydra hold an informative position on the phylogenetic tree as sister group to bilaterians (16) and largely have the same complement of gene families found in vertebrates (17–19). Thus, this data set, in combination with the existing cnidarian single-cell data set for Nematostella (20), offers the opportunity to identify conserved developmental mechanisms.
Results
Single-cell RNA sequencing of whole Hydra reveals cell state transitions
Thirteen Drop-seq libraries were prepared from dissociated whole adult Hydra polyps and two neuron-enriched libraries were prepared using FACS-enriched GFP-positive neurons from transgenic Hydra (figs. S1,S2, tables S1,S2). We mapped sequencing reads to a reference transcriptome and filtered for cells with 300–7,000 detected genes and 500–50,000 Unique Molecular Identifiers (UMIs), resulting in a data set with a detected median of 1,936 genes and 5,672 UMIs per cell (table S3). We clustered the cells and annotated cluster identity using published gene expression patterns (Fig. 1E,F, fig. S3) and further validated identities by performing RNA in situ hybridization experiments (fig. S4). In the clustering, cells separated according to cell lineage (Fig. 1E), and within each lineage, we observed the expected cell populations (Fig. 1F). We captured cells in a wide range of differentiation states and several differentiation trajectories are evident even in the t-SNE representation, similar to findings in scRNA-seq studies performed in planarians (21, 22). For example, clusters that correspond to differentiated head and foot epithelial cells are connected to their respective body column stem cell clusters (Fig. 1F). Additionally, the interstitial stem cell clusters are connected to both neuronal and nematocyte progenitors (nematoblasts). We also identified distinct clusters for differentiated cells of the interstitial lineage — neurons, gland cells, nematocytes, and germ cells (Fig. 1F). We applied non-negative matrix factorization (NMF) to the full dataset and subsequently to all lineage subsets to identify modules of genes that are co-expressed within cell populations (fig. S5) (23, 24). As described below and in the supplementary material, the recovered gene modules were used for doublet identification (see supplementary methods for discussion of doublet categories, figs. S2,S6–S9), trajectory characterization, and the identification of transcription factor binding sites enriched in the cis-regulatory elements of co-regulated genes.
Trajectory reconstruction of epithelial cells reveals position-dependent gene expression
Epithelial cells constantly adjust their gene expression relative to their position as they divide in the body column and are displaced toward the extremities (Fig. 1A). To identify these position-dependent gene expression patterns, we performed trajectory analyses on subsets of endodermal and ectodermal epithelial cells (Fig. 2A,B, fig. S10A–C). We ordered cells along the oral–aboral axis by using the R package URD to generate branching trajectories for each lineage spanning from the foot (aboral) to the hypostome and tentacle (oral) as two separate endpoints (24). URD connects cells with similar gene expression and uses simulated random walks to find gene expression trajectories between terminal cell populations and a starting progenitor cell population. This required removing biological and technical doublets from the epithelial cell subsets, which we accomplished by implementing a novel approach using NMF co-expression modules to identify doublet signatures (see methods, fig. S7). To validate these differentiation trajectories, we visualized the spatial expression of several previously characterized genes and validated the expression of several uncharacterized genes by RNA in situ hybridization (Fig. 2C–M, figs. S11–13).
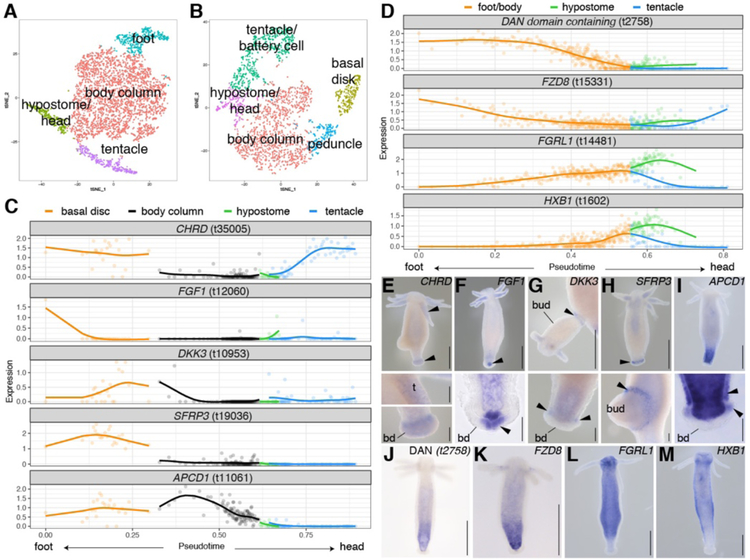
Figure 2. Identification of genes with differential expression along the oral-aboral axis.
A) t-SNE representation of subclustered endodermal epithelial cells and B) subclustered ectodermal epithelial cells. C-D) Epithelial cells were ordered using URD to reconstruct a trajectory where pseudotime represents spatial position. Scaled and log-transformed expression is visualized. C) Trajectory plots for previously uncharacterized putative signaling genes expressed in ectodermal epithelial cells of foot and tentacles. BMP antagonist CHRD (t35005), fibroblast growth factor FGF1 (t12060); Wnt antagonists DKK3 (t10953), SFRP3 (t19036) and APCD1 (t11061). D) Trajectory plots for genes expressed in a graded manner in endodermal epithelial cells. BMP antagonist “DAN domain containing gene” t2758, secreted Wnt antagonist FZD8 (t15331), fibroblast growth factor receptor FGRL1 (t14481), homeobox protein HXB1 (t1602). E-M) Epithelial expression patterns obtained using RNA in situ hybridization consistent with predicted patterns. Whole mounts and selected close-ups. Arrows indicate ectodermal signal. t: tentacle, bd: basal disk. Scale bars: whole mounts (including G): 500 μm, close-ups: 100 μm.
We identified epithelial genes with variable expression along the oral-aboral axis, including differentially expressed gene modules identified by NMF (figs. S14–17). These spatially and temporally resolved gene expression profiles for body column epithelial cells provide access to putative regulators of epithelial cell terminal differentiation at the oral and aboral ends, such as transcription factors and signaling molecules (Fig. 2, figs. S12,S15). For example, we find differential expression along the body axis of previously uncharacterized genes in the Wnt, BMP, and FGF signaling pathways (Fig. 2). Therefore, these data suggest candidate genes and pathways for functional testing to better understand oral-aboral patterning in Hydra.
Identification of multipotent interstitial stem cells and trajectory reconstruction of the interstitial lineage
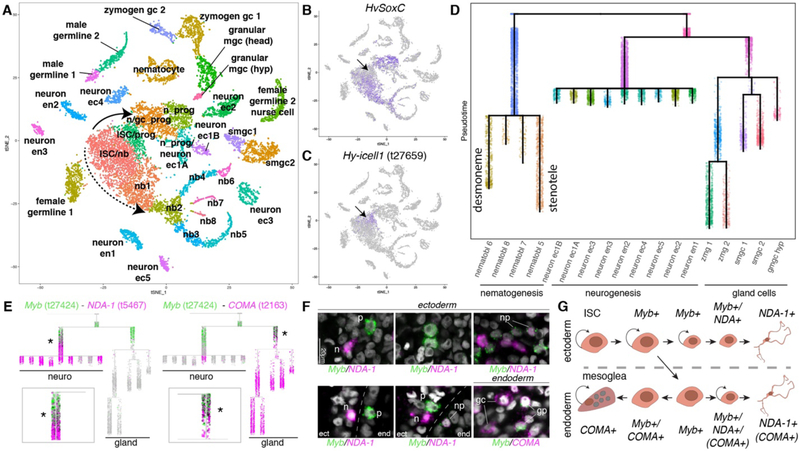
We extracted 12,470 interstitial cells from the whole data set (Fig. 1E, fig. S10D), performed subclustering, and annotated the clusters through the expression of known and new markers (Fig. 3A, fig. S18). The t-SNE representation of interstitial cells showed evidence for ISC differentiation (Fig. 3A, fig. S18A–H). NMF analysis was used to identify gene modules associated with interstitial lineage differentiation pathways (fig. S19). We identified a population of cells that largely lack expression of differentiation gene modules (i.e. the putative multipotent ISCs) and used this cell population as the root in a trajectory reconstruction using URD (fig. S19). HvSoxC (25) was found to be expressed in transition states between candidate ISCs and differentiated neurons and nematoblasts, suggesting that expression of this gene marks cells undergoing differentiation (Fig. 3B, fig. S20). We attempted to identify transcripts specific to the putative ISC population and found only a single marker with no shared similarities to known proteins (Fig. 3C, fig. S21A). ISCs may therefore be largely defined by an absence of cell type specific markers similar to planarian cNeoblasts (21). The URD reconstruction recovered a branching tree of interstitial stem cell differentiation that resolves neurogenesis, nematogenesis, and gland cell differentiation (Fig. 3D). We performed double fluorescent in situ hybridization (FISH) to validate predicted transition states (Fig. 3E–F, figs. S20–S22).
Figure 3. Trajectory reconstruction for cells of the interstitial lineage suggests a cell state common to neurogenesis and gland cell differentiation.
A) t-SNE representation of interstitial cells with clusters labeled by cell state. Solid arrow: neurogenesis/gland cell differentiation. Dashed arrow: nematogenesis. B) HvSoxC expression in progenitor cells. Arrow indicates putative ISC population, which is negative for HvSoxC and positive for biomarker Hy-icell1 expression (C). D) URD differentiation tree of the interstitial lineage. Color represents URD segments and do not correspond to the colors in the t-SNE (see fig. S19. E)) Myb (green) is expressed in the neuron/gland cell progenitor state and during early neurogenesis/gland cell differentiation. Expression of Myb (green, > 0) partially overlaps with high expression of the neuronal gene NDA-1 (magenta, > 3) and the gland cell gene COMA (t2163) (magenta, > 0); COMA is also expressed in a subset of endodermal neurons. Co-expressing cells are black. Star and close-up highlights cell states with co-expression. F) Double labeling using fluorescent RNA in situ hybridization is consistent with neuron differentiation in the endodermal and ectodermal epithelial layers and demonstrates the existence of transition states observed in the trajectory analysis. Additionally, endodermal gland cell differentiation transition states were observed in the endodermal epithelial layer (see also fig. S22). gc: gland cell, gp: gland cell progenitor, n: neuron, np: neuron progenitor, p: Myb positive progenitor. G) Model for progenitor specification. Ectodermal ISCs give rise to a progenitor that can give rise to ectodermal neurons. Progenitors that translocate to the endoderm are able to give rise to glands or neurons. ec: ectoderm, en: endoderm, gmgc: granular mucous gland cell, gc: gland cell, hyp: hypostome, ISC: interstitial multipotent stem cell, mgc: mucous gland cell, nb: nematoblast, smgc: spumous mucous gland cell, prog: progenitor, zmg: zymogen gland cell.
The trajectory analysis of the interstitial lineage suggests that neuron and gland cell differentiation transit through a previously undescribed shared cell state (Fig. 3D), whereas nematogenesis is distinct. To test this result, we identified genes that are expressed in the progenitor state common to neural and gland cell differentiation, including Myc3 (t18095) (26) and Myb (t27424) (Fig. 3D and E, fig. S21). We identified the spatial location of Myb-positive cells using FISH and found positive cells in both the endodermal and ectodermal layers (Figs. 3F, fig. S22). A subset of Myb positive cells co-express the neuronal marker NDA-1 (27), consistent with Myb-positive cells giving rise to neurons in both epithelial layers (Fig. 3E,F, fig. S22). Furthermore, we found endodermal Myb-positive cells that co-express COMA (t2163), a gene expressed during gland cell differentiation and in all gland cell states (Fig. 3E,F, fig. S22). ISCs reside in the ectodermal layer, but are the source of both new gland cells and neurons in the endodermal layer (12, 28). The data suggest the existence of two Myb-positive progenitor populations, one that stays in the ectoderm to give rise to neurons and one that crosses the mesoglea to the endodermal layer and subsequently gives rise to endodermal neurons and gland cells (Fig. 3G). Finally, we find that many of the gene modules identified by NMF analysis were specific to each differentiation pathway with ordered expression in pseudotime (fig. S23), thus revealing gene expression changes that underlie differentiation in the interstitial lineage.
Subtrajectory analyses of interstitial cell types
We next explored specification of different cell types within the interstitial lineage (Fig. 1D). First, we examined nematocytes, which contain one of the most complex eukaryotic organelles, nematocysts (29); these are used to sting and immobilize prey. Hydra nematocytes each have one of four types of nematocysts: desmonemes, holotrichous or atrichous isorhizas, and stenoteles (30, 31). We identified one cluster of differentiated nematocytes, which contains nematocytes that harbor either desmonemes or stenoteles (Fig. 3A, cluster “nematocyte”, fig. S24A–F). In addition, we annotated the differentiation trajectories of nematoblasts and identified gene modules that are expressed as they produce these two types of nematocytes (Figs. 3A,D, 4A, figs. S23–25). While extensive work on nematocyst diversity has been facilitated by their extreme morphological and functional differentiation, little is known about nematocyte molecular diversity. The identification of genes that are differentially expressed between nematocytes harboring different nematocyst types (Fig. 4A, figs. S23–25) provides a basis for understanding the specification and construction of these extraordinary organelles, which are the defining feature of Cnidaria.
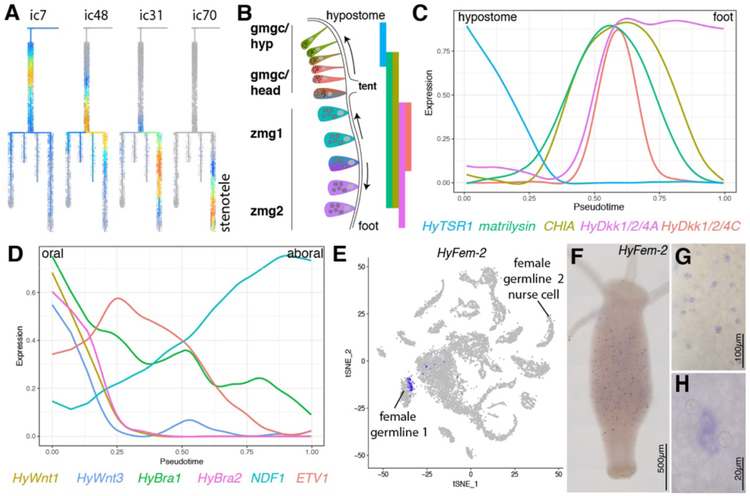
Figure 4. Subtrajectory analyses of interstitial cell types.
A) Interstitial gene modules successively expressed in nematocytes forming a stenotele (ic: interstitial gene module). B) Model for gland cell (ZMG/gMGC) location dependent changes. Gland cells integrated in the endodermal epithelium get displaced towards the extremities and undergo changes in expression and morphology. Bars show known expression domains for genes depicted in (C). gmgc: granular mucous gland cell, hyp: hypostome, tent: tentacle, zmg: zymogen gland cell. C) URD linear ZMG/gMGC trajectory recapitulates known position dependent gene expression in gland cells along the body column. HyTSR1 (15), HyDkk1/2/4 A/C (51, 52), matrilysin-like (t32151)/CHIA (t18356) (fig. S26B–E). D) URD linear sMGC trajectory plot for HyWnt1 (53), HyWnt3 (54), Hybra1 (55), Hybra2 (56), ETV1 (t22116) and NDF1 (t21810) showing expression changes in pseudotime that correlate to position along the oral-aboral axis. Cells are ordered according to pseudotime with putative hypostomal cell states to the left and putative lower head cell states to the right. E) Plot showing HyFem-2 expression in a subset of cells in the early female cluster. F-H) HyFem-2 is expressed in single cells or pairs scattered within the body column.
Second, we analyzed gland cells, which are interspersed between endodermal epithelial cells. Gland cell numbers are maintained both by specification of new gland cells from ISCs and by mitotic divisions of differentiated gland cells (7). We were able to capture ISC differentiation into gland cells in the trajectory analysis (Fig. 3D, fig. S26). Zymogen gland cells (ZMGs) are found throughout the body and transdifferentiate into granular mucous gland cells (gMGCs) when they are displaced into the head (Fig. 4B). Both of these cell types exhibit location-dependent changes in gene expression, and we captured these by building linear trajectories along the oral-aboral axis (Fig. 4C, figs. S14,S27). We hypothesized that spumous mucous gland cells (sMGCs), a separate type of gland cells present in the head, may exhibit similar location-dependent gene expression profiles that were previously unappreciated. Indeed, reconstruction of a linear trajectory uncovered oral–aboral organization of gene expression in this cell type, including several oral organizer genes (such as HyWnt1, HyWnt3, Hybra1 and Hybra2) in the orally located sMGCs (Fig. 4D, figs. S14, S28). This raises the possibility that these cells participate in patterning the head. Overall, our analysis reveals a broad range of gland cell states in Hydra that can be achieved through multiple paths.
Finally, we explored the germ cell clusters recovered in the data set. We excluded germline cells from the interstitial lineage tree reconstruction because differentiation of GSCs from ISCs does not typically occur in a homeostatic animal (11); thus we did not expect to observe transition states linking ISCs to GSCs. However, we did elucidate the spermatogenesis trajectory by analyzing the progression of cell states found in the two male germline clusters that were recovered in the subclustering of interstitial cells and used these data to identify and confirm several new male germline genes (Fig. 4D, figs. S14, S29–30). We identified two female germ cell clusters, which likely correspond to early and late female germ cell development (Fig. 3A). We performed in situ hybridizations for two genes (HyFem-1, HyFem-2) expressed in a subset of cells found in the early female germline cluster and found positive cells scattered throughout the body column, which we hypothesize are GSCs (Fig. 4E–H, fig. S30). If so, this would be the first report of gene expression in Hydra that is specific to GSCs and would allow for the study of GSCs in Hydra through the construction of GSC reporter transgenic lines.
Identification of putative transcriptional regulators of cell state specific regulatory modules
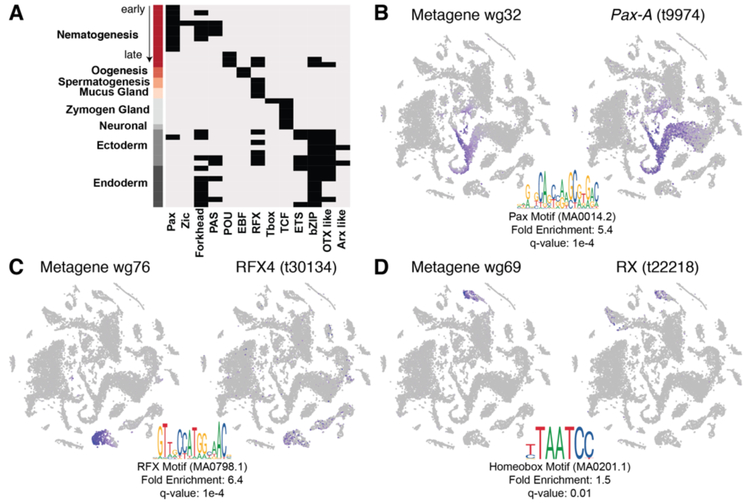
The construction of differentiation trajectories allows us to determine the spatial and temporal expression patterns of transcription factors, and thus gain insight into the gene regulatory networks that control cell type specification. We aimed to identify the transcription factor binding sites shared by co-expressed genes and candidate transcription factors that may bind these sites. To identify co-expressed genes, we used NMF to interrogate a genome-mapped data set and found 58 metagenes (i.e. sets of co-expressed genes) (figs. S31–32). To identify the putative regulatory regions of these co-expressed gene sets, we performed ATAC-seq on whole Hydra (fig. S33). We identified regions of locally enriched ATAC-seq read density (peaks) – signifying regions of open chromatin – and restricted the analysis to peaks within 5 kb upstream of the start codon of the genes in each NMF metagene (figs. S32–34). We then performed motif enrichment analysis to identify transcription factor binding sites that may control the expression of genes belonging to a metagene. We found at least one significantly enriched motif for each of 39 metagenes. These metagenes had distinct sets of enriched motifs, suggesting differences in the transcription factor classes underlying various cell states (Fig. 5A, fig. S35). For example, the paired box (Pax) motif is enriched in regulatory regions of genes expressed during early and mid-stages of nematogenesis, the forkhead (Fox) motif is enriched at mid- and late stages, and the POU motif is enriched only in late stages. The B-cell factor (EBF) motif is enriched in the female germline and the TCF motif is enriched in neurons and gland cells. Among epithelial cell states, motif enrichment is less tightly restricted to particular cell states. However, the ETS domain binding motif is enriched in metagenes expressed in endodermal and ectodermal epithelial cells in the extremities (tentacles and foot). Additionally, homeodomain (Otx and Arx) and bZip motifs are enriched throughout both epithelial lineages, and forkhead motifs appeared associated with genes expressed in endodermal epithelial cells (Fig. 5A). The enrichment of forkhead motifs in Hydra endoderm and Nematostella digestive filaments is consistent with a conserved function for forkhead transcription factors in cnidarian endodermal fate specification that is also found across bilaterians (20, 32).
Figure 5. Motif enrichment analysis for gene modules and identification of candidate regulators.
A) Enriched motifs (columns) found in open chromatin of putative 5’ cis-regulatory regions of co-expressed gene sets (metagenes) for listed cell states (rows). B-D) Metagene scores visualized on the t-SNE representation (left), significantly enriched motif found in putative 5’ cis-regulatory regions (bottom) and candidate regulators likely to bind identified motif with correlated expression (right). B) Metagene expressed during nematogenesis and putative PAX regulator. C) Metagene expressed in gland cells and putative RFX regulator. D) Metagene expressed in ectodermal epithelial cells of the foot and putative homeobox regulator.
To determine the regulatory factors that may be coordinating gene co-expression modules, we identified transcription factors within each metagene that are predicted to interact with the binding site(s) enriched in that metagene using a combination of Pfam domain annotation and profile inference (JASPAR) (fig. S34). For 25 of the 39 metagenes with enriched binding motifs, we found one or multiple candidate transcription factors with putative function in cell fate specification (table S5). For example, we find a metagene (wg32) that consists of 73 genes co-expressed during nematogenesis. A Pax transcription factor binding motif was significantly enriched in the potential regulatory regions near those genes, and the Pax-A transcription factor (t9974) is part of the metagene (Fig. 5A,B, fig. S35). The results therefore strongly suggest that Pax-A functions during early nematogenesis. This is concordant with a recent finding that Pax-A is required for nematogenesis during Nematostella development (20, 33). Similarly, we found evidence that an RX homeobox transcription factor (t22218) functions in basal disk development and an RFX transcription factor (t30134) functions in gland cell specification; the latter was also reported for Nematostella (Fig. 5C,D) (20). Homeodomain transcription factor binding motifs are enriched in ectoderm tentacle genes (metagene wg71) and the analysis recovered aristaless-related homeobox gene HyAlx (t16456) as a regulator (table S5, fig. S11A). A role for HyAlx during tentacle formation has been established previously (34). We provide all transcription factors that met our criteria as candidate regulators, including cases such as the basal disk where multiple TFs are both expressed in the proper context and predicted to bind an enriched motif (table S5). Overall, we identified several candidates for regulators of Hydra cell fate specification.
A molecular map of the Hydra nervous system
The Hydra nervous system consists of two nerve nets, one embedded in the ectodermal epithelial layer and one embedded in the endodermal epithelial layer. Neurons are concentrated at the oral and aboral ends of the polyp (35). To determine the molecular nature of neuronal subtypes, we extracted neural progenitors and differentiated neurons from the dataset for subcluster analysis (figs. S10E–F, S36). We identified 15 clusters: three clusters consist of neuronal progenitor cells, expressing progenitor genes like Myb/Myc3, and the remaining 12 clusters are differentiated neuronal subtypes (Fig. 6A, figs. S37–S40). To place these 12 neuronal subtypes into the ectodermal or endodermal nerve net, we performed TagSeq (36) on separated tissue layers and conducted differential gene expression (DGE) analysis to identify genes with significantly higher expression in the endodermal or ectodermal epithelial layer (fig. S37, see methods). Since the neurons remained attached to the epithelia, differentially expressed genes included neuron-specific genes, which allowed us to score the neuronal clusters as ectodermal or endodermal (Fig. 6A, fig. S37).
Figure 6. Molecular map of the Hydra nervous system with spatial resolution.
A) Subclustering of neurons and neuronal progenitors. Cell states are annotated with cell layer, localization along the body column, tentative neuronal subtype category — sensory (S) or ganglion (G), and gene markers used in annotations. B) Heatmap shows top twelve markers for neuronal cell states. C-E) First molecular markers for endodermal neurons. C) Transgenic line (NDF1(t14976)::GFP) expressing GFP in endodermal ganglion neurons along the body column (cluster “en1”). D-E) Body column cross-section of transgenic line (Alpha-LTX-Lhe1a-like(t33301)::GFP) expressing GFP in putative sensory neurons (cluster “en2”). Phalloidin staining (red) marks ECM and Hoechst (blue) marks nuclei. en: endoderm, ec: ectoderm.
To determine the location of the ectodermal neuronal subtypes along the oral-aboral axis, we generated a list of neuronal markers and selected genes to test spatial location using a combination of new and previously published in situ expression patterns (Fig. 6A,B, figs. S38–S 41). To test the endodermal identity of clusters “en1” and “en2”, we examined NDF1 (t14976, specific to cluster “en1”) and Alpha-LTX-Lhe1a-like (t33301, specific to cluster “en2”) expression by generating GFP reporter lines. For NDF1, GFP is expressed in endodermal ganglion neurons in the entire body except tentacles (Fig. 6C, fig. S40N–O). For Alpha-LTX-Lhe1a-like, GFP is expressed in sensory neurons along the body column in the endoderm (Fig. 6D,E). Therefore the transgenic reporter lines confirm endodermal localization of clusters “en1” and “en2” and demonstrate our ability to identify specific biomarkers for each neuronal subtype. In summary, we have produced a molecular map of the Hydra nervous system that describes 12 molecularly distinct neuronal subtypes and their in situ locations.
Discussion
We present an extensively validated gene expression map of Hydra cell states and differentiation trajectories, thus providing access to transcription factors expressed at key developmental decision points. Several recent studies have similarly demonstrated the value of conducting whole animal (20–22) or whole embryo scRNA-seq (20, 24, 37–39) to uncover cell type diversity and the regulatory programs that drive cell type specification. Conducting scRNA-seq on a diversity of organisms will provide insights into the core regulatory modules underling cell type specification and the evolution of cellular diversity (40). Thus, our Hydra data set provides an additional opportunity for comparisons to be made in an evolutionary context.
Analysis of Hydra by scRNA-seq uncovered new technical challenges, and we provide solutions to these challenges that will likely be applicable to many systems. For example, Hydra epithelial cells are highly phagocytic (41), a phenomenon that has been observed in a variety of systems, and thus will likely present a challenge for interpretation of scRNA-seq results in future studies (42–44). We implemented an approach that has been incorporated into URD, in which we use NMF as an unbiased method to identify anomalies in the data that likely represent cell doublets or phagocytic events. We envision that our approach could be applied to other systems and will be particularly useful in animals where existing expression data are limiting.
We present a gene expression map of a dynamic and regenerative nervous system, which opens the door to understanding the molecular basis of neuronal plasticity and regeneration. Of the twelve neuronal subtypes we have identified, three (the endodermal neurons) were previously uncharacterized molecularly. Three distinct neuronal circuits have been described in Hydra; two in the ectoderm — rhythmic potential 1 (RP1) and contraction burst (CB) — and one in the endoderm — rhythmic potential 2 (RP2) (45). These circuits are likely composed of ganglion neurons connected throughout the body. The characteristic localization of these circuits, combined with the in situ locations of the ganglion neuron molecular subtypes we identified (Fig. 6A), suggest the molecular identities of the neurons that comprise these distinct circuits. We propose the following: 1) the endodermal neurons of cluster “en1” (Fig. 6A,B) make up the RP2 circuit, 2) the neurons of clusters “ec3A”, “ec3B”, and “ec3C” make up the RP1 circuit, and 3) the neurons of clusters “ec1A”, “ec1B”, and “ec5” make up the ectodermal CB circuit. This is supported by the observation that the RP1 circuit is active in the basal disk (cluster ec3A), whereas the CB circuit extends aborally only to the peduncle (cluster “ec5”) (45). Neuron subtype-specific transgenes, such as the two examples presented here, will provide powerful tools for experimental perturbations to test neuronal function and nervous system regeneration by enabling precise alterations to these neural circuits. Nervous system function in such engineered animals can be tested using newly developed microfluidic tools that allow for simultaneous electrical and optical recordings in behaving animals (46).
The interstitial lineage differentiation trajectories provided several new insights. First, we identified a marker that may be specific to the multipotent stem cell population, which could provide a powerful tool for understanding stem cell function and fate decisions. Second, our data suggest the existence of a cell state that is shared by the neuron and gland cell trajectory (Fig. 3D). This interpretation is supported by the co-localization of neural and gland cell progenitors in several independent clustering analyses (Fig. 1F, 3A, fig. S31) that consider different sets of variable genes and sets of cells, and by the overlap of gene modules for neurogenesis and gland cell differentiation (fig. S42). The shared stem cell of gland cells, neurons and nematocytes suggests a shared evolutionary history of these cell types. The data further suggest that the evolution of nematocytes coincided with the emergence of a distinct progenitor. We thus propose a model in which multipotent ISCs first decide between a nematocyte or gland/neuron fate and then a second decision is made by the common gland/neuron progenitor. This contrasts with previous models that posit a common neuron/nematocyte progenitor (47). However, an alternative explanation is that gland and neuronal progenitors are separate populations that share early transcriptional events, thus future fate mapping experiments will be crucial. Additionally, our data suggest a model in which a bipotential gland/neuron progenitor born in the ectodermal layer, where multipotent ISCs reside, traverses the extracellular matrix to provide the endodermal layer with both gland cells and neurons (Fig. 3G).
Adult Hydra polyps, which are in a constant state of development, enable the capture of all states of cellular differentiation using scRNA-seq. An important future goal is to use scRNA-seq to rapidly assess the effect of mutations on all cell types (24, 39, 48). Hydra has a diversity of fate specifications from multiple stem cell types, yet is simple enough to be completely captured by a relatively small number of sequenced single cells from one life stage. Thus we are now able to study organism-wide changes at a single-cell level in response to perturbations. The transcription factors that we identified at key developmental decision points are exciting candidates to test using this approach. In conclusion, this resource and the experimental approaches we describe open doors in multiple fields including developmental biology, evolutionary biology, and neurobiology.
Methods
Hydra vulgaris AEP and Hydra vulgaris transgenic lines were dissociated into single cells and were prepared for Drop-seq (49); FACS was used to enrich for neurons. Sequencing reads were mapped to a de novo assembled transcriptome and a Hydra genome reference and clustering was performed. Subclustering was performed on the following subsets of the data: epithelial ectodermal cells, epithelial endodermal cells, interstitial cells, and neurons and neuronal progenitors. The in situ location of neuron subclusters was determined using in situ hybridization and differential gene expression analysis of separated epithelial layers. URD (24) was used to build differentiation trajectories for the interstitial and male germline lineages and to analyze the spatial expression of genes in the ectodermal, endodermal, and gland lineages. To analyze regulatory regions, co-expression modules were identified using NMF, ATAC-seq was performed to identify regions of open chromatin, and motif enrichment analysis was used to identify candidate regulators of the gene modules. Colorimetric in situ hybridization, fluorescent in situ hybridization, immunohistochemistry, and generation of transgenic lines was performed and used to validate biomarkers and cell states. For complete methods see supplementary material and methods.
Supplementary Material
Acknowledgements
We thank Robert Monroy for collection of separated endoderm and ectoderm epithelial layers for TagSeq and Ram Abhineet, Jamie Ho, Qianyan Li, Noemi Sierra and Adrienne Cho for wet lab validations by RNA in situ hybridization. We thank Yiqun Wang for help with NMF analysis and helpful discussion. We thank Rob Steele, Catherine Dana and Kristine Glauber for kindly providing transgenic lines PT1, enGreen1 and nGreen, and Thomas C. G. Bosch for kindly providing Hydra vulgaris AEP. We thank Lutz Froenicke, Vanessa Rashbrook, Siranoosh Ashtari, Emily Kumimoto of the UC Davis DNA Technologies Core for expert advice and assistance with sequencing. The Olympus FV1000 confocal used in this study was purchased using NIH Shared Instrumentation Grant 1S10RR019266-01. We thank the MCB Light Microscopy Imaging Facility, which is a UC Davis Campus Core Research Facility, for the use of this microscope and Michael Paddy for excellent support. Many thanks to Charlie David for providing numerous and plentiful insights on Hydra biology that helped with the interpretation of the data. We thank Bryan Teefy for helpful discussion and moral support. We thank the following people for critical reading of the manuscript: Alex Schier, Rob Steele, Gary Wessel, Bruce Draper, Stefan Materna, Jacob Robinson, and Casey Dunn.
Funding: This study was supported by DARPA Contract # HR0011–17-C-0026 (C.E.J.), UC Davis Start-up funding (C.E.J.), and by the NIH (J.A.F, K99HD091291).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: The raw data reported in this paper are archived at NCBI GEO (accession no. GSE121617) and in processed and interactively browsable forms in the Broad Single-Cell Portal (https://portals.broadinstitute.org/single_cell/study/SCP260/stem-cell-differentiation-trajectories-in-hydra-resolved-at-single-cell-resolution). RNA-seq data: Bioproject PRJNA497966. Transcriptome Shotgun Assembly project: GHHG00000000. Transcriptome Blast and ATACseq read/peak display: (https://research.nhgri.nih.gov/hydra/). Analysis code and objects: github https://github.com/cejuliano/hydra_single_cell and Dryad https://doi.org/10.5061/dryad.v5r6077.
References
- 1.Trembley A, Mémoires Pour Servirà l’Histoire d’un Genre de Polypes d’Eau Douce, à Bras en Forme de Cornes (Chez Jean & Herman Verbeek, Leiden, 1744). [Google Scholar]
- 2.Weismann A, Die Entstehung der Sexualzellen bei den Hydromedusen: Zugleich ein Beitrag zur Kenntniss des Baues und der Lebenserscheinungen dieser Gruppe (Gustav Fischer Verlag, Jena, 1883). [Google Scholar]
- 3.Campbell RD, Development of Hydra Lacking Interstitial and Nerve Cells (“Epithelial Hydra”) in Determinants of Spatial Organization (Elsevier, 1979), pp. 267–293. [Google Scholar]
- 4.Sugiyama T, Fujisawa T, Genetic analysis of developmental mechanisms in Hydra. II. Isolation and characterization of an interstitial cell-deficient strain. J. Cell. Sci 29, 35–52 (1978). [DOI] [PubMed] [Google Scholar]
- 5.David CN, Murphy S, Characterization of interstitial stem cells in Hydra by cloning. Dev. Biol 58, 372–383 (1977). [DOI] [PubMed] [Google Scholar]
- 6.Bosch TCG, David CN, Stem cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. 121, 182–191 (1987). [Google Scholar]
- 7.Bode HR, Heimfeld S, Chow MA, Huang LW, Gland cells arise by differentiation from interstitial cells in Hydra attenuata. Dev. Biol 122, 577–585 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Campbell RD, Tissue dynamics of steady state growth in Hydra littoralis. II. Patterns of tissue movement. J. Morphol 121, 19–28 (1967). [DOI] [PubMed] [Google Scholar]
- 9.Bosch TCG, Anton-Erxleben F, Hemmrich G, Khalturin K, The Hydra polyp: Nothing but an active stem cell community. Dev. Growth Differ 52, 15–25 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Holstein TW, Hobmayer E, David CN, Pattern of epithelial cell cycling in Hydra. Dev. Biol 148, 602–611 (1991). [DOI] [PubMed] [Google Scholar]
- 11.Nishimiya-Fujisawa C, Kobayashi S, Germline stem cells and sex determination in Hydra. Int. J. Dev. Biol 56, 499–508 (2012). [DOI] [PubMed] [Google Scholar]
- 12.David CN, Interstitial stem cells in Hydra: multipotency and decision-making. Int. J. Dev. Biol 56, 489–497 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Campbell RD, Cell movements in Hydra. Integr. Comp. Biol 14, 523–535 (1974). [Google Scholar]
- 14.Koizumi O, Bode HR, Plasticity in the nervous system of adult Hydra. I. The position-dependent expression of FMRFamide-like immunoreactivity. Dev. Bio 116, 407–421 (1986). [DOI] [PubMed] [Google Scholar]
- 15.Siebert S, Anton-Erxleben F, Bosch TCG, Cell type complexity in the basal metazoan Hydra is maintained by both stem cell based mechanisms and transdifferentiation. Dev. Biol 313, 13–24 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Dunn CW, Giribet G, Edgecombe GD, Hejnol A, Animal phylogeny and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst 45, 371–395 (2014). [Google Scholar]
- 17.Erwin DH, Early origin of the bilaterian developmental toolkit. Philos. Trans. R. Soc. Lond., B, Biol. Sci 364, 2253–2261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Technau U et al. , Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 21, 633–639 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Kortschak RD, Samuel G, Saint R, Miller DJ, EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr. Biol 13, 2190–2195 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Sebé-Pedrós A et al. , Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell. 173, 1520–1534.e20 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW, Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science. 360, eaaq1736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plass M et al. , Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science. 360, eaaq1723 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Brunet JP, Tamayo P, Golub TR, Mesirov JP, Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. U.S.A 101, 4164–4169 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrell JA et al. , Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. 360, eaar3131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemmrich G et al. , Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol. Biol. Evol 29, 3267–3280 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Hobmayer B et al. , Stemness in Hydra - a current perspective. Int. J. Dev. Biol 56, 509–517 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Augustin R et al. , A secreted antibacterial neuropeptide shapes the microbiome of Hydra. Nat. Commun 8, 698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bode HR, The interstitial cell lineage of Hydra: a stem cell system that arose early in evolution. J. Cell. Sci 109(Pt 6), 1155–1164 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Özbek S, The cnidarian nematocyst: a miniature extracellular matrix within a secretory vesicle. Protoplasma. 248, 635–640 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Bode HR, Flick KM, Distribution and dynamics of nematocyte populations in Hydra attenuata. J. Cell. Sci 21, 15–34 (1976). [DOI] [PubMed] [Google Scholar]
- 31.Holstein T, The morphogenesis of nematocytes in Hydra and Forskålia: an ultrastructural study. J. Ultrastruct. Res 75, 276–290 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Grapin-Botton A, Constam D, Evolution of the mechanisms and molecular control of endoderm formation. Mech. Dev 124, 253–278 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Babonis LS, Martindale MQ, PaxA, but not PaxC, is required for cnidocyte development in the sea anemone Nematostella vectensis. Evodevo. 8, 431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith KM, Gee L, Bode HR, HyAlx, an aristaless-related gene, is involved in tentacle formation in hydra. Development. 127, 4743–4752 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Bode H et al. , Quantitative analysis of cell types during growth and morphogenesis in Hydra. Roux. Arch. Dev. Biol 171, 269–285 (1973). [DOI] [PubMed] [Google Scholar]
- 36.Lohman BK, Weber JN, Bolnick DI, Evaluation of TagSeq, a reliable low-cost alternative for RNAseq. Mol. Ecol. Resour 16, 1315–1321 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Karaiskos N et al. , The Drosophila embryo at single-cell transcriptome resolution. Science. 358, 194–199 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Briggs JA et al. , The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science. 360, eaar5780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner DE et al. , Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science. 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marioni JC, Arendt D, How single-cell genomics is changing evolutionary and developmental Biology. Annu. Rev. Cell Dev. Biol 33, 537–553 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Campbell RD, Elimination by Hydra interstitial and nerve cells by means of colchicine. J. Cell. Sci 21, 1–13 (1976). [DOI] [PubMed] [Google Scholar]
- 42.Schafer DP et al. , Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Z et al. , Phagocytic activity of neuronal progenitors regulates adult neurogenesis. Nat. Cell. Biol 13, 1076–1083 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakanishi Y, Shiratsuchi A, Phagocytic removal of apoptotic spermatogenic cells by sertoli cells: Mechanisms and consequences. Biol. Pharm. Bull 27, 13–16 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Dupre C, Yuste R, Non-overlapping neural networks in Hydra vulgaris. Curr. Biol 27, 1085–1097 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badhiwala KN, Gonzales DL, Vercosa DG, Avants BW, Robinson JT, Microfluidics for electrophysiology, imaging, and behavioral analysis of Hydra. Lab Chip. 18, 2523–2539 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Miljkovic-Licina M, Chera S, Ghila L, Galliot B, Head regeneration in wild-type hydra requires de novo neurogenesis. Development. 134, 1191–1201 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Harland RM, A new view of embryo development and regeneration. Science. 360, 967–968 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Macosko EZ et al. , Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siebert S, Hydra cell lineages (http://commons.wikimedia.org/wiki/File:Hydra_cell_lineages.svg) (2018). [Google Scholar]
- 51.Guder C et al. , An ancient Wnt-Dickkopf antagonism in Hydra. Development. 133, 901–911 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Augustin R et al. , Dickkopf related genes are components of the positional value gradient in Hydra. Dev. Biol 296, 62–70 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Lengfeld T et al. , Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Bio 330, 186–199 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Hobmayer B et al. , WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature. 407, 186–189 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Technau U, Bode HR, HyBra1, a Brachyury homologue, acts during head formation in Hydra. Development. 126, 999–1010 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Bielen H et al. , Divergent functions of two ancient Hydra Brachyury paralogues suggest specific roles for their C-terminal domains in tissue fate induction. Development. 134, 4187–4197 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Greber MJ, David CN, Holstein TW, A quantitative method for separation of living Hydra cells. Roux. Arch. Dev. Biol 201, 296–300 (1992). [DOI] [PubMed] [Google Scholar]
- 58.Gierer A et al. , Regeneration of Hydra from reaggregated cells. Nat. New Biol. 239, 98–101 (1972). [DOI] [PubMed] [Google Scholar]
- 59.Dunn CW, Howison M, Zapata F, Agalma: an automated phylogenomics workflow. BMC Bioinformatics. 14, 330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gnerre S et al. , High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U.S.A 108, 1513–1518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grabherr MG et al. , Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen KD, Brenner SE, Dudoit S, Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic. Acids. Res 38, e131–e131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilbert D, Gene-omes built from mRNA seq not genome DNA. In: 7th Annual Arthropod Genomics Symposium. Notre Dame (2013). [Google Scholar]
- 64.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Stover NA, Steele RE, Trans-spliced leader addition to mRNAs in a cnidarian. Proc. Natl. Acad. Sci. U.S.A 98, 5693–5698 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011). [Google Scholar]
- 67.Finn RD et al. , The Pfam protein families database: towards a more sustainable future. Nucleic. Acids. Res 44, D279–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voigt O, Erpenbeck D, Wörheide G, A fragmented metazoan organellar genome: the two mitochondrial chromosomes of Hydra magnipapillata. BMC Genomics 9, 350 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chapman JA et al. , The dynamic genome of Hydra. Nature. 464, 592–596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu SM, Westfall JA, Dunne JF, Light and electron microscopic localization of a monoclonal antibody in neurons in situ in the head region of Hydra. J. Morphol 184, 183–193 (1985). [DOI] [PubMed] [Google Scholar]
- 72.Hufnagel LA, Kass-Simon G, Lyon MK, Functional organization of battery cell complexes in tentacles of Hydra attenuata. J. Morphol 184, 323–341 (1985). [DOI] [PubMed] [Google Scholar]
- 73.Hobmayer E, David CN, Differentiation of a Nerve Cell-Battery Cell Complex in Hydra in Evolution of the First Nervous Systems (Springer US, Boston, MA, 1989), pp. 71–80. [Google Scholar]
- 74.Dübel S, Cell differentiation in the head of Hydra. Differentiation. 41, 99–109 (1989). [Google Scholar]
- 75.Lyon MK, Kass-Simon G, Hufnagel LA, Ultrastructural analysis of nematocyte removal in Hydra. Tissue Cell. 14, 415–424 (1982). [DOI] [PubMed] [Google Scholar]
- 76.McNeil PL, Mechanisms of nutritive endocytosis. I. Phagocytic versatility and cellular recognition in Chlorohydra digestive cells, a scanning electron microscope study. J. Cell. Sci 49, 311–339 (1981). [DOI] [PubMed] [Google Scholar]
- 77.Seybold A, Salvenmoser W, Hobmayer B, Sequential development of apical-basal and planar polarities in aggregating epitheliomuscular cells of Hydra. Dev. Bio 412, 148–159 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Satija R, Farrell JA, Gennert D, Schier AF, Regev A, Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitgutsch C, Hauser F, Grimmelikhuijzen CJP, Expression and developmental regulation of the Hydra-RFamide and Hydra-LWamide preprohormone genes in Hydra: Evidence for transient phases of head formation. Dev. Biol 207, 189–203 (1999). [DOI] [PubMed] [Google Scholar]
- 80.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corces MR et al. , An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol 109, 21.29.1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Q, Brown JB, Huang H, Bickel PJ, Measuring reproducibility of high-throughput experiments. Annals of Applied Statistics. 5, 1752–1779 (2011). [Google Scholar]
- 86.de Mendoza A et al. , Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl. Acad. Sci. U.S.A 110, E4858–66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khan A, Mathelier A, JASPAR RESTful API: accessing JASPAR data from any programming language. Bioinformatics. 34, 1612–1614 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Khan A et al. , JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic. Acids. Res 46, D260–D266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heinz S et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haghverdi L, Buettner F, Theis FJ, Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 31, 2989–2998 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ, Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Hartigan JA, Hartigan PM, The Dip Test of Unimodality. Ann. Stat 13, 70–84 (1985). [Google Scholar]
- 93.Grens A, Gee L, Fisher DA, Bode HR, CnNK-2, an NK-2 homeobox gene, has a role in patterning the basal end of the axis in Hydra. Dev. Biol 180, 473–488 (1996). [DOI] [PubMed] [Google Scholar]
- 94.Lesh-Laurie GE, Separating Viable Tissue Layers in Hydra: Research Methods (Springer US, Boston, MA, 1983), pp. 267–271. [Google Scholar]
- 95.Glauber KM et al. , A small molecule screen identifies a novel compound that induces a homeotic transformation in Hydra. Development. 140, 4788–4796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muscatine L, in The Biology of Hydra, Lenhoff HM, Loomis WF, Eds. (Coral Gables, Fla: ), pp. 255–268. [Google Scholar]
- 97.Davis LE, Burnett AL, Haynes JF, Mumaw VR, A histological and ultrastructural study of dedifferentiation and redifferentiation of digestive and gland cells in Hydra viridis. Dev. Biol 14, 307–329 (1966). [Google Scholar]
- 98.Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Juliano CE, Lin H, Steele RE, Generation of transgenic Hydra by embryo microinjection. J. Vis. Exp, 51888 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TCG, Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. U.S.A 103, 6208–6211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schindelin J et al. , Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thomsen S, Bosch TCG, Foot differentiation and genomic plasticity in Hydra: lessons from the PPOD gene family. Dev. Genes Evol. 216, 57–68 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Endl I, Lohmann JU, Bosch TCG, Head-specific gene expression in Hydra: complexity of DNA- protein interactions at the promoter of ks1 is inversely correlated to the head activation potential. Proc. Natl. Acad. Sci. U.S.A 96, 1445–1450 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T, Expression and evolutionary conservation of nanos-related genes in Hydra. Roux. Arch. Dev. Biol 210, 591–602 (2000). [DOI] [PubMed] [Google Scholar]
- 107.Hwang JS et al. , Nematogalectin, a nematocyst protein with GlyXY and galectin domains, demonstrates nematocyte-specific alternative splicing in Hydra. Proc. Natl. Acad. Sci. U.S.A 107, 18539–18544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hwang JS et al. , Cilium evolution: identification of a novel protein, nematocilin, in the mechanosensory cilium of Hydra nematocytes. Mol. Biol. Evol 25, 2009–2017 (2008). [DOI] [PubMed] [Google Scholar]
- 109.Fraune S et al. , In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc. Natl. Acad. Sci. U.S.A 107, 18067–18072 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alexopoulos H et al. , Evolution of gap junctions: the missing link? Curr. Biol 14, R879–80 (2004). [DOI] [PubMed] [Google Scholar]
- 111.Darmer D et al. , Three different prohormones yield a variety of Hydra-RFamide (Arg-Phe-NH2) neuropeptides in Hydra magnipapillata. Biochem. J 332 ( Pt 2), 403–412 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Noro Y et al. , Regionalized nervous system in Hydra and the mechanism of its development. Gene Expr. Patterns 31, 42–59 (2019). [DOI] [PubMed] [Google Scholar]
- 113.Yum S et al. , A novel neuropeptide, Hym-176, induces contraction of the ectodermal muscle in Hydra. Biochem. Biophys. Res. Commun 248, 584–590 (1998). [DOI] [PubMed] [Google Scholar]
- 114.Takahashi T et al. , Hym-301, a novel peptide, regulates the number of tentacles formed in hydra. Development. 132, 2225–2234 (2005). [DOI] [PubMed] [Google Scholar]
- 115.Lommel M et al. , Hydra mesoglea proteome identifies thrombospondin as a conserved component active in head organizer restriction. Sci Rep. 8, 11753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weinziger R, Salgado LM, David CN, Bosch TCG, Ks1, an epithelial cell-specific gene, responds to early signals of head formation in Hydra. Development. 120, 2511–2517 (1994). [DOI] [PubMed] [Google Scholar]
- 117.Smith KM, Gee L, Blitz IL, Bode HR, CnOtx, a member of the Otx gene family, has a role in cell movement in Hydra. Dev. Biol 212, 392–404 (1999). [DOI] [PubMed] [Google Scholar]
- 118.Martinez DE et al. , Budhead, a Fork Head/HNF-3 homologue, is expressed during axis formation and head specification in Hydra. Dev. Biol 192, 523–536 (1997). [DOI] [PubMed] [Google Scholar]
- 119.Watanabe H et al. , Nodal signalling determines biradial asymmetry in Hydra. Nature. 515, 112–115 (2014). [DOI] [PubMed] [Google Scholar]
- 120.Philipp I et al. , Wnt/beta-catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra. Proc. Natl. Acad. Sci. U.S.A 106, 4290–4295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bosch TCG et al. , Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev. Comp. Immunol 33, 559–569 (2009). [DOI] [PubMed] [Google Scholar]
- 122.Juliano CE et al. , PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc. Natl. Acad. Sci. U.S.A 111, 337–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lim RSM, Anand A, Nishimiya-Fujisawa C, Kobayashi S, Kai T, Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev. Biol 386, 237–251 (2014). [DOI] [PubMed] [Google Scholar]
- 124.Engel U et al. , Nowa, a novel protein with minicollagen Cys-rich domains, is involved in nematocyst formation in Hydra. J. Cell. Sci 115, 3923–3934 (2002). [DOI] [PubMed] [Google Scholar]
- 125.Fedders H, Augustin R, Bosch TCG, A Dickkopf- 3-related gene is expressed in differentiating nematocytes in the basal metazoan Hydra. Roux. Arch. Dev. Biol 214, 72–80 (2004). [DOI] [PubMed] [Google Scholar]
- 126.Fujisawa T, David CN, Commitment during nematocyte differentiation in Hydra. J. Cell. Sci 48, 207–222 (1981). [DOI] [PubMed] [Google Scholar]
- 127.Fraune J et al. , Hydra meiosis reveals unexpected conservation of structural synaptonemal complex proteins across metazoans. Proc. Natl. Acad. Sci. U.S.A 109, 16588–16593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hayakawa E, Fujisawa C, Fujisawa T, Involvement of Hydra achaete-scute gene CnASH in the differentiation pathway of sensory neurons in the tentacles. Roux. Arch. Dev. Biol (2004). [DOI] [PubMed] [Google Scholar]
- 129.Takaku Y et al. , Innexin gap junctions in nerve cells coordinate spontaneous contractile behavior in Hydra polyps. Sci Rep. 4, 3573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Takahashi T et al. , A novel neuropeptide, Hym-355, positively regulates neuron differentiation in Hydra. Comp. Biochem. Physiol. A. Mol. Integr. Physiol 124, S93 (1999). [DOI] [PubMed] [Google Scholar]
- 131.Galliot B et al. , Origins of neurogenesis, a cnidarian view. Dev. Biol 332, 2–24 (2009). [DOI] [PubMed] [Google Scholar]
- 132.Miljkovic-Licina M, Gauchat D, Galliot B, Neuronal evolution: analysis of regulatory genes in a first-evolved nervous system, the Hydra nervous system. Biosystems. 76, 75–87 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.