ABSTRACT
Cleft lip and palate are common birth defects resulting from failure of the facial processes to fuse during development. The mammalian grainyhead-like (Grhl1-3) genes play key roles in a number of tissue fusion processes including neurulation, epidermal wound healing and eyelid fusion. One family member, Grhl2, is expressed in the epithelial lining of the first pharyngeal arch in mice at embryonic day (E)10.5, prompting analysis of the role of this factor in palatogenesis. Grhl2-null mice die at E11.5 with neural tube defects and a cleft face phenotype, precluding analysis of palatal fusion at a later stage of development. However, in the first pharyngeal arch of Grhl2-null embryos, dysregulation of transcription factors that drive epithelial-mesenchymal transition (EMT) occurs. The aberrant expression of these genes is associated with a shift in RNA-splicing patterns that favours the generation of mesenchymal isoforms of numerous regulators. Driving the EMT perturbation is loss of expression of the EMT-suppressing transcription factors Ovol1 and Ovol2, which are direct GRHL2 targets. The expression of the miR-200 family of microRNAs, also GRHL2 targets, is similarly reduced, resulting in a 56-fold upregulation of Zeb1 expression, a major driver of mesenchymal cellular identity. The critical role of GRHL2 in mediating cleft palate in Zeb1−/− mice is evident, with rescue of both palatal and facial fusion seen in Grhl2−/−;Zeb1−/− embryos. These findings highlight the delicate balance between GRHL2/ZEB1 and epithelial/mesenchymal cellular identity that is essential for normal closure of the palate and face. Perturbation of this pathway may underlie cleft palate in some patients.
KEY WORDS: Grhl2, Zeb1, Palate, EMT, Explant
Summary: Epithelial transcription factor GRHL2 is required for face closure while mesenchymal transcription factor ZEB1 is required for palate closure. Surprisingly, animals lacking both factors close their face and secondary palate.
INTRODUCTION
Cleft lip and/or palate affects approximately 1 in 700 live births (Dixon et al., 2011) and is associated with significant morbidity. This birth defect arises when facial processes fail to fuse during embryonic development. The upper lip and anterior region of the palate, termed primary palate, has a separate ontogeny from the posterior region, termed secondary palate. The upper lip and primary palate are derived from fusion between the medial nasal processes (MNPs) and maxillary processes (MXPs). The secondary palate is formed when palatal shelves outgrow from the MXPs, initially downwards adjacent to the tongue, then reorientating to grow towards the midline. Fusion between the palatal shelves creates a midline seam of epithelium that dissolves to allow mesenchymal confluence. In the mouse, MXP-MNP fusion occurs at embryonic day (E)10.5, while palatal shelf outgrowth and fusion occur between E11.5 and E15.5, resulting in palate formation being complete by E17 (Bush and Jiang, 2012). In the sixth week of human gestation, the MXPs, MNPs and lateral nasal process (LNP) fuse, and the palatal shelves begin to grow from the oral side of the MXPs. In the eighth week, the palatal shelves elevate above the tongue and merge in the midline. They also fuse with primary palate and nasal septum, with fusion complete by the twelfth week of gestation (Lan et al., 2015).
Numerous genes have been implicated in the regulation of mammalian palatogenesis (Bush and Jiang, 2012), including the grainyhead-like 3 (GRHL3) gene (Peyrard-Janvid et al., 2014), a member of a large family of highly conserved developmental transcription factors (Wilanowski et al., 2002). The first-discovered member of this family, Drosophila grainyhead (grh), is required for formation of the head skeleton, dorsal hole closure, integrity of the cuticle, and other cellular polarity and migration events during fly development (Bray and Kafatos, 1991). Truncating mutations in human GRHL3 cause cleft palate (Mangold et al., 2016), while a missense variant is associated with increased risk of cleft palate at the population level (Leslie et al., 2016). The congenital disorder Van Der Woude syndrome, involving cleft lip and/or palate with lower lip pits, can also be caused by dominant mutations in GRHL3 (Peyrard-Janvid et al., 2014). Grhl3 is also critical for neural tube closure, a role it shares with its nearest mammalian paralog, Grhl2. We and others have shown that Grhl2−/− mouse embryos exhibit a cleft face, cranioschisis and an open posterior neuropore at E10.5 (Rifat et al., 2010; Werth et al., 2010). Embryos carrying N-ethyl-N-nitrosourea (ENU)-induced mutations in Grhl2 can survive until advanced stages of development, at which point they display a cleft upper jaw (Menke et al., 2015; Pyrgaki et al., 2011). The non-neural ectoderm adjacent to the neural plate displays mesenchymal characteristics in Grhl2 mutant embryos, including aberrant vimentin expression and increased cell motility (Ray and Niswander, 2016). The resulting lack of epithelial integrity prevents apposition of the neural folds, resulting in failed neural tube closure.
ZEB1 is another transcription factor implicated in palatogenesis. Zeb1−/− mouse embryos die neonatally with a cleft palate and widespread skeletal abnormalities (Takagi et al., 1998). Zeb1 is expressed in the mesenchyme but not the epithelium during palatal shelf outgrowth (Liu et al., 2008; Shin et al., 2012). Zeb1−/− palatal mesenchyme fails to express the mesenchymal marker vimentin but ectopically expresses the epithelial marker E-cadherin at E16.5 (Liu et al., 2016). A spontaneously arising mutant, the Twirler mouse, displays cleft lip and secondary palate associated with failure of palatal shelf outgrowth (Gong et al., 2000). This phenotype is due to a point mutation in intron 1 of Zeb1 that dysregulates its expression (Kurima et al., 2011). Although mutations in ZEB1 are not associated with cleft palate in humans, haploinsufficiency causes posterior polymorphic corneal dystrophy, a characteristic feature of which is acquisition of epithelial characteristics by corneal endothelium (Krafchak et al., 2005; Liskova et al., 2016).
The developing palate is composed of epithelial and mesenchymal cell types. Epithelial cells are immotile and characterized by their apical-basal polarity, attachment to a basal lamina, cortical ring of actin and expression of intercellular tight and adherens junctions. In contrast, mesenchymal cells exhibit anteroposterior polarity and actin stress fibres, are not attached to a basal lamina and can be motile. The epithelial-mesenchymal transition (EMT) is a phenotype shift that occurs during embryonic development and cancer metastasis. Where a cell sits on the epithelial-mesenchymal phenotypic spectrum at any given time is determined by expression of microRNAs, transcription factors and splicing regulators (Nieto et al., 2016). The miR-200 and miR-34 family microRNAs, GRHL2, OVOL1 and OVOL2 transcription factors, and ESRP1 and ESRP2 splicing regulators promote epithelial cellular identity. The ZEB1, ZEB2, SNAI1, PRRX1 and TWIST1 transcription factors and the QKI, SRSF1 and RBFOX2 splicing factors promote mesenchymal cellular identity. A number of observations link GRHL2 to regulation of EMT, particularly the strong promotion and maintenance of epithelial identity, albeit largely in non-developmental contexts. GRHL2 suppresses EMT in a breast cancer cell line by directly repressing the ZEB1 promoter (Cieply et al., 2013, 2012), while key EMT suppressors are directly transactivated by GRHL2, including MIR200B, MIR200A, MIR429 (Chung et al., 2016) and Ovol2 (Aue et al., 2015).
In this paper, we demonstrate that GRHL2 maintains the cellular identity of palatal epithelium by transactivating EMT-suppressing microRNAs, transcription factors and splicing regulators. These findings indicate that the pathway by which GRHL2 suppresses EMT in cancer is also crucial for palatogenesis.
RESULTS
The MXPs of Grhl2−/− embryos are smaller than those of wild-type littermates at E10.5
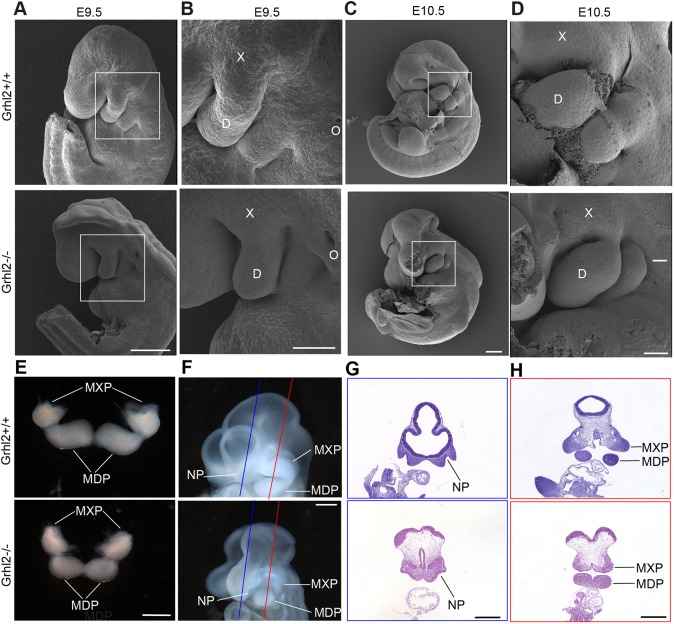
Grhl2 is expressed in the epithelium lining the maxillary and nasal processes at E10.5 (Brouns et al., 2011), with expression continuing in oral epithelium until E17.5 (Auden et al., 2006). This expression pattern is consistent with Grhl2 playing a role in closure of the palate. As Grhl2−/− embryos have a cleft face, we first determined whether their maxillary and nasal processes formed normally between E9.5 and E10.5. Scanning electron micrographs revealed that the MXPs were abnormally small in Grhl2−/− embryos at E10.5 (Fig. 1A-D). The mean ratio of MXP/MDP area was 0.96 for wild-type embryos and 0.78 for Grhl2−/− embryos, although this difference was not statistically significant. Next, we dissected the first pharyngeal arch (PA1) at E10.5, and noted again that the Grhl2−/− MXPs were reduced in size (Fig. 1E). Images of the head of E10.5 embryos revealed that the nasal processes were malformed in Grhl2−/− embryos, making it infeasible to determine whether the lambdoid junction had formed (Fig. 1F). We cut defined section planes through the nasal and maxillary processes of E10.5 embryos and determined that the nasal pits were present in Grhl2−/− embryos, as were the MXPs, which stained darkly with Haematoxylin and Eosin (H&E) (Fig. 1G,H). These findings showed that Grhl2−/− embryos have small MXPs at E10.5. In order to determine whether Grhl2−/− embryos had a deficiency of cranial neural crest cells, we stained coronal sections through E10.5 PA1 for SOX9. Grhl2−/− embryos exhibited normal numbers of SOX9-positive neural crest cells in their MXP and mandibular process (MDP) mesenchyme at E10.5 (Fig. S1A,B). In order to determine whether GRHL2 is required in neural crest cells for palate closure, we conditionally inactivated Grhl2 using Wnt1Cre (Danielian et al., 1998). Grhl2+/−;Wnt1Cre+×Grhl2fl/fl timed matings were performed and the embryos harvested at E17.5. Of 55 embryos harvested, 16 (29%) were Grhl2fl/−;Wnt1Cre+. This was not significantly different from the Mendelian expectation of 25%, indicating that embryos lacking Grhl2 in neural crest cells were viable to E17.5. Furthermore, skeletal preparations stained with Alizarin Red and Alcian Blue revealed normal palate closure in Grhl2fl/−;Wnt1Cre+ conditional knockout embryos at E17.5 (Fig. S1C). These findings indicate that GRHL2 is not required for neural crest cells to populate the first pharyngeal arch or required in neural crest cells for palate closure.
Fig. 1.
Grhl2−/− embryos have small maxillary processes. (A-D) Scanning electron micrographs of Grhl2+/+ and Grhl2−/− E9.5 (A,B) and E10.5 (C,D) embryos. Boxes indicate the location of adjacent higher-magnification images. D, mandibular process; O, otic pit; X, maxillary process. (E) PA1 dissected from E10.5 embryos. (F) Planes of sections through E10.5 embryos shown in G (blue line) and H (red line). (G,H) Coronal sections through the nasal process (G) and maxillary process (H) stained with Haematoxylin and Eosin (H&E). MDP, mandibular process; MXP, maxillary process; NP, nasal pit. Images are representative of four embryos of each genotype. Scale bars: 250 μm (A,C), 100 μm (B,D) and 500 μm (E-H).
Grhl2−/− first pharyngeal arch epithelium displays mesenchymal characteristics
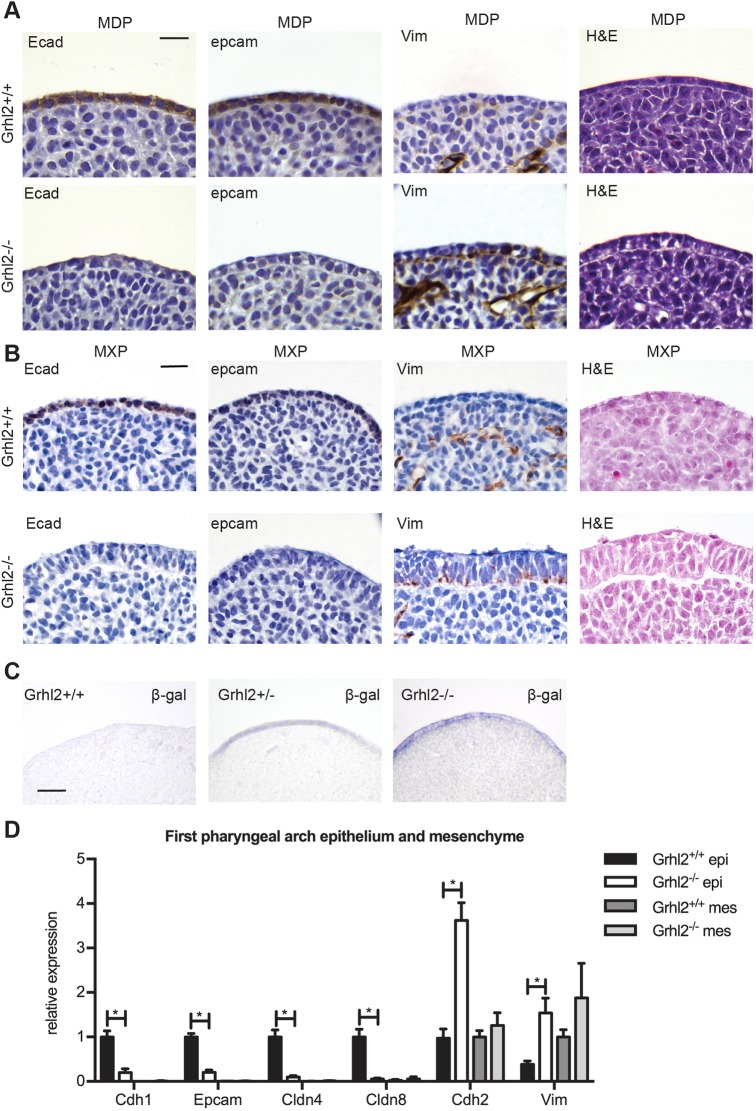
As GRHL2 is required to maintain the epithelial integrity of non-neural ectoderm at E8.5 (Ray and Niswander, 2016), we determined whether absence of this transcription factor perturbs E-cadherin and vimentin expression at E10.5. The first pharyngeal arch (PA1) comprises the MXPs, which form the palate, and the MDPs, which form the lower jaw. We collected sections through the PA1 of Grhl2+/+ and Grhl2−/− embryos at E10.5, and performed immunohistochemical staining on the MDPs and MXPs (Fig. 2A,B). The epithelial markers E-cadherin and Epcam were downregulated in Grhl2−/− epithelium. Conversely, the mesenchymal marker vimentin was ectopically expressed in Grhl2−/− epithelium in this system. Staining for β-galactosidase (which reports expression from the Grhl2 gene-targeted allele) confirmed that Grhl2 was expressed in the epithelial but not mesenchymal compartments of the PA1 (Fig. 2C). We isolated the epithelial and mesenchymal compartments of PA1 and performed quantitative reverse transcription PCR (Q-RT-PCR) using primers for epithelial (Cdh1, Epcam, Cldn4, Cldn8) and mesenchymal (Cdh2, Vim) gene transcripts. Epithelial genes were downregulated and mesenchymal genes were upregulated in Grhl2−/− PA1 epithelium compared to that of wild-type controls (Fig. 2D). In contrast, expression of these genes was unaltered in Grhl2−/− PA1 mesenchyme, in keeping with the lack of Grhl2 expression in this tissue in wild-type mice.
Fig. 2.
Grhl2−/− PA1 epithelium has mesenchymal characteristics. (A) Transverse sections through the E10.5 PA1 MDP stained by immunohistochemistry for E-cadherin (Ecad), Epcam or vimentin (Vim), or with H&E. Images are representative of four embryos. (B) As for panel A, except images are of coronal sections through the MXP. (C) Transverse sections through the MDP of E10.5 embryos stained for β-galactosidase (β-gal) activity. (D) Q-RT-PCR on E10.5 PA1 epithelium (epi) and mesenchyme (mes). Graph shows mean±s.d. n=6 embryos. Grhl2+/+ and Grhl2−/− epithelium were compared using Student's t-tests corrected for multiple comparisons with the Holm-Sidak method. *P<0.0001. Scale bars: 20 μm (A,B) and 40 μm (C).
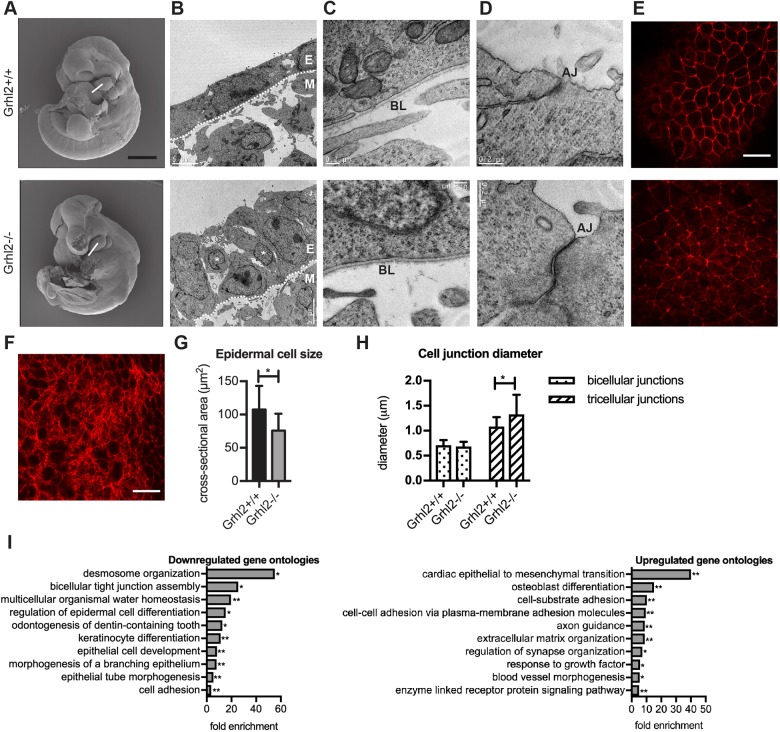
To determine which epithelial cellular characteristics were retained in the absence of GRHL2, we imaged the PA1 of E10.5 embryos using cryo-electron microscopy. Images through the MDP revealed that the wild-type arch was lined by an epithelium of single-cell thickness, with all cells in contact with the basal lamina (Fig. 3A,B). In contrast, the Grhl2−/− epithelium was thicker and presented with a disorderly cellular arrangement, with some cells not contacting the basal lamina, characteristic of ‘mesenchymal’ cell behaviour. Higher-magnification images revealed that the basal lamina was intact in Grhl2−/− embryos (Fig. 3C), and apical junctions were readily identifiable at the epithelial surface (Fig. 3D), indicating some maintenance of apico-basal polarity. Confocal microscope optical slices at various depths through the MDPs of embryos stained for filamentous actin revealed that cortical actin was retained in Grhl2−/− surface epithelium (Fig. 3E) but was not apparent in the underlying mesenchyme (Fig. 3F). Furthermore, the cortical area of the Grhl2−/− epithelial cells was reduced (Fig. 3G). Cortical actin width was normal at bicellular junctions but increased at tricellular junctions in Grhl2−/− epithelium (Fig. 3H). These data suggest that Grhl2−/− PA1 epithelium has a cellular phenotype intermediate between that of wild-type epithelium and mesenchyme.
Fig. 3.
Grhl2-/- epidermis retains some epithelial characteristics. (A) Scanning electron microscopy (SEM) images of E10.5 Grhl2+/+ and Grhl2-/- embryos. Images are repeated from Fig. 1C to illustrate the plane of sections used for transmission electron microscopy (TEM; white line). (B) 5000× TEM image of Grhl2−/− PA1 epithelium (E) and underlying mesenchyme (M). Asterisks indicate epithelial cells not contacting the basal lamina (dotted line). (C) 70,000× TEM image of the basal end of a PA1 epithelial cell. BL, basal lamina. (D) 120,000× TEM image of the apical end of a PA1 epithelial cell. AJ, apical junction. (E) Confocal microscopy slice through MDP epithelium after staining with rhodamine-phalloidin. Red staining shows the cortical actin ring that surrounds each epithelial cell. (F) An orthoslice through the mesenchymal region of a MDP showing actin stress fibres. SEM, TEM and confocal images are representative of four embryos. (G) PA1 epithelial cell size measured from images in E and similar images of three other Grhl2+/+ and three other Grhl2−/− embryos. Cross-sectional area of 20 MDP epithelial cells was measured in each embryo. *P<0.0001 using Mann–Whitney test. (H) PA1 epithelial junction diameter measured from images similar to those in E. 110-163 bicellular junctions and 96-104 tricellular junctions were measured. *P<0.0001 using Mann–Whitney test. (I) Most strongly downregulated and upregulated gene ontologies in Grhl2−/− PA1. *P<0.05, **P<0.01 using binomial test with Bonferroni correction. Graphs in G and H show mean±s.d. Scale bars: 500 μm (A), 5 μm (B), 0.2 μm (C,D) and 20 μm (E,F).
To further examine the shift towards mesenchymal gene expression in the Grhl2−/− epithelium, we performed RNA sequencing (RNAseq) on the dissected PA1 at E10.5. We identified 163 genes that were downregulated (Table S1) and 117 genes that were upregulated (Table S2) in Grhl2−/− PA1 compared to wild-type PA1. Given that the PA1 is composed of epithelial and mesenchymal cells, we expect some of the observed gene expression changes to have arisen from each compartment. We next searched for gene ontologies that were enriched in the downregulated and upregulated gene lists. The gene ontologies most strongly down-regulated in the absence of GRHL2 were integral to epithelial functions, including ‘desmosome organization’, ‘bicellular tight junction assembly’, ‘multicellular organismal water homeostasis’ and ‘regulation of epidermal cell differentiation’ (Fig. 3I). In contrast, the most significantly upregulated gene ontology in Grhl2−/− PA1 was ‘cardiac epithelial to mesenchymal transition’, due to upregulation of Has2, Tmem100, Tgfbr3, Bmp2 and Hey1. Remodelling of the primitive heart into a mature, four-chambered heart requires the formation of endocardial cushion tissue in the atrioventricular canal and outflow tract. Endocardial cells undergo EMT to delaminate and repopulate underlying extracellular matrix to form the cushion tissues. HAS2 synthesizes the extracellular matrix component hyaluronan, required for endocardial cell EMT (Camenisch et al., 2000). TMEM100 is a transmembrane protein and HEY1 is a transcription factor also required for endocardial cell EMT (Fischer et al., 2007; Mizuta et al., 2015). BMP2 binds to the TGFBR3 cell surface receptor to mediate EMT in chick endocardial cushion explants (Kirkbride et al., 2008). This indicated that a widespread shift in gene expression from epithelial to mesenchymal occurs in the absence of GRHL2. Table S3 lists genes from key ontologies that were downregulated in Grhl2−/− PA1. Table S4 lists genes from key ontologies that were upregulated in Grhl2−/− PA1. We next performed gene set enrichment analysis to determine whether defined sets of genes showed significant, concordant differences between wild-type and Grhl2−/− PA1 (Subramanian et al., 2005). By interrogating the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp), we found a significant correlation with a set of epithelial genes that are repressed by ZEB1 (Aigner et al., 2007) (Fig. S2). These genes were upregulated by knockdown of ZEB1 in a metastatic breast cancer cell line and are thus likely to be targets of ZEB1-mediated transcriptional repression. Of the 26 mouse orthologues, 17 displayed core enrichment, meaning that this subset of genes contributes most to the enrichment result. Furthermore, 11 of these 26 mouse orthologues were significantly downregulated in Grhl2−/− PA1 compared to wild-type PA1, indicating a strong inverse correlation between these datasets.
EMT transcription factors, microRNAs and splicing patterns are dysregulated in the epithelial lining of the first pharyngeal arch in Grhl2−/− embryos
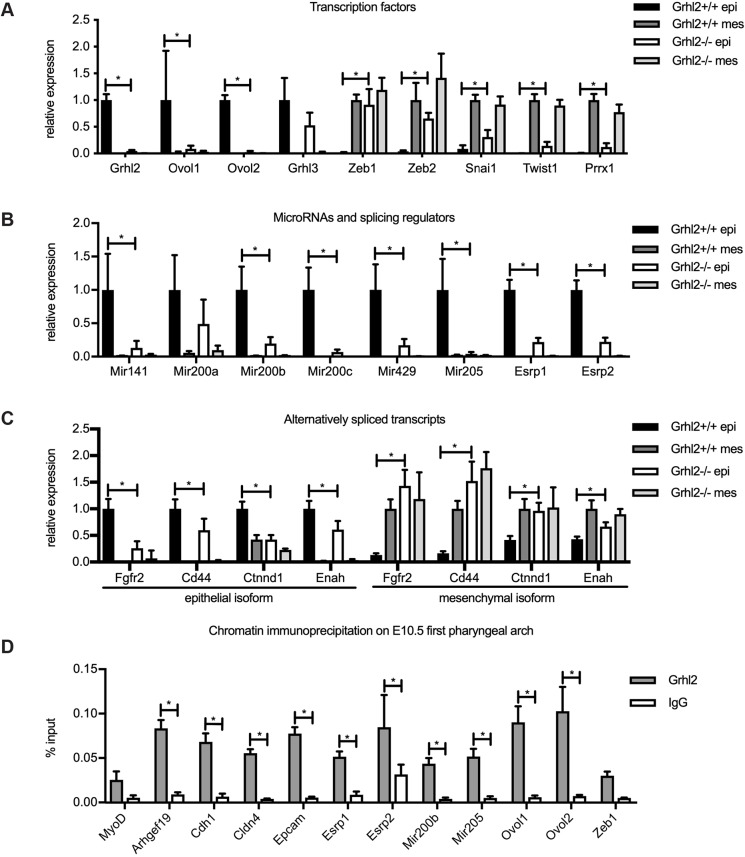
To determine the mechanism underpinning the shift towards mesenchymal phenotype in the absence of GRHL2, we quantified the expression of the transcription factors and microRNAs that regulate EMT. We performed Q-RT-PCR on PA1 epithelium and mesenchyme of Grhl2−/− and wild-type embryos at E10.5. This revealed that the Ovol1 and Ovol2 transcriptional repressors, which suppress EMT, were downregulated, whereas the Zeb1, Zeb2, Snai1, Twist1 and Prrx1 master regulators, which promote EMT, were upregulated (Fig. 4A). Of note, Zeb1 and Zeb2 were expressed at equivalent levels in Grhl2−/− epithelium as in wild-type mesenchyme, implying that they were completely dysregulated in the absence of GRHL2. In contrast, although Snai1, Twist1 and Prrx1 were also upregulated, they were still expressed at submesenchymal levels. Interestingly, of these genes, only the epithelial transcription factors were detected as differentially expressed in RNAseq (Table S1). In that dataset, Grhl2 was downregulated 8-fold, Ovol1 was downregulated 14-fold and Ovol2 was downregulated 16-fold in Grhl2−/− PA1 compared to wild-type PA1. Likely, we failed to detect misexpression of mesenchymal transcription factors in the RNAseq experiment because the bulk of PA1 is composed of mesenchyme, and hence the ectopic expression of these factors in epithelium did not significantly increase their overall expression level. Because of its known role in palate closure, we also measured the expression of Grhl3. This transcription factor displayed epithelial-specific expression but was not significantly downregulated in Grhl2−/− PA1 epithelium. Similarly, this factor was not misexpressed in the RNAseq dataset. As miR-200 microRNAs and miR-205 inhibit the expression of Zeb1 and Zeb2 (Gregory et al., 2008), we postulated that these might be downregulated in Grhl2−/− epithelium. Measurement of these microRNAs by Q-RT-PCR confirmed this, with miR-200 family microRNAs Mir141, Mir200b, Mir200c and Mir429, along with Mir205, significantly downregulated in Grhl2−/− PA1 epithelium compared to wild-type PA1 (Fig. 4B). MicroRNAs were not captured in the RNAseq experiment.
Fig. 4.
GRHL2 maintains epithelial cellular identity via multiple direct target genes. Q-RT-PCR on E10.5 PA1 epithelium (epi) and mesenchyme (mes). (A) Epithelial and mesenchymal transcription factors. (B) Zeb1-repressing microRNAs and epithelial splicing regulators. (C) Isoform-specific Q-RT-PCR for Fgfr2, Cd44, Ctnnd1 and Enah. Grhl2+/+ and Grhl2−/− epithelium were compared using Student's t-tests corrected for multiple comparisons with the Holm-Sidak method. *P<0.05. n=5-6 embryos. (D) ChIP on E10.5 PA1 with Grhl2 antibody or normal rabbit immunoglobulin (IgG) antibody, followed by PCR with primers spanning predicted GRHL2 binding sites. Results are representative of three independent experiments. *P<0.0001 by two-way ANOVA with Sidak's multiple comparison test. Graphs show mean±s.d of quadruplicate Q-RT-PCRs.
As patterns of pre-mRNA splicing change during EMT (Nieto et al., 2016), we next measured expression of the epithelial splicing regulator genes Esrp1 and Esrp2. These factors promote splicing into the epithelial isoform over the mesenchymal isoform for numerous transcripts (Warzecha et al., 2009a,b). Q-RT-PCR revealed that Esrp1 and Esrp2 were both downregulated in Grhl2−/− epithelium compared to wild-type epithelium (Fig. 4B). Similarly, Esrp1 was downregulated 4-fold and Esrp2 was downregulated 3-fold in Grhl2−/− PA1, according to RNAseq. This implied that the patterns of splicing might also be perturbed in Grhl2−/− PA1 epithelium. We explored this by designing Q-RT-PCR assays specific for the epithelial and mesenchymal isoforms of four transcripts subject to ESRP-mediated splicing – Fgfr2, Cd44, Ctnnd1 and Enah. In all four cases, the epithelial isoform was downregulated and the mesenchymal isoform upregulated in Grhl2−/− compared to wild-type epithelium (Fig. 4C). These results indicated that Grhl2−/− epithelium displays a partial shift towards a mesenchymal pattern of splicing.
To identify the target genes by which GRHL2 maintains the epithelial phenotype, we initially analysed four published GRHL2 chromatin immunoprecipitation (ChIP) massively parallel sequencing (ChIP-seq) datasets (Aue et al., 2015; Chung et al., 2016; Gao et al., 2013; Walentin et al., 2015), and identified regions that showed peaks in multiple datasets and contained evolutionarily conserved GRHL recognition motifs (AACCGGTT) (Ting et al., 2005). Putative GRHL2 binding sites were found in the Mir200b/Mir200a/Mir429 promoter, Mir205 enhancer, Ovol1 promoter, Ovol2 promoter, Esrp1 enhancer and Esrp2 intron 2. Notably, there were no peaks in the promoter or enhancer regions of Zeb1 and Zeb2, suggesting that these are not direct targets of GRHL2 in the tissues used for ChIP-seq. We designed primers to span these putative binding sites and performed ChIP on chromatin pooled from 25 E10.5 PA1 samples. We also designed primers to amplify a predicted GRHL2 binding site in the Zeb1 promoter, previously reported but not seen in ChIP datasets (Cieply et al., 2013). In comparison with the IgG control, the anti-GRHL2 antibody enriched precipitated chromatin for the well-defined binding sites in the known targets Arhgef19, Cdh1, Cldn4 and Epcam (Fig. 4D). We also observed specific binding of GRHL2 to the predicted sites in the Esrp1, Esrp2, Mir200b, Mir205, Ovol1 and Ovol2 loci, but not to the Zeb1 promoter or the negative control Myod promoter. Furthermore, significant GRHL2 enrichment at the binding sites near Esrp1, Esrp2, Mir205, Ovol1 and Ovol2 did not extend to loci spaced 1-1.5 kb away (Fig. S3). This suggests that GRHL2 maintains the epithelial phenotype by transactivating Ovol1, Ovol2, Mir200b, Mir205, Esrp1 and Esrp2, but not by directly repressing Zeb1. Rather, our data suggest that GRHL2 may indirectly repress Zeb1 by transactivating miR-200 microRNAs.
Palate closure is restored in Grhl2−/−;Zeb1−/− embryos
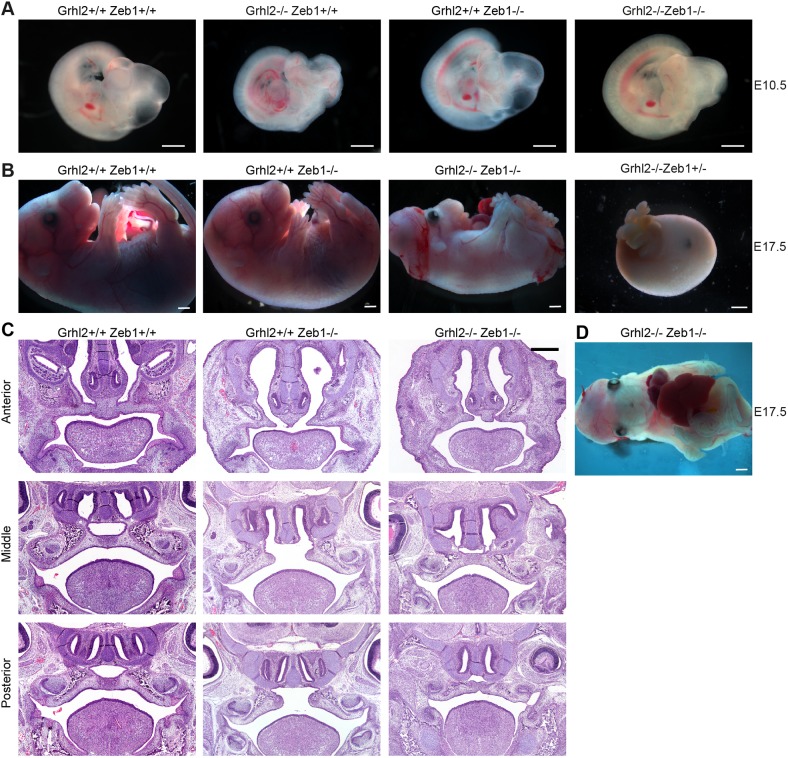
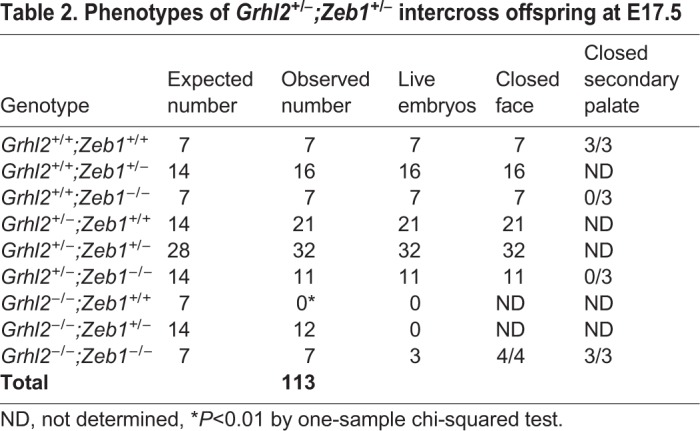
Given the central role of Zeb1 in the regulation of EMT, we asked whether inactivation of that gene could restore epithelial identity in the absence of GRHL2. Previous studies have shown that inactivation of Zeb1 in mice leads to cleft palate associated with ectopic expression of E-cadherin and loss of vimentin expression in the palatal mesenchyme (Liu et al., 2008; Takagi et al., 1998). We reasoned that the balance between epithelial and mesenchymal gene expression may be restored sufficiently in Grhl2−/−;Zeb1−/− embryos for palate closure to occur. In order to answer this question, we intercrossed the Grhl2− and Zeb1− mouse lines. Importantly, both were on the C57BL/6J background, indicating that any phenotypic rescue that occurred would not be due to mixed genetic background. We first analysed offspring of Grhl2+/−;Zeb1+/−×Grhl2+/−;Zeb1+/− timed matings at E10.5 (Fig. 5A). Chi-squared tests revealed that all genotypes were present at expected Mendelian ratios at this stage of development (Table 1). Due to Grhl2−/−;Zeb1−/− embryos occurring at 1/16 in this cross, we were not able to determine whether their facial closure at E10.5 was significantly rescued. We next analysed offspring of these timed matings at E17.5 (Table 2). Chi-squared tests revealed that Grhl2−/−;Zeb1+/+ embryos were absent by this stage of development, while all other embryos were present at expected Mendelian ratios (Table 2). Of the seven Grhl2−/−;Zeb1−/− embryos we collected at this stage of development, three were alive, 6 days after Grhl2-null mice succumb (Rifat et al., 2010). The seven Grhl2−/−;Zeb1−/− embryos displayed a range of phenotypes, with live embryos displaying fully penetrant exencephaly and open posterior neuropore, as seen in Grhl2-null embryos (Fig. 5B). They also displayed thoracogastroschisis (Fig. 5D), a phenotype observed in a previously described ENU-generated Grhl2 mutant line that also exhibits extended survival compared to the null line (Pyrgaki et al., 2011). However, major phenotypic differences were also observed, with loss of ZEB1 allowing complete facial fusion in the four embryos that developed sufficiently for this to be determined. Twelve Grhl2−/−;Zeb1+/− embryos were present at E17.5, but all were dead and displaying a distinct phenotype from Grhl2−/−;Zeb1−/− embryos. These embryos had a ‘boiled egg’ appearance, with their only recognizable features being gastroschisis and an eye. No Grhl2−/−;Zeb1+/+ live or dead embryos were detected at E17.5. This gene dosage effect confirmed that ZEB1 levels mediated rescue of Grhl2−/− lethality.
Fig. 5.
Secondary palate closure is rescued in Grhl2−/−;Zeb1−/− embryos. (A) Images at E10.5 showing cleft face in Grhl2−/− embryo and closed face in Grhl2−/−;Zeb1−/− embryo. (B) Images at E17.5 showing viable Grhl2−/−;Zeb1−/− embryo with closed face, gastroschisis, uncovered eyes, exencephaly and spina bifida, and a dead Grhl2−/−;Zeb1+/− embryo with a round, smooth appearance and gastroschisis. (C) Coronal sections through the palate of E17.5 embryos at anterior, middle and posterior levels. (D) Ventral view of Grhl2−/−;Zeb1−/− embryo at E17.5. Scale bars: 1 mm (A,B,D) and 500 μm (C).
Table 1.
Phenotypes of Grhl2+/−;Zeb1+/− intercross offspring at E10.5
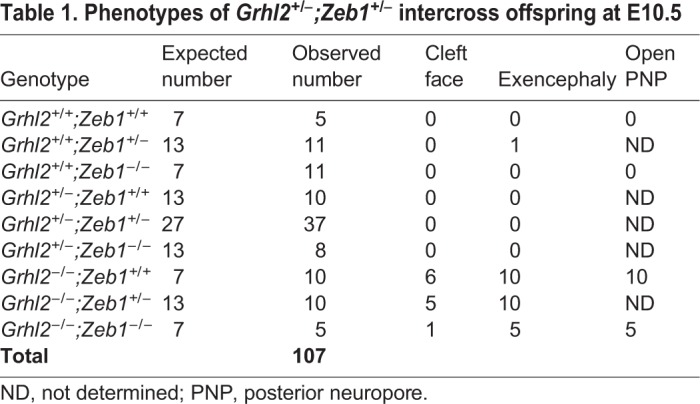
Table 2.
Phenotypes of Grhl2+/−;Zeb1+/− intercross offspring at E17.5
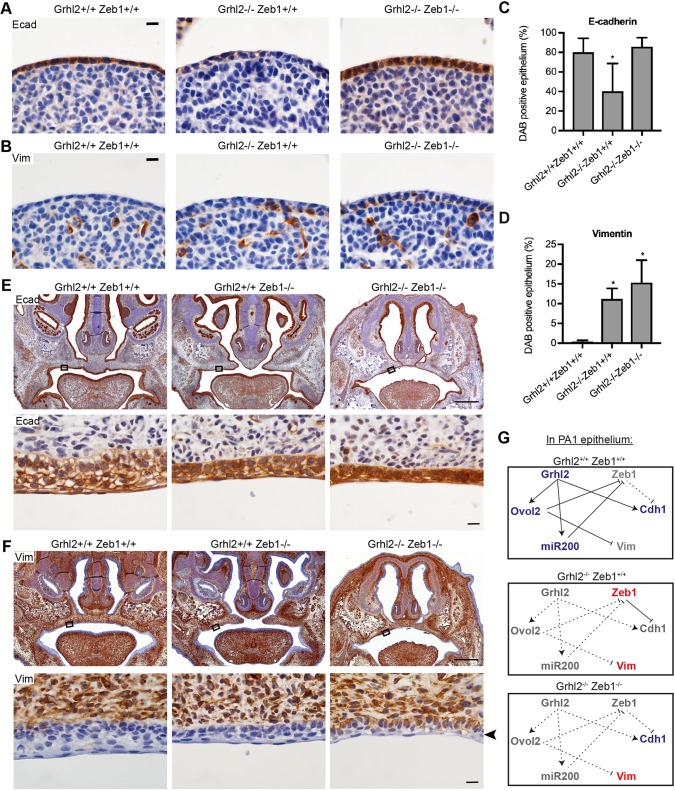
Live Grhl2−/−;Zeb1−/− embryos also displayed closure of the secondary palate, although the primary palate did not fully close (Fig. 5C; Fig. S4). The palate of Grhl2+/−;Zeb1−/− embryos remained fully cleft, indicating that rescue of secondary palate closure requires inactivation of both alleles of Grhl2. Transverse sections through E10.5 PA1 revealed that E-cadherin expression was restored to normal in Grhl2−/−;Zeb1−/− PA1 epithelium, although ectopic vimentin expression was still observed (Fig. 6A-D). Similarly, Grhl2−/−;Zeb1−/− palate epithelium co-expressed E-cadherin and vimentin at E17.5 (Fig. 6E,F). This indicated that ZEB1 likely represses the Cdh1 locus in Grhl2−/− embryos. However, it is not clear that de-repression of this locus contributes to normalization of palate closure. Ectopic expression of E-cadherin in palatal mesenchyme was not observed in E17.5 Zeb1−/− embryos. Also, Grhl2 mRNA was not elevated in PA1 mesenchyme of E10.5 Zeb1−/− embryos (Fig. S5). These results indicate that GRHL2 activity underlies failed closure of Zeb1−/− secondary palate.
Fig. 6.
E-cadherin expression is rescued in Grhl2−/−;Zeb1−/− palatal epithelium. (A,B) Immunohistochemistry on transverse sections through E10.5 PA1 for epithelial marker E-cadherin (Ecad; A) or mesenchymal marker vimentin (Vim; B). Images are representative of four embryos. (C,D) Quantification of the proportion of E10.5 PA1 epithelium staining for Ecad (C) or Vim (D). n=4 embryos. Graphs show mean±s.d. *P<0.05 versus wild type by one-way ANOVA with Tukey's multiple comparison test. (E,F) Coronal sections of E17.5 embryo heads stained with immunohistochemistry for Ecad (E) or Vim (F). Bottom rows show high-magnification images of regions of the palatal epithelium indicated by boxes. Arrowhead indicates vimentin-positive Grhl2−/−;Zeb1−/− palatal epithelium. Images are representative of three embryos of each genotype. (G) Schematic showing the pathways by which GRHL2 maintains epithelial cellular identity in PA1 epithelium. Blue genes promote the epithelial phenotype, red genes promote the mesenchymal phenotype and grey genes are not expressed. In wild-type embryos (top), GRHL2 directly transactivates Cdh1, Ovol2 and miR-200 microRNAs. The latter two factors repress Zeb1 and Vim. In the absence of GRHL2 (middle), ectopically expressed ZEB1 represses Cdh1, and the lack of OVOL2-mediated repression leads to ectopic Vim expression. In the absence of both GRHL2 and ZEB1 (bottom), Cdh1 and Vim are free from transcriptional repression so are both expressed. Scale bars: 500 μm (top rows in E,F) and 10 μm (A,B, bottom rows in E,F).
DISCUSSION
We have shown that the epithelial abnormalities observed in Grhl2−/− non-neural ectoderm at E8.5 (Ray and Niswander, 2016) also occur in PA1 epithelium at E10.5. This implies that other aspects of the Grhl2 mutant mouse phenotype, such as thoracogastroschisis (Pyrgaki et al., 2011), are also a consequence of epithelial dysfunction. Furthermore, we have demonstrated that numerous key regulators of EMT are perturbed in Grhl2−/− embryos, and that the miR-200 family microRNAs, Ovol1/Ovol2 transcription repressors and Esrp1/Esrp2 splicing regulators are direct targets of GRHL2 in PA1 epithelium. Zeb1/Zeb2 are de-repressed in Grhl2−/− PA1 to equivalent expression levels as in mesenchyme, while Snai1, Twist1 and Prrx1 are expressed at submesenchymal levels. Therefore, we hypothesize that upregulation of Zeb1/Zeb2 is a driving factor in the shift from epithelial to mesenchymal phenotype in the absence of GRHL2. Our ChIP analysis of E10.5 PA1 shows that GRHL2 does not bind a previously identified binding site in the Zeb1 promoter (Cieply et al., 2012). Rather, Zeb1 is likely suppressed in PA1 epithelium by OVOL1/OVOL2 at the transcriptional level and by miR-200 microRNAs at the post-transcriptional level. Therefore, we hypothesize that failure to transactivate the Ovol1, Ovol2 and Mir200b/Mir200a/Mir429 loci are the key events leading to EMT in Grhl2−/− epithelium. This is consistent with the observation that GRHL2 functions as both an activator and a repressor of transcription (Aue et al., 2015). Esrp1, Sostdc1, Fermt1, Tmprss2 and Lamc2 have been previously identified as key GRHL2 target genes that suppress EMT in non-neural ectoderm (Ray and Niswander, 2016). Although Snai1 and Zeb2 were not detected in Grhl2−/− non-neural ectoderm (Ray and Niswander, 2016), it would be interesting to determine if Zeb1, Twist1, Prrx1, Ovol1, Ovol2, Esrp2 and miR-200 microRNAs are expressed in these cells. The similarity in cellular phenotype between Grhl2−/− non-neural ectoderm at E8.5 and PA1 epithelium at E10.5 suggests that the same EMT pathways are likely perturbed in the two developmental contexts.
GRHL2 suppresses EMT and has an expression pattern inversely correlated with that of ZEB1 in numerous cancers (Chen et al., 2016; Chung et al., 2016; Nishino et al., 2017; Paltoglou et al., 2017; Pan et al., 2017; Quan et al., 2014; Werner et al., 2013; Xiang et al., 2017). The miR-200 family of microRNAs, including MIR141, MIR200A, MIR200B, MIR200C and MIR429, along with MIR205, bind to the ZEB1 and ZEB2 mRNAs and inhibit their translation (Gregory et al., 2008). GRHL2 upregulates expression of the MIR141, MIR200A, MIR200B, MIR200C and MIR429 microRNAs by binding to their regulatory elements in a number of cancers (Chen et al., 2016; Chung et al., 2016; Cieply et al., 2012). Furthermore, ZEB1 directly represses the MIR200C/MIR141 locus in colorectal cancer cells (Burk et al., 2008). Although GRHL2 directly represses the ZEB1 promoter in breast cancer (Cieply et al., 2012), our findings indicate that GRHL2 does not bind the Zeb1 promoter in E10.5 PA1 epithelium. Similarly, ChIP-seq experiments showed no binding of GRHL2 to the Zeb1 or Zeb2 promoters in kidney, placenta, ovarian cancer or lung epithelial cells (Aue et al., 2015; Chung et al., 2016; Gao et al., 2013; Walentin et al., 2015). Interestingly, ZEB1 binds and represses the GRHL2 promoter in breast cancer (Cieply et al., 2013; Werner et al., 2013). These observations indicate that the GRHL2/miR-200 and ZEB1/ZEB2 factors antagonize one another's expression.
Perturbation of other transcription factors in Grhl2−/− PA1 epithelium likely contributes to the mesenchymal phenotype. Like Grhl2−/− mouse embryos, Ovol2−/− embryos die mid-gestation with an open cranial neural tube (Mackay et al., 2006; Rifat et al., 2010). The phenotypic similarity between these knockout embryos suggests that Ovol2 may be a key GRHL2 target during embryogenesis. Ovol1−/− mice present with subtle epidermal defects including abnormal hairs, expansion of epidermal progenitors and delayed acquisition of the skin barrier (Dai et al., 1998; Nair et al., 2006; Teng et al., 2007). As Ovol2 is upregulated in Ovol1−/− epidermis, mice that lack expression of both Ovol1 and Ovol2 in epidermis were created (Lee et al., 2014). Keratinocytes derived from these embryos had mesenchymal characteristics including a stress fibre type of actin cytoskeleton, upregulation of ZEB1, SNAI2, vimentin, fibronectin, smooth muscle actin and N-cadherin, and downregulation of α-catenin. The epithelial phenotype of these keratinocytes was restored by knockdown of Zeb1. Furthermore, OVOL2 directly repressed the Zeb1 promoter in keratinocytes. These observations support the idea that OVOL2-mediated repression of Zeb1 is key to maintenance of the epithelial phenotype of PA1 epithelium.
The epithelial splicing regulatory proteins ESRP1 and ESRP2 have an epithelial-specific pattern of expression and are downregulated during EMT (Warzecha et al., 2009a). These factors promote splicing of pre-mRNAs into the epithelial isoform for Fgfr2, Ctnnd1, Enah, Cd44 and other genes (Warzecha et al., 2009b). Interestingly, ZEB1 directly represses the ESRP1 locus in breast cancer cells (Preca et al., 2015). Esrp1−/− mice have bilateral cleft lip and palate and die neonatally, whereas Esrp2−/− mice have no overt phenotype (Bebee et al., 2015). Esrp1−/−;Esrp2−/− mice also display agenesis of lungs and salivary glands, but survive until E18.5. This indicates that absence of ESRP1/ESRP2 does not reconstitute all aspects of the Grhl2−/− embryo phenotype.
The secondary palate closed in viable Grhl2−/−;Zeb1−/− embryos but remained cleft in Zeb1−/− littermates at E17.5. This implies that GRHL2 activity in Zeb1−/− embryos disrupts closure. It is not possible to deduce the precise mechanism underlying the rescue from our work. However, the lack of ectopic Grhl2 expression in Zeb1−/− PA1 mesenchyme at E10.5 and the absence of ectopic E-cadherin expression in Zeb1−/− palatal mesenchyme at E17.5 argue against a simple restoration of mesenchymal cell identity. Rather, the most plausible explanation is restoration of an epithelial-mesenchymal signalling pathway that promotes proliferation of palatal shelf mesenchyme. By comparison with Grhl2NiswNisw embryos that display cleft upper jaw at E18.5 (Pyrgaki et al., 2011), Grhl2−/−;Zeb1−/− embryos successfully closed their face and secondary palate at E17.5. This implies that ectopic expression of ZEB1 underlies failure of Grhl2Nisw/Nisw secondary palate to close. More broadly, our results imply that both epithelial and mesenchymal transcription factors are required for palate closure and that they cooperatively effect morphogenesis.
Ectopic expression of ZEB1 has been hypothesized to underlie the neural tube defects in Grhl2 mutant mouse embryos (Ray and Niswander, 2016). We have demonstrated that this is not the case in the hindbrain and tail regions, as Grhl2−/−;Zeb1−/− embryos displayed fully penetrant exencephaly and open posterior neuropore at E10.5. Grhl2−/−;Zeb1−/− embryos sometimes survive until E17.5, while Grhl2−/− embryos die at E11.5. This indicates that ZEB1 expression in Grhl2−/− embryos underlies mid-gestation lethality. Although the cause of this early lethality is unknown, cardiac defects are a likely candidate as heart development is abnormal in Grhl2Nisw/Nisw embryos (Pyrgaki et al., 2011) and numerous fusion events occur during heart development (Ray and Niswander, 2012).
In conclusion, we have demonstrated that GRHL2 maintains epithelial cellular identity in PA1 epithelium via multiple pathways (Fig. 6G). Restoration of E-cadherin expression in Grhl2−/−;Zeb1−/− epithelium implies that ZEB1-mediated repression has a dominant effect on Cdh1 expression over GRHL2-mediated transactivation. Similarly, ectopic expression of vimentin in Grhl2−/−;Zeb1−/− epithelium implies that OVOL2-mediated repression has a dominant effect on Vim expression. Grhl2−/− maxillary epithelium maintains a cellular phenotype and gene expression patterns intermediate between that of wild-type epithelium and mesenchyme. This implies that either a co-expressed transcription factor, or epigenetic marks laid down at an earlier stage of development, maintain some epithelial gene expression in these cells. It would be of interest to identify this factor, which reveals that GRHL2 is not the sole driver of epithelial phenotype during palate closure.
MATERIALS AND METHODS
Mouse lines
Use of animals conformed to the Australian code for the care and use of animals for scientific purposes. Experiments involving animals were approved by the Alfred Research Alliance Animal Ethics Committee (application number E/1200/2012/M). Mice carrying the Grhl2-null allele were maintained by heterozygote intercrosses and genotyped as described (Rifat et al., 2010). To collect embryos, mice were mated in the afternoon and females checked for a vaginal plug the following morning. Embryos were harvested at 12:00 and yolk sac DNA used for genotyping. Mice carrying a conditional allele of Zeb1 (Brabletz et al., 2017) were crossed with mice carrying a CMV-Cre transgene (Schwenk et al., 1995). This deletes exon 6 of Zeb1, leading to a premature termination of translation and resulting in a phenotype equivalent to that of other Zeb1−/− mice (Brabletz et al., 2017). Mice carrying this Zeb1Δ allele, referred to as Zeb1− in this paper, were maintained by heterozygote intercrosses and genotyped as described (Brabletz et al., 2017). Grhl2− and Zeb1− mouse lines were both back-crossed ten times to C57BL/6J before this study commenced. Zeb1+/− mice were mated with Grhl2+/− mice and the resulting Grhl2+/−;Zeb1+/− offspring intercrossed as time matings. Sex of embryos was not determined. Mice carrying the Wnt1Cre transgene have previously been described (Danielian et al., 1998). Mice carrying the Grhl2fl allele were maintained as a homozygous line and genotyped as described (Kersbergen et al., 2018).
Immunohistochemistry
E10.5 embryos were fixed in 4% paraformaldehyde (PFA) at 4°C overnight and then orientated in 2% low-melting-point agarose in PBS. E17.5 embryo heads were fixed similarly then processed without agarose orientation. Samples were processed using a Leica ASP300S and embedded into paraffin. Embryo heads were sectioned in a coronal orientation, while E10.5 embryos were sectioned in a transverse or coronal orientation. Five-micrometre sections were cut onto Superfrost plus slides, and immunohistochemistry was performed using standard 3,3′-diaminobenzidine (DAB) protocols. The antibodies used were anti-E-cadherin (3195, batch 02/2017, Cell Signaling Technology; 1:100), anti-vimentin (5741S, batch 04/2017, Cell Signaling Technology; 1:100) and anti-Epcam (ab71916, batch GR231753-3, Abcam; 1:1200). Validation profiles for these antibodies are available on the supplier websites. H&E staining was performed using standard protocols. Immunohistochemical staining was quantified with ImageJ software using image deconvolution and a mask for DAB-positive areas.
Immunofluorescence
E10.5 embryos were fixed in 4% PFA, cryoprotected in 30% sucrose and embedded in OCT. Then, 10 μm sections were permeabilized with 0.25% Triton X-100 in PBS and blocked with 20% normal goat serum, 0.3% Triton X-100 in PBS. Sections were stained with rabbit monoclonal antibody against SOX9 (ab185966, Abcam) at 1:500 in 2% bovine serum albumin, 0.3% Triton X-100 in PBS overnight. After washing, sections were stained with goat polyclonal antibody against rabbit IgG conjugated to Alexa Fluor 594 (ab150080, Abcam; 1:1000) for 2 h. Sections were counterstained with 1 μg/ml Hoechst 33342 for 15 min, mounted in Vectastain mounting medium for fluorescence (H-1000, Vector Laboratories), coverslipped and cured at 4°C overnight. Sections were imaged with a Nikon A1r confocal microscope using a 20× multi-immersion objective and SOX9-positive cells were quantified using ImageJ software.
Phalloidin staining
Embryos stored in 4% PFA were washed in PBS 3×5 min, permeabilized in 0.5% Triton X in PBS for 15 min, washed in PBS 2×2 min and blocked in 10% normal goat serum, 0.1% Triton X in PBS for 1 h. Samples were incubated in block containing rhodamine-phalloidin (1:1000; Invitrogen R415) and 1 μM Hoechst 33342 (Sigma-Aldrich) at 4°C overnight then washed with 0.1% Triton X in PBS 3×10 min. Samples were orientated onto 0.17±0.01 mm thick coverslips in 1% low-melting-point agarose in PBS and imaged using a Nikon A1r confocal microscope (Tokyo, Japan). Samples were imaged with a 60× 1.27 WI Plan Apo water immersion objective with 2× zoom. Five slices spaced 0.5 μm apart were collected using a channel series, with the depth set to image through the epithelial layer. Cell cross-sectional area and diameter of bicellular and tricellular junctions were measured using ImageJ software.
Electron microscopy
For scanning electron microscopy (SEM), embryos were fixed overnight in 2.5% glutaraldehyde in PBS at 4°C, then rinsed in PBS for 2×10 min. Embryos were treated with 1-2.5% osmium tetroxide for 1 h then washed 2× in 0.1 M sodium cacodylate, followed by dehydration in the following washes: 50% ethanol (30 min), 70% ethanol (30 min), 95% ethanol (30 min) and 100% ethanol (2×30 min). Embryos were incubated in 50% ethanol 50% hexamethyldisilazane (HMDS; Pro Sci Tech) for 15 min, followed by 2×15 min washes in 100% HMDS. Excess HMDS was removed, and the embryos were left to dehydrate in a fume hood overnight. Embryos were mounted onto stubs with carbon tabs, sputter coated with gold using a Bal-Tec SCD 005 sputter coater and visualized using a Hitachi S-570 scanning electron microscope at 10 kV.
For transmission electron microscopy (TEM), the hind limbs and tail were removed and embryos fixed for 2 h at room temperature in 2% paraformaldehyde, 2.5% glutaraldehyde, 0.1 M sodium cacodylate, 250 μM CaCl2, 500 μM MgCl2. Embryos were then rinsed in 0.1 M sodium cacodylate and placed in 1% OsO4, 1.5% K3Fe(III)CN6, 0.065 M Na-cacodylate buffer (pH 7.4) for 1 h. Samples were rinsed in deionized water then dehydrated through a graded ethanol and propylene oxide series into epon resin. Samples were polymerized for 48 h at 60°C and 80 nm transverse sections through the MDP were mounted onto metal grids. Sections were imaged using a Hitachi H7500 microscope with a Gatan Multiscan 791 CCD camera between 5000× and 120,000× magnification.
RNAseq
The PA1s of E10.5 mouse embryos were snap frozen on liquid nitrogen. Yolk sac DNA was used to determine embryo sex as described (Lambert et al., 2000). Only male samples were used for RNAseq to minimize gene expression differences due to sex. RNA from five wild-type and five Grhl2−/− PA1s was isolated using a QIAGEN micro RNeasy kit with on column DNase digestion. RNA integrity was determined using a Qubit and bioanalyser (Agilent). Libraries were constructed using the Illumina TruSeq Stranded Total RNA Library preparation (RiboZero) protocol. Sequencing was run over three HiSeq lanes using 50 bp single-end reads on HiSeq1500. Reads were aligned to the GRCm38 mouse reference genome using STAR aligner (Dobin et al., 2013), and reads were counted using featureCounts (Liao et al., 2014). Differential gene expression analysis was performed using limma-voom through the Degust interface (David Powell, Monash University, Melbourne, Australia) and trimmed mean of M-values (TMM) normalization was performed (Robinson and Oshlack, 2010). Degust software can be downloaded from http://degust.erc.monash.edu. FDR values were assigned to each gene and a cut-off of FDR<0.01 was used to select differentially expressed genes. Gene ontology analysis was performed using the Panther Overrepresentation Test and the Mus musculus reference list 2019-07-03 release (Ashburner et al., 2000; Mi et al., 2017) on two lists of genes: the 163 genes downregulated and the 117 genes upregulated in Grhl2−/− PA1. Bonferroni correction for multiple testing was applied. Gene set enrichment analysis was performed using GSEA v3.0 software and the molecular signatures database v5.2 (Subramanian et al., 2005).
Q-RT-PCR
PA1 epithelium and mesenchyme were isolated from E10.5 embryos as described (Li and Williams, 2013). RNA was isolated with Trisure (Bioline) according to the manufacturer's instructions, except that precipitation was performed at −30°C overnight. RNA was reverse transcribed with the transcriptor first strand cDNA synthesis kit (Roche) for mRNA or the Quantimir RT kit (#RA420A-1, Systems Biosciences) for microRNA. Quantitative PCR (qPCR) was performed with GoTaq qPCR master mix (Promega) as duplicate 20 μl reactions or triplicate 10 μl reactions. Relative expression values were calculated using the ΔΔCT method with the normalization controls Actb for mRNA or Rnu6 for microRNA. Isoform-specific Q-RT-PCR primers for Fgfr2, Cd44, Ctnnd1 and Enah spanned an exon-exon junction exclusive to either the epithelial or mesenchymal isoform. Oligonucleotide primer sequences are listed in Table S5.
ChIP
ChIP was performed essentially as described (Voss et al., 2012) on chromatin pooled from 25 E10.5 PA1s. Cross-linked chromatin was incubated with 5 μg Grhl2 antibody (HPA004820, batch B96161, Sigma-Aldrich) or 5 μg normal rabbit IgG (2729, batch 09/2009, Cell Signaling Technology). Dynabeads protein A (#10001D, Invitrogen) were used to precipitate the antibody-bound chromatin. Samples were eluted in 60 μl volume and assayed in quadruplicate Q-RT-PCR reactions using the primers listed in Table S5. The percentage input was determined by the ΔΔCt method.
Skeletal preparations
Mouse embryo heads were fixed in 80% ethanol for 1 day, dehydrated in 96% ethanol for 1 day and incubated in 75 mM Alcian Blue, 16.5 M ethanol, 3.2 M acetic acid for 3 days. Samples were rehydrated in 70% ethanol, 40% ethanol and 15% ethanol for 2 h each then in water for 1 day. Samples were cleared in 1% KOH for 2 days, stained in 15 mM Alizarin Red, 1% KOH for 4 h then washed for 2 h three times in 1% KOH. Samples were immersed in 20% glycerol 1% KOH, 50% glycerol 1% KOH then 80% glycerol 1% KOH for 1 day each prior to imaging.
Statistics
A sample size of four to six embryos was deemed sufficient to detect differences between genotypes based on previous experience using inbred mouse strains. No animals were excluded from the analyses. Randomization of animals to experimental groups was not performed and littermate controls were used wherever possible. Investigators were blinded to genotype when imaging embryos and histological sections. Statistical analyses were performed using Prism 7 for Mac OS X software. When comparing epidermal cell size, the Mann–Whitney t-test was used, as this non-parametric test does not assume a Gaussian distribution of the data. For Q-RT-PCR data, two genotypes were compared using Student's t-tests, not assuming consistent standard deviation, with P-values adjusted for multiple testing with the Holm-Sidak method. For ChIP-qPCR in Fig. 4D, two-way ANOVA was used to compare 11 different loci, with P-values adjusted for multiple comparisons using Sidak's method. In this case, a statistical significance cut-off of P<0.01 was deemed appropriate due to the slightly higher Grhl2 than IgG signal at the Myod locus. For ChIP-qPCR in Fig. S3, Grhl2 was compared to IgG using an unpaired Student's t-test. For comparison of immunohistochemical staining between three different genotypes, ordinary one-way ANOVA with Tukey's multiple comparison test was used. Chi-squared values were calculated as (observed-expected)2/expected and P-values determined for 1 degree of freedom.
Supplementary Material
Acknowledgements
We thank Kylie Spark for excellent animal husbandry. We also acknowledge the support of the Monash Histology Platform, Monash Micro Imaging, the Monash Cryo-Electron Microscopy Facility, the Monash Animal Research Platform, the Monash Medical Genomics Facility and the Monash Bioinformatics Platform.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.R.C., S.D., S.M.J.; Methodology: M.R.C., M.E.d.V., S.D., S.M.J.; Software: M.R.C., S.D., S.M.J.; Validation: M.R.C., S.D., S.M.J.; Formal analysis: M.R.C., S.D., S.M.J.; Investigation: M.R.C., A.A., T. Butt, Z.D., D.D.P., L.B.M., S.R.G., S.D., S.M.J.; Resources: M.R.C., J.J.H., C.D., S.B., T. Brabletz, M.P.S., S.D., S.M.J.; Data curation: M.R.C., S.D., S.M.J.; Writing - original draft: M.R.C., S.D., S.M.J.; Writing - review & editing: M.R.C., S.D., S.M.J.; Visualization: M.R.C., S.D., S.M.J.; Supervision: M.R.C., S.D., S.M.J.; Project administration: M.R.C., S.D., S.M.J.; Funding acquisition: S.D., S.M.J.
Funding
This work was supported by the Australian National Health and Medical Research Council (project grant 1063837) and the Australian Research Council (Discovery Early Career Researcher Award 140100500 to S.D.).
Data availability
RNAseq data are available from the Gene Expression Omnibus under accession number GSE106130.
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.042218.supplemental
References
- Aigner K., Dampier B., Descovich L., Mikula M., Sultan A., Schreiber M., Mikulits W., Brabletz T., Strand D., Obrist P. et al. (2007). The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 26, 6979-6988. 10.1038/sj.onc.1210508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T. et al. (2000). Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 25, 25-29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auden A., Caddy J., Wilanowski T., Ting S. B., Cunningham J. M. and Jane S. M. (2006). Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr. Patterns 6, 964-970. 10.1016/j.modgep.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Aue A., Hinze C., Walentin K., Ruffert J., Yurtdas Y., Werth M., Chen W., Rabien A., Kilic E., Schulzke J.-D. et al. (2015). A grainyhead-like 2/ovo-like 2 pathway regulates renal epithelial barrier function and lumen expansion. J. Am. Soc. Nephrol. 26, 2704-2715. 10.1681/ASN.2014080759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebee T. W., Park J. W., Sheridan K. I., Warzecha C. C., Cieply B. W., Rohacek A. M., Xing Y. and Carstens R. P. (2015). The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife 4, e08954 10.7554/eLife.08954.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz S., Lasierra Losada M., Schmalhofer O., Mitschke J., Krebs A., Brabletz T. and Stemmler M. P. (2017). Generation and characterization of mice for conditional inactivation of Zeb1. Genesis 55, e23024 10.1002/dvg.23024 [DOI] [PubMed] [Google Scholar]
- Bray S. J. and Kafatos F. C. (1991). Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 5, 1672-1683. 10.1101/gad.5.9.1672 [DOI] [PubMed] [Google Scholar]
- Brouns M. R., De Castro S. C. P., Terwindt-Rouwenhorst E. A., Massa V., Hekking J. W., Hirst C. S., Savery D., Munts C., Partridge D., Lamers W. et al. (2011). Over-expression of Grhl2 causes spina bifida in the Axial defects mutant mouse. Hum. Mol. Genet. 20, 1536-1546. 10.1093/hmg/ddr031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S. and Brabletz T. (2008). A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582-589. 10.1038/embor.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush J. O. and Jiang R. (2012). Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 139, 231-243. 10.1242/dev.067082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch T. D., Spicer A. P., Brehm-Gibson T., Biesterfeldt J., Augustine M. L., Calabro A. Jr., Kubalak S., Klewer S. E. and McDonald J. A. (2000). Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J. Clin. Invest. 106, 349-360. 10.1172/JCI10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Yi J. K., Shimane T., Mehrazarin S., Lin Y.-L., Shin K. H., Kim R. H., Park N.-H. and Kang M. K. (2016). Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis 37, 500-510. 10.1093/carcin/bgw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung V. Y., Tan T. Z., Tan M., Wong M. K., Kuay K. T., Yang Z., Ye J., Muller J., Koh C. M., Guccione E. et al. (2016). GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep. 6, 19943 10.1038/srep19943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B., Riley P. T., Pifer P. M., Widmeyer J., Addison J. B., Ivanov A. V., Denvir J. and Frisch S. M. (2012). Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res. 72, 2440-2453. 10.1158/0008-5472.CAN-11-4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B., Farris J., Denvir J., Ford H. L. and Frisch S. M. (2013). Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res. 73, 6299-6309. 10.1158/0008-5472.CAN-12-4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Schonbaum C., Degenstein L., Bai W., Mahowald A. and Fuchs E. (1998). The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 12, 3452-3463. 10.1101/gad.12.21.3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian P. S., Muccino D., Rowitch D. H., Michael S. K. and McMahon A. P. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323-1326. 10.1016/S0960-9822(07)00562-3 [DOI] [PubMed] [Google Scholar]
- Dixon M. J., Marazita M. L., Beaty T. H. and Murray J. C. (2011). Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet. 12, 167-178. 10.1038/nrg2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Steidl C., Wagner T. U., Lang E., Jakob P. M., Friedl P., Knobeloch K.-P. and Gessler M. (2007). Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ. Res. 100, 856-863. 10.1161/01.RES.0000260913.95642.3b [DOI] [PubMed] [Google Scholar]
- Gao X., Vockley C. M., Pauli F., Newberry K. M., Xue Y., Randell S. H., Reddy T. E. and Hogan B. L. M. (2013). Evidence for multiple roles for grainyhead-like 2 in the establishment and maintenance of human mucociliary airway epithelium.[corrected]. Proc. Natl. Acad. Sci. USA 110, 9356-9361. 10.1073/pnas.1307589110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S.-G., White N. J. and Sakasegawa A. Y. (2000). The Twirler mouse, a model for the study of cleft lip and palate. Arch. Oral Biol. 45, 87-94. 10.1016/S0003-9969(99)00101-6 [DOI] [PubMed] [Google Scholar]
- Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y. and Goodall G. J. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593-601. 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Kersbergen A., Best S. A., Dworkin S., Ah-Cann C., de Vries M. E., Asselin-Labat M.-L., Ritchie M. E., Jane S. M. and Sutherland K. D. (2018). Lung morphogenesis is orchestrated through Grainyhead-like 2 (Grhl2) transcriptional programs. Dev. Biol. 443, 1-9. 10.1016/j.ydbio.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Kirkbride K. C., Townsend T. A., Bruinsma M. W., Barnett J. V. and Blobe G. C. (2008). Bone morphogenetic proteins signal through the transforming growth factor-beta type III receptor. J. Biol. Chem. 283, 7628-7637. 10.1074/jbc.M704883200 [DOI] [PubMed] [Google Scholar]
- Krafchak C. M., Pawar H., Moroi S. E., Sugar A., Lichter P. R., Mackey D. A., Mian S., Nairus T., Elner V., Schteingart M. T. et al. (2005). Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am. J. Hum. Genet. 77, 694-708. 10.1086/497348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurima K., Hertzano R., Gavrilova O., Monahan K., Shpargel K. B., Nadaraja G., Kawashima Y., Lee K. Y., Ito T., Higashi Y. et al. (2011). A noncoding point mutation of Zeb1 causes multiple developmental malformations and obesity in Twirler mice. PLoS Genet. 7, e1002307 10.1371/journal.pgen.1002307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.-F., Benoit B. O., Colvin G. A., Carlson J., Delville Y. and Quesenberry P. J. (2000). Quick sex determination of mouse fetuses. J. Neurosci. Methods 95, 127-132. 10.1016/S0165-0270(99)00157-0 [DOI] [PubMed] [Google Scholar]
- Lan Y., Xu J. and Jiang R. (2015). Cellular and molecular mechanisms of palatogenesis. Curr. Top. Dev. Biol. 115, 59-84. 10.1016/bs.ctdb.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Villarreal-Ponce A., Fallahi M., Ovadia J., Sun P., Yu Q.-C., Ito S., Sinha S., Nie Q. and Dai X. (2014). Transcriptional mechanisms link epithelial plasticity to adhesion and differentiation of epidermal progenitor cells. Dev. Cell 29, 47-58. 10.1016/j.devcel.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie E. J., Liu H., Carlson J. C., Shaffer J. R., Feingold E., Wehby G., Laurie C. A., Jain D., Laurie C. C., Doheny K. F. et al. (2016). A genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3. Am. J. Hum. Genet. 98, 744-754. 10.1016/j.ajhg.2016.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. and Williams T. (2013). Separation of mouse embryonic facial ectoderm and mesenchyme. J. Vis. Exp. e50248 10.3791/50248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liskova P., Evans C. J., Davidson A. E., Zaliova M., Dudakova L., Trkova M., Stranecky V., Carnt N., Plagnol V., Vincent A. L. et al. (2016). Heterozygous deletions at the ZEB1 locus verify haploinsufficiency as the mechanism of disease for posterior polymorphous corneal dystrophy type 3. Eur. J. Hum. Genet. 24, 985-991. 10.1038/ejhg.2015.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., El-Naggar S., Darling D. S., Higashi Y. and Dean D. C. (2008). Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 135, 579-588. 10.1242/dev.007047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Leslie E. J., Jia Z., Smith T., Eshete M., Butali A., Dunnwald M., Murray J. and Cornell R. A. (2016). Irf6 directly regulates Klf17 in zebrafish periderm and Klf4 in murine oral epithelium, and dominant-negative KLF4 variants are present in patients with cleft lip and palate. Hum. Mol. Genet. 25, 766-776. 10.1093/hmg/ddv614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D. R., Hu M., Li B., Rhéaume C. and Dai X. (2006). The mouse Ovol2 gene is required for cranial neural tube development. Dev. Biol. 291, 38-52. 10.1016/j.ydbio.2005.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E., Bohmer A. C., Ishorst N., Hoebel A. K., Gültepe P., Schuenke H., Klamt J., Hofmann A., Gölz L., Raff R. et al. (2016). Sequencing the GRHL3 coding region reveals rare truncating mutations and a common susceptibility variant for nonsyndromic cleft palate. Am. J. Hum. Genet. 98, 755-762. 10.1016/j.ajhg.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke C., Cionni M., Siggers T., Bulyk M. L., Beier D. R. and Stottmann R. W. (2015). Grhl2 is required in nonneural tissues for neural progenitor survival and forebrain development. Genesis 53, 573-582. 10.1002/dvg.22875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D. and Thomas P. D. (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183-d189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K., Sakabe M., Hashimoto A., Ioka T., Sakai C., Okumura K., Hattammaru M., Fujita M., Araki M., Somekawa S. et al. (2015). Impairment of endothelial-mesenchymal transformation during atrioventricular cushion formation in Tmem100 null embryos. Dev. Dyn. 244, 31-42. 10.1002/dvdy.24216 [DOI] [PubMed] [Google Scholar]
- Nair M., Teng A., Bilanchone V., Agrawal A., Li B. and Dai X. (2006). Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J. Cell Biol. 173, 253-264. 10.1083/jcb.200508196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M. A., Huang R. Y.-J., Jackson R. A. and Thiery J. P. (2016). EMT: 2016. Cell 166, 21-45. 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Nishino H., Takano S., Yoshitomi H., Suzuki K., Kagawa S., Shimazaki R., Shimizu H., Furukawa K., Miyazaki M. and Ohtsuka M. (2017). Grainyhead-like 2 (GRHL2) regulates epithelial plasticity in pancreatic cancer progression. Cancer Med. 6, 2686-2696. 10.1002/cam4.1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltoglou S., Das R., Townley S. L., Hickey T. E., Tarulli G. A., Coutinho I., Fernandes R., Hanson A. R., Denis I., Carroll J. S. et al. (2017). Novel androgen receptor coregulator GRHL2 exerts both oncogenic and antimetastatic functions in prostate cancer. Cancer Res. 77, 3417-3430. 10.1158/0008-5472.CAN-16-1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Zhang R., Xie C., Gan M., Yao S., Yao Y., Jin J., Han T., Huang Y., Gong Y. et al. (2017). GRHL2 suppresses tumor metastasis via regulation of transcriptional activity of RhoG in non-small cell lung cancer. Am. J. Transl. Res. 9, 4217-4226. [PMC free article] [PubMed] [Google Scholar]
- Peyrard-Janvid M., Leslie E. J., Kousa Y. A., Smith T. L., Dunnwald M., Magnusson M., Lentz B. A., Unneberg P., Fransson I., Koillinen H. K. et al. (2014). Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am. J. Hum. Genet. 94, 23-32. 10.1016/j.ajhg.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preca B.-T., Bajdak K., Mock K., Sundararajan V., Pfannstiel J., Maurer J., Wellner U., Hopt U. T., Brummer T., Brabletz S. et al. (2015). A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int. J. Cancer 137, 2566-2577. 10.1002/ijc.29642 [DOI] [PubMed] [Google Scholar]
- Pyrgaki C., Liu A. and Niswander L. (2011). Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev. Biol. 353, 38-49. 10.1016/j.ydbio.2011.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Y., Jin R., Huang A., Zhao H., Feng B., Zang L. and Zheng M. (2014). Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer Biol. Ther. 15, 878-887. 10.4161/cbt.28877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray H. J. and Niswander L. (2012). Mechanisms of tissue fusion during development. Development 139, 1701-1711. 10.1242/dev.068338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray H. J. and Niswander L. A. (2016). Grainyhead-like 2 downstream targets act to suppress epithelial-to-mesenchymal transition during neural tube closure. Development 143, 1192-1204. 10.1242/dev.129825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat Y., Parekh V., Wilanowski T., Hislop N. R., Auden A., Ting S. B., Cunningham J. M. and Jane S. M. (2010). Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 345, 237-245. 10.1016/j.ydbio.2010.07.017 [DOI] [PubMed] [Google Scholar]
- Robinson M. D. and Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F., Baron U. and Rajewsky K. (1995). A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080-5081. 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.-O., Nakagawa E., Kim E.-J., Cho K.-W., Lee J.-M., Cho S.-W. and Jung H.-S. (2012). miR-200b regulates cell migration via Zeb family during mouse palate development. Histochem. Cell Biol. 137, 459-470. 10.1007/s00418-012-0915-6 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T., Moribe H., Kondoh H. and Higashi Y. (1998). DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development 125, 21-31. [DOI] [PubMed] [Google Scholar]
- Teng A., Nair M., Wells J., Segre J. A. and Dai X. (2007). Strain-dependent perinatal lethality of Ovol1-deficient mice and identification of Ovol2 as a downstream target of Ovol1 in skin epidermis. Biochim. Biophys. Acta 1772, 89-95. 10.1016/j.bbadis.2006.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S. B., Caddy J., Hislop N., Wilanowski T., Auden A., Zhao L. L., Ellis S., Kaur P., Uchida Y., Holleran W. M. et al. (2005). A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308, 411-413. 10.1126/science.1107511 [DOI] [PubMed] [Google Scholar]
- Voss A. K., Dixon M. P., McLennan T., Kueh A. J. and Thomas T. (2012). Chromatin immunoprecipitation of mouse embryos. Methods Mol. Biol. 809, 335-352. 10.1007/978-1-61779-376-9_23 [DOI] [PubMed] [Google Scholar]
- Walentin K., Hinze C., Werth M., Haase N., Varma S., Morell R., Aue A., Potschke E., Warburton D., Qiu A. et al. (2015). A Grhl2-dependent gene network controls trophoblast branching morphogenesis. Development 142, 1125-1136. 10.1242/dev.113829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha C. C., Sato T. K., Nabet B., Hogenesch J. B. and Carstens R. P. (2009a). ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell 33, 591-601. 10.1016/j.molcel.2009.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha C. C., Shen S., Xing Y. and Carstens R. P. (2009b). The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 6, 546-562. 10.4161/rna.6.5.9606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S., Frey S., Riethdorf S., Schulze C., Alawi M., Kling L., Vafaizadeh V., Sauter G., Terracciano L., Schumacher U. et al. (2013). Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J. Biol. Chem. 288, 22993-23008. 10.1074/jbc.M113.456293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth M., Walentin K., Aue A., Schonheit J., Wuebken A., Pode-Shakked N., Vilianovitch L., Erdmann B., Dekel B., Bader M. et al. (2010). The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 137, 3835-3845. 10.1242/dev.055483 [DOI] [PubMed] [Google Scholar]
- Wilanowski T., Tuckfield A., Cerruti L., O'Connell S., Saint R., Parekh V., Tao J., Cunningham J. M. and Jane S. M. (2002). A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech. Dev. 114, 37-50. 10.1016/S0925-4773(02)00046-1 [DOI] [PubMed] [Google Scholar]
- Xiang J., Fu X., Ran W. and Wang Z. (2017). Grhl2 reduces invasion and migration through inhibition of TGFbeta-induced EMT in gastric cancer. Oncogenesis 6, e284 10.1038/oncsis.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.