Summary
Par-3 regulates animal cell polarity by targeting the Par complex proteins Par-6 and atypical Protein Kinase C (aPKC) to specific cortical sites. Although numerous physical interactions between Par-3 and the Par complex have been identified [1–6], we discovered a novel interaction between Par-3’s second PDZ domain and a highly conserved aPKC PDZ binding motif (PBM) that is required in the context of the full-length, purified Par-6/aPKC complex. We also found that Par-3 is phosphorylated by the full Par complex and phosphorylation induces dissociation of the Par-3 phosphorylation site from aPKC’s kinase domain but does not disrupt the Par-3 PDZ2 – aPKC PBM interaction. In asymmetrically dividing Drosophila neuroblasts, the aPKC PBM is required for cortical targeting, consistent with its role in mediating a persistent interaction with Par-3. Our results define a physical connection that targets the Par complex to polarized sites on the cell membrane.
eTOC blurb
Holly et al. discover an interaction between Par-3’s second PDZ domain and a newly identified PDZ Binding Motif in aPKC that is conserved across metazoa and required for cortical polarity in Drosophila neuroblasts
Graphical Abstract
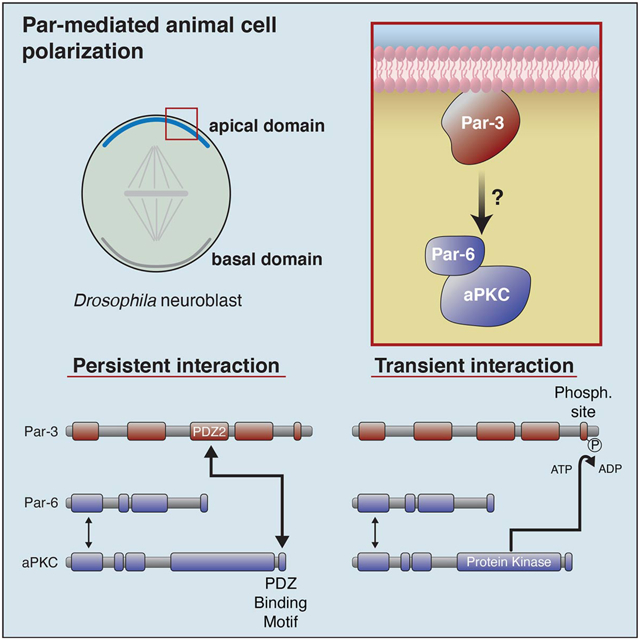
Results and Discussion
Par complex phosphorylation of Par-3
The catalytic activity of aPKC defines mutually exclusive cortical domains in diverse animal cells [7,8]. Par-6 and aPKC are recruited to specific cellular sites where aPKC phosphorylation polarizes downstream factors by displacing them from the Par cortical domain. For example, in Drosophila neural stem cells or neuroblasts, the Par complex localizes to an apical cortical domain during mitosis where it excludes neuronal differentiation factors [9–11]. Apical exclusion separates these factors into a distinct cortical domain at the basal cortex, which is segregated into the basal daughter cell following cytokinesis [8,12]. Par polarized factors such as Miranda and Numb contain sequences that bind the membrane but are also phosphorylation motifs for aPKC [13]. The direct connection of aPKC’s catalytic activity to the polarization of downstream factors makes the regulatory pathways that control its cortical targeting critical to animal cell polarity.
In many cellular contexts, Par-3 (Bazooka in flies) is essential for recruitment of Par-6 and aPKC to specific cortical sites [7,8,14]. Par-3’s role in regulating Par complex cortical recruitment is thought to be direct because five physical interactions have been discovered with both Par-6 and aPKC [1–6] (Figure 1A). Four of the interactions involve at least one of Par-3’s three PDZ protein interaction domains: Par-3 PDZ1 binding to the Par-6 PDZ domain [1,5,15], Par-3 PDZ1 and PDZ3 domain interactions with Par-6’s PDZ Binding Motif (PBM) [6], and an interaction with an undefined region of aPKC that requires both Par-3 PDZ2 and PDZ3 [2]. Additionally, because Par-3 is an aPKC substrate, the aPKC kinase domain interacts with Par-3’s aPKC Phosphorylation Motif (APM aka CR3). Although protein kinases are typically thought to interact transiently with their substrates, the interaction with the APM has been proposed to mediate complex assembly [3,4,16]. Previous investigations used small fragments of the Par complex that did not contain all potential binding motifs, such that it was not possible to assess whether any of the interactions are required for binding in the context of the purified, full-length Par-6/aPKC complex. Furthermore, none of the interactions have been shown to be required for cortical targeting of aPKC in a functional context.
Figure 1. Par-6/aPKC binds and phosphorylates Par-3.
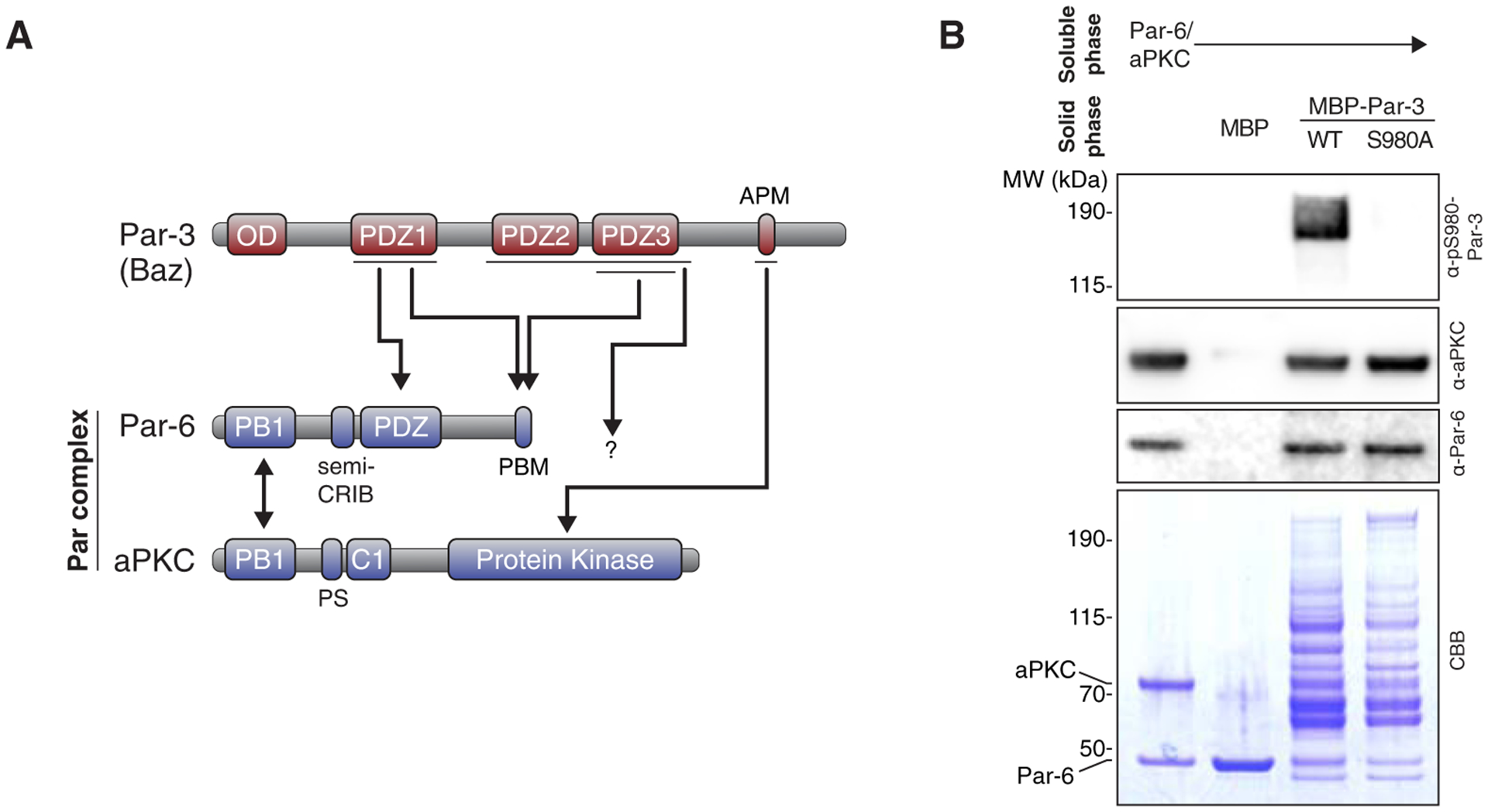
(A) Domain structure (not to scale) and previously described Par-3 interactions with the Par complex. Single direction arrows define the five previously identified Par-3 interactions with Par-6/aPKC. PBM, PDZ Binding Motif; APM, aPKC Phosphorylation Motif (aka CR3); PS, pseudosubstrate. Double headed arrow denotes the interaction between Par-6 and aPKC. (B) Par-3 interaction with and phosphorylation by the Par complex. Solid phase (amylose resin) bound Maltose Binding Protein (MBP) fused Par-3 (MBP-Par-3 has an expected mass of 199.9 kDa) with the Par-6/aPKC complex. CBB, Coomassie Brilliant Blue; α-pS980-Par-3, antibody specific to the S980 site within the APM [17]. Shaded region indicates fraction applied to gel (soluble phase or solid phase components after mixing with soluble phase components and washing).
We investigated Par-3 interactions with the Par-6/aPKC complex by reconstituting full-length Drosophila Par-6 and aPKC. While we were able to purify the Par-6/aPKC complex to a high degree (Figure 1B), Par-3 is very large (157.4 kDa) and the Maltose Binding Protein fused Par-3 (MBP-Par-3; total mass 199.9 kDa) we were able to obtain included significant amounts of degradation products in addition to full-length protein. Nevertheless, using this preparation we were able to detect an interaction with reconstituted Par complex using a qualitative affinity chromatography (i.e. “pull-down”) assay (Figure 1B). Additionally, we detected phosphate transfer to full-length Par-3 (and some smaller fragments with masses consistent with COOH-terminal truncations that contain the APM) using an antibody specific to the phosphorylated APM [17] (Figure 1B). Phosphorylation of Par-3 by aPKC has been controversial [4,18,19]. This result contributes to our understanding of the process by demonstrating that aPKC phosphorylates Par-3 in the context of the full-length, purified Par complex in addition to the isolated catalytic domain and APM peptide [19].
Par-3 PDZ2 is required for interaction with the Par complex
Using the system of purified Par complex and MBP-fusions of full-length Par-3 and its degradation products, we attempted to identify Par-3 domains required for interaction with the full Par complex (Figure 2). We also tested the Par-6 PBM within the Par complex as it has been reported to bind both the Par-3 PDZ1 and PDZ3 domains [6] (Figure 2B). We included ATP in binding experiments since Par-3 is a substrate in the context of the full Par complex (Figure 1B). Using this experimental setup, we identified Par-3 PDZ2 as a required interaction domain for binding to the full Par complex (Figure 2).
Figure 2. Par-3 PDZ2 is required for Par complex binding.
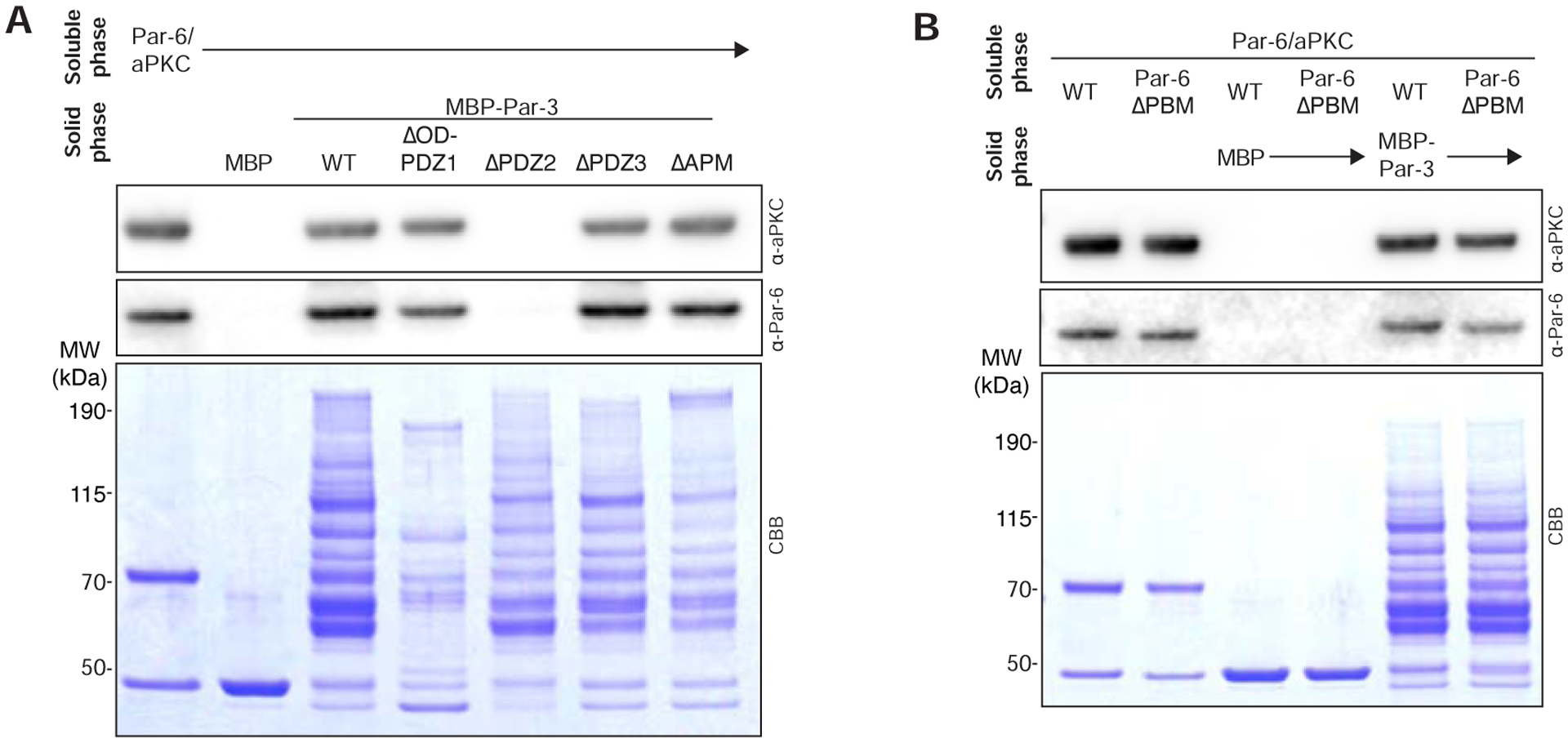
(A) Effect of removing Par-3 domains on its interaction with the Par complex. Solid phase (amylose resin) bound Maltose Binding Protein (MBP) fused Par-3 (full length or the PDZ1-APM fragment) was incubated with soluble Par-6/aPKC complex. CBB, Coomassie Brilliant Blue. Shaded region indicates fraction applied to gel (soluble phase or solid phase components after mixing with soluble phase components and washing).
(B) Effect of removing the Par-6 PDZ Binding Motif on the Par complex interaction with Par-3. Labeling as in A.
A conserved aPKC PDZ Binding Motif is required for interaction with Par-3
To determine the mechanism by which Par-3 PDZ2 mediates binding to the Par complex, we first sought to identify the recognition site on the complex. The Drosophila aPKC COOH-terminal sequence has the characteristics of a “class 3” PDZ Binding Motif (PBM) (Figure 3A) and is consistent with the binding specificity of the PDZ2 domain as assessed using a phage display assay [20]. The aPKC COOH-terminal sequence is also highly conserved among metazoan orthologues (Figure 3A), the same evolutionary interval in which Par-3 is found [21]. We tested whether the aPKC COOH-terminus is required for the interaction with Par-3 by purifying Par complex lacking aPKC’s final six residues. As shown in Figure 3B, the aPKC COOH-terminus is required for Par-3’s interaction with the Par complex.
Figure 3. A conserved aPKC PDZ Binding Motif required for Par-3’s interaction with the Par complex.
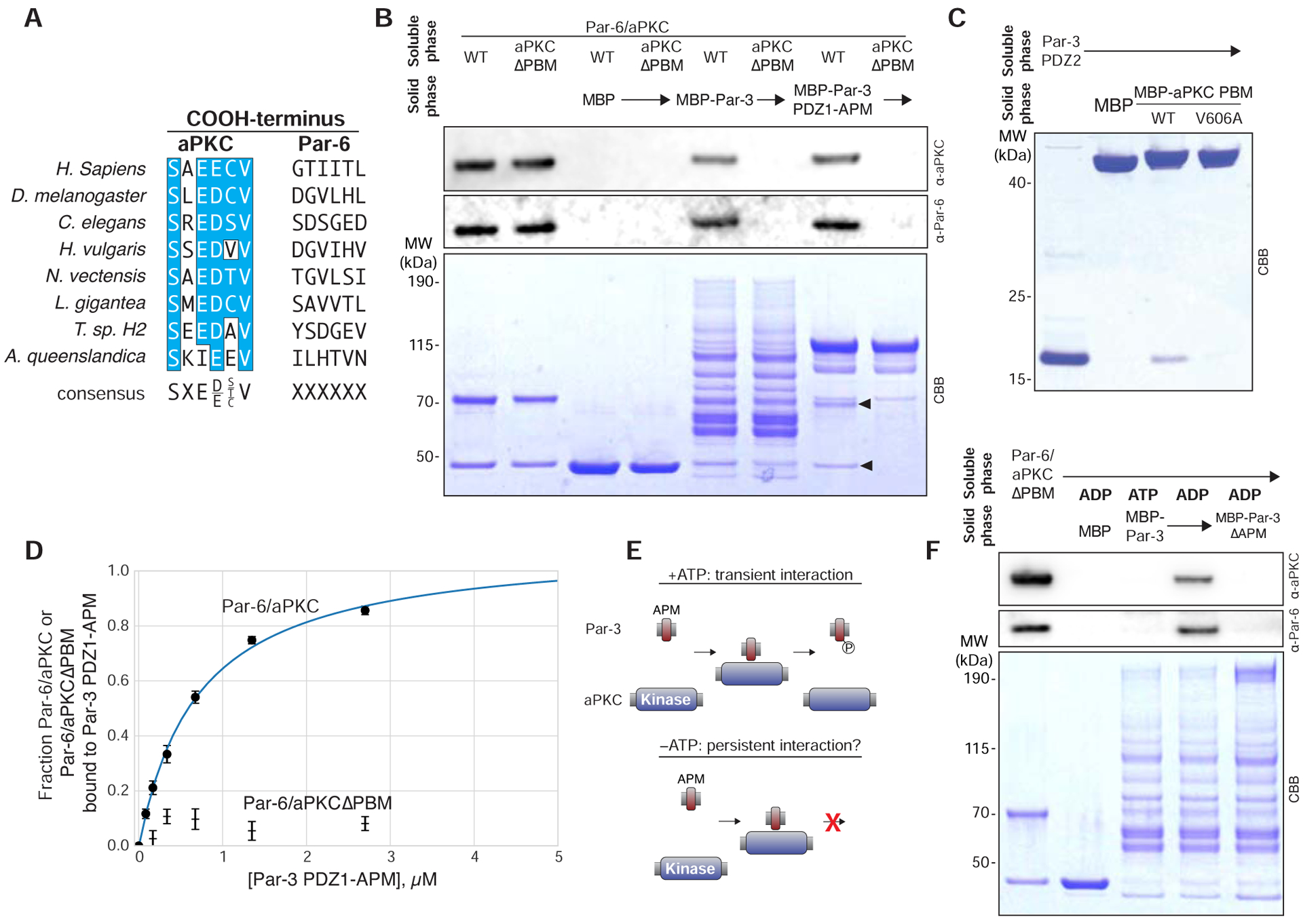
(A) Sequence alignment of the aPKC and Par-6 COOH-terminuses from diverse metazoan organisms.
(B) Effect of removing the aPKC PDZ Binding Motif on the Par complex interaction with Par-3. Solid phase (amylose resin) bound Maltose Binding Protein (MBP) fused Par-3 (full length or the PDZ1-APM fragment that contains all known interaction domains) was incubated with wild type Par-6/aPKC or the complex lacking aPKC’s PBM (Par-6/aPKCΔPBM). Arrowheads indicate Par-6 and aPKC. CBB, Coomassie Brilliant Blue. Shaded region indicates fraction applied to gel (soluble phase or solid phase components after mixing with soluble phase components and washing).
(C) Interaction of Par-3 PDZ2 with the aPKC PBM. Labeling as in B. For other organisms, see Figure S1A.
(D) Comparison of Par-6/aPKC and Par-6/aPKCΔPBM binding to Par-3 PDZ1-APM using an equilibrium supernatant depletion assay [22]. Mean values of the fraction of the Par complex bound, as determined by both anti-Par-6 and anti-aPKC western blot analysis (see Figure S1B), from two experimental replicates are shown along with standard error at each concentration.
(E) Schematic depicting how the lack of ATP may convert a transient interaction between the Par-3 APM and the aPKC kinase domain into a persistent one.
(F) Effect of nucleotide on the interaction of Par-3 with Par-6/aPKCΔPBM. Solid phase (amylose resin) bound Maltose Binding Protein (MBP) fused Par-3 (full length and with the specified interaction domains removed) was incubated with Par-6/aPKC lacking aPKC’s PBM (Par-6/aPKCΔPBM). “ADP” or “ATP” indicates which nucleotide was present in the binding reaction. CBB, Coomassie Brilliant Blue. Shaded region indicates fraction applied to gel (soluble phase or solid phase components after mixing with soluble phase components and washing).
To test whether the Par-3 PDZ2 and aPKC COOH-terminus are sufficient for binding, we examined the interaction of the isolated motifs. As shown in Figure 3C, the isolated proteins are sufficient for complex assembly. In general, PDZ–PBM interactions are strongly dependent on the identity of the terminal residue and we found that Par-3 PDZ2 failed to bind the aPKC COOH-terminus when the final residue was mutated from valine to alanine (aPKC V606A; Figure 3C). We conclude that the aPKC COOH-terminus is a bona fide PDZ Binding Motif (PBM). We also confirmed that the aPKC PBM and Par-3 PDZ2 interaction is broadly conserved across metazoans by examining orthologues from a chordate (human), a placozoan (Trichoplax), and a cnidarian (Hydra), in addition to the arthropod Drosophila (Figure S1A). We observed binding for each of the orthologous pairs indicating that the interaction is conserved across diverse metazoan organisms. Together, these results indicate that the Par-3 PDZ2 and aPKC PBM are sufficient for binding and their interaction is conserved across metazoa.
To assess the role of the Par-3 PDZ2–aPKC PBM interaction quantitatively, we implemented an equilibrium supernatant depletion assay [22]. We measured the affinity of the Par-3 PDZ1-APM for the Par complex, as this region could be purified to a level suitable for quantitative measurements (Figure 3B). Addition of Par-3 PDZ1-APM depleted Par-6 and aPKC from the supernatant consistent with a Kd of 0.7 μM (95% confidence interval of 0.5 – 0.9 μM; Figures 3D, S2B). To determine the effect of disrupting the Par-3 PDZ2–aPKC PBM interaction on binding affinity, we examined Par-3 PDZ1-APM binding to Par-6/aPKCΔPBM. We did not observe sufficient depletion of Par-6 and aPKCΔPBM by PDZ1-APM to allow fitting to a binding isotherm (Figures 3D, S2B), indicating the absence of the aPKC PBM substantially decreases the affinity of the Par-3 interaction with the Par complex, consistent with the results of qualitative measurements (Figure 3B).
The role of Par-3 phosphorylation in its interaction with the Par complex
Our results indicate that the Par-3 PDZ2 and aPKC PBM are required for Par-3’s interaction with the Par complex (Figures 2A, 3B). The requirement for these domains suggests that the Par-3 phosphorylation site (i.e. APM) does not form a persistent interaction with the Par complex. However, this conclusion appears to be in conflict with previous work showing that the Par-3 APM is sufficient for binding to the aPKC kinase domain, both with binding assays and structure determination using x-ray crystallography [4,23]. Furthermore, a stable APM-kinase interaction forms the basis of a model in which the unphosphorylated Par-3 APM forms a stable, persistent interaction with the aPKC kinase domain that is not phosphorylated until an unknown activating event occurs [4]. The finding that Par-3 is phosphorylated by the full Par complex (Figure 1B) is inconsistent with this model, but it does not fully resolve whether the Par-3 APM is sufficient for forming a stable, persistent interaction with the Par complex [4,23].
We hypothesized that the presence of ATP could influence the binding behavior of the Par-3 APM with the Par complex. A key difference between our experiments and previous reports is that our experiments included ATP, whereas previous binding experiments and structural analysis lacked ATP [4,23]. Without ATP, completion of the protein kinase catalytic cycle is not possible, and interactions that would otherwise form transiently could persist (Figure 3E).
We tested whether the Par-3 APM forms a stable, persistent interaction with the Par complex in the absence of ATP. We were able to detect binding between Par-3 and the Par complex after replacing ATP with ADP in a context where the Par-3 PDZ2 – aPKC PBM interaction is disrupted (Figure 3F). This interaction requires the APM, leading us to conclude that the Par-3 APM can form a persistent interaction with the Par complex, but only in the absence of ATP. When ATP is present the APM interacts transiently with the Par complex, because it is phosphorylated (Figure 1B) and subsequently dissociates (Figures 3B,D,F). Under the same conditions, the Par-3 PDZ2 interaction with the Par complex is not disrupted, however (e.g. Figure 3B,D). Although it is possible to form a stalled complex between the Par-3 APM and the Par complex in the absence of ATP, we propose that persistent binding of the APM to the kinase domain due to the lack of ATP is unlikely in vivo because ATP concentrations are high under normal cellular conditions.
The aPKC PDZ Binding Motif is required for neuroblast polarization
Although numerous interactions have been identified between Par-3 and Par-6/aPKC [1–6], none have been demonstrated to be required for cortical targeting of the Par complex. In fact, the interactions of Par-6 with Par-3 have been shown to be dispensable for function [6,15,24]. To determine if the Par-3 PDZ2 – aPKC PBM interaction is required for Par complex polarization, we investigated the localization of aPKC harboring the V606A PBM point mutation during neuroblast asymmetric division by expressing aPKC-V606A in larval brain neuroblasts and comparing its localization to that of wild-type aPKC. Consistent with previous observations [9,25], we found that wild-type aPKC is polarized to a cortical crescent around the apical pole at metaphase (Figures 4A, B). In contrast, aPKC-V606A remained in the cytoplasm and was not recruited to the cortex, even though the localization of Par-3 was unaffected (Figures 4A–D). The aPKC-V606A protein also failed to be recruited to the apical cortex in neuroblasts lacking endogenous aPKC (Figure S2). We conclude that the Par-3 PDZ2-aPKC PBM interaction is required for cortical recruitment and polarization of aPKC in neuroblasts.
Figure 4. aPKC polarization requires its PDZ Binding Motif.
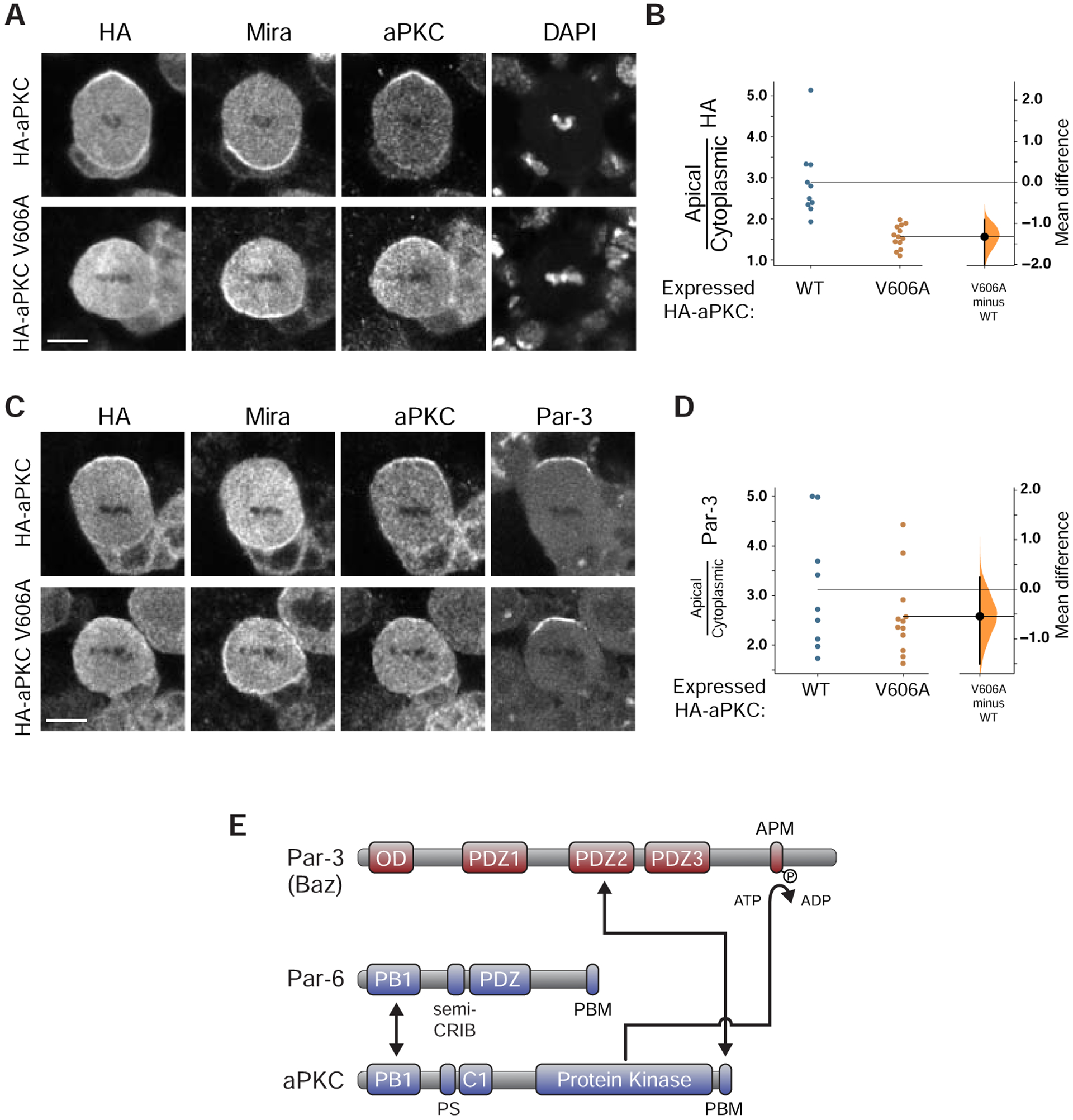
(A) Protein localization in metaphase neuroblasts expressing WT or V606A aPKC. The localization of HA-tagged WT or V606A aPKC, expressed using Worniu-GAL4/UAS, is shown with the basal marker Miranda, total aPKC (transgenically expressed and endogenous) using an anti-aPKC antibody, and DNA (DAPI). Scale bar is 5 μm. A similar analysis in aPKC mutant neuroblasts is shown in Figure S2.
(B) Gardner-Altman estimation plot of effect of the aPKC V606A mutation on aPKC cortical localization. Ratios of apical cortical to cytoplasmic anti-HA signal intensities are shown for individual metaphase neuroblasts expressing either HA-WT or HA-V606A aPKC. Statistics: Bootstrap 95% confidence interval (bar in “V606A minus WT” column).
(C) Localization of Par-3 in metaphase neuroblasts expressing WT or V606A aPKC, as in panel d.
(D) Gardner-Altman estimation plot of effect of the aPKC V606A mutation on Par-3 cortical localization. Apical cortical to cytoplasmic signal intensities of anti-Par-3 signals are shown for individual metaphase neuroblasts expressing either HA-WT or HA-V606A aPKC. Statistics: Statistics: Bootstrap 95% confidence interval (bar in “V606A minus WT” column).
(E) Par-3 interactions with Par-6/aPKC analyzed in this study. The Par-3 PDZ2–aPKC PBM interaction forms a persistent connection while the aPKC kinase domain interacts transiently with the Par-3 APM when ATP is present.
We have examined the interaction of Par-3 with the full-length Par complex and found that Par-3 PDZ2 and a previously unrecognized PBM at the COOH-terminus of aPKC are required for complex assembly (Figure 4E). The Par-3 phosphorylation site (APM) can also form a persistent interaction with the aPKC kinase domain, but only if phosphorylation is not allowed to occur due to the absence of ATP. Unlike the APM–kinase domain interaction, the Par-3 PDZ2 interaction with the aPKC PBM is not influenced by the presence of ATP, suggesting that additional mechanisms besides APM phosphorylation must exist to dissociate Par-3 from the Par complex, an important component of current polarity models [17,26,27].
The identification of Par-3 PDZ2 domain as a key factor in recruiting the Par complex to the cortex during animal cell polarization is consistent with previous work demonstrating that while Par-3 PDZ1 and 3 are dispensable in C. elegans, PDZ2 is required for cortical recruitment of Par-6 and aPKC [24]. It is also consistent with work in both C. elegans and Drosophila showing that the interaction of Par-6 with Par-3 is not required [6,15]. In Drosophila, the role of PDZ2 is less clear but is known to be required for downstream effects on epithelial structure [28]. We suggest that the Par-3 PDZ2 – aPKC PBM interaction represents an important physical connection for animal cell polarity and that the reconstitution approach used to identify this interaction will likely be useful for understanding how other regulatory molecules, such as Cdc42, control polarity.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
All DNA constructs generated in this study and the UAS-HA-aPKC and UAS-HA-aPKC-V606A fly lines are available from the Lead Contact (Ken Prehoda; prehoda@uoregon.edu) without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals: Drosophila
Flies were reared on cornmeal/yeast/dextrose culture medium at the specific temperatures specified below. A mix of male and female larvae were used for all experiments. The strains used in this study were: ;Worniu-Gal4 (BDSC_56553), ; FRT-G13, aPKCK06403/CyO (Gift from C.Q. Doe), and elav-Gal4, UAS-mCD8:GFP, hs:flp; FRT-G13, tubPGal80 (BDSC_5145).
In addition, two fly lines were created for this study, 3xHA-aPKC 1–606 and 3xHA-aPKC V606A. These were made using Phi-C31 integration. In brief, the coding region of aPKC wild-type (1–606) (A1Z9X0) or aPKC PBM mutant (V606A) were cloned into a pUAST vector (GenBank: EF362409.1) modified to contain an N-terminal 3xHA tag. Vector was injected into attP2 containing flies in a y, w background (BDSC_8622) and integrated using PhiC31 (BestGene, Inc.). F1 generation progeny were backcrossesd to y, w adults and F2 progeny were screened for the presence of red eyes.
METHOD DETAILS
Expression and Purification of Par-complex
Plasmids (pCMV) containing the coding sequences for aPKC and His-Par-6 including the mutants aPKCΔPBM or His-Par-6ΔPBM were co-transfected into HEK293-F suspension cells using 293fectin (Thermo Fisher) and grown in shaker flasks for 60h at 37°C. Cells were collected by centrifugation at 1000 RPM × 3 minutes, resuspended in lysis buffer (50 mM NH3PO4, 300 mM NaCl, 10 mM Imidazole, pH 8.0), and lysed by probe sonication at 50% amplitude, 0.3s/0.7s pulse on/off, 3 × 1 minute. After clearing cellular debris by centrifugation at 15k RPM × 20 minutes, protein was gently mixed with 4mL HisPur Cobalt (ThermoFisher) resin for 45 minutes. Resin was washed twice with 20mL lysis buffer supplemented with 5 mM MgCl2 and 100 μM ATP followed by a final wash with 20mL nickel lysis buffer. Protein was eluted with elution buffer (50 mM NH3PO4, 300 mM NaCl, 300 mM Imidazole, pH 8.0) by gravity and buffer exchanged with 20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM DTT, 5 mM MgCl2, and 100 μM ATP using a PD10 desalting column (GE Healthcare). Finally, protein was purified by Source Q anion exchange chromatography with a salt gradient from 100 mM to 550 mM. Fractions from the elution peak containing aPKC phosphorylated at the activation loop and turn motif, as verified by reactivity with phosphospecific antibodies (Rabbit α-PKCζ p-410 Santa Cruz Biotech, sc-12894-R; Rabbit α-PKC p560 Abcam ab62372), were pooled concentrated to 2.5mL, and buffer shifted to 20 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 100 μM ATP, and 1 mM DTT, followed by concentration to ~400 μL. Protein was aliquoted, flash frozen, and stored at −80°C until use. Par-complex was quantified by western blot analysis using a standard curve generated with a sample of known concentration using an anti-aPKC antibody (Mouse α-PKCζ H-1 Santa Cruz Biotech sc-17781).
Expression and Purification of Par-3.
Par-3 PDZ1 – APM (aa 309 – 991) was cloned (Gibson Cloning) into the pMal expression vector allowing for an NH2-terminal MBP tag as well as a COOH-terminal His6 tag to allow for dual affinity purification. Full length Par-3 (aa 1 – 1464) and all full length Par-3 domain deletion mutants were cloned into the pMal expression vector allowing for an NH2-terminal MBP tag. Plasmids were transformed into E. coli BL21 (DE3) cells, plated on LB + AMP and allowed to grow for 18h at 37 C. Single colonies were picked to inoculate 100 mL of LB + AMP starter culture and grown for ~4h. Starter culture was used to inoculate 2L LB + AMP and cultures were grown to an OD600 of 0.8 – 1, followed by a 3h induction with 500 μM IPTG. Cell pellets were collected at 5000 RPM × 20 minutes and resuspended in lysis buffer (50 mM NH3PO4, 300 mM NaCl, 10 mM Imidazole, pH 8.0). Cell extracts were thawed under running water and lysed by probe sonication at 70% amplitude, 0.3s/0.7s pulse on/off, 3 × 1 minute. Cellular debris was cleared at 15,000 RPM × 20 minutes and supernatant was added to 5mL HisPur cobalt resin and incubated for 30 minutes at 4 C. Resin was washed 3 times with lysis buffer followed by elution with nickel elution buffer (50 mM NH3PO4, 300 mM NaCl, 300 mM Imidazole, pH 8.0). Fractions containing protein were pooled and concentrated to 2.5mL. Proteins were buffer shifted over PD10 desalting columns (GE Healthcare) to 20 mM Tris, pH 7.5, 200 mM NaCl, 1 mM DTT followed by flash freezing and storage at −80°C. Protein was quantified by Bradford and purity was assessed by SDS-Page. All other Par-3 constructs were cloned into pMal, pGex, or pET expression vectors allowing for a single NH2-terminal MBP, GST, or His tag. Proteins were expressed in transformed BL21 (DE3) competent cells as above. MBP-protein cell extracts were resuspended in MBP lysis buffer (20mM Tris, pH 7.5, 200mM NaCl, 1mM EDTA, 1mM DTT). GST-protein cell extracts were resuspended in GST lysis buffer (1x PBS, pH 7.4, 1mM DTT). His-protein cell extracts were resuspended in nickel lysis buffer (50 mM NH3PO4, 300 mM NaCl, 10 mM Imidazole, pH 8.0). Cells were lysed and cleared as above. For proteins used as bait in pull down assays, cleared lysate was aliquoted, flash frozen, and stored at −80 C. Purified proteins were prepared by incubating cleared lysates with 5mL of amylose, glutathione, or HisPur cobalt resin for 30 minutes at 4 C. Resin was washed 3x with either GST, MBP, or nickel lysis buffer. Proteins were then eluted with MBP, GST, or Nickel lysis buffer supplemented with 10 mM maltose, 10mM glutathione, or 300mM Imidizole, respectively. Fractions containing protein were pooled and concentrated to 2.5mL with Vivaspin 20 centrifugal concentrators. Proteins were buffer shifted to 20mM HEPES, pH 7.5, 100mM NaCl, 1mM DTT. Finally, proteins were concentrated to 500 μL, aliquoted, flash frozen, and stored at −80 C.
Affinity chromatography interaction assay
Amylose or glutathione resin was loaded with bacterial lysate (or his purification elutions in the case of Par-3 PDZ1-APM or Par-3 PDZ1-PDZ3 as these proteins contain COOH-terminal his tags) containing MBP-or GST-fusion protein for 30 minutes at 4° C and then washed with wash buffer three times (20 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 0.5% Tween 20, and 1 mM DTT). Par-complex was then added to a concentration of 0.5 μM and incubated for 10 minutes at room temperature. In the case where ATP is present, ATP was used at a final concentration of 200 μM in all buffers throughout the pull down experiment and and binding reactions were carried out for 30 minutes at room temperature. Finally, beads were washed two times briefly to remove unbound Par-complex and beads were resuspended in loading dye. Samples were analyzed by SDS-Page and stained by Coomassie as well as Western Blot using α-aPKC (Mouse α-PKCζ H-1 Santa Cruz Biotech sc-17781) and rat α-Par-6.
Supernatant depletion interaction assay
Amylose resin was loaded with bacterial lysate containing MBP – Par-3 PDZ1 – APM for 30 minutes at 4° C and then washed with wash buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 200 μM ATP, 0.5% Tween 20, and 1 mM DTT). 2-fold serial dilutions of beads were prepared from 20 μL to 0.625 μL in a total volume of 200 μL. Par-complex was added to a final concentration of 40 nM diluted in wash buffer. After incubation for 30 minutes, beads were collected by centrifugation and an aliquot of supernatant was diluted in loading dye for western blot analysis using α-aPKC (Mouse α-PKCζ H-1 Santa Cruz Biotech sc-17781) and rat α-Par-6. The concentration of protein loaded on the beads was verified by SDS-Page using a standard curve generated with known concentrations of BSA.
Drosophila Neuroblast Immunofluoresence
Flies were allowed to lay in vials for 24 hours at room temperature, after which the flies were removed, and the vial was moved to 30°C. During wandering third instar larval stage (5–6 days later), larvae were dissected within 20 minutes into Schneider’s Insect Medium (Sigma, S0146). Brains were fixed using 4% PFA in PBS for 20 minutes followed by 3 washes in PBST (1xPBS + 0.3% Triton-X 100, Sigma-Aldrich). Note that during all fixing, blocking, and washing steps, brains were kept moving on a nutator. At this stage brains could be kept for up to 3 days nutating at 4˚C in PBST or washed an additional 20 minutes in PBST before blocking and staining. Brains were blocked for 30 minutes in PBSBT (PBST + 1% BSA, Fisher Scientific). Incubation in primary antibodies occurred overnight at 4°C. Primary antibodies: Rat α-Mira (1:500; Abcam, ab197788), Mouse α-PKCζ H-1(1:1,000; Santa Cruz Biotech sc-17781), Rabbit α-PKCζ C-20 (1:1,000; Santa Cruz Biotech sc-216), Rabbit α-HA C29F4(1:1,000; Cell Signaling Technologies, 3724), Mouse α-HA (1:500; Covance, MMS-101P), and Guinea Pig α-Baz(1:2,000; gift from C.Doe). Brains were then washed quickly followed by 3×15 minutes washes in PBSBT. Incubation in secondary antibodies occurred at room temperature protected from light for 2 hours followed by a quick wash and 3×15 minutes washes in PBST. Secondary antibodies used were from Jackson Immunoresearch Laboratories, Inc.: Dk α-Rt Cy3 (712-165-153; 1:500), Dk α-Rb 647 (711-605-152; 1:500), Dk α-Ms 488 (715-545-151), Dk α-Gp 405 (706-475-148; 1:500). Brains were stored in SlowFade Diamond with DAPI (Invitrogen, S36964) for at least 24 hours before imaging. Brains were imaged using a Leica TCS SPE upright confocal microscope using an ACS APO 40× 1.15 NA Oil CS objective.
Drosophila Neuroblast MARCM Clones
To generate Drosophila neuroblast MARCM clones, FRT-G13, aPKCK06403/CyO Virgin flies were crossed to ;;3xHA-aPKC V606A male flies. The resulting non-Cyo male progeny were crossed to elav-Gal4, UAS-mCD8:GFP, hs:flp; FRT-G13, tubPGal80 Virgins and allowed to lay for 24 hours at room temperature. The vials were then allowed to stay at room temperature for an additional 24 hours at which time they were heat-shocked @37°C for 90 minutes. This was followed by a possible second 90 minute heat-shock within 18 hours. Vials were raised at 18°C or room temperature until wandering third instar stage when they were dissected and stained as described above with the following antibodies: Primary antibodies: Rat α-Mira (1:500; Abcam, ab197788), Rabbit α-HA C29F4(1:1,000; Cell Signaling Technologies, 3724) or Mouse α-HA (1:500; Covance, MMS-101P), and Chicken α-GFP (1:500; Abcam, ab13970). Secondary antibodies: Dk α-Rt Cy3 (712-165-153; 1:500), Dk α-Rb 647 (711-605-152; 1:500) or Dk α-Ms 647 (715-605-151; 1:500), and Dk α-Ck 488 (703-545-155). Brains were imaged using a Leica TCS SPE upright confocal microscope using an ACS APO 40× 1.15 NA Oil CS objective or an Olympus Fluoview FV1000 upright confocal microscope using a PlanApo N 60× 1.42 NA Oil objective.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of equilibrium dissociation constants
The equilibrium dissociation constant for Par-3 PDZ1-APM binding to Par-6/aPKC was calculated by measuring both Par-6 and aPKC western signals from the supernatant of solutions containing increasing concentrations of amylose resin-bound MBP-Par-3 PDZ1-APM and fitting to the following equation:
“fb” was calculated from the experimentally measured Par-6 and aPKC western signals using the equation 1 – Wx/W0 where Wx is the western signal at Par-3 concentration × and W0 is the western signal in the absence of Par-3 “free” and “bound” are the fraction of Par-6/aPKC bound when saturated and in the absence of Par-3, respectively. These parameters were allowed to float Kd is the equilibrium dissociation constant.
Fitting and calculation of 95% confidence intervals were done using the LMFIT python package.
Drosophila Neuroblast Quantification
All images were analyzed using Fiji. For quantification of apical cortical to cytoplasmic signal intensity ratios, corresponding signals were measured from an intensity profile averaged from 10 pixels across the apical portion of the cell parallel with the polarity axis in a central optical section. The apical value was taken as the highest peak data point corresponding with the apical domain of the cell, the cytoplasmic value was an average of 20 data points that were a distance of 10 points away from the apical value.
DATA AND SOFTWARE AVAILABILITY
The study did not generate any unique datasets or code.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-PKC zeta (H1) | SCBT | SC-17781 |
| Rabbit Anti-PKC zeta (C-20) | SCBT | SC-216 |
| Rabbit Anti-PKC zeta p-410 | SCBT | SC-12894-R |
| Rabbit Anti-PKC zeta p560 | Abcam | Ab62372 |
| Mouse Anti-His (AD1.1.10 monoclonal) | SCBT | SC-53073 |
| Rat Anti-Par-6 (polyclonal custom antibody) | Alpha diagnostic | N/A |
| Rabbit Anti- Par-3 S980 phosphospecific | Daniel St. Johnston Lab | |
| Rat Anti-Mira | Abcam | Ab197788 |
| Rabbit Anti-HA (C29F4) | Cell Signaling Tech. | 3724 |
| Mouse Anti-HA | Covance | MMS-101P |
| Guinea Pig Anti-Baz (polyclonal custom antibody) | C.Q.Doe Lab | N/A |
| Chicken Anti-GFP | Abcam | Ab13970 |
| Donkey Anti-Rat Cy3 | Jackson Immuoresearch Lab. | 712-165-153 |
| Donkey Anti-Rabbit 647 | Jackson Immunoresearch Lab. | 711-605-152 |
| Donkey Anti-Mouse 647 | Jackson Immunoresearch Lab. | 715-605-151 |
| Donkey Anti-Mouse 488 | Jackson Immunoresearch Lab. | 715-545-151 |
| Donkey Anti-Chicken 488 | Jackson Immunoresearch Lab. | 703-545-155 |
| Donkey Anti-Guinea Pig 405 | Jackson Immunoresearch Lab. | 706-475-148 |
| IRDye 680RD Donkey anti-mouse secondary | Licor | 926–68072 |
| IRDye 800CW Goat anti-rat secondary | Licor | 926–32219 |
| Bacterial and Virus Strains | ||
| BL21-DE3 | ||
| TG1 | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| All Par-3 constructs prepared from Drosophila melanogaster transcript variant A: NM_001347807.1 | ||
| MBP – Par-3 (1 – 1464) | This Paper | MBP – Par-3 - WT |
| MBP – Par-3 -His (309 – 991) | This Paper | MBP – Par-3 – PDZ1 – APM – His |
| MBP – Par-3 (393 – 1464) | This Paper | MBP – Par-3 – ΔOD -PDZ1 |
| MBP – Par-3 (1 – 1464 Δ 437–533) | This Paper | MBP – Par-3 – ΔPDZ2 |
| MBP – Par-3 (1 – 1464 Δ 616 – 741) | This Paper | MBP – Par-3 – ΔPDZ3 |
| MBP – Par-3 (1 – 1464 Δ 969 – 987) | This Paper | MBP – Par-3 – ΔAPM |
| MBP – Par-3 (309 – 1464) | This Paper | MBP – Par-3 – PDZ1 – COOH |
| MBP – Par-3 (1– 987) | This Paper | MBP – Par-3 – NH2 term – APM |
| MBP – Par-3 (1–1464) (S980A) | This Paper |
MBP-Par-3
S980A |
| His – Dr - Par-3 (437 – 741) | This Paper | His – DrPar-3 – PDZ2 |
| His – Human-Par-3 (454 – 549) (Q9BYG5) | This Paper | His – HuPar-3 – PDZ2 |
| His – Hydra - Par-3 (370 – 463) (T2M6J3) | This Paper | His – HyPar-3 – PDZ2 |
| His – Trichoplax - Par-3 (250–340) (A0A369RWF0) | This Paper | His – TrPar-3 – PDZ2 |
| GST-Dr – aPKC (601 – 606) (A1Z9X0) | This Paper | GST-DraPKC – PBM |
| GST-Dr – aPKC (601 – 606 -V606A) (A1Z9X0) | This Paper | GST-DraPKC – PBM (V606A) |
| GST-Human – aPKC (591–596) (P41743) | This Paper | GST-HuaPKC – PBM |
| GST-Hydra – aPKC (574–579) (T2MGA0) | This Paper | GST-HyaPKC – PBM |
| GST-Trichoplax – aPKC (404 – 410)(A0A369S098) | This Paper | GST-TraPKC – PBM |
| aPKC (1 – 606) | This Paper | aPKC |
| aPKC (1 – 606) (V606A) | This Paper | aPKC (V606A) |
| aPKC ΔPBM (1 – 600) | This Paper | aPKC ΔPBM |
| His – Par-6 (1 – 351) | This Paper | His – Par-6 |
| His – Par-6 ΔPBM (1 – 343) | This Paper | His – Par-6 ΔPBM |
| 293fectin | ThermoFisher | 12347019 |
| Freestyle 293 expression Medium | ThermoFisher | 12338026 |
| Opti-Mem | ThermoFisher | 3198588 |
| HisPur cobalt resin | ThermoFisher | 89965 |
| Amylose Resin | NEB | E8021L |
| Glutathione Resin | GoldBio | G250–100 |
| Source Q anion exchange resin | GE Healthcare | 17-1275-01 |
| Bolt 12% Bis-Tris Gels | ThermoFisher | NW00125BOX |
| TMB Substrate | ThermoFisher | N301 |
| Odyssey blocking buffer (TBS) | LiCor | 927–50000 |
| Schneider’s Insect Medium (SIM) | Sigma | S0146 |
| SlowFade Diamond Antifade Mountant with DAPI | Invitrogen | S36964 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Human Embryonic Kindey cells (HEK293) | ThermoFisher | R79007 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: ;Worniu-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_56553 |
| D. melanogaster: ;;3xHA-aPKC 1–606 | This Paper | N/A |
| D. melanogaster: ;;3xHA-aPKC V606A | This Paper | N/A |
| D. melanogaster: ; FRT-G13, aPKCK06403/CyO | C.Q. Doe Lab | N/A |
| D. melanogaster: elav-Gal4, UAS-mCD8:GFP, hs:flp; FRT-G13, tubPGal80 | Bloomington Drosophila Stock Center | RRID:BDSC_5145 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| pCMV mammalian expression plasmid | ThermoFisher | 10586014 |
| pMal C4X bacterial expression plasmid | Addgene | 75288 |
| pGex 4Ti bacterial expression plasmid | Amersham | 27458001 |
| pUASTattB fly cloning and transformation plasmid | Addgene | EF362409.1 |
| Software and Algorithms | ||
| ImageJ | ||
| GraphPad | ||
| Other | ||
| PD10 Desalting columns | 95017–001 | |
| VivaSpin 20 sample concentrators MWCO 30kD | VWR | 95056–130 |
| VivaSpin 20 sample concentrators MWCO 10kD | VWR | 95056–128 |
| Shaker Flasks – 125mL | VWR | 89095–258 |
Highlights.
Par-3 and Par-6/aPKC interact through a PDZ2 – PDZ Binding Motif interaction
The Par-3 PDZ2 interaction with the aPKC PBM is conserved across metazoan
The aPKC PBM is required for cortical polarity in Drosophila neuroblasts
Acknowledgments
The authors thank Mike Drummond for help at the inception of the project and Brad Nolen and members of the Prehoda Lab for helpful comments. This work was supported by NIH grant GM127092 (K.E.P.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Joberty G, Petersen C, Gao L, and Macara IG (2000). The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol 2, 531–539. [DOI] [PubMed] [Google Scholar]
- 2.Wodarz A, Ramrath A, Grimm A, and Knust E (2000). Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol 150, 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, Fujimoto T, Tabuse Y, Kemphues KJ, and Ohno S (1998). An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J. Cell Biol 143, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soriano EV, Ivanova ME, Fletcher G, Riou P, Knowles PP, Barnouin K, Purkiss A, Kostelecky B, Saiu P, Linch M, et al. (2016). aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization. Dev. Cell 38, 384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, and Pawson T (2000). A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol 2, 540–547. [DOI] [PubMed] [Google Scholar]
- 6.Renschler FA, Bruekner SR, Salomon PL, Mukherjee A, Kullmann L, Schütz-Stoffregen MC, Henzler C, Pawson T, Krahn MP, and Wiesner S (2018). Structural basis for the interaction between the cell polarity proteins Par3 and Par6. Sci. Signal 11. [DOI] [PubMed] [Google Scholar]
- 7.Lang CF, and Munro E (2017). The PAR proteins: from molecular circuits to dynamic self-stabilizing cell polarity. Dev. Camb. Engl 144, 3405–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkei ZG, and Yamashita YM (2018). Emerging mechanisms of asymmetric stem cell division. J. Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolls MM, Albertson R, Shih H-P, Lee C-Y, and Doe CQ (2003). Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol 163, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wodarz A, Ramrath A, Kuchinke U, and Knust E (1999). Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402, 544–547. [DOI] [PubMed] [Google Scholar]
- 11.Atwood SX, and Prehoda KE (2009). aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. CB 19, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knoblich JA (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol 11, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey MJ, and Prehoda KE (2015). Establishment of Par-Polarized Cortical Domains via Phosphoregulated Membrane Motifs. Dev. Cell 35, 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen W, and Zhang M (2018). Protein Complex Assemblies in Epithelial Cell Polarity and Asymmetric Cell Division. J. Mol. Biol 430, 3504–3520. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Kim H, Aceto DG, Hung J, Aono S, and Kemphues KJ (2010). Binding to PKC-3, but not to PAR-3 or to a conventional PDZ domain ligand, is required for PAR-6 function in C. elegans. Dev. Biol 340, 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, and Ohno S (2002). Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells Devoted Mol. Cell. Mech 7, 1161–1171. [DOI] [PubMed] [Google Scholar]
- 17.Morais-de-Sá E, Mirouse V, and St Johnston D (2010). aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell 141, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson BJ, and McDonald NQ (2019). Competitive Inhibition of aPKC by Par-3/Bazooka and Other Substrates. Dev. Cell 49, 680. [DOI] [PubMed] [Google Scholar]
- 19.Holly RW, and Prehoda KE (2019). Phosphorylation of Par-3 by Atypical Protein Kinase C and Competition between Its Substrates. Dev. Cell 49, 678–679. [DOI] [PubMed] [Google Scholar]
- 20.Yu CG, Tonikian R, Felsensteiner C, Jhingree JR, Desveaux D, Sidhu SS, and Harris TJC (2014). Peptide binding properties of the three PDZ domains of Bazooka (Drosophila Par-3). PloS One 9, e86412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahey B, and Degnan BM (2010). Origin of animal epithelia: insights from the sponge genome. Evol. Dev 12, 601–617. [DOI] [PubMed] [Google Scholar]
- 22.Pollard TD (2010). A guide to simple and informative binding assays. Mol. Biol. Cell 21, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Shang Y, Yu J, and Zhang M (2012). Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKCι in complex with a substrate peptide from Par-3. Structure 20, 791–801. [DOI] [PubMed] [Google Scholar]
- 24.Li B, Kim H, Beers M, and Kemphues K (2010). Different domains of C. elegans PAR-3 are required at different times in development. Dev. Biol 344, 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oon CH, and Prehoda KE (2019). Asymmetric recruitment and actin-dependent cortical flows drive the neuroblast polarity cycle. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S-C, Low TYF, Nishimura Y, Gole L, Yu W, and Motegi F (2017). Cortical forces and CDC-42 control clustering of PAR proteins for Caenorhabditis elegans embryonic polarization. Nat. Cell Biol 19, 988–995. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez J, Peglion F, Martin J, Hubatsch L, Reich J, Hirani N, Gubieda AG, Roffey J, Fernandes AR, St Johnston D, et al. (2017). aPKC Cycles between Functionally Distinct PAR Protein Assemblies to Drive Cell Polarity. Dev. Cell 42, 400–415.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKinley RFA, Yu CG, and Harris TJC (2012). Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J. Cell Sci 125, 1177–1190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The study did not generate any unique datasets or code.


