Abstract
Noroviruses are the main cause of viral gastroenteritis with new variants emerging frequently. There are three norovirus genogroups infecting humans. These genogroups are divided based on the sequence of their major capsid protein, which is able to form virus-like particles (VLPs) when expressed recombinantly. VLPs of the prototypical GI.1 Norwalk virus are known to disassemble into specific capsid protein oligomers upon alkaline treatment. Here, native mass spectrometry and electron microscopy on variants of GI.1 and of GII.17 were performed, revealing differences in terms of stability between these groups. Beyond that, these experiments indicate differences even between variants within a genotype. The capsid stability was monitored in different ammonium acetate solutions varying both in ionic strength and pH. The investigated GI.1 West Chester isolate showed comparable disassembly profiles to the previously studied GI.1 Norwalk virus isolate. However, differences were observed with the West Chester being more sensitive to alkaline pH. In stark contrast to that, capsids of the variant belonging to the currently prevalent genogroup GII were stable in all tested conditions. Both variants formed smaller capsid particles already at neutral pH. Certain amino acid substitutions in the S domain of West Chester relative to the Norwalk virus potentially result in the formation of these T = 1 capsids.
Keywords: virus assembly, native mass spectrometry, structural virology, capsid stability, VLP size
1. Introduction
Human noroviruses cause the majority of nonbacterial gastroenteritis outbreaks worldwide (Ahmed et al 2014). Symptoms include fever, vomiting, and diarrhea that usually last for several days (Kaplan et al 1982). In addition to the surface of fecal contaminated food, noroviruses can be found in environmental samples such as sewage and spread through person-to-person contact (Kaplan et al 1982, Hedberg and Osterholm 1993, Ozawa et al 2007). A small number of particles of this pathogen is sufficient for infection. Severe disease is mostly observed in the elderly, children and immunocompromised people (Teunis et al 2008).
Noroviruses belong to the Caliciviridae family and are non-enveloped, single-stranded, positive-sense ribonucleic acid (RNA) viruses. Human norovirus genomes are 7.4–7.7 kb in length and have three open reading frames (ORFs). The non-structural proteins, such as the RNA dependent polymerase (RdRp), are encoded by ORF1, while the structural proteins are encoded by ORF2 (VP1) and ORF3 (VP2) (Xi et al 1990, Jiang et al 1993). The major capsid protein VP1 forms dimers, which assemble into the viral capsid with T = 3 icosahedral symmetry of approximately 36–42 nm diameter. VP2 is considered as minor capsid protein and carries multiple functions, which are still not completely deciphered (Vongpunsawad et al 2013). VP1 can be divided into a shell (S) domain and a protruding (P) domain, which is further subdivided into P1 and P2. The S domain forms a scaffold surrounding the viral RNA with a diameter of 30 nm, whereas the genetically more variable P domain builds the viral spikes and facilitates cell attachment (Prasad et al 1994, 1999). Research on human noroviruses has been limited by the lack of a robust cell culture system. Nevertheless, production of virus-like VP1 particles (VLPs) show comparable characteristics to the native virion (Jiang et al 1992). Electron microscopy (EM) and x-ray crystallography studies have deepened the understanding of capsid assembly and interactions with potential receptors like human histo-blood group antigens (Harrington et al 2002, Tan and Jiang 2005).
Native mass spectrometry (MS) has further elucidated these processes (Shoemaker et al 2010, Uetrecht and Heck 2011, Mallagaray et al 2015). Here, intact non-covalent protein complexes are transferred into the gas phase from an aqueous solution of a volatile buffer surrogate, such as ammonium acetate, via nano-electrospray ionization (ESI). All coexisting oligomers can be detected with high sensitivity, allowing to monitor assembly processes. Recently, charge detection MS has been introduced, enabling simultaneous charge and mass-to-charge (m/z) ratio determination and therefore mass assignment of single ions (Keifer et al 2017). VLPs can also be analyzed by mobility-based approaches (Weiss et al 2015). However, ESI with time-of-flight (ToF) detectors are mostly employed, which rely on charge state resolution for exact mass assignment of m/z signals.
Remarkably, Shoemaker et al demonstrated the alkaline and ionic strength sensitivity of norovirus VLPs, and showed that capsid disassembly was reversible (Shoemaker et al 2010). VLPs completely disassembled at low ionic strength and alkaline pH, whereas smaller particles formed at high ionic strength in alkaline conditions. Most importantly, they showed that small VLPs, which were previously observed in EM under alkaline conditions, corresponded to T = 1 capsids. The dynamic behavior of VP1 upon changing pH may have implications for the infection process as the virus passes through very different environments in the intestinal tract. Using ion mobility, pH dependent intermediates were shown to be consistent with partial capsid structures and an assembly pathway was proposed (Uetrecht et al 2011). These findings confirmed previous studies, which combined EM with spectroscopic techniques like circular dichroism and high resolution second-derivative UV absorption spectroscopy (Ausar et al 2006).
Importantly, MS studies focused on the prototypical Norwalk virus first isolated in 1972 in Ohio (Kapikian et al 1972), disregarding the emergence of new variants, which could exhibit altered assembly behavior. Noroviruses can be divided into at least seven genogroups (GI-GVII), with GI, GII, and GIV causing infections in human. Each genogroup can be further subdivided into numerous genotypes based on the capsid amino acid sequences (Kageyama et al 2004, Hansman et al 2006). The prototypical Norwalk virus (GI.1 Norwalk) is rarely found to cause outbreaks nowadays. It is replaced by strains of the genogroup II, where GII.4 noroviruses have been prevalent (Eden et al 2013). A reemerging genotype of this genogroup, GII.17, seems to become a major threat nowadays in East Asia. First isolated in 1978, they were the cause of several outbreaks in Asia in 2014 to 2015 (Chan et al 2015, Lee et al 2015, Zhang et al 2015, Singh et al 2016).
Here, GI.1 West Chester noroviruses closely related to and isolated three decades after the Norwalk strain and a newly emerging GII.17 variant isolated 2015 in Kawasaki, Japan, are compared. The VLP stability in different pH and ionic strength conditions of these genotypes was examined using native MS and negative stain EM. Surprisingly, the outbreak-causing isolate showed a much stronger resistance to pH. Furthermore, both variants form smaller T = 1 capsids at neutral pH, which points to amino acid substitutions potentially influencing the particle size. Differences between variants in terms of mass and stability hint towards new approaches for virus identification.
2. Experimental details
2.1. VLP production and preparation
In order to produce norovirus VLPs, recombinant VP1 protein was expressed in insect cells as described previously (Hansman et al 2005, 2007). Genebank accession numbers are AY602016.1 and Kawasaki308 for the GI.1 and GII.17 variant, respectively. Briefly, recombinant VP1 bacmid was transfected into Spodoptera frugiperda (Sf9) cells using Effectene. After incubation for five days at 27 °C, the cell culture medium was clarified by low-speed centrifugation. The supernatant containing the baculovirus was used to infect high five (H5) insect cells for 6 d at 27 °C. To separate secreted VLPs from the cell medium and cells, the solution was centrifuged at low speed and then concentrated by ultracentrifugation at 35 000 rpm for 2 h at 4 °C. VLPs were purified using a caesium chloride (CsCl) equilibrium gradient ultracentrifugation at 35 000 rpm for 18 h at 4 °C and then stored in phosphate buffered saline at 4 °C at a concentration of 3–5 mg ml−l.
Prior to MS and EM analysis, the VLPs were exchanged into 50 and 250 mM ammonium acetate solutions at different pH using Vivaspin 500 centrifugal concentrators (10 000 MWCO, Sartorius, Göttingen, Germany). Zeba micro spin™ desalting columns 0.5 ml (7000 MWCO, Thermo Fisher Scientific, Massachusetts, USA) were used if protein yields were too low for analysis using Vivaspin.
2.2. Mass spectrometry
Native MS measurements on VLPs were performed on a Q-Tof 2 instrument (Waters, Manchester, UK and MS Vision, Almere, the Netherlands) with modifications enabling high mass experiments (van den Heuvel et al 2006). All spectra were recorded in positive ion mode. Generally, samples were measured at a VP1 concentration of 10 µM. Ions were introduced via a nano-ESI source into the vacuum at a source pressure of 10 mbar using both premade (Waters, Manchester, UK) and handmade capillaries. For the in-house production, borosilicate glass tubes (inner diameter 0.68 mm, outer diameter 1.2 mm with filament; World Precision Instruments, Sarasota, USA) were pulled using a two-step program in a micropipette puller (Sutter Instruments, Novato, USA) with a squared box filament (2.5 × 2.5 mm). The capillaries were gold-coated using a sputter coater (Quorum Technologies Ltd., East Sussex, UK, 40 mA, 200 s, tooling factor of 2.3 and end bleed vacuum of 8 × 10−2 mbar argon) and opened directly on the sample cone of the mass spectrometer. Voltages of 1.55–1.65 kV and 145–155 V were applied to the capillary and cone, respectively. Xenon was used as a collision gas at a pressure of 2.0 × 10−2 mbar to improve the transmission of large ions (Lorenzen et al 2007). Starting from 10 V, collision energies were increased up to 200 V in steps of 25 V. The presented spectra were generally recorded at 50 V collision energy. MS profile and pusher settings were kept constant for all measurements. A caesium iodide spectrum was recorded the same day in order to test the performance of the instrument and to calibrate spectra where necessary. Analysis was performed using MassLynx™ (Waters, Manchester, UK). In order to generate intensity fraction plots, signals were accumulated over 100 s and in a respective m/z range all signals belonging to the broad ion distributions of a given oligomer were summed. The range was kept constant for both variants to simplify comparison, although ion distribution widths varied. Tables S2 and S3 (stacks.iop.org/JPhysCM/30/064006/mmedia) list respective m/z ranges that correspond to different VP1 oligomers and their intensity fractions.
2.3. Electron microscopy
After buffer exchange to ammonium acetate solutions, the norovirus VLPs were stored at 4 °C. The samples were adsorbed onto glow discharge-activated carbon coated grids (Science Services, Munich, Germany) after storage in ammonium acetate solution. The sample coated grids were washed three times with distilled water, following a negative staining with 1% uranyl acetate. Images were acquired using the FEI Tecnai™ G2 transmission electron microscope and the wide angle Veleta CCD camera (FEI, Thermo Fisher Scientific, USA and Olympus, Tokyo, Japan) at 80 kV.
3. Results and discussion
3.1. Altered capsid stability and size distribution in GI.1 isolates
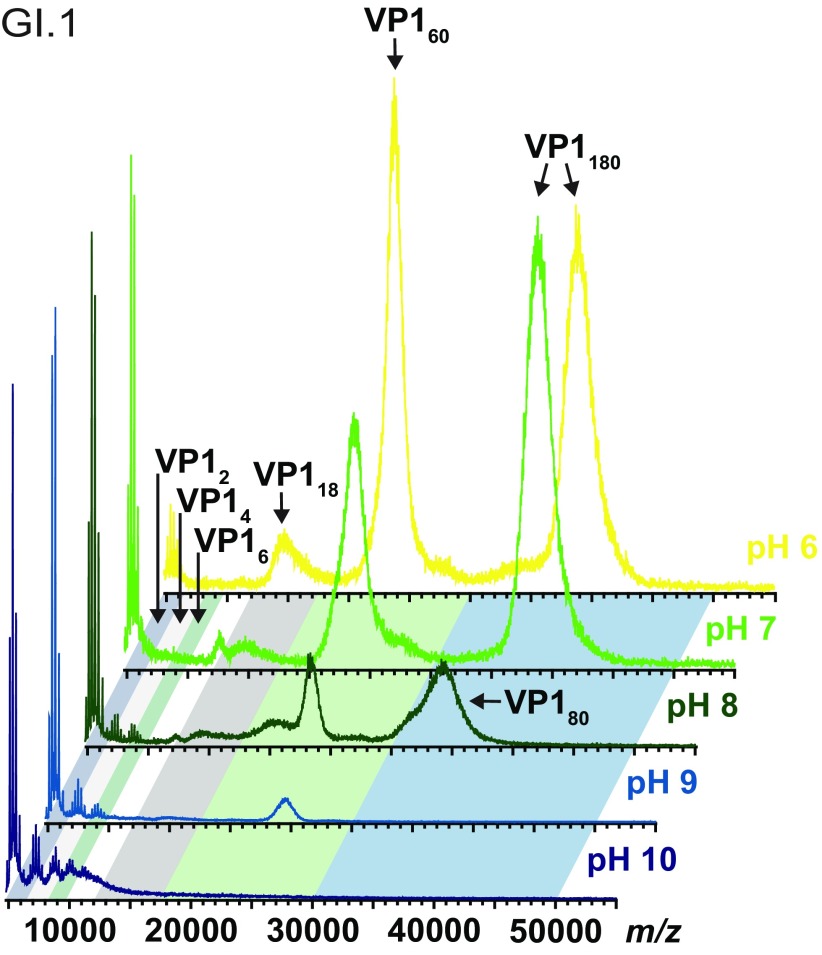
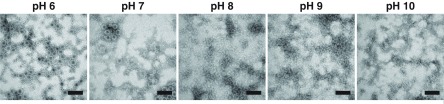
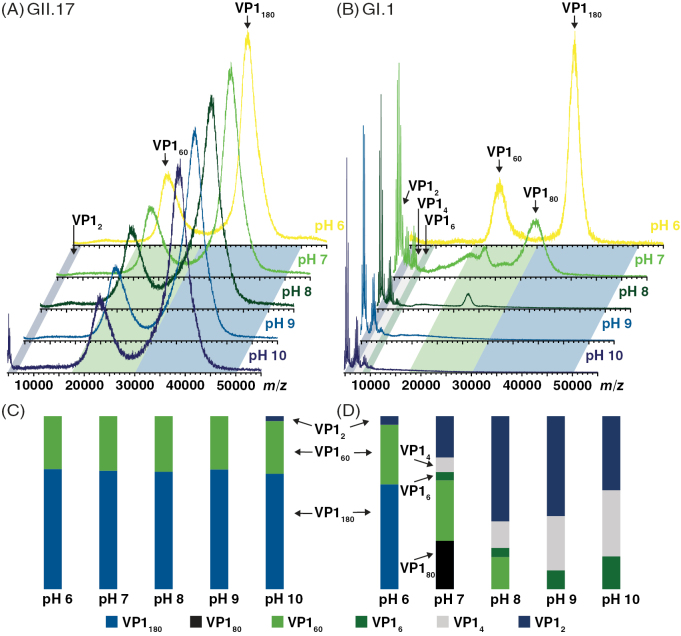
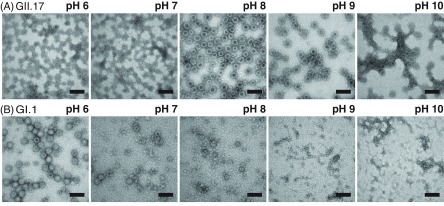
In order to compare the stability of VLPs within a genotype, native MS measurements in varying solution conditions were performed according to Shoemaker et al (2010). They observed the intact T = 3 capsid, at neutral and near neutral pH, which disassembled upon alkaline treatment into VP1 oligomers. Whereas the prototypical GI.1 Norwalk virus was investigated previously, the GI.1 West Chester variant is studied here. Our assignment of ion distributions is analogous to the Norwalk results (Shoemaker et al 2010), since VP1 heterogeneity again precluded charge state resolution (figure S1). The broad ion distribution around m/z 40 000 indicates the presence of intact VLPs consisting of 180 copies of the VP1 protein (figure 1). The observed m/z is in line with the linear relation of charge state to the square root of the mass (Heck and van den Heuvel 2004) and is therefore assigned to T = 3 particles. Following this scheme, further VP1 oligomers, namely VP1 80 mers and 60 mers (T = 1), can be assigned in high ionic strength solutions (250 mM ammonium acetate) at pH ranging from 6–10 (figure 1). Intact T = 3 is observed at pH 6 and pH 7. A shift of the corresponding ion distribution at pH 8 indicates that T = 3 disassembles and instead VP1 80 mers form. A second dominant ion distribution is located around m/z 20 000. Ions in this m/z region can be assigned to the T = 1 capsid. These ions are detected at pH 6 up to pH 9 and high ionic strength, although with strongly decreasing signals at alkaline pH. Several smaller oligomers are detected up to pH 8 between m/z 10 000–20 000 in line with disassembly of T = 3 and reformation of 80 mer particles. The dominant signal around m/z 15 000 corresponds to VP1 18 mers already detected at pH 6. VP1 6 mers and 4 mers are observed in solutions at pH 7 and higher. VP1 dimers are present in all tested solutions with strongly increasing signals upon alkaline treatment. To further illustrate and confirm the different VP1 oligomers, the ammonium acetate VLP preparations were imaged in negative stain EM (figure 2). At pH 6 and pH 7, round structures with a diameter of about 40 nm corresponding to intact T = 3 capsids can be found. Smaller particles with a diameter of about 20 nm are also seen in pH 6–8, likely the detected T = 1 particles. Size estimation is affected by aggregation. However, a clear trend in line with the MS measurements is observed as bigger spherical structures are no longer visible at pH 9 and 10.
Figure 1.
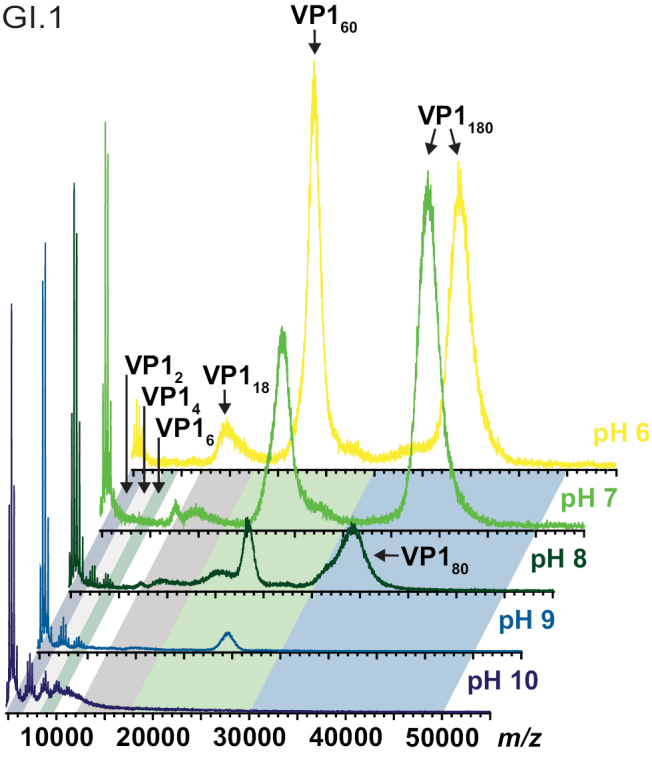
Norovirus GI.1 West Chester VLP stability monitored by native MS. Representative mass spectra of norovirus VLPs (10 µM VP1) at pH values indicated (from top to bottom pH 6–10) in 250 mM ammonium acetate. Data are normalized to the highest peak.
Figure 2.
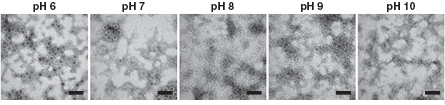
EM images of norovirus GI.1 West Chester VLPs. VLPs are in 250 mM ammonium acetate solutions with increasing pH from left to right. The bar represents 100 nm. All images are taken at the same magnification.
Notably, West Chester VP1 60 mers are present in ammonium acetate solutions from pH 6 up to pH 9, whereas 60 mers were only observed at pH 8 and pH 9 for the Norwalk virus despite comparable measurement conditions and very similar purification protocols. In both protocols a baculovirus expression system was used and VLPs were purified via CsCl gradient centrifugation. The T = 1 particles with a diameter of approximately 21 nm are a known byproduct in VLP production and also found in stool samples (Taniguchi et al 1981, White et al 1997). Truncated VP1 proteins were described as a possible reason for formation of smaller particles (Huo et al 2015). The versions of West Chester VP1 observed in the low m/z region after particle disassembly show differences in methionine processing (Δm ~ 270 Da per monomer) as was previously described for Norwalk VP1 (Shoemaker et al 2010). Therefore, the T = 1 formation likely represents an intrinsic property of the VP1 protein. This is also in agreement with the observation of T = 3 particles at neutral pH of the Norwalk virus S domain (Baclayon et al 2011). Nevertheless, it cannot be ruled out that small particles in stool samples are T = 3 particles lacking the P domain or other aberrant assemblies. Furthermore, West Chester VLPs are more sensitive to alkaline treatment as they disassemble at even lower pH than the previously described Norwalk virus (Shoemaker et al 2010). Smaller oligomers, such as the VP1 18 mer, are also detected at lower pH values for the West Chester. Importantly like Norwalk, the West Chester virus retained the ability to reversibly assemble when diluted from a high pH solution into a low pH solution including reformation of T = 1 particles (figure S2).
3.2. GII.17 Kawasaki norovirus shows vast stability at alkaline pH
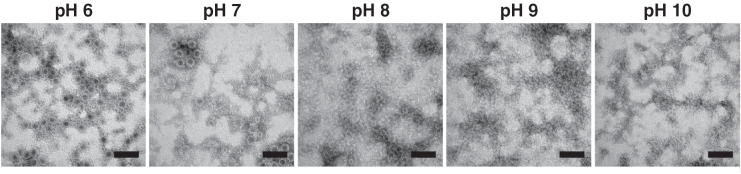
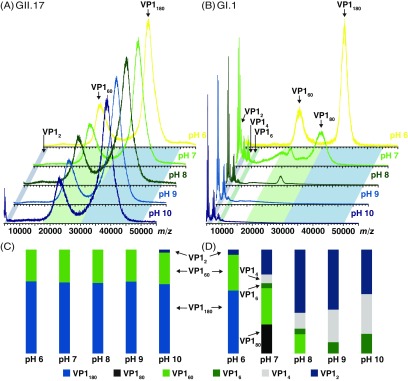
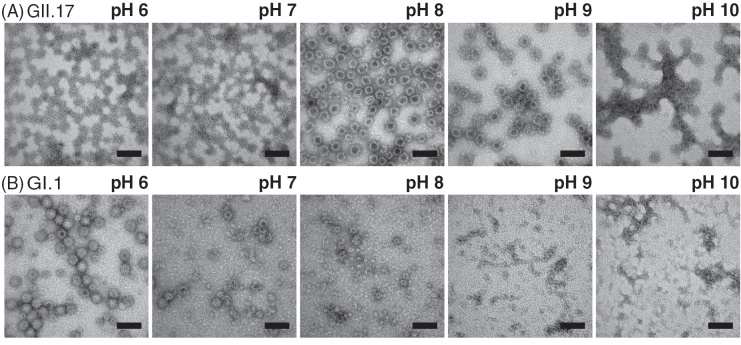
Although the Norwalk virus is known as the prototype strain, GI noroviruses have been replaced by emerging GII noroviruses (Blanton et al 2006). It is therefore of interest, whether these still exhibit a similar stability profile as GI noroviruses, in other words whether particle stability is a conserved feature in noroviruses. Therefore, GII.17 Kawasaki VLPs are also analyzed with native MS (figure 3). Broad ion distributions corresponding to the T = 3 and T = 1 intact capsids are detected throughout the whole pH range (pH 6–10) both at low (50 mM ammonium acetate, figure 3) and high ionic strength (250 mM ammonium acetate, figure S3(A)). The ratios of the capsid sizes remain unchanged up to pH 9. At pH 10, mild disassembly is observed, as is evident from the arising dimer signal. Based on the intensity fractions, no preference for disassembly of either capsid form is observed. Disulfide bonds as cause of this extraordinary stabilization can be excluded, as no additional bands corresponding to oligomers are observed under non-reducing conditions in SDS-PAGE (figure S4).
Figure 3.
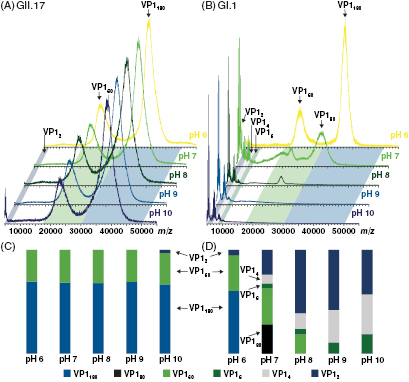
Comparison of norovirus VLP stability monitored by native MS at low ionic strength. Representative mass spectra of norovirus VLPs (10 µM VP1) in 50 mM ammonium acetate at pH values indicated (from top to bottom pH 6–10) (A) mass spectra obtained for the GII.17 Kawasaki virus and (B) for the GI.1 West Chester virus. Data are normalized to the highest peak. Colored inlets indicate the respective m/z range, where intensities were summed for the respective VP1 oligomers. Due to overlapping distributions, the VP1 80 mer falls into the same m/z range as VP1 180 mers, the descriptor therefore changes at pH 7 in (D). The intensity fractions of oligomers are shown for GII.17 Kawasaki (C) and for GI.1 West Chester (D) viruses. Spectra for measurements in 250 mM ammonium acetate can be found in figure S3 of both variants.
Ions in the m/z range of 30 000–50 000 m/z assigned to T = 3 signal cannot be detected above pH 7 for the GI.1 West Chester in 50 mM ammonium acetate (figure 3(B)). At pH 7 however, the signal already shifts to lower m/z as it does in 250 mM ammonium acetate, pH 8; therefore it likely corresponds to VP1 80 mers and not complete T = 3 particles. At pH 9, also T = 1 particles have disappeared. Thus, West Chester VLPs maintain a lower stability at lower ionic strength as was shown for Norwalk VLPs (Shoemaker et al 2010). Conversely, the Kawasaki VLPs show a more intense VP1 dimer signal at pH 10 at higher ionic strength indicative of decreased stability (figures 3(C) and S3(C)). VLP solutions were further imaged in negative stain EM (figure 4). Spherical structures with a diameter of about 40 nm can be found in all conditions for GII.17 Kawasaki VLPs. For GI.1 West Chester VLPs, the T = 3 is most obvious at pH 6. Consistent with the findings at high ionic strength, several smaller West Chester VP1 oligomers arise with alkaline treatment albeit with less defined size (figures 1 and 3).
Figure 4.
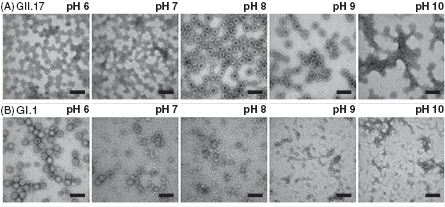
EM images of norovirus VLPs in 50 mM ammonium acetate. Images of VLPs of GII.17 Kawasaki virus (A) and of GI.1 West Chester virus (B) in increasing pH solutions from left to right (pH 6–10, respectively). The bar represents 100 nm. Images were taken at the same magnification.
The mass spectra and intensity fractions are normalized and therefore do not include information on the absolute amount of assemblies or altered electrospray ionization due to pH induced changes. To further illustrate differences between the strains, absolute intensities for m/z ranges corresponding to T = 3, T = 1 and a combined lower m/z range (corresponding to VP1 oligomers between 4800 to 9600 m/z at 50 mM and 4800–17 500 m/z at 250 mM) are plotted (figure S5). The GII.17 ion signals for both capsid forms are increasing up to pH 9 until they drop again at pH 10 in 250 mM ammonium acetate. The ion counts for both forms are overall lower at 50 mM ammonium acetate but less affected by pH. Furthermore, the spray is more stable at low ionic strength as is evident from smaller error bars. In contrast, ion counts for GI.1 West Chester capsids are always lower but reproducible. High intensity counts can only be obtained for the signal corresponding to low mass VP1 oligomers upon VLP dissociation.
4. Discussion
The two human norovirus strains GI.1 West Chester and GII.17 Kawasaki show a completely different pattern when monitored in ammonium acetate solutions of increasing pH. The GI.1 West Chester virus behaves similar to Norwalk virus (Shoemaker et al 2010) and disassembles upon alkaline treatment into capsid protein oligomers. The alkaline sensitivity is furthermore dependent on ionic strength, as higher ionic strength favors higher-mass oligomers. In stark contrast, GII.17 Kawasaki VLPs exhibit immense stability throughout the tested pH range up to pH 10. Capsid destabilization is indicated only at pH 10 as VP1 dimer peaks arise in low abundance. For GII.17 Kawasaki VLPs, the influence of ionic strength is low but inversed compared to GI.1 West Chester and Norwalk viruses.
Previous studies using various techniques on norovirus VLP stability were able to show alkaline sensitivity for the Norwalk virus (Ausar et al 2006, Shoemaker et al 2010, da Silva et al 2011). Stability studies on noroviruses are of special interest, as these are foodborne and need to persist in various environmental settings. The influence of temperature, ionic strength and pH on the adhesion of VLPs to surfaces also revealed higher stability and more adhesion for GII.4 viruses than GI.1 (da Silva et al 2011, Samandoulgou et al 2015). However, the investigated pH range was limited. In 2010, Donath and coworkers investigated the norovirus genotype GII.7 with atomic force microscopy (AFM) in a pH range of 2–10 (Cuellar et al 2010). Despite belonging to GII genogroup, VLPs were found to be sensitive to alkaline treatment. GII.7 noroviruses were reported as slowly evolving genotypes with decreasing prevalence (Hoffmann et al 2012). Based on these and our own findings on GI.1 and emerging GII.17 viruses, it is tempting to speculate whether increased VLP stability could be related to prevalence. However, other causes cannot be excluded and genome containing viruses likely have altered stability profiles.
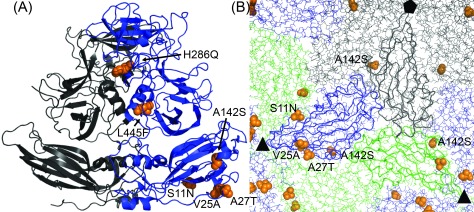
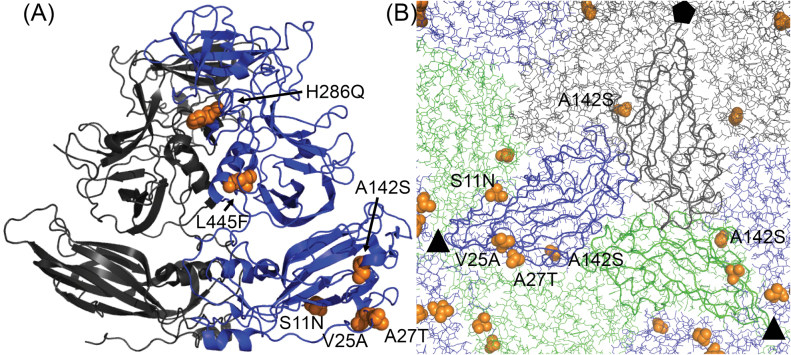
A sequence alignment of VP1 of GI.1 Norwalk and West Chester virus reveals 13 substitutions (figure S6), with seven amino acids exchanged for similar residues. Four out of the six dissimilar residues are located in the S domain (N-terminal 225 amino acids); all of them are located at the dimer surface and potentially involved in inter-dimer contacts (figure 5). Substitutions for similar amino acids are exclusively found in the P domain together with two dissimilar residues close to the intra-dimer interface and therefore unlikely to cause the differences in assembly behavior. The P domain also contains the only substitution involving charged residues (H286Q), therefore altered electrostatic interactions are an unlikely cause of the observed changes. In light of the Norwalk S domain, which assembles into T = 3 particles like the full length protein VP1 albeit with lower stability (Baclayon et al 2011), the amino acid exchanges in the S domain most likely cause the occurrence of the smaller West Chester T = 1 at neutral pH. Which amino acids contribute to stability or alteration of capsid size requires further investigations by mutagenesis. Nevertheless, the quasi-equivalent B subunit shows a serine to asparagine substitution at residue 11 in the N-terminal arm, which directly interfaces with the β-sheet of subunit C. A/B and C/C dimers show a different arrangement of the S domain, bent and flat, respectively (Prasad et al 1999). The substitution at residue 11 may alter the capability of the VP1 protein to switch to the alternate C conformation and therefore also small particles can form at neutral pH. The GI.1 Norwalk and GII.17 Kawasaki sequences are 46% identical (figure S6). The substitutions are located across the entire sequence but with higher frequency in the P domain. However, in all three variants the pI remains similar (Norwalk virus pI 5.64, West Chester isolate pI 5.57 and Kawasaki isolate pI 5.24). Given the number of substitutions, it is impossible to link the increased stability to any specific residues. Interestingly, residue 11 is not substituted in Kawasaki virus but flanked by two substitutions, S10P and V12N, which could again be an explanation for the increased T = 1 amount compared to Norwalk virus. Again, further studies are required to pinpoint the residues responsible for the observed stability and size differences.
Figure 5.
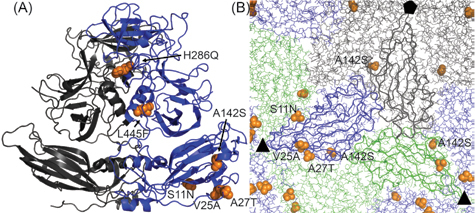
Norovirus GI.1 Norwalk and GI.1 West Chester capsid protein VP1 show six dissimilar substitutions. (A) Cartoon structure of a Norwalk VP1 dimer (PDB: 1IHM, residues 10–520) with quasi-equivalent monomers A (grey) and B (blue). Amino acids substituted for similar residues are not shown. Only the B subunit has a sufficiently resolved N-terminal arm to observe all four dissimilar amino acids in the S domain (bottom). The upper P domain only contains two major substitutions at the intra-dimer interface. All dissimilar residues are depicted as orange spheres and the substitution from Norwalk to West Chester is annotated. (B) S domains in the capsid context viewed from the interior with one subunit each (A—grey, B—blue, C—green) highlighted as ribbon. All substituted residues locate to the inter-dimer interface. Threefold and fivefold symmetry axes are labelled with black symbols.
This study demonstrates that native MS is the method of choice to investigate genotype and even isolate specific stability differences as it can resolve the different assembly states. The combination with negative stain EM confirmed the observed differences between norovirus-like particles of different genogroups. Notably, also isolates from the same genotype show distinct profiles and can pinpoint residues altering the assembly. The results are therefore informative for designing capsids with specific properties for nanotechnological applications (Worsdorfer et al 2011). It is of interest to test whether ligand binding to e.g. glycans alters the stability profiles, which would be indicative of structural changes. Furthermore, if stability differences persist at the virion level, mass determination in conjunction with determination of mechanical properties from AFM or similar methods could have potential as a diagnostic tool with the capability to distinguish specific isolates. Especially for rapidly evolving RNA viruses like noroviruses, this is a promising approach to identify new variants without the need for prior knowledge.
Supplementary Material
Acknowledgments
The Heinrich-Pette-Institute, Leibniz Institute for Experimental Virology is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit (BMG). CU acknowledges funding from the Leibniz Association through SAW-2014-HPI-4 grant. RP acknowledges funding from EU Horizon 2020 project VIRUSCAN 731868. CU, RP and GH further thank the DFG FOR2327 Virocarb for support. We thank the HPI microscopy technology platform for help acquiring the electron microscopy images.
References
- Ahmed S M, Hall A J, Robinson A E, Verhoef L, Premkumar P, Parashar U D, Koopmans M, Lopman B A. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:725–30. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausar S F, Foubert T R, Hudson M H, Vedvick T S, Middaugh C R. Conformational stability and disassembly of Norwalk virus-like particles. Effect of pH and temperature. J. Biol. Chem. 2006;281:19478–88. doi: 10.1074/jbc.M603313200. [DOI] [PubMed] [Google Scholar]
- Baclayon M, Shoemaker G K, Uetrecht C, Crawford S E, Estes M K, Prasad B V, Heck A J, Wuite G J, Roos W H. Prestress strengthens the shell of Norwalk virus nanoparticles. Nano Lett. 2011;11:4865–9. doi: 10.1021/nl202699r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton L H, Adams S M, Beard R S, Wei G, Bulens S N, Widdowson M-A, Glass R I, Monroe S S. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000–2004. J. Infect. Dis. 2006;193:413–21. doi: 10.1086/499315. [DOI] [PubMed] [Google Scholar]
- Chan M C, Lee N, Hung T N, Kwok K, Cheung K, Tin E K, Lai R W, Nelson E A, Leung T F, Chan P K. Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014. Nat. Commun. 2015;6:10061. doi: 10.1038/ncomms10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar J L, Meinhoevel F, Hoehne M, Donath E. Size and mechanical stability of norovirus capsids depend on pH: a nanoindentation study. J. Gen. Virol. 2010;91:2449–56. doi: 10.1099/vir.0.021212-0. [DOI] [PubMed] [Google Scholar]
- da Silva A K, Kavanagh O V, Estes M K, Elimelech M. Adsorption and aggregation properties of norovirus GI and GII virus-like particles demonstrate differing responses to solution chemistry. Environ. Sci. Technol. 2011;45:520–6. doi: 10.1021/es102368d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden J S, Tanaka M M, Boni M F, Rawlinson W D, White P A. Recombination within the pandemic norovirus GII.4 lineage. J. Virol. 2013;87:6270–82. doi: 10.1128/JVI.03464-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman G S, et al. Genetic and antigenic diversity among noroviruses. J. Gen. Virol. 2006;87:909–19. doi: 10.1099/vir.0.81532-0. [DOI] [PubMed] [Google Scholar]
- Hansman G S, Natori K, Oka T, Ogawa S, Tanaka K, Nagata N, Ushijima H, Takeda N, Katayama K. Cross-reactivity among sapovirus recombinant capsid proteins. Arch. Virol. 2005;150:21–36. doi: 10.1007/s00705-004-0406-8. [DOI] [PubMed] [Google Scholar]
- Hansman G S, Saito H, Shibata C, Ishizuka S, Oseto M, Oka T, Takeda N. Outbreak of gastroenteritis due to sapovirus. J. Clin. Microbiol. 2007;45:1347–9. doi: 10.1128/JCM.01854-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington P R, Lindesmith L, Yount B, Moe C L, Baric R S. Binding of norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 2002;76:12335–43. doi: 10.1128/JVI.76.23.12335-12343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck A J R, van den Heuvel R H H. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 2004;23:368–89. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- Hedberg C W, Osterholm M T. Outbreaks of food-borne and waterborne viral gastroenteritis. Clin. Microbiol. Rev. 1993;6:199–210. doi: 10.1128/CMR.6.3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann D, Hutzenthaler M, Seebach J, Panning M, Umgelter A, Menzel H, Protzer U, Metzler D. Norovirus GII.4 and GII.7 capsid sequences undergo positive selection in chronically infected patients. Infect. Genet. Evol. 2012;12:461–6. doi: 10.1016/j.meegid.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Huo Y, Wan X, Wang Z, Meng S, Shen S. Production of norovirus VLPs to size homogeneity. Virus Res. 2015;204:1–5. doi: 10.1016/j.virusres.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang M, Graham D Y, Estes M K. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 1992;66:6527–32. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang M, Wang K, Estes M K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- Kageyama T, Shinohara M, Uchida K, Fukushi S, Hoshino F B, Kojima S, Takai R, Oka T, Takeda N, Katayama K. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to Norovirus in Japan. J. Clin. Microbiol. 2004;42:2988–95. doi: 10.1128/JCM.42.7.2988-2995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapikian A Z, Wyatt R G, Dolin R, Thornhill T S, Kalica A R, Chanock R M. Visualization by immune electron microscopy of a 27 nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 1972;10:1075–81. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J E, Feldman R, Campbell D S, Lookabaugh C, Gary G W. The frequency of a Norwalk-like pattern of illness in outbreaks of acute gastroenteritis. Am. J. Public Health. 1982;72:1329–32. doi: 10.2105/AJPH.72.12.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keifer D Z, Pierson E E, Jarrold M F. Charge detection mass spectrometry: weighing heavier things. Analyst. 2017;142:1654–71. doi: 10.1039/C7AN00277G. [DOI] [PubMed] [Google Scholar]
- Lee C-C, Feng Y, Chen S-Y, Tsai C-N, Lai M-W, Chiu C-H. Emerging Norovirus GII.17 in Taiwan. Clin. Infect. Dis. 2015;61:1762–4. doi: 10.1093/cid/civ647. [DOI] [PubMed] [Google Scholar]
- Lorenzen K, Versluis C, Van Duijn E, Van Den Heuvel R H H, Heck A J R. Optimizing macromolecular tandem mass spectrometry of large non-covalent complexes using heavy collision gases. Int. J. Mass Spectrom. 2007;268:198–206. doi: 10.1016/j.ijms.2007.06.012. [DOI] [Google Scholar]
- Mallagaray A, Lockhauserbaumer J, Hansman G, Uetrecht C, Peters T. Attachment of norovirus to histo blood group antigens: a cooperative multistep process. Angew. Chem., Int. Ed. Engl. 2015;54:12014–9. doi: 10.1002/anie.201505672. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Oka T, Takeda N, Hansman G S. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 2007;45:3996–4005. doi: 10.1128/JCM.01516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B V, Hardy M E, Dokland T, Bella J, Rossmann M G, Estes M K. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–90. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- Prasad B V, Rothnagel R, Jiang X, Estes M K. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J. Virol. 1994;68:5117–25. doi: 10.1128/jvi.68.8.5117-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samandoulgou I, Hammami R, Morales Rayas R, Fliss I, Jean J. Stability of secondary and tertiary structures of virus-like particles representing noroviruses: effects of pH, ionic strength, and temperature and implications for adhesion to surfaces. Appl. Environ. Microbiol. 2015;81:7680–6. doi: 10.1128/AEM.01278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker G K, Van Duijn E, Crawford S E, Uetrecht C, Baclayon M, Roos W H, Wuite G J, Estes M K, Prasad B V, Heck A J. Norwalk virus assembly and stability monitored by mass spectrometry. Mol. Cell. Proteomics. 2010;9:1742–51. doi: 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B K, Koromyslova A, Hefele L, Gurth C, Hansman G S. Structural evolution of the emerging 2014-2015 GII.17 noroviruses. J. Virol. 2016;90:2710–5. doi: 10.1128/JVI.03119-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Jiang X. The p domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 2005;79:14017–30. doi: 10.1128/JVI.79.22.14017-14030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Urasawa S, Urasawa T. Further studies of 35–40 nm virus-like particles associated with outbreaks of acute gastroenteritis. J. Med. Microbiol. 1981;14:107–18. doi: 10.1099/00222615-14-1-107. [DOI] [PubMed] [Google Scholar]
- Teunis P F M, Moe C L, Liu P, Miller S E, Lindesmith L, Baric R S, Le Pendu J, Calderon R L. Norwalk virus: how infectious is it? J. Med. Virol. 2008;80:1468–76. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- Uetrecht C, Barbu I M, Shoemaker G K, Van Duijn E, Heck A J. Interrogating viral capsid assembly with ion mobility-mass spectrometry. Nat. Chem. 2011;3:126–32. doi: 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- Uetrecht C, Heck A J. Modern biomolecular mass spectrometry and its role in studying virus structure, dynamics, and assembly. Angew. Chem., Int. Ed. Engl. 2011;50:8248–62. doi: 10.1002/anie.201008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Heuvel R H, et al. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal. Chem. 2006;78:7473–83. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- Vongpunsawad S, Venkataram Prasad B V, Estes M K. Norwalk virus minor capsid protein VP2 associates within the VP1 shell domain. J. Virol. 2013;87:4818–25. doi: 10.1128/JVI.03508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss V U, et al. Analysis of a common cold virus and its subviral particles by gas-phase electrophoretic mobility molecular analysis and native mass spectrometry. Anal. Chem. 2015;87:8709–17. doi: 10.1021/acs.analchem.5b01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White L J, Hardy M E, Estes M K. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J. Virol. 1997;71:8066–72. doi: 10.1128/jvi.71.10.8066-8072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsdorfer B, Woycechowsky K J, Hilvert D. Directed evolution of a protein container. Science. 2011;331:589–92. doi: 10.1126/science.1199081. [DOI] [PubMed] [Google Scholar]
- Xi J, Graham D, Wang K, Estes M. Norwalk virus genome cloning and characterization. Science. 1990;250:1580–3. doi: 10.1126/science.2177224. [DOI] [PubMed] [Google Scholar]
- Zhang X F, et al. An outbreak caused by GII.17 norovirus with a wide spectrum of HBGA-associated susceptibility. Sci. Rep. 2015;5:17687. doi: 10.1038/srep17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.