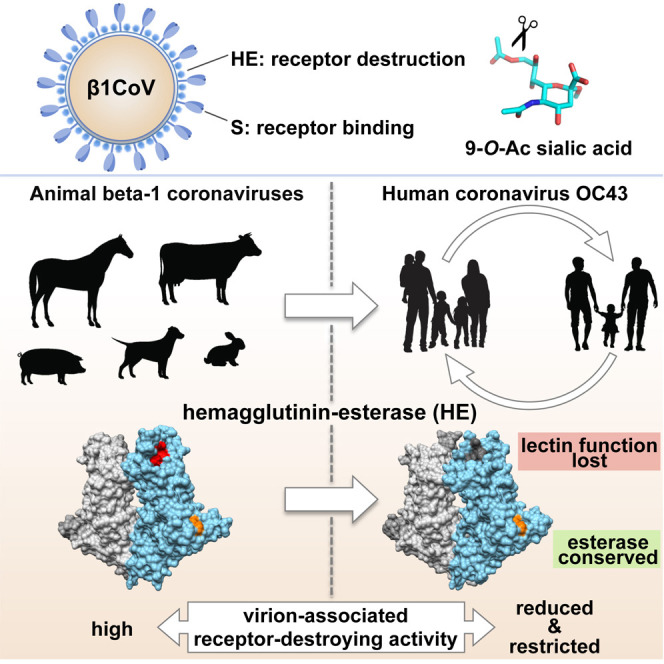
Summary
Human beta1-coronavirus (β1CoV) OC43 emerged relatively recently through a single zoonotic introduction. Like related animal β1CoVs, OC43 uses 9-O-acetylated sialic acid as receptor determinant. β1CoV receptor binding is typically controlled by attachment/fusion spike protein S and receptor-binding/receptor-destroying hemagglutinin-esterase protein HE. We show that following OC43’s introduction into humans, HE-mediated receptor binding was selected against and ultimately lost through progressive accumulation of mutations in the HE lectin domain. Consequently, virion-associated receptor-destroying activity toward multivalent glycoconjugates was reduced and altered such that some clustered receptor populations are no longer cleaved. Loss of HE lectin function was also observed for another respiratory human coronavirus, HKU1. This thus appears to be an adaptation to the sialoglycome of the human respiratory tract and for replication in human airways. The findings suggest that the dynamics of virion-glycan interactions contribute to host tropism. Our observations are relevant also to other human respiratory viruses of zoonotic origin, particularly influenza A virus.
Graphical Abstract
Highlights
-
•
Adaption of coronaviruses OC43 and HKU1 to humans involved loss of HE lectin function
-
•
OC43 HE receptor binding site was lost via progressive accumulation of mutations
-
•
Loss of HE receptor binding alters sialate-9-O-acetylesterase receptor destroying activity
-
•
Balance of receptor binding and receptor destruction contributes to host tropism
Coronavirus OC43 entered the human population relatively recently. Bakkers et al. report that as an adaptation to replication in human airways, the OC43 hemagglutinin-esterase lost its receptor-binding function. Consequently, virion-associated receptor-destroying activity toward clustered sialoglycan-based receptor determinants was reduced. Suggestive of convergent evolution, human respiratory coronavirus HKU1 underwent similar changes.
Introduction
Coronaviruses (CoVs), long considered of high veterinary impact exclusively, are now generally recognized as zoonotic threats of pandemic potential in consequence of the 2002/2003 outbreak of severe acute respiratory syndrome (SARS) and the emergence of Middle East respiratory syndrome (MERS) in 2012 (de Wit et al., 2016). SARS-CoV was contained within 3 months after its discovery, while MERS-CoV causes a classical zoonotic infection with limited human-to-human spread and, as yet, is incapable of sustained community transmission (Reusken et al., 2016, Zumla et al., 2015). However, four other respiratory coronaviruses—alphacoronaviruses NL63 and 229E and lineage A betacoronaviruses OC43 and HKU1—successfully breached the species barrier and are currently maintained in the human population worldwide through continuous circulation (Su et al., 2016, de Groot et al., 2011). Conceivably, the study of these genuine human coronaviruses (HCoVs) may yield clues to what is required for viral adaptation to the human host and thereby increase our understanding of the probabilities and risks of coronavirus cross-species transmission.
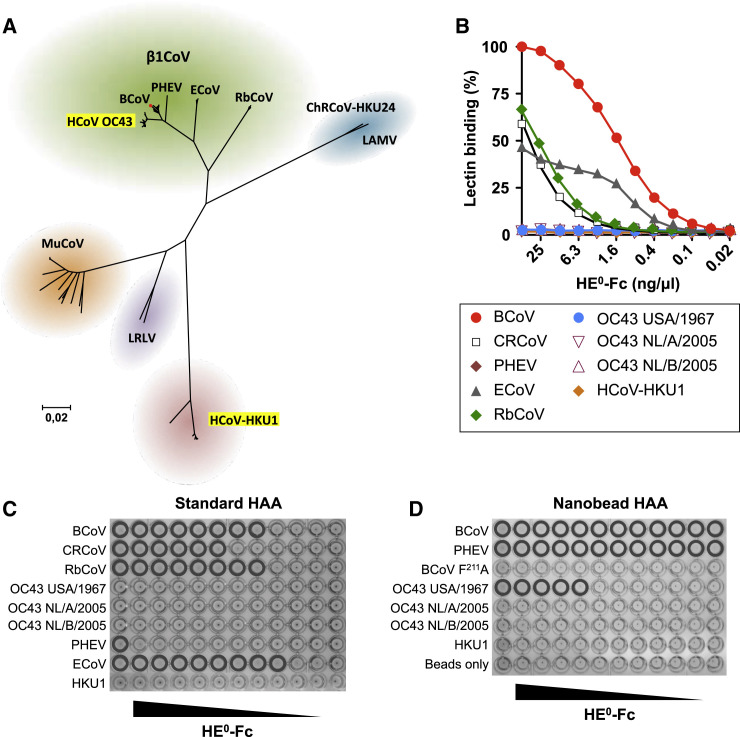
OC43 and HKU1, while generally associated with benign common colds in healthy immunocompetent individuals, may cause significant morbidity and even mortality in the frail (Morfopoulou et al., 2016, Woo et al., 2005a). OC43, the best-studied HCoV, apparently arose relatively recently, 120 to 70 years ago, with the most recent common ancestor of all extant OC43 variants dating to the 1950s (Lau et al., 2011, Vijgen et al., 2005, Vijgen et al., 2006). OC43 groups in the species Betacoronavirus-1 (β1CoV), together with highly related viruses from ruminants (bovine coronavirus, BCoV), swine (porcine hemagglutinating encephalomyelitis virus, PHEV), equines (equine coronavirus, ECoV), leporids (rabbit coronavirus HKU14, RbCoV), and canines (canine respiratory coronavirus, CRCoV) (de Groot et al., 2011; Figure 1 A). The extraordinary radiation of β1CoVs might be explained from their receptor usage, as they attach to 9-O-acetylated sialic acids (9-O-Ac-Sias) (Matrosovich et al., 2015), i.e., glycan components common in mammals and birds (Traving and Schauer, 1998). Paradoxically, however, most β1CoVs, including OC43, have very narrow host ranges. They form distinct monophyletic clades congruent with host selectivity, and phylogenetic evidence strongly argues against recurrent inter-species transmissions (Lau et al., 2011, Vijgen et al., 2006; see also Figure 1A). With the emergence of OC43 seemingly sparked by a singular one-time zoonotic event, the founder virus likely possessed unique traits already to allow efficient infection of, and transmission among, humans. In turn, these traits and subsequent adaptations to the new niche must have closed the door on reintroduction of the virus into animals. Despite the close genetic relationship between OC43 and BCoV-Mebus, particularly (96.6% identity across their entire genomes), their high-prevalence endemicity/enzooticity, and the scale and frequency of interactions between their host species, there is no evidence of OC43 spreading to cattle nor of BCoV (or any other β1CoV) efficiently spreading into humans (Figure 1A).
Figure 1.
Loss of HE-Mediated Receptor Binding in Human Betacoronaviruses
(A) Evolutionary relationships among lineage A betacoronaviruses. Neighbor-joining tree based on βCoV lineage A ORF1b sequences in the NCBI database (n = 206), with 100% bootstrap support for all major branches. Evolutionary distances were computed using the Maximum Composite Likelihood method in MEGA6 (Tamura et al., 2013). The positions of human coronaviruses OC43 and HKU1 are highlighted relative to those of animal lineage A betacoronaviruses, including the various β1CoV subspecies and classical mouse hepatitis virus-type murine coronavirus (MuCoV). ChRCoV-HKU24, Chinese rat coronavirus HKU24 (Lau et al., 2015); LAMV, Longquan Aa mouse coronavirus (Wang et al., 2015); LRLV, Longquan RI rat coronavirus (Wang et al., 2015). The position of CRCoV, a recent split-off of BCoV, is indicated by a red dot. (See also Figure S1.)
(B) HE0-Fc lectins (2-fold serial dilutions, starting at 50 ng/μL) were compared by sp-LBA for relative binding to BSM (with 50 ng/μL BCoV-HE0-Fc set at 100%).
(C) Conventional HAA with 2-fold serial dilutions of HE0-Fc fusion proteins of β1CoV members and HCoV-HKU1 (starting at 25 ng/well). Wells positive for hemagglutination are encircled.
(D) High-sensitivity nanobead HAA. Non-complexed nanobeads (“beads only”) and nanobeads complexed with lectin-inactive mutant BCoV HE0 F211A were included as negative controls.
β1CoV attachment involves two distinct types of surface projections: large 20-nm “spikes” comprised of homotrimers of the class I fusion protein S and stubby 8-nm protrusions that are homodimeric assemblies of the hemagglutinin-esterase protein HE (de Groot et al., 2011). The S protein, common to all coronaviruses, mediates receptor binding and fusion of the viral and host membranes (Heald-Sargent and Gallagher, 2012). HEs are found in toroviruses and in influenza C and D viruses, but among coronaviruses, only in lineage A betacoronaviruses (de Groot, 2006, de Groot et al., 2011, Hause et al., 2014, Matrosovich et al., 2015). HE monomers have a bimodular structure with a carbohydrate-binding (“lectin”) domain appended to an enzymatically active sialate-O-acetylesterase (“esterase”) domain (Langereis et al., 2009, Rosenthal et al., 1998, Zeng et al., 2008). Typically, in β1CoVs, both the spikes and HEs bind 9-O-Ac-Sias, while the HE esterase domain promotes virus elution through receptor destruction. In consequence of the opposing activities of receptor binding and receptor destruction, β1CoV attachment to sialoglycans is dynamic and reversible. Thus, dead-end binding of virions to decoy receptors in oropharyngeal, respiratory, and gastrointestinal mucus may be prevented. Moreover, in infected host cells, HE-mediated receptor destruction is essential for efficient release of viral progeny (Desforges et al., 2013).
Here we present a comprehensive structure-function study of OC43 HE. We demonstrate that over decades after OC43’s introduction, its evolution was marked by a progressive loss of HE receptor-binding activity through the accumulation of select mutations in the HE lectin domain. The effect of these mutations on the organization of the carbohydrate-binding site (CBS) and on receptor binding is explained from the crystal structure of the BCoV HE-receptor complex (Zeng et al., 2008) and visualized directly by the structure of a contemporary OC43 HE solved to 2.45Å resolution. Evidence is provided that loss of HE receptor-binding activity resulted in a reduction of virion-associated esterase activity toward multivalent clustered substrates. We propose that inactivation of the HE lectin domain altered the balance between virion binding and esterase-mediated virion elution, apparently as an adaptation to the sialoglycome of the human respiratory tract, and we speculate that this may be a contributing factor to host selectivity. This view is supported by our observation that the HE of HCoV-HKU1 also lost its receptor-binding properties. A mechanism is proposed in which accessibility of receptors to destruction depends on HE lectin function in relation to S-HE size differences and in which the balance between attachment and catalysis-driven virion elution is a determinant of host tropism.
Results and Discussion
Lectin Properties of β1CoV HEs
A comprehensive set of β1CoV HEs, expressed as esterase-inactive Fc-fusion proteins (HE0-Fc), was compared for their Sia-binding properties. In accordance with previous findings (Langereis et al., 2015), the HEs of most animal β1CoVs bound to bovine submaxillary mucin (BSM) in a 9-O-Ac-Sia-dependent fashion in solid-phase lectin-binding assays (sp-LBA; Figure 1B). Remarkably, however, those of PHEV strain VW572 and prototypic OC43 strain USA/1967 (also known as ATCC-VR759; McIntosh et al., 1967) showed no detectable binding. In hemagglutination assays (HAA), more sensitive than sp-LBA, PHEV HE0 tested positive, albeit weakly (Figure 1C). OC43 USA/1967 HE0, however, did not hemagglutinate. To augment HAA sensitivity through multivalency-driven high-avidity binding, we complexed HE0-Fc chimeras to protein A-coated nanobeads (Figure 1D). For PHEV HE0, sensitivity was increased 250-fold as compared to the standard assay. Under these conditions, modest, but reproducible, hemagglutination by OC43 USA/1967 HE0 was detected (Figure 1D). Apparently, it has lost most, though not all, of its lectin function.
Our observations prompted the question of whether loss of HE lectin activity is a strain-specific trait resulting from adaptation to in vitro propagation or a characteristic also shared by naturally occurring OC43 viruses. We therefore RT-PCR-amplified HE genes from more recent sputum-derived OC43 strains (respiratory season 2005). The encoded proteins OC43/NL/A/2005 and OC43/NL/B/2005 HE0, representative for the two major HE lineages in OC43 (designated “A” and “B,” Figure S1 and vide infra), did not show any detectable binding by sp-LBA or nanobead HAA (Figures 1B–1D). Apparently, loss of affinity for 9-O-Ac-Sias as seen for OC43/USA/1967 HE is not an artifact. In fact, the data suggest that HEs of contemporary OC43 variants have lost receptor-binding activity altogether.
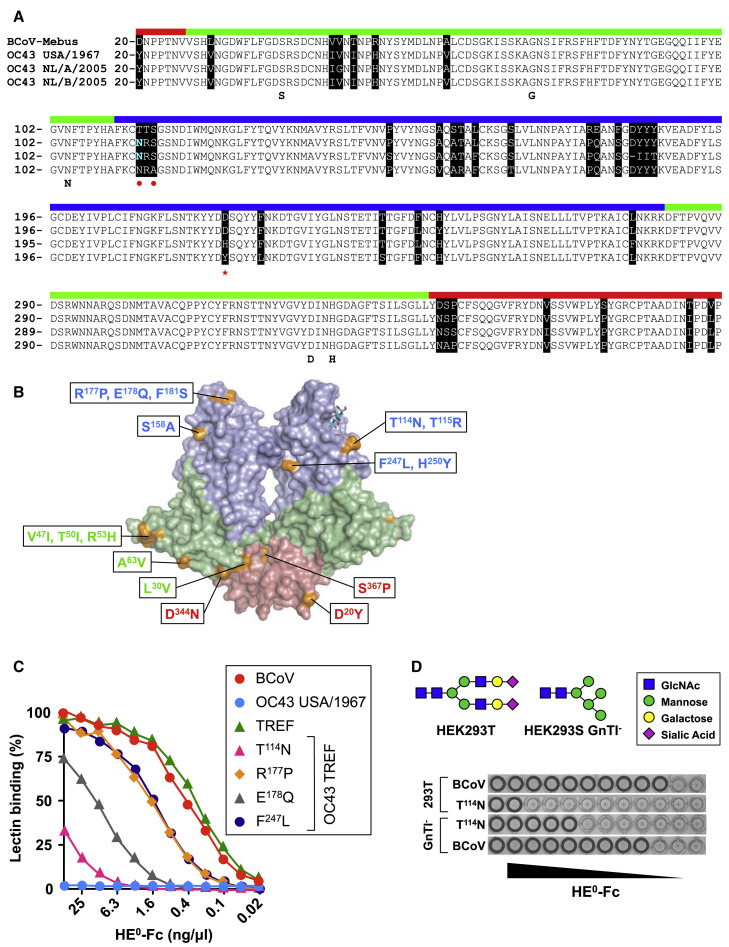
Lectin Function of OC43 HE Impeded by a Combination of Mutations
The HE ectodomains of BCoV strain Mebus and OC43 USA/1967, each 365 residues in length, differ at 18 positions only (Figures 2A and 2B). To identify the differences accounting for loss of lectin activity in OC43 USA/1967 HE, we introduced OC43-specific substitutions in BCoV-Mebus HE0-Fc, in sets and individually, and tested for a reduction in lectin activity by HAA (Figure S2A). In a complementary approach, OC43 HE residues were systematically replaced by BCoV HE orthologs (Figures S2B–S2D). The results show that the mutations in the esterase and MP domains of OC43 USA/1967 HE (Figures 2A–2C; Figure S2A) do not affect ligand binding. In fact, loss of 9-O-Ac-Sia binding can be attributed to four out of eight substitutions in the lectin domain (T114N, R177P, E178Q, and F247L; Figures S2A–S2D). Combined replacement of these residues in OC43 USA/1967 HE0 by BCoV orthologs restored binding affinity to that of BCoV-Mebus HE (Figure 2C). Individual replacement of these residues in the context of a restored OC43 USA/1967 HE (“TREF”) showed that each mutation affects receptor binding to more (T114N, E178Q) or lesser (R177P, F247L) extent (Figure 2C).
Figure 2.
Loss of OC43 USA/1967 HE0 Lectin Affinity Attributed to a Combination of Four Mutations
(A) Alignment of BCoV, OC43 USA/1967, OC43 NL/A/2005, and OC43 NL/B/2005 HE with sequences color coded by domain (membrane-proximal domain, red; esterase domain, green; lectin domain, blue). Residues crucial for esterase activity (SGNDH) are annotated. Amino acid differences are marked in black. N114 substituting for Thr is colored in cyan, and the resulting N-linked glycosylation site (NRS) marked with red dots. D220 is marked with a red asterisk.
(B) Overall structure of BCoV HE (PDB: 3CL5) with mutations in OC43 USA/1967 HE indicated. Domain coloring as in (A).
(C) Comparison of binding affinities of BCoV HE0, OC43 USA/1967 HE0, and derivatives by sp-LBA. BCoV, BCoV-Mebus HE0; OC43 TREF, OC43 USA/1967 HE0 with N114, P177, Q178 and L247 replaced by BCoV-Mebus HE orthologs; T114N, R177P, E178Q, and F247L, TREF derivatives with indicated residues re-converted to the autologous OC43 orthologs.
(D) Conventional HAA with BCoV-Mebus HE0 T114N expressed in HEK293T or HEK293S GnTI− cells. (See also Figure S2.)
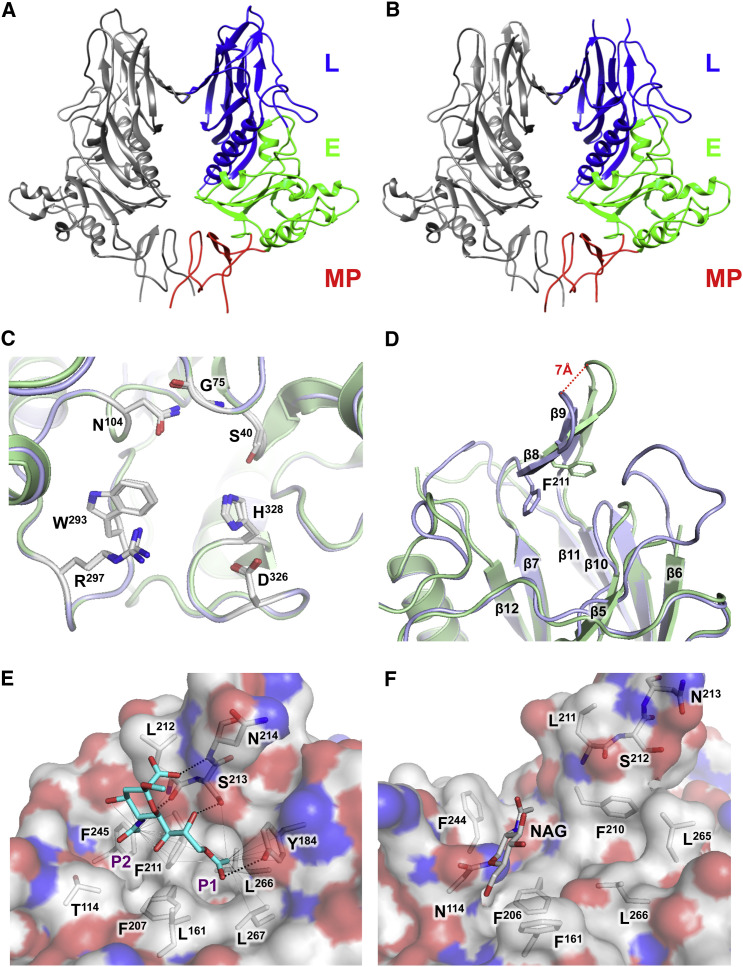
The impact of these mutations on 9-O-Ac-Sia binding can be understood from the crystal structure of the BCoV-Mebus HE-receptor complex (Zeng et al., 2008). The BCoV HE receptor-binding region is comprised of six surface loops, five grafted on the beta-sandwich core of the lectin domain and one emanating from the esterase domain (Figure S3A). The actual carbohydrate-binding site consists of a deep hydrophobic pocket P1 and a more shallow hydrophobic depression P2 that accommodate the methyl groups of the Sia-9-O- and −5-N-acetyl moieties, respectively (Figure 4E). The side chain of F211 (a residue in the β12/β13 β-hairpin) separates P1 from P2 and, in the HE-receptor complex, intercalates between the Sia-acetyl groups. Ligand binding, thus largely based on shape complementarity and hydrophobic interactions, is stabilized by extensive protein-sugar hydrogen bonding, particularly involving the β7–β10 β-hairpin, through main chain atoms of L212 and N214 to the sialate-5-N-acyl amide and the Sia carboxylate, respectively, and through the side-chain hydroxyl of S213 to the sialate-C8-hydroxyl and -C9-acyl oxygen. Y184 in the β5-β6 loop (residues 176–185) is particularly important for ligand binding as its side-chain hydroxyl group hydrogen bonds with the sialate-9-O-acetyl carbonyl, while its aromatic ring together with the side chains of F211, L266, and L267 walls the P1 pocket that is key to receptor recognition (Figure 4E).
Figure 4.
Crystal Structure of OC43 NL/A/2005 HE
(A and B) Side-by-side cartoon representations of the overall crystal structures of BCoV-Mebus HE (A) and OC43 NL/A/2005 HE (B). HE monomers are colored gray or by domain (as in Figure 2A).
(C) BCoV and OC43 HE have identical esterase catalytic sites. Overlay of BCoV (blue) and OC43 NL/A/2005 (green) HE esterase domains. Cartoon representations with residues crucial for activity indicated as sticks.
(D) Overlay of BCoV and OC43 NL/A/2005 HE lectin domains. F211 indicated with sticks.
(E) 9-O-Ac-Sia (with carbon atoms in cyan) binding in the BCoV-Mebus HE lectin CBS as observed in the crystal complex (PDB: 3CL5). Close up with contacting amino acid side chains shown in stick representation. Hydrogen bonds are shown as black dashed lines, and hydrophobic interactions with the Sia-9-O- and −5-N-methyl groups as thin gray lines. P1 and P2 indicate the pocket and hydrophobic depression, which accommodate the methyl groups of the Sia-9-O- and −5-N-acetyls, respectively.
(F) Close up of the inactivated CBS of OC43 NL/A/2005 HE. Residues corresponding to those in (E) are in stick representation and colored by atom type. NAG, N-acetylglucosamine attached to N114, is shown in stick representation. (See also Figure S3 and Table S1.)
Residues of the α5-α6 loop (residues 111–118) do not directly contact the ligand. However, the T114N substitution in OC43 HE created a novel N-linked glycosylation site (TTS→NRS) (Figure 2A). A glycan attached to this site would extend into the CBS and potentially reduce receptor binding through steric hindrance. Indeed, the introduction of this glycosylation site in BCoV HE0 resulted in an almost complete loss of binding as detected by sp-LBA (Figure 2C) and a 250-fold loss of apparent binding affinity as measured by HAA (Figure 2D; Figure S2A). The same mutant protein, but now expressed in N-acetyl glucosamine transferase I (GnTI)-deficient HEK293S GnTI− cells instead of HEK293T cells, showed a less pronounced loss of binding affinity (30-fold in HAA; Figure 2D). The data show that glycosylation at N114 indeed reduces the affinity of the HE lectin domain for 9-O-Ac-Sias and that this reduction is positively correlated with glycan size/complexity.
In BCoV HE, E178 fixates the β5-β6 loop by engaging in double hydrogen bonds with the backbone nitrogen atoms of S155 and A156 in the β4-β5 loop (Figure S3B). We offer that in OC43 HE, the replacement by Gln and consequential loss of inter-loop hydrogen bonding destabilizes the β5-β6 loop and that its increased flexibility impairs the Sia-binding site, presumably by affecting the critical P1 pocket. The consequences of the other two substitutions in OC43 USA/1967 HE are less evident, although it can be envisaged that the R177P replacement might again affect folding and/or stability of the β5-β6 loop and thereby indirectly the positioning of the critical Y184 residue. The F247L substitution is more difficult to explain as the affected residue locates to the core of the HE lectin domain, distal to the CBS, and the mutation apparently decreases ligand binding through indirect long-range effects. The combined data show for the early OC43 isolate USA/1967 that the reduced lectin activity of its HE resulted not from a single but from a combination of mutations.
Progressive Loss of HE Lectin Function during OC43 Evolution
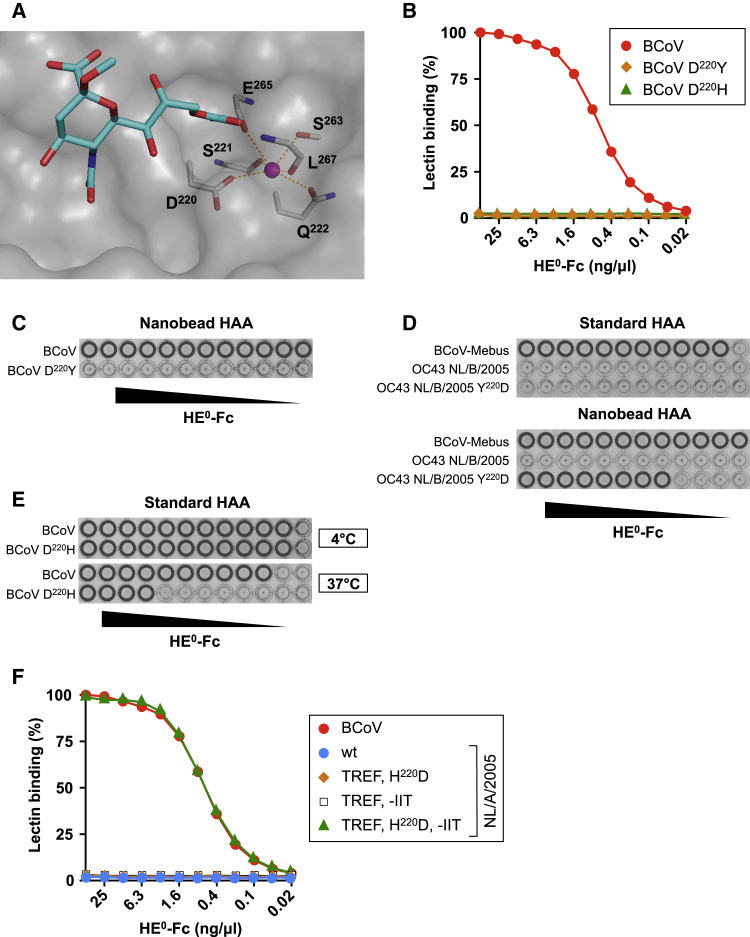
In the course of this study, many full genome sequences from OC43 field variants from the US, Western Europe, and China, well documented with respect to place and date of virus sampling, became available in GenBank. This wealth of information allowed us to study the recent evolution of the OC43 HE protein. In phylograms, in accordance with the presumptive date of OC43 emergence, the HE of the USA/1967 strain is placed close to the root (Figure S1). In the decades thereafter, the HE proteins divided into three clades. Clade C, the least populous, is comprised of HEs from OC43 strains sampled in the early 1990s with no recent representatives and may have been replaced by clades A and B. Comparative sequence analysis in combination with structure-function analysis showed that both type A and B HEs acquired several mutations in the lectin domain additional to those already present in OC43 USA/1967, which explains the loss of HE receptor binding in extant OC43 variants. Unexpectedly, in B-type HEs, but not in those of types A and C, glycosylation at N114 was lost again due to a S116A substitution (NRS→NRA). Further inspection revealed the replacement in B-type OC43 HEs of D220 by Y. D220 is part of a potassium ion-coordinating metal-binding site (MBS), a signature element of coronavirus HE lectin domains involved in the organization of the Sia-binding site (Figure 3 A) (Zeng et al., 2008). The MBS is formed by main-chain carbonyl atoms of S221, E265, and L267, together with the side chains of Q222, S263, and D220, the latter of which is particularly important as it balances the positive charge of the metal ion. In BCoV HE0, the D220Y mutation resulted in loss of all detectable binding to 9-O-Ac-Sia (Figures 3B and 3C). In turn, a Y220D back mutation in the type B HE of OC43 NL/B/2005 restored Sia-binding activity to levels detectable by high-sensitivity nanobead HAA (Figure 3D). Again, the structure of the BCoV HE-receptor complex provides an explanation for the detrimental effect of the mutation. As L266 and L267 are in a loop that is stabilized through the coordination of the potassium ion, disruption of the MBS might affect the folding of this loop, the positioning of the Leu residues, and thereby the structure of receptor-binding pocket P1.
Figure 3.
Complete Loss of HE Lectin Function during OC43 Evolution due to Progressive Accumulation of Mutations
(A) Close up of the BCoV-Mebus HE CBS in complex with α-Neu5,9Ac22Me (in sticks). Residues comprising the metal-binding site (MBS) are also shown in sticks, the potassium ion is shown as a magenta sphere, and interactions between the metal ion and coordinating amino acid residues as red dashed lines.
(B) Disruption of the MBS in BCoV HE leads to loss of lectin affinity. sp-LBA as in Figure 1B.
(C) Disruption of the BCoV HE lectin MBS through D220Y substitution renders receptor binding non-detectable even by high-sensitivity nanobead HAA.
(D) The Y220D mutation partially restores lectin affinity of OC43 NL/B/2005 HE0.
(E) D220H substitution in BCoV HE results in thermolability of receptor binding. Conventional HAA before (4°C) and after (37°C) a temperature shift up.
(F) Receptor binding of OC43 NL/A/2005 HE0 and derivatives as determined by sp-LBA. In OC43 NL/A/2005 HE0, N114, P177, Q178, and L247 were replaced by BCoV-Mebus HE orthologs (TREF), in combination with (1) H220D substitution (TREF, H220D), (2) repair of the β5-β6 loop by substituting D183YYY186 for IIT (TREF, −IIT), or (3) H220D and repair of the β5-β6 loop (TREF, H220D, −IIT).
Remarkably, also in A-type OC43 HEs, the MBS was targeted by substitution of D220, now by His (Figure 2A). As evident from comparative sequence and phylogenetic analysis, the substitutions in A- and B-type HEs were independent events, and their selection must be ascribed to convergent evolution. Again, D220H mutation in BCoV HE resulted in loss of detectable Sia binding as measured by sp-LBA (Figure 3B). Unexpectedly, however, in conventional HAA routinely performed at 4°C, wild-type and mutant BCoV HEs hemagglutinated to similar extent (Figure 3E). The effect of the mutation became evident after a shift up to 37°C, upon which hemagglutination with wild-type BCoV HE remained stable while that of the mutant protein resolved (Figure 3E). Apparently, substitution of D220 by His is less disruptive than by Tyr, but results in thermolability of HE receptor binding, likely to cause reduced binding affinity under physiological conditions. Repair of the OC43-specific substitutions already present in the HE of the 1967 strain (Figure 2A), in conjunction with a H220D back substitution, did not restore lectin activity of OC43 NL/A/2005 HE (Figure 3F), suggesting the presence of at least one additional mutation to prevent receptor binding. Comparative sequence analysis revealed a major change uniformly shared by A-type HEs again in the β5-β6 loop, with four adjacent residues 183–186, Asp-Tyr-Tyr-Tyr, replaced by Ile-Ile-Thr (Figure 2A), apparently as a result of a double frameshift mutation (Figure S3C). Repair of this mutation, in combination with restorative changes at the other five sites, was required for OC43 NL/A/2005 HE to regain receptor-binding activity (Figure 3F). The impact of the β5-β6 loop mutation is easily understood from the structure of the BCoV HE-receptor complex, as it involved loss of Y184, an essential residue for ligand binding.
Crystal Structure of an OC43A-type HE
To assess the consequences of the combined mutations in the type A HE lectin domain, we determined the crystal structure of OC43 NL/A/2005 HE at 2.45Å resolution (for crystallographic details, see Table S1; PDB: 5N11). In overall fold, OC43 HE is highly similar to other coronavirus HEs (Figure 4 ). Differences in the esterase domain are limited to substitution of five surface residues, none likely to impair esterase function or activity. OC43 HE is also identical to BCoV HE in most of its lectin domain, particularly in the beta-sandwich core, but the loops shaping the receptor-binding site have changed dramatically (Figure 4D; Figure S3D). The β11-β12 and β5-β6 loops are disordered, suggestive of extensive flexibility, in the case of the latter loop in accordance with loss of E178-mediated inter-loop hydrogen bonding. Those (parts of the) loops that can be visualized are reoriented and displaced with respect to their original position in BCoV HE, utterly destroying the CBS. For example the β7/β10 β-hairpin, comprising F211, L212, S213, and N214 that are key to protein-ligand interaction in BCoV HE (Zeng et al., 2008), has shifted by approximately 7Å (Figure 4D). The P1 pocket, arguably the most critical element of the CBS, no longer exists as (1) F211 is no longer at its original position and its side chain is rotated by 90°, (2) Y184 was lost as a result of the frameshift mutation in the β5-β6 loop, and (3) the side chains of L266 and L267 are reoriented apparently due to the loss of the MBS (Figures 4E and 4F). Finally, attached to N114 is an N-acetylglucosamine, providing formal evidence that the newly introduced glycosylation site in OC43 HEs is functional. Although the remaining residues of the sugar chain could not be visualized, the glycan would stretch across the remains of the CBS (Figure 4F).
Loss of HE Receptor Binding in HCoV-HKU1: A Case of Convergent Evolution?
OC43 and HKU1 occupy the same niche, prompting the question whether they have been subject to similar selective constraints. HCoV-HKU1 is also a lineage A betacoronavirus but distantly enough related to the β1CoVs to be assigned to a separate species (de Groot et al., 2011) (Figure 1A). Like OC43, HCoV-HKU1 binds to 9-O-Ac-Sia receptor determinants via its spike (Huang et al., 2015), and it possesses an HE (Woo et al., 2005b). The HE of the prototypical HCoV-HKU1 2005 Hong Kong genotype A strain (Woo et al., 2006) tested negative for 9-O-Ac-Sia binding by sp-LBA, standard HAA, and nanobead HAA (Figures 1B–1D). Comparative sequence analysis, including genotypes A and B, of HCoV-HKU1 (Woo et al., 2006) shows that the HKU1 HE has undergone massive deletions, as a result of which most of the lectin domain was lost (Figure S4).
Loss of HE Lectin Function Reduces Receptor-Destroying Enzyme Activity toward Multivalent Substrates
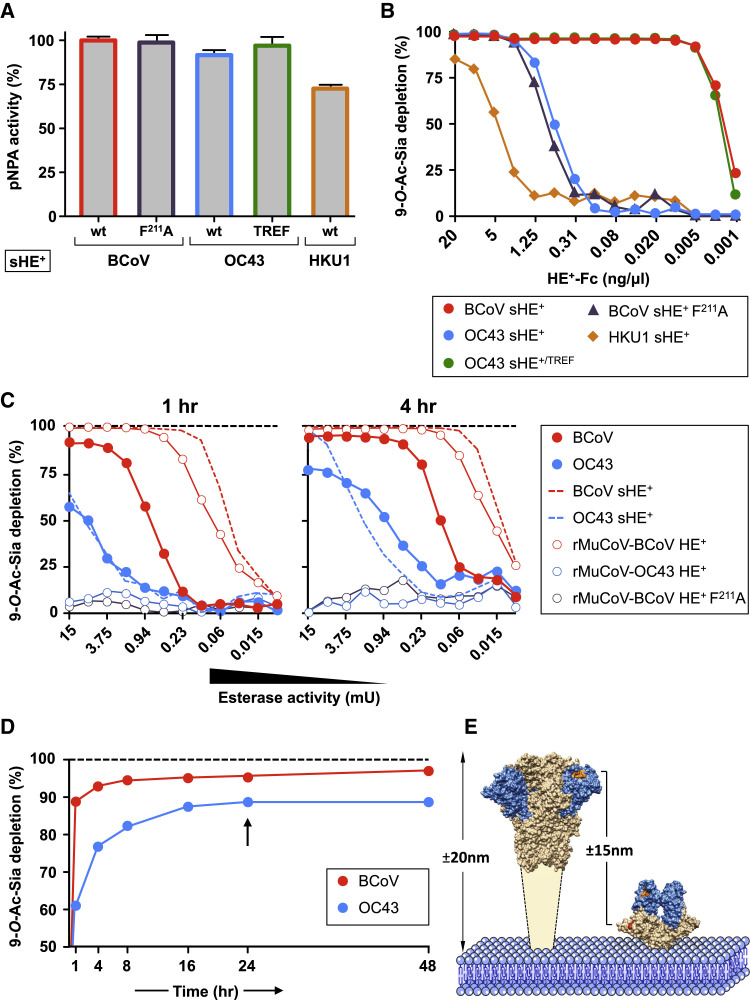
In influenza C and D viruses, the hemagglutinin-esterase fusion protein is uniquely responsible for viral attachment and entry, and hence HE function is receptor binding first and foremost. The same holds for the HEs of MHV-type murine betacoronaviruses (MuCoV) that mediate virion binding to O-Ac-sialoglycan-based attachment factors (Langereis et al., 2010), while S binds to the proteinaceous entry receptor CEACAM-1 (Peng et al., 2011). At the other end of the spectrum, in OC43 and HKU1, HE’s sole remaining function is receptor destruction, with virion attachment to 9-O-Ac-sialoglycans exclusively assigned to S. For the animal β1CoVs that retain a functional HE lectin domain, however, the role of HE is less obvious. On the one hand, HE might participate in attachment to increase virion binding avidity. On the other, its lectin domain may serve primarily to promote catalytic activity toward high-multivalency substrates by bringing the esterase in close proximity to and prolonged association with clustered glycotopes, in analogy with the carbohydrate-binding modules of cellular glycoside hydrolases (Boraston et al., 2004). In the latter case, the HE lectin CBS in the human lineage A βCoVs may have been lost to downregulate receptor-destroying activity. To study this, we compared soluble recombinant HEs of BCoV, OC43, and HKU1 for their relative enzymatic activity toward the monovalent substrate p-nitrophenol acetate (pNPA) and the multivalent substrate BSM. The latter glycoconjugate carries hundreds of O-linked glycans clustered in such densities to give the protein a filamentous bottlebrush appearance typical for mucins (Zappone et al., 2015). Whereas all three HEs showed comparable specific activities when assayed with pNPA (Figure 5 A), those of OC43 and HKU1 were more than 250-fold less active than BCoV HE in receptor-destruction assays with BSM (Figure 5B). Inactivation of the BCoV HE lectin CBS through a F211A substitution reduced its esterase activity toward BSM to that of wild-type OC43 HE. Conversely, restoration of the OC43 HE lectin domain increased esterase activity to that of BCoV HE (see “TREF,” Figure 5B). Thus, BCoV and HCoV HEs indeed differ in their reactivity toward clustered receptors in consequence of the respective conservation or loss of lectin function.
Figure 5.
Loss of HE Lectin-Mediated Receptor Binding Alters Sialate-9-O-Acetylesterase Receptor, Destroying Activity toward Multivalent Substrates
(A) Esterase activity of soluble recombinant HE+-Fc fusion proteins (sHE+) toward monovalent substrate pNPA. Enzymatic activity is shown as percentage of BCoV HE wild-type activity. WT, sHE+ with wild-type HE ectodomains of BCoV-Mebus, OC43 USA/1967, or HCoV-HKU1 as indicated; F211A, BCoV-Mebus sHE+ derivative with the lectin CBS inactivated through a F211A substitution (Zeng et al., 2008); TREF, OC43 USA/1967 HE with the lectin CBS repaired (as in Figure 2C). Data are represented as mean ± SEM.
(B) Loss of sHE+-mediated receptor binding results in reduced esterase activity toward high-multivalency substrates as determined by on-the-plate O-Ac-Sia depletion assay. BSM, coated on maxisorp plates, was incubated for 1 hr with 2-fold serial dilutions of sHE+. Receptor destruction was assessed by sp-LBA with BCoV HE0-Fc at fixed concentration (2 ng/μL).
(C) Whole-virion receptor-destruction assays with 2-fold serial dilutions of purified viruses (starting at 15 mU). BCoV and OC43 sHE+s were included for comparison. Receptor destruction was measured after 1 hr or 4 hr incubation. Black dotted lines indicate 100% receptor destruction as determined with excess amounts of BCoV or OC43 sHE+.
(D) Whole-virus-mediated receptor destruction over time with fixed amounts of BCoV-Mebus and OC43 USA/1967 virions (15 mU). To exclude “exhaustion” (inactivation of OC43 esterase and/or changes in the physicochemical properties of the virions over time), we removed BCoV and OC43 at t = 24 and replaced them with equal amounts of freshly thawed aliquots of the virus stocks (black arrow).
(E) Scaled side-by-side representations of the structures of coronavirus S (Walls et al., 2016) and HE proteins. Indicated are the estimated heights of S and HE and the approximate distance separating S receptor-binding sites and HE catalytic sites. (See also Figure S4 and S5 and Table S2.)
Consequences of Loss of Lectin Function for Virion-Mediated Receptor Destruction
OC43 HE efficiently depletes endogenous pools of O-Ac-Sias within infected cells (Figure S5) as is critical for the release of viral progeny (Desforges et al., 2013). The loss of HE lectin function therefore is more likely to affect virus biology at another stage, namely that of virion (pre)attachment. Of note, the observations for recombinant soluble HEs cannot be extrapolated one-to-one to virion-associated HEs, because in virus particles, HE esterase activity will be affected by S-mediated attachment to 9-O-Ac-Sias as well as by the close packing of HE molecules in the viral envelope. Serial dilutions of intact BCoV and OC43 virions, adjusted for pNPA esterase activity (Table S2), were therefore compared in solid-phase receptor-destruction assays with BSM (Figures 5C and 5D). Remarkably, the difference between the two viruses as measured by the onset of detectable receptor destruction was smaller than for their respective soluble HEs.
The activity of virion-associated BCoV HE relative to that of soluble BCoV HE was reduced 8- to 16-fold. Conversely, the relative activity of virion-associated OC43 was increased at least in that destruction of receptors already became apparent at lower enzyme concentrations. (Figure 5C). To study whether these phenomena relate to the S protein and to S-mediated virion attachment to 9-O-Ac-Sias, we constructed recombinant murine coronaviruses (rMuCoVs) that express MuCoV S, which does not bind 9-O-Ac-Sias, in combination with either BCoV or OC43 HE. rMuCoV virions studded with OC43 HE lacked detectable esterase activity toward BSM. In contrast, the esterase activity of rMuCoVs expressing BCoV HE was 6- to 8-fold higher than that of wild-type BCoV virions (Figure 5C). Apparently, β1CoV S proteins, by binding to 9-O-Ac-Sias negatively or positively, affect HE esterase activity in BCoV and OC43 virions, respectively, depending on whether or not the HE lectin domain is functional. Most strikingly, BCoV and OC43 virions differed in the extent to which receptors were depleted. The soluble HEs of OC43 and BCoV, despite their difference in enzyme activity, destroyed all O-Ac-Sias as detectable by sp-LBA. BCoV virions destroyed 95% of the receptors after 24 hr incubation, which increased to 98% after 48 hr (Figure 5D). For OC43 virions, the effect was more pronounced such that after 24 hr only 85% of the receptors were destroyed. The remaining receptors were resistant even to continued incubation (Figure 5D). The data suggest that, in the context of the virion, the absence of a functional HE lectin domain results not only in reduced enzyme activity toward clustered receptors, but also prohibits cleavage of particular receptor populations. As these “protected” receptor populations are readily depleted by soluble OC43 HE (Figures 5B–5D), their resistance to cleavage by OC43 virions suggests that they are inaccessible to HEs when embedded in the viral envelope and in the presence of S.
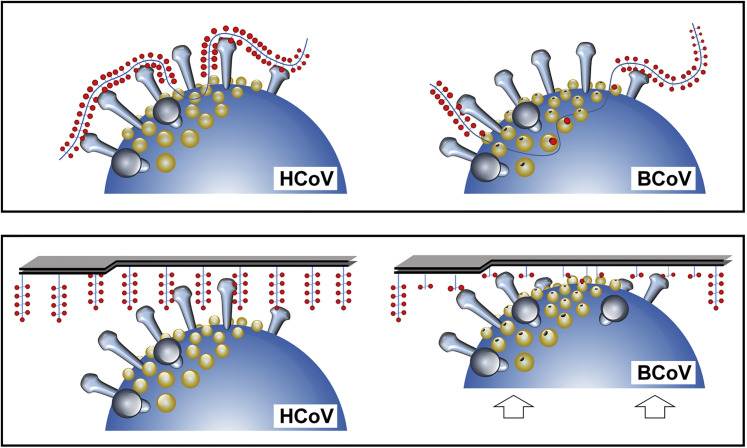
The data prompt a model in which the susceptibility of populations of clustered receptors to cleavage depends on (1) their accessibility, as determined by the distance by which they extend from a fixed surface—the bottom of the ELISA well in our artificial system and, for example, the host cell membrane in vivo—and (2) the considerable difference in height between S and HE (20 and 8 nm, respectively), with the CBSs of S and those of HE esterase domains separated by a distance of up to 15 nm (Figure 5E). S-mediated virion attachment to a 9-O-Ac-sialylated surface would bring the HE catalytic site in proximity of some clustered receptors (which would explain the stimulating effect of S on HE esterase activity in the case of OC43 virions, but not observed for rMuCoV-OC43 HE+) but at the same time would keep the virus-associated enzyme at “arm’s length” from other receptor populations, readily accessible to soluble HEs (which would explain the inhibiting effect of S on esterase activity in the case of BCoV virions and the apparent lack thereof in the case of rMuCoV-BCoV HE+) (Figures 5C and 5D). Thus, a distinction arises between clustered receptor populations that come within immediate reach of the HE esterase and that therefore would be cleaved by virion-associated HE as efficiently as by soluble HE and those that are kept out of reach of virion-associated HE as a result of S-mediated receptor-binding and that are therefore cleaved at strongly reduced rates or even rendered non-cleavable unless HE is provided with a functional lectin domain (Figure 6 ). By virions grasping on to multivalent sialoglycoconjugates via the HE receptor-binding sites and “drawing them in” closer to the surface of the envelope, clustered receptors would yet become available to the HE esterase domain (Figure 6). Such a mechanism may be promoted by cooperativity among adjacent HE molecules and, membrane fluidity permitting, possibly even lead to redistribution of surface projections in the viral envelope, resulting in displacement of spikes, local recruitment of HEs, and formation of a “receptor-destruction patch.” Saliently, some receptor populations may be completely resistant to destruction even by virion-associated HEs with a functional lectin domain, such as the 2% of detectable O-Ac-Sias remaining after 48 hr of incubation with BCoV (Figure 5D). It is tempting to speculate that under natural conditions, the distinction between decoy and entry receptors is made by default on basis of their accessibility to cleavage by virion-associated HE.
Figure 6.
Hypothetical Model for the Interaction of HCoV and BCoV Virions with Multivalent Glycoconjugates
Schematically depicted are portions of HCoV (OC43 or HKU1) and BCoV virions, with large spikes comprised of S (in gray) and smaller protrusions comprised of HE (yellow) extending from the viral membrane (blue). Functional HE lectin CBSs are indicated by black holes (only one shown per HE dimer for reasons of simplicity). Also shown schematically are membrane-anchored (bottom) and non-anchored (top) mucin-type glycoconjugates of bottle-brush filamentous appearance with clustered receptors (9-O-Ac-Sias, red dots) arranged in linear arrays and with absence of red dots indicating receptor-destruction. The model, based on the size difference between S and HE, visualizes how loss of HE lectin function might alter virion-associated receptor-destroying activity, reducing the specific activity of virions as well as the rate and selectivity of receptor destruction. In virions with lectin-deficient HEs, clustered substrates will be largely kept at a distance from the HE esterase catalytic pocket as a result of S-glycoconjugate interaction. In contrast, HEs with intact lectin CBS may draw in portions of the glycoconjugates (or will draw the virion-associated HEs toward them), aided by cooperativity of binding between adjacent HEs within the viral envelope. Thus, clustered glycotopes become fixed within reach of the esterase catalytic sites and receptor destruction is promoted.
Conclusion
We showed that OC43—after its zoonotic introduction—has been under incessant selective pressure to adapt to the sialoglycome of the human host. HE-associated receptor binding was selected against, largely lost early on, and ultimately lost altogether through an accumulation of mutations in the lectin domain over a period of decades. In result, the balance between virion attachment and catalytically driven virion elution was reset, apparently to meet the specific requirements for optimal replication in human airways and to allow the virus in this particular niche to distinguish between decoys marked for destruction and functional receptors that should be preserved for cell entry. This view is reinforced by our observation that HCoV-HKU1, another respiratory lineage A βCoV of humans, followed a convergent evolutionary path and also lost HE-mediated receptor binding. We offer that differences in the sialoglycomes of bovids and humans, such as in the local expression, structure, and density of 9-O-acetylated sialoglycoconjugates, may pose incompatibilities during cross-species transmission of OC43 and BCoV and that the respective loss or preservation of HE-mediated receptor binding contributes to the host tropism of these viruses. This said, comparative sialoglycomics is still in its infancy, humans and bovids have not been studied for differences in O-Ac-sialoglycan expression in any detail, and the precise sialoglycan-based constraints that selected for the particular traits of the animal and human βCoVs therefore remain to be identified. At any rate, our findings do reveal an as yet unappreciated aspect of lineage A βCoV adaptation to humans. Of broader relevance, they provide a general paradigm for dynamic, catalysis-driven virus-sialoglycan interactions in relation to host selectivity. This notion is supported by observations for influenza A viruses, where neuraminidase (NA) activity toward multivalent substrates varies with differences in the length of the stalk domain and positively correlates with NA size (Castrucci and Kawaoka, 1993, Els et al., 1985), where NA catalytic activity is modulated by the absence or presence of a second Sia-binding site (Uhlendorff et al., 2009, Varghese et al., 1997) and where changes in NA stalk length and NA lectin affinity have been implicated in host specificity (Blumenkrantz et al., 2013, Castrucci and Kawaoka, 1993, Lai et al., 2012, Li et al., 2014, Matrosovich et al., 1999, Munier et al., 2010, Uhlendorff et al., 2009, Varghese et al., 1997). These observations can be readily interpreted in the context of our data and model proposed.
Experimental Procedures
Materials and experimental procedures are detailed in the Supplemental Experimental Procedures.
Expression and Purification of HEs
Human codon-optimized sequences of the HEs of BCoV-Mebus, ECoV-NC99, PHEV-VW572, RbCoV-HKU14-1, CRCoV-240/05, HCoV-HKU1, and OC43 USA/1967 (ATCC VR-759) were cloned in expression plasmid pCD5-T-Fc (Zeng et al., 2008). The resulting constructs encode chimeric proteins comprised of the HE ectodomain fused to the human IgG1 Fc domain, with the domains separated by a thrombin cleavage site. The fusion proteins were either expressed in an enzymatically active form (HE+) or rendered inactive through a catalytic Ser-to-Ala substitution (HE0). Plasmids coding for OC43 NL/A/2005 and OC43 NL/B/2005 HE were constructed through site-directed mutagenesis of the OC43 USA/1967 HE expression vector using the Q5 kit (New England Biolabs). For lectin affinity and esterase activity assays, HE-Fcs were produced by transient expression in HEK293T cells and purified from cell culture supernatants by protein A affinity chromatography and low pH elution as described (Zeng et al., 2008). For crystallization, OC43 NL/A/2005 HE+-Fc was expressed in HEK293 GnTI(−) cells and purified by protein A affinity chromatography, followed by on-the-beads thrombin cleavage as described (Zeng et al., 2008). Beads were pelleted, and the HE ectodomain in the supernatant was concentrated to ∼15 mg/mL. HE genes from OC43 field strains were RT-PCR amplified with OC43 viral RNA, directly isolated from nasopharyngeal aspirates (van de Pol et al., 2006) as a template, and their sequences were used to construct codon-optimized versions.
Purification of Virions for Whole-Virus Receptor-Destruction Assays
Cell monolayers were infected at MOI 0.01. Virions were purified by ultracentrifugation through 20% (w/v) sucrose cushions and resuspended in PBS. Virus preparations were analyzed for particle content by qPCR, plaque assay, quantitative latex bead ratio EM, and pNPA esterase activity assay (Table S2).
Hemagglutination Assay
HAA was performed with rat erythrocytes (Rattus norvegicus strain Wistar; 50% suspension in PBS) and, in standard tests, with 2-fold serial dilutions of HE0-Fc proteins (starting at 25 ng/well) as described (Zeng et al., 2008). For high-sensitivity HAA, based on multivalency-driven high-avidity binding, 5 μg HE0-Fc was complexed to 2 × 109 protein A-coated 100 nm nanobeads (Chemicell) in 100 μL PBS for 45 min at 4°C prior to 2-fold serial dilution. Hemagglutination was for 2 hr at 4°C unless stated otherwise.
Solid-Phase Lectin-Binding Assay and On-the-Plate O-Ac-Sia Depletion Assay
sp-LBAs were performed with bovine submaxillary mucin (BSM) and 2-fold serial dilutions of HE0-Fc proteins as described (Langereis et al., 2012, Langereis et al., 2015).
Receptor-destroying esterase activities of soluble recombinant HEs (sHE+) and whole viruses were measured by on-the-plate O-Ac-Sia depletion assay, performed essentially as described (Langereis et al., 2015). BSM, coated on Maxisorp flat-bottom 96-well plates, was (mock-)treated with 2-fold serial dilutions of sHE+ in PBS (100 μL/well) starting at 20 ng/μL for 1 hr, or, for whole-virion receptor-destruction assays, with virions or sHE+ starting at 15 mU esterase activity for up to 48 hr at 37°C. Depletion of O-Ac-Sia receptors was detected by sp-LBA with BCoV-LUN HE0 (2 ng/μL; 100 μL/well). Esterase activity was plotted as the inverse of lectin-binding measurements, expressed in percentages.
Author Contributions
M.J.G.B. designed and performed experiments, analyzed the data, and wrote the paper. Y.L., L.J.F., R.J.G.H., and M.A.L. designed and performed experiments and analyzed data. S.A.H.d.P., A.L.W.v.V., and I.M. performed experiments. J.D.F.d.G.-M. provided materials, analyzed data, and advised on data interpretation. F.J.M.v.K. and E.G.H. provided funding, analyzed data, and advised on data interpretation. R.J.d.G. conceived and supervised the study, analyzed data, provided funding, and wrote the paper. All authors commented upon/edited the manuscript.
Acknowledgments
We acknowledge the Paul Scherrer Institut, Villigen, Switzerland for provision of synchrotron radiation beam time at beamline PX of the Swiss Light Source (SLS) and its beamline scientists for assistance. We thank Jean-Luc Murk and Marco Viveen from the Diagnostic Electron Microscopy Unit, Medical Microbiology, University Medical Center Utrecht for the titration of virus stocks and Richard Wubbolts from the Center for Cell Imaging of the Utrecht Faculty of Veterinary Medicine for advice and assistance. This work was supported by ECHO grant 711.011.006 of the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW).
Published: March 8, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2017.02.008.
Accession Numbers
The accession number for the crystal structure of HCoV-OC43 NL/A/2005 HE reported in this paper is PDB: 5N11.
Supplemental Information
References
- Blumenkrantz D., Roberts K.L., Shelton H., Lycett S., Barclay W.S. The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J. Virol. 2013;87:10539–10551. doi: 10.1128/JVI.00967-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraston A.B., Bolam D.N., Gilbert H.J., Davies G.J. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrucci M.R., Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J. Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj. J. 2006;23:59–72. doi: 10.1007/s10719-006-5438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J. Family Coronaviridae. In: King A., Lefkowitz E., Adams M.J., Carstens E.B., editors. Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; 2011. pp. 806–828. [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desforges M., Desjardins J., Zhang C., Talbot P.J. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J. Virol. 2013;87:3097–3107. doi: 10.1128/JVI.02699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Els M.C., Air G.M., Murti K.G., Webster R.G., Laver W.G. An 18-amino acid deletion in an influenza neuraminidase. Virology. 1985;142:241–247. doi: 10.1016/0042-6822(85)90332-0. [DOI] [PubMed] [Google Scholar]
- Hause B.M., Collin E.A., Liu R., Huang B., Sheng Z., Lu W., Wang D., Nelson E.A., Li F. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio. 2014;5 doi: 10.1128/mBio.00031-14. e00031–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q.K., Marasco W.A., Baric R.S., Sims A.C., Pyrc K. Human Coronavirus HKU1 spike protein Uses O-acetylated sialic Acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J.C.C., Garcia J.M., Dyason J.C., Böhm R., Madge P.D., Rose F.J., Nicholls J.M., Peiris J.S.M., Haselhorst T., von Itzstein M. A secondary sialic acid binding site on influenza virus neuraminidase: fact or fiction? Angew. Chem. Int. Ed. Engl. 2012;51:2221–2224. doi: 10.1002/anie.201108245. [DOI] [PubMed] [Google Scholar]
- Langereis M.A., Zeng Q., Gerwig G.J., Frey B., von Itzstein M., Kamerling J.P., de Groot R.J., Huizinga E.G. Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases. Proc. Natl. Acad. Sci. USA. 2009;106:15897–15902. doi: 10.1073/pnas.0904266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis M.A., van Vliet A.L.W., Boot W., de Groot R.J. Attachment of mouse hepatitis virus to O-acetylated sialic acid is mediated by hemagglutinin-esterase and not by the spike protein. J. Virol. 2010;84:8970–8974. doi: 10.1128/JVI.00566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis M.A., Zeng Q., Heesters B.A., Huizinga E.G., de Groot R.J. The murine coronavirus hemagglutinin-esterase receptor-binding site: a major shift in ligand specificity through modest changes in architecture. PLoS Pathog. 2012;8:e1002492. doi: 10.1371/journal.ppat.1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis M.A., Bakkers M.J.G., Deng L., Padler-Karavani V., Vervoort S.J., Hulswit R.J.G., van Vliet A.L.W., Gerwig G.J., de Poot S.A.H., Boot W. Complexity and diversity of the mammalian sialome revealed by nidovirus virolectins. Cell Rep. 2015;11:1966–1978. doi: 10.1016/j.celrep.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Lee P., Tsang A.K.L., Yip C.C.Y., Tse H., Lee R.A., So L.-Y., Lau Y.-L., Chan K.-H., Woo P.C.Y., Yuen K.Y. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Tsang A.K.L., Fan R.Y.Y., Luk H.K.H., Cai J.-P., Chan K.-H., Zheng B.-J., Wang M., Yuen K.Y. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J. Virol. 2015;89:3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen S., Zhang X., Fu Q., Zhang Z., Shi S., Zhu Y., Gu M., Peng D., Liu X. A 20-amino-acid deletion in the neuraminidase stalk and a five-amino-acid deletion in the NS1 protein both contribute to the pathogenicity of H5N1 avian influenza viruses in mallard ducks. PLoS ONE. 2014;9:e95539. doi: 10.1371/journal.pone.0095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M., Zhou N., Kawaoka Y., Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M., Herrler G., Klenk H.D. Sialic acid receptors of viruses. Top. Curr. Chem. 2015;367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfopoulou S., Brown J.R., Davies E.G., Anderson G., Virasami A., Qasim W., Chong W.K., Hubank M., Plagnol V., Desforges M. Human coronavirus OC43 associated with fatal encephalitis. N. Engl. J. Med. 2016;375:497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- Munier S., Larcher T., Cormier-Aline F., Soubieux D., Su B., Guigand L., Labrosse B., Cherel Y., Quéré P., Marc D., Naffakh N. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J. Virol. 2010;84:940–952. doi: 10.1128/JVI.01581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. USA. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B.E.M., Raj V.S., Koopmans M.P., Haagmans B.L. Cross host transmission in the emergence of MERS coronavirus. Curr. Opin. Virol. 2016;16:55–62. doi: 10.1016/j.coviro.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal P.B., Zhang X., Formanowski F., Fitz W., Wong C.-H., Meier-Ewert H., Skehel J.J., Wiley D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traving C., Schauer R. Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. 1998;54:1330–1349. doi: 10.1007/s000180050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlendorff J., Matrosovich T., Klenk H.D., Matrosovich M. Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses. Arch. Virol. 2009;154:945–957. doi: 10.1007/s00705-009-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pol A.C., Wolfs T.F.W., Jansen N.J.G., van Loon A.M., Rossen J.W.A. Diagnostic value of real-time polymerase chain reaction to detect viruses in young children admitted to the paediatric intensive care unit with lower respiratory tract infection. Crit. Care. 2006;10:R61. doi: 10.1186/cc4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J.N., Colman P.M., van Donkelaar A., Blick T.J., Sahasrabudhe A., McKimm-Breschkin J.L. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc. Natl. Acad. Sci. USA. 1997;94:11808–11812. doi: 10.1073/pnas.94.22.11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moës E., Thoelen I., Wollants E., Lemey P., Vandamme A.-M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X.-D., Guo W.-P., Zhou R.-H., Wang M.-R., Wang C.-Q., Ge S., Mei S.-H., Li M.-H., Shi M. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Huang Y., Poon R.W., Chu C.M., Lee R.A., Luk W.K., Wong G.K., Wong B.H. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect. Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.K. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Yip C.C.Y., Huang Y., Tsoi H.-W., Chan K.-H., Yuen K.-Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappone B., Patil N.J., Madsen J.B., Pakkanen K.I., Lee S. Molecular structure and equilibrium forces of bovine submaxillary mucin adsorbed at a solid−liquid interface. Langmuir. 2015;31:4524–4533. doi: 10.1021/acs.langmuir.5b00548. [DOI] [PubMed] [Google Scholar]
- Zeng Q., Langereis M.A., van Vliet A.L.W., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. USA. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.