Abstract
Channel-forming proteins are found in a number of viral genomes. In some cases, their role in the viral life cycle is well understood, in some cases it needs still to be elucidated. A common theme is that their mode of action involves a change of electrochemical or proton gradient across the lipid membrane which modulates the viral or cellular activity. Blocking these proteins can be a suitable therapeutic strategy as for some viruses this may be “lethal.” Besides the many biological relevant questions still to be answered, there are also many open questions concerning the biophysical side as well as structural information and the mechanism of function on a molecular level. The immanent biophysical issues are addressed and the work in the field is summarized.
Key Words: Viruses, Viral channel proteins, Antiviral therapy, Protein assembly
1. Introduction
Genomes of viruses encode a series of proteins which span the membrane and alter the permeability of viral and cellular membranes (Fischer, 2005, Gonzales and Carrasco, 2003). In combination with in vitro experiments, it has been proposed that these proteins need to form pores or some kind of channel since small molecules as well as ions cross a lipid bilayer in the presence of these proteins. Due to their size, it is evident that these proteins have to self-assemble to form a gateway across the membrane. Self-assembly and formation of homo- or hetero-oligomers is a strategy which is also commonly used by the host cells to manufacture functional units such as ion channels that are usually about five times larger (Karlin, 2002). And on an even larger scale, porins (e.g., from Rhodobacter) are found to assemble in trimeric units even though their activity as a pore is due to the individual protein subunit (Danese and Silhavy, 1998, Schirmer, 1998, Yoshihara et al., 1991).
Today, it is unquestionably accepted that M2 from influenza A is a channel-forming protein enabling the flux of protons (Lin and Schroeder, 2001, Mould et al., 2000). M2 has also been the first ion channel target in antiviral therapy (Davies et al., 1964). For others, such as Vpu from HIV-1 and p7 from HCV, there is evidence that they form channels in vitro. Recently, more proteins have been suggested to act as channels or pores such as 3a from SARS-Co (Lu et al., 2006), BM2 from influenza B (Mould et al., 2003), 2B from polio virus (van Kuppeveld et al., 1997a), and others.
This review presents an update of the research field of viral channel-forming proteins guided by biophysical questions along the lines of a putative mechanism of function: how we can identify a putative channel in the genome of the virus, how these proteins are assembled, what their structure is, and finally what their mode of action is in terms of gating and selectivity.
In asking for the role of these proteins within the cellular life cycle of the viruses, one might speculate that they modulate cellular environment (1) via inducing changes of electrochemical or proton gradients and/or (2) via direct interaction between viral and host membrane proteins.
2. Channel-Forming Proteins
2.1. M2 from influenza A
M2 (Allen et al., 1980, Winter and Fields, 1980) consists of 97 amino acids (aa), has a single transmembrane (TM) domain (Fig. 2.1A ), and forms homotetramers from dimers covalently linked via disulfide bridges (Sakaguchi et al., 1997, Sugrue and Hay, 1991). It has a phosphorylation site in the cytoplasmic domain (Holsinger et al., 1995) and is palmitoylated (Sugrue et al., 1990b, Veit et al., 1991). The protein is involved in the entry of the virus into the host cell via the endocytosis pathway which occurs by membrane fusion at a specific low pH (Ciampor et al., 1992, Sugrue et al., 1990a). This event takes place when the virion is entrapped in the endosome. Sensing the low pH in the endosome, M2 conducts protons into the interior of the virion thereby enabling conformational changes of viral proteins such as the matrix protein M1 and the fusion protein hemagglutinin (HA) eventually leading to fusion of the viral and endosomal membrane and uncoating of the viral genome. In addition, during the late stage of the infectious cycle of certain influenza A strains, M2 maintains a near-neutral pH in the Golgi, preventing HA from adopting the low-pH structure which would cause the assembly of nonfunctional HA in the virion. It seems also that M2 facilitates the budding process (McCown and Pekosz, 2006). For this role the cytoplasmic domain of M2 is responsible. This mode of action is not affected by the antiviral drug amantadine which otherwise blocks M2 channel activity.
Figure 2.1.
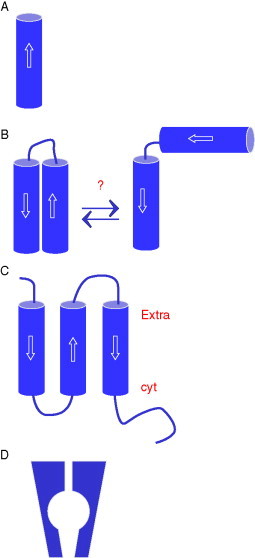
Topologies of the TM region of the to-date known viral channel-forming proteins. (A) A single TM domain [e.g., M2 (influenza A), Vpu (HIV-1), 6K (alphavirus)]. The proteins have a parallel alignment. (B) Two TM domains as proposed for p7 (HCV) or 2B (Polio). (C) Three TM domains proposed for 3a (SARS-CoV). (D) Kcv model based on sequence homology (Plugge et al., 2000) to KcsA channel (Doyle et al., 1998).
A series of investigations have been done focusing on channel activity of M2 in dependence of lipid composition. It has been reported that the association of peptides corresponding to the TM domain of M2 is promoted in the presence of cholesterol as it is found in the Golgi (Cristian et al., 2003). NMR spectroscopic investigations reveal that the tilt of such peptides varies according to membrane thickness (Nishimura et al., 2002, Wang et al., 2001).
M2 has little association with rafts (Leser and Lamb, 2005, Zhang et al., 2000). Recently, a model for raft association has been suggested (Schroeder and Lin, 2005, Schroeder et al., 2005). M2 from infected cells and eukaryotic expression systems copurifies with cholesterol (Schroeder et al., 2005). However, channel activity in liposomes seems to be independent of cholesterol or raft lipids (Lin and Schroeder, 2001). Based on M2′ palmitoylation site Cys-50 and other motifs such as a cholesterol recognition/interaction amino consensus motif (CRAC) including a phosphatidylinositol phosphate (PIP) recognition site, it is therefore proposed that M2 can be raft anchored with its cytoplasmic domain. The actual TM domain, however, remains in a raft-free lipid environment. This mechanism may explain the incorporation of M2 into the rafts of the virion and also may be the reason for the low amount of M2 in the virion since it needs also raft-free areas in the membrane. In the light of the early discoveries, it has been speculated that other viruses, at least the enveloped viruses, should encode for proteins with similar properties.
2.2. Vpu from HIV-1
Vpu has been detected in the mid-1980s independently by two groups (Cohen et al., 1988, Strebel et al., 1988). Their results indicated a virus protein “U” of 81 aa with a single TM domain (Fig. 2.1A) and specific for human immunodeficiency virus type-1 (HIV-1) and related simian immunodeficiency virus (SIV) isolates (Courgnaud et al., 2002, Huet et al., 1990). Its presence in T lymphocytes leads to a 5- to 10-fold increase in the release of progeny virions (Strebel et al., 1988). However, in the absence of the Vpu open reading frame, viruses can still replicate and the protein then has been classified as an auxiliary protein (Cullen, 1998). Investigations using confocal microscopy have shown that Vpu is localized to the recycling endosome (Varthakavi et al., 2006). With its retention signal to the Golgi (Pacyniak et al., 2005), Stephens and coworkers have been able to visualize Vpu in the Golgi apparatus and the ER also using confocal microscopy (Hout et al., 2006). A most recent study shows evidence of Vpu assembling as a pentamer (Hussain et al., 2007).
Vpu is supposed to influence HIV egress from the host cell by three distinct mechanisms:
-
(1)
Vpu is involved in the enhanced release of progeny virions (Klimkait et al., 1990) by directing the HIV receptor CD4 to the ubiquitin-dependent proteasome degradation pathway in the ER (Bour et al., 1995, Chen et al., 1993, Friborg et al., 1995, Margottin et al., 1996, Tiganos et al., 1997). The CD4–Vpu interaction is determined by the Vpu cytoplasmic domain (Chen et al., 1993, Kimura et al., 1994, Margottin et al., 1996, Schubert et al., 1996a, Willey et al., 1992).
-
(2)
It is assumed that Vpu forms ion-conducting channels with a positive effect on the budding process. This is supposed to be due to the oligomerization of the protein (Maldarelli et al., 1993) and consequently the ability to form a channel through which ions can pass (Ewart et al., 1996, Schubert et al., 1996b). The finding that Vpu forms ion channels emerged from experiments with peptides corresponding to the TM domain and with full-length protein, both of which exhibit similar channel activity when reconstituted into artificial lipid bilayers. Oocytes expressed Vpu has shown channel activity in whole cell current recordings. Applying a range of −60 to −130 mV holding potentials generates currents in the nanoampere range (Schubert et al., 1996b). In a similar experiment in which the sequence of amino acids of the TM domain has been randomly chosen, no significant current has been detected. Similarly, a 27-aa synthetic peptide corresponding with the same scrambled sequence TM domain of Vpu reconstituted into artificial bilayer does not generate any current (Schubert et al., 1996b). The data can be taken as a proof that the wild type amino acid sequence of the TM domain is responsible for channel activity and that the TM domain and with it the channel activity may be functionally associated with enhanced particle release. The data above have been thoroughly discussed in the literature (Lamb and Pinto, 1997). At that state of investigations, it has been concluded that it still may be possible that Vpu modulates another cation channel and that there is the need of a selective Vpu blocker to further investigate the issue while using whole cell recordings.
In a more recent bilayer study (peptide or protein reconstituted into a lipid bilayer according to Montal and Müller, 1972), it has been reported that a peptide corresponding to the first 32-aa of Vpu shows channel activity even at lowest or zero current (Mehnert et al., 2007). Together with the finding of an asymmetric voltage-dependent open rate, it has been suggested that the closure of the bundle is voltage independent and due to other factors such as the lateral pressure profile of the bilayer. Mutant studies based on the synthetic peptides have shown that presumably pore-lining serines are essential for channel activity and that the tryptophan is involved in the gating kinetics of the bundle (Mehnert et al., 2008).
-
(3)
There is increasing evidence of Vpu interacting with other cellular factors. It has been reported that action of Vpu to enhance particle release is dependent on the cells which are invaded by HIV-1 (Varthakavi et al., 2003). Most recently, it has been found that Vpu interacts with a potassium channel (TASK-1) of the host cell, thereby blocking its channel activity in a dose-dependent manner and thus indirectly affecting the release of progeny virions (Hsu et al., 2004). Interesting to note is the sequence similarity between Vpu and the first TM domain (TM-1) of the TASK channel. Expressing just the TM-1 domain, a similar effect of lowering TASK channel activity occurs in the host cell. This has been taken as an indication that an alteration of channel activity may be important for viral release, but it may not necessarily be due to channel activity of Vpu itself. In addition, Vpu interacts with the cellular protein BST-2 which would otherwise prevent the pinching off of the virus particle (Neil et al., 2008, van Damme et al., 2008). For this mode of action, both the TM and the cytoplasmic domain of Vpu are necessary. BST-2 has a TM domain and a luminal GPI anchor and is colocalized with structural gag proteins of HIV-1 in the endosome and the plasma membrane. Vpu deficient HIV-1 will be retained at the plasma membrane. Unlike for M2, research on Vpu lacks still essential experiments like those involving escape mutants to assess the intracellular role of this protein.
2.3. p7 from HCV
The genome of HCV encodes approximately 10 proteins which are expressed as a large “polyprotein” and cleaved into single proteins by viral and host proteases and peptidases (Penin et al., 2004). One of this protein is p7, a 63-aa TM protein (Lin et al., 1994). It sites between E2 and NS2 and all three proteins represent the precursor before final cleavage. Thus, occasionally E2-p7 remains uncleaved. Up to now it is not yet clear whether p7 belongs to the structural proteins, such as E1 and E2, or the nonstructural proteins, such as NS3, NS4A, NS4B, NS5A, and NS5B. It is also not yet confirmed whether p7 is part of the virion. Deletion of p7 leads to noninfectious viruses, suggesting that p7 is essential for the production of the virus (Harada et al., 2000). The protein is detected on the cell surface and with a large portion retained in the endoplasmic reticulum (Carrère-Kremer et al., 2002). It is a polytopic membrane protein with its N- and C-termini exposed to the extracellular environment (Carrère-Kremer et al., 2002) (Fig. 2.1B). The translocation experiments have been done with CD4-p7 chimeras detected by immunofluorescence in nonpermeabilized cells while the studies on the topology have been based on Myc epitope mapping. Secondary structure prediction proposes two TM domains connected by a short basic segment. Both ends are found to face the interior of the endoplasmic reticulum. Protein p7 is suggested to be less immunogenic, since the generation of antisera has failed so far. The exact role of this protein remains to be elucidated. The protein seems to be important for infectivity of the virion (Sakai et al., 2003, Steinmann et al., 2007a) but not essential for the replication of viral RNA in hepatoma cell lines (Lohmann et al., 1999). It is assumed that p7 has a role within the compartments of the secretory pathway (Carrère-Kremer et al., 2002). Recent modeling analysis suggests that a histidine residue is pointing into a putative pore, formed by the assembly of the protein (Patargias et al., 2006). Transmission electron microscopy (TEM) data of HIS-p7 reconstituted into unilamellar vesicles indicate that the protein assembly is a hexamer with a putative diameter of 3–5 nm (Griffin et al., 2003). In a more recent study, it is suggested that p7 forms also homoheptamers when analyzed as a GST-FLAG-p7 protein using the same technique (Clarke et al., 2006). Full-length p7 reconstituted into lipid bilayer shows channel activity which can be blocked by several drugs (Griffin et al., 2003, Pavlovic et al., 2003, Premkumar et al., 2004). Structural data is lacking to date.
2.4. 2B from polio virus
2B is part of nonstructural proteins encoded by enteroviruses (e.g., poliovirus, coxsackie virus, and ECHO virus) (de Jong et al., 2003, Nieva et al., 2003). The protein (coxsackie) is localized in the Golgi to affect the permeability of secretory membranes and the plasma membrane (de Jong et al., 2003) but is not transported outside the Golgi. The plasma membrane has been found in experiments to be more permeable to calcium, thereby modulating apoptosis (Campanella et al., 2004), and to low molecular weight compounds (Aldabe et al., 1996, Doedens and Kirkegaard, 1995, van Kuppeveld et al., 1997a). How the retention in the Golgi affects the plasma membrane is unclear. Two proposals have been made to explain the retention of 2B in the Golgi: (1) the oligomers are too large (“kin-recognition” model: Nilsson et al., 1993, Nilsson et al., 1996) and (2) lack of specific signals (Munro, 1998). 2B is supposed to consist of two hydrophobic transmembrane stretches (van Kuppeveld et al., 1995, van Kuppeveld et al., 1997b) (Fig. 2.1B). Similar to the other channel-forming proteins, 2B forms multimers (Cuconati et al., 1998, de Jong et al., 2002, van Kuppeveld et al., 2002). The roles of several amino acids on oligomerization, permeabilization of the membrane, and the localization within the cell have been identified (de Jong et al., 2004). Mutations of hydrophilic residues within the short linker region between the two TM domains readily impair the multimerization and also decrease membrane permeability. Mutating residues (tryptophans) toward the C-terminus of the second TM domain also shows an abrogation of membrane permeability without affecting multimerization of the protein. The results are based on a hygromycin B assay. In this assay, protein activity is monitored under the suppressing affect of hygromycin B. Hygromycin B is supposed to enter the cell through leakages or pores formed in the cell membrane. Permeability tests have also been done with unilamellar vesicles in which a 2B maltose-binding protein (MBP) fusion construct has been reconstituted (Agirre et al., 2002). With this test, membrane permeability was shown to be increased for molecules with a molecular weight of up to 660 kDa. It has been shown that the fusion construct forms SDS-resistant tetramers with an approximate pore size of 6 Å. The topology is suggested to consist of two TM domains, one of which is lysine rich, a short linker region followed by another TM domain which is folded back into the membrane. The suggestion is based on the defined cutoff in size for the permeating molecules. A carpet-like mechanism in which one of the TM domains may lie on the surface of the membrane while the other forms the pore, should lead to pores which allow the permeation of molecules of a large variation of size, which is in contrast to their findings. Channel conductance has not yet been reported for 2B.
2.5. 6K from alphavirus
Members of the alphavirus genus express 6K proteins with a sequence in the range of 58–61 aa (Welch and Sefton, 1980) (Fig. 2.1A). The role in the life cycle of the viruses is not yet known. Expression of 6K proteins in Escherichia coli increases membrane permeability fostering the budding process when expressed in eukaryotic cells (Liljestrom et al., 1991, Loewy et al., 1995, Sanz et al., 1994). Its topology is assumed to be a single helix according to theoretical secondary structure prediction programs (Sonnhammer et al., 1998). Using in vitro translation-translocation assays, it is suggested that 6K consists of two helices (Liljestrom and Garoff, 1991). Full-length protein expressed in E. coli, purified and reconstituted into lipid bilayers, has shown channel activity with a preference for cations (Melton et al., 2002).
2.6. 3a and E proteins from SARS coronavirus
Recently, a 274-aa protein called 3a and located in an open reading frame between the S and E protein loci in the genome of the severe acute respiratory syndrome-associated coronavirus (SARS-CoV) has been reported to modulate membrane currents when expressed in Xenopus oocytes (Lu et al., 2006). Its topology has been identified to consist of three TM domains with the C-terminus in the cytoplasm (Fig. 2.1C). The protein forms homodimers and tetramers and is located in the plasma membrane of infected cells.
Another short 9–12 kDa protein (Siddell, 1995), called E protein, with a short 10-aa hydrophobic region (Shen et al., 2003) is encoded by SARS-CoV. The protein is nonessential for the survival of the virus, and its exact function remains to be elucidated. A synthetic 76-aa construct of the full-length protein has been identified to show channel activity (Wilson et al., 2004). Structural analysis using Fourier-transform infrared (FTIR) spectroscopy, X-ray scattering, and electron microscopy together with global search molecular dynamics (MD) simulations reveals a hairpin like motif for the proposed TM region of the protein (Arbely et al., 2004).
Other proteins from the genome of SARS-CoV are currently under investigation and show promising results that they my form channels (C.-C. Chen, J. Krüger, P. Henklein, Y.-M. Chen, and W. B. Fischer, unpublished results).
2.7. Kcv from PBCV-1
Chlorella virus PBCV-1 (family Phycodnaviridae) encodes a 94-aa membrane protein (Plugge et al., 2000). Hydropathy analysis reveals two putative hydrophobic stretches which are separated by about 44-aa. This part of the sequence follows the same sequence as found for the pore region of the KcsA channel (Doyle et al., 1998). The protein when expressed in Xenopus oocytes exhibits K+ selective channel activity (Plugge et al., 2000) and is suggested to be active during viral entry (Mehmel et al., 2003) (Fig. 2.1D). It is expected that the number of channels from plant viruses may increase in the near future, since their genomes are larger than those from animal viruses.
2.8. NB and BM2
Like influenza A, influenza B and C encode for small transmembrane proteins, NB (Fischer et al., 2000b, Sunstrom et al., 1996), BM2 (Mould et al., 2003, Paterson et al., 2003), and CM2 (Hongo et al., 1997, Kukol and Arkin, 2000). NB and BM2 have been shown channel activity in reconstitution experiments. These proteins are type III integral membrane proteins with an NoutCin motif which are expressed at the surface of virus-infected cells (Paterson et al., 2003) and in the virion (NB: Betakova et al., 1996; BM2: Odagiri et al., 1999; CM2: Hongo et al., 1997). In both cases, proton selectivity has been reported. BM2 is active in the Golgi (Mould et al., 2003) equilibrating the pH between the Golgi and the cytoplasm, a function similar to M2 from influenza A. Also, the TM topology with a HxxxW motif is similar to M2 and strengthens the experimental findings. Influenza M2 protein appears to play an additional role during virus assembly, which is not dependent on the channel function (Schroeder and Lin, 2005, Schroeder et al., 2005). Recent findings on BM2 indicate that the protein captures the M1–vRNP complex during the budding process (Imai et al., 2004, Imai et al., 2008). Following the recognition that BM2 is the ion channel responsible for biological functions, the role of NB is now obscure (Hatta and Kawaoka, 2003).
3. Detecting a Channel-Forming Protein
3.1. Sequence identification
Discovery of a putative channel-forming protein is driven by the detection of hydrophobic stretches within the amino acid sequence of a viral protein. Specific amino acids have a certain probability to reside within the TM stretch. The probability is based on investigations of known membrane proteins, available in protein databases.
Using secondary structure prediction tools as well as tools for TM segment prediction, it is possible to identify suitable candidates. These tools usually assume an α-helical or β-barrel motif for the membrane spanning part. So far only helical motifs have been found for most of the membrane proteins (Doyle et al., 1998, Kuo et al., 2003, Miyazawa et al., 2003, Stroud et al., 2003). These predictions can, of cause, only suggest a certain idealized hydrophobic stretch; kinks or bends within the transmembrane domain which may be meaningful for the mechanism of function cannot be predicted.
An example of the prediction of the TM domain of 3a protein from SARS-CoV is shown in Fig. 2.2 . A series of programs such as Membrane Protein Explorer (MPEx, http://blanco.biomol.uci.edu/mpex/), Dense Alignment Surface prediction of transmembrane regions in proteins (DAS, http://www.enzim.hu/DAS/DAS.html), TMpred (prediction of transmembrane regions and orientations, http://www.ch.embnet.org/software/TMPRED_form.html), TMHMM (prediction of transmembrane helices, http://www.cbs.dtu.dk/services/TMHMM/), and HMMTop (prediction of TM helices and topology of proteins, http://www.enzim.hu/hmmtop/index.html) are commonly used. The programs try to identify the membrane spanning part based on sequence homology with well characterized proteins and on lipophilicity of individual amino acids derived from octanol/water distribution coefficients. The prediction from all of these programs does not always overlap perfectly well. Whilst the core region is usually well defined, the residues presumably located within the lipid head group region are harder to grasp. Therefore, it is recommended to use not only one prediction program but to work with a consensus based on different methods. Using multiple secondary structure prediction tools three TM domains of SARS-CoV 3a have been identified with a longer linker region between TM1 and TM2 and a shorter linker region between TM2 and TM3. Also for Vpu (Fischer et al., 2000a) and p7 (Patargias et al., 2006), the predictions reveal one and two TM domains, respectively.
Figure 2.2.

Prediction programs used to assess the topology of the channel-forming protein 3a from SARS-CoV. The asterisks indicate a presumably helical TM conformation. The consensus among the programs is highlighted in light grey, but would need experimental confirmation.
The analysis mentioned is then the starting point for further prediction of the tertiary structure of the assembled proteins within the TM region. Sequence homology with other proteins may also give a clue about the structure and role of these proteins and consequently about their mechanism of function.
3.2. Detection of channel activity
Channel activity of all of these proteins has so far been verified experimentally with whole cell recordings in which the particular viral protein is overexpressed in either Xenopus oocytes or other cell lines (Fischer, 2005, Fischer and Sansom, 2002, Gonzales and Carrasco, 2003). Another source of evidence for channel-forming properties is derived from reconstitution of the proteins and parts of them in artificial membranes (bilayer technique: Montal and Müller, 1972). In this respect, data from Vpu from HIV-1 (Ewart et al., 1996, Mehnert et al., 2007, Schubert et al., 1996a) (Fig. 2.3 ), p7 from HCV (Griffin et al., 2003, Pavlovic et al., 2003), and other proteins (Melton et al., 2002, Piller et al., 1996, Wilson et al., 2004) have been accumulated. For Vpu, a scrambled sequence of the TM domain, which is thought to be solely responsible for channel function, has shown no channel activity and thus confirms the credibility of the technique (Schubert et al., 1996b). Based on the use of this technique, the statement can be made that these proteins self-assemble in lipid membranes and display channel characteristics. Thus, within the cellular membranes they are expected to behave in a similar way.
Figure 2.3.
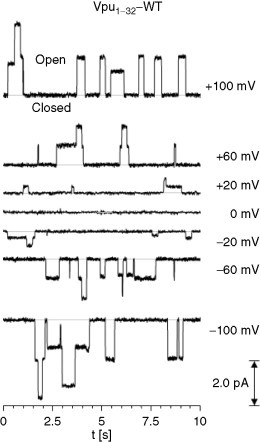
Typical channel recordings of a peptide corresponding to the first 32-aa of Vpu (HIV-1) reconstituted into lipid bilayers [POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine) and DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) 1:4] at various holding potentials. The system was hold in 300 mM KCl, 5 mM K-HEPES at pH 7.0.
However, there is increasing evidence at least for Vpu that the protein interferes with several host membrane proteins at various locations within the cell. Since no feedback mechanism regulating Vpu production is known to date, it is reasonable to assume that Vpu can engage in these protein–protein interactions (CD4: Bour et al., 1995, Chen et al., 1993, Friborg et al., 1995, Kimura et al., 1994, Margottin et al., 1996, Schubert et al., 1996a, Tiganos et al., 1997, Willey et al., 1992; TASK: Hsu et al., 2004; BST-2: Neil et al., 2008, van Damme et al., 2008) but also can self-assemble forming channels or pores. The same situation counts for the other channel-forming proteins and needs to be elucidated in the future.
4. Functional and Structural Characterization
4.1. Biophysics of the channel-forming proteins and channel–pore dualism
In models of ion-conducting peptide assemblies hydrophilic residues are usually proposed to face the lumen of the pores (Dieckmann et al., 1999, Tieleman et al., 1999). In recent investigations on viral ion channels, the orientation of hydrophilic residues has been done accordingly (Fischer and Sansom, 2002). The requirement of an adequate counterplay between hydrophilic and hydrophobic residues for a TM peptide to show ion channel function is outlined by the synthetic peptides consisting of specific Leu-Ser repeats (Lear et al., 1988). Modeling these peptides in a pore, the hydrophilic residues are oriented toward the lumen (Dieckmann et al., 1999). The position of these hydrophilic residues in conjunction with their hydrophobic counterparts makes the channel selective for protons and other ions. It has been found that a serine side chains in 6K protein from alphavirus affects ion selectivity if mutated (Melton et al., 2002). Computational studies on the assembly of two TM helices from p7 from HCV indicate that hydrophilic residues such as serines and threonines, but also histidine, align within a putative pore (Patargias et al., 2006). For M2, histidines and tryptophans are lining the pore and are important for the proton-conducting properties of the channel (Hu et al., 2006).
Thus, from the topological point of view, the viral channels fulfill the criteria of being able to show ion selectivity. They have a helical TM motif with hydrophilic pore facing residues and there are indications that side chains create a selectivity filter (Wilson et al., 2004) as well as hydrophic stretches (Mehnert et al., 2008). Recent crystallographic data of a pentameric ligand-gated ion channel from bacteria belonging to the cys-loop family support considerations on the orientation of pore-lining residues (Hilf and Dutzler, 2008). With an increasing number of channel-forming viral proteins with several TM domains, it may be possible that also other channel architectures similar to those of voltage-gated channels are adopted, as in the case of the Kcv channel from chlorella virus (Plugge et al., 2000).
Whilst M2 is held together at least partially by covalent links (Cys-Cys), the other channels are solely held together by noncovalent protein–protein interactions. Thus, in the latter case and in comparison of the pore-lining motifs of larger channels, the viral channel-forming proteins comprise a minimalist approach towards channel architecture. The hydrophilic residues may be just enough to attract and accumulate the ions to be conducted.
Any mechanism of gating may involve minimal conformational changes. The mechanism may also be driven by the lipid environment. Lipid composition and thickness in the Golgi network, for example, could induce altered conformations which allow an ion flux. For M2, a tendency for associating with rafts in the membrane is outlined (Schroeder et al., 2005). For Vpu, a lipid dependency for closure is also suggested on the basis of channel recordings in artificial bilayers reflecting small conformational changes (Mehnert et al., 2007, Mehnert et al., 2008). The lateral pressure profile is proposed to induce the closure. Consequently, channel activity may depend also on membrane thickness, another indication for activity which may depend on lipids in the Golgi and the cell membrane.
In general, evidence is emerging that for a gating mechanism only subtle conformational change are necessary, a suggestion based on computational studies on nanopores (Beckstein et al., 2001, Beckstein et al., 2004) and the nAChR structure (Beckstein and Sansom, 2006). This suggestion is confirmed by experimental studies on the nAChR (Cymes and Grosman, 2008). However, these suggestions still remain controversial since large-scale conformational changes have also been proposed for the larger channels (Cymes and Grosman, 2008) and also M2 from influenza A based on X-ray (Stouffer et al., 2008) and NMR studies (Schnell and Chou, 2008).
Experiments have shown that some of the channels, such as Vpu, alter membrane permeability for small molecules (Gonzales and Carrasco, 2003). In line with the finding that, apart from the influenza M2 family, most of the viral ion-conducting channels are only weakly ion selective it is suggested that these proteins may embody a “channel–pore dualism”. The term is coined in analogy to the particle–wave dualism of light: depending on the experimental conditions they exhibit channel characteristics or pore (low or no ion selectivity) characteristics.
4.2. Structural information
There is a large amount of structural information available for M2 and Vpu (see Fischer, 2005, references therein). The information is based from early studies using CD spectroscopy (M2: Duff et al., 1992; Vpu: Wray et al., 1999), NMR spectroscopy (M2: Kovacs and Cross, 1997, Kovacs et al., 2000; Vpu: Ma et al., 2002, Park et al., 2003), FTIR spectroscopy (M2: Kukol et al., 1999; Vpu: Kukol and Arkin, 1999), and X-ray reflectivity (Zheng et al., 2001). Most of the experiments have used parts of the TM domain or parts of the cytoplasmic domains in the case of Vpu. Recently for M2 the first crystal structure has been published showing a TM domain assembly in absence of a hydrophobic lipid environment in a state at low-pH conditions cocrystallized with amantadine (Stouffer et al., 2008). Without the drug in the pore the conformation seems to represent a solvent accessible pore and therefore an open state of the bundle. This is in contrast of findings from solution NMR spectroscopic studies (Schnell and Chou, 2008). The data show the tetrameric assembly with the tryptophans (Trp-41) occluding the putative pore and interpreted this as the closed state of the channel. In the study, amantadine is found in binding pockets outside the pore.
Vpu is the only protein for which the cytoplasmic domain structure has been characterized by NMR spectroscopy. The spectra reveal either two (Federau et al., 1996) or three helices (Willbold et al., 1997) connected via short loops. In addition, interactions of the cytoplasmic domain with cellular proteins have also been reported for Vpu (Coadou et al., 2003, Gharbi-Benarous et al., 2004).
Depending on the experimental conditions, TEM data of p7 reveal a hexameric (Griffin et al., 2003) or heptameric assembly (Clarke et al., 2006).
4.3. Modeling the mechanism of function
Modeling the mechanism of function has two aspects: experimental data need to be derived and the data have to be converted into theoretical models which then have to be filled with atomistic details.
The working hypothesis is that the viral proteins are formed in the ER and exist as monomeric units prior to any assembly. Assembling is a common feature for membrane proteins in general (Engelman, 2005, Popot and Engelman, 2000) as well as for other channel or pore-forming proteins such as alamethicin and melittin (Bechinger, 1997, Tossi et al., 2000). For these proteins, the topology of the pore once formed is also an assembly of helical TM domains (Boheim et al., 1983). Up to now only gramicidin channel is known to consist of two β-barrel units forming hydrogen bonds in a head-to-head formation at their N-termini (Woolley and Wallace, 1992).
To model the pore or channel some care need to be taken to obtain structural details of the monomeric protein. After careful assessment of the conformational space of the monomeric TM domain, the domain may then be replicated and reoriented to form the appropriate bundle. This has now been done for Vpu (Krüger and Fischer, 2008). At this stage also the effect of lipid composition and thickness on the structure of the TM domain of Vpu can be addressed. It has been reported that Vpu adopts to the environment via kinking rather than forcing the lipid environment to adjust (Krüger and Fischer, 2008). This is in contrast to experimental findings of X-ray reflectivity data but may be due to the high protein to lipid ratio (0.02) used in the experiments where already assemblies may have formed (Khattari et al., 2006). None of the sequences of the putative channel-forming proteins contain a Leu heptad (Gurezka et al., 1999) or GxxxG motives (Russ and Engelman, 2000) with the latter especially known to promote close helix–helix packing.
For a functional and selective pore, it is required to have an ingenious balance of hydrophilic and hydrophobic pore-lining residues. Purely hydrophobic pores would pose a barrier for the permeating ion similar to the lipid environment. At contrast a highly hydrophilic motif would trap the passing ion. Modeling a hydrophobic pore, Beckstein and Sansom (2003) have shown that the amount of water within the pore changes continuously between “water filled” and “empty” (termed by the authors as “vapor” phase) depending on the pore diameter. Therefore, the term “oscillating liquid/vapor phase” within the pore has been created. Conductance can only be observed when a sufficient amount of water molecules is present within the gorge portion of the pore. The observed oscillation between the liquid and the vapor state within the pore is just one aspect which addresses the role of a specific type of wall within a channel. The lumen of a biological ion channels consists of a complex alignment of polar and nonpolar residues. Many more aspects have to be considered for a proper description of the ion permeation such as its interaction with the residues at the entrance/lumen of the pore and the behavior of the hardly removable first tetrameric hydration shell within the pore.
Recently, the TM domain of Vpu (Ulmschneider and Ulmschneider, 2007) has been folded within a hydrophobic slab mimicking a lipid bilayer. In this approach, the slab shows a gradual change of the dielectric constant toward the center of the membranes. Monte Carlo simulations have been used in combination with replica exchange methods to assess the fold of the domains. This is another approach toward a fully in silico modeling of membrane protein structure.
Several approaches on the level of bundles have been tried for Vpu to elucidate the mechanism of function once the pore is formed. Molecular dynamics simulations in combination with physical models of ion permeation have been undertaken. In all of these studies solely the TM domain of Vpu has been used. The TM domain was oligomerized into tetrameric, pentameric, and hexameric bundles. In one of the studies, after using MD simulations to achieve a reasonably stable structural model, the energy profiles for Na+ and Cl- along the pore have been calculated by placing them at various positions within the pore (Grice et al., 1997). Recently, steered MD simulations were used to assess the potential of mean force of the ions across the lumen of the pore (Patargias et al., 2009). Based on the energy profile the pentameric model supports the experimental findings of a weak cation-specific channel. In a combined computational and experimental approach, structural features of the Vpu bundle, such as the tilt of the helices in the mechanism of function and the role of a putative selectivity filter based on a EYR-motif at the C-terminus, has been demonstrated (Cordes et al., 2002). Together with other MD simulation work in which the instability of the hexameric bundle has been explicitly shown (Moore et al., 1998), the pentameric bundle model is now fairly established until further proof to the contrary. The essential question remains, what happens between the stage of the monomer and the functional bundle? Two kinetic pathways are possible, either the assembly happens in an all or nothing step with minor adjustments or more likely the monomers assemble sequentially.
For the influenza A M2 channel, proton conductance has been modeled using MD simulations (Chen et al., 2007, Smondyrev and Voth, 2002). Based on theoretical considerations, there are two possible scenarios for the proton to pass the pore and specifically the histidines (His-37): either the protonated histidine flips and carries the proton to the other side, or all histidines undergo a “swing motion” due to electrostatic repulsion and thereby open the pore accordingly (Schweighofer and Pohorille, 2000).
5. Channel-Forming Proteins in Antiviral Therapy
Inhibition of proteins which are essential in the virus life cycle is a basic approach in antiviral therapy. In the case of channels and pores, there are in principle two strategies (1) to inhibit the channel itself by a suitable inhibitor, either small organic molecule or peptide drug and (2) to prevent the formation of the channel by hindering the assembly of the monomers, where custom designed “anti-oligomerization” peptides appear to be most promising.
5.1. Small molecule drugs
There are several small molecule drugs under investigation which inhibit some of the channel proteins. A derivative of amiloride such as hexamethylene amiloride has been shown to be effective against HIV-1 and in particular against Vpu (Ewart et al., 2002). Experiments have been done in HELA cells which coexpressed Vpu and Gag proteins as well as with pure Vpu reconstituted into artificial membranes from proteoliposomes. Also p7 from HCV, synthesized by solid phase peptide synthesis and reconstituted into lipid bilayers, is affected in the presence of hexamethylene amiloride (Ewart et al., 2002, Premkumar et al., 2004). M protein from Dengue virus-1 also forms channels when reconstituted into artificial membranes and can be blocked by hexamethylene amiloride and amantadine (Premkumar et al., 2005).
Amantadine, known as one of the first drugs against influenza A, blocks the proton channel M2 in a concentration of up to 5 μM (Chizhmakov et al., 1996, Hay et al., 1985, Tu et al., 1996) by changing M2 dynamics (Hu et al., 2007). Depending on the technique used, the drug may either enter the pore (X-ray crystallography: Stouffer et al., 2008) or diffuses into hydrophilic/hydrophobic pockets in the outer rim of the bundle (NMR spectroscopy: Schnell and Chou, 2008). Amantadine/rimantadine resistance is on the rise because most mutations determining drug resistance do not impair channel function. New derivatives of these drugs are being developed in the hope of overcoming drug resistance (Kolocouris et al., 1996). Amantadine is also under investigation for treatment of chronic hepatitis C virus infection (Lim et al., 2005) and seems to affect p7 when this protein is used within a GST-His-p7 or GST-p7 construct (Griffin et al., 2003, Griffin et al., 2004). In a recent study with full-length p7, amantadine shows no affect on the channel activity of p7 in a range of up to 10 μg/ml (Steinmann et al., 2007b). In a computational study using global search algorithms, a hexameric model of p7 has been suggested (Patargias et al., 2006). Amantadine has been docked into the pore and its residence is suggested to be in the vicinity of the histidine in the first TM domain at position 17. A clinical study has shown that amantadine does not lead to a mutation at the position of His-17 (Castelain et al., 2007). The attribute affects the pH-dependent entry pathway of HCV. One still can argue that the residence of amantadine at the site of His-17 is possible, but as can be seen in the study of Patargias et al. (2006), it does not necessarily occlude the pore. As a consequence, it may not to be a potent candidate for blocking of p7. Kcv from PBCV-1 (Plugge et al., 2000) and Vpu also respond on amantadine and derivatives (Kolocouris et al., 1996), but only to fairly high doses.
It has been shown that iminosugar derivatives are affective against bovine viral diarrhea virus (Durantel et al., 2001). In follow-up studies, it could be shown that p7 from HCV is a possible target of these small molecules as well (Pavlovic et al., 2003). With a liposome assay for p7 activity, large-scale drug screening for p7 comes into reach (StGelais et al., 2007). Investigations on the interaction of p7 with iminosugars indicate an effect in the range of up to 10 μg/ml (Steinmann et al., 2007b). Especially derivatives such as N-nonyl-deoxynojirimycin (NN-DNJ) and N-nonyl-deoxygalactonojirimycin (NN-DGJ) are potential candidates.
5.2. Peptide drugs
Peptide drugs have several advantages over small molecule drugs (1) they are highly active and specific and (2) they accumulate in tissue to a lower extent and therefore exhibit lower toxicity. There is a major drawback such as the difficulty to deliver the drug to the target site. Usually peptide drugs have very low oral bioavailability and need to be injected (Hamman et al., 2005). They are commonly used for severe chronic diseases. Peptide drugs have a low uptake by the gastrointestinal tract and experience rapid presystemic enzymatic degradation. However, advances in peptide manufacturing and improvements in peptide drug delivery systems have and will further rejuvenate this field.
The concept of interfering with the molecular action by using parts of protein itself has been shown in HIV research. It has been found that Vpu, a channel-forming protein of HIV-1, interferes with the function of the acid-sensitive leakage K+ channel (TASK) (Hsu et al., 2004). Vpu has high sequence homology with parts of the TASK protein. The authors have additionally shown that parts of TASK can also hamper channel activity of TASK. The mechanism of function is proposed to be a disturbed assembly of the protein in the presence of the protein fractions.
The concept of interfering with protein assembly within the aqueous phase has already been used to interrupt the fusion mechanism of gp41 from HIV-1 (Root et al., 2001). The interaction of the trimeric gp41 with the cellular membrane induces conformational change into a bundle of six helices which is called the “trimer-of-hairpins.” This intermediate catalyzes the fusion of the viral with the cellular membrane. A protein which corresponds to only five out of these six helices has been synthesized, called “five-helix” protein. The gap generated by the missing sixth helix in this assembly acts as a high-affinity target site for gp41.
6. Concluding Remarks
After almost 20 years of research viral channel-forming proteins still leave a lot of open questions. Elucidation of their modes of action should be accelerated since for some viruses these proteins are essential and consequently a valuable drug target. Several questions remain to be answered: what is the relation of channel and pore functionality; how do they assemble and which oligomeric structure do they adopt; and finally, which sequence elements and interactions govern the fate of nascent viral channel monomers to form either homo-oligomeric channels or instead interact with host cell proteins?
For a full understanding of the mode of action, the full-length proteins need to be considered more thoroughly. Up to now all functional analysis is based on looking solely on the TM domains. For Vpu, bilayer experiments and assays indicate that the protein can be seen as a unification of two domains, the TM and the cytoplasmic domain, each of which can act independently from the other. It may be due to future research whether this is also the case for the other proteins.
Acknowledgments
WBF thanks the NSC, NYMU, and the Government of Taiwan for financial support. JK acknowledges the receipt of an Alexander von Humboldt—NSC fellowship. We thank Cornelia Schroeder (Dresden) for valuable discussions and critically reading the manuscript.
References
- Agirre A., Barco A., Carrasco L., Nieva J.L. Viroporin-mediated membrane permeabilization. Pore formation by nanostructural poliovirus 2B protein. J. Biol. Chem. 2002;277:40434–40441. doi: 10.1074/jbc.M205393200. [DOI] [PubMed] [Google Scholar]
- Aldabe R., Barco A., Carrasco L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 1996;271:23134–23137. doi: 10.1074/jbc.271.38.23134. [DOI] [PubMed] [Google Scholar]
- Allen H., McCauley J., Waterfield M., Gethering M. Influenza virus RNA segment 7 has the coding capacity for two polypeptides. Virology. 1980;107:548–551. doi: 10.1016/0042-6822(80)90324-4. [DOI] [PubMed] [Google Scholar]
- Arbely E., Khattari Z., Brotons G., Akkawi M., Salditt T., Arkin I.T. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 2004;341:769–779. doi: 10.1016/j.jmb.2004.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger B. Structure and functions of channel-forming peptides: Magainins, Cecropins, Melittin and Alamethicin. J. Membr. Biol. 1997;156:197–211. doi: 10.1007/s002329900201. [DOI] [PubMed] [Google Scholar]
- Beckstein O., Sansom M.S.P. Liquid-vapor oscillations of water in hydrophobic nanopores. Proc. Natl. Acad. Sci. USA. 2003;100:7063–7068. doi: 10.1073/pnas.1136844100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstein O., Sansom M.S.P. A hydrophobic gate in an ion channel: The closed state of the nicotinic acetylcholine receptor. Phys. Biol. 2006;3:147–159. doi: 10.1088/1478-3975/3/2/007. [DOI] [PubMed] [Google Scholar]
- Beckstein O., Biggin P.C., Sansom M.S.P. A hydrophobic gating mechanism for nanopores. J. Phys. Chem. B. 2001;105:12902–12905. [Google Scholar]
- Beckstein O., Tai K., Sansom M.S. Not ions alone: Barriers to ion permeation in nanopores and channels. J. Am. Chem. Soc. 2004;126:14694–14695. doi: 10.1021/ja045271e. [DOI] [PubMed] [Google Scholar]
- Betakova T., Nermut M.V., Hay A.J. The NB protein is an integral component of the membrane of influenza B virus. J. Gen. Virol. 1996;77:2689–2694. doi: 10.1099/0022-1317-77-11-2689. [DOI] [PubMed] [Google Scholar]
- Boheim G., Hanke W., Jung G. Alamethicin pore formation: Voltage-dependent flip-flop of alpha-helix dipoles. Biophys. Struct. Mech. 1983;9:181–191. [Google Scholar]
- Bour S., Schubert U., Strebel K. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: Implications for the mechanism of degradation. J. Virol. 1995;69:1510–1520. doi: 10.1128/jvi.69.3.1510-1520.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella M., de Jong A.S., Lanke K.W., Melchers W.J., Willems P.H., Pinton P., Rizzuto R., van Kuppeveld F.J. The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J. Biol. Chem. 2004;279:18440–18450. doi: 10.1074/jbc.M309494200. [DOI] [PubMed] [Google Scholar]
- Carrère-Kremer S., Montpellier-Pala C., Cocquerel L., Wychowski C., Penin F., Dubuisson J. Subcellular localization and topology of the p7 polypeptide of Hepatitis C virus. J. Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelain S., Bonte D., Penin F., François C., Capron D., Dedeurwaerder S., Zawadzki P., Morel V., Wychowski C., Deuverlie G. Hepatitis C virus p7 membrane protein quasispecies variability in chronically infected patients treated with interferon and ribavirin, with or without amantadine. J. Med. Virol. 2007;79:144–154. doi: 10.1002/jmv.20772. [DOI] [PubMed] [Google Scholar]
- Chen M.Y., Maldarelli F., Martin M.A., Strebel K. Human immunodeficiency virus type 1 Vpu protein induces degradation of CD4 in vitro: The cytoplasmic domain contributes to Vpu sensitivity. J. Virol. 1993;67:3877–3884. doi: 10.1128/jvi.67.7.3877-3884.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wu Y., Voth G.A. Proton transport behaviour through the Influenza A M2 channel: Insights from molecular simulations. Biophys. J. 2007;93:3470–3479. doi: 10.1529/biophysj.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhmakov I.V., Geraghty F.M., Ogden D.C., Hayhurst A., Antoniou M., Hay A.J. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J. Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampor F., Bayley P.M., Nermut M.V., Hirst E.M., Sugrue R.J., Hay A.J. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans Golgi compartment. Virology. 1992;188:14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- Clarke D., Griffin S., Beales L., Gelais C.S., Burgess S., Harris M., Rowlands D. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 2006;281:37057–37068. doi: 10.1074/jbc.M602434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coadou G., Gharbi-Benarous J., Megy S., Bertho G., Evrard-Todeschi N., Segeral E., Benarous R., Giraudat J.P. NMR studies of the phosphorylation motif of the HIV-1 protein Vpu bound to the F-box protein beta-TrCP. Biochemistry. 2003;42:14741–14751. doi: 10.1021/bi035207u. [DOI] [PubMed] [Google Scholar]
- Cohen E.A., Terwilliger E.F., Sodroski J.G., Haseltine W.A. Identification of a protein encoded by the vpu gene of HIV-1. Nature. 1988;334:532–534. doi: 10.1038/334532a0. [DOI] [PubMed] [Google Scholar]
- Cordes F.S., Tustian A., Sansom M.S.P., Watts A., Fischer W.B. Bundles consisting of extended transmembrane segments of Vpu from HIV-1: Computer simulations and conductance measurements. Biochemistry. 2002;41:7359–7365. doi: 10.1021/bi025518p. [DOI] [PubMed] [Google Scholar]
- Courgnaud V., Salemi M., Pourrut X., Mpoudi-Ngole E., Abela B., Auzel P., Bibollet-Ruche F., Hahn B., Vandamme A.M., Delaporte E., Peeters M. Characterization of a novel Simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into Simian/Human immunodeficiency virus phylogeny. J. Virol. 2002;76:8298–8309. doi: 10.1128/JVI.76.16.8298-8309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristian L., Lear J.D., DeGrado W.F. Use of thiol disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 2003;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuconati A., Xiang W., Lahser F., Pfister T., Wimmer E. A protein linkage map of the P2 nonstructural proteins of poliovirus. J. Virol. 1998;72:1297–1307. doi: 10.1128/jvi.72.2.1297-1307.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B.R. HIV-1 auxiliary proteins: Making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- Cymes G.D., Grosman C. Pore-opening mechanism of the nicotinic acetylcholine receptor evinced by proton transfer. Nat. Struct. Mol. Biol. 2008;15:389–396. doi: 10.1038/nsmb.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese P.N., Silhavy T.J. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- Davies W.L., Grunert R.R., Haff R.F., McGahen J.W., Neumayer E.M., Paulshock M., Watts J.C., Wood T.R., Hermann E.C., Hoffmann C.E. Antiviral activity of 1-adamantanamine (amantadine) Science. 1964;144:862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- de Jong A.S., Schrama I.W., Willems P.H., Galama J.M., Melchers W.J., van Kuppeveld F.J. Multimerization reactions of coxsackievirus proteins 2B, 2C and 2BC: A mammalian two-hybrid analysis. J. Gen. Virol. 2002;83:783–793. doi: 10.1099/0022-1317-83-4-783. [DOI] [PubMed] [Google Scholar]
- de Jong A.S., Wessels E., Dijkman H.B.P.M., Galama J.M.D., Melchers W.J.G., Willems P.H.G.M., van Kuppeveld F.J.M. Determinants for membrane association and permeabilization of the coxsackievirus 2B protein and the identification of the Golgi complex as the target organelle. J. Biol. Chem. 2003;278:1012–1021. doi: 10.1074/jbc.M207745200. [DOI] [PubMed] [Google Scholar]
- de Jong A.S., Melchers W.J., Glaudemans D.H., Willems P.H., van Kuppeveld F.J. Mutational analysis of different regions in the coxsackievirus 2B protein: Requirements for homo-multimerization, membrane permeabilization, subcellular localization, and virus replication. J. Biol. Chem. 2004;279:19924–19935. doi: 10.1074/jbc.M314094200. [DOI] [PubMed] [Google Scholar]
- Dieckmann G.R., Lear J.D., Zhong Q., Klein M.L., DeGrado W.F., Sharp K.A. Exploration of the structural features defining the conduction properties of a synthetic ion channel. Biophys. J. 1999;76:618–630. doi: 10.1016/S0006-3495(99)77230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedens J.R., Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Cabral J.M., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Duff K.C., Kelly S.M., Price N.C., Bradshaw J.P. The secondary structure of influenza A M2 transmembrane domain. A circular dichroism study. FEBS Lett. 1992;311:256–258. doi: 10.1016/0014-5793(92)81114-2. [DOI] [PubMed] [Google Scholar]
- Durantel D., Branza-Nichita N., Carrouée-Durantel S., Butters T.D., Dwek R.A., Zitzmann N. Study of the mechanism of antiviral action of iminosugar derivatives against bovine viral diarrhea virus. J. Virol. 2001;75:8987–8998. doi: 10.1128/JVI.75.19.8987-8998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D.M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- Ewart G.D., Sutherland T., Gage P.W., Cox G.B. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol. 1996;70:7108–7115. doi: 10.1128/jvi.70.10.7108-7115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart G.D., Mills K., Cox G.B., Gage P.W. Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur. Biophys. J. 2002;31:26–35. doi: 10.1007/s002490100177. [DOI] [PubMed] [Google Scholar]
- Federau T., Schubert U., Floßdorf J., Henklein P., Schomburg D., Wray V. Solution structure of the cytoplasmic domain of the human immunodeficiency virus type 1 encoded virus protein U (Vpu) Int. J. Peptide Protein Res. 1996;47:297–310. doi: 10.1111/j.1399-3011.1996.tb01359.x. [DOI] [PubMed] [Google Scholar]
- Fischer W.B. Viral membrane proteins: Structure, function and drug design. In: Atassi M.Z., editor. Protein Reviews. Kluwer Academic/Plenum Publisher; New York: 2005. [Google Scholar]
- Fischer W.B., Sansom M.S.P. Viral ion channels: Structure and function. Biochim. Biophys. Acta. 2002;1561:27–45. doi: 10.1016/s0304-4157(01)00009-0. [DOI] [PubMed] [Google Scholar]
- Fischer W.B., Forrest L.R., Smith G.R., Sansom M.S.P. Transmembrane domains of viral ion channel proteins: A molecular dynamics simulation study. Biopolymers. 2000;53:529–538. doi: 10.1002/(SICI)1097-0282(200006)53:7<529::AID-BIP1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Fischer W.B., Pitkeathly M., Wallace B.A., Forrest L.R., Smith G.R., Sansom M.S.P. Transmembrane peptide NB of influenza B: A simulation, structure, and conductance study. Biochemistry. 2000;39:12708–12716. doi: 10.1021/bi001000e. [DOI] [PubMed] [Google Scholar]
- Friborg J., Ladha A., Göttlinger H., Haseltine W.A., Cohen E.A. Functional analysis of the phosphorylation sites on the human immunodeficiency virus type-1 Vpu protein. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995;8:10–22. [PubMed] [Google Scholar]
- Gharbi-Benarous J., Bertho G., Evrard-Todeschi N., Coadou G., Megy S., Delaunay T., Benarous R., Girault J.P. Epitope mapping of the phosphorylation motif of the HIV-1 protein Vpu bound to the selective monoclonal antibody using TRNOESY and STD NMR spectroscopy. Biochemistry. 2004;43:14555–14565. doi: 10.1021/bi0492861. [DOI] [PubMed] [Google Scholar]
- Gonzales M.E., Carrasco L. Viroporins. FEBS Lett. 2003;552:28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- Grice A.L., Kerr I.D., Sansom M.S.P. Ion channels formed by HIV-1 Vpu: A modelling and simulation study. FEBS Lett. 1997;405:299–304. doi: 10.1016/s0014-5793(97)00198-1. [DOI] [PubMed] [Google Scholar]
- Griffin S.D.C., Beales L.P., Clarke D.S., Worsfold O., Evans S.D., Jäger J., Harris M.P.G., Rowlands D.J. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- Griffin S.D.C., Harvey R., Clarke D.S., Barclay W.S., Harris M., Rowlands D.J. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J. Gen. Virol. 2004;85:451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- Gurezka R., Laage R., Brosig B., Langosch D. A heptad motif of leucine residues found in membrane proteins can drive self-assembly of artificial transmembrane segments. J. Biol. Chem. 1999;274:9265–9270. doi: 10.1074/jbc.274.14.9265. [DOI] [PubMed] [Google Scholar]
- Hamman J.H., Enslin G.M., Kotze A.F. Oral delivery of peptide drugs: Barriers and developments. Biodrugs. 2005;19:165–177. doi: 10.2165/00063030-200519030-00003. [DOI] [PubMed] [Google Scholar]
- Harada T., Tautz N., Thiel H.J. E2-p7 region of the bovine viral diarrhea virus polyprotein: Processing and functional studies. J. Virol. 2000;74:9498–9506. doi: 10.1128/jvi.74.20.9498-9506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M., Kawaoka Y. The NB protein of Influenza B virus is not necessary for virus replication in vitro. J. Virol. 2003;77:6050–6054. doi: 10.1128/JVI.77.10.6050-6054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A.J., Wolstenholme A.J., Skehel J.J., Smith M.H. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf R.J.C., Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- Holsinger L.J., Shaughnessy A., Micko A., Pinto L.H., Lamb R.A. Analysis of the posttranslational modifications of the influenza virus M2 protein. J. Virol. 1995;69:1219–1225. doi: 10.1128/jvi.69.2.1219-1225.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongo S., Sugawara K., Muraki Y., Kitame F., Nakamura K. Characterization of a second protein (CM2) encoded by RNA segment 6 of influenza C virus. J. Virol. 1997;71:2786–2792. doi: 10.1128/jvi.71.4.2786-2792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hout D.R., Gomez L.M., Pacyniak E., Miller J.M., Hill M.S., Stephens E.B. A single amino acid substitution within the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein renders simian-human immunodeficiency virus (SHIV(KU-1bMC33)) susceptible to rimantadine. Virology. 2006;348:449–461. doi: 10.1016/j.virol.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Hsu K., Seharaseyon J., Dong P., Bour S., Marbán E. Mutual functional destruction of HIV-1 Vpu and host TASK-1 channel. Mol. Cell. 2004;14:259–267. doi: 10.1016/s1097-2765(04)00183-2. [DOI] [PubMed] [Google Scholar]
- Hu J., Fu R., Nishimura K., Zhang L., Zhou H.X., Busath D.D., Vijayvergiya V., Cross T.A. Histidines, heart of the hydrogen ion channel from influenza A virus: Towards an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. USA. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Fu R., Cross T.A. The chemical and dynamical influence of the anti-viral drug amantadine on the M2 proton channel transmembrane domain. Biophys. J. 2007;93:276–283. doi: 10.1529/biophysj.106.102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T., Cheynier R., Meyerhans A., Roelants G. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- Hussain A., Das S.R., Tanwar C., Jameel S. Oligomerization of the human immunodeficiency virus type I (HIV-1) Vpu protein—A genetic, biochemical and biophysical analysis. Virol. J. 2007;4:1–11. doi: 10.1186/1743-422X-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Watanabe S., Ninomiya A., Obuchi M., Odagiri T. Influenza B virus BM2 protein is a crucial component for incorporation of viral ribonucleoprotein complex into virions during virus assembly. J. Virol. 2004;78:11007–11015. doi: 10.1128/JVI.78.20.11007-11015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Kawasaki K., Odagiri T. Cytoplasmic domain of Influenza B virus BM2 protein plays critical roles in production of infectious virus. J. Virol. 2008;82:728–739. doi: 10.1128/JVI.01752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin A. Emerging structure of the nicotinic acetylcholine receptor. Nat. Rev. Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- Khattari Z., Arbely E., Arkin I.T., Salditt T. Viral ion channel proteins in model membranes: A comparative study by X-ray reflectivity. Eur. Biophys. J. 2006;36:45–55. doi: 10.1007/s00249-006-0099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Nishikawa M., Ohyama A. Intracellular membrane traffic of human immunodeficiency virus type 1 envelope glycoproteins: Vpu liberates Golgi-targeted gp160 from CD4-dependent retention in the endoplasmic reticulum. J. Biochem. 1994;115:1010–1020. doi: 10.1093/oxfordjournals.jbchem.a124414. [DOI] [PubMed] [Google Scholar]
- Klimkait T., Strebel K., Hoggan M.D., Martin M.A., Orenstein J.M. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolocouris N., Kolocouris A., Foscolos G.B., Fytas G., Neyts J., Padalko E., Balzarini J., Snoeck R., Andrei G., De Clercq E. Synthesis and antiviral activity evaluation of some new aminoadamantane derivatives. 2. J. Med. Chem. 1996;39:3307–3318. doi: 10.1021/jm950891z. [DOI] [PubMed] [Google Scholar]
- Kovacs F.A., Cross T.A. Transmembrane four-helix bundle of influenza A M2 protein channel: Structural implications from helix tilt and orientation. Biophys. J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs F.A., Denny J.K., Song Z., Quine J.R., Cross T.A. Helix tilt of the M2 transmembrane peptide from influenza A virus: An intrinsic property. J. Mol. Biol. 2000;295:117–125. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- Krüger J., Fischer W.B. Exploring the conformational space of Vpu from HIV-1: A versatile and adaptable protein. J. Comput. Chem. 2008;29:2416–2424. doi: 10.1002/jcc.20986. [DOI] [PubMed] [Google Scholar]
- Kukol A., Arkin I.T. Vpu transmembrane peptide structure obtained by site-specific Fourier transform infrared dichroism and global molecular dynamics searching. Biophys. J. 1999;77:1594–1601. doi: 10.1016/S0006-3495(99)77007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukol A., Arkin I.T. Structure of the influenza C virus CM2 protein transmembrane domain obtained by site-specific infrared dichroism and global molecular dynamics searching. J. Biol. Chem. 2000;275:4225–4229. doi: 10.1074/jbc.275.6.4225. [DOI] [PubMed] [Google Scholar]
- Kukol A., Adams P.D., Rice L.M., Brunger A.T., Arkin I.T. Experimentally based orientational refinement of membrane protein models: A structure for the influenza A M2 H+ channel. J. Mol. Biol. 1999;286:951–962. doi: 10.1006/jmbi.1998.2512. [DOI] [PubMed] [Google Scholar]
- Kuo A., Gulbis J.M., Antcliff J.F., Rahman T., Lowe E.D., Zimmer J., Cuthbertson J., Ashcroft F.M., Ezaki T., Doyle D.A. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- Lamb R.A., Pinto L.H. Do Vpu and Vpr of human immunodeficiency virus type 1 and NB of influenza B virus have ion channel activities in the viral life cycles? Virology. 1997;229:1–11. doi: 10.1006/viro.1997.8451. [DOI] [PubMed] [Google Scholar]
- Lear J.D., Wasserman Z.R., DeGrado W.F. Synthetic amphiphilic peptide models for proteins ion channels. Science. 1988;240:1177–1181. doi: 10.1126/science.2453923. [DOI] [PubMed] [Google Scholar]
- Leser G.P., Lamb R.A. Influenza virus assembly and budding in raft-derived microdomains: A quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Liljestrom P., Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 1991;65:147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljestrom P., Lusa S., Huylebroeck D., Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: The small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.K., Wooten D., Siegel R., Cheung R.C. Amantadine in treatment of chronic hepatitis C virus infection? J. Viral Hepatitis. 2005;12:445–455. doi: 10.1111/j.1365-2893.2005.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J. Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Lindenbach B.D., Pragai B.M., McCourt D.W., Rice C.M. Processing in the hepatitis C virus E2-NS2 region: Identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewy A., Smyth J., von Bonsdorff C.H., Liljestrom P., Schlesinger M.J. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J. Virol. 1995;69:469–475. doi: 10.1128/jvi.69.1.469-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V., Korner F., Koch J., Herian U., Theilmann L., Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K.L., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Marassi F.M., Jones D.H., Straus S.K., Bour S., Strebel K., Schubert U., Oblatt-Montal M., Montal M., Opella S.J. Expression, purification, and activities of full-length and truncated versions of the integral membrane protein Vpu from HIV-1. Protein Sci. 2002;11:546–557. doi: 10.1110/ps.37302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F., Chen M.Y., Willey R.L., Strebel K. Human-immunodeficiency-virus type-1 Vpu protein is an oligomeric type-I integral membrane protein. J. Virol. 1993;67:5056–5061. doi: 10.1128/jvi.67.8.5056-5061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin F., Benichou S., Durand H., Richard V., Liu L.X., Benarous R. Interaction between the cytoplasmic domains of HIV-1 Vpu and CD4: Role of Vpu residues involved in CD4 interaction and in vitro CD4 degradation. Virology. 1996;223:381–386. doi: 10.1006/viro.1996.0491. [DOI] [PubMed] [Google Scholar]
- McCown M.F., Pekosz A. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 2006;80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmel M., Rothermel M., Meckel T., Van Etten J.L., Moroni A., Thiel G. Possible function for virus encoded K+ channel Kcv in the replication of chlorella virus PBCV-1. FEBS Lett. 2003;552:7–11. doi: 10.1016/s0014-5793(03)00776-2. [DOI] [PubMed] [Google Scholar]
- Mehnert T., Lam Y.H., Judge P.J., Routh A., Fischer D., Watts A., Fischer W.B. Towards a mechanism of function of the viral ion channel Vpu from HIV-1. J. Biomol. Struct. Dyn. 2007;24:589–596. doi: 10.1080/07391102.2007.10507148. [DOI] [PubMed] [Google Scholar]
- Mehnert T., Routh A., Judge P.J., Lam Y.H., Fischer D., Watts A., Fischer W.B. Biophysical characterisation of Vpu from HIV-1 suggests a channel–pore dualism. Proteins. 2008;70:1488–1497. doi: 10.1002/prot.21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton J.V., Ewart G.D., Weir R.C., Board P.G., Lee E., Gage P.W. Alphavirus 6K proteins form ion channels. J. Biol. Chem. 2002;277:46923–46931. doi: 10.1074/jbc.M207847200. [DOI] [PubMed] [Google Scholar]
- Miyazawa A., Fujiyoshi Y., Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Montal M., Müller P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. USA. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.B., Zhong Q., Husslein T., Klein M.L. Simulation of the HIV-1 Vpu transmembrane domain as a pentameric bundle. FEBS Lett. 1998;431:143–148. doi: 10.1016/s0014-5793(98)00714-5. [DOI] [PubMed] [Google Scholar]
- Mould J.A., Li H.C., Dudlak C.S., Lear J.D., Pekosz A., Lamb R.A., Pinto L.H. Mechanism for proton conduction of the M2 ion channel of influenza A virus. J. Biol. Chem. 2000;275:8592–8599. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- Mould J.A., Paterson R.G., Takeda M., Ohigashi Y., Venkataraman P., Lamb R.A., Pinto L.H. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Munro S. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S.J.D., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–431. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Nieva J.L., Agirre A., Nir S., Carrasco L. Mechanism of membrane permeabilization by picornavirus 2B viroporin. FEBS Lett. 2003;552:68–73. doi: 10.1016/s0014-5793(03)00852-4. [DOI] [PubMed] [Google Scholar]
- Nilsson T., Slusarewicz P., Hoe M.H., Warren G. Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- Nilsson T., Rabouille C., Hui N., Watson R., Warren G. The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase I in retention, kin recognition and structural maintenance of the Golgi apparatus in HeLa cells. J. Cell Sci. 1996;109:1975–1989. doi: 10.1242/jcs.109.7.1975. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Kim S., Zhang L., Cross T.A. The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- Odagiri T., Hong J., Ohara Y. The BM2 protein of influenza B virus is synthesized in the late phase of infection and incorporated into virions as a subviral component. J. Gen. Virol. 1999;80:2573–2581. doi: 10.1099/0022-1317-80-10-2573. [DOI] [PubMed] [Google Scholar]
- Pacyniak E., Gomez M.L., Gomez L.M., Mulcahy E.R., Jackson M., Hout D.R., Wisdom B.J., Stephens E.B. Identification of a region within the cytoplasmic domain of the subtype B Vpu protein of human immunodeficiency virus type 1 (HIV-1) that is responsible for retention in the Golgi complex and its absence in the Vpu protein from a subtype C HIV-1. AIDS Res. Hum. Retrovir. 2005;21:379–394. doi: 10.1089/aid.2005.21.379. [DOI] [PubMed] [Google Scholar]
- Park S.H., Mrse A.A., Nevzorov A.A., Mesleh M.F., Oblatt-Montal M., Montal M., Opella S.J. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J. Mol. Biol. 2003;333:409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- Patargias G., Martay H., Fischer W.B. Reconstructing potentials of mean force from short steered molecular dynamics simulations of Vpu from HIV-1. J. Biomol. Struc. Dyn. 2009 doi: 10.1080/07391102.2009.10507291. in print. [DOI] [PubMed] [Google Scholar]
- Patargias G., Zitzmann N., Dwek R., Fischer W.B. Protein–protein interactions: Modeling the hepatitis C virus ion channel p7. J. Med. Chem. 2006;49:648–655. doi: 10.1021/jm050721e. [DOI] [PubMed] [Google Scholar]
- Paterson R.G., Takeda M., Ohigashi Y., Pinto L.H., Lamb R.A. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology. 2003;306:7–17. doi: 10.1016/s0042-6822(02)00083-1. [DOI] [PubMed] [Google Scholar]
- Pavlovic D., Neville D.C.A., Argaud O., Blumberg B., Dwek R.A., Fischer W.B., Zitzmann N. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penin F., Dubuisson J., Rey F.A., Moradpour D., Pawlotsky J.M. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- Piller S.C., Ewart G.D., Premkumar A., Cox G.B., Gage P.W. Vpr protein of human immunodeficiency virus type 1 forms cation-selective channels in planar lipid bilayers. Proc. Natl. Acad. Sci. USA. 1996;93:111–115. doi: 10.1073/pnas.93.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plugge B., Gazzarrini S., Nelson M., Cerana R., Van Etten J.L., Derst C., DiFrancesco D., Moroni A., Thiel G. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287:1641–1644. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- Popot J.L., Engelman D.M. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- Premkumar A., Wilson L., Ewart G.D., Gage P.W. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- Premkumar A., Horan C.R., Gage P.W. Dengue virus M protein C-terminal peptide (DVM-C) forms ion channels. J. Membr. Biol. 2005;204:33–38. doi: 10.1007/s00232-005-0744-9. [DOI] [PubMed] [Google Scholar]
- Root M.J., Kay M.S., Kim P.S. Protein design of an HIV-1 entry inhibitor. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- Russ W.P., Engelman D.M. The GxxxG motif: A framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T., Tu Q.A., Pinto L.H., Lamb R.A. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc. Natl. Acad. Sci. USA. 1997;94:5000–5005. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A., St. Claire M., Faulk K., Govindarajan S., Emerson S.U., Purcell R.H., Bukh J. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc. Natl. Acad. Sci. USA. 2003;100:11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz M.A., Perez L., Carrasco L. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J. Biol. Chem. 1994;269:12106–12110. [PubMed] [Google Scholar]
- Schirmer T. General and specific porins from bacterial outer membranes. J. Struct. Biol. 1998;121:101–109. doi: 10.1006/jsbi.1997.3946. [DOI] [PubMed] [Google Scholar]
- Schnell J.R., Chou J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C., Lin T.I. Influenza A virus M2 protein: Proton selectivity of the ion channel, cytotoxicity, and a hypothesis on peripheral raft association and virus budding. In: Fischer W.B., editor. Viral Membrane Proteins: Structure, Function, and Drug Design. Kluwer Academic; New York: 2005. pp. 113–130. [Google Scholar]
- Schroeder C., Heider H., Moncke-Buchner E., Lin T.I. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur. Biophys. J. 2005;34:52–66. doi: 10.1007/s00249-004-0424-1. [DOI] [PubMed] [Google Scholar]
- Schubert U., Bour S., Ferrer-Montiel A.V., Montal M., Maldarelli F., Strebel K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 1996;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U., Ferrer-Montiel A.V., Oblatt-Montal M., Henklein P., Strebel K., Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- Schweighofer K.J., Pohorille A. Computer simulation of ion channel gating: The M2 channel of influenza A virus in a lipid bilayer. Biophys. J. 2000;78:150–163. doi: 10.1016/S0006-3495(00)76581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Xue J.H., Yu C.Y., Luo H.B., Qin L., Yu X.J., Chen J., Chen L.L., Xiong B., Yue L.D., Cai J.H., Shen J.H. Small envelope protein E of SARS: Cloning, expression, purification, CD determination, and bioinformatics analysis. Acta Pharmacol. Sin. 2003;24:505–511. [PubMed] [Google Scholar]
- Siddell S. The small-membrane protein. In: Siddell S., editor. The Coronaviridae. Plenum Press; New York: 1995. [Google Scholar]
- Smondyrev A.M., Voth G.A. Molecular dynamics simulation of proton transport through the influenza A virus M2 channel. Biophys. J. 2002;83:1987–1996. doi: 10.1016/S0006-3495(02)73960-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Steinmann E., Penin F., Kallis S., Patel A.H., Bartenschlager R., Pietschmann T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathogens. 2007;3:962–971. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann E., Whitfield T., Kallis S., Dwek R., Zitzmann N., Pietschmann T., Bartenschlager R. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology. 2007;46:330–338. doi: 10.1002/hep.21686. [DOI] [PubMed] [Google Scholar]
- StGelais C., Tuthill T.J., Clarke D.S., Rowlands D.J., Harris M., Griffin S. Inhibition of hepatitis C virus p7 membrane channels in a liposome-based assay system. Antiviral Res. 2007;76:48–58. doi: 10.1016/j.antiviral.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer A.L., Acharya R., Salom D., Levine A.S., Di Constanzo L., Soto C.S., Tereshko V., Nanda V., Stayrook S., DeGrado W.F. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K., Klimkait T., Martin M.A. Novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- Stroud R.M., Savage D., Miercke L.J.W., Lee J.K., Khademi S., Harries W. Selectivity and conductance among the glycerol and water conducting aquaporin family of channels. FEBS Lett. 2003;555:79–84. doi: 10.1016/s0014-5793(03)01195-5. [DOI] [PubMed] [Google Scholar]
- Sugrue R.J., Hay A.J. Structural characteristics of the M2 protein of influenza A viruses: Evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue R.J., Bahadur G., Zambon M.C., Hall-Smith M., Douglas A.R., Hay A.J. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990;9:3469–3476. doi: 10.1002/j.1460-2075.1990.tb07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue R.J., Belshe R.B., Hay A.J. Palmitoylation of the influenza A virus M2 protein. Virology. 1990;179:51–56. doi: 10.1016/0042-6822(90)90272-s. [DOI] [PubMed] [Google Scholar]
- Sunstrom N.A., Prekumar L.S., Prekumar A., Ewart G., Cox G.B., Gage P.W. Ion channels formed by NB, an influenza B virus protein. J. Membr. Biol. 1996;150:127–132. doi: 10.1007/s002329900037. [DOI] [PubMed] [Google Scholar]
- Tieleman D.P., Berendsen H.J.C., Sansom M.S.P. An alamethicin channel in a lipid bilayer: Molecular dynamics simulations. Biophys. J. 1999;76:1757–1769. doi: 10.1016/s0006-3495(99)77337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiganos E., Yao X.J., Friborg J., Daniel N., Cohen E.A. Putative a-helical structures in the human immunodeficiency virus type 1 Vpu protein and CD4 are involved in binding and degradation of the CD4 molecule. J. Virol. 1997;71:4452–4460. doi: 10.1128/jvi.71.6.4452-4460.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi A., Sandri L., Giangaspero A. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Tu Q., Pinto L.H., Luo G., Shaughnessy M.A., Mullaney D., Kurtz S., Krystal M., Lamb R.A. Characterization of inhibition of M2 ion channel activity by BL-1743, an inhibitor of Influenza A virus. J. Virol. 1996;70:4246–4252. doi: 10.1128/jvi.70.7.4246-4252.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmschneider J.P., Ulmschneider M. Folding simulations of the transmembrane helix of virus protein u in an implicit membrane model. J. Chem. Theory Comput. 2007;3:2335–2346. doi: 10.1021/ct700103k. [DOI] [PubMed] [Google Scholar]
- van Damme N., Goff D., Katsura C., Jorgensen R.L., Mitchell R., Johnson M.C., Stephens E.B., Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:1–8. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuppeveld F.J., Galama J.M., Zoll J., Melchers W.J. Genetic analysis of a hydrophobic domain of coxsackie B3 virus protein 2B: A moderate degree of hydrophobicity is required for a cis-acting function in viral RNA synthesis. J. Virol. 1995;69:7782–7790. doi: 10.1128/jvi.69.12.7782-7790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuppeveld F.J.M., Hoenderop J.G.J., Smeets R.L.L., Willems P.H.G.M., Dijkman H.B.P.M., Galama J.M.D., Melchers W.J.G. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997;16:3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuppeveld F.J.M., Melchers W.J.G., Kirkegaard K., Doedens J.R. Structure-function analysis of coxsackie B3 virus protein 2B. Virology. 1997;227:111–118. doi: 10.1006/viro.1996.8320. [DOI] [PubMed] [Google Scholar]
- van Kuppeveld F.J.M., Melchers W.J.G., Willems P.H.G.M., Gadella T.W.J., Jr. Homomultimerization of the coxsackievirus 2B protein in living cells visualized by fluorescence resonance energy transfer microscopy. J. Virol. 2002;76:9446–9456. doi: 10.1128/JVI.76.18.9446-9456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varthakavi V., Smith R.M., Bour S.P., Strebel K., Spearman P. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA. 2003;100:15154–15159. doi: 10.1073/pnas.2433165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varthakavi V., Smith R.M., Martin K.L., Derdowski A., Lapierre L.A., Goldenring J.R., Spearman P. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic. 2006;7:298–307. doi: 10.1111/j.1600-0854.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- Veit M., Klenk H.D., Kendal A., Rott R. The M2 protein of influenza A virus is acylated. Virology. 1991;184:227–234. doi: 10.1099/0022-1317-72-6-1461. [DOI] [PubMed] [Google Scholar]
- Wang J., Kim S., Kovacs F., Cross T.A. Structure of the transmembrane region of the M2 protein H+ channel. Protein Sci. 2001;10:2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch W.J., Sefton B.M. Characterization of a small, nonstructural viral polypeptide present late during infection of BHK cells by Semliki Forest virus. J. Virol. 1980;33:230–237. doi: 10.1128/jvi.33.1.230-237.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willbold D., Hoffmann S., Rösch P. Secondary structure and tertiary fold of the human immunodeficiency virus protein U (Vpu) cytoplasmatic domain in solution. Eur. J. Biochem. 1997;245:581–588. doi: 10.1111/j.1432-1033.1997.t01-1-00581.x. [DOI] [PubMed] [Google Scholar]
- Willey R.L., Maldarelli F., Martin M.A., Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., McKinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330:322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G., Fields S. Cloning of influenza cDNA into M13: The sequence of the RNA segment encoding the A/PR/8/34 matrix protein. Nucleic Acids Res. 1980;8:1965–1974. doi: 10.1093/nar/8.9.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley G.A., Wallace B.A. Model ion channels: Gramicidin and alamethicin. J. Membr. Biol. 1992;129:109–136. doi: 10.1007/BF00219508. [DOI] [PubMed] [Google Scholar]
- Wray V., Kinder R., Federau T., Henklein P., Bechinger B., Schubert U. Solution structure and orientation of the transmembrane anchor domain of the HIV-1-encoded virus protein U by high resolution and solid-state NMR spectroscopy. Biochemistry. 1999;38:5272–5282. doi: 10.1021/bi982755c. [DOI] [PubMed] [Google Scholar]
- Yoshihara E., Yoneyama H., Nakae T. In vitro assembly of the functional porin trimer from dissociated monomers in Pseudomonas aeruginosa. J. Biol. Chem. 1991;226:952–957. [PubMed] [Google Scholar]
- Zhang J., Pekosz A., Lamb R.A. Influenza virus assembly and lipid raft microdomains: A role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 2000;74:4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Strzalka J., Ma C., Opella S.J., Ocko B.M., Blasie J.K. Structural studies of the HIV-1 accessory protein Vpu in Langmuir monolayers: Synchrotron X-ray reflectivity. Biophys. J. 2001;80:1837–1850. doi: 10.1016/S0006-3495(01)76154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


