Abstract
Compared to classical epidemiologic methods, genomics can be used to precisely monitor virus evolution and transmission in real time across large, diverse populations. Integration of pathogen genomics with data about host genetics and global transcriptional responses to infection allows for comprehensive studies of population-level responses to infection and provides novel methods for predicting clinical outcomes. As genomic technologies become more accessible, these methods will redefine how emerging viruses are studied and outbreaks are contained. Here we review the existing and emerging genomic technologies that are enabling systems epidemiology and systems virology and making it possible to respond rapidly to emerging viruses such as Zika.
The advent of new genomic technologies has enabled unprecedented speed and precision in understanding human viral pathogens. Rasmussen and Katze review the existing and emerging genomic technologies that are enabling systems epidemiology and systems virology and making it possible to respond rapidly to emerging viruses such as Zika.
Main Text
Introduction
The development of genomics technologies has heralded a new era in epidemiology, one in which viral epidemics can be detected and monitored in real time (Rasmussen, 2015). Historical efforts to track and contain outbreaks have relied on classical methods to study disease incidence at the population level. Outbreak containment was achieved primarily through isolating infected patients and contact tracing to establish appropriate quarantine procedures. While often effective, these methods are time consuming and imprecise, particularly when diagnosis is based exclusively on clinical data. Until the advent and implementation of polymerase chain reaction (PCR)-based methods in the early 2000s, laboratory diagnostics were heavily reliant on serology. Unfortunately, serological assays are typically not useful during an acute virus infection, as they cannot detect exposure to a particular pathogen until the patient has mounted a detectable neutralizing antibody response. Even then, they cannot distinguish between different viral strains within a given serotype and are not useful for monitoring virus evolution. While PCR and reverse-transcriptase PCR (RT-PCR) have enabled direct early detection of virus genomes, these amplification-based methods are unfortunately heavily biased and dependent on the sequence of oligonucleotide primers. PCR-based methodologies are not useful for screening or surveillance of unknown pathogens or those that diverge highly from known prototypic strains, nor can they monitor virus evolution in real time during an outbreak. Thus, methods that can provide detailed information about an emerging virus at the sequence level are proving essential to realizing the promise of genomics as an epidemiologic tool (Barzon et al., 2011, Quiñones-Mateu et al., 2014, Sloots et al., 2015). Combining experimental, clinical, and computational approaches with pathogen genomics will give us a systems-level understanding of how viruses cause illness (Figure 1 ). Moving forward, we will also be able to more confidently identify the etiologic agents associated with pathogenic virus infections utilizing host transcriptional signatures. Thus, rather than having to fulfill Koch’s postulates to be certain of the cause of disease, scientists and clinicians will be able to use genomic information about the virus combined with the host response transcriptional signature to make more confident associations and diagnoses. In this review, we examine the existing and emerging genomic technologies that are being employed for the analysis of viruses and the infections they cause. Throughout, we include examples of the effective use of these technologies for epidemiological, virological, and medical applications. Finally, we provide some thoughts looking forward toward emerging viruses, such as Zika.
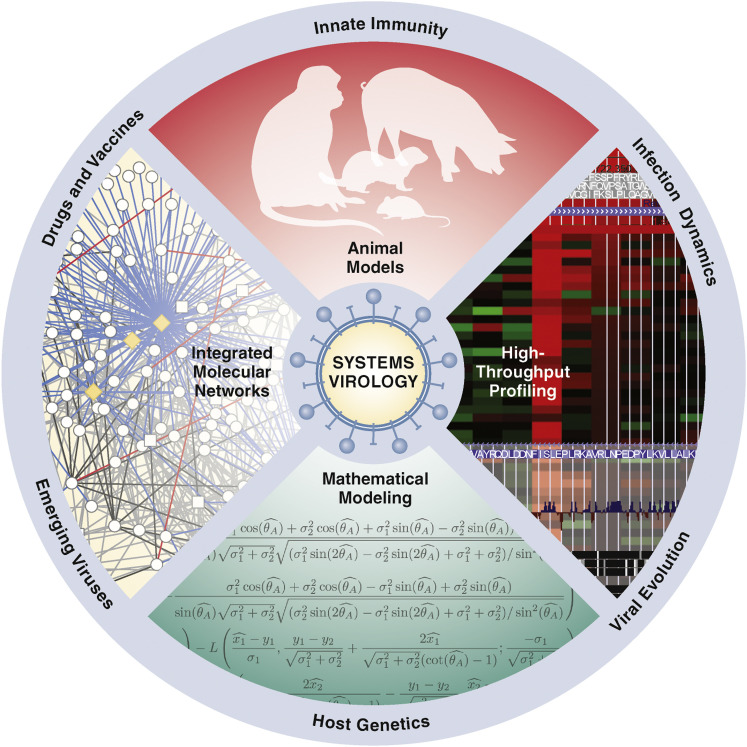
Figure 1.
Systems Virology Model for Genomic Characterization, Host Response, and Surveillance for Emerging Viral Pathogens
Future epidemiology and virology research will use genomics in concert with a variety of clinical, experimental, and bioinformatics approaches to rapidly facilitate a more comprehensive understanding of emergent viruses and the host response to the pathogen. One usually starts with a reproducible animal or tissue culture virus infection model, followed by high-throughput profiling utilizing microarrays and/or RNA-seq. The next steps involve defining the host responses using mathematical, computational, and bioinformatics tools. Predictions are made, followed by the iterative process of validating the predictions experimentally. These approaches are powerful tools for drug repurposing, correlating the host response with discovery of novel virus pathogens and defining mechanisms underlying innate immunity.
Technologies Enabling Molecular Epidemiology
The currently available genomic technologies that have been used for analyzing viruses and their relative advantages and disadvantages are summarized in Table 1 . Initial efforts to conduct broad-spectrum screening for emerging viruses from clinical specimens were based primarily on RT-PCR panels. While most PCR-based assays can be performed on standard laboratory equipment, these assays do not provide genetic information beyond the virus species or strain and usually do not cover the entire pathogen genome. The RT-PCR assays used in diagnostics today are based on older studies using validated primers capable of amplifying emerging pathogens of interest (Casas et al., 1997, Drosten et al., 2002, Valassina et al., 1997). Multiplex PCR technology was subsequently combined with primer tagging (MassTag PCR) to survey a wide range of viral sequences used as diagnostic panels for hemorrhagic fever viruses and respiratory viruses (Palacios et al., 2006). This method was used to identify a novel rhinovirus clade associated with severe influenza-like illness (Lamson et al., 2006), although the MassTag platform is restricted by primer bias. Gradually, multiplex PCR primer panels were extended to RT-PCR arrays. These arrays of multiplexed RT-PCR probe sets enable multiple PCR amplifications at once and typically cover groups of related viruses rather than the entire virome (Renois et al., 2010, Sanghavi et al., 2012).
Table 1.
Genomic Technologies Available for Virus Analysis
| Method | Sample Requirements | Advantages | Disadvantages |
|---|---|---|---|
| qPCR/RT-PCR | Purified RNA/DNA | Highly sensitive; used for absolute or relative quantitation | Requires prior knowledge of sequence of interest |
| Microarray-ViroChip | Purified RNA | Requires low input; well-developed technology; developed for Agilent platform | Requires high-quality input RNA; requires prior knowledge of sequences of interest and may not detect novel viruses or viral variants; only useful for relative quantitation; non-specific hybridization can confound results |
| Microarray-host gene expression | Purified RNA | Requires low input; well-developed technology; multiple platforms with numerous product lines | Requires high-quality input RNA; off-the-shelf products do not detect viral sequences; only useful for relative quantitation; non-specific hybridization can confound results |
| Sanger sequencing | Purified RNA/DNA | Low cost; uses equipment available to most institutions; numerous commercial entities offer these services | Requires prior knowledge of sequence of interest; low throughput; not applicable to global transcriptomic applications |
| Next-gen sequencing-mRNA-seq | Purified mRNA | Does not require prior knowledge of sequence of interest; provides unbiased global view of the full coding transcriptome | Requires high-quality input RNA; difficult to distinguish splice isoforms using short-read platform, especially at lower read depths; does not detect transcripts that are not polyadenylated (some virus genomes and non-coding RNA); fragmentation and amplification during library preparation can introduce bias; large data output requires substantial storage and computational power to manage and analyze; short-read platform makes absolute quantification difficult; viral reads are often undetectable due to high host transcript background |
| Next-gen sequencing-total RNA-seq | Purified RNA | Does not require prior knowledge of sequence of interest; can be used with fragmented or degraded RNA; includes sequences from non-coding and non-polyadenylated transcripts | Difficult to distinguish splice isoforms using short-read platform, especially at lower read depths; fragmentation and amplification during library preparation can introduce bias; large data output requires substantial storage and computational power to manage and analyze; short-read platform makes absolute quantification difficult; viral reads are often undetectable due to high host transcript background |
| Virome capture sequencing—VirCapSeq-VERT and ViroCap | Purified RNA | Covers the entire known human virome; significantly enriches virus reads from complex materials; can improve read coverage | Can only detect uncharacterized viruses based on conserved sequences capable of hybridizing with the capture oligos |
| Nanopore sequencing | Purified RNA | Long reads; single molecule detection; does not require amplification or labeling; miniaturizable; cloud-based analysis possible on laptop computers for deployment in field | Many technical difficulties remain for deploying hand-held systems in remote areas (data too large to upload to cloud-based base callers); primer bias may still impact sample preparation methods requiring amplification |
Other groups developed oligonucleotide microarrays (the Virochip) that could identify and relatively quantify the presence of genomes from known, sequenced viruses. The Virochip system is capable of simultaneous parallel screening for multiple viruses and as such is useful for detecting pathogens from clinical specimens in the absence of a confirmed diagnosis (Wang et al., 2002). The Virochip method has subsequently been used to identify viral pathogens from clinical specimens, including both human and porcine respiratory pathogens such as rhinoviruses, circoviruses, and coronaviruses (Kistler et al., 2007, Nicholson et al., 2011). Unfortunately, the Virochip requires high-quality input material and is not suitable for samples with fragmented or degraded nucleic acids from aldehyde-based fixatives, temperature fluctuations or extensive freeze-thawing, or nuclease activity. Furthermore, while they are becoming more common, microarray scanners are not standard equipment in many diagnostic laboratories, limiting the clinical sites where this technology can be used. Carbon nanotube field effect transistor technology under development enables label-free hybridization and eliminates the need for a microarray fluorescence scanner (Martinez et al., 2009, Sorgenfrei et al., 2011, Star et al., 2006); however, these are still largely at the prototype stage and are not widely available.
As next-generation sequencing has become more prevalent, accessible, and affordable, virus surveillance and diagnosis are increasingly performed by directly monitoring viral genomes. However, detecting sufficient viral reads at conventional read depths is a significant problem. High background host gene expression in complex clinical specimens (such as blood or tissue biopsies) interferes with complete coverage of viral genomes, thus reducing the reliability of this method. Therefore, new methods to enrich viral sequences have been developed to minimize bias and enhance coverage. Initial efforts relied on depletion of ribosomal RNAs, which typically comprise at least 50% of the host transcriptome. However, in the last year, systems were developed using targeted probe-based selection methods to enrich viral sequences from complex specimens. The virome capture sequencing platform for vertebrate viruses (VirCapSeq-VERT) can enrich viral reads by up to 10,000-fold and results in near-total genome coverage for all known viruses. Furthermore, based on highly conserved sequences, this platform could also identify highly divergent, completely uncharacterized viruses, suggesting its utility for virus discovery (Briese et al., 2015). An alternative targeted enrichment panel, ViroCap, enriches virus reads up to 700-fold and provides up to ∼80% genome coverage (Wylie et al., 2015). These enrichment methods offer great promise for rapid diagnosis of a virus infection from clinical specimens, metagenomic virome characterization, and monitoring virus evolution in real time. Unfortunately, although decreasing, the cost of next-generation sequencing instruments remains high, and this technology is not widely available in most clinical diagnostic laboratories. Short-read sequencers are large, cumbersome instruments, and the computational infrastructure necessary for robust sequence analysis is not conducive to work requiring mobility and real-time data collection, such as outbreak response or field surveillance.
The first instance of using sequence data to characterize a novel emerging virus was the complete mapping of the severe acute respiratory syndrome coronavirus (SARS-CoV) genome as it emerged in 2003 (Marra et al., 2003). This was an especially remarkable achievement, considering the genome was sequenced using Sanger dideoxy chain termination sequencing technology. Rapid sequencing of the SARS-CoV genome during the epidemic demonstrated the great potential for applying genomic analysis to the epidemiological toolbox. Recent efforts to bring sequencing technology into the field or the clinic focus on improving portability and access to cloud-based data storage and analysis. Recent work to develop analytical software that can be run on a laptop in the field has circumvented issues with web access in areas with limited connectivity (Quick et al., 2016). The MinION system is a small long-read nanopore sequencing device capable of assembling complete pathogen genomes (Madoui et al., 2015). In conjunction with a web-based bioinformatics pipeline, nanopore sequencing has been used to detect chikungunya virus (CHIKV), Ebola virus (EBOV), hepatitis B virus (HBV), hepatitis C virus (HCV), and influenza virus from clinical specimens (Greninger et al., 2015, Hoenen et al., 2015, Hoenen et al., 2016, Wang et al., 2015, Yao et al., 2014). Increased availability of the MinION system and online bioinformatics resources represent the future of molecular epidemiology, allowing rapid deployment of cutting-edge sequencing technology even in remote or resource-poor environments.
Tracking Ebola in West Africa
An unexpected outbreak of EBOV in Guinea in late 2013 sparked an epidemic that blazed through West Africa throughout 2014, killing thousands. Virus detection for most patients was limited to PCR-based assays, as the affected areas were already lacking necessary infrastructure for laboratory diagnostics. However, samples collected for research were also used to rapidly generate a wealth of genetic sequence data concerning the origins and evolution of the Makona variant causing the 2014 West African EBOV epidemic (Gire et al., 2014). This is the first example of effectively using sequencing technologies in the field to monitor a large epidemic caused by an emerging virus in real time as it unfolded. A critical question concerned the origins of this epidemic, as all known previous outbreaks of Zaire ebolaviruses occurred thousands of miles away in Central Africa. Genome assemblies from samples collected early in the outbreak from 78 patients in Sierra Leone suggested initially that the Makona variant diverged from central African strains in 2004 (Gire et al., 2014). Analysis of these early samples suggested that the substitution rate of the Makona variant was higher than those observed during any previous EBOV outbreak. Subsequent sequencing studies from other clinical populations throughout Guinea, Sierra Leone, Liberia, and Mali demonstrated that the nucleotide substitution rate associated was roughly equivalent to central African Zaire ebolaviruses, suggesting that this was more likely the result of different analytical approaches (Hoenen et al., 2015, Kugelman et al., 2015, Ladner et al., 2015, Quick et al., 2016, Scarpino et al., 2015, Stadler et al., 2014, Tong et al., 2015). However, sequence analysis of many viruses throughout this outbreak provided an unprecedented opportunity to understand how purifying selection drives the emergence of different phylogenetic lineages as well as to identify specific mutations in viral proteins that may have functional significance (Park et al., 2015, Simon-Loriere et al., 2015). The University of California, Santa Cruz has developed a genome browser to facilitate continued bioinformatics studies investigating EBOV phylogeny (Haeussler et al., 2014), enabling large-scale genome studies that will improve our understanding of viral evolution.
Virus sequencing has also demonstrated at least two instances of sexual transmission by persistent EBOV in semen from a disease survivor (Mate et al., 2015). Unlike PCR-based methods that are capable of simply detecting virus in the EBOV disease survivor’s semen, sequencing allowed the determination that the survivors were infected with the same virus by sexual transmission.
In addition to the EBOV genomes sequenced primarily by short-read sequencers as described above, nanopore sequencing was used to sequence clinical Makona variant isolates on the ground in both Guinea and Liberia (Hoenen et al., 2016, Quick et al., 2016). This represented a proof-of-concept use of this technology to conduct molecular epidemiology in the field and clearly demonstrated the power of portable sequencing devices. This technology has been hindered by difficulties performing sequence analysis in the field, such as uploading large amounts of sequence data for analysis by cloud-based base-calling software. In particular, the development of customized, laptop-based base-callers enabled the first completely in-field demonstration of portable nanopore technology for epidemiology (Quick et al., 2016). Future virus epidemics can be easily monitored and studied by placing this technology in the hands of those responding to an epidemic.
As portable sequencers become more common, they can be used for simultaneous pathogen detection and genomic analysis. This will permit identification of a virus in the context of an outbreak but allow for virtually real-time analysis of virus evolution and transmission. This would also allow monitoring EBOV prevalence in different species of bats thought to be reservoirs, as well as disease outbreaks in wild populations of non-human primates. Portable sequencers have the potential to integrate detection or diagnosis with genomic methods to study virus transmission and evolution. This is invaluable in understanding the origin and epidemiology of disease outbreaks and virus ecology in the wild.
Tracking Viruses with Host Response Data
The increased use of genomics technology has created opportunities for monitoring epidemics in the absence of samples containing detectable virus by assaying the host response. Identifying and characterizing gene expression profiles associated with viral disease can detect cases for illnesses not associated with high viremia or in cases in which the infection has been cleared. Prior efforts to study epidemic survivors retrospectively have primarily been confined to serologic surveys. Positive serology can identify individuals who have been exposed to a particular pathogen, and antibody titers can be used to assess adaptive immunity, but these are not highly precise measures of the host response (MacNeil et al., 2014). Serotypic cross-reactivity between structurally similar epitopes conserved among or within viral taxa can make it difficult to distinguish specific pathogen strains or species, and antibody titers can vary dramatically between individuals and decrease over time. Other functional immune assays, such as multiplexed cytokine analysis to look at global inflammatory responses, or ELISpot assays or MHC tetramers to quantify and phenotype antigen-specific T cells, have also been used in many studies (Edwards, 2014, Luebke et al., 2004, McElhaney et al., 2016). However, these require fresh, temperature-controlled samples and specialized equipment and are generally not available to epidemiologists or researchers in the field. Measures of host responses across many different parameters thus are an appealing alternative means of studying virus emergence or prevalence in the absence of detectable virus, in the presence of a completely novel virus about which little is known, or after an infection has been cleared.
As the mobility of sequencing or microarray technology improves, opportunities have arisen to use global host transcriptomic responses for epidemiology or diagnostics. Myriad studies demonstrate that very specific host response profiles can be associated with viral disease at early stages and late stages and can be closely linked to disease severity and treatment response (Dong et al., 2015, Jansen et al., 2015, Rasmussen et al., 2012, Sun et al., 2013). These responses are not limited solely to antiviral immunity or inflammation, but also identify host pathways that are induced by viral infection and provide insight into how these pathways are involved in pathogenesis. In some cases, these have been linked closely to human cohorts of disease in both acute and chronic virus infections (Dong et al., 2015, Rasmussen et al., 2012). Host transcriptional responses to rhinovirus in primary cells ex vivo have also been shown to be concordant with those in vivo (Gardeux et al., 2015), indicating the potential for high-throughput development of broadly applicable diagnostic signatures, including those that can differentiate viral and bacterial pathogens (Huang et al., 2011, Zaas et al., 2013). As both microarray and sequencing technologies for collecting global transcriptomic data become more affordable and portable, increased surveys of host responses in diverse human cohorts during and after outbreaks of viral disease will facilitate the development of host response signatures that can be used for diagnosis or surveillance (Figure 2 ).
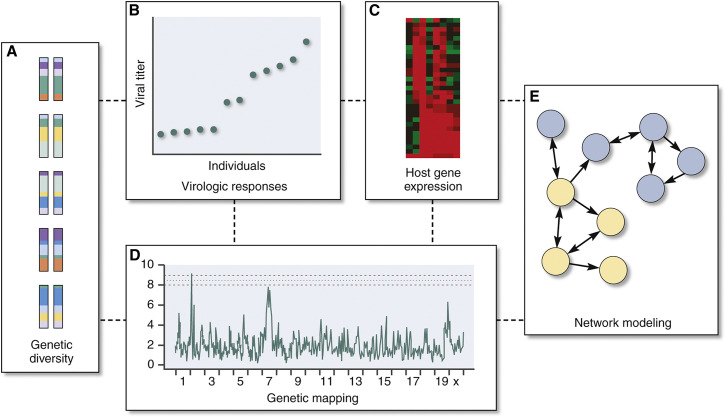
Figure 2.
Systems Genetics Incorporates Complexity to Explain Differential Disease Responses
(A–E) An advantage of genetically tractable yet complex experimental systems such as the Collaborative Cross Recombinant Inbred panels is the ability to explicitly integrate (A) host genetics and (B) virologic and (C) transcriptional responses to identify (D) polymorphic genetic loci that contribute to differential virologic responses and to develop (E) transcriptional networks that shed mechanistic insight into these polymorphic responses (see Rasmussen et al., 2014) (reprinted with permission from Katze et al., 2016).
Transcriptomic data allow for the most comprehensive look at host response dynamics during a viral infection. Gene expression profiles are regulated in part by genetic elements, such as particular alleles, transposons, or single nucleotide polymorphisms (SNPs) (Elbarbary et al., 2016, Haraksingh and Snyder, 2013, Huang, 2015). For many years, genome-wide association studies (GWAS) have been subject to considerable criticism (Huang, 2015), in part because of practical and logistic challenges associated with fully sequencing genomes from sufficient individuals to associate a given genetic element with a disease outcome in a diverse population. However, the increasing availability and decreasing cost of human genome sequencing is rapidly changing that paradigm. Genome sequencing and GWAS across a large, diverse cohort have revealed a link between SNPs in the interleukin-28B (IL-28B) promoter and hepatitis C virus (HCV) clearance and response to treatment (Ge et al., 2009, Thomas et al., 2009). As the overall body of human genome data becomes increasingly available, our understanding of particular genetic features that increase epidemic risk or predispose a population to specific pathogen susceptibilities or disease outcomes will grow.
Host Genetics and the Collaborative Cross
Genetic variation in humans poses a major challenge in assessing the host signature in response to virus infection. A relatively new resource, the Collaborative Cross (CC) mouse model, has helped define the role of host genetics and its contribution to determining the outcome of a viral infection (Bottomly et al., 2012, Ferris et al., 2013, Graham et al., 2015, Gralinski et al., 2015, Josset et al., 2014, Rasmussen et al., 2014, Xiong et al., 2014). These mice, due to their novel breeding scheme, resemble humans in terms of the complexity of their genetic variation. Not only does this ensure that CC mice are more reliable indicators of human disease, but it also allows investigators to identify novel alleles associated with disease resistance and susceptibility. Indeed, this model has been used to study a range of phenotypes recapitulating those occurring in human patients infected with influenza virus, West Nile virus, SARS-CoV, and EBOV (Ferris et al., 2013, Graham et al., 2015, Gralinski et al., 2015, Josset et al., 2014, Rasmussen et al., 2014, Xiong et al., 2014). By utilizing a combination of transcriptomic and genetic approaches, we will be able to computationally decipher which immune cell populations are responsible for the differential host cell transcriptional signatures (Figure 2). This, in turn, should help us understand the response characteristic of certain viral infections in more detail. The CC mouse complements other animal models of viral pathogenesis.
Final Thoughts
When considering how genomics can revolutionize epidemiology, it is worth remembering the postulates established by the great 19th century microbiologist Robert Koch. Koch’s postulates dictate that a given pathogen must be isolated from a diseased organism, grown in pure culture, inoculated in a healthy organism, and re-isolated from that organism after disease onset. These criteria definitively show that a pathogen is the causative agent of a disease. However, they cannot be easily extrapolated for human pathogens that cannot be cultured or do not cause disease in available model organisms.
In the era of modern molecular biology, Fredricks and Relman (1996) have proposed a series of updated criteria using genomic technology to identify pathogens that are the causative agents of a disease. These allow for pathogens to be detected by PCR or sequencing from clinical specimens, rather than requiring isolation in pure culture and subsequent challenge experiments in a well-developed animal model (Fredricks and Relman, 1996). These “molecular Koch’s postulates” were further refined by Falkow, who proposed demonstrating virulence factor function by deletion and complementation experiments (Falkow, 2004). These methods for establishing disease etiology as applied to present next-generation sequencing technology demonstrate how much pathogen discovery has transformed within the past two centuries.
Clearly we are in a new renaissance of pathogen discovery: as genomics technology becomes more widespread, the response to future outbreaks will be increasingly reliant on it to conduct effective diagnosis and surveillance and to use genetic and transcriptomic data to effect truly personalized precision medicine. The sudden and shocking emergence of Zika virus (ZIKV) in the Americas is proving to be another example of how the virus can be monitored in real time, and genomics can be used to understand how the virus is transmitted, how it is evolving, how it interacts with the host, and whether or not it can be linked to other conditions (Faria et al., 2016, Wang et al., 2016). Already, genomics technologies have enabled the establishment of the link between ZIKV infection and infants born with microcephaly (Driggers et al., 2016), although it’s important to note that detection of virus sequence alone is not sufficient to prove a causative link. However, this link provided fundamental early information that led to further work conclusively linking ZIKV to abnormalities in brain development (Garcez et al., 2016, Rasmussen et al., 2016, Tang et al., 2016). Genomics technologies have also provided evidence of sexual transmission (D’Ortenzio et al., 2016). This represents a paradigm shift in how epidemiology and surveillance are conducted and will help us respond rapidly and effectively to new emerging infectious challenges that we will surely face.
References
- Barzon L., Lavezzo E., Militello V., Toppo S., Palù G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011;12:7861–7884. doi: 10.3390/ijms12117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomly D., Ferris M.T., Aicher L.D., Rosenzweig E., Whitmore A., Aylor D.L., Haagmans B.L., Gralinski L.E., Bradel-Tretheway B.G., Bryan J.T. Expression quantitative trait Loci for extreme host response to influenza a in pre-collaborative cross mice. G3 (Bethesda) 2012;2:213–221. doi: 10.1534/g3.111.001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Kapoor A., Mishra N., Jain K., Kumar A., Jabado O.J., Lipkin W.I. Virome Capture Sequencing Enables Sensitive Viral Diagnosis and Comprehensive Virome Analysis. MBio. 2015;6 doi: 10.1128/mBio.01491-15. e01491–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas I., Tenorio A., Echevarría J.M., Klapper P.E., Cleator G.M. Detection of enteroviral RNA and specific DNA of herpesviruses by multiplex genome amplification. J. Virol. Methods. 1997;66:39–50. doi: 10.1016/s0166-0934(97)00035-9. [DOI] [PubMed] [Google Scholar]
- D’Ortenzio E., Matheron S., de Lamballerie X., Hubert B., Piorkowski G., Maquart M., Descamps D., Damond F., Yazdanpanah Y., Leparc-Goffart I. Evidence of Sexual Transmission of Zika Virus. N. Engl. J. Med. 2016 doi: 10.1056/NEJMc1604449. Published online April 13, 2016. [DOI] [PubMed] [Google Scholar]
- Dong H., Qian Z., Zhang L., Chen Y., Ren Z., Ji Q. Genomic and transcriptome profiling identified both human and HBV genetic variations and their interactions in Chinese hepatocellular carcinoma. Genom. Data. 2015;6:1–3. doi: 10.1016/j.gdata.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers R.W., Ho C.Y., Korhonen E.M., Kuivanen S., Jääskeläinen A.J., Smura T., Rosenberg A., Hill D.A., DeBiasi R.L., Vezina G. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N. Engl. J. Med. 2016 doi: 10.1056/NEJMoa1601824. Published online March 30, 2016. [DOI] [PubMed] [Google Scholar]
- Drosten C., Göttig S., Schilling S., Asper M., Panning M., Schmitz H., Günther S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K.M. Review of the laboratory approaches to the detection of antibody and cell-mediated immunity to pertussis disease and vaccine. Expert Rev. Vaccines. 2014;13:1183–1190. doi: 10.1586/14760584.2014.946015. [DOI] [PubMed] [Google Scholar]
- Elbarbary R.A., Lucas B.A., Maquat L.E. Retrotransposons as regulators of gene expression. Science. 2016;351:aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkow S. Molecular Koch’s postulates applied to bacterial pathogenicity--a personal recollection 15 years later. Nat. Rev. Microbiol. 2004;2:67–72. doi: 10.1038/nrmicro799. [DOI] [PubMed] [Google Scholar]
- Faria N.R., Azevedo Rdo.S., Kraemer M.U., Souza R., Cunha M.S., Hill S.C., Thézé J., Bonsall M.B., Bowden T.A., Rissanen I. Zika virus in the Americas: Early epidemiological and genetic findings. Science. 2016;352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris M.T., Aylor D.L., Bottomly D., Whitmore A.C., Aicher L.D., Bell T.A., Bradel-Tretheway B., Bryan J.T., Buus R.J., Gralinski L.E. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2013;9:e1003196. doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks D.N., Relman D.A. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin. Microbiol. Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez P.P., Loiola E.C., Madeiro da Costa R., Higa L.M., Trindade P., Delvecchio R., Nascimento J.M., Brindeiro R., Tanuri A., Rehen S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016 doi: 10.1126/science.aaf6116. Published online April 10, 2016. [DOI] [PubMed] [Google Scholar]
- Gardeux V., Bosco A., Li J., Halonen M.J., Jackson D., Martinez F.D., Lussier Y.A. Towards a PBMC “virogram assay” for precision medicine: Concordance between ex vivo and in vivo viral infection transcriptomes. J. Biomed. Inform. 2015;55:94–103. doi: 10.1016/j.jbi.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Gire S.K., Goba A., Andersen K.G., Sealfon R.S., Park D.J., Kanneh L., Jalloh S., Momoh M., Fullah M., Dudas G. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.B., Thomas S., Swarts J., McMillan A.A., Ferris M.T., Suthar M.S., Treuting P.M., Ireton R., Gale M., Jr., Lund J.M. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. MBio. 2015;6 doi: 10.1128/mBio.00493-15. e00493–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Ferris M.T., Aylor D.L., Whitmore A.C., Green R., Frieman M.B., Deming D., Menachery V.D., Miller D.R., Buus R.J. Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross. PLoS Genet. 2015;11:e1005504. doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Naccache S.N., Federman S., Yu G., Mbala P., Bres V., Stryke D., Bouquet J., Somasekar S., Linnen J.M. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M., Karolchik D., Clawson H., Raney B.J., Rosenbloom K.R., Fujita P.A., Hinrichs A.S., Speir M.L., Eisenhart C., Zweig A.S. The UCSC Ebola Genome Portal. PLoS Curr. 2014;6:6. doi: 10.1371/currents.outbreaks.386ab0964ab4d6c8cb550bfb6071d822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraksingh R.R., Snyder M.P. Impacts of variation in the human genome on gene regulation. J. Mol. Biol. 2013;425:3970–3977. doi: 10.1016/j.jmb.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Hoenen T., Safronetz D., Groseth A., Wollenberg K.R., Koita O.A., Diarra B., Fall I.S., Haidara F.C., Diallo F., Sanogo M. Virology. Mutation rate and genotype variation of Ebola virus from Mali case sequences. Science. 2015;348:117–119. doi: 10.1126/science.aaa5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T., Groseth A., Rosenke K., Fischer R.J., Hoenen A., Judson S.D., Martellaro C., Falzarano D., Marzi A., Squires R.B. Nanopore Sequencing as a Rapidly Deployable Ebola Outbreak Tool. Emerg. Infect. Dis. 2016;22:22. doi: 10.3201/eid2202.151796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q. Genetic study of complex diseases in the post-GWAS era. J. Genet. Genomics. 2015;42:87–98. doi: 10.1016/j.jgg.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Huang Y., Zaas A.K., Rao A., Dobigeon N., Woolf P.J., Veldman T., Øien N.C., McClain M.T., Varkey J.B., Nicholson B. Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza a infection. PLoS Genet. 2011;7:e1002234. doi: 10.1371/journal.pgen.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L., de Niet A., Makowska Z., Dill M.T., van Dort K.A., Terpstra V., Bart Takkenberg R., Janssen H.L., Heim M.H., Kootstra N.A., Reesink H.W. An intrahepatic transcriptional signature of enhanced immune activity predicts response to peginterferon in chronic hepatitis B. Liver Int. 2015;35:1824–1832. doi: 10.1111/liv.12768. [DOI] [PubMed] [Google Scholar]
- Josset L., Tchitchek N., Gralinski L.E., Ferris M.T., Eisfeld A.J., Green R.R., Thomas M.J., Tisoncik-Go J., Schroth G.P., Kawaoka Y. Annotation of long non-coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014;11:875–890. doi: 10.4161/rna.29442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze M., Korth M., Law G.L., Nathanson N. Third Edition. Academic Press; 2016. Viral Pathogenesis: From Essentials to Systems Biology. [Google Scholar]
- Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S., Schnurr D., Ganem D., DeRisi J.L., Boushey H.A. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J. Infect. Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelman J.R., Wiley M.R., Mate S., Ladner J.T., Beitzel B., Fakoli L., Taweh F., Prieto K., Diclaro J.W., Minogue T., US Army Medical Research Institute of Infectious Diseases. National Institutes of Health. Integrated Research Facility–Frederick Ebola Response Team 2014–2015 Monitoring of Ebola Virus Makona Evolution through Establishment of Advanced Genomic Capability in Liberia. Emerg. Infect. Dis. 2015;21:1135–1143. doi: 10.3201/eid2107.150522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Wiley M.R., Mate S., Dudas G., Prieto K., Lovett S., Nagle E.R., Beitzel B., Gilbert M.L., Fakoli L. Evolution and Spread of Ebola Virus in Liberia, 2014-2015. Cell Host Microbe. 2015;18:659–669. doi: 10.1016/j.chom.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J., Dean A., St George K., Briese T., Lipkin W.I. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J. Infect. Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebke R.W., Parks C., Luster M.I. Suppression of immune function and susceptibility to infections in humans: association of immune function with clinical disease. J. Immunotoxicol. 2004;1:15–24. doi: 10.1080/15476910490438342. [DOI] [PubMed] [Google Scholar]
- MacNeil A., Lee C.W., Dietz V. Issues and considerations in the use of serologic biomarkers for classifying vaccination history in household surveys. Vaccine. 2014;32:4893–4900. doi: 10.1016/j.vaccine.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madoui M.A., Engelen S., Cruaud C., Belser C., Bertrand L., Alberti A., Lemainque A., Wincker P., Aury J.M. Genome assembly using Nanopore-guided long and error-free DNA reads. BMC Genomics. 2015;16:327. doi: 10.1186/s12864-015-1519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Martinez M.T., Tseng Y.C., Ormategui N., Loinaz I., Eritja R., Bokor J. Label-free DNA biosensors based on functionalized carbon nanotube field effect transistors. Nano Lett. 2009;9:530–536. doi: 10.1021/nl8025604. [DOI] [PubMed] [Google Scholar]
- Mate S.E., Kugelman J.R., Nyenswah T.G., Ladner J.T., Wiley M.R., Cordier-Lassalle T., Christie A., Schroth G.P., Gross S.M., Davies-Wayne G.J. Molecular Evidence of Sexual Transmission of Ebola Virus. N. Engl. J. Med. 2015;373:2448–2454. doi: 10.1056/NEJMoa1509773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhaney J.E., Kuchel G.A., Zhou X., Swain S.L., Haynes L. T-Cell Immunity to Influenza in Older Adults: A Pathophysiological Framework for Development of More Effective Vaccines. Front. Immunol. 2016;7:41. doi: 10.3389/fimmu.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson T.L., Kukielka D., Vincent A.L., Brockmeier S.L., Miller L.C., Faaberg K.S. Utility of a panviral microarray for detection of swine respiratory viruses in clinical samples. J. Clin. Microbiol. 2011;49:1542–1548. doi: 10.1128/JCM.01876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Briese T., Kapoor V., Jabado O., Liu Z., Venter M., Zhai J., Renwick N., Grolla A., Geisbert T.W. MassTag polymerase chain reaction for differential diagnosis of viral hemorrhagic fever. Emerg. Infect. Dis. 2006;12:692–695. doi: 10.3201/eid1204.051515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.J., Dudas G., Wohl S., Goba A., Whitmer S.L., Andersen K.G., Sealfon R.S., Ladner J.T., Kugelman J.R., Matranga C.B. Ebola Virus Epidemiology, Transmission, and Evolution during Seven Months in Sierra Leone. Cell. 2015;161:1516–1526. doi: 10.1016/j.cell.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L., Bore J.A., Koundouno R., Dudas G., Mikhail A. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones-Mateu M.E., Avila S., Reyes-Teran G., Martinez M.A. Deep sequencing: becoming a critical tool in clinical virology. J. Clin. Virol. 2014;61:9–19. doi: 10.1016/j.jcv.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.L. Probing the Viromic Frontiers. MBio. 2015;6 doi: 10.1128/mBio.01767-15. e01767–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.L., Tchitchek N., Susnow N.J., Krasnoselsky A.L., Diamond D.L., Yeh M.M., Proll S.C., Korth M.J., Walters K.A., Lederer S. Early transcriptional programming links progression to hepatitis C virus-induced severe liver disease in transplant patients. Hepatology. 2012;56:17–27. doi: 10.1002/hep.25612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.L., Okumura A., Ferris M.T., Green R., Feldmann F., Kelly S.M., Scott D.P., Safronetz D., Haddock E., LaCasse R. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.A., Jamieson D.J., Honein M.A., Petersen L.R. Zika Virus and Birth Defects - Reviewing the Evidence for Causality. N. Engl. J. Med. 2016 doi: 10.1056/NEJMsr1604338. Published online April 13, 2016. [DOI] [PubMed] [Google Scholar]
- Renois F., Talmud D., Huguenin A., Moutte L., Strady C., Cousson J., Lévêque N., Andréoletti L. Rapid detection of respiratory tract viral infections and coinfections in patients with influenza-like illnesses by use of reverse transcription-PCR DNA microarray systems. J. Clin. Microbiol. 2010;48:3836–3842. doi: 10.1128/JCM.00733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghavi S.K., Bullotta A., Husain S., Rinaldo C.R. Clinical evaluation of multiplex real-time PCR panels for rapid detection of respiratory viral infections. J. Med. Virol. 2012;84:162–169. doi: 10.1002/jmv.22186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpino S.V., Iamarino A., Wells C., Yamin D., Ndeffo-Mbah M., Wenzel N.S., Fox S.J., Nyenswah T., Altice F.L., Galvani A.P. Epidemiological and viral genomic sequence analysis of the 2014 ebola outbreak reveals clustered transmission. Clin. Infect. Dis. 2015;60:1079–1082. doi: 10.1093/cid/ciu1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Faye O., Faye O., Koivogui L., Magassouba N., Keita S., Thiberge J.M., Diancourt L., Bouchier C., Vandenbogaert M. Distinct lineages of Ebola virus in Guinea during the 2014 West African epidemic. Nature. 2015;524:102–104. doi: 10.1038/nature14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloots T.P., Nissen M.D., Ginn A.N., Iredell J.R. Rapid identification of pathogens using molecular techniques. Pathology. 2015;47:191–198. doi: 10.1097/PAT.0000000000000241. [DOI] [PubMed] [Google Scholar]
- Sorgenfrei S., Chiu C.Y., Gonzalez R.L., Jr., Yu Y.J., Kim P., Nuckolls C., Shepard K.L. Label-free single-molecule detection of DNA-hybridization kinetics with a carbon nanotube field-effect transistor. Nat. Nanotechnol. 2011;6:126–132. doi: 10.1038/nnano.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler T., Kühnert D., Rasmussen D.A., du Plessis L. Insights into the early epidemic spread of ebola in sierra leone provided by viral sequence data. PLoS Curr. 2014;6:6. doi: 10.1371/currents.outbreaks.02bc6d927ecee7bbd33532ec8ba6a25f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star A., Tu E., Niemann J., Gabriel J.C., Joiner C.S., Valcke C. Label-free detection of DNA hybridization using carbon nanotube network field-effect transistors. Proc. Natl. Acad. Sci. USA. 2006;103:921–926. doi: 10.1073/pnas.0504146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., García J., Comach G., Vahey M.T., Wang Z., Forshey B.M., Morrison A.C., Sierra G., Bazan I., Rocha C. Sequential waves of gene expression in patients with clinically defined dengue illnesses reveal subtle disease phases and predict disease severity. PLoS Negl. Trop. Dis. 2013;7:e2298. doi: 10.1371/journal.pntd.0002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.02.016. Published online March 4, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O’Huigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y.G., Shi W.F., Liu D., Qian J., Liang L., Bo X.C., Liu J., Ren H.G., Fan H., Ni M., China Mobile Laboratory Testing Team in Sierra Leone Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature. 2015;524:93–96. doi: 10.1038/nature14490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valassina M., Cuppone A.M., Cusi M.G., Valensin P.E. Rapid detection of different RNA respiratory virus species by multiplex RT-PCR: application to clinical specimens. Clin. Diagn. Virol. 1997;8:227–232. doi: 10.1016/s0928-0197(97)10001-0. [DOI] [PubMed] [Google Scholar]
- Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Moore N.E., Deng Y.M., Eccles D.A., Hall R.J. MinION nanopore sequencing of an influenza genome. Front Microbiol. 2015;6:766. doi: 10.3389/fmicb.2015.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Valderramos S.G., Wu A., Ouyang S., Li C., Brasil P., Bonaldo M., Coates T., Nielsen-Saines K., Jiang T. From Mosquitos to Humans: Genetic Evolution of Zika Virus. Cell Host Microbe. 2016;19:561–565. doi: 10.1016/j.chom.2016.04.006. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie T.N., Wylie K.M., Herter B.N., Storch G.A. Enhanced virome sequencing using targeted sequence capture. Genome Res. 2015;25:1910–1920. doi: 10.1101/gr.191049.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Morrison J., Ferris M.T., Gralinski L.E., Whitmore A.C., Green R., Thomas M.J., Tisoncik-Go J., Schroth G.P., Pardo-Manuel de Villena F. Genomic profiling of collaborative cross founder mice infected with respiratory viruses reveals novel transcripts and infection-related strain-specific gene and isoform expression. G3 (Bethesda) 2014;4:1429–1444. doi: 10.1534/g3.114.011759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F., Zhang Y., Wei Y., Kang X. A rapid and sensitive detection of HBV DNA using a nanopore sensor. Chem. Commun. (Camb.) 2014;50:13853–13856. doi: 10.1039/c4cc06135g. [DOI] [PubMed] [Google Scholar]
- Zaas A.K., Burke T., Chen M., McClain M., Nicholson B., Veldman T., Tsalik E.L., Fowler V., Rivers E.P., Otero R. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci. Transl. Med. 2013;5:203ra126. doi: 10.1126/scitranslmed.3006280. [DOI] [PMC free article] [PubMed] [Google Scholar]