Abstract
Type I interferons (IFNs) comprise of pro-inflammatory cytokines created, as well as sensed, by all nucleated cells with the main objective of blocking pathogens-driven infections. Owing to this broad range of influence, type I IFNs also exhibit critical functions in many sterile inflammatory diseases and immunopathologies, especially those associated with endoplasmic reticulum (ER) stress-driven signaling pathways. Indeed, over the years accumulating evidence has indicated that the presence of ER stress can influence the production, or sensing of, type I IFNs induced by perturbations like pattern recognition receptor (PRR) agonists, infections (bacterial, viral or parasitic) or autoimmunity. In this article we discuss the link between type I IFNs and ER stress in various diseased contexts. We describe how ER stress regulates type I IFNs production or sensing, or how type I IFNs may induce ER stress, in various circumstances like microbial infections, autoimmunity, diabetes, cancer and other ER stress-related contexts.
Keywords: Toll-like receptors (TLRs), STING, Interferon-stimulated genes (ISGs), Unfolded protein response (UPR), PERK, IRE1, Inflammation, Oncolytic viruses, Danger signals, NF-κB, Chemokine
1. Introduction
Type I interferons (IFNs) are a family of pro-inflammatory cytokines (especially IFNα or IFNβ) created by nucleated cells with the main objective of blocking (pathogenic) infections (Makris et al., 2017; McNab et al., 2015; Negishi et al., 2018; Tailor et al., 2006). A number of non-sterile or sterile factors associated with stress, injury, or trauma, are all sensed by our cells through a combination of pattern recognition receptors (PRRs) that, in reaction, enhance the production of immunological factors like type I IFNs (Bianchi, 2007; Garg and Agostinis, 2017; Tang et al., 2012; Tatematsu et al., 2018; van de Veerdonk et al., 2008). Due to the very extensive presence of type I IFN-binding receptor complex composed of IFN-α/β receptor-1/2 (IFNAR1/2), on almost all cells in an organism's body, type I IFNs exert widespread cellular or immunological functions (Bald et al., 2014; de Weerd et al., 2013; Taft and Bogunovic, 2018). Additionally, type I IFNs are decisive for the maturation and differentiation of assorted innate immune cells (Longhi et al., 2009; Nardelli et al., 2002). Owing to this broad range of influence, type I IFNs exhibit critical functions in many inflammatory diseases thereby positioning them at the core of almost all functional immunological paradigms (Bracci et al., 2017; Parker et al., 2016; Sprooten et al., 2019; Wu et al., 2016; Zitvogel et al., 2015).
Like a number of cytokines and their cognate receptors, IFNα/β and IFNAR1/2 are considerably glycosylated (especially N-glycosylated but also O-glycosylated) (Ling et al., 1995; Sommereyns and Michiels, 2006) and hence the endoplasmic reticulum (ER)/Golgi apparatus system, where such protein glycosylations are executed (Smirle et al., 2013), is critical for the proper functioning of the type I IFNs signaling. Indeed, the ER is an important organelle that is accountable for the production, maturation and post-translational modifications of a number of proteins (Garg et al., 2015d; Kang and Hegde, 2008; Sovolyova et al., 2014). Moreover, a number of pathological perturbations that can elicit type I IFNs also tend to affect ER homeostasis, e.g., chemotherapeutic or radiotherapeutic stress, oxidative injury, and microbial infections (especially of viral origin) (Garg et al., 2012a, Garg et al., 2014; Kang and Hegde, 2008; Tardif et al., 2005). Such interference with ER homeostasis compromises ER-associated proteostasis thereby causing ER stress (Cabral-Miranda and Hetz, 2018). The ER typically responds to such insults by stimulating a complex signaling cascade, called the unfolded protein response (UPR) (Ron and Walter, 2007; Rutkowski and Kaufman, 2007; Schröder and Kaufman, 2005). Herein, the primary aim of the UPR is to overcome “proteotoxicity” and rescue ER homeostasis thereby prolonging cellular survival (Gorman et al., 2012; Hendershot, 2004). However this primary aim is only met in conditions of low-to-medium ER stress such that severe ER stress almost always steers the UPR to engage pro-death signaling that tends to culminate into intrinsic (mitochondria-driven) apoptosis (mostly but not always) (Estornes et al., 2014; Urra et al., 2013; Verfaillie et al., 2012). Due to the overlaps between type I IFNs production or sensing and the ER, not surprisingly, ER stress has critical consequences for type I IFNs-associated functions. Moreover, even type I IFNs themselves are capable of eliciting ER stress in target cells, given the right conditions (Mollereau et al., 2014; Rahmati et al., 2018; Yoshida, 2007).
In this article we discuss the link between type I IFNs and ER stress in various pathological contexts. Initially, we introduce the basic concepts underlying type I IFNs production and sensing, ER stress and ER stress-associated inflammatory signaling pathways. Thereafter we describe how ER stress regulates type I IFNs production or sensing, or how type I IFNs may induce ER stress, in various pathological contexts like microbial infections (viral, bacterial or parasitic), diabetes, cancer and general ER stress-related contexts.
2. Type I interferons: An introduction
2.1. Mechanisms underlying type I IFNs production
Type I IFNs, as mentioned above, evolved to hinder pathogenic diseases (Hron and Peng, 2004; Ivashkiv and Donlin, 2014), which they accomplish by stimulating innate as well as adaptive immune cells, and balancing this with the blunting of any unfavorable immune reactions, thereby ensuring rapid purging of microbial infections coupled with eventual resolution of inflammation (Ivashkiv and Donlin, 2014; Iwasaki and Medzhitov, 2015). A large majority of cells are highly competent at producing IFNβ, however only particular immune cells have evolved to proficiently secrete IFNα, e.g., particular dendritic cell (DC) subsets like plasmacytoid DCs (Björck et al., 2011; Diebold et al., 2003; Garg et al., 2017a, Garg et al., 2017e; Ivashkiv and Donlin, 2014). Despite this, production of type I IFNs is not specific for non-sterile pathologies. In various sterile pathologies, type I IFNs play either a disease ameliorating (e.g., cancer) or even a disease exacerbating role (e.g., autoimmune diseases especially interferonopathies) (Arimoto et al., 2018; Burdick et al., 2009; Durbin et al., 2000; Palucka et al., 2005; Rizzo et al., 2014). This is made possible by a major overlap (in terms of shared cognate PRRs) between danger signals derived from pathogens (together classified under the term, pathogen-associated molecular patterns or PAMPs) and those of “self” or endogenous origin (together classified under the term, damage-associated molecular patterns or DAMPs); as well as the ubiquitous expression of IFNAR1/2 (de Weerd et al., 2013; Garg and Agostinis, 2017; Garg et al., 2017c; Ragimbeau et al., 2003; Rubartelli and Lotze, 2007).
Binding of PAMPs (during microbial invasion) or DAMPs (during tissue injury, trauma, or due to toxic insults) to cellular PRRs drives the production of type I IFNs (Fig. 1 ) (Janeway's, 2016; Janeway and Medzhitov, 2002; Zhang et al., 2010). PRRs tend to have diverse subcellular localizations (e.g., surface, cytosol and endosomes/phagosomes) in order to effectively sense microbial threats capable of entering the cells from disparate routes (Hoffmann et al., 2015; Liu et al., 2011; Thompson et al., 2011; Wu and Chen, 2014). As far as induction of type I IFNs is concerned, nucleic acids of either microbial or endogenous origin are the most potent inducers engaging either cytosolic or endosomal PRRs (Fig. 1) (Roers et al., 2016; Tan et al., 2018; Thompson et al., 2011; Wu and Chen, 2014; Yang et al., 2010). On the cell surface, type I IFNs are majorly elicited through agonists of TLR4, e.g., bacterial lipopolysaccharide (LPS) (Leu et al., 2011; Ohto et al., 2012; Walker and Tough, 2006). Similarly, major endosomal PRRs crucial for type I IFN induction include TLR9, TLR3 or TLR7/8, that are frequently engaged by danger signals like unmethylated CpG DNA, double-stranded RNA (dsRNA) or single-stranded RNA (ssRNA), respectively (Moynagh, 2005; Stetson and Medzhitov, 2006; Sykes et al., 2013; Thompson and Locarnini, 2007). Lastly, major cytosolic PRRs crucial for type I IFN induction include, cytosolic cGAMP-AMP synthase (cGAS)/absent in melanoma 2 (AIM2)-like receptors (ALRs) or, melanoma differentiation-associated gene 5 (MDA5)/RNA helicases retinoic acid-inducible gene I (RIG-I), that are frequently engaged by danger signals like cytosolic DNA species or microbial RNA species (e.g., dsRNA or 5′ppp-RNA), respectively (Fig. 1) (Elion and Cook, 2018; Iurescia et al., 2018; Lee et al., 2019; Saito and Gale, 2008; Tan et al., 2018; Vanpouille-Box et al., 2018).
Fig. 1.
A schematic depiction of production and sensing mechanisms for type I interferons (IFNs). Danger signals like lipopolysaccharide (LPS) or different nucleic acid species (single-stranded RNA or ssRNA, double-stranded RNA or dsRNA, CpG DNA) bind their cognate receptors like, Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated protein 5 (MDA5), or cyclic GMP-AMP (cGAMP) synthase (cGAS). These receptors execute a complex intracellular pathway that ultimately leads to the activation of transcription factors like IFN regulatory factor (IRF)-3/7 that eventually transcribe IFNα/β, which are thereafter translated and secreted. Upon extracellular emergence, IFNα/β bind to the IFNα receptor (IFNAR) complex, which causes activation of tyrosine kinase 2 (TYK2) and receptor-associated janus kinase 1 (JAK1). This eventually activates various downstream pathways that primarily aim to induce production of various interferon-stimulated genes (ISGs) derived protein products but also modulate cellular homeostasis. Of note, the inflammatory potential of IFNα/β is often limited by ubiquitination (Ub)-driven, lysosomal degradation of the IFNAR complex. AKT, protein kinase B alpha; ER, endoplasmic reticulum; IKKβ, I-kappa-B kinase beta; IKKɛ, I-kappa-B kinase epsilon; IRS1/2, insulin receptor substrate 1; ISRE, interferon-stimulated response element; MAVS, mitochondrial antiviral signaling protein; mTOR, mechanistic target of rapamycin; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; PI-3K, phosphoinositide-3-kinase; STAT1/2/3/4/5/6, signal transducer and activator of transcription 1/2/2/4/5/6; STING, stimulator of interferon genes; TBK1, TANK-binding kinase 1.
A diversity of adaptor proteins or downstream signaling modules function beyond the above mentioned PRRs to tightly regulate the production of type I IFNs (Battesti and Gottesman, 2013; Koretzky and Myung, 2001) (Fig. 1). For instance, TLRs, cGAS or RIG-I/MDA5 mostly function through cytosolic proteins like TIR-domain containing adaptor-inducing interferon-β (TRIF)/myeloid differentiation primary response 88 (MyD88), stimulator of interferon genes (STING) or mitochondrial antiviral-signaling protein (MAVS), respectively (Liu et al., 2015; Yamamoto et al., 2003). Mostly “standard” biochemical events like protein-protein binding based activation, phosphorylation or modification of protein structure/conformations underlie the mobilization of the above signaling pathways; however in some cases, this downstream signaling might be conveyed through intermediate metabolites, e.g., stimulation of cGAS tends to generate 2′3′-cGAMP, a potent agonist of STING (Ahn et al., 2018; Khoo and Chen, 2018; Kranzusch et al., 2015). Beyond the standard PRR agonists, specific inflammatory factors like cytokines can also elicit type I IFNs production, e.g., receptor activator of NF-κB ligand (RANKL), tumor-necrosis factor (TNF) or, macrophage colony-stimulating factor (M-CSF) (Ivashkiv and Donlin, 2014; McMahon and Ueki, 2009; Venkatesh et al., 2013; Yarilina et al., 2008; Zauli et al., 2004). These signaling pathways ultimately pave way for the activation of specific kinases like the kinases IκB kinase-ɛ (IKKɛ) and TANK-binding kinase 1 (TBK1), which thereafter recruit the nuclear activity of particular transcription factors belonging to the IFN-regulatory factor (IRF) family, such as IRF1/3/7, for the transcription of type I IFNs (Fig. 1) (Ikeda et al., 2007; Lei et al., 2010; Li et al., 2017; Taft and Bogunovic, 2018; Tanaka and Chen, 2012; Vanpouille-Box et al., 2018). Type I IFNs transcription is often induced in a wave-like format such that TLRs tend to be relatively faster than other PRRs in inducing type I IFN production (Katakura et al., 2005; Ma et al., 2015; Perkins and Vogel, 2015; Sato et al., 1998).
Like many other cytokines, IFNα/β transcripts are translated and folded at the ER and thereafter transported from the ER to the cis-Golgi (Kurokawa et al., 2014; Oku et al., 2011; Stow and Murray, 2013). While in the ER/Golgi, IFNα/β are also glycosylated (mostly N-glycosylation which happens in the Golgi but also some degree of O-glycosylation that happens in the ER) (Ling et al., 1995; Smirle et al., 2013; Sommereyns and Michiels, 2006). Thereafter within the Golgi complex, these cytokines are directed toward the trans-Golgi network (TGN) and sorted into the post-Golgi secretory vesicles that transport them toward the cell surface for eventual extracellular secretion (Gu et al., 2001; Stow and Murray, 2013). It is still debatable to what extent the type I IFNs trafficking pathway deviates from this conventional secretory pathway (Stow and Murray, 2013). There are some unconventional secretory routes observed for various cytokines including IFNγ but it remains unclear whether type I IFNs are also sorted into these pathways, e.g., recycling endosomes-plasma membrane pathway or the Golgi-early endosome pathway (Short, 2015; Zemskov et al., 2011). The role of these pathways in type I IFNs is also unclear because it is not known whether it matters how type I IFNs are secreted, i.e., is it enough that they are secreted via multiple points on the cell surface or do they also need to be secreted in a directionally-defined manner (Reefman et al., 2010; Stow and Murray, 2013).
2.2. Sensing of type I IFNs
Upon release, extracellular IFNα/β engage their cognate IFNα receptor (IFNAR1/2), which activates receptor-associated tyrosine kinases like tyrosine kinase 2 (TYK2) and Janus kinase 1/2 (JAK1/2) (Fig. 1) (Ragimbeau et al., 2003; Uzé et al., 2007; Wallweber et al., 2014). In basal conditions the IFNAR1/2 cytoplasmic tails remain bound, in an inactive conformation, by the JAK proteins (Gui et al., 2017; Ma et al., 2019; Ragimbeau et al., 2003; Teijaro et al., 2016). Engagement by IFNα/β causes dimerization of IFNAR1/2 receptor chains thereby triggering the JAK/TYK2 kinase domains to undertake activation directed through their trans-phosphorylation (Pfeffer et al., 1997; Schneider et al., 2014). Consequently, JAK/TYK2 kinases phosphorylate Signal transducer and activator of transcription 1/2 (STAT1/STAT2) proteins causing their dimerization, nuclear translocation, and binding to the IRF9 protein thereby creating the IFN-stimulated gene factor 3 (ISGF3) complex (Ivashkiv and Donlin, 2014; Liu et al., 2018; Schneider et al., 2014; Tenoever et al., 2007). Thereafter, ISGF3 engages the IFN-stimulated response elements (ISREs) thus guiding the transcription of throngs of interferon-stimulated genes or ISGs (Fig. 1) (Ivashkiv and Donlin, 2014; Liu et al., 2018; Schneider et al., 2014; Tenoever et al., 2007). In principle the above described pathway is considered to be “canonical”; however some variants of this pathway can also be activated and these are mediated by different STAT1/2 homodimers, some of which are generally linked with IFNγ-associated signaling (Ivashkiv and Donlin, 2014; Ramana et al., 2002). Interestingly, even diverse STAT isomers such as STAT3/4/5A and 5B can direct the production of ISGs (Blaszczyk et al., 2015; Rauch et al., 2013). Such diversity, in terms of downstream mediators activated following engagement of IFNAR1/2 by IFNα/β, account for the broad immunological effects of type I IFNs in both health and disease (de Weerd et al., 2013; Mizutani et al., 2012).
Protein products resulting from translation of ISGs (i.e., ISG-products) chiefly strive to minimize the injury instigated by contagions through, disturbing the processes enabling their growth, and proliferation (Fig. 1) (Borden, 2019; Cheon et al., 2014; Lai et al., 2019; Schneider et al., 2014). The best known ISG-products comprise of (Parker et al., 2016): proteins functioning in antigen processing and presentation cascades, T cells or inflammatory myeloid cells-attracting chemokines (e.g., CXCL10 or CXCL9), proteins or micro-RNAs that impede or inhibit angiogenesis, factors capable of inducing or modulating cell death (e.g., ligands that engage death receptors-driven apoptosis or necroptosis) or immunological ligands crucial for myeloid or lymphocytic inflammatory sensitization (e.g., co-stimulatory proteins like CD40 or CD80) (Borden, 2019; Schoggins, 2014, Schoggins, 2018; Schoggins and Rice, 2011; Schneider et al., 2014; van der Vorst et al., 2019; Wang et al., 2017). Type I IFNs and its ISG-products function together to enhance the cell-to-cell communication (in both tissue and immune cells context) so as to eliminate the pathogenic or sterile threat challenging the organismal homeostasis and to resolve inflammation (Borden, 2019; Cheon et al., 2014; Divangahi et al., 2015; Kono et al., 2014; Schneider et al., 2014).
It is clear from the above discussion that type I IFNs are extremely crucial for maintaining tissue homeostasis and impeding progression of diseases (Chovatiya and Medzhitov, 2014; Divangahi et al., 2015; Goldmann et al., 2016). Thus not surprisingly, cellular processes crucial for type I IFNs production are also considered to be important (Ludigs et al., 2012; Mancuso et al., 2007; Schmeisser et al., 2014). Although type I IFN-aiding cellular processes directly associated with immunological signaling have received considerable attention yet some others haven't received as much attention despite being foundational for type I IFNs production (Bogdan et al., 2004; Haque et al., 2014; Steinman, 2009). One such process is ER stress or UPR signaling. In the following sections, we introduce the basic concepts underlying ER stress/UPR signaling before discussing the broad trends within the current literature connecting it with the regulation of type I IFN responses.
3. ER stress and inflammation: An introduction
3.1. ER stress-associated UPR signaling
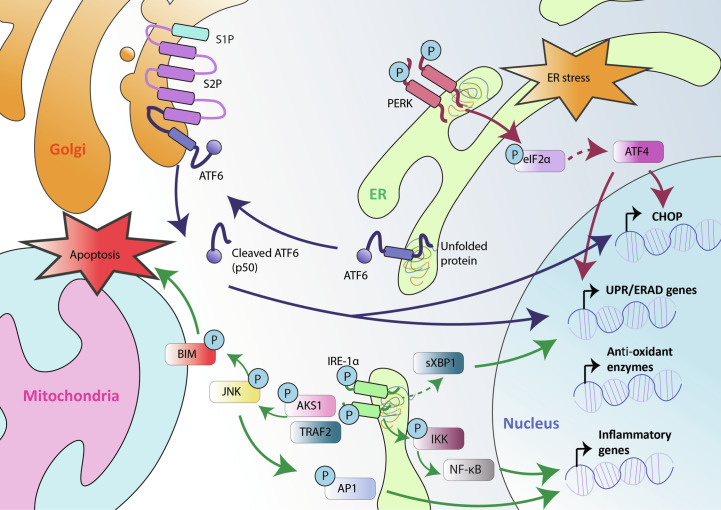
A stressed ER, as mentioned earlier, aims to overcome the disturbance in its homeostasis by activating a complex set of signaling pathways together representing the UPR signaling (Imanikia et al., 2018; Menu et al., 2012; Oliveira et al., 2011; Szegezdi et al., 2009). The UPR involves a multifaceted interaction amongst three main signaling pathways, each of which is activated by three different ER-sessile sensors of ER stress, i.e., pancreatic ER kinase (PKR)-like ER kinase (PERK), inositol requiring enzyme 1 (IRE1), and activating transcription factor 6 (ATF6) (Fig. 2 ) (Chen and Brandizzi, 2013; Glembotski et al., 2019; Hetz, 2012; Mori, 2010; Pytel et al., 2016; Verfaillie et al., 2012). PERK, IRE1 and ATF6 are ER-transmembrane proteins that, in basal or homeostatic conditions, exist in an inactive conformation enforced by the direct binding of the chaperone, 78 kDa glucose-regulated protein (GRP78), to their luminal domains (Carrara et al., 2015; Pincus et al., 2010; Sepulveda et al., 2018; Sundaram et al., 2018). Upon increased accumulation of unfolded or misfolded proteins that create ER-localized proteotoxicity, GRP78 seizes to bind these ER stress sensors in order to prioritize its main function of protein chaperone so it can amplify ER's protein re-folding capacity (Christis et al., 2010; Das et al., 2013; Ma and Hendershot, 2004; Määttänen et al., 2010; Pincus et al., 2010; Schröder and Kaufman, 2005). This stoppage in GRP78 binding paves way for the activation of PERK, IRE1 and ATF6 (Gardner et al., 2013; Sundaram et al., 2018). In the following sub-sections, we give an overview of mechanisms underlying ER stress sensors' activation (Fig. 2) however it is worth noting that for the sake of focus and clarity we present a relatively simplified overview of these mechanisms although these are much more complex and multi-factorial. For those who want to gain a deeper understanding of this subject, we refer to other more comprehensive reviews focussing on ER stress-associated mechanisms.
Fig. 2.
A broad depiction of the endoplasmic reticulum (ER) stress-driven unfolded protein response (UPR) signaling. See the text for further details on this pathway. S1P or S2P, site-1 or site-2 protease; eIF2α, eukaryotic translation initiation factor 2 alpha; PERK, protein kinase RNA-like ER kinase; ERAD, ER-associated protein degradation; CHOP, C/EBP homologous protein; XBP1, X-box binding protein 1; IRE1, inositol requiring enzyme 1; AP1, activator protein 1; TRAF2, TNF receptor-associated factor 2; ASK1, apoptosis signal-regulating kinase 1; JNK, c-Jun N-terminal kinase; BIM, BCL2-like protein 11; ATF4/6, activating transcription factor 4/6.
Molecularly speaking, PERK is a Ser/Thr kinase that conveys signaling through at least two known downstream substrates, i.e., eukaryotic initiation factor 2-alpha (eIF2α) or nuclear factor-E2-related factor 2 (NRF2) (Fig. 2) (Atkins et al., 2013; Harding et al., 2000; Marciniak et al., 2006). Upon ER stress, PERK orchestrates the phosphorylation of eIF2α on its Ser51 amino acid residue thereby restraining Cap-dependent protein translation (Balsa et al., 2019; Gonen et al., 2019; Gupta et al., 2012). Of note, Cap-dependent translation is distinguished by the obligatory requirement for the ribosomes to first engage the mRNA transcript at its 5′ end (i.e., 5′ cap sequence) before moving toward the start codon whereas the process when a ribosome is “directly” transported to the start codon site thereby bypassing this obligation is simply termed as cap-independent protein translation. The PERK-eIF2α phosphorylation driven impediment of protein translation helps in dropping the amount of new proteins arriving at the ER for maturation (Araki and Nagata, 2011; Ellgaard et al., 2016; Naidoo, 2009; Nehls et al., 2000; Pedrazzini and Vitale, 2018). Of note, this inhibition of Cap-dependent translation is sometimes termed as global translational shut-down which may not be entirely correct since under eIF2α inhibitory conditions, Cap-independent protein translation can still continue and may in fact be crucial for UPR functioning itself (Kim et al., 2011; Knutsen et al., 2015; Thakor and Holcik, 2012; Van Der Kelen et al., 2009; Walters and Thompson, 2016). More specifically the transcription factor, activating transcription factor 4 (ATF4), that is crucial for the downstream deliverables of the PERK signaling pathway, is one of the products of Cap-independent translation (Ameri and Harris, 2008; Saito et al., 2011; Wang et al., 2012). This PERK-eIF2α-ATF4 signaling module controls the expression of numerous genes necessary for restoration of the ER homeostasis including, those relevant for regulating ordered synthesis of amino acids, protein or peptide transportation, and antioxidant responses (amongst others) (Grootjans et al., 2016; Todd et al., 2008; Wang and Kaufman, 2014) (Fig. 2). Similarly, PERK also phosphorylates NRF2 which encourages NRF2’s detachment from its cytosolic repressor protein, Kelch-like Ech associated protein 1 (KEAP1) (Chien et al., 2015; Cullinan et al., 2003; Del Vecchio et al., 2014; Holland and Fishbein, 2010; Uruno et al., 2015). This allows NRF2 to transit into the nucleus where it directs transcription of several genes involved in the antioxidant responses (Faraonio et al., 2006; Levings et al., 2018; Sun et al., 2009; Xue et al., 2015). Thus, through coordination of these two (parallel) signaling modules, the PERK arm of the UPR strives to help increase the survival of ER stressed cells by facilitating ER's adaptation to stress, especially oxidative stress (Dadey et al., 2018; Karali et al., 2014; Verfaillie et al., 2012; Walter et al., 2015; Zhang et al., 2013).
In comparison to PERK, that operates primarily through phosphorylation of its substrates, IRE1 arm of the UPR orchestrates a relatively more complex set of biological processes (Fig. 2) (Jwa and Chang, 2012; Reverendo et al., 2019; Rubio-Patiño et al., 2018; Sundaram et al., 2018). On one hand, activated IRE1 functions as an endoribonuclease or RNase to splice X-Box binding protein 1 (XBP1) mRNA (or XBP1u), and cause the emergence of a fully functional form of it, i.e., spliced XBP1 (XBP1s) (Hassler et al., 2015; Lee et al., 2002; Mimura et al., 2012; Yanagitani et al., 2011; Yoshida et al., 2006). This XBP1s functions as a transcription factor and directs the transcription of several crucial genes involved in quality control within the ER, redox homeostasis, biogenesis of ER/Golgi structures, antioxidant response, as well as proteins involved in the ER-associated protein degradation (ERAD; a process that helps in degradation of misfolded proteins accumulating in the ER via cytosolic proteasomes) (Almanza et al., 2019; Hiller et al., 1996; Lhomond et al., 2018; Logue et al., 2018; Ruggiano et al., 2014; Wu and Rapoport, 2018). Of note, beyond XBP1 splicing, IRE1 also undertakes another RNase activity involving degradation of ER membrane-associated mRNAs, a process termed as the IRE1-dependent decay of mRNA (RIDD) activity (Blazanin et al., 2017; Guydosh et al., 2017; Hollien et al., 2009; Maurel et al., 2014; So et al., 2015). IRE1’s RIDD activity has a highly context-dependent role in facilitating stress adaptive responses, or cell death (Bouchecareilh et al., 2011; Coelho and Domingos, 2014; Hassler et al., 2012; Lerner et al., 2012; Maurel et al., 2014; Rubio et al., 2011). On the other hand, activated IRE1 also displays some degree of kinase activity, which it exploits for trans-autophosphorylation; although the exact identity of other downstream substrates targeted for phosphorylation by IRE1 remains enigmatic (Ali et al., 2011; Feldman and Maly, 2017; Korennykh et al., 2009; Lee et al., 2008; Prischi et al., 2014). In comparison to PERK and IRE1, ATF6 activation mechanism is highly distinct in the UPR setting (Jerry Chiang and Lin, 2014; Sundaram et al., 2018; Tam et al., 2018; Teske et al., 2011; Yoshida et al., 2001). The titration of GRP78 away from ATF6 encourages its transportation toward the Golgi complex wherein it is sliced by particular Golgi-resident proteases (Fig. 2) (Papaioannou et al., 2018; Schindler and Schekman, 2009; Shen and Prywes, 2005; Shen et al., 2002; Sommer and Jarosch, 2002). This processing of ATF6 liberates it as an active transcription factor that not only transcribes specific ER chaperones, or proteins involved in the ERAD, but also contributes toward adaptive lipid metabolism, and expansion of the ER to better cope with the proteotoxic stress (Oka et al., 2019; Shen et al., 2002; Tam et al., 2018; Ye et al., 2000; Zeng et al., 2004).
The above pathway mainly represents the attempt of the UPR to re-establish ER homeostasis and facilitate the survival of the stressed cell. However this cascade is best executed only in condition of low or mild ER stress where the ER-associated signaling still has enough time and resources to tackle the proteotoxic insult (Binet and Sapieha, 2015; Gorman et al., 2012; Hendershot, 2004; Lindholm et al., 2006; Yoshida, 2007). However in some situations, prolonged stress or very intense stress in a short amount of time, can both overwhelm the UPR's ability to restore ER homeostasis (Chevet et al., 2015; McGrath et al., 2018; Senft and Ronai, 2015; Urra and Hetz, 2014). In such a situation, the UPR is inclined to engage the pro-death signaling pathways. The molecular switching between these two crucial cell fates (i.e., cell survival versus cell death) in the context of ER stress is complex and still poorly understood (Chevet et al., 2015; Maurel et al., 2015; Szegezdi et al., 2006). However this is facilitated by the ability of the main ER stress sensors to control a dualistic signaling cascade that balances stress adaptation with pro-death signaling (Rutkowski et al., 2006; Sano and Reed, 2013; Sovolyova et al., 2014). A possible contrivance that may account for this dichotomy is the differential stability of mRNAs coding for proteins regulating pro-survival or pro-death signaling wherein this stability might depend upon the overall intensity of ER stress (Haynes et al., 2004; Kaneko and Nomura, 2003; Kondo et al., 2012; Rutkowski et al., 2006; Urra et al., 2013). So for instance, a transiently activated PERK will likely direct the ATF4-driven transcription program to undertake pro-survival signaling (Dadey et al., 2018; Rozpedek et al., 2016). This program will not readily engage pro-death signaling via downstream mediators, like C/EBP homologous protein (CHOP), largely because of the intrinsic instability of mRNAs coding for pro-apoptotic proteins (Cubillos-Ruiz et al., 2017; Tavernier et al., 2017). These mRNA species probably require a stronger and sustained PERK activation (as in the case of severe or prolonged ER stress) to allow commencement of an apoptotic program driven through ATF4-directed activation of CHOP (Balsa et al., 2019; Wang et al., 2019). In case of the IRE1 arm of the UPR, published research indicates that the IRE1 → XBP1 signaling elicited by mild ER stress plays a predominantly pro-survival role whereas in case of severe ER stress, IRE1 tends to orchestrate a XBP1-independent scaffold signaling characteristics to commence apoptotic signaling (Back et al., 2005, Back et al., 2006; Cubillos-Ruiz et al., 2016; Gupta et al., 2010; Heindryckx et al., 2016; Tang et al., 2014; Yoshida et al., 2001). More specifically, upon exposure to severe ER stress, IRE1 can recruit pro-death signaling proteins like TNF-receptor-associated receptor 2 (TRAF2) (Hu et al., 2006; Pincus et al., 2010; Qiu et al., 2013; Salzberg et al., 2017; Takeda et al., 2019; Urano et al., 2000), which is an E3 ubiquitin ligase that can facilitate IRE1’s ability to engage activation of other signaling cascades like those involving, apoptosis signal-regulating kinase 1 (ASK1), c-Jun N-terminal kinase (JNK), and/or p38 mitogen-activated protein kinases (MAPK) cascades, to modulate cellular fate (Covino et al., 2018; Fey et al., 2012; Gallo and Johnson, 2002; Madden et al., 2019; Mitra and Ryoo, 2019; Preissler and Ron, 2019; Urano et al., 2000; Yang et al., 2009) (Fig. 2).
Beyond the above disparate routes connecting ER stress with pro-death signaling, it is clear from a series of studies that the transcription factor CHOP plays a dominant role in driving ER stress-induced apoptosis (Chiribau et al., 2010; Mihailidou et al., 2010; Teske et al., 2013). In fact, activation of CHOP is not exclusive for the PERK arm of the UPR, since its formation or activation is shared between transcription factors downstream of all UPR arms, i.e., ATF4, XBP1 and ATF6 (Hirsch et al., 2014; Masuda et al., 2013; Shoulders et al., 2013; Takayanagi et al., 2013; Teske et al., 2011; Tsuru et al., 2016). CHOP facilitates apoptosis via transcription of various pro-apoptotic genes like, growth arrest and DNA-damage inducible protein (GADD34), and ER oxidoreductase 1 alpha (ERO1α) (amongst others) (Brush et al., 2003; Fransson et al., 2014; Hollander et al., 2003; Iwasaki et al., 2015; Kojima et al., 2003; Novoa et al., 2001). For instance, GADD34 directs protein phosphatase 1 (PP1) to de-phosphorylate the eIF2α protein thereby causing recommencement of (Cap-dependent) protein synthesis (Choy et al., 2015; Perego et al., 2018; Rojas et al., 2015). This resumption of protein synthesis overwhelms the protein-folding capacity of the ER, which is already dealing with proteotoxicity, thereby amplifying ER stress and pushing the cell toward cell death (Han et al., 2013; Haynes et al., 2004; Malhotra and Kaufman, 2011; Sano and Reed, 2013; Sovolyova et al., 2014; Thomas et al., 2013). Similarly, ERO1α activation can severely disturb the redox homeostasis of the ER, thereby also facilitating proteotoxicity-driven lethality (Appenzeller-Herzog et al., 2008; Araki and Nagata, 2011; Gomez et al., 2014; Kim et al., 2012; Sevier and Kaiser, 2008; Sevier et al., 2007). Thus, the UPR signaling, that strives to maintain the redox homeostasis of the ER during stressful conditions, can instead facilitate oxidation within the ER via CHOP activity, thereby paving way for apoptosis (Menu et al., 2012; Song et al., 2008; Urra et al., 2013; Verfaillie et al., 2012; Xu et al., 2017). Besides these proteostasis and redox homeostasis regulating proteins, CHOP can also control the expression of specific proteins belonging to the B-cell lymphoma 2 (BCL2) family, which can not only directly regulate mitochondrial or intrinsic apoptosis but can also modify UPR signaling cascades (Hollville et al., 2014; Szegezdi et al., 2009; Zagorodna et al., 2012; Zheng et al., 2016). For example, the pro-apoptotic BCL2 proteins, BCL2-associated X (BAX) and BCL2 homologous antagonist/killer (BAK), can directly regulate the release of Ca2+ from the ER by forming membrane pores there (a potent initiator event for intrinsic apoptosis) (Andreu-Fernández et al., 2017; Jones et al., 2007; Wang et al., 2011; Willis et al., 2007); but, BAX/BAK can also modulate IRE1 activity through a direct interaction to facilitate intrinsic apoptosis (Castillo et al., 2011; Hetz et al., 2006; Hollville and Martin, 2012; Lisbona et al., 2009; Peña-Blanco and García-Sáez, 2018). Moreover, CHOP can also encourage the transcription of pro-apoptotic proteins like BCL2-like protein 11 (BIM), while simultaneously subduing the stimulation of anti-apoptotic BCL2 proteins (Chipuk et al., 2008; John et al., 2011; Zhu et al., 2004). Of note, ER-associated BAK can also activate an atypical pathway involving IRE1 → TRAF2 cascade, which mobilizes ER Ca2+ and promotes persistent activation of the multi-functional protein, JNK, all of which further facilitates mitochondrial apoptosis (Chen and Brandizzi, 2013; Chen et al., 2014; Hu et al., 2006; Kato et al., 2012; Pincus et al., 2010; Urano et al., 2000; Xu et al., 2018; Zhu et al., 2014). Thus, a dynamic but complex cross-talk between BCL2 proteins and the UPR signaling axes underlies the cellular response to ER stress, and this interplay also regulates the fate of an ER stressed cell (Chevet et al., 2015; Pihán et al., 2017; Rodriguez et al., 2011; Senft and Ronai, 2015; Szegezdi et al., 2009).
3.2. ER stress-induced inflammation
Stress adaptive processes co-ordinated by ER stress are not limited to taking only life or death decisions in a cellular context (Imanikia et al., 2018; Malhotra and Kaufman, 2011; Sano and Reed, 2013). Along with these cell fate-related resolutions, ER stress in a particular cell can also establish cross-talk of these stressed cells with the rest of the tissue, in which they are located, by modulating inflammation (Dandekar et al., 2015; Reverendo et al., 2019; Yoshida, 2007). ER stress or UPR-associated modulation of inflammation generally strives to limit tissue damage, thereby allowing tissue-level adaptation to extrinsic or intrinsic stress (aided by the immune system) (Garg et al., 2012a; Reverendo et al., 2019; Rodvold et al., 2016). But, in line with the multifaceted characteristics of ER stress, in specific settings ER stress-induced inflammation aggravates circumstances (Garg et al., 2012a; Lindholm et al., 2006; McGuckin et al., 2011; Scull and Tabas, 2011; Yoshida, 2007; Zhang et al., 2016). Such ER stress-associated worsening of pathologies is especially applicable to diseases like, atherosclerosis, cancer, diabetes, and obesity (Binet and Sapieha, 2015; Garg et al., 2012a; McAlpine et al., 2010; Mollereau et al., 2014; Morita et al., 2017; Yoshida, 2007). In such inflammatory situations, ER stress/UPR can strongly modulate the durability, strength, and the nature of immune reactions (Fung and Liu, 2014; Imanikia et al., 2018; Reverendo et al., 2019; Rodvold et al., 2016). However, the final immunological outcome of ER stress-induced inflammation tends to be harmful or protective depending on various factors consisting of (but not limited to): the cellular or tissue context, types of inflammatory signals generated by ER stress/UPR (i.e., pro-inflammatory or anti-inflammatory factors), type of immune cells interacting with ER stressed cells, the intensity of ER stress, and the overall immunological or health status of the organism (Cubillos-Ruiz et al., 2015; Dandekar et al., 2015; Garg et al., 2015d; Lindholm et al., 2006; Minamino and Kitakaze, 2010; Morito and Nagata, 2012; Verfaillie et al., 2013; Zhang, 2010).
Indeed, UPR signaling is frequently connected to the creation of numerous pro-inflammatory cytokines or chemokines (Dandekar et al., 2015; Fung and Liu, 2014; Garg et al., 2012a; Morito and Nagata, 2012; Rodvold et al., 2016; Turner et al., 2014). In fact, all three UPR axes discussed above can facilitate inflammatory transcriptional programs (Guo and Li, 2014; Medzhitov and Horng, 2009; Mogilenko et al., 2019). Such ER stress/UPR-directed inflammatory program is usually executed by notorious inflammatory transcription factors like, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) or activator protein 1 (AP1) (Erbay et al., 2009; Garg et al., 2012a; Jorgensen et al., 2008; Lenna et al., 2014; Meyerovich et al., 2016; Mozzini et al., 2017; Zhang et al., 2008). It is indeed common knowledge that NF-κB is one of the most dominant regulators of pro-inflammatory signaling since, it transcribes for a plethora of cytokines, immunological enzymes, or chemokines crucial for deciding both the amplitude and the duration of inflammation (Bottero et al., 2006; Hanada and Yoshimura, 2002; Hayden and Ghosh, 2011; Hayden et al., 2006; Lawrence, 2009; Novack, 2011; Pasparakis, 2009; Sun, 2012). NF-κB driven inflammation is amongst the most fastest immunological processes and can be elicited by a large array of microbial products, as well as endogenous factors released or secreted during tissue injury, or damage (Baldwin, 1996; Dolcet et al., 2005; Laskin and Pendino, 1995; Scott and Roifman, 2019). NF-κB is also downstream of almost all immune receptors (Chalmers et al., 2019; Dev et al., 2011; Ju Hwang et al., 2019). Interestingly, NF-κB can influence type I IFN responses. For instance, NF-κB can suppress expression of specific ISGs like guanylate-binding protein 1 (GBP1), interferon gamma inducible protein 47 (IFI47), myxovirus resistance 1/2 (MX1 or MX2), transporter associated with antigen processing (TAP1), while enhancing expression of other ISGs like, GBP2 or, TAP2 (Pfeffer, 2011).
PERK, IRE1 and ATF6 axes are all capable of activating or modulating the NF-κB signaling cascade, although each of these UPR arms may differ in terms of qualitative and quantitative NF-κB activity (Sundaram et al., 2018; Tam et al., 2012; Teske et al., 2011; Wang et al., 2014; Wolfson et al., 2008). Usually NF-κB is preserved as an inactive cytosolic form by the IκB proteins, thereby avoiding its instigation and nuclear transportation (Capece et al., 2018; May and Ghosh, 1997; Mussbacher et al., 2019; Sun, 2017). However, inflammatory factors engage IκB kinase (IKK) complex to enforce proteasomes-mediated IκB protein degradation, thereby unleashing NF-κB's (nucleus-associated) inflammatory transcriptional activity (Capece et al., 2018; May and Ghosh, 1997; Mussbacher et al., 2019; Sun, 2017). Similarly, the IRE1 axis also stimulates NF-κB activity by enforcing proteasomal degradation of IκB (Duran-Aniotz et al., 2017; Rubio et al., 2011; Tam et al., 2012). PERK commands a slightly different pathway for this purpose, it enforces translational inhibition of IκB thereby freeing NF-κB for engaging its transcriptional program (Blais et al., 2006; Halliday et al., 2017; Pytel et al., 2016; Qiao et al., 2017; Tam et al., 2012). Lastly, although ATF6 axis has been shown to influence NF-κB activation yet, the exact mechanism underlying this cascade has not been established (Tam et al., 2018; Yamazaki et al., 2009). Similar to NF-κB, AP1 is an important inflammatory transcription factor composed of monomers of a variety of proteins, like Jun proto-oncogene or JUN, Fos proto-oncogene or FOS, ATF and/or, musculoaponeurotic fibrosarcoma (MAF) (Uluçkan et al., 2015; van Dam and Castellazzi, 2001; Wagner, 2010). Different combinations of these proteins give rise to functional AP1 homo/hetero-dimers, which in turn influence the nature of the transcriptional program (Macián et al., 2001; Shaulian and Karin, 2001; Vesely et al., 2009). TNF, granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin 8 (IL8), and various cytokine receptors, are some of the most notorious genes within the transcriptional control of AP1 (Adcock, 1997; Hamilton, 2002; Hansen et al., 2008; Kyriakis, 1999; Thomas et al., 1997; Wu et al., 2001). Herein, mainly the IRE1 axis has been documented to activate the AP1-driven transcriptional program whereas the direct role of other UPR signaling arms is still controversial (Rubio et al., 2011; Shah et al., 2016).
Apart from the specific inflammatory transcriptional programs, ER stress/UPR can also initiate the more multifarious inflammatory process of acute-phase response (APR) (Morito and Nagata, 2012; Reverendo et al., 2019; Zhang, 2010; Zhang et al., 2006). APR is typically elicited during the very early phases of an innate immune reaction, driven primarily by a set of pro-inflammatory acute-phase proteins (APPs) consisting of cytokines, like IL1, IL6 and/or TNF (Banks et al., 1995; Bode et al., 2012; Slaats et al., 2016). However APR is not a solitary (coherent) reaction but, rather a cluster of cell, tissue, and organ-level stress adaptive processes, aimed at activating the immune system so it can prepare to tackle an impending pathological, or sterile injury (Joung et al., 2019; Kubes and Mehal, 2012; McDonald and Kubes, 2016; Natoli and Ostuni, 2019; Zelenay and Reis e Sousa, 2013). APR is largely responsible for various early physiological signs of injury or trauma, like fever, increased “permeability” of the vascular network, shift in the metabolites profile (often detectable via serum diagnostic methods), modified neurological reactions, and other similar (macroscopic) pathological symptoms (Joung et al., 2019; Kubes and Mehal, 2012; McDonald and Kubes, 2016; Natoli and Ostuni, 2019; Zelenay and Reis e Sousa, 2013). This is typically accompanied by a sharp increase in serum levels of nearly 200 different APPs including pro-inflammatory cytokines, factors modulating the pituitary-adrenal hormonal axis or cardiovascular system (e.g., C-reactive protein or CRP), and factors eliciting localized or systemic neutrophilia (mainly contributed by hepatocytes, and supported by few other cell-types) (Dinarello, 2006; Hribal et al., 2014; Kim and Moudgil, 2017; Li and Li, 2019; Paik et al., 2007; Tilg et al., 1997; Walsh et al., 2007). Even though APR aims to control damage to the tissue yet, very prolonged persistence of APPs can be the reason behind APR-associated adverse immunological events (Joung et al., 2019; Kubes and Mehal, 2012; McDonald and Kubes, 2016; Natoli and Ostuni, 2019; Zelenay and Reis e Sousa, 2013). In contrast to activation of NF-κB or AP1, where PERK/IRE1 axes of UPR dominate, in case of APR the ATF6 axis of the UPR seems to play a more important role (Duvigneau et al., 2018; Lehotský et al., 2009). In some cases, the cyclic-AMP-responsive-element-binding protein H (CREBH) may support the pro-APR activity of the ATF6 (Zhang et al., 2006, Zhang et al., 2012).
Last but not least, ER stress/UPR can also modulate danger signaling mediated by damage-associated molecular patterns (DAMPs) acting as danger signals (Brenner et al., 2013; Fucikova et al., 2015; Hou et al., 2013; Krysko et al., 2012; Land et al., 2016a, Land et al., 2016b; Ravichandran, 2011; Reverendo et al., 2019; Rufo et al., 2017; Vénéreau et al., 2015). Danger signaling is a context-dependent inflammatory process, that can aid or impede canonical inflammatory reactions, depending upon the type of DAMPs emitted and cognate receptors engaged, thereby either ameliorating or exacerbating a particular disease (de Haan et al., 2013; Brenner et al., 2013; Foell et al., 2007; Garg and Agostinis, 2017; Garg et al., 2016a; O'Shea and Murray, 2008; Rock and Kono, 2008). For instance, in case of diseases with dysregulated inflammatory milieu (e.g., cancer), ordered exposure of pro-phagocytic, chemotactic and immune cell activating DAMPs can be crucial for induction of anticancer immunity (Bracci et al., 2017; Cronin and Penninger, 2007; Fang et al., 2019; Galluzzi et al., 2017; He et al., 2013; Iurescia et al., 2018; Lee et al., 2016; Ma et al., 2011; Ravichandran, 2011; Seong and Matzinger, 2004). In contrast, DAMPs can not only support autoimmune diseases (e.g., GRP78 in case of autoimmune diabetes) but, they can also sometime be the primary pathological factors underlying their progression (e.g., nucleic acids during lupus) (Anaya et al., 2007; Anders, 2005; Cheng and Anderson, 2012; Cunha-Neto et al., 2011; Pedersen et al., 2011; Rizzo et al., 2014; Rondas et al., 2015). Nevertheless, in case of cancer it has been shown that the PERK axis can facilitate the active secretion or, surface exposure of danger signals, like calreticulin (a well-established “eat me” signal) or, ATP (a short-range “find me” signal) during the process of apoptotic immunogenic cell death (ICD) (Garg et al., 2012b, Garg et al., 2015c, Garg et al., 2017b; Kepp et al., 2014; Tesniere et al., 2008). However it is apparent that this function is sometimes contingent on PERK's UPR-related role (in case chemotherapy-induced surface-calreticulin) but, on other occasions it involves an as-yet-uncharacterized direct role of PERK in regulating the expansion of the secretory apparatus (in case of surface-calreticulin or secreted ATP elicited by ER-directed therapeutic oxidative stress achieved by hypericin-based photodynamic therapy) (Galluzzi et al., 2018; Garg et al., 2012b, Garg et al., 2015a, Garg et al., 2015b, Garg et al., 2015c, Garg et al., 2016b; Kroemer et al., 2013; Liu et al., 2019; Martins et al., 2014; Moserova et al., 2017; Panaretakis et al., 2009).
From the above sections it is clear that ER stress/UPR can influence both qualitative and quantitative aspects of inflammation. Indeed, type I IFN response is also a crucial inflammatory process (Mancuso et al., 2007; Uccellini and García-Sastre, 2018); however, the connection of ER stress/UPR with type I IFN responses has received comparatively much less attention than its connection with canonical transcriptional processes, like those mediated by NF-κB/AP1 or, underlying APR and danger signaling. In the next sections, we elaborate and attempt to summarize the broad interplay between type I IFNs and ER stress.
4. ER stress and type I IFNs: There and back again
4.1. Impact of ER stress on production or sensing of type I IFNs
A direct link between ER stress/UPR and type I IFNs has always been obscure because most chemical or pharmacological ER stress inducers do not (autonomously) cause considerable production of type I IFNs (Smith, 2014). However over the years accumulating evidence has indicated that the presence of ER stress can influence the production or sensing of type I IFNs induced by other perturbations like PRR agonists, infections (bacterial, viral or parasitic), autoimmune reactions or sterile injury-related factors (Smith, 2014). To understand the exact impact of ER stress/UPR on production or sensing of type I IFNs, we carried out a systematic literature analyses of publications indexed in PubMed database (Table 1 ). Remarkably it is clear that ER stress never directly engages type I IFNs production (Table 1). However, in presence of other inducers of type I IFNs (like infectious agents, PRR agonists, pharmacological ER stressors), ER stress plays an important role in modulating type I IFNs production, although whether it augments or suppresses it depends strongly on the context (Table 1).
Table 1.
Regulation of type I interferon (IFN) production or sensing by endoplasmic reticulum (ER) stress and unfolded protein response (UPR)
| Context | UPR or ISR entity involved | Effect of ER stress on production of type I IFNs or their sensing by IFNα-receptor (IFNAR) | ER stressor or UPR activator | Cell type | Refs. |
|---|---|---|---|---|---|
| Bacterial infection | IRE1α → XBP1 → ATG9A | Suppression of production; bacteria-induced UPR limits STING-dependent IFNβ production via ATG9A | Streptococcus pneumoniae | Murine BMDMs | Mitzel et al. (2014) |
| IRE1α → PKR | Augmentation of production; TLR4-mediated IRE1α activation causes PKR-driven stimulation of IFNβ production | Chlamydia trachomatis | Human mDCs and murine BMDMs | Webster et al. (2016) | |
| STING → XBP1 | Augmentation of production; bacteria-elicited, STING-dependent, XBP1 activation enhances IFNβ production | Brucella abortus | Murine BMDMs | Guimarães et al. (2019) | |
| DCs responding to poly(I:C) or VSV | IRE1α → XBP1 | Augmentation of production; XBP1s enhances poly(I:C)-induced IFNβ | Tunicamycin, thapsigargin and VSV | Murine BMDCs | Hu et al. (2011) |
| Diabetes | PERK | Suppression of sensing; whole-body or pancreas-specific ablation of PERK increases IFNAR1 levels thereby causing pancreatic toxicity-driven diabetes in mice | PERK knock-out phenotype? | Mouse derived pancreatic islets | Yu et al. (2015) |
| General ER stress | ATF6 or STING | Augmentation of production; Ca2 + mobilizing ER stressors caused STING-dependent & IRF3-induced IFNβ; other ER stressors exploited ATF6 for IRF3-induced IFNβ | Thapsigargin, oxygen-glucose deprivation, tunicamycin and 2-DE | RAW264.7 cells and murine BMDMs | Liu et al. (2012) |
| Hepatocellular steatosis during HCV infection | Not tested | Suppression of sensing; FFAs-based UPR causes phosphorylation-dependent and ubiquitination-driven IFNAR1 degradation | Saturated and unsaturated FFAs | S3-GFP replicon cell line (HCV2a) | Gunduz et al. (2012) |
| Macrophages responding to LPS | XBP1 | Augmentation of production; XBP1 together with IRF3, CBP and p300, enhances LPS-induced IFNβ | Thapsigargin | RAW264.7 cells and murine BMDMs | Zeng et al. (2010) |
| Parasitic infection | IRE1α → XBP1 | Augmentation of production; XBP1s enhances IFNβ production (and shows increased binding to IFNβ enhancer and promoter sequences) | Leishmania amazonensis and thapsigargin | RAW264.7 cells and murine BMDMs | Dias-Teixeira et al. (2016) |
| PBMCs and pDCs responding to PRR agonists (TLR2/4/9) | XBP1 | Augmentation of production; XBP1s enhances PRR agonists-induced IFNα | PRR agonists | PBMCs and pDCs | Beisel et al. (2017) |
| PRR agonist and chemical ER stress | IRE1α | Augmentation of production; In cells depleted of SKIV2L or XRN1, UPR-associated IRE1α generates endogenous RLR ligands that stimulate type I IFNs | Thapsigargin and tunicamycin | BMDMs | Eckard et al. (2014) |
| PRR agonist (TLR3) and viral infection | PKR → phospho-eIF2α → ATF4 → GADD34 | Augmentation of production; GADD34 activity enhanced IFNβ production | Chikungunya virus and poly(I:C) | Mouse embryonic fibroblasts | Clavarino et al. (2012) |
| PRR agonists (TLR4/3, MDA5) | XBP1 | Augmentation of production; XBP1 (but not PERK or ATF6) enhances TLR4/3 or MDA5 agonists-induced IFNβ | Tunicamycin and thapsigargin | RAW264.7 cells and murine BMDMs | Smith et al. (2008) |
| Viral infection | PERK → phospho-eIF2α → ATF4 → CHOP | Suppression of sensing; 3a protein & UPR causes phosphorylation-dependent and ubiquitination-driven IFNAR1 degradation | 3a protein of SARS-CoV | Huh7 | Minakshi et al. (2009) |
| CHOP | Suppression of production; knocking-down CHOP activates IFNβ production | HCV and Dengue virus | Huh7 | Ke and Chen (2011) | |
| IRE1α → XBP1; ATF6 | Suppression of sensing; IRE1α-XBP1 and ATF6 together inhibit JAK-STAT signaling thereby obstructing responses to IFNα | WNV | Mouse embryonic fibroblasts | Ambrose and Mackenzie, 2011, Ambrose and Mackenzie, 2013 | |
| PERK | Suppression of sensing; PERK and UPR-induced autophagy caused IFNAR1 degradation | HCV and thapsigargin | Huh7 | Chandra et al. (2014) | |
| PERK → phospho-eIF2α | Augmentation of production; TGEV-induced UPR enhances type I IFN production via the PERK arm | TGEV | ST cells | Xue et al. (2018) | |
| Viral infection and chemical ER stress | Not tested | Suppression of sensing; During UPR, CK1α kinase causes S535 phosphorylation-dependent and ubiquitination-driven IFNAR1 degradation | VSV and thapsigargin | Mouse embryonic fibroblasts, HeLa and 2fTGH | Bhattacharya et al. (2010) and Liu et al. (2009a) |
| PERK | Suppression of sensing; PERK promotes phosphorylation-dependent and ubiquitination-driven IFNAR1 degradation | VSV, HCV and thapsigargin | Mouse embryonic fibroblasts | Liu et al. (2009b) | |
| PERK → p38 | Suppression of sensing; p38 protein kinase causes phosphorylation-dependent and ubiquitination-driven IFNAR1 degradation | VSV and thapsigargin | Mouse embryonic fibroblasts, HeLa and 2fTGH | Bhattacharya et al. (2011) |
Abbreviations: 2-DE, 2-deoxyglucose; ATF4/6, activating transcription factor 4 or 6; ATG9, autophagy related 9; BMDC, bone marrow derived dendritic cells; BMDM, bone marrow derived macrophages; CBP, cAMP response element binding protein (CREB)-binding protein; CHOP, CCAAT-enhancer-binding protein homologous protein; CK1α, casein kinase 1-alpha; DCs, dendritic cells; ER, endoplasmic reticulum; FFAs, free fatty acids; GADD34, growth-arrest and DNA damage-inducible protein 34; HCV, hepatitis C virus; IFN, interferon; IFNAR, interferon alpha/beta receptor; IRE1α, inositol-requiring enzyme 1 alpha; IRF3, interferon regulatory factor 3; ISR, integrated stress response; JAK, janus kinase; LPS, lipopolysaccharide; MDA5, melanoma differentiation-associated protein 5; mDCs, monocyte-derived dendritic cells; PBMCs, peripheral blood mononuclear cells; pDCs, plasmacytoid dendritic cells; PERK, PKR-like endoplasmic reticulum kinase; Phospho-eIF2α, phosphorylated eukaryotic translation initiation factor 2A; PKR, protein kinase R; Poly(I:C), polyinosinic:polycytidylic acid; PRR, pattern recognition receptor; RLR, retinoic acid-inducible gene I (RIG-I)-like receptors; SARS-CoV, severe acute respiratory syndrome-related coronavirus; SKIV2L, superkiller viralicidic activity 2-like RNA helicase; STAT, signal transducer and activator of transcription; STING, stimulator of interferon genes; TGEV, transmissible gastroenteritis virus; TLR, toll-like receptor; UPR, unfolded protein response; VSV, vesicular stomatitis virus; WNV, West Nile virus; XBP1, X-box binding protein 1; XBP1s, spliced XBP1 isoform; XRN1, 5′–3′ exoribonuclease 1.
Mechanistically speaking, the IRE1/XBP1 arm of the UPR mostly augments type I IFN production (Table 1), a function that seems to be aided by other stress-responsive proteins (protein kinase R or PKR, CREB-binding protein or CBP, p300), interferon-pathway related proteins (STING, IRF3) or even UPR-related proteins (ATF6). Interestingly this pro-type I IFNs activity of the IRE1 arm often operates through XBP1s' ability to bind the IFNβ enhancer and promoter sequences (Table 1). Type I IFNs induced by agonists of PRRs, especially TLR2/3/4/9, cGAS/STING, melanoma differentiation-associated protein 5 (MDA5), seem to gain most advantage from the enhancing effects of the IRE1/XBP1 arm of the UPR (Table 1). In couple of instances, phosphorylation of eIF2α (either by PERK or PKR) also seems to support type I IFNs production, however this occurs much less often when compared to the IRE1/XBP1 arm (Table 1).
Nonetheless in line with the complex nature of UPR's functional effects (Hazari et al., 2016; Milev and Gatfield, 2018), UPR signaling also plays a detrimental role by suppressing type I IFN responses. Based on our survey (Table 1), two main type I IFN impeding pathways emerged: (1) in most cases, UPR was found to disrupt the sensing of type I IFNs by reducing the overall expression of its cognate receptor IFNAR1; (2) in other cases, ER stress-autophagy interplay (Klionsky et al., 2016) was found to target particular components of the type I IFNs signaling pathway, thereby disrupting either the type I IFNs production itself or its IFNAR1-dependent sensing. Interestingly, the PERK arm of the UPR was mostly reported to play a role in suppressing type I IFNs production or sensing (Table 1). In most cases the major pathway PERK was enforcing for suppressing type I IFN responses involved, IFNAR1 phosphorylation that drove the ubiquitination-based degradation of IFNAR1, which compromised the sensitization of target cells toward type I IFNs (Table 1; Fig. 1). Of note, degradation of IFNAR1 is one of the most prevalent phenomena for compromising responsiveness to type I IFNs in various diseased contexts (Araya and Goldszmid, 2017; Bhattacharya et al., 2010, Bhattacharya et al., 2014). It involves TYK2-dependent phosphorylation of serine residues in IFNAR1 (S526 in mice or S535 in humans) that attracts βTrcp E3 ubiquitin ligase-dependent IFNAR1 ubiquitination. Thereafter the ubiquitinated IFNAR1 is internalized and degraded via the lysosomes (Liu et al., 2009b) (Fig. 1). Signaling modules mostly reported to cause IFNAR1 degradation include those operated by the p38 protein kinase pathway (Araya and Goldszmid, 2017; Bhattacharya et al., 2011), casein kinase 1α (CK1alpha) (Liu et al., 2009a), and sphingosine 1-phosphate receptor (S1PR1)-associated pathway (Teijaro et al., 2016).
In terms of types of physicochemical ER stressors (Couve and Hetz, 2014), a wide-range of ER stress inducers including oxygen-glucose deprivation and pharmacological agents (tunicamycin, thapsigargin) can modulate type I IFNs induced by PRR agonists (Liu et al., 2012) (Table 1). However the mechanisms behind this modulation, as well as, its ultimate outcome (i.e., augmentation or deprivation of type I IFNs production or response) varies significantly from one ER stress-inducing agent to another (Liu et al., 2012). Such variation is even applicable to the context of viral infections despite the fact that, unlike the above physicochemical ER stressors, viruses are capable of eliciting both ER stress and type I IFNs (Jheng et al., 2014; Tardif et al., 2005; Yoshida, 2007). As evident from Table 1, whereas viral induction of ER stress-driven UPR signaling is indeed a ubiquitous phenomenon yet, the qualitative characteristics of virus-induced UPR do differ depending on the type of virus (Liu et al., 2009b; Shinjo et al., 2013). More specifically, depending on the virus, the UPR signaling might be driven by either the IRE1 arm or, PERK arm (Lin et al., 2009; Song et al., 2019); and this differential dominance of UPR arms may eventually govern the effects of UPR on production or sensing of type I IFNs (Table 1). Of note, ATF6 activation is not always analyzed with as much consistency as IRE1 or PERK (Sundaram et al., 2018; Tay et al., 2014); however in few studies where this arm was investigated, it seemed that ATF6 tended to synergize more with IRE1’s contribution in regulating type I IFN production or sensing (Table 1).
Finally, although none of the studies mentioned in Table 1 were directly dealing with cancer, yet several utilized cancer cells lines (e.g., HeLa cells). Thus, these results may also have direct implications for cancer immunology, at least in the context of oncolytic virotherapy.
4.2. Induction of ER stress by type I IFNs
Type I IFNs, as mentioned previously, do not always have a disease-impeding effect (Burdick et al., 2009). In the context of specific autoimmune pathologies like type 1 diabetes or interferonopathies, type I IFNs can play a disease-supportive, or even a disease-initiating role (Burdick et al., 2009; Lee-Kirsch et al., 2015). Research has shown that persistent presence of type I IFNs that fails to resolve in time supports chronic inflammation that may cause worsening of some pathologies (Akbar et al., 2000; Lee-Kirsch et al., 2015). This scenario is particularly applicable to type 1 diabetes wherein inflammation and/or autoimmunity causes apoptosis of pancreatic beta cells thereby impairing secretion of insulin (Eizirik et al., 2009). For instance, children with genetic predisposition toward type 1 diabetes often exhibit an augmented type I IFN-associated genetic signature, preceding the emerge of auto-antibodies (Marroqui et al., 2017). Moreover, antibody-based blockade of IFNAR1 has been often shown to ameliorate type 1 diabetes, at least pre-clinically (Marroqui et al., 2017). It is known for a long time that ER stress has an important role in regulating beta cell apoptosis during type 1 diabetes, thereby leading to prioritization of preclinical research on ER stress modulators for treating type 1 diabetes (Brozzi and Eizirik, 2016; Garg et al., 2012a; Rondas et al., 2015). Interestingly, recently a link between type I IFNs and ER stress is starting to emerge in this context. Apparently IFNα, along with IFNβ, can augment apoptosis of beta cells in-part via ER stress thereby facilitating type 1 diabetes (Marroqui et al., 2017). In fact, even without beta cell apoptosis, IFNα-induced ER stress can create ER-associated proteotoxicity that can impede the maturation and secretion of insulin (Lombardi and Tomer, 2017). Thus not surprisingly, pre-treatment of beta cells with ER stress-impeding chemical chaperones can almost completely prevent the negative effects of IFNα on beta cells (Lombardi and Tomer, 2017). Nevertheless, apoptosis induced by type I IFNs, while detrimental in contexts like type 1 diabetes, can also be beneficial in case of other pathologies. For example, during infections (to eliminate an infected cell, and along with it, the endogenous microbes) and cancer (to eliminate cancer cells via apoptosis), type I IFN-induced elimination of diseased cells is highly desirable. Interestingly, type I IFNs can indeed cause apoptosis in cancer cells or virally infected cells at least partially via ER stress induction (Mihailidou et al., 2017; Shi et al., 2016; Smith, 2014). In case of cancer however, from the available data it is not conclusive whether this ER stress induction by type I IFNs is as potent as pharmacological ER stressors in inducing high levels of apoptosis. Moreover, most studies reporting this phenomenon achieved this effect in vitro, and hence it remains unclear whether type I IFNs can reach high enough concentrations in vivo to elicit ER stress-driven apoptosis consistently.
4.3. ER stress and STING-based type I IFN signaling
Besides serving as a biosynthetic “factory” for type I IFNs production, ER can also provide a certain degree of structural support to type I IFN response. More specifically STING, which is part of an important PRR complex (i.e., cGAS/STING, as discussed above), localizes to the ER membrane in basal conditions (Ishikawa and Barber, 2008). When cGAS encounters its cognate PAMPs or DAMPs, it elicits (via cGAMP moieties-driven activation) translocation of STING toward the ER-Golgi intermediate compartment (ERGIC) wherein STING engages TBK1 and IRF3 to eventually orchestrate type I IFNs production (Barber, 2015). Of note in basal conditions, STING might also be present in mitochondria-associated ER membrane (MAMs), a site of physical association between mitochondria and ER (Ishikawa et al., 2009). However, this partial MAMs localization is of no palpable functional significance since upon activation, STING is only found in the ERGIC (Ishikawa et al., 2009). Interestingly, several lines of evidence have linked STING with ER stress-linked pathologies (Garg et al., 2012a; Wu et al., 2019). For instance, Gram-positive bacteria based elicitation of STING causes ER stress, which can engage an autophagic pathway to carryout ER-phagy (in order to ameliorate bacteria-induced ER stress) (Moretti et al., 2017). Interestingly, this ER-phagy stimulates an interferon response by transferring STING to the autophagosomes (Moretti et al., 2017). Overall, this pathway seems to play an important role in ensuring the post-infection viability of cells (Moretti et al., 2017). On the flip side, STING-mediated disruption of Ca2+ homeostasis can end up activating ER stress that can cause apoptosis of various normal cells thereby leading to STING-linked immunopathologies (Wu et al., 2019). Similarly STING and IRF3 can together drive alcoholic liver disease by linking ER stress with apoptosis (Petrasek et al., 2013). More specifically, alcoholic insult can elicit ER stress accompanied by IRF3-STING interaction that paves way for the activation of pro-apoptotic machinery (Petrasek et al., 2013). Interestingly, deletion of STING overcomes these disparities and also reduces ER stress thereby substantiating a direct connection in this context (Petrasek et al., 2013). Mechanistically it seems that ER might be restraining over-activation of STING in basal conditions via the Ca2+-binding protein, Stromal Interaction Molecule 1 (STIM1); since, absence of STIM1 causes spontaneous activation of the STING-TBK1-IRF3 pathway thereby leading to type I IFN-mediated autoimmune reactions (Srikanth et al., 2019).
Although these cross-talks between ER stress and STING is largely independent of the cGAS/STING-based type I IFNs production pathway yet it is of critical importance considering the attention STING agonists (for cancer immunotherapy) or antagonists (for autoimmune diseases) have recently received in preclinical and clinical studies (Berger et al., 2019; Corrales and Gajewski, 2015; Garg et al., 2017d; Lemos et al., 2015; McCaffary, 2017; Ramanjulu et al., 2018). In case these STING-targeting therapeutics cause adverse side-effects like autoimmune reactions in the clinic, then this may create a precedent to explore the application of ER stress ameliorating therapeutics, in combinatorial manner.
5. Conclusion
Type I IFNs exhibit critical functions in many sterile as well as non-sterile inflammatory diseases or immunopathologies, thereby positioning them at the core of almost all therapeutic paradigms. Like several other cytokine families, there also exist extensive overlaps between type I IFNs production or sensing, and the ER, which positions ER stress at a critical cross-road with the type I IFNs-associated functions. Indeed, stress adaptive processes co-ordinated by ER stress are not limited to taking only life or death decisions in a cellular context. Along with these, ER stress also modulates inflammation in order to recruit the services of immune cells to maintain tissue homeostasis. Thus, it is not surprising that ER stress is frequently connected to the creation of numerous pro-inflammatory cytokines. However, a direct link between ER stress and type I IFNs has always been obscure because, unlike many other cytokines, most ER stress inducers do not (autonomously) cause considerable production of type I IFNs. However, the presence of ER stress can in fact influence the production or sensing of type I IFNs induced by other perturbations like PRR agonists, infections (bacterial, viral or parasitic), interferonopathies, autoimmune reactions, or trauma/injury-related factors. Despite this crucial interplay between ER stress and type I IFNs that can have overarching implications for several human diseases, this connection has received comparatively much less attention than the connection of ER stress with, canonical transcriptional processes (like those mediated by NF-κB/AP1), APR or danger signaling. Thus, in future it is crucial to carryout both systematic as well as large-scale explorative studies to comprehensively understand how ER stress influences type I IFNs. This is especially important for disease contexts where these two play an important role however their cross-talk is not completely understood, e.g., cancer, respiratory pathologies, Crohn's disease or ulcerative colitis (amongst others).
Acknowledgments
We would also like to acknowledge the support of Research Foundation Flanders' (FWO) Excellence of Science (EOS) grant (30837538) for the “DECODE” consortium to A.D.G.; and KU Leuven support to A.D.G. via C1 grant (C14/19/098) and POR award funds (POR/16/040). A.D.G. is also a recipient of the FWO Postdoctoral Mandate (2016–2019).
References
- Adcock I.M. Transcription factors as activators of gene transcription: AP-1 and NF-kappa B. Monaldi Arch. Chest Dis. 1997;52:178–186. [PubMed] [Google Scholar]
- Ahn J., Xia T., Rabasa Capote A., Betancourt D., Barber G.N. Extrinsic phagocyte-dependent STING signaling dictates the immunogenicity of dying cells. Cancer Cell. 2018;33 doi: 10.1016/j.ccell.2018.03.027. 862–873.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar A.N., Lord J.M., Salmon M. IFN-alpha and IFN-beta: a link between immune memory and chronic inflammation. Immunol. Today. 2000;21:337–342. doi: 10.1016/s0167-5699(00)01652-2. [DOI] [PubMed] [Google Scholar]
- Ali M.M.U., Bagratuni T., Davenport E.L., Nowak P.R., Silva-Santisteban M.C., Hardcastle A., McAndrews C., Rowlands M.G., Morgan G.J., Aherne W., Collins I., Davies F.E., Pearl L.H. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almanza A., Carlesso A., Chintha C., Creedican S., Doultsinos D., Leuzzi B., Luís A., McCarthy N., Montibeller L., More S., Papaioannou A., Püschel F., Sassano M.L., Skoko J., Agostinis P., de Belleroche J., Eriksson L.A., Fulda S., Gorman A.M., Healy S., Kozlov A., Muñoz-Pinedo C., Rehm M., Chevet E., Samali A. Endoplasmic reticulum stress signalling—from basic mechanisms to clinical applications. FEBS J. 2019;286:241–278. doi: 10.1111/febs.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose R.L., Mackenzie J.M. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 2011;85:2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose R.L., Mackenzie J.M. ATF6 signaling is required for efficient West Nile virus replication by promoting cell survival and inhibition of innate immune responses. J. Virol. 2013;87:2206–2214. doi: 10.1128/JVI.02097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameri K., Harris A.L. Activating transcription factor 4. Int. J. Biochem. Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Anaya J.-M., Corena R., Castiblanco J., Rojas-Villarraga A., Shoenfeld Y. The kaleidoscope of autoimmunity: multiple autoimmune syndromes and familial autoimmunity. Expert. Rev. Clin. Immunol. 2007;3:623–635. doi: 10.1586/1744666X.3.4.623. [DOI] [PubMed] [Google Scholar]
- Anders H.J. A Toll for lupus. Lupus. 2005;14:417–422. doi: 10.1191/0961203305lu2102rr. [DOI] [PubMed] [Google Scholar]
- Andreu-Fernández V., Sancho M., Genovés A., Lucendo E., Todt F., Lauterwasser J., Funk K., Jahreis G., Pérez-Payá E., Mingarro I., Edlich F., Orzáez M. Bax transmembrane domain interacts with prosurvival Bcl-2 proteins in biological membranes. Proc. Natl. Acad. Sci. U. S. A. 2017;114:310–315. doi: 10.1073/pnas.1612322114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Riemer J., Christensen B., Sørensen E.S., Ellgaard L. A novel disulphide switch mechanism in Ero1alpha balances ER oxidation in human cells. EMBO J. 2008;27:2977–2987. doi: 10.1038/emboj.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K., Nagata K. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya R.E., Goldszmid R.S. IFNAR1 degradation: a new mechanism for tumor immune evasion? Cancer Cell. 2017;31:161–163. doi: 10.1016/j.ccell.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K.-I., Miyauchi S., Stoner S.A., Fan J.-B., Zhang D.-E. Negative regulation of type I IFN signaling. J. Leukoc. Biol. 2018;103:1099–1116. doi: 10.1002/JLB.2MIR0817-342R. [DOI] [PubMed] [Google Scholar]
- Atkins C., Liu Q., Minthorn E., Zhang S.-Y., Figueroa D.J., Moss K., Stanley T.B., Sanders B., Goetz A., Gaul N., Choudhry A.E., Alsaid H., Jucker B.M., Axten J.M., Kumar R. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- Back S.H., Schröder M., Lee K., Zhang K., Kaufman R.J. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35:395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Back S.H., Lee K., Vink E., Kaufman R.J. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J. Biol. Chem. 2006;281:18691–18706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- Bald T., Landsberg J., Lopez-Ramos D., Renn M., Glodde N., Jansen P., Gaffal E., Steitz J., Tolba R., Kalinke U., Limmer A., Jönsson G., Hölzel M., Tüting T. Immune cell-poor melanomas benefit from PD-1 blockade after targeted type I IFN activation. Cancer Discov. 2014;4:674–687. doi: 10.1158/2159-8290.CD-13-0458. [DOI] [PubMed] [Google Scholar]
- Baldwin A.S. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Balsa E., Soustek M.S., Thomas A., Cogliati S., García-Poyatos C., Martín-García E., Jedrychowski M., Gygi S.P., Enriquez J.A., Puigserver P. ER and nutrient stress promote assembly of respiratory chain supercomplexes through the PERK-eIF2α axis. Mol. Cell. 2019;74 doi: 10.1016/j.molcel.2019.03.031. 877–890.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks R.E., Forbes M.A., Storr M., Higginson J., Thompson D., Raynes J., Illingworth J.M., Perren T.J., Selby P.J., Whicher J.T. The acute phase protein response in patients receiving subcutaneous IL-6. Clin. Exp. Immunol. 1995;102:217–223. doi: 10.1111/j.1365-2249.1995.tb06659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber G.N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A., Gottesman S. Roles of adaptor proteins in regulation of bacterial proteolysis. Curr. Opin. Microbiol. 2013;16:140–147. doi: 10.1016/j.mib.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C., Ziegler S., Martrus Zapater G., Chapel A., Griesbeck M., Hildebrandt H., Lohse A.W., Altfeld M. TLR7-mediated activation of XBP1 correlates with the IFNα production in humans. Cytokine. 2017;94:55–58. doi: 10.1016/j.cyto.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Berger G., Marloye M., Lawler S.E. Pharmacological modulation of the STING pathway for cancer immunotherapy. Trends Mol. Med. 2019;25:412–427. doi: 10.1016/j.molmed.2019.02.007. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., HuangFu W.-C., Liu J., Veeranki S., Baker D.P., Koumenis C., Diehl J.A., Fuchs S.Y. Inducible priming phosphorylation promotes ligand-independent degradation of the IFNAR1 chain of type I interferon receptor. J. Biol. Chem. 2010;285:2318–2325. doi: 10.1074/jbc.M109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Qian J., Tzimas C., Baker D.P., Koumenis C., Diehl J.A., Fuchs S.Y. Role of p38 protein kinase in the ligand-independent ubiquitination and down-regulation of the IFNAR1 chain of type I interferon receptor. J. Biol. Chem. 2011;286:22069–22076. doi: 10.1074/jbc.M111.238766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Katlinski K.V., Reichert M., Takano S., Brice A., Zhao B., Yu Q., Zheng H., Carbone C.J., Katlinskaya Y.V., Leu N.A., McCorkell K.A., Srinivasan S., Girondo M., Rui H., May M.J., Avadhani N.G., Rustgi A.K., Fuchs S.Y. Triggering ubiquitination of IFNAR1 protects tissues from inflammatory injury. EMBO Mol. Med. 2014;6:384–397. doi: 10.1002/emmm.201303236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M.E. DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukoc. Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- Binet F., Sapieha P. ER stress and angiogenesis. Cell Metab. 2015;22:560–575. doi: 10.1016/j.cmet.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Björck P., Leong H.X., Engleman E.G. Plasmacytoid dendritic cell dichotomy: identification of IFN-α producing cells as a phenotypically and functionally distinct subset. J. Immunol. 2011;186:1477–1485. doi: 10.4049/jimmunol.1000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais J.D., Addison C.L., Edge R., Falls T., Zhao H., Wary K., Koumenis C., Harding H.P., Ron D., Holcik M., Bell J.C. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol. Cell. Biol. 2006;26:9517–9532. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk K., Olejnik A., Nowicka H., Ozgyin L., Chen Y.-L., Chmielewski S., Kostyrko K., Wesoly J., Balint B.L., Lee C.-K., Bluyssen H.A.R. STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1. Biochem. J. 2015;466:511–524. doi: 10.1042/BJ20140644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazanin N., Son J., Craig-Lucas A.B., John C.L., Breech K.J., Podolsky M.A., Glick A.B. ER stress and distinct outputs of the IRE1α RNase control proliferation and senescence in response to oncogenic Ras. Proc. Natl. Acad. Sci. U. S. A. 2017;114:9900–9905. doi: 10.1073/pnas.1701757114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J.G., Albrecht U., Häussinger D., Heinrich P.C., Schaper F. Hepatic acute phase proteins—regulation by IL-6- and IL-1-type cytokines involving STAT3 and its crosstalk with NF-κB-dependent signaling. Eur. J. Cell Biol. 2012;91:496–505. doi: 10.1016/j.ejcb.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Bogdan C., Mattner J., Schleicher U. The role of type I interferons in non-viral infections. Immunol. Rev. 2004;202:33–48. doi: 10.1111/j.0105-2896.2004.00207.x. [DOI] [PubMed] [Google Scholar]
- Borden E.C. Interferons α and β in cancer: therapeutic opportunities from new insights. Nat. Rev. Drug Discov. 2019;18:219–234. doi: 10.1038/s41573-018-0011-2. [DOI] [PubMed] [Google Scholar]
- Bottero V., Withoff S., Verma I.M. NF-kappaB and the regulation of hematopoiesis. Cell Death Differ. 2006;13:785–797. doi: 10.1038/sj.cdd.4401888. [DOI] [PubMed] [Google Scholar]
- Bouchecareilh M., Higa A., Fribourg S., Moenner M., Chevet E. Peptides derived from the bifunctional kinase/RNase enzyme IRE1α modulate IRE1α activity and protect cells from endoplasmic reticulum stress. FASEB J. 2011;25:3115–3129. doi: 10.1096/fj.11-182931. [DOI] [PubMed] [Google Scholar]
- Bracci L., Sistigu A., Proietti E., Moschella F. The added value of type I interferons to cytotoxic treatments of cancer. Cytokine Growth Factor Rev. 2017;36:89–97. doi: 10.1016/j.cytogfr.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Brenner C., Galluzzi L., Kepp O., Kroemer G. Decoding cell death signals in liver inflammation. J. Hepatol. 2013;59:583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- Brozzi F., Eizirik D.L. ER stress and the decline and fall of pancreatic beta cells in type 1 diabetes. Ups. J. Med. Sci. 2016;121:133–139. doi: 10.3109/03009734.2015.1135217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush M.H., Weiser D.C., Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick L.M., Somani N., Somani A.-K. Type I IFNs and their role in the development of autoimmune diseases. Expert Opin. Drug Saf. 2009;8:459–472. doi: 10.1517/14740330903066726. [DOI] [PubMed] [Google Scholar]
- Cabral-Miranda F., Hetz C. ER stress and neurodegenerative disease: a cause or effect relationship? Curr. Top. Microbiol. Immunol. 2018;414:131–157. doi: 10.1007/82_2017_52. [DOI] [PubMed] [Google Scholar]
- Capece D., Verzella D., Tessitore A., Alesse E., Capalbo C., Zazzeroni F. Cancer secretome and inflammation: the bright and the dark sides of NF-κB. Semin. Cell Dev. Biol. 2018;78:51–61. doi: 10.1016/j.semcdb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Carrara M., Prischi F., Nowak P.R., Kopp M.C., Ali M.M. Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. eLife. 2015;4 doi: 10.7554/eLife.03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo K., Rojas-Rivera D., Lisbona F., Caballero B., Nassif M., Court F.A., Schuck S., Ibar C., Walter P., Sierralta J., Glavic A., Hetz C. BAX inhibitor-1 regulates autophagy by controlling the IRE1α branch of the unfolded protein response. EMBO J. 2011;30:4465–4478. doi: 10.1038/emboj.2011.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers S.A., Garcia S.J., Reynolds J.A., Herlitz L., Putterman C. NF-kB signaling in myeloid cells mediates the pathogenesis of immune-mediated nephritis. J. Autoimmun. 2019;98:33–43. doi: 10.1016/j.jaut.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra P.K., Bao L., Song K., Aboulnasr F.M., Baker D.P., Shores N., Wimley W.C., Liu S., Hagedorn C.H., Fuchs S.Y., Wu T., Balart L.A., Dash S. HCV infection selectively impairs type I but not type III IFN signaling. Am. J. Pathol. 2014;184:214–229. doi: 10.1016/j.ajpath.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Brandizzi F. IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 2013;23:547–555. doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xu S., Liu L., Wen X., Xu Y., Chen J., Teng J. Cab45S inhibits the ER stress-induced IRE1-JNK pathway and apoptosis via GRP78/BiP. Cell Death Dis. 2014;5:e1219. doi: 10.1038/cddis.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.H., Anderson M.S. Monogenic autoimmunity. Annu. Rev. Immunol. 2012;30:393–427. doi: 10.1146/annurev-immunol-020711-074953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon H., Borden E.C., Stark G.R. Interferons and their stimulated genes in the tumor microenvironment. Semin. Oncol. 2014;41:156–173. doi: 10.1053/j.seminoncol.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevet E., Hetz C., Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. 2015;5:586–597. doi: 10.1158/2159-8290.CD-14-1490. [DOI] [PubMed] [Google Scholar]
- Chien M.-H., Lee W.-J., Hsieh F.-K., Li C.-F., Cheng T.-Y., Wang M.-Y., Chen J.-S., Chow J.-M., Jan Y.-H., Hsiao M., Hua K.-T., Kuo M.-L. Keap1-Nrf2 interaction suppresses cell motility in lung adenocarcinomas by targeting the S100P protein. Clin. Cancer Res. 2015;21:4719–4732. doi: 10.1158/1078-0432.CCR-14-2880. [DOI] [PubMed] [Google Scholar]
- Chipuk J.E., Fisher J.C., Dillon C.P., Kriwacki R.W., Kuwana T., Green D.R. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20327–20332. doi: 10.1073/pnas.0808036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiribau C.-B., Gaccioli F., Huang C.C., Yuan C.L., Hatzoglou M. Molecular symbiosis of CHOP and C/EBP beta isoform LIP contributes to endoplasmic reticulum stress-induced apoptosis. Mol. Cell. Biol. 2010;30:3722–3731. doi: 10.1128/MCB.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovatiya R., Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol. Cell. 2014;54:281–288. doi: 10.1016/j.molcel.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy M.S., Yusoff P., Lee I.C., Newton J.C., Goh C.W., Page R., Shenolikar S., Peti W. Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep. 2015;11:1885–1891. doi: 10.1016/j.celrep.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christis C., Fullaondo A., Schildknegt D., Mkrtchian S., Heck A.J.R., Braakman I. Regulated increase in folding capacity prevents unfolded protein stress in the ER. J. Cell Sci. 2010;123:787–794. doi: 10.1242/jcs.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavarino G., Cláudio N., Couderc T., Dalet A., Judith D., Camosseto V., Schmidt E.K., Wenger T., Lecuit M., Gatti E., Pierre P. Induction of GADD34 is necessary for dsRNA-dependent interferon-β production and participates in the control of Chikungunya virus infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho D.S., Domingos P.M. Physiological roles of regulated Ire1 dependent decay. Front. Genet. 2014;5:76. doi: 10.3389/fgene.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L., Gajewski T.F. Molecular pathways: targeting the stimulator of interferon genes (STING) in the immunotherapy of cancer. Clin. Cancer Res. 2015;21:4774–4779. doi: 10.1158/1078-0432.CCR-15-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couve A., Hetz C. RESETing ER proteostasis: selective stress pathway hidden in the secretory route. EMBO J. 2014;33:2444–2446. doi: 10.15252/embj.201489845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covino R., Hummer G., Ernst R. Integrated functions of membrane property sensors and a hidden side of the unfolded protein response. Mol. Cell. 2018;71:458–467. doi: 10.1016/j.molcel.2018.07.019. [DOI] [PubMed] [Google Scholar]
- Cronin S.J.F., Penninger J.M. From T-cell activation signals to signaling control of anti-cancer immunity. Immunol. Rev. 2007;220:151–168. doi: 10.1111/j.1600-065X.2007.00570.x. [DOI] [PubMed] [Google Scholar]
- Cubillos-Ruiz J.R., Silberman P.C., Rutkowski M.R., Chopra S., Perales-Puchalt A., Song M., Zhang S., Bettigole S.E., Gupta D., Holcomb K., Ellenson L.H., Caputo T., Lee A.-H., Conejo-Garcia J.R., Glimcher L.H. ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell. 2015;161:1527–1538. doi: 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz J.R., Bettigole S.E., Glimcher L.H. Molecular pathways: immunosuppressive roles of IRE1α-XBP1 signaling in dendritic cells of the tumor microenvironment. Clin. Cancer Res. 2016;22:2121–2126. doi: 10.1158/1078-0432.CCR-15-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz J.R., Bettigole S.E., Glimcher L.H. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan S.B., Zhang D., Hannink M., Arvisais E., Kaufman R.J., Diehl J.A. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Neto E., Teixeira P.C., Nogueira L.G., Kalil J. Autoimmunity. Adv. Parasitol. 2011;76:129–152. doi: 10.1016/B978-0-12-385895-5.00006-2. [DOI] [PubMed] [Google Scholar]
- Dadey D.Y.A., Kapoor V., Khudanyan A., Thotala D., Hallahan D.E. PERK regulates glioblastoma sensitivity to ER stress although promoting radiation resistance. Mol. Cancer Res. 2018;16:1447–1453. doi: 10.1158/1541-7786.MCR-18-0224. [DOI] [PubMed] [Google Scholar]
- Dandekar A., Mendez R., Zhang K. Cross talk between ER stress, oxidative stress, and inflammation in health and disease. Methods Mol. Biol. 2015;1292:205–214. doi: 10.1007/978-1-4939-2522-3_15. [DOI] [PubMed] [Google Scholar]
- Das I., Png C.W., Oancea I., Hasnain S.Z., Lourie R., Proctor M., Eri R.D., Sheng Y., Crane D.I., Florin T.H., McGuckin M.A. Glucocorticoids alleviate intestinal ER stress by enhancing protein folding and degradation of misfolded proteins. J. Exp. Med. 2013;210:1201–1216. doi: 10.1084/jem.20121268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan J.J., Smeets M.B., Pasterkamp G., Arslan F. Danger signals in the initiation of the inflammatory response after myocardial infarction. Mediat. Inflamm. 2013;2013:206039. doi: 10.1155/2013/206039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weerd N.A., Vivian J.P., Nguyen T.K., Mangan N.E., Gould J.A., Braniff S.-J., Zaker-Tabrizi L., Fung K.Y., Forster S.C., Beddoe T., Reid H.H., Rossjohn J., Hertzog P.J. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 2013;14:901–907. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- Del Vecchio C.A., Feng Y., Sokol E.S., Tillman E.J., Sanduja S., Reinhardt F., Gupta P.B. De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev A., Iyer S., Razani B., Cheng G. NF-κB and innate immunity. Curr. Top. Microbiol. Immunol. 2011;349:115–143. doi: 10.1007/82_2010_102. [DOI] [PubMed] [Google Scholar]
- Dias-Teixeira K.L., Calegari-Silva T.C., dos Santos G.R.R.M., Vitorino Dos Santos J., Lima C., Medina J.M., Aktas B.H., Lopes U.G. The integrated endoplasmic reticulum stress response in Leishmania amazonensis macrophage infection: the role of X-box binding protein 1 transcription factor. FASEB J. 2016;30:1557–1565. doi: 10.1096/fj.15-281550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold S.S., Montoya M., Unger H., Alexopoulou L., Roy P., Haswell L.E., Al-Shamkhani A., Flavell R., Borrow P., Reis e Sousa C. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- Dinarello C.A. The paradox of pro-inflammatory cytokines in cancer. Cancer Metastasis Rev. 2006;25:307–313. doi: 10.1007/s10555-006-9000-8. [DOI] [PubMed] [Google Scholar]
- Divangahi M., King I.L., Pernet E. Alveolar macrophages and type I IFN in airway homeostasis and immunity. Trends Immunol. 2015;36:307–314. doi: 10.1016/j.it.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Dolcet X., Llobet D., Pallares J., Matias-Guiu X. NF-kB in development and progression of human cancer. Virchows Arch. 2005;446:475–482. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- Duran-Aniotz C., Cornejo V.H., Espinoza S., Ardiles Á.O., Medinas D.B., Salazar C., Foley A., Gajardo I., Thielen P., Iwawaki T., Scheper W., Soto C., Palacios A.G., Hoozemans J.J.M., Hetz C. IRE1 signaling exacerbates Alzheimer's disease pathogenesis. Acta Neuropathol. 2017;134:489–506. doi: 10.1007/s00401-017-1694-x. [DOI] [PubMed] [Google Scholar]
- Durbin J.E., Fernandez-Sesma A., Lee C.K., Rao T.D., Frey A.B., Moran T.M., Vukmanovic S., García-Sastre A., Levy D.E. Type I IFN modulates innate and specific antiviral immunity. J. Immunol. 2000;164:4220–4228. doi: 10.4049/jimmunol.164.8.4220. [DOI] [PubMed] [Google Scholar]
- Duvigneau J.C., Luís A., Gorman A.M., Samali A., Kaltenecker D., Moriggl R., Kozlov A.V. Crosstalk between inflammatory mediators and endoplasmic reticulum stress in liver diseases. Cytokine. 2018;124:154577. doi: 10.1016/j.cyto.2018.10.018. [DOI] [PubMed] [Google Scholar]
- Eckard S.C., Rice G.I., Fabre A., Badens C., Gray E.E., Hartley J.L., Crow Y.J., Stetson D.B. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat. Immunol. 2014;15:839–845. doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik D.L., Colli M.L., Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat. Rev. Endocrinol. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- Elion D.L., Cook R.S. Harnessing RIG-I and intrinsic immunity in the tumor microenvironment for therapeutic cancer treatment. Oncotarget. 2018;9:29007–29017. doi: 10.18632/oncotarget.25626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., McCaul N., Chatsisvili A., Braakman I. Co- and post-translational protein folding in the ER. Traffic. 2016;17:615–638. doi: 10.1111/tra.12392. [DOI] [PubMed] [Google Scholar]
- Erbay E., Babaev V.R., Mayers J.R., Makowski L., Charles K.N., Snitow M.E., Fazio S., Wiest M.M., Watkins S.M., Linton M.F., Hotamisligil G.S. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat. Med. 2009;15:1383–1391. doi: 10.1038/nm.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornes Y., Aguileta M.A., Dubuisson C., De Keyser J., Goossens V., Kersse K., Samali A., Vandenabeele P., Bertrand M.J.M. RIPK1 promotes death receptor-independent caspase-8-mediated apoptosis under unresolved ER stress conditions. Cell Death Dis. 2014;5:e1555. doi: 10.1038/cddis.2014.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Agostinis P., Salven P., Garg A.D. Decoding cancer cell death-driven immune cell recruitment: an in vivo method for site-of-vaccination analyses. In: Galluzzi L., Rudqvist N.-P., editors. Methods in Enzymology. Elsevier; 2019. [DOI] [PubMed] [Google Scholar]
- Faraonio R., Vergara P., Di Marzo D., Pierantoni M.G., Napolitano M., Russo T., Cimino F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J. Biol. Chem. 2006;281:39776–39784. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- Feldman H.C., Maly D.J. Profiling the dual enzymatic activities of the serine/threonine kinase ire1α. Methods Mol. Biol. 2017;1513:233–242. doi: 10.1007/978-1-4939-6539-7_17. [DOI] [PubMed] [Google Scholar]
- Fey D., Croucher D.R., Kolch W., Kholodenko B.N. Crosstalk and signaling switches in mitogen-activated protein kinase cascades. Front. Physiol. 2012;3:355. doi: 10.3389/fphys.2012.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foell D., Wittkowski H., Roth J. Mechanisms of disease: a “DAMP” view of inflammatory arthritis. Nat. Clin. Pract. Rheumatol. 2007;3:382–390. doi: 10.1038/ncprheum0531. [DOI] [PubMed] [Google Scholar]
- Fransson L., Sjöholm A., Ortsäter H. Inhibition of palmitate-induced GADD34 expression augments apoptosis in mouse insulinoma cells (MIN6) Cell Biochem. Funct. 2014;32:445–452. doi: 10.1002/cbf.3036. [DOI] [PubMed] [Google Scholar]
- Fucikova J., Moserova I., Urbanova L., Bezu L., Kepp O., Cremer I., Salek C., Strnad P., Kroemer G., Galluzzi L., Spisek R. Prognostic and predictive value of DAMPs and DAMP-associated processes in cancer. Front. Immunol. 2015;6:402. doi: 10.3389/fimmu.2015.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo K.A., Johnson G.L. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat. Rev. Mol. Cell Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Buqué A., Kepp O., Zitvogel L., Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner B.M., Pincus D., Gotthardt K., Gallagher C.M., Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Agostinis P. Cell death and immunity in cancer: from danger signals to mimicry of pathogen defense responses. Immunol. Rev. 2017;280:126–148. doi: 10.1111/imr.12574. [DOI] [PubMed] [Google Scholar]
- Garg A.D., Kaczmarek A., Krysko O., Vandenabeele P., Krysko D.V., Agostinis P. ER stress-induced inflammation: does it aid or impede disease progression? Trends Mol. Med. 2012;18:589–598. doi: 10.1016/j.molmed.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Garg A.D., Krysko D.V., Verfaillie T., Kaczmarek A., Ferreira G.B., Marysael T., Rubio N., Firczuk M., Mathieu C., Roebroek A.J.M., Annaert W., Golab J., de Witte P., Vandenabeele P., Agostinis P. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J. 2012;31:1062–1079. doi: 10.1038/emboj.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Martin S., Golab J., Agostinis P. Danger signalling during cancer cell death: origins, plasticity and regulation. Cell Death Differ. 2014;21:26–38. doi: 10.1038/cdd.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Dudek-Peric A.M., Romano E., Agostinis P. Immunogenic cell death. Int. J. Dev. Biol. 2015;59:131–140. doi: 10.1387/ijdb.150061pa. [DOI] [PubMed] [Google Scholar]
- Garg A.D., Elsen S., Krysko D.V., Vandenabeele P., de Witte P., Agostinis P. Resistance to anticancer vaccination effect is controlled by a cancer cell-autonomous phenotype that disrupts immunogenic phagocytic removal. Oncotarget. 2015;6:26841–26860. doi: 10.18632/oncotarget.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Galluzzi L., Apetoh L., Baert T., Birge R.B. Molecular and translational classifications of damps in immunogenic cell death. Front. Immunol. 2015;6:588. doi: 10.3389/fimmu.2015.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Maes H., van Vliet A.R., Agostinis P. Targeting the hallmarks of cancer with therapy-induced endoplasmic reticulum (ER) stress. Mol. Cell. Oncol. 2015;2 doi: 10.4161/23723556.2014.975089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., De Ruysscher D., Agostinis P. Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: a large-scale meta-analysis. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2015.1069938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Vandenberk L., Koks C., Verschuere T., Boon L., Van Gool S.W., Agostinis P. Dendritic cell vaccines based on immunogenic cell death elicit danger signals and T cell-driven rejection of high-grade glioma. Sci. Transl. Med. 2016;8:328ra27. doi: 10.1126/scitranslmed.aae0105. [DOI] [PubMed] [Google Scholar]
- Garg A.D., Coulie P.G., Van den Eynde B.J., Agostinis P. Integrating next-generation dendritic cell vaccines into the current cancer immunotherapy landscape. Trends Immunol. 2017;38:577–593. doi: 10.1016/j.it.2017.05.006. [DOI] [PubMed] [Google Scholar]
- Garg A.D., More S., Rufo N., Mece O., Sassano M.L., Agostinis P., Zitvogel L., Kroemer G., Galluzzi L. Trial watch: immunogenic cell death induction by anticancer chemotherapeutics. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2017.1386829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Vandenberk L., Fang S., Fasche T., Van Eygen S., Maes J., Van Woensel M., Koks C., Vanthillo N., Graf N., de Witte P., Van Gool S., Salven P., Agostinis P. Pathogen response-like recruitment and activation of neutrophils by sterile immunogenic dying cells drives neutrophil-mediated residual cell killing. Cell Death Differ. 2017;24:832–843. doi: 10.1038/cdd.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Vandenberk L., Van Woensel M., Belmans J., Schaaf M., Boon L., De Vleeschouwer S., Agostinis P. Preclinical efficacy of immune-checkpoint monotherapy does not recapitulate corresponding biomarkers-based clinical predictions in glioblastoma. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2017.1295903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.D., Vara Perez M., Schaaf M., Agostinis P., Zitvogel L., Kroemer G., Galluzzi L. Trial watch: dendritic cell-based anticancer immunotherapy. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2017.1328341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glembotski C.C., Rosarda J.D., Wiseman R.L. Proteostasis and beyond: ATF6 in ischemic disease. Trends Mol. Med. 2019;25:538–550. doi: 10.1016/j.molmed.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T., Blank T., Prinz M. Fine-tuning of type I IFN-signaling in microglia—implications for homeostasis, CNS autoimmunity and interferonopathies. Curr. Opin. Neurobiol. 2016;36:38–42. doi: 10.1016/j.conb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J.A., Tyra H.M., DeZwaan-McCabe D., Olivier A.K., Rutkowski D.T. Synthetic embryonic lethality upon deletion of the ER cochaperone p58(IPK) and the ER stress sensor ATF6α. Biochem. Biophys. Res. Commun. 2014;443:115–119. doi: 10.1016/j.bbrc.2013.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen N., Sabath N., Burge C.B., Shalgi R. Widespread PERK-dependent repression of ER targets in response to ER stress. Sci. Rep. 2019;9:4330. doi: 10.1038/s41598-019-38705-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman A.M., Healy S.J.M., Jäger R., Samali A. Stress management at the ER: regulators of ER stress-induced apoptosis. Pharmacol. Ther. 2012;134:306–316. doi: 10.1016/j.pharmthera.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Grootjans J., Kaser A., Kaufman R.J., Blumberg R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016;16:469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F., Crump C.M., Thomas G. Trans-Golgi network sorting. Cell. Mol. Life Sci. 2001;58:1067–1084. doi: 10.1007/PL00000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J., Zhao B., Lyu K., Tong W., Fuchs S.Y. Downregulation of the IFNAR1 chain of type 1 interferon receptor contributes to the maintenance of the haematopoietic stem cells. Cancer Biol. Ther. 2017;18:534–543. doi: 10.1080/15384047.2017.1345395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães E.S., Gomes M.T.R., Campos P.C., Mansur D.S., Dos Santos A.A., Harms J., Splitter G., Smith J.A., Barber G.N., Oliveira S.C. Brucella abortus cyclic dinucleotides trigger STING-dependent unfolded protein response that favors bacterial replication. J. Immunol. 2019;202:2671–2681. doi: 10.4049/jimmunol.1801233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunduz F., Aboulnasr F.M., Chandra P.K., Hazari S., Poat B., Baker D.P., Balart L.A., Dash S. Free fatty acids induce ER stress and block antiviral activity of interferon alpha against hepatitis C virus in cell culture. Virol. J. 2012;9:143. doi: 10.1186/1743-422X-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Li Z. Endoplasmic reticulum stress in hepatic steatosis and inflammatory bowel diseases. Front. Genet. 2014;5:242. doi: 10.3389/fgene.2014.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Deepti A., Deegan S., Lisbona F., Hetz C., Samali A. HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1alpha-XBP1 signaling through a physical interaction. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Giricz Z., Natoni A., Donnelly N., Deegan S., Szegezdi E., Samali A. NOXA contributes to the sensitivity of PERK-deficient cells to ER stress. FEBS Lett. 2012;586:4023–4030. doi: 10.1016/j.febslet.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Guydosh N.R., Kimmig P., Walter P., Green R. Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. pombe. eLife. 2017;6 doi: 10.7554/eLife.29216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday M., Hughes D., Mallucci G.R. Fine-tuning PERK signaling for neuroprotection. J. Neurochem. 2017;142:812–826. doi: 10.1111/jnc.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J.A. GM-CSF in inflammation and autoimmunity. Trends Immunol. 2002;23:403–408. doi: 10.1016/s1471-4906(02)02260-3. [DOI] [PubMed] [Google Scholar]
- Han J., Back S.H., Hur J., Lin Y.-H., Gildersleeve R., Shan J., Yuan C.L., Krokowski D., Wang S., Hatzoglou M., Kilberg M.S., Sartor M.A., Kaufman R.J. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T., Yoshimura A. Regulation of cytokine signaling and inflammation. Cytokine Growth Factor Rev. 2002;13:413–421. doi: 10.1016/s1359-6101(02)00026-6. [DOI] [PubMed] [Google Scholar]
- Hansen G., Hercus T.R., McClure B.J., Stomski F.C., Dottore M., Powell J., Ramshaw H., Woodcock J.M., Xu Y., Guthridge M., McKinstry W.J., Lopez A.F., Parker M.W. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134:496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- Haque A., Best S.E., Montes de Oca M., James K.R., Ammerdorffer A., Edwards C.L., de Labastida Rivera F., Amante F.H., Bunn P.T., Sheel M., Sebina I., Koyama M., Varelias A., Hertzog P.J., Kalinke U., Gun S.Y., Rénia L., Ruedl C., MacDonald K.P.A., Hill G.R., Engwerda C.R. Type I IFN signaling in CD8− DCs impairs Th1-dependent malaria immunity. J. Clin. Invest. 2014;124:2483–2496. doi: 10.1172/JCI70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Hassler J., Cao S.S., Kaufman R.J. IRE1, a double-edged sword in pre-miRNA slicing and cell death. Dev. Cell. 2012;23:921–923. doi: 10.1016/j.devcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler J.R., Scheuner D.L., Wang S., Han J., Kodali V.K., Li P., Nguyen J., George J.S., Davis C., Wu S.P., Bai Y., Sartor M., Cavalcoli J., Malhi H., Baudouin G., Zhang Y., Yates J.R., Itkin-Ansari P., Volkmann N., Kaufman R.J. The ire1α/xbp1s pathway is essential for the glucose response and protection of β cells. PLoS Biol. 2015;13 doi: 10.1371/journal.pbio.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., West A.P., Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- Haynes C.M., Titus E.A., Cooper A.A. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell. 2004;15:767–776. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Hazari Y.M., Bashir A., Haq E.U., Fazili K.M. Emerging tale of UPR and cancer: an essentiality for malignancy. Tumour Biol. 2016;37:14381–14390. doi: 10.1007/s13277-016-5343-0. [DOI] [PubMed] [Google Scholar]
- He Y., Zha J., Wang Y., Liu W., Yang X., Yu P. Tissue damage-associated “danger signals” influence T-cell responses that promote the progression of preneoplasia to cancer. Cancer Res. 2013;73:629–639. doi: 10.1158/0008-5472.CAN-12-2704. [DOI] [PubMed] [Google Scholar]
- Heindryckx F., Binet F., Ponticos M., Rombouts K., Lau J., Kreuger J., Gerwins P. Endoplasmic reticulum stress enhances fibrosis through IRE1α-mediated degradation of miR-150 and XBP-1 splicing. EMBO Mol. Med. 2016;8:729–744. doi: 10.15252/emmm.201505925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L.M. The ER function BiP is a master regulator of ER function. Mt Sinai J. Med. 2004;71:289–297. [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Hetz C., Bernasconi P., Fisher J., Lee A.-H., Bassik M.C., Antonsson B., Brandt G.S., Iwakoshi N.N., Schinzel A., Glimcher L.H., Korsmeyer S.J. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- Hiller M.M., Finger A., Schweiger M., Wolf D.H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hirsch I., Weiwad M., Prell E., Ferrari D.M. ERp29 deficiency affects sensitivity to apoptosis via impairment of the ATF6-CHOP pathway of stress response. Apoptosis. 2014;19:801–815. doi: 10.1007/s10495-013-0961-0. [DOI] [PubMed] [Google Scholar]
- Hoffmann H.-H., Schneider W.M., Rice C.M. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland R., Fishbein J.C. Chemistry of the cysteine sensors in Kelch-like ECH-associated protein 1. Antioxid. Redox Signal. 2010;13:1749–1761. doi: 10.1089/ars.2010.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M.C., Poola-Kella S., Fornace A.J. Gadd34 functional domains involved in growth suppression and apoptosis. Oncogene. 2003;22:3827–3832. doi: 10.1038/sj.onc.1206567. [DOI] [PubMed] [Google Scholar]
- Hollien J., Lin J.H., Li H., Stevens N., Walter P., Weissman J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollville E., Martin S.J. Greasing the path to BAX/BAK activation. Cell. 2012;148:845–846. doi: 10.1016/j.cell.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Hollville E., Carroll R.G., Cullen S.P., Martin S.J. Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol. Cell. 2014;55:451–466. doi: 10.1016/j.molcel.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Hou W., Zhang Q., Yan Z., Chen R., Zeh Iii H.J., Kang R., Lotze M.T., Tang D. Strange attractors: DAMPs and autophagy link tumor cell death and immunity. Cell Death Dis. 2013;4:e966. doi: 10.1038/cddis.2013.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hribal M.L., Fiorentino T.V., Sesti G. Role of C reactive protein (CRP) in leptin resistance. Curr. Pharm. Des. 2014;20:609–615. doi: 10.2174/13816128113199990016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hron J.D., Peng S.L. Type I IFN protects against murine lupus. J. Immunol. 2004;173:2134–2142. doi: 10.4049/jimmunol.173.3.2134. [DOI] [PubMed] [Google Scholar]
- Hu P., Han Z., Couvillon A.D., Kaufman R.J., Exton J.H. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol. Cell. Biol. 2006;26:3071–3084. doi: 10.1128/MCB.26.8.3071-3084.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Yu X., Wang H., Zuo D., Guo C., Yi H., Tirosh B., Subjeck J.R., Qiu X., Wang X.-Y. ER stress and its regulator X-box-binding protein-1 enhance polyIC-induced innate immune response in dendritic cells. Eur. J. Immunol. 2011;41:1086–1097. doi: 10.1002/eji.201040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F., Hecker C.M., Rozenknop A., Nordmeier R.D., Rogov V., Hofmann K., Akira S., Dötsch V., Dikic I. Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. EMBO J. 2007;26:3451–3462. doi: 10.1038/sj.emboj.7601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanikia S., Sheng M., Taylor R.C. Cell non-autonomous UPRER signaling. Curr. Top. Microbiol. Immunol. 2018;414:27–43. doi: 10.1007/82_2017_38. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurescia S., Fioretti D., Rinaldi M. Targeting cytosolic nucleic acid-sensing pathways for cancer immunotherapies. Front. Immunol. 2018;9:711. doi: 10.3389/fimmu.2018.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki N., Sugiyama Y., Miyazaki S., Nakagawa H., Nishimura K., Matsuo S. An ATF4-signal-modulating machine other than GADD34 acts in ATF4-to-CHOP signaling to block CHOP expression in ER-stress-related autophagy. J. Cell. Biochem. 2015;116:1300–1309. doi: 10.1002/jcb.25085. [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Janeway’s . ninth ed. Garland Science/Taylor & Francis; New York, NY: 2016. Janeway's Immunobiology. [Google Scholar]
- Jerry Chiang W.-C., Lin J.H. The effects of IRE1, ATF6, and PERK signaling on adRP-linked rhodopsins. Adv. Exp. Med. Biol. 2014;801:661–667. doi: 10.1007/978-1-4614-3209-8_83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheng J.-R., Ho J.-Y., Horng J.-T. ER stress, autophagy, and RNA viruses. Front. Microbiol. 2014;5:388. doi: 10.3389/fmicb.2014.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John L., Thomas S., Herchenröder O., Pützer B.M., Schaefer S. Hepatitis E virus ORF2 protein activates the pro-apoptotic gene CHOP and anti-apoptotic heat shock proteins. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.G., Bui T., White C., Madesh M., Krawczyk C.M., Lindsten T., Hawkins B.J., Kubek S., Frauwirth K.A., Wang Y.L., Conway S.J., Roderick H.L., Bootman M.D., Shen H., Foskett J.K., Thompson C.B. The proapoptotic factors Bax and Bak regulate T cell proliferation through control of endoplasmic reticulum Ca(2 +) homeostasis. Immunity. 2007;27:268–280. doi: 10.1016/j.immuni.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen E., Stinson A., Shan L., Yang J., Gietl D., Albino A.P. Cigarette smoke induces endoplasmic reticulum stress and the unfolded protein response in normal and malignant human lung cells. BMC Cancer. 2008;8:229. doi: 10.1186/1471-2407-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J.-Y., Cho J.-H., Kim Y.-H., Choi S.-H., Son C.-G. A literature review for the mechanisms of stress-induced liver injury. Brain Behav. 2019;9 doi: 10.1002/brb3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Hwang C., Choi D.-Y., Park M.H., Hong J.T. NF-κB as a key mediator of brain inflammation in Alzheimer's disease. CNS Neurol. Disord. Drug Targets. 2019;18:3–10. doi: 10.2174/1871527316666170807130011. [DOI] [PubMed] [Google Scholar]
- Jwa M., Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nat. Cell Biol. 2012;14:1223–1230. doi: 10.1038/ncb2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Nomura Y. ER signaling in unfolded protein response. Life Sci. 2003;74:199–205. doi: 10.1016/j.lfs.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Kang S.-W., Hegde R.S. Lighting up the stressed ER. Cell. 2008;135:787–789. doi: 10.1016/j.cell.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali E., Bellou S., Stellas D., Klinakis A., Murphy C., Fotsis T. VEGF Signals through ATF6 and PERK to promote endothelial cell survival and angiogenesis in the absence of ER stress. Mol. Cell. 2014;54:559–572. doi: 10.1016/j.molcel.2014.03.022. [DOI] [PubMed] [Google Scholar]
- Katakura K., Lee J., Rachmilewitz D., Li G., Eckmann L., Raz E. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J. Clin. Invest. 2005;115:695–702. doi: 10.1172/JCI22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Nakajima S., Saito Y., Takahashi S., Katoh R., Kitamura M. mTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1-JNK pathway. Cell Death Differ. 2012;19:310–320. doi: 10.1038/cdd.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P.-Y., Chen S.S.-L. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepp O., Senovilla L., Vitale I., Vacchelli E., Adjemian S. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology. 2014;3 doi: 10.4161/21624011.2014.955691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo L.T., Chen L.-Y. Role of the cGAS-STING pathway in cancer development and oncotherapeutic approaches. EMBO Rep. 2018;19 doi: 10.15252/embr.201846935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.Y., Moudgil K.D. Immunomodulation of autoimmune arthritis by pro-inflammatory cytokines. Cytokine. 2017;98:87–96. doi: 10.1016/j.cyto.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Park S.M., Park J.H., Keum S.J., Jang S.K. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Sideris D.P., Sevier C.S., Kaiser C.A. Balanced Ero1 activation and inactivation establishes ER redox homeostasis. J. Cell Biol. 2012;196:713–725. doi: 10.1083/jcb.201110090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen J.H.J., Rødland G.E., Bøe C.A., Håland T.W., Sunnerhagen P., Grallert B., Boye E. Stress-induced inhibition of translation independently of eIF2α phosphorylation. J. Cell Sci. 2015;128:4420–4427. doi: 10.1242/jcs.176545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima E., Takeuchi A., Haneda M., Yagi A., Hasegawa T., Yamaki K., Takeda K., Akira S., Shimokata K., Isobe K. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB J. 2003;17:1573–1575. doi: 10.1096/fj.02-1184fje. [DOI] [PubMed] [Google Scholar]
- Kondo S., Hino S.I., Saito A., Kanemoto S., Kawasaki N., Asada R., Izumi S., Iwamoto H., Oki M., Miyagi H., Kaneko M., Nomura Y., Urano F., Imaizumi K. Activation of OASIS family, ER stress transducers, is dependent on its stabilization. Cell Death Differ. 2012;19:1939–1949. doi: 10.1038/cdd.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H., Onda A., Yanagida T. Molecular determinants of sterile inflammation. Curr. Opin. Immunol. 2014;26:147–156. doi: 10.1016/j.coi.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Korennykh A.V., Egea P.F., Korostelev A.A., Finer-Moore J., Zhang C., Shokat K.M., Stroud R.M., Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koretzky G.A., Myung P.S. Positive and negative regulation of T-cell activation by adaptor proteins. Nat. Rev. Immunol. 2001;1:95–107. doi: 10.1038/35100523. [DOI] [PubMed] [Google Scholar]
- Kranzusch P.J., Wilson S.C., Lee A.S.Y., Berger J.M., Doudna J.A., Vance R.E. Ancient origin of cGAS-STING reveals mechanism of universal 2’,3' cGAMP signaling. Mol. Cell. 2015;59:891–903. doi: 10.1016/j.molcel.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Galluzzi L., Kepp O., Zitvogel L. Immunogenic cell death in cancer therapy. Annu. Rev. Immunol. 2013;31:51–72. doi: 10.1146/annurev-immunol-032712-100008. [DOI] [PubMed] [Google Scholar]
- Krysko D.V., Garg A.D., Kaczmarek A., Krysko O., Agostinis P., Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer. 2012;12:860–875. doi: 10.1038/nrc3380. [DOI] [PubMed] [Google Scholar]
- Kubes P., Mehal W.Z. Sterile inflammation in the liver. Gastroenterology. 2012;143:1158–1172. doi: 10.1053/j.gastro.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Kurokawa K., Okamoto M., Nakano A. Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat. Commun. 2014;5:3653. doi: 10.1038/ncomms4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis J.M. Activation of the AP-1 transcription factor by inflammatory cytokines of the TNF family. Gene Expr. 1999;7:217–231. [PMC free article] [PubMed] [Google Scholar]
- Lai J.K.H., Gagalova K.K., Kuenne C., El-Brolosy M.A., Stainier D.Y.R. Induction of interferon-stimulated genes and cellular stress pathways by morpholinos in zebrafish. Dev. Biol. 2019;454:21–28. doi: 10.1016/j.ydbio.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land W.G., Agostinis P., Gasser S., Garg A.D., Linkermann A. DAMP-induced allograft and tumor rejection: the circle is closing. Am. J. Transplant. 2016;16:3322–3337. doi: 10.1111/ajt.14012. [DOI] [PubMed] [Google Scholar]
- Land W.G., Agostinis P., Gasser S., Garg A.D., Linkermann A. Transplantation and damage-associated molecular patterns (DAMPs) Am. J. Transplant. 2016;16:3338–3361. doi: 10.1111/ajt.13963. [DOI] [PubMed] [Google Scholar]
- Laskin D.L., Pendino K.J. Macrophages and inflammatory mediators in tissue injury. Annu. Rev. Pharmacol. Toxicol. 1995;35:655–677. doi: 10.1146/annurev.pa.35.040195.003255. [DOI] [PubMed] [Google Scholar]
- Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1 doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.P.K., Dey M., Neculai D., Cao C., Dever T.E., Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Penberthy K.K., Wheeler K.M., Juncadella I.J., Vandenabeele P., Lysiak J.J., Ravichandran K.S. Boosting apoptotic cell clearance by colonic epithelial cells attenuates inflammation in vivo. Immunity. 2016;44:807–820. doi: 10.1016/j.immuni.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Chiang C., Gack M.U. Endogenous nucleic acid recognition by RIG-I-like receptors and cGAS. J. Interf. Cytokine Res. 2019;39:450–458. doi: 10.1089/jir.2019.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Kirsch M.A., Wolf C., Kretschmer S., Roers A. Type I interferonopathies—an expanding disease spectrum of immunodysregulation. Semin. Immunopathol. 2015;37:349–357. doi: 10.1007/s00281-015-0500-x. [DOI] [PubMed] [Google Scholar]
- Lehotský J., Urban P., Pavlíková M., Tatarková Z., Kaminska B., Kaplán P. Molecular mechanisms leading to neuroprotection/ischemic tolerance: effect of preconditioning on the stress reaction of endoplasmic reticulum. Cell. Mol. Neurobiol. 2009;29:917–925. doi: 10.1007/s10571-009-9376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C.-Q., Zhong B., Zhang Y., Zhang J., Wang S., Shu H.-B. Glycogen synthase kinase 3β regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity. 2010;33:878–889. doi: 10.1016/j.immuni.2010.11.021. [DOI] [PubMed] [Google Scholar]
- Lemos H., Huang L., McGaha T., Mellor A.L. STING, nanoparticles, autoimmune disease and cancer: a novel paradigm for immunotherapy? Expert. Rev. Clin. Immunol. 2015;11:155–165. doi: 10.1586/1744666X.2015.995097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenna S., Han R., Trojanowska M. Endoplasmic reticulum stress and endothelial dysfunction. IUBMB Life. 2014;66:530–537. doi: 10.1002/iub.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner A.G., Upton J.-P., Praveen P.V.K., Ghosh R., Nakagawa Y., Igbaria A., Shen S., Nguyen V., Backes B.J., Heiman M., Heintz N., Greengard P., Hui S., Tang Q., Trusina A., Oakes S.A., Papa F.R. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu S.-W., Shi L., Xu C., Zhao Y., Liu B., Li Y., Shiedlin A., Xiang C., Shen H., Quinn D.A., Hales C.A., Zhao H. TLR4 through IFN-β promotes low molecular mass hyaluronan-induced neutrophil apoptosis. J. Immunol. 2011;186:556–562. doi: 10.4049/jimmunol.1001630. [DOI] [PubMed] [Google Scholar]
- Levings D.C., Wang X., Kohlhase D., Bell D.A., Slattery M. A distinct class of antioxidant response elements is consistently activated in tumors with NRF2 mutations. Redox Biol. 2018;19:235–249. doi: 10.1016/j.redox.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhomond S., Avril T., Dejeans N., Voutetakis K., Doultsinos D., McMahon M., Pineau R., Obacz J., Papadodima O., Jouan F., Bourien H., Logotheti M., Jégou G., Pallares-Lupon N., Schmit K., Le Reste P.-J., Etcheverry A., Mosser J., Barroso K., Vauléon E., Maurel M., Samali A., Patterson J.B., Pluquet O., Hetz C., Quillien V., Chatziioannou A., Chevet E. Dual IRE1 RNase functions dictate glioblastoma development. EMBO Mol. Med. 2018;10 doi: 10.15252/emmm.201707929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.-Y., Li H.-Y. Determination of CRP autoantibodies by SPR immunoassay. Methods Mol. Biol. 2019;1901:205–219. doi: 10.1007/978-1-4939-8949-2_17. [DOI] [PubMed] [Google Scholar]
- Li K., Qu S., Chen X., Wu Q., Shi M. Promising targets for cancer immunotherapy: TLRs, RLRs, and STING-mediated innate immune pathways. Int. J. Mol. Sci. 2017;18:404. doi: 10.3390/ijms18020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.H., Li H., Zhang Y., Ron D., Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D., Wootz H., Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- Ling L.E., Zafari M., Reardon D., Brickelmeier M., Goelz S.E., Benjamin C.D. Human type I interferon receptor, IFNAR, is a heavily glycosylated 120-130 kD membrane protein. J. Interf. Cytokine Res. 1995;15:55–61. doi: 10.1089/jir.1995.15.55. [DOI] [PubMed] [Google Scholar]
- Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J.H., Walter P., Reed J.C., Glimcher L.H., Hetz C. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol. Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Carvalho L.P., Bhattacharya S., Carbone C.J., Kumar K.G.S., Leu N.A., Yau P.M., Donald R.G.K., Weiss M.J., Baker D.P., McLaughlin K.J., Scott P., Fuchs S.Y. Mammalian casein kinase 1alpha and its leishmanial ortholog regulate stability of IFNAR1 and type I interferon signaling. Mol. Cell. Biol. 2009;29:6401–6412. doi: 10.1128/MCB.00478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., HuangFu W.-C., Kumar K.G.S., Qian J., Casey J.P., Hamanaka R.B., Grigoriadou C., Aldabe R., Diehl J.A., Fuchs S.Y. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lu L., Yang Z., Palaniyandi S., Zeng R., Gao L.-Y., Mosser D.M., Roopenian D.C., Zhu X. The neonatal FcR-mediated presentation of immune-complexed antigen is associated with endosomal and phagosomal pH and antigen stability in macrophages and dendritic cells. J. Immunol. 2011;186:4674–4686. doi: 10.4049/jimmunol.1003584. [DOI] [PubMed] [Google Scholar]
- Liu Y.-P., Zeng L., Tian A., Bomkamp A., Rivera D., Gutman D., Barber G.N., Olson J.K., Smith J.A. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J. Immunol. 2012;189:4630–4639. doi: 10.4049/jimmunol.1102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.-T., Grishin N.V., Chen Z.J. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu B.C., Sarhan J., Poltorak A. Host-intrinsic interferon status in infection and immunity. Trends Mol. Med. 2018;24:658–668. doi: 10.1016/j.molmed.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Zhao L., Pol J., Levesque S., Petrazzuolo A., Pfirschke C., Engblom C., Rickelt S., Yamazaki T., Iribarren K., Senovilla L., Bezu L., Vacchelli E., Sica V., Melis A., Martin T., Xia L., Yang H., Li Q., Chen J., Durand S., Aprahamian F., Lefevre D., Broutin S., Paci A., Bongers A., Minard-Colin V., Tartour E., Zitvogel L., Apetoh L., Ma Y., Pittet M.J., Kepp O., Kroemer G. Crizotinib-induced immunogenic cell death in non-small cell lung cancer. Nat. Commun. 2019;10:1486. doi: 10.1038/s41467-019-09415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue S.E., McGrath E.P., Cleary P., Greene S., Mnich K., Almanza A., Chevet E., Dwyer R.M., Oommen A., Legembre P., Godey F., Madden E.C., Leuzzi B., Obacz J., Zeng Q., Patterson J.B., Jäger R., Gorman A.M., Samali A. Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat. Commun. 2018;9:3267. doi: 10.1038/s41467-018-05763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi A., Tomer Y. Interferon alpha impairs insulin production in human beta cells via endoplasmic reticulum stress. J. Autoimmun. 2017;80:48–55. doi: 10.1016/j.jaut.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi M.P., Trumpfheller C., Idoyaga J., Caskey M., Matos I., Kluger C., Salazar A.M., Colonna M., Steinman R.M. Dendritic cells require a systemic type I interferon response to mature and induce CD4 + Th1 immunity with poly IC as adjuvant. J. Exp. Med. 2009;206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludigs K., Parfenov V., Du Pasquier R.A., Guarda G. Type I IFN-mediated regulation of IL-1 production in inflammatory disorders. Cell. Mol. Life Sci. 2012;69:3395–3418. doi: 10.1007/s00018-012-0989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Hendershot L.M. ER chaperone functions during normal and stress conditions. J. Chem. Neuroanat. 2004;28:51–65. doi: 10.1016/j.jchemneu.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Ma Y., Conforti R., Aymeric L., Locher C., Kepp O., Kroemer G., Zitvogel L. How to improve the immunogenicity of chemotherapy and radiotherapy. Cancer Metastasis Rev. 2011;30:71–82. doi: 10.1007/s10555-011-9283-2. [DOI] [PubMed] [Google Scholar]
- Ma F., Li B., Liu S., Iyer S.S., Yu Y., Wu A., Cheng G. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J. Immunol. 2015;194:1545–1554. doi: 10.4049/jimmunol.1402066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Yang W., Zhang L., Liu S., Zhao M., Zhou G., Wang L., Jin S., Zhang Z., Hu J. Interferon-alpha promotes immunosuppression through IFNAR1/STAT1 signalling in head and neck squamous cell carcinoma. Br. J. Cancer. 2019;120:317–330. doi: 10.1038/s41416-018-0352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Määttänen P., Gehring K., Bergeron J.J.M., Thomas D.Y. Protein quality control in the ER: the recognition of misfolded proteins. Semin. Cell Dev. Biol. 2010;21:500–511. doi: 10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Macián F., López-Rodríguez C., Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- Madden E., Logue S.E., Healy S.J., Manie S., Samali A. The role of the unfolded protein response in cancer progression: from oncogenesis to chemoresistance. Biol. Cell. 2019;111:1–17. doi: 10.1111/boc.201800050. [DOI] [PubMed] [Google Scholar]
- Makris S., Paulsen M., Johansson C. Type I interferons as regulators of lung inflammation. Front. Immunol. 2017;8:259. doi: 10.3389/fimmu.2017.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra J.D., Kaufman R.J. ER stress and its functional link to mitochondria: role in cell survival and death. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G., Midiri A., Biondo C., Beninati C., Zummo S., Galbo R., Tomasello F., Gambuzza M., Macrì G., Ruggeri A., Leanderson T., Teti G. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J. Immunol. 2007;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- Marciniak S.J., Garcia-Bonilla L., Hu J., Harding H.P., Ron D. Activation-dependent substrate recruitment by the eukaryotic translation initiation factor 2 kinase PERK. J. Cell Biol. 2006;172:201–209. doi: 10.1083/jcb.200508099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroqui L., Dos Santos R.S., Op de Beeck A., Coomans de Brachène A., Marselli L., Marchetti P., Eizirik D.L. Interferon-α mediates human beta cell HLA class I overexpression, endoplasmic reticulum stress and apoptosis, three hallmarks of early human type 1 diabetes. Diabetologia. 2017;60:656–667. doi: 10.1007/s00125-016-4201-3. [DOI] [PubMed] [Google Scholar]
- Martins I., Wang Y., Michaud M., Ma Y., Sukkurwala A.Q., Shen S., Kepp O., Métivier D., Galluzzi L., Perfettini J.L., Zitvogel L., Kroemer G. Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ. 2014;21:79–91. doi: 10.1038/cdd.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M., Miyazaki-Anzai S., Levi M., Ting T.C., Miyazaki M. PERK-eIF2α-ATF4-CHOP signaling contributes to TNFα-induced vascular calcification. J. Am. Heart Assoc. 2013;2 doi: 10.1161/JAHA.113.000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel M., Chevet E., Tavernier J., Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Maurel M., McGrath E.P., Mnich K., Healy S., Chevet E., Samali A. Controlling the unfolded protein response-mediated life and death decisions in cancer. Semin. Cancer Biol. 2015;33:57–66. doi: 10.1016/j.semcancer.2015.03.003. [DOI] [PubMed] [Google Scholar]
- May M.J., Ghosh S. Rel/NF-kappa B and I kappa B proteins: an overview. Semin. Cancer Biol. 1997;8:63–73. doi: 10.1006/scbi.1997.0057. [DOI] [PubMed] [Google Scholar]
- McAlpine C.S., Bowes A.J., Werstuck G.H. Diabetes, hyperglycemia and accelerated atherosclerosis: evidence supporting a role for endoplasmic reticulum (ER) stress signaling. Cardiovasc. Hematol. Disord. Drug Targets. 2010;10:151–157. doi: 10.2174/187152910791292529. [DOI] [PubMed] [Google Scholar]
- McCaffary D. STING signalling: an emerging common pathway in autoimmunity and cancer. Immunopharmacol. Immunotoxicol. 2017;39:253–258. doi: 10.1080/08923973.2017.1350704. [DOI] [PubMed] [Google Scholar]
- McDonald B., Kubes P. Innate immune cell trafficking and function during sterile inflammation of the liver. Gastroenterology. 2016;151:1087–1095. doi: 10.1053/j.gastro.2016.09.048. [DOI] [PubMed] [Google Scholar]
- McGrath E.P., Logue S.E., Mnich K., Deegan S., Jäger R., Gorman A.M., Samali A. The unfolded protein response in breast cancer. Cancers (Basel) 2018;10:344. doi: 10.3390/cancers10100344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin M.A., Eri R.D., Das I., Lourie R., Florin T.H. Intestinal secretory cell ER stress and inflammation. Biochem. Soc. Trans. 2011;39:1081–1085. doi: 10.1042/BST0391081. [DOI] [PubMed] [Google Scholar]
- McMahon M.S., Ueki Y. SH3BP2 is a critical regulator of macrophage and osteoclast response to M-CSF and RANKL stimulation. HSS J. 2009;5:49–50. doi: 10.1007/s11420-008-9091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Horng T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Menu P., Mayor A., Zhou R., Tardivel A., Ichijo H., Mori K., Tschopp J. ER stress activates the NLRP3 inflammasome via an UPR-independent pathway. Cell Death Dis. 2012;3:e261. doi: 10.1038/cddis.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerovich K., Ortis F., Allagnat F., Cardozo A.K. Endoplasmic reticulum stress and the unfolded protein response in pancreatic islet inflammation. J. Mol. Endocrinol. 2016;57:R1–R17. doi: 10.1530/JME-15-0306. [DOI] [PubMed] [Google Scholar]
- Mihailidou C., Papazian I., Papavassiliou A.G., Kiaris H. CHOP-dependent regulation of p21/waf1 during ER stress. Cell. Physiol. Biochem. 2010;25:761–766. doi: 10.1159/000315096. [DOI] [PubMed] [Google Scholar]
- Mihailidou C., Papavassiliou A.G., Kiaris H. Cell-autonomous cytotoxicity of type I interferon response via induction of endoplasmic reticulum stress. FASEB J. 2017;31:5432–5439. doi: 10.1096/fj.201700152R. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Milev N.B., Gatfield D. Circadian clocks and UPR: new twists as the story unfolds. Dev. Cell. 2018;44:7–9. doi: 10.1016/j.devcel.2017.12.018. [DOI] [PubMed] [Google Scholar]
- Mimura N., Fulciniti M., Gorgun G., Tai Y.-T., Cirstea D., Santo L., Hu Y., Fabre C., Minami J., Ohguchi H., Kiziltepe T., Ikeda H., Kawano Y., French M., Blumenthal M., Tam V., Kertesz N.L., Malyankar U.M., Hokenson M., Pham T., Zeng Q., Patterson J.B., Richardson P.G., Munshi N.C., Anderson K.C. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–5781. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Kitakaze M. ER stress in cardiovascular disease. J. Mol. Cell. Cardiol. 2010;48:1105–1110. doi: 10.1016/j.yjmcc.2009.10.026. [DOI] [PubMed] [Google Scholar]
- Mitra S., Ryoo H.D. The unfolded protein response in metazoan development. J. Cell Sci. 2019;132 doi: 10.1242/jcs.217216. jcs217216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitzel D.N., Lowry V., Shirali A.C., Liu Y., Stout-Delgado H.W. Age-enhanced endoplasmic reticulum stress contributes to increased Atg9A inhibition of STING-mediated IFN-β production during Streptococcus pneumoniae infection. J. Immunol. 2014;192:4273–4283. doi: 10.4049/jimmunol.1303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T., Neugebauer N., Putz E.M., Moritz N., Simma O., Zebedin-Brandl E., Gotthardt D., Warsch W., Eckelhart E., Kantner H.-P., Kalinke U., Lienenklaus S., Weiss S., Strobl B., Müller M., Sexl V., Stoiber D. Conditional IFNAR1 ablation reveals distinct requirements of Type I IFN signaling for NK cell maturation and tumor surveillance. Oncoimmunology. 2012;1:1027–1037. doi: 10.4161/onci.21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilenko D.A., Haas J.T., L’homme L., Fleury S., Quemener S., Levavasseur M., Becquart C., Wartelle J., Bogomolova A., Pineau L., Molendi-Coste O., Lancel S., Dehondt H., Gheeraert C., Melchior A., Dewas C., Nikitin A., Pic S., Rabhi N., Annicotte J.-S., Oyadomari S., Velasco-Hernandez T., Cammenga J., Foretz M., Viollet B., Vukovic M., Villacreces A., Kranc K., Carmeliet P., Marot G., Boulter A., Tavernier S., Berod L., Longhi M.P., Paget C., Janssens S., Staumont-Sallé D., Aksoy E., Staels B., Dombrowicz D. Metabolic and innate immune cues merge into a specific inflammatory response via the UPR. Cell. 2019;177 doi: 10.1016/j.cell.2019.03.018. 1201–1216.e19. [DOI] [PubMed] [Google Scholar]
- Mollereau B., Manié S., Napoletano F. Getting the better of ER stress. J. Cell Commun. Signal. 2014;8:311–321. doi: 10.1007/s12079-014-0251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti J., Roy S., Bozec D., Martinez J., Chapman J.R., Ueberheide B., Lamming D.W., Chen Z.J., Horng T., Yeretssian G., Green D.R., Blander J.M. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 2017;171 doi: 10.1016/j.cell.2017.09.034. 809–823.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. Divest yourself of a preconceived idea: transcription factor ATF6 is not a soluble protein! Mol. Biol. Cell. 2010;21:1435–1438. doi: 10.1091/mbc.E09-07-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S., Villalta S.A., Feldman H.C., Register A.C., Rosenthal W., Hoffmann-Petersen I.T., Mehdizadeh M., Ghosh R., Wang L., Colon-Negron K., Meza-Acevedo R., Backes B.J., Maly D.J., Bluestone J.A., Papa F.R. Targeting ABL-IRE1α signaling spares ER-stressed pancreatic β cells to reverse autoimmune diabetes. Cell Metab. 2017;25 doi: 10.1016/j.cmet.2017.04.026. 883–897.e8. [DOI] [PubMed] [Google Scholar]
- Morito D., Nagata K. ER stress proteins in autoimmune and inflammatory diseases. Front. Immunol. 2012;3:48. doi: 10.3389/fimmu.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moserova I., Truxova I., Garg A.D., Tomala J., Agostinis P., Cartron P.F., Vosahlikova S., Kovar M., Spisek R., Fucikova J. Caspase-2 and oxidative stress underlie the immunogenic potential of high hydrostatic pressure-induced cancer cell death. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2016.1258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynagh P.N. TLR signalling and activation of IRFs: revisiting old friends from the NF-kappaB pathway. Trends Immunol. 2005;26:469–476. doi: 10.1016/j.it.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Mozzini C., Cominacini L., Garbin U., Fratta Pasini A.M. Endoplasmic reticulum stress, NRF2 signalling and cardiovascular diseases in a nutshell. Curr. Atheroscler. Rep. 2017;19:33. doi: 10.1007/s11883-017-0669-7. [DOI] [PubMed] [Google Scholar]
- Mussbacher M., Salzmann M., Brostjan C., Hoesel B., Schoergenhofer C., Datler H., Hohensinner P., Basílio J., Petzelbauer P., Assinger A., Schmid J.A. Cell type-specific roles of NF-κB linking inflammation and thrombosis. Front. Immunol. 2019;10:85. doi: 10.3389/fimmu.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N. ER and aging-protein folding and the ER stress response. Ageing Res. Rev. 2009;8:150–159. doi: 10.1016/j.arr.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Nardelli B., Zaritskaya L., Semenuk M., Cho Y.H., LaFleur D.W., Shah D., Ullrich S., Girolomoni G., Albanesi C., Moore P.A. Regulatory effect of IFN-kappa, a novel type I IFN, on cytokine production by cells of the innate immune system. J. Immunol. 2002;169:4822–4830. doi: 10.4049/jimmunol.169.9.4822. [DOI] [PubMed] [Google Scholar]
- Natoli G., Ostuni R. Adaptation and memory in immune responses. Nat. Immunol. 2019;20:783–792. doi: 10.1038/s41590-019-0399-9. [DOI] [PubMed] [Google Scholar]
- Negishi H., Taniguchi T., Yanai H. The interferon (IFN) class of cytokines and the IFN regulatory factor (IRF) transcription factor family. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls S., Snapp E.L., Cole N.B., Zaal K.J., Kenworthy A.K., Roberts T.H., Ellenberg J., Presley J.F., Siggia E., Lippincott-Schwartz J. Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- Novack D.V. Role of NF-κB in the skeleton. Cell Res. 2011;21:169–182. doi: 10.1038/cr.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I., Zeng H., Harding H.P., Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J.J., Murray P.J. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U., Yamakawa N., Akashi-Takamura S., Miyake K., Shimizu T. Structural analyses of human Toll-like receptor 4 polymorphisms D299G and T399I. J. Biol. Chem. 2012;287:40611–40617. doi: 10.1074/jbc.M112.404608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka O.B., van Lith M., Rudolf J., Tungkum W., Pringle M.A., Bulleid N.J. ERp18 regulates activation of ATF6α during unfolded protein response. EMBO J. 2019;38 doi: 10.15252/embj.2018100990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M., Tanakura S., Uemura A., Sohda M., Misumi Y., Taniguchi M., Wakabayashi S., Yoshida H. Novel cis-acting element GASE regulates transcriptional induction by the Golgi stress response. Cell Struct. Funct. 2011;36:1–12. doi: 10.1247/csf.10014. [DOI] [PubMed] [Google Scholar]
- Oliveira S.J., de Sousa M., Pinto J.P. ER stress and iron homeostasis: a new frontier for the UPR. Biochem. Res. Int. 2011;2011:896474. doi: 10.1155/2011/896474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik J.K., Kim O.Y., Koh S.J., Jang Y., Chae J.S., Kim J.Y., Kim H.J., Hyun Y.J., Cho J.R., Lee J.H. Additive effect of interleukin-6 and C-reactive protein (CRP) single nucleotide polymorphism on serum CRP concentration and other cardiovascular risk factors. Clin. Chim. Acta. 2007;380:68–74. doi: 10.1016/j.cca.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Palucka A.K., Blanck J.-P., Bennett L., Pascual V., Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3372–3377. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretakis T., Kepp O., Brockmeier U., Tesniere A., Bjorklund A.-C., Chapman D.C., Durchschlag M., Joza N., Pierron G., van Endert P., Yuan J., Zitvogel L., Madeo F., Williams D.B., Kroemer G. Mechanisms of pre-apoptotic calreticulin exposure in immunogenic cell death. EMBO J. 2009;28:578–590. doi: 10.1038/emboj.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou A., Higa A., Jégou G., Jouan F., Pineau R., Saas L., Avril T., Pluquet O., Chevet E. Alterations of EDEM1 functions enhance ATF6 pro-survival signaling. FEBS J. 2018;285:4146–4164. doi: 10.1111/febs.14669. [DOI] [PubMed] [Google Scholar]
- Parker B.S., Rautela J., Hertzog P.J. Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat. Rev. Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- Pedersen E.B., Nielsen J.T., Nielsen C., Filichev V.V. Enhanced anti-HIV-1 activity of G-quadruplexes comprising locked nucleic acids and intercalating nucleic acids. Nucleic Acids Res. 2011;39:2470–2481. doi: 10.1093/nar/gkq1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini E., Vitale A. Protein biosynthesis and maturation in the ER. Methods Mol. Biol. 2018;1691:179–189. doi: 10.1007/978-1-4939-7389-7_14. [DOI] [PubMed] [Google Scholar]
- Peña-Blanco A., García-Sáez A.J. Bax, Bak and beyond—mitochondrial performance in apoptosis. FEBS J. 2018;285:416–431. doi: 10.1111/febs.14186. [DOI] [PubMed] [Google Scholar]
- Perego J., Mendes A., Bourbon C., Camosseto V., Combes A., Liu H., Manh T.-P.V., Dalet A., Chasson L., Spinelli L., Bardin N., Chiche L., Santos M.A.S., Gatti E., Pierre P. Guanabenz inhibits TLR9 signaling through a pathway that is independent of eIF2α dephosphorylation by the GADD34/PP1c complex. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aam8104. eaam8104. [DOI] [PubMed] [Google Scholar]
- Perkins D.J., Vogel S.N. Space and time: new considerations about the relationship between Toll-like receptors (TLRs) and type I interferons (IFNs) Cytokine. 2015;74:171–174. doi: 10.1016/j.cyto.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J., Iracheta-Vellve A., Csak T., Satishchandran A., Kodys K., Kurt-Jones E.A., Fitzgerald K.A., Szabo G. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer L.M. The role of nuclear factor κB in the interferon response. J. Interf. Cytokine Res. 2011;31:553–559. doi: 10.1089/jir.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer L.M., Mullersman J.E., Pfeffer S.R., Murti A., Shi W., Yang C.H. STAT3 as an adapter to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science. 1997;276:1418–1420. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- Pihán P., Carreras-Sureda A., Hetz C. BCL-2 family: integrating stress responses at the ER to control cell demise. Cell Death Differ. 2017;24:1478–1487. doi: 10.1038/cdd.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D., Chevalier M.W., Aragón T., van Anken E., Vidal S.E., El-Samad H., Walter P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S., Ron D. Early events in the endoplasmic reticulum unfolded protein response. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prischi F., Nowak P.R., Carrara M., Ali M.M.U. Phosphoregulation of Ire1 RNase splicing activity. Nat. Commun. 2014;5:3554. doi: 10.1038/ncomms4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pytel D., Majsterek I., Diehl J.A. Tumor progression and the different faces of the PERK kinase. Oncogene. 2016;35:1207–1215. doi: 10.1038/onc.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q., Sun C., Han C., Han N., Zhang M., Li G. Endoplasmic reticulum stress pathway PERK-eIF2α confers radioresistance in oropharyngeal carcinoma by activating NF-κB. Cancer Sci. 2017;108:1421–1431. doi: 10.1111/cas.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q., Zheng Z., Chang L., Zhao Y.-S., Tan C., Dandekar A., Zhang Z., Lin Z., Gui M., Li X., Zhang T., Kong Q., Li H., Chen S., Chen A., Kaufman R.J., Yang W.-L., Lin H.-K., Zhang D., Perlman H., Thorp E., Zhang K., Fang D. Toll-like receptor-mediated IRE1α activation as a therapeutic target for inflammatory arthritis. EMBO J. 2013;32:2477–2490. doi: 10.1038/emboj.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragimbeau J., Dondi E., Alcover A., Eid P., Uzé G., Pellegrini S. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 2003;22:537–547. doi: 10.1093/emboj/cdg038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmati M., Moosavi M.A., McDermott M.F. ER stress: a therapeutic target in rheumatoid arthritis? Trends Pharmacol. Sci. 2018;39:610–623. doi: 10.1016/j.tips.2018.03.010. [DOI] [PubMed] [Google Scholar]
- Ramana C.V., Gil M.P., Schreiber R.D., Stark G.R. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- Ramanjulu J.M., Pesiridis G.S., Yang J., Concha N., Singhaus R., Zhang S.-Y., Tran J.-L., Moore P., Lehmann S., Eberl H.C., Muelbaier M., Schneck J.L., Clemens J., Adam M., Mehlmann J., Romano J., Morales A., Kang J., Leister L., Graybill T.L., Charnley A.K., Ye G., Nevins N., Behnia K., Wolf A.I., Kasparcova V., Nurse K., Wang L., Puhl A.C., Li Y., Klein M., Hopson C.B., Guss J., Bantscheff M., Bergamini G., Reilly M.A., Lian Y., Duffy K.J., Adams J., Foley K.P., Gough P.J., Marquis R.W., Smothers J., Hoos A., Bertin J. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature. 2018;564:439–443. doi: 10.1038/s41586-018-0705-y. [DOI] [PubMed] [Google Scholar]
- Rauch I., Müller M., Decker T. The regulation of inflammation by interferons and their STATs. JAKSTAT. 2013;2 doi: 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran K.S. Beginnings of a good apoptotic meal: the find-me and eat-me signaling pathways. Immunity. 2011;35:445–455. doi: 10.1016/j.immuni.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reefman E., Kay J.G., Wood S.M., Offenhäuser C., Brown D.L., Roy S., Stanley A.C., Low P.C., Manderson A.P., Stow J.L. Cytokine secretion is distinct from secretion of cytotoxic granules in NK cells. J. Immunol. 2010;184:4852–4862. doi: 10.4049/jimmunol.0803954. [DOI] [PubMed] [Google Scholar]
- Reverendo M., Mendes A., Argüello R.J., Gatti E., Pierre P. At the crossway of ER-stress and proinflammatory responses. FEBS J. 2019;286:297–310. doi: 10.1111/febs.14391. [DOI] [PubMed] [Google Scholar]
- Rizzo R., Bortolotti D., Bolzani S., Fainardi E. HLA-G molecules in autoimmune diseases and infections. Front. Immunol. 2014;5:592. doi: 10.3389/fimmu.2014.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K.L., Kono H. The inflammatory response to cell death. Annu. Rev. Pathol. 2008;3:99–126. doi: 10.1146/annurev.pathmechdis.3.121806.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez D., Rojas-Rivera D., Hetz C. Integrating stress signals at the endoplasmic reticulum: the BCL-2 protein family rheostat. Biochim. Biophys. Acta. 2011;1813:564–574. doi: 10.1016/j.bbamcr.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Rodvold J.J., Mahadevan N.R., Zanetti M. Immune modulation by ER stress and inflammation in the tumor microenvironment. Cancer Lett. 2016;380:227–236. doi: 10.1016/j.canlet.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Roers A., Hiller B., Hornung V. Recognition of endogenous nucleic acids by the innate immune system. Immunity. 2016;44:739–754. doi: 10.1016/j.immuni.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Rojas M., Vasconcelos G., Dever T.E. An eIF2α-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E3466–E3475. doi: 10.1073/pnas.1501557112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rondas D., Crèvecoeur I., D’Hertog W., Ferreira G.B., Staes A., Garg A.D., Eizirik D.L., Agostinis P., Gevaert K., Overbergh L., Mathieu C. Citrullinated glucose-regulated protein 78 is an autoantigen in type 1 diabetes. Diabetes. 2015;64:573–586. doi: 10.2337/db14-0621. [DOI] [PubMed] [Google Scholar]
- Rozpedek W., Pytel D., Mucha B., Leszczynska H., Diehl J.A., Majsterek I. The role of the perk/eif2α/atf4/chop signaling pathway in tumor progression during endoplasmic reticulum stress. Curr. Mol. Med. 2016;16:533–544. doi: 10.2174/1566524016666160523143937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubartelli A., Lotze M.T. Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 2007;28:429–436. doi: 10.1016/j.it.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Rubio C., Pincus D., Korennykh A., Schuck S., El-Samad H., Walter P. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J. Cell Biol. 2011;193:171–184. doi: 10.1083/jcb.201007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Patiño C., Bossowski J.P., Chevet E., Ricci J.-E. Reshaping the immune tumor microenvironment through IRE1 signaling. Trends Mol. Med. 2018;24:607–614. doi: 10.1016/j.molmed.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Rufo N., Garg A.D., Agostinis P. The unfolded protein response in immunogenic cell death and cancer immunotherapy. Trends Cancer. 2017;3:643–658. doi: 10.1016/j.trecan.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Ruggiano A., Foresti O., Carvalho P. ER-associated degradation: protein quality control and beyond. J. Cell Biol. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D.T., Kaufman R.J. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem. Sci. 2007;32:469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Rutkowski D.T., Arnold S.M., Miller C.N., Wu J., Li J., Gunnison K.M., Mori K., Sadighi Akha A.A., Raden D., Kaufman R.J. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Gale M. Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J. Exp. Med. 2008;205:1523–1527. doi: 10.1084/jem.20081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Ochiai K., Kondo S., Tsumagari K., Murakami T., Cavener D.R., Imaizumi K. Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J. Biol. Chem. 2011;286:4809–4818. doi: 10.1074/jbc.M110.152900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg Y., Coleman A.J., Celestrin K., Cohen-Berkman M., Biederer T., Henis-Korenblit S., Bülow H.E. Reduced insulin/insulin-like growth factor receptor signaling mitigates defective dendrite morphogenesis in mutants of the ER stress sensor IRE-1. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano R., Reed J.C. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Hata N., Asagiri M., Nakaya T., Taniguchi T., Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- Schindler A.J., Schekman R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17775–17780. doi: 10.1073/pnas.0910342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser H., Bekisz J., Zoon K.C. New function of type I IFN: induction of autophagy. J. Interf. Cytokine Res. 2014;34:71–78. doi: 10.1089/jir.2013.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W. Interferon-stimulated genes: roles in viral pathogenesis. Curr. Opin. Virol. 2014;6:40–46. doi: 10.1016/j.coviro.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W. Recent advances in antiviral interferon-stimulated gene biology. F1000Res. 2018;7:309. doi: 10.12688/f1000research.12450.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M., Kaufman R.J. ER stress and the unfolded protein response. Mutat. Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Scott O., Roifman C.M. NF-κB pathway and the Goldilocks principle: lessons from human disorders of immunity and inflammation. J. Allergy Clin. Immunol. 2019;143:1688–1701. doi: 10.1016/j.jaci.2019.03.016. [DOI] [PubMed] [Google Scholar]
- Scull C.M., Tabas I. Mechanisms of ER stress-induced apoptosis in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2011;31:2792–2797. doi: 10.1161/ATVBAHA.111.224881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft D., Ronai Z.A. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 2015;40:141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong S.-Y., Matzinger P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 2004;4:469–478. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
- Sepulveda D., Rojas-Rivera D., Rodríguez D.A., Groenendyk J., Köhler A., Lebeaupin C., Ito S., Urra H., Carreras-Sureda A., Hazari Y., Vasseur-Cognet M., Ali M.M.U., Chevet E., Campos G., Godoy P., Vaisar T., Bailly-Maitre B., Nagata K., Michalak M., Sierralta J., Hetz C. Interactome screening identifies the ER luminal chaperone hsp47 as a regulator of the unfolded protein response transducer ire1α. Mol. Cell. 2018;69 doi: 10.1016/j.molcel.2017.12.028. 238–252.e7. [DOI] [PubMed] [Google Scholar]
- Sevier C.S., Kaiser C.A. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim. Biophys. Acta. 2008;1783:549–556. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Sevier C.S., Qu H., Heldman N., Gross E., Fass D., Kaiser C.A. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell. 2007;129:333–344. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Shah A., Vaidya N.K., Bhat H.K., Kumar A. HIV-1 gp120 induces type-1 programmed cell death through ER stress employing IRE1α, JNK and AP-1 pathway. Sci. Rep. 2016;6:18929. doi: 10.1038/srep18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E., Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Shen J., Prywes R. ER stress signaling by regulated proteolysis of ATF6. Methods. 2005;35:382–389. doi: 10.1016/j.ymeth.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Shen J., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shi W.-Y., Cao C., Liu L. Interferon α induces the apoptosis of cervical cancer HeLa cells by activating both the intrinsic mitochondrial pathway and endoplasmic reticulum stress-induced pathway. Int. J. Mol. Sci. 2016;17:1832. doi: 10.3390/ijms17111832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinjo S., Mizotani Y., Tashiro E., Imoto M. Comparative analysis of the expression patterns of UPR-target genes caused by UPR-inducing compounds. Biosci. Biotechnol. Biochem. 2013;77:729–735. doi: 10.1271/bbb.120812. [DOI] [PubMed] [Google Scholar]
- Short B. Sorting out endosome form and function. J. Cell Biol. 2015;210:870. doi: 10.1083/jcb.2106fta. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders M.D., Ryno L.M., Genereux J.C., Moresco J.J., Tu P.G., Wu C., Yates J.R., Su A.I., Kelly J.W., Wiseman R.L. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaats J., Ten Oever J., van de Veerdonk F.L., Netea M.G. IL-1β/IL-6/CRP and IL-18/ferritin: distinct inflammatory programs in infections. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirle J., Au C.E., Jain M., Dejgaard K., Nilsson T., Bergeron J. Cell biology of the endoplasmic reticulum and the Golgi apparatus through proteomics. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.A. A new paradigm: innate immune sensing of viruses via the unfolded protein response. Front. Microbiol. 2014;5:222. doi: 10.3389/fmicb.2014.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.A., Turner M.J., DeLay M.L., Klenk E.I., Sowders D.P., Colbert R.A. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur. J. Immunol. 2008;38:1194–1203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So J.-S., Cho S., Min S.-H., Kimball S.R., Lee A.-H. IRE1α-dependent decay of CReP/Ppp1r15b mRNA increases eukaryotic initiation factor 2α phosphorylation and suppresses protein synthesis. Mol. Cell. Biol. 2015;35:2761–2770. doi: 10.1128/MCB.00215-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T., Jarosch E. BiP binding keeps ATF6 at bay. Dev. Cell. 2002;3:1–2. doi: 10.1016/s1534-5807(02)00210-1. [DOI] [PubMed] [Google Scholar]
- Sommereyns C., Michiels T. N-glycosylation of murine IFN-beta in a putative receptor-binding region. J. Interf. Cytokine Res. 2006;26:406–413. doi: 10.1089/jir.2006.26.406. [DOI] [PubMed] [Google Scholar]
- Song B., Scheuner D., Ron D., Pennathur S., Kaufman R.J. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 2008;118:3378–3389. doi: 10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Chi M., Luo X., Song Q., Xia D., Shi B., Han J. Non-structural protein 2B of human rhinovirus 16 activates both PERK and ATF6 rather than IRE1 to trigger ER stress. Viruses. 2019;11:133. doi: 10.3390/v11020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovolyova N., Healy S., Samali A., Logue S.E. Stressed to death—mechanisms of ER stress-induced cell death. Biol. Chem. 2014;395:1–13. doi: 10.1515/hsz-2013-0174. [DOI] [PubMed] [Google Scholar]
- Sprooten J., Agostinis P., Garg A.D. Type I interferons and dendritic cells in cancer immunotherapy. In: Galluzzi L., Lhuillier C., editors. International Review of Cell and Molecular Biology. Elsevier; 2019. [DOI] [PubMed] [Google Scholar]
- Srikanth S., Woo J.S., Wu B., El-Sherbiny Y.M., Leung J., Chupradit K., Rice L., Seo G.J., Calmettes G., Ramakrishna C., Cantin E., An D.S., Sun R., Wu T.-T., Jung J.U., Savic S., Gwack Y. The Ca2 + sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 2019;20:152–162. doi: 10.1038/s41590-018-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman L. Shifting therapeutic attention in MS to osteopontin, type 1 and type 2 IFN. Eur. J. Immunol. 2009;39:2358–2360. doi: 10.1002/eji.200939814. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Stow J.L., Murray R.Z. Intracellular trafficking and secretion of inflammatory cytokines. Cytokine Growth Factor Rev. 2013;24:227–239. doi: 10.1016/j.cytogfr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Sun S.-C. The noncanonical NF-κB pathway. Immunol. Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Chin Y.E., Zhang D.D. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol. Cell. Biol. 2009;29:2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram A., Appathurai S., Plumb R., Mariappan M. Dynamic changes in complexes of IRE1α, PERK, and ATF6α during endoplasmic reticulum stress. Mol. Biol. Cell. 2018;29:1376–1388. doi: 10.1091/mbc.E17-10-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes A., Edwards M.R., Macintyre J., Del Rosario A., Gielen V., Haas J., Kon O.M., McHale M., Johnston S.L. TLR3, TLR4 and TLRs7-9 induced interferons are not impaired in airway and blood cells in well controlled asthma. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegezdi E., Logue S.E., Gorman A.M., Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegezdi E., Macdonald D.C., Ní Chonghaile T., Gupta S., Samali A. Bcl-2 family on guard at the ER. Am. J. Phys. Cell Physiol. 2009;296:C941–C953. doi: 10.1152/ajpcell.00612.2008. [DOI] [PubMed] [Google Scholar]
- Taft J., Bogunovic D. The Goldilocks zone of type I IFNs: lessons from human genetics. J. Immunol. 2018;201:3479–3485. doi: 10.4049/jimmunol.1800764. [DOI] [PubMed] [Google Scholar]
- Tailor P., Tamura T., Ozato K. IRF family proteins and type I interferon induction in dendritic cells. Cell Res. 2006;16:134–140. doi: 10.1038/sj.cr.7310018. [DOI] [PubMed] [Google Scholar]
- Takayanagi S., Fukuda R., Takeuchi Y., Tsukada S., Yoshida K. Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress. Cell Stress Chaperones. 2013;18:11–23. doi: 10.1007/s12192-012-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Nagashima S., Shiiba I., Uda A., Tokuyama T., Ito N., Fukuda T., Matsushita N., Ishido S., Iwawaki T., Uehara T., Inatome R., Yanagi S. MITOL prevents ER stress-induced apoptosis by IRE1α ubiquitylation at ER-mitochondria contact sites. EMBO J. 2019;38 doi: 10.15252/embj.2018100999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A.B., Mercado E.L., Hoffmann A., Niwa M. ER stress activates NF-κB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A.B., Roberts L.S., Chandra V., Rivera I.G., Nomura D.K., Forbes D.J., Niwa M. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev. Cell. 2018;46 doi: 10.1016/j.devcel.2018.04.023. 327–343.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Sun L., Chen J., Chen Z.J. Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 2018;72:447–478. doi: 10.1146/annurev-micro-102215-095605. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Chen Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Kang R., Coyne C.B., Zeh H.J., Lotze M.T. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol. Rev. 2012;249:158–175. doi: 10.1111/j.1600-065X.2012.01146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.-H.A., Ranatunga S., Kriss C.L., Cubitt C.L., Tao J., Pinilla-Ibarz J.A., Del Valle J.R., Hu C.-C.A. Inhibition of ER stress-associated IRE-1/XBP-1 pathway reduces leukemic cell survival. J. Clin. Invest. 2014;124:2585–2598. doi: 10.1172/JCI73448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif K.D., Waris G., Siddiqui A. Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 2005;13:159–163. doi: 10.1016/j.tim.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Tatematsu M., Funami K., Seya T., Matsumoto M. Extracellular RNA sensing by pattern recognition receptors. J. Innate Immun. 2018;10:398–406. doi: 10.1159/000494034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernier S.J., Osorio F., Vandersarren L., Vetters J., Vanlangenakker N., Van Isterdael G., Vergote K., De Rycke R., Parthoens E., van de Laar L., Iwawaki T., Del Valle J.R., Hu C.-C.A., Lambrecht B.N., Janssens S. Regulated IRE1-dependent mRNA decay sets the threshold for dendritic cell survival. Nat. Cell Biol. 2017;19:698–710. doi: 10.1038/ncb3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay K.H., Luan Q., Croft A., Jiang C.C., Jin L., Zhang X.D., Tseng H.-Y. Sustained IRE1 and ATF6 signaling is important for survival of melanoma cells undergoing ER stress. Cell. Signal. 2014;26:287–294. doi: 10.1016/j.cellsig.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Teijaro J.R., Studer S., Leaf N., Kiosses W.B., Nguyen N., Matsuki K., Negishi H., Taniguchi T., Oldstone M.B.A., Rosen H. S1PR1-mediated IFNAR1 degradation modulates plasmacytoid dendritic cell interferon-α autoamplification. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1351–1356. doi: 10.1073/pnas.1525356113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenoever B.R., Ng S.-L., Chua M.A., McWhirter S.M., García-Sastre A., Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- Teske B.F., Wek S.A., Bunpo P., Cundiff J.K., McClintick J.N., Anthony T.G., Wek R.C. The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol. Biol. Cell. 2011;22:4390–4405. doi: 10.1091/mbc.E11-06-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske B.F., Fusakio M.E., Zhou D., Shan J., McClintick J.N., Kilberg M.S., Wek R.C. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol. Biol. Cell. 2013;24:2477–2490. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesniere A., Panaretakis T., Kepp O., Apetoh L., Ghiringhelli F., Zitvogel L., Kroemer G. Molecular characteristics of immunogenic cancer cell death. Cell Death Differ. 2008;15:3–12. doi: 10.1038/sj.cdd.4402269. [DOI] [PubMed] [Google Scholar]
- Thakor N., Holcik M. IRES-mediated translation of cellular messenger RNA operates in eIF2α-independent manner during stress. Nucleic Acids Res. 2012;40:541–552. doi: 10.1093/nar/gkr701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R.S., Tymms M.J., McKinlay L.H., Shannon M.F., Seth A., Kola I. ETS1, NFkappaB and AP1 synergistically transactivate the human GM-CSF promoter. Oncogene. 1997;14:2845–2855. doi: 10.1038/sj.onc.1201125. [DOI] [PubMed] [Google Scholar]
- Thomas S.E., Malzer E., Ordóñez A., Dalton L.E., van ’t Wout E.F.A., Liniker E., Crowther D.C., Lomas D.A., Marciniak S.J. p53 and translation attenuation regulate distinct cell cycle checkpoints during endoplasmic reticulum (ER) stress. J. Biol. Chem. 2013;288:7606–7617. doi: 10.1074/jbc.M112.424655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J.V., Locarnini S.A. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 2007;85:435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- Thompson M.R., Kaminski J.J., Kurt-Jones E.A., Fitzgerald K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H., Dinarello C.A., Mier J.W. IL-6 and APPs: anti-inflammatory and immunosuppressive mediators. Immunol. Today. 1997;18:428–432. doi: 10.1016/s0167-5699(97)01103-1. [DOI] [PubMed] [Google Scholar]
- Todd D.J., Lee A.-H., Glimcher L.H. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat. Rev. Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- Tsuru A., Imai Y., Saito M., Kohno K. Novel mechanism of enhancing IRE1α-XBP1 signalling via the PERK-ATF4 pathway. Sci. Rep. 2016;6:24217. doi: 10.1038/srep24217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M.D., Nedjai B., Hurst T., Pennington D.J. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta. 2014;1843:2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- Uccellini M.B., García-Sastre A. ISRE-reporter mouse reveals high basal and induced type I IFN responses in inflammatory monocytes. Cell Rep. 2018;25 doi: 10.1016/j.celrep.2018.11.030. 2784–2796.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uluçkan Ö., Guinea-Viniegra J., Jimenez M., Wagner E.F. Signalling in inflammatory skin disease by AP-1 (Fos/Jun) Clin. Exp. Rheumatol. 2015;33:S44–S49. [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Urra H., Hetz C. A novel ER stress-independent function of the UPR in angiogenesis. Mol. Cell. 2014;54:542–544. doi: 10.1016/j.molcel.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Urra H., Dufey E., Lisbona F., Rojas-Rivera D., Hetz C. When ER stress reaches a dead end. Biochim. Biophys. Acta. 2013;1833:3507–3517. doi: 10.1016/j.bbamcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Uruno A., Yagishita Y., Yamamoto M. The Keap1-Nrf2 system and diabetes mellitus. Arch. Biochem. Biophys. 2015;566:76–84. doi: 10.1016/j.abb.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Uzé G., Schreiber G., Piehler J., Pellegrini S. The receptor of the type I interferon family. Curr. Top. Microbiol. Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- van Dam H., Castellazzi M. Distinct roles of Jun : Fos and Jun : ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- van de Veerdonk F.L., Kullberg B.J., van der Meer J.W.M., Gow N.A.R., Netea M.G. Host-microbe interactions: innate pattern recognition of fungal pathogens. Curr. Opin. Microbiol. 2008;11:305–312. doi: 10.1016/j.mib.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Van Der Kelen K., Beyaert R., Inzé D., De Veylder L. Translational control of eukaryotic gene expression. Crit. Rev. Biochem. Mol. Biol. 2009;44:143–168. doi: 10.1080/10409230902882090. [DOI] [PubMed] [Google Scholar]
- van der Vorst E.P.C., Peters L.J.F., Müller M., Gencer S., Yan Y., Weber C., Döring Y. G-protein coupled receptor targeting on myeloid cells in atherosclerosis. Front. Pharmacol. 2019;10:531. doi: 10.3389/fphar.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpouille-Box C., Demaria S., Formenti S.C., Galluzzi L. Cytosolic DNA sensing in organismal tumor control. Cancer Cell. 2018;34:361–378. doi: 10.1016/j.ccell.2018.05.013. [DOI] [PubMed] [Google Scholar]
- Vénéreau E., Ceriotti C., Bianchi M.E. DAMPs from cell death to new life. Front. Immunol. 2015;6:422. doi: 10.3389/fimmu.2015.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh D., Ernandez T., Rosetti F., Batal I., Cullere X., Luscinskas F.W., Zhang Y., Stavrakis G., García-Cardeña G., Horwitz B.H., Mayadas T.N. Endothelial TNF receptor 2 induces IRF1 transcription factor-dependent interferon-β autocrine signaling to promote monocyte recruitment. Immunity. 2013;38:1025–1037. doi: 10.1016/j.immuni.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verfaillie T., Rubio N., Garg A.D., Bultynck G., Rizzuto R., Decuypere J.P., Piette J., Linehan C., Gupta S., Samali A., Agostinis P. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012;19:1880–1891. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verfaillie T., Garg A.D., Agostinis P. Targeting ER stress induced apoptosis and inflammation in cancer. Cancer Lett. 2013;332:249–264. doi: 10.1016/j.canlet.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Vesely P.W., Staber P.B., Hoefler G., Kenner L. Translational regulation mechanisms of AP-1 proteins. Mutat. Res. 2009;682:7–12. doi: 10.1016/j.mrrev.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Wagner E.F. Bone development and inflammatory disease is regulated by AP-1 (Fos/Jun) Ann. Rheum. Dis. 2010;69(Suppl. 1):i86–i88. doi: 10.1136/ard.2009.119396. [DOI] [PubMed] [Google Scholar]
- Walker J., Tough D.F. Modification of TLR-induced activation of human dendritic cells by type I IFN: synergistic interaction with TLR4 but not TLR3 agonists. Eur. J. Immunol. 2006;36:1827–1836. doi: 10.1002/eji.200635854. [DOI] [PubMed] [Google Scholar]
- Wallweber H.J.A., Tam C., Franke Y., Starovasnik M.A., Lupardus P.J. Structural basis of recognition of interferon-α receptor by tyrosine kinase 2. Nat. Struct. Mol. Biol. 2014;21:443–448. doi: 10.1038/nsmb.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D.M., Minogue A.M., Sala Frigerio C., Fadeeva J.V., Wasco W., Selkoe D.J. The APP family of proteins: similarities and differences. Biochem. Soc. Trans. 2007;35:416–420. doi: 10.1042/BST0350416. [DOI] [PubMed] [Google Scholar]
- Walter F., Schmid J., Düssmann H., Concannon C.G., Prehn J.H.M. Imaging of single cell responses to ER stress indicates that the relative dynamics of IRE1/XBP1 and PERK/ATF4 signalling rather than a switch between signalling branches determine cell survival. Cell Death Differ. 2015;22:1502–1516. doi: 10.1038/cdd.2014.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters B., Thompson S.R. Cap-independent translational control of carcinogenesis. Front. Oncol. 2016;6:128. doi: 10.3389/fonc.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Kaufman R.J. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer. 2014;14:581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- Wang X., Olberding K.E., White C., Li C. Bcl-2 proteins regulate ER membrane permeability to luminal proteins during ER stress-induced apoptosis. Cell Death Differ. 2011;18:38–47. doi: 10.1038/cdd.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Alam G.N., Ning Y., Visioli F., Dong Z., Nör J.E., Polverini P.J. The unfolded protein response induces the angiogenic switch in human tumor cells through the PERK/ATF4 pathway. Cancer Res. 2012;72:5396–5406. doi: 10.1158/0008-5472.CAN-12-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cheng Q., Wang X., Zu B., Xu J., Xu Y., Zuo X., Shen Y., Wang J., Shen Y. Deficiency of IRE1 and PERK signal pathways in systemic lupus erythematosus. Am. J. Med. Sci. 2014;348:465–473. doi: 10.1097/MAJ.0000000000000328. [DOI] [PubMed] [Google Scholar]
- Wang W., Xu L., Su J., Peppelenbosch M.P., Pan Q. Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol. 2017;25:573–584. doi: 10.1016/j.tim.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.-G., Fan R.-F., Li W.-H., Zhang D., Yang D.-B., Wang Z.-Y., Wang L. Activation of PERK-eIF2α-ATF4-CHOP axis triggered by excessive ER stress contributes to lead-induced nephrotoxicity. Biochim. Biophys. Acta, Mol. Cell Res. 2019;1866:713–726. doi: 10.1016/j.bbamcr.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Webster S.J., Ellis L., O’Brien L.M., Tyrrell B., Fitzmaurice T.J., Elder M.J., Clare S., Chee R., Gaston J.S.H., Goodall J.C. IRE1α mediates PKR activation in response to Chlamydia trachomatis infection. Microbes Infect. 2016;18:472–483. doi: 10.1016/j.micinf.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis S.N., Fletcher J.I., Kaufmann T., van Delft M.F., Chen L., Czabotar P.E., Ierino H., Lee E.F., Fairlie W.D., Bouillet P., Strasser A., Kluck R.M., Adams J.M., Huang D.C.S. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Wolfson J.J., May K.L., Thorpe C.M., Jandhyala D.M., Paton J.C., Paton A.W. Subtilase cytotoxin activates PERK, IRE1 and ATF6 endoplasmic reticulum stress-signalling pathways. Cell. Microbiol. 2008;10:1775–1786. doi: 10.1111/j.1462-5822.2008.01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- Wu X., Rapoport T.A. Mechanistic insights into ER-associated protein degradation. Curr. Opin. Cell Biol. 2018;53:22–28. doi: 10.1016/j.ceb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Tanimoto A., Murata Y., Fan J., Sasaguri Y., Watanabe T. Induction of human matrix metalloproteinase-12 gene transcriptional activity by GM-CSF requires the AP-1 binding site in human U937 monocytic cells. Biochem. Biophys. Res. Commun. 2001;285:300–307. doi: 10.1006/bbrc.2001.5161. [DOI] [PubMed] [Google Scholar]
- Wu D., Sanin D.E., Everts B., Chen Q., Qiu J., Buck M.D., Patterson A., Smith A.M., Chang C.-H., Liu Z., Artyomov M.N., Pearce E.L., Cella M., Pearce E.J. Type 1 interferons induce changes in core metabolism that are critical for immune function. Immunity. 2016;44:1325–1336. doi: 10.1016/j.immuni.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Chen Y.-J., Dobbs N., Sakai T., Liou J., Miner J.J., Yan N. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 2019;216:867–883. doi: 10.1084/jem.20182192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Xu Y., Chen L., Fang Q., Song S., Chen J., Teng J. RCN1 suppresses ER stress-induced apoptosis via calcium homeostasis and PERK-CHOP signaling. Oncogenesis. 2017;6:e304. doi: 10.1038/oncsis.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wang H., Wei S., Wang Z., Ji G. Inhibition of ER stress-related IRE1α/CREB/NLRP1 pathway promotes the apoptosis of human chronic myelogenous leukemia cell. Mol. Immunol. 2018;101:377–385. doi: 10.1016/j.molimm.2018.07.002. [DOI] [PubMed] [Google Scholar]
- Xue M., Momiji H., Rabbani N., Barker G., Bretschneider T., Shmygol A., Rand D.A., Thornalley P.J. Frequency modulated translocational oscillations of nrf2 mediate the antioxidant response element cytoprotective transcriptional response. Antioxid. Redox Signal. 2015;23:613–629. doi: 10.1089/ars.2014.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Fu F., Ma Y., Zhang X., Li L., Feng L., Liu P. The PERK arm of the unfolded protein response negatively regulates transmissible gastroenteritis virus replication by suppressing protein translation and promoting type I interferon production. J. Virol. 2018;92 doi: 10.1128/JVI.00431-18. e00431-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Yamazaki H., Hiramatsu N., Hayakawa K., Tagawa Y., Okamura M., Ogata R., Huang T., Nakajima S., Yao J., Paton A.W., Paton J.C., Kitamura M. Activation of the Akt-NF-kappaB pathway by subtilase cytotoxin through the ATF6 branch of the unfolded protein response. J. Immunol. 2009;183:1480–1487. doi: 10.4049/jimmunol.0900017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K., Kimata Y., Kadokura H., Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- Yang W., Tiffany-Castiglioni E., Koh H.C., Son I.-H. Paraquat activates the IRE1/ASK1/JNK cascade associated with apoptosis in human neuroblastoma SH-SY5Y cells. Toxicol. Lett. 2009;191:203–210. doi: 10.1016/j.toxlet.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Yang P., An H., Liu X., Wen M., Zheng Y., Rui Y., Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Yarilina A., Park-Min K.-H., Antoniv T., Hu X., Ivashkiv L.B. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat. Immunol. 2008;9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- Ye J., Rawson R.B., Komuro R., Chen X., Davé U.P., Prywes R., Brown M.S., Goldstein J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Oku M., Suzuki M., Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Zhao B., Gui J., Katlinski K.V., Brice A., Gao Y., Li C., Kushner J.A., Koumenis C., Diehl J.A., Fuchs S.Y. Type I interferons mediate pancreatic toxicities of PERK inhibition. Proc. Natl. Acad. Sci. U. S. A. 2015;112:15420–15425. doi: 10.1073/pnas.1516362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorodna O., Martin S.M., Rutkowski D.T., Kuwana T., Spitz D.R., Knudson C.M. 2-deoxyglucose-induced toxicity is regulated by Bcl-2 family members and is enhanced by antagonizing Bcl-2 in lymphoma cell lines. Oncogene. 2012;31:2738–2749. doi: 10.1038/onc.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauli G., Rimondi E., Nicolin V., Melloni E., Celeghini C., Secchiero P. TNF-related apoptosis-inducing ligand (TRAIL) blocks osteoclastic differentiation induced by RANKL plus M-CSF. Blood. 2004;104:2044–2050. doi: 10.1182/blood-2004-03-1196. [DOI] [PubMed] [Google Scholar]
- Zelenay S., Reis e Sousa C. Adaptive immunity after cell death. Trends Immunol. 2013;34:329–335. doi: 10.1016/j.it.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Zemskov E.A., Mikhailenko I., Hsia R.-C., Zaritskaya L., Belkin A.M. Unconventional secretion of tissue transglutaminase involves phospholipid-dependent delivery into recycling endosomes. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Lu M., Mori K., Luo S., Lee A.S., Zhu Y., Shyy J.Y.-J. ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 2004;23:950–958. doi: 10.1038/sj.emboj.7600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Liu Y.-P., Sha H., Chen H., Qi L., Smith J.A. XBP-1 couples endoplasmic reticulum stress to augmented IFN-beta induction via a cis-acting enhancer in macrophages. J. Immunol. 2010;185:2324–2330. doi: 10.4049/jimmunol.0903052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Integration of ER stress, oxidative stress and the inflammatory response in health and disease. Int. J. Clin. Exp. Med. 2010;3:33–40. [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D.T., Back S.H., Kaufman R.J. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang G., Zhang H., Karin M., Bai H., Cai D. Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell. 2008;135:61–73. doi: 10.1016/j.cell.2008.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., Hauser C.J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang G., Zheng Z., Maddipati K.R., Zhang X., Dyson G., Williams P., Duncan S.A., Kaufman R.J., Zhang K. Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology. 2012;55:1070–1082. doi: 10.1002/hep.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Hietakangas V., Wee S., Lim S.C., Gunaratne J., Cohen S.M. ER stress potentiates insulin resistance through PERK-mediated FOXO phosphorylation. Genes Dev. 2013;27:441–449. doi: 10.1101/gad.201731.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang K., Li Z., Guo B. ER Stress-induced inflammasome activation contributes to hepatic inflammation and steatosis. J. Clin. Cell Immunol. 2016;7:457. doi: 10.4172/2155-9899.1000457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.H., Viacava Follis A., Kriwacki R.W., Moldoveanu T. Discoveries and controversies in BCL-2 protein-mediated apoptosis. FEBS J. 2016;283:2690–2700. doi: 10.1111/febs.13527. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Swanson B.J., Wang M., Hildeman D.A., Schaefer B.C., Liu X., Suzuki H., Mihara K., Kappler J., Marrack P. Constitutive association of the proapoptotic protein Bim with Bcl-2-related proteins on mitochondria in T cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7681–7686. doi: 10.1073/pnas.0402293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Zhang J., Sun H., Jiang C., Dong Y., Shan Q., Su S., Xie Y., Xu N., Lou X., Liu S. Ubiquitination of inositol-requiring enzyme 1 (IRE1) by the E3 ligase CHIP mediates the IRE1/TRAF2/JNK pathway. J. Biol. Chem. 2014;289:30567–30577. doi: 10.1074/jbc.M114.562868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Galluzzi L., Kepp O., Smyth M.J., Kroemer G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015;15:405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]