Abstract
By controlling gene expression at the level of mRNA translation, organisms temporally and spatially respond swiftly to an ever-changing array of environmental conditions. This capacity for rapid response is ideally suited for mobilizing host defenses and coordinating innate responses to infection. Not surprisingly, a growing list of pathogenic microbes target host mRNA translation for inhibition. Infection with bacteria, protozoa, viruses, and fungi has the capacity to interfere with ongoing host protein synthesis and thereby trigger and/or suppress powerful innate responses. This review discusses how diverse pathogens manipulate the host translation machinery and the impact of these interactions on infection biology and the immune response.
Main Text
A cardinal means by which microbes influence host physiology is through the cellular translation machinery. Indeed, microbes and their hosts strategically maneuver to control cellular messenger RNA (mRNA) translation for their own benefit, illustrating a spectrum of host-pathogen interactions. On one extreme, replication of obligate intracellular parasites like viruses is completely reliant on the host translational apparatus. Not only must viruses commandeer this machinery to translate their own mRNAs and ensure viral mRNAs compete with cellular counterparts, but they also thwart host defenses aimed at inactivating the cellular translation apparatus. Even bacteria and protozoa, which are not dependent upon host translational components for their protein synthetic needs, manipulate the cellular translation machinery. Pathogen-encoded effectors, like bacterial toxins, inhibit host translation. As virulence factors associated with severe infections, toxins kill eukaryotic cells and facilitate tissue invasion. While interfering with host mRNA translation limits production of host defenses, it can also generate a danger signal capable of triggering a conserved innate immune response. Sensing of pathogens indirectly by monitoring changes in cell homeostasis is used extensively by plants, where it is called effector-triggered immunity (Jones and Dangl, 2006). Alternatively, changes in cellular protein synthesis can indicate host nutritional status and its relationship to a dynamic environment, illustrating how pathways that control mRNA translation are responsive to physiological stress and metabolic status. For persistent pathogens, monitoring of ongoing mRNA translation represents a vital window into the host metabolic state and prospects for future homeostasis. In addition, many cellular innate immune effectors are important regulators of global and mRNA specific translation. Finally, adaptive immunity requires presentation of protein antigens complexed with cell surface MHC molecules for immune surveillance. Thus, the cell protein repertoire can signal infection and is a key determinant discriminating self from nonself. In this review, we focus on how different pathogens induce changes to ongoing cellular protein synthesis and consider the varied strategies by which host translation impacts infection and immunity.
A Primer on Translation and Its Control in Eukaryotic Host Cells
Regulated mRNA translation is a powerful posttranscriptional means to control gene expression spatially and temporally in eukaryotes. In addition to fundamental roles in cell growth and proliferation, development, learning, memory, and synaptic plasticity, translational control plays a major role in host stress responses, including pathogenic infection, and defenses by enabling rapid responses in surroundings that abound with microbes. Indeed, the role of translation in infection and host immunity to viruses is aptly illustrated in crop plant species, where all known recessive resistance genes encode translation initiation factors (Truniger and Aranda, 2009). With respect to infection and immune defenses, host translation is regulated primarily at the initiation and elongation steps (Figure 1 ).
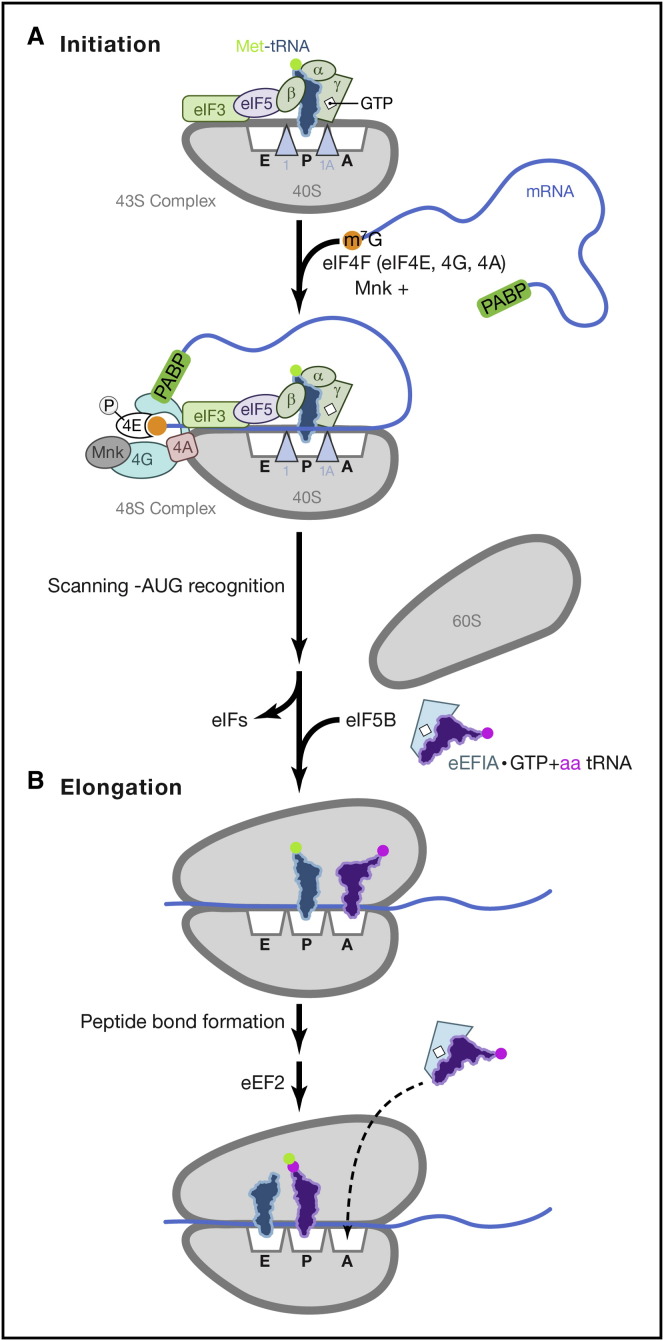
Figure 1.
Translation Initiation and Elongation in Eukaryotes
Initiation and elongation steps each require discrete translation factors.
(A) Initiation. Formation of the 43S preinitiation complex involves loading the initiator-methionine tRNA (Met-tRNAi) into the ribosomal P site as a complex with eIF2·GTP. The 40S subunit is also associated with eIF1 (blue triangle, 1), eIF1A (blue triangle, 1A), eIF3, and eIF5. The 43S complex is next positioned onto the 5′ end of a capped (orange circle, m7G), polyadenylated mRNA by eIF4F, a three-subunit complex composed of the cap-binding protein eIF4E, eIF4G, and eIF4A (shown as 4E, 4G, and 4A). Poly(A)-binding protein (PABP) bound to the polyadenylated 3′ mRNA also associates with eIF4G bound to the 5′ end. This forms a “closed-loop” topology, linking 5′ and 3′ mRNA ends. The eIF4E kinase Mnk is shown associated with eIF4G where it phosphorylates eIF4E (circle, P). The mRNA-bound 48S complex scans the mRNA to locate the AUG start codon, whose recognition is facilitated by eIF3, eIF1, and 1A. Initiation factor release follows 60S subunit joining, which requires eIF5B.
(B) Elongation. Loading of a charged tRNA into the 80S ribosome A site requires eEF1A·GTP. Ribosome-catalyzed peptide bond formation ensues. 80S translocation requires eukaryotic elongation factor 2 (eEF2), which moves the deacetylated tRNA to the E site, positioned the peptidyl-tRNA in the P site and re-exposes the A site.
Before mRNA loading, the 40S ribosomal subunit forms a 43S complex with translation initiation factors and the methionine-charged initiator tRNA (Met-tRNAi; Figure 1). This requires eIF2, a heterotrimeric guanine nucleotide binding protein that in its active state forms a ternary complex (TC) containing GTP and the initiator tRNA, Met-tRNAi. The TC loads 40S ribosomes with Met-tRNAi and is required to initiate protein synthesis at open reading frames (ORFs) commencing with an AUG codon or near-cognate AUGs, with one exception described later (reviewed by Hinnebusch and Lorsch, 2012). Unlike the prokaryotic 30S ribosomal subunit that directly recognizes the Shine-Dalgarno RNA sequence upstream of the initiator AUG, eukaryotic 40S subunits require assistance from the heterotrimeric translation initiation factor eIF4F (reviewed by Hinnebusch and Lorsch, 2012). By binding to the 7-methylguanosine (m7G) cap structure that is present at the 5′ terminus of all nuclear transcribed eukaryotic mRNAs, eIF4F positions the ribosome onto the mRNA 5′ end and facilitates an ATP-dependent movement termed “scanning” that locates the 5′ proximal AUG on the majority of mRNAs (reviewed by Parsyan et al., 2011). eIF4F is comprised of the cap-binding protein eIF4E, the DEAD box RNA helicase eIF4A, and the large molecular scaffold eIF4G. While eIF4E binds the cap, eIF4G associates with ribosome-bound eIF3 to tether the 43S complex to the mRNA 5′ end. Once bound to eIF4G, eIF4E can be phosphorylated by an eIF4G-associated kinase, Mnk1 or Mnk2. Unlike Mnk2, which confers basal eIF4E phosphorylation, Mnk1 mediates inducible p38 and ERK-responsive eIF4E phosphorylation and links eIF4E phosphorylation to MAP kinase signaling. Finally, eIF4F assembly juxtaposes the mRNA polyadenylated 3′ end, bound by PABP, with the 5′ end through an interaction between eIF4G and PABP (reviewed by Hinnebusch and Lorsch, 2012). This probably limits 40S subunit recruitment to mRNAs containing intact 5′ and 3′ ends. Significantly, initiation is thought to be the rate-limiting step under most circumstances (reviewed by Hinnebusch and Lorsch, 2012). The extent to which translation of individual mRNAs varies with respect to eIF4F levels is heavily influenced by the degree of secondary structure in the 5′ untranslated region (UTR) (reviewed by Parsyan et al., 2011).
Importantly, regulated binding of eIF4E to eIF4G represents a key point whereby physiological signals control initiation through assembly of a functional eIF4F complex (reviewed by Hinnebusch and Lorsch, 2012). This is mediated by the host ser/thr kinase mTOR complex 1 (mTORC1; Figure 2 ), which responds to changes in physiologic homeostasis, including growth factors, nutrients, amino acids, oxygen, and energy availability (reviewed by Laplante and Sabatini, 2012) and controls activity of the eIF4E-binding protein family of translation repressors (4E-BP1,2,3). While hypophosphorylated 4E-BPs bind to eIF4E, prevent its assembly into the eIF4F complex, and suppress cap-dependent translation, hyperphosphorylation of 4E-BP by activated mTORC1 releases eIF4E, promotes eIF4E binding to eIF4G, and stimulates cap-dependent translation.
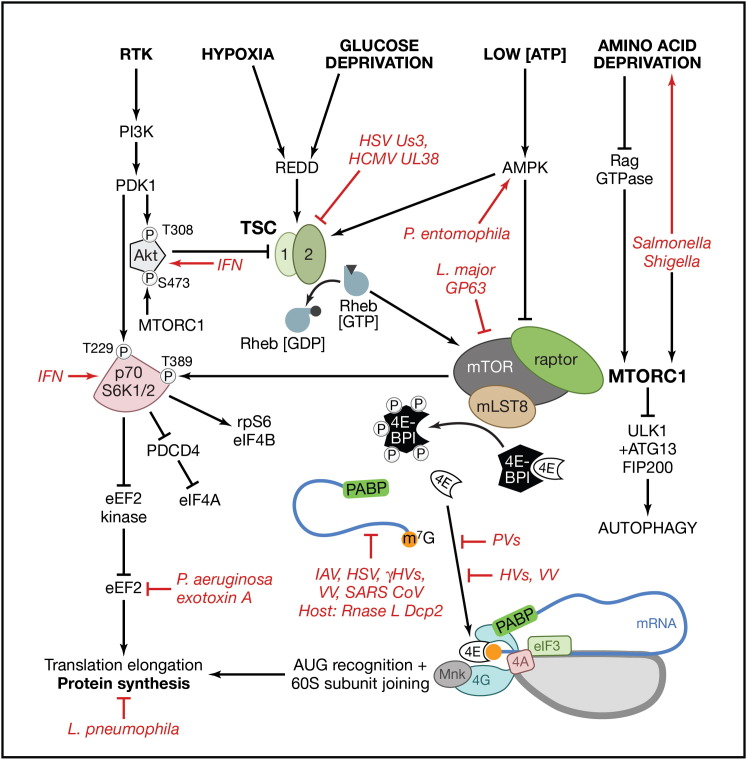
Figure 2.
Integration of Signals by mTORC1 Controls Cap-Dependent Translation
Availability of growth factors (receptor tyrosine kinase [RTK] signaling), oxygen, glucose, and energy regulate the GTPase activating protein TSC (composed of subunits hamartin [TSC1] and tuberin [TSC2]), which represses mTOR complex 1 (mTORC1) by promoting Rheb·GDP accumulation. Amino acid availability controls mTORC1 through the Rag GTPases. Inhibition of TSC allows Rheb·GTP accumulation and mTORC1 activation and results in p70 ribosomal protein S6 kinase (p70 S6K) and 4E-BP1 phosphorylation. 4E-BP1 hyperphosphorylation relieves translational repression and releases eIF4E (labeled 4E), allowing eIF4E to bind eIF4G (labeled 4G) and assemble the heterotrimer subunit initiation factor eIF4F (composed of eIF4E, eIF4A [labeled 4A], and eIF4G) on 7-methylguanosine (m7G; orange circle)-capped mRNA (see Figure 1A). Poly(A)-binding protein (PABP) is depicted bound to the 3′ poly(A) tail and also associates with eIF4G to stimulate translation. For simplicity, only eIF3, but not other 43S complex components (eIF1, eIF1A, eIF2, eIF5, and Met-tRNAi as shown in Figure 1), is depicted. In addition to stimulating ribosomal protein S6 (RPS6) phosphorylation, p70 S6K activation by mTORC1 stimulates the eIF4A accessory factor eIF4B, relieves PDCD4-mediated repression of eIF4A, and inhibits eukaryotic elongation factor 2 (eEF2) kinase to stimulate elongation. Pathogen strategies for activating and inhibiting steps within this pathway to control host translation are indicated in red; see the main text for details. IFN, interferon; HVs, herpesviruses; VV, vaccinia virus; PVs, picornaviruses (poliovirus, EMCV, rhinovirus); SARS coV, SARS coronavirus; IAV, influenza A virus; γHVs, gamma herpesviruses (EBV, KSHV, MHV68).
Upon AUG recognition, GTP hydrolysis is stimulated by eIF5, an eIF2 GTPase-activating protein (GAP) that also stabilizes GDP binding to eIF2 and functions as a guanine nucleotide dissociation inhibitor (Hinnebusch and Lorsch, 2012). While AUG recognition promotes inorganic phosphate release from eIF2, eIF5B-dependent 60S subunit joining promotes initiation factor release and allows inactive eIF2⋅GDP to access its heteropentameric subunit (α-ε) recycling factor, eIF2B, which exchanges eIF2-bound GDP for GTP (Figure 3 ; reviewed by Hinnebusch and Lorsch, 2012).
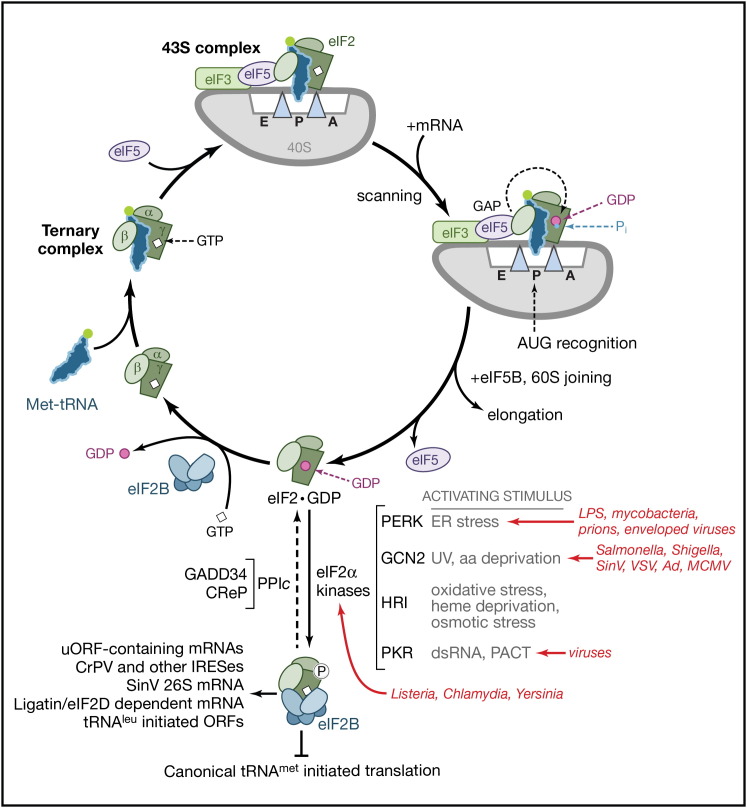
Figure 3.
Control of Translation Initiation by eIF2 and Host Regulatory Kinases that Phosphorylate Its α subunit
A ternary complex, comprised of eIF2 (α, β, and γ subunits, depicted) and GTP bound to the initiator-methionine tRNA (Met-tRNAi), loads Met-tRNAi into the ribosomal P site and forms a 43S preinitiation complex. After recruitment of eIF4F- and poly(A)-binding protein (PABP)-bound mRNA and recognition of the AUG start codon by scanning ribosomes, the GTPase-activating protein eIF5 stimulates GTP hydrolysis, and 60S subunit joining triggers the release of eIF2·GDP and inorganic phosphate (Pi). The resulting 80S ribosome carries out the elongation phase (Figure 1B). Inactive, GDP-bound eIF2 (eIF2·GDP) is recycled to the active GTP-bound form by the five-subunit guanine nucleotide exchange factor eIF2B.
Four different cellular eIF2α kinases (described in the main text), each of which is activated by a discrete stress, phosphorylate eIF2α, and prevent eIF2 recycling. Phosphorylated eIF2 binds tightly to and inhibits eIF2B, blocking canonical tRNAmet-initiated translation. When bound to either the inducible (growth arrest and DNA damage-inducible protein 34 [GADD34]) or constitutively active (CReP) regulatory subunit, the host protein phosphatase 1 catalytic (PP1c) subunit dephosphorylates eIF2. Pathogens shown in red and the different eIF2α kinases they activate are indicated. Viral (CrPV and other IRESs, Sindbis 26S mRNA) and host (uORF-containing, eIF2D-dependent, tRNAleu-initiated) mRNAs that are translated in the presence of phosphorylated eIF2 are indicated. SinV, Sindbis virus; VSV, vesicular stomatitis virus; Ad, adenovirus; CrPV, cricket paralysis virus; uORF, upstream open reading frame; aa, amino acid.
After AUG initiation codon recognition and 60S subunit joining, the 80S ribosome commences polypeptide chain elongation (Figure 1; reviewed in Dever and Green, 2012). This requires elongation factor eEF1A, a G protein that delivers aminoacylated tRNA to the ribosome and whose activity is regulated by the guanine nucleotide exchange factor (GEF) eEF1B, and eEF2, which promotes ribosome translocation after peptide bond formation. While casein kinase II and PKC stimulate eEF1A activity, eEF2 phosphorylation by eEF2 kinase inhibits elongation (Figure 2). Elongation continues until a termination codon is encountered.
Infection and Its Bearing on Host mRNA Translation
Microbial infection impacts host translation in a variety of ways. Not only can infection modify the translational capacity of the host, but regulated mRNA translation can influence microbial pathogenesis by limiting or promoting translation of host mRNAs encoding the effector proteins that mediate innate responses. Damage to epithelia by bacteria can suppress host translation via cell signaling pathways, alerting the host to danger. By introducing effectors directly into the cytoplasm, bacteria or protozoa can directly suppress host translation. Finally, viruses are absolutely dependent on the host translation machinery to produce their proteins, which are required for their replication, and must effectively seize control of translation factors and their extensive regulatory network. Some RNA viruses like poliovirus inactivate factors required for canonical cap-dependent translation of host mRNAs to favor noncanonical, cap-independent mechanisms that allow 40S ribosomes to be recruited to viral mRNA containing an internal ribosome entry site (IRES) (reviewed by Doudna and Sarnow, 2007). DNA viruses produce capped and polyadenylated mRNAs similar to the host and must effectively recruit limiting translation components to viral mRNAs. For a more-detailed discussion of the varied tactics that viruses use to interact with the host to promote viral mRNA translation, the reader is referred to two recent reviews (Walsh and Mohr, 2011; Walsh et al., 2012).
Translation Initiation Factor eIF2 and Its Regulatory Kinases: Innate Sensors and Direct Effectors at the Front Lines of Host Defense
Their exquisite sensitivity to environmental factors, metabolic status, and stress coupled with their capacity to rapidly change host gene expression renders translation factors ideal innate immune effectors to directly limit pathogen replication. A seminal innate immune guardian is eIF2. Host translation initiation can be impaired by four cellular kinases that phosphorylate the α regulatory subunit of eIF2 on S51, each of which is triggered by a discrete environmental or metabolic stress (Figure 3). Double-strand RNA (dsRNA), a pathogen-associated molecular pattern (PAMP) indicative of virus infection, and the host protein PACT can each activate the dsRNA-dependent protein kinase PKR, an interferon (IFN)-induced eIF2α kinase that inhibits protein synthesis upon activation in virus-infected cells (Walsh et al., 2012). In this manner, IFN production by infected cells induces the accumulation of antiviral host defense molecules in uninfected neighboring cells to restrict viral replication and spread (Walsh et al., 2012). Exceeding the ER protein folding capacity engenders activation of PERK, an ER membrane spanning protein with a luminal sensor domain and a cytoplasmic eIF2α kinase domain (Ron and Walter, 2007). Heme depletion, arsenite-induced oxidative stress, heat shock, and osmotic stress activate the eIF2α kinase HRI, while amino acid deprivation or UV light activates GCN2 (Lu et al., 2001; Deng et al., 2002). Relative to its unphosphorylated form, eIF2 containing a phosphorylated α subunit exhibits greater affinity for the recycling factor eIF2B and inhibits its guanine nucleotide exchange factor (GEF) activity. Importantly, since eIF2B is limiting, small changes in phosphorylated eIF2α concentration have dramatic effects on translation initiation (Hinnebusch and Lorsch, 2012). The consequences for viruses can be dire unless they have a strategy to prevent accumulation of phosphorylated eIF2, which can also result in autophagy (Tallóczy et al., 2002). Indeed, the ability to antagonize PKR, either via virus-encoded dsRNA decoy molecules, dsRNA-binding proteins, or PKR-binding proteins, each of which prevent PKR activation, or via induction of cellular PKR antagonists, represents a major strategy used by viruses (Table 1 ) to resist the potent antiviral effects of IFN (Walsh and Mohr, 2011). Among the least sensitive to IFNs, large DNA viruses including adenoviruses (Ad), poxviruses, and herpesviruses encode multiple, independent effectors to prevent eIF2 phosphorylation, some of which are specific antagonists of different eIF2α kinases or globally suppress eIF2α phosphorylation (reviewed in Walsh et al., 2012). This latter class of effectors, including virus-encoded eIF2α pseudosubstrates and phosphatase regulatory components, potentially counteract PERK and GCN2, both of which also have antiviral activity (reviewed in Walsh and Mohr, 2011; Won et al., 2012). Viral genes that inhibit eIF2α phosphorylation are virulence determinants, as their deletion results in attenuated strains that grow poorly (Chou et al., 1990; Beattie et al., 1995; Mohr and Gluzman, 1996; Mulvey et al., 2004). While this has likely driven viruses to acquire functions that prevent eIF2α phosphorylation, the host pkr gene has also undergone rapid evolutionary change to evade virus-encoded inhibitors (Elde et al., 2009; Rothenburg et al., 2009), illustrating how viruses and their hosts continuously maneuver to control eIF2. Recently, eIF2α phosphorylation has been reported after infection with Listeria monocytogenes, Chlamydia trachomatis, or Yersinia pseudotuberculosis, all intracellular bacteria (Shrestha et al., 2012). Cells unable to phosphorylate eIF2α were more susceptible to bacterial invasion, illustrating a role for eIF2α phosphorylation in activating NF-κB and proinflammatory cytokine production. In the case of Yersinia, the bacterial YopJ protein impairs eIF2α phosphorylation by an unknown mechanism, hinting that bacteria and viruses both counteract host defenses that operate through eIF2 (Shrestha et al., 2012).
Table 1.
Viral Strategies to Prevent or Counteract eIF2α Phosphorylation
| Mechanism | Virus (Gene Product) |
|---|---|
| dsRNA BPs | Influenza (NS1), reovirus (σ3), HSV (Us11), HCMV (TRS1, IRS1), MCMV (m142, m143), EBV (SM), Vaccinia (E3L) |
| RNA antagonists of PKR | Adenovirus (VA RNA), HCV (IRES), EBV (EBERs) |
| Protein antagonists of PKR | Sendai (C), HCV (NS5A), Adenovirus (E1b 55k, E4 orf6), KSHV (vIRF2), PRV (IE 180) |
| PERK antagonists | HCV (E2), Influenza, TEV, TMV (activate host p58IPK), HSV1 (gB) |
| eIF2α kinase pseudosubstrate | Vaccinia (K3L), Ranavirus (vIF2) |
| eIF2α phosphatase regulatory subunit | HSV (γ34.5), ASFV (DP71L), HPV (E6) |
| eIF2-independent initiation | CrPV, Sindbis virus, poliovirus |
TEV, tobacco etch virus; TMV, tobacco mosaic virus; ASFV, African swine fever virus; PRV, pseudorabies virus; MCMV, murine cytomegalovirus.
Not all viruses prevent eIF2α phosphorylation. While many RNA viruses like vesicular stomatitis virus (VSV) and encephalomyocarditis virus (EMCV) are exquisitely IFN sensitive, others use uncommon, alternative translation initiation strategies that do not require eIF2-mediated Met-tRNAi loading. Insect viruses like cricket paralysis virus (CrPV) and Plautia stali intestine virus represent the most extreme examples and contain an IRES that directly recruits 40S subunits without needing any initiation factors, bypassing tRNAi loading altogether (Wilson et al., 2000; Spahn et al., 2004; Pfingsten et al., 2010). Sindbis virus, however, uses an alternative cellular factor distinct from eIF2, eIF2D, to recruit Met-tRNAi to the P site of 40S-mRNA complexes in a GTP-independent manner and subsequently initiates translation of the viral subgenomic 26S mRNA in the presence of phosphorylated eIF2α (Ventoso et al., 2006; Dmitriev et al., 2010; Skabkin et al., 2010). While the hepatitis C virus (HCV) IRES also initiates translation in an eIF2-independent manner, elegant studies with an in vitro reconstituted system showed that eIF2D is required to recruit Met-tRNAi to 40S-mRNA complexes, whereas studies in cultured cells implicated a different eIF2-independent factor called eIF2A (Dmitriev et al., 2010; Skabkin et al., 2010, Kim et al., 2011).
In some cases, chronic pathogen infection engenders persistent, unresolved ER stress. Exposure of cells to LPS, a PAMP produced by gram-negative bacteria that activates TLR4 signaling, stimulates proinflammatory cytokine and antimicrobial protein production causing ER stress (Woo et al., 2009). PERK, which is activated by ER stress and phosphorylates eIF2α, suppresses global translation but promotes the translation of a subset of cellular mRNAs harboring upstream ORFs (uORFs). This mechanism is based on the property of eukaryotic ribosomes to infrequently reinitiate translation and on the time they transit through the 5′ UTR to reacquire an active TC before encountering the next AUG (Hinnebusch, 2011). One such uORF-containing mRNA encodes the transcription factor ATF4, which in turn induces the transcription factors ATF-3 and CHOP (C/EBP homologous protein). While transient CHOP induction has a salubrious effect on cells, sustained production is harmful and causes apoptosis. During ER stress, macrophage TLR4 signaling stimulates eIF2B GEF activity, which enables continued synthesis of essential proteins without activating ATF4 and CHOP (Woo et al., 2012). Similarly, activation of ER stress pathways may also be important for Mycobacterium tuberculosis survival in macrophages (Seimon et al., 2010; Lim et al., 2011). In contrast, sustained eIF2α phosphorylation can have severe pathogenic consequences. Bacterial components including LPS, cytolysins, and intracellular-acting toxins all induce ER stress (Zhang et al., 2006; Wolfson et al., 2008; Pillich et al., 2012). Misfolded prion protein PrPSc accumulation during prion replication triggers the unfolded protein response (UPR), resulting in eIF2α phosphorylation and global protein synthesis suppression, followed by synaptic failure and neurodegeneration in mice (Moreno et al., 2012). Importantly, prevention of phosphorylated eIF2α accumulation rescued synaptic failure and neuronal loss, suggesting that newly described PERK inhibitors may have efficacy for ameliorating prion diseases like Alzheimer’s Disease (Axten et al., 2012).
Sculpting the Innate Immune Response through Translational Regulation of Host Defense Effector Molecules
In addition to the direct role in host defense played by initiation factors like eIF2 that control translation, production of cellular innate immune effectors is subject to translation control. Notably, eIF4F and its regulators control translation of mRNAs encoding innate immune effectors through multiple, independent signaling pathways that probably act synergistically to regulate different components of the host innate response. One arm of this response involves mTOR signaling, which in addition to regulating translation in response to metabolic status and physiological stress also controls type I IFN production (Cao et al., 2008; Colina et al., 2008). RNA virus replication (VSV, influenza, sindbis) is markedly suppressed in mouse embryonic fibroblasts (MEFs) lacking the mTORC1 substrates 4E-BP1 and 4E-BP2, both of which repress cap-dependent mRNA translation (Colina et al., 2008). Moreover, plasmacytoid dendritic cells (pDCs), the main type I IFN producers in response to virus infection, that were derived from 4E-BP1/2-deficient mice produced more IFN in response to infection. In addition, mice lacking 4E-BP1/2 were resistant to a lethal VSV challenge. The molecular mechanism underlying these findings is the upregulation of IRF-7 (interferon regulatory factor-7, a transcription factor) mRNA translation in 4E-BP1/2-deficient MEFs, leading to higher basal type I IFN production (Colina et al., 2008). mTORC1 and its upstream regulator TSC1/2 also regulate inflammatory mediators in response to bacterial stimulation of mononuclear phagocytes by L. monocytogenes, as mTORC1 activation limits inflammation by blocking NF-κB and enhancing STAT3 (Weichhart et al., 2008). Consistent with this, inhibition of mTORC1 by rapamycin protects mice from a lethal L. monocytogenes infection (Weichhart et al., 2008).
While the mTORC1 substrates 4E-BP1/2 regulate translation by controlling eIF4E binding to eIF4G, phosphorylation of the cap-binding protein eIF4E on S209 by the eIF4G-associated kinase Mnk1 independently controls translation of the mRNA encoding the NF-κB inhibitor IκBα. MEFs from knockin mice, in which the wild-type eif4e gene was replaced with an S209A allele, produced more type I IFN and were less susceptible to infection with a variety of RNA (EMCV, VSV, sindbis) and DNA viruses (vaccinia, herpes simplex virus [HSV]) (Herdy et al., 2012). These DNA viruses produce capped, polyadenylated mRNAs that rely on eIF4F and promote eIF4F assembly in infected cells (Walsh and Mohr, 2006; Walsh et al., 2008). They also stimulate eIF4E phosphorylation, which can be viewed as a viral strategy to antagonize NF-κB activation. Importantly, not all viruses stimulate eIF4E phosphorylation. In adenovirus and influenza virus-infected cells, unphosphorylated eIF4E accumulates (Feigenblum and Schneider, 1993; Cuesta et al., 2000). These viruses use other strategies to counter host innate defenses, including suppressing host protein synthesis and preventing eIF2α phosphorylation. While eIF2α phosphorylation globally inhibits translation, regulated eIF4E phosphorylation contributes to antiviral host defense by selectively controlling translation of IκBα mRNA, encoding a critical suppressor of the innate antiviral response (Herdy et al., 2012).
IFN signaling through specific cell surface receptors also activates the AKT/mTOR pathway and stimulates phosphorylation of downstream effectors, including 4E-BP1, p70S6K, and PDCD4, which control translation of IFN-induced mRNAs (Alain et al., 2010; Cao et al., 2008; Kroczynska et al., 2012). Likewise, activation of MEK-ERK MAPK signaling by IFN stimulates the eIF4G-associated, eIF4E kinase Mnk1, which in turn promotes eIF4E phosphorylation and has mRNA-specific effects on translation (Joshi et al., 2009; Furic et al., 2010).
Type I IFN is also important for host defense against parasite infections, such as leishmaniasis (Bogdan et al., 2004). Although little is known regarding how protozoan parasite infection affects host translation, Leishmania promotes survival within macrophage by downregulating host protein synthesis. This requires a key Leishmania virulence gene, the surface glycoprotein GP63 (Joshi et al., 2002). In addition to cleaving and thereby activating host phosphatases (Gomez et al., 2009), GP63 also causes cleavage of mTOR, the catalytic kinase component of both mTORC1 and mTORC2 (Jaramillo et al., 2011). This activates the 4E-BP1/2 translation repressors and promotes Leishmania survival within macrophages. Importantly, 4E-BP1/2-deficient mice are less susceptible to cutaneous leishmaniasis, in part due to their constitutive type I IFN response, which leads to increased iNOS levels and nitric oxide production. Thus, repression of host cap-dependent mRNA translation by L. major probably limits the synthesis of host antimicrobial and host defense proteins, thereby contributing to the pathogenesis of Leishmaniasis (Jaramillo et al., 2011). In an analogous manner, inhibition of mTORC1 with rapamycin enhances the replication of poliovirus and EMCV (Beretta et al., 1996).
Two Sides to Disrupting Host mRNA Translation
Antagonizing Production of Host Defense Proteins Can Activate a Potent Innate Response
Changes in ongoing host protein synthesis alert the host to dangerous microbes, helping distinguish innocuous commensals from pathogens. In an infection model using the nematode C. elegans, which normally feeds on nonpathogenic E. coli, ingestion of a virulent P. aeruginosa strain causes a lethal intestinal infection that requires some of the same virulence factors needed in mammals (Tan et al., 1999). The virulence of P. aeruginosa is due in part to exotoxin A (ToxA), which ADP-ribosylates eEF2 to inhibit host translation (McEwan et al., 2012). Indeed, host sensing of the resulting translation inhibition leads to the activation of the cellular zip-2/irg-1 pathway, which induces production of host defense factors, including transporters and UDP-glucuronosyltrasferases, that remedy toxin damage by pumping it out or inactivating it (Estes et al., 2010). Importantly, inhibition of translation provides the sensor that alerts the host to the pathogen’s presence (Dunbar et al., 2012). This in turn activates translation of a select set of host mRNAs and requires a region of the zip-2 5′ UTR that contains several small uORFs. While uORFs are important for stress-induced responses including amino acid sensing in yeast and the UPR, their effects on translation typically involve eIF2 phosphorylation (described earlier). How uORFs might stimulate translation in which eEF2 is made limiting by ToxA requires further study. Nevertheless, translation is a core cellular process monitored intracellularly in real time to mount a potent innate response against a pathogen.
Similarly, infection of Drosophila with virulent strains of P. entomophila, a bacterial pathogen that disrupts gut homeostasis and is lethal at high does, induces protective immune and repair pathways at the transcriptional level (Chakrabarti et al., 2012). However, ROS accumulation in the gut activates both GCN2, which phosphorylates eIF2α, and AMP-activated protein kinase (AMPK), which inhibits mTORC1 via activating TSC-GAP and phosphorylating the mTORC1 raptor subunit. This was not observed in flies infected with an avirulent strain or nonlethal bacteria. While the precise mechanism of GCN2 and AMPK activation remains unresolved, excessive stress pathway activation by pathogenic bacteria can severely restrict the host response to infection by impairing cellular protein synthesis. Importantly, GCN2-depletion by RNA interference (RNAi) or chemical inhibitors of AMPK allowed gut repair and limited pathogenesis in infected animals. AMPK hypomorphic flies were also relatively resistant to infection.
Finally, secretion of five effector proteins by virulent L. pneumophila also decreased global host translation in macrophages (Fontana et al., 2011). The lgt1,2,3 genes encode related glucosyltransferases that modify eEF1A, sidI encodes a protein that binds to eEF1A and eEF1βγ, and sidL is toxic to mammalian cells and inhibits translation in vitro by an unknown mechanism. The resulting suppression of ongoing host mRNA translation depletes IκB and activates NF-κB responsive genes, many of which encode innate immune effectors. How these mRNAs are translated in L. pneumophila-infected macrophage while global translation is suppressed is not clear. Nevertheless, global suppression of host mRNA translation triggers a potent innate response and allows the host to distinguish pathogenic from nonpathogenic strains.
Coincidental, collateral cellular damage during bacterial infection also impacts the host translation machinery. Pathogen-induced membrane damage resulting from Shigella or Salmonella epithelia cell infection depletes intracellular amino acids. Starving for amino acids suppresses mTORC1 activity and induces autophagy by inhibiting the Rag GTPases, which are regulated by amino acid availability, reside on lysosomal membranes, and dock mTORC1 to its activator rheb (reviewed in Laplante and Sabatini, 2012). A full-blown general amino acid starvation response follows, evidenced by activation of the eIF2α kinase GCN2 (Tattoli et al., 2012). This is accompanied by an integrated stress response initiated by ATF-4 mRNA translation, which is stimulated upon eIF2α phosphorylation and in turn activates transcription of a second transcription factor, ATF-3, which promotes expression of stress responsive genes (Tattoli et al., 2012). Shigella itself remains camouflaged from host autophagic defenses due to secreted bacterial effectors. In Salmonella-infected cells, mTORC1 activity is initially inhibited and amino acid starvation results due to pathogen-induced membrane damage. However, mTOR is progressively relocalized from the surface of lysosomal membranes to Salmonella cytoplasmic-containing vacuoles (SCVs). This coincides with restoration of normal amino acid levels and reduced GCN2 activation (Tattoli et al., 2012). Thus, Salmonella escapes autophagy by redistributing mTORC1 to SCVs and ensuring that this host kinase remains active. In a related strategy to maintain activated mTORC1, the kinase and its activator rheb are redistributed to a perinuclear location in a dynein-dependent manner by human cytomegalovirus (HCMV) (Clippinger and Alwine, 2012).
Altering Host Translation Can Facilitate Virus Replication
Inhibition of host translation is not limited to protozoa and bacteria that have their own translation machinery. Many viruses also impair cellular protein synthesis, typically referred to as host shutoff, and this plays a significant role in pathogenesis (Walsh et al., 2012). While suppression of bulk host protein synthesis in cells infected with bacterial pathogens can activate translation of select cellular mRNAs important for host defense as described earlier, a more-severe inhibition of host translation resulting from the concerted action of multiple pathogen-encoded effectors can typically facilitate replication of numerous viruses (Walsh et al., 2012). Besides reducing overall host mRNA complexity and allowing viral mRNAs to compete for limiting initiation factors and ribosomes, it severely restricts the ability of the host to translate mRNAs encoding inflammatory cytokines, HLA class I and class II molecules required for antigen presentation, or proteins with antiviral activity, including those specified by interferon-stimulated genes (ISGs). Indeed, antiviral immune response gene expression is selectively elevated in cells deficient for the decapping enzyme Dcp2, which impairs mRNA turnover (Li et al., 2012). The resulting sustained type I IFN production renders these cells more refractory to virus infection.
While many viruses induce host shutoff, different underlying molecular mechanisms prevail that are presumably tailored to the biological needs of each pathogen. The simplest involves inactivating host translation factors by direct proteolysis as seen in picornavirus-infected cells (reviewed in Walsh et al., 2012). By cleaving eIF4G, or interfering with eIF4F assembly by promoting 4E-BP hypophosphorylation, some picornaviruses (poliovirus, coxsakievirus, rhinovirus) suppress cap-dependent translation, the major means whereby host proteins are produced. The related picornavirus EMCV induces nuclear accumulation of the cap-binding protein eIF4E (Groppo et al., 2011), and enterovirus 73 stimulates the production of miR141, which reduces eIF4E levels and promotes the switch from cap-dependent to cap-independent mRNA translation (Ho et al., 2011). A newly identified X-ORF within genomic RNA segment 3 of influenza A virus generated by ribosomal frameshifting contains an endonuclease domain and is required for global host shutoff via an unknown mechanism that may involve accelerated mRNA decay (Jagger et al., 2012). In the absence of X, the magnitude of host response to infection, notably inflammatory, apoptotic, and T-lymphocyte signaling, were all elevated. Finally, the nsp1 protein encoded by SARS coronavirus suppresses host type I IFN production by (1) binding to and inactivating 40S ribosome subunits and (2) promoting selective degradation of host, but not viral mRNAs (Kamitani et al., 2009; Huang et al., 2011).
A common means by which complex DNA viruses suppress host mRNA translation is via interfering with mRNA metabolism, splicing, and nucleocytoplasmic trafficking (reviewed in Walsh and Mohr, 2011). This can be supplemented by global increase of mRNA turnover through virus-encoded decapping or endonuclease functions that effectively subvert rate-limiting steps in the host mRNA quality-control pathway (Feng et al., 2005; Parrish et al., 2007, 2009; Kronstad and Glaunsinger, 2012; Gaglia et al., 2012). While viral mRNAs are not spared in this process, their sheer abundance allows them to engage the translational machinery. mRNA turnover also allows viruses to adjust their gene expression profile as their developmental replicative cycle progresses (Read and Frenkel, 1983; Parrish and Moss, 2006). Importantly, HSV mutants deficient in their ability to promote mRNA turnover are attenuated and sensitive to interferon (Pasieka et al., 2008; Murphy et al., 2003). Finally, host shutoff can impact a virus’s ability to establish long-term latent infections. Murine gamma herpesvirus MHV68 mutants unable to induce host shutoff replicate to near-WT levels in the lung during the acute infection phase but fail to traffic to the lymph system and establish latency in splenocytes at reduced levels (Richner et al., 2011). While more work is needed to understand how host shutoff contributes to latency establishment, failure to suppress host response genes during acute infection could play a contributing role.
Unlike many viruses, HCMV does not suppress ongoing host protein synthesis in productively infected cells (Stinski, 1977). This renders translation of viral mRNAs difficult, as they must compete with host mRNAs. In a surprising twist, CMV induces its host to make more translation factors, raising the intracellular concentration of eIF4E, eIF4G, eIF2, and PABP (Isler et al., 2005; Perez et al., 2011). Importantly, the increase in PABP abundance is required to stimulate assembly of the cap-recognition complex eIF4F. Failure to increase PABP reduces viral replication and spread. Virus-induced new PABP synthesis is translationally controlled by the HCMV UL38 mTORC1 activator (McKinney et al., 2012). Thus, because HCMV does not induce host cell shutoff, it exploits host mRNA translation to support its productive replication. This comes at a potential huge liability, however, as indiscriminate translation of host mRNAs may also interfere with viral productive growth. Whether or not the virus is capable of globally controlling which host mRNAs engage ribosomes and are translated and how these contribute to viral replication remain to be determined.
While viruses stimulate mRNA degradation to antagonize the host, mRNA turnover directed by the cellular interferon-induced 2′,5′-oligoadenylate synthetase (OAS)-RNase L system effectively limits virus replication. In response to the virus dsRNA PAMP, OAS generates oligoadenylate (OA) chains with a distinct 2′,5′ linkage that activate the latent ribonuclease RNase L (Sadler and Williams, 2008). Viral and cellular mRNA cleavage inhibits protein synthesis and generates more short RNAs that amplify the type I IFN response via other pattern recognition receptors (RIGI and MDA5) (Malathi et al., 2007). Many dsRNA BPs that prevent PKR activation also inhibit OAS. In a unique strategy, the murine hepatitis virus ns2 gene encodes a phosphodiesterase that cleaves 2′,5′-OA and reduces its intracellular concentration to limit the potent OAS activator. While the ns2-deficient virus was not pathogenic in WT mice, its virulence was restored in RNase L-deficient mice, illustrating the importance of host defenses that target mRNA decay in coronavirus pathogenesis (Zhao et al., 2012).
Infections and Stress Granules
Viral infection diverts the subcellular compartmentalization of mRNAs into discrete structures called stress granules (SGs). These granules are dynamic aggregates detected when translation initiation is impaired and are comprised of nontranslating mRNAs, several cellular mRNA binding proteins (PABP, TIA-1, TIAR, and G3BP), 40S ribosome subunits, and a subset of initiation factors including eIF4E, eIF4G, eIF4A, eIF3, and eIF2 (Decker and Parker, 2012). Thus, SGs contain stalled, aggregated translation initiation complexes and may be an intermediary repository between mRNAs destined for polysomes and active translation and those slated for mRNA decay in P bodies. Many viruses impair SG formation. Viral factors that promote mRNA decay deplete SG RNA components, whereas those that suppress eIF2α phosphorylation limit stalled initiation complex accumulation (White and Lloyd, 2012). The G3BP stress granule subunit is directly cleaved by the poliovirus 2A proteinase (White et al., 2007; Piotrowska et al., 2010). Other viruses, however, stimulate SG assembly (White and Lloyd, 2012). While these findings are largely descriptive and more work is required to understand SG function, changes to these dynamic structures may represent another strategy whereby viruses manipulate cytoplasmic mRNA organization to control host translation. Indeed, cycles of SG assembly/disassembly have been observed in HCV-infected cells (Ruggieri et al., 2012). Manipulation of SG dynamics by opposing actions of PKR and protein phosphatase 1 acting on eIF2 may prevent sustained translational repression and be a conserved response to viral infection that can be exploited for persistence (Ruggieri et al., 2012). Bacterial infection can likewise induce SG formation, as the amino acid depletion, GCN2 activation, and eIF2α phosphorylation triggered by Shigella or Salmonella results in TIA-1-containing cytoplasmic SGs (Tattoli et al., 2012).
Fueling Adaptive Host Immune Responses: Translation and Antigen Production
The translational machinery is the major source of peptide antigens recognized as self or nonself by producing proteins that are processed into peptides and presented for adaptive immune surveillance in association with MHC class I on the host cell surface. Not only are these peptides encoded by conventional ORFs that begin with methionine, they are also encoded by alternate reading frames, some of which do not begin with a canonical AUG start codon or lie in 5′ or 3′ UTRs distinct from the major protein encoding ORF (Starck and Shastri, 2011). Indeed, introduction of a frameshift mutation after codon 4 of an influenza virus NP transgene prevents synthesis of the full-length protein antigen without substantial effects on antigen presentation to CTLs (Fetten et al., 1991). Peptides for MHC class I antigen presentation derived from alternative ORFs have been termed defective ribosomal products or DRiPs, as they are distinct from peptides resulting from normal turnover of full-length proteins (Yewdell, 2011). DRiPs are rapidly synthesized and have short half-lives, helping to explain the swiftness with which peptides derived from stable viral proteins can be presented by cell surface MHC molecules. How DRiPs are produced is only beginning to be understood. Importantly, translation initiation at alternate ORFs that commence with CUG does not require eIF2 and instead requires one of the tRNALeu isoforms capable of initiating protein synthesis at CUG (Starck et al., 2012). While most antigenic peptides are derived from the standard reading frame of proteins, some antigenic peptide production can take place from ORFs commencing with CUG when eIF2 is inactivated. In conclusion, alternative ORFs in mRNAs and the mode with which their translation is controlled contribute to regulate the adaptive immune response.
Translational Control Strategies for Persistence and Immune Avoidance
The host translational machinery also provides opportunities for microbes to avoid adaptive immune surveillance. Herpesviruses use a strategy termed latency to persist indefinitely in specialized host cell types. The neurotrophic alpha herpesvirus, HSV, avoids producing proteins during latency (Wilson and Mohr, 2012). Instead, a non-protein-coding, latency-associated transcript (LAT) accumulates and is processed into microRNAs. MicroRNAs (miRs) act posttranscriptionally, either by inhibiting translation or promoting mRNA decay, to repress gene expression by binding cognate sites typically located within 3′ UTRs of mRNA targets (Fabian and Sonenberg, 2012). Presumably, these virus-encoded miRs act as buffers preventing translation of lytic mRNAs that may spontaneously accumulate below a critical threshold in latently infected neurons (Skalsky and Cullen, 2010). While the virus influences ganglia gene expression in latently infected mice (Kramer et al., 2003), more work is needed to determine how virus-encoded miRs affect neuronal mRNA translation. Studies on related beta and gamma herpesviruses, however, have shown that virus-encoded miRs can indeed influence host gene expression (Gottwein et al., 2011; Riley et al., 2012; Skalsky et al., 2012; Jurak et al., 2012).
Sustained PI3K-Akt signaling is required to maintain the HSV latent state in a neuron cell culture model of latency, hinting at virus-neuron interactions fundamental to latency (Camarena et al., 2010). Persistent mTORC1 activation is required to inactivate the 4E-BP translation repressor and maintain latency, while environmental cues like hypoxia, which suppresses protein synthesis by inhibiting mTORC1 activation, trigger reactivation (Kobayashi et al., 2012). This suggests that latency in neurons requires active signaling that controls cap-dependent mRNA translation. Having these signals integrated through mTORC1 allows the virus to change its lifecycle in response to fundamental indicators of cell homeostasis including neurotrophic factor support, energy levels, amino acid sufficiency, and oxygen levels in a neuron cell-autonomous manner. While it clearly involves changes to ongoing host mRNA translation, it differs from the global inhibition in response to Pseudomonas exotoxin as discussed above and highlights the more subtle changes resulting from altering the population of mRNAs that are translated in a cap-dependent manner on polyribosomes. Remarkably, transient changes in mTOR signaling can be sensed locally in axons and transmitted to latent viral genomes resident in nuclei, offering a glimpse of how axons receive signals from the periphery and transmit them long distances to latent viral genomes in distant ganglia (Kobayashi et al., 2012).
Unlike alpha herpesviruses that colonize nondividing cells, the gamma herpesviruses Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) establish life-long latency in B cells and encode functions to ensure persistence of their genomes within a dividing cell compartment (Speck and Ganem, 2010). EBV EBNA1 is important in maintaining the viral minichromosome during latency in B cells and is a prime target for CTL recognition. To counter the host’s capacity to sample EBNA1-derived antigens and antigens encoded by ORFs other than the primary ORF on a given mRNA, the EBNA1 Gly/Ala repeat (GAr) region suppresses translation in cis. In this manner, GAr reduces the amount of not only the full-length antigen, but also any translation products derived from that mRNA, and thereby decreases their likelihood of being presented (Yin et al., 2003). An unstable mRNA secondary structure resulting from a purine bias within the EBNA1 mRNA GAr-coding sequences is thought to account for the reduced translational efficiency, illustrating how viral mRNA structure impacts immune surveillance (Tellam et al., 2008). The KSHV LANA protein plays a similar role in viral episome maintenance and also contains a segment that inhibits translation in cis. However, LANA’s ability to inhibit translation in cis can be separated from its impact on antigen presentation (Kwun et al., 2011). Further work is required to resolve these questions.
Fine Tuning Inflammatory and Immune Responses via MicroRNAs
Many cellular mRNAs that control innate inflammatory responses and host defenses are subject to miR regulation, enabling cells to balance their response to pathogenic microbes with an additional layer of control (O’Connell et al., 2012). Infection with viruses, bacteria, and protozoa perturb the host miR-ome. In some cases, infection with pathogenic bacteria potentiates innate responses, as L. monocytogenes and M. bovis bacillus Calmette-Guérin (BCG) decrease levels of miR29, which represses IFNγ mRNA expression, in NK, CD4+, and CD8+ T cells (Ma et al., 2011). Alternatively, M. leprae-infected monocytes were found to upregulate hsa-miR-21, allowing the pathogen to inhibit antimicrobial peptide expression (Liu et al., 2012). In response to the invasive intracellular bacteria Salmonella, downregulation of the let-7 miR family derepresses the production of IL-10, which attenuates proinflammatory cytokines and prevents excessive immune activation (Schulte et al., 2011). Likewise, PAMPs sensed by the host can differentially regulate miR expression. Growth of a Pseudomonas syringae mutant defective in type III secretion, which elicits a PAMP-triggered response but cannot suppress it, is partially restored in Arabidopsis mutants deficient in siRNA or miR accumulation (Navarro et al., 2008). In human peripheral blood mononuclear cells (PBMCs), bacterial LPS induces miR21 expression, which in turn decreases the abundance of PDCD4, an inhibitor of eIF4A (Figure 2). By depleting PDCD4, IL-10 mRNA translation is stimulated, limiting excessive immune activation (Sheedy et al., 2010). Even more striking, TLR-4-dependent induction of miR-146a, which dampens levels of the essential TLR-signaling molecule IRAK1, is triggered in neonates by PAMPs when the intestinal mucosa transitions from a sterile, protected site to an environmentally exposed, permanently colonized surface (Chassin et al., 2010). This facilitates protective innate immune tolerance in the neonate intestine. To counter RNAi-mediated host defenses, the Turnip crinkle virus (TCV) capsid, the P38 protein, physically interacts with Arabidopsis Ago1 to suppress RNA silencing (Azevedo et al., 2010). It recognizes Ago1 by mimicking host GW repeat proteins, which bind Ago and promote gene silencing. P38 mutant viruses abrogate TCV virulence, which is partially restored in Arabidopsis Ago1 hypomorphic strains. Similarly, abrogation of miR biogenesis by gut-specific depletion of the miR processing component dicer rendered mice more susceptible to infection with helminthes, indicating a role of miRs in intestinal homeostasis and mucosal immunity (Biton et al., 2011). Apicomplexan parasites, including P. falciparum, P vivax, C. parvum, and T. gondii also change the host miR-ome (Hakimi and Cannella, 2011). Finally, while the poxvirus-encoded poly A polymerase polyadenylates the 3′ end of cellular miRs and stimulates their degradation, endogenous siRNAs and mIRs modified with a terminal 2′O-methyl group persisted, consistent with a role for this RNA modification in host defense (Backes et al., 2012).
Besides influencing the cellular miR-ome, some viruses encode miRs that target both virus and host genes. HCMV establishes latency in monocyte-macrophage precursors. When it initiates lytic replication, HCMV subverts antigen presentation on the cell surface via multiple strategies. While most involve manipulating MHC class I peptide loading, one in particular relies on miR regulation of ERAP1, a host amino peptidase required for MHC class I-presented peptide maturation (Kim et al., 2011). By targeting ERAP1 with the virus miR US4-1, HCMV suppresses the CD8+ T cell response in a distinctive manner involving translational regulation that does not involve antigenic protein production and instead relies on a small non-protein-coding miR. Several herpesviruses (HCMV, EBV, and KSHV) also produce miRs that repress expression of MICB, a stress-induced host cell ligand of the NK cell activating receptor NKG2D NK ligand, to escape NK cell recognition and killing (Stern-Ginossar et al., 2007; Nachmani et al., 2009). Polyomaviruses JC and BK encode a miR that targets ULBP3, a different stress-induced NKG2D ligand for similar purposes (Bauman et al., 2011). Viral infection can likewise suppress cellular miR biogenesis, as Ad VA RNA impairs dicer function (reviewed in Skalsky and Cullen, 2010). Using a more-specific method, MCMV and herpesvirus saimiri (HVS) downregulate the host antiviral miR27 via a virus-encoded transcript. Whereas MCMV utilizes a viral mRNA (m169) that binds the host miR and promotes its degradation (Libri et al., 2012), HVS relies on a small, viral noncoding RNA (Cazalla et al., 2010).
Do Fungal Infections Interfere with Host Translation?
While little is known regarding how fungal infection influences host translation, parallels have been drawn between intracellular fungi that escape phagosomes and bacteria (Bliska and Casadevall, 2009). Exposure of a murine macrophpage cell line to C. albicans decreased abundance of a host eIF3 subunit and the appearance of an acidic, modified form of eEF2, consistent with eEF2 phosphorylation (Martínez-Solano et al., 2006). Transcriptional profiling of Drosophila cells after Candida infection revealed that the fly 4E-BP transcript was among those upregulated, and 4E-BP-deficient flies were approximately 50% less resistant to infection than controls (Levitin et al., 2007). Furthermore, RNAi screens for host factors important for C. neoformans intracellular growth in Drosophila cells identified a potential role for the mTORC1 regulators rheb and TSC together with host autophagy genes (Qin et al., 2011). Finally, commensal fungi can be detected via the C-type lectin receptor Dectin-1, a host pattern recognition receptor important for inducing innate immune responses to fungi, and Dectin-1-deficient mice are more susceptible to colitis (Iliev et al., 2012). Perhaps pathogenic fungi may, like bacteria, be sensed by their hosts as a consequence of their ability to suppress ongoing host translation. Further investigation and experimental validation in human cells is required to test this hypothesis.
Concluding Remarks and Future Perspectives
Infection with a diverse assortment of pathogenic microbes impacts ongoing protein synthesis in the host. This is not limited to viruses, which are known to commandeer the host protein synthesis machinery, but holds true for many bacteria, protozoa, and perhaps fungi. Indeed, innate defense functions performed by host proteins allow host organisms to deftly respond to microbial infection, changes in metabolic status, and environmental or physiologic stress. While control of which mRNAs and ORFs are translated shapes innate and adaptive immune response, the underlying basis for these distinctions remains unknown, although it probably relies on recognition of mRNA structural and sequence determinants. Interference with host mRNA translation likewise provides a powerful indicator of host cell distress. By integrating signals from intracellular and extracellular cues through key regulatory circuits, the host translation machinery can alter initiation and elongation rates on select mRNAs and discrete mRNA subpopulations or suppress global translation through modification of discrete factors. Thus, eIF2 and its inducible regulatory kinases, one of which is an ISG, function as innate immune sentinels, suppressing global translation in infected cells in response to different metabolic, physiologic, or environmental stresses while concomitantly activating translation of host defense mRNAs that contain uORFs. Translation initiation and elongation are controlled for mRNAs that encode innate immune effectors by TSC and mTORC1, which integrate fundamental indicators of cell homeostasis including the availability of energy, nutrients, oxygen, and growth/trophic factors and coordinate translational output. In response to MAP kinase signaling, eIF4E phosphorylation by Mnk prevents induction of NF-κB-responsive genes by stimulating translation of IκB mRNA. Indeed, the extent to which translational control circuits are interwoven into vital cell signaling pathway that monitor fundamental indicators of metabolic and environmental stress makes them ideal sensors to detect pathogen-induced changes or collateral damage. In addition, changes to ongoing host mRNA translation can be harnessed to distinguish harmful organisms from commensals and play a role in determining how persistent infections are maintained through the life of an organism. Importantly, cellular responses to stimulate translation of host defense-related mRNAs are balanced by microbial strategies designed to blunt them. These include global mechanisms to enhance host mRNA turnover and alter mRNP compartmentalization. More selective strategies also operate to subvert signaling to the translational machinery and suppress translation of host defense-related mRNAs, limit microbial antigen production in persistently infected cells, or selectively silence discrete transcripts via miRs. The impact of microbe-induced changes to ongoing host protein synthesis on pathogenesis and the diversity of microbial countermeasures that suppress translation of host defense-related mRNAs illustrates how host mRNA translation helps effectively shape infection and immunity.
Acknowledgments
The authors apologize to colleagues whose contributions were not cited owing to space restrictions. We thank Maritza Jaramillo for critical comments on the manuscript and Andrew Darwin, Victor Torres, and Heran Darwin for helpful discussions. Work in the authors’ laboratories is supported by grants from the National Institutes of Health (AI073898 and GM056927 to I.M.) and the Canadian Institutes of Health Research (MOP93607 to N.S.).
Contributor Information
Ian Mohr, Email: ian.mohr@med.nyu.edu.
Nahum Sonenberg, Email: nahum.sonenberg@mcgill.ca.
References
- Alain T., Lun X., Martineau Y., Sean P., Pulendran B., Petroulakis E., Zemp F.J., Lemay C.G., Roy D., Bell J.C. Vesicular stomatitis virus oncolysis is potentiated by impairing mTORC1-dependent type I IFN production. Proc. Natl. Acad. Sci. USA. 2010;107:1576–1581. doi: 10.1073/pnas.0912344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axten J.M., Medina J.R., Feng Y., Shu A., Romeril S.P., Grant S.W., Li W.H., Heerding D.A., Minthorn E., Mencken T. Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK) J. Med. Chem. 2012;55:7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- Azevedo J., Garcia D., Pontier D., Ohnesorge S., Yu A., Garcia S., Braun L., Bergdoll M., Hakimi M.A., Lagrange T., Voinnet O. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes S., Shapiro J.S., Sabin L.R., Pham A.M., Reyes I., Moss B., Cherry S., Tenoever B.R. Degradation of Host MicroRNAs by Poxvirus Poly(A) Polymerase Reveals Terminal RNA Methylation as a Protective Antiviral Mechanism. Cell Host Microbe. 2012;12:200–210. doi: 10.1016/j.chom.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman Y., Nachmani D., Vitenshtein A., Tsukerman P., Drayman N., Stern-Ginossar N., Lankry D., Gruda R., Mandelboim O. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9:93–102. doi: 10.1016/j.chom.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Beattie E., Paoletti E., Tartaglia J. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L- and E3L- mutant viruses. Virology. 1995;210:254–263. doi: 10.1006/viro.1995.1342. [DOI] [PubMed] [Google Scholar]
- Beretta L., Svitkin Y.V., Sonenberg N. Rapamycin stimulates viral protein synthesis and augments the shutoff of host protein synthesis upon picornavirus infection. J. Virol. 1996;70:8993–8996. doi: 10.1128/jvi.70.12.8993-8996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton M., Levin A., Slyper M., Alkalay I., Horwitz E., Mor H., Kredo-Russo S., Avnit-Sagi T., Cojocaru G., Zreik F. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat. Immunol. 2011;12:239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- Bliska J.B., Casadevall A. Intracellular pathogenic bacteria and fungi—a case of convergent evolution? Nat. Rev. Microbiol. 2009;7:165–171. doi: 10.1038/nrmicro2049. [DOI] [PubMed] [Google Scholar]
- Bogdan C., Mattner J., Schleicher U. The role of type I interferons in non-viral infections. Immunol. Rev. 2004;202:33–48. doi: 10.1111/j.0105-2896.2004.00207.x. [DOI] [PubMed] [Google Scholar]
- Camarena V., Kobayashi M., Kim J.Y., Roehm P., Perez R., Gardner J., Wilson A.C., Mohr I., Chao M.V. Nature and duration of growth factor signaling through receptor tyrosine kinases regulates HSV-1 latency in neurons. Cell Host Microbe. 2010;8:320–330. doi: 10.1016/j.chom.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Manicassamy S., Tang H., Kasturi S.P., Pirani A., Murthy N., Pulendran B. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D., Yario T., Steitz J.A. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Liehl P., Buchon N., Lemaitre B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe. 2012;12:60–70. doi: 10.1016/j.chom.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Chassin C., Kocur M., Pott J., Duerr C.U., Gütle D., Lotz M., Hornef M.W. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. 2010;8:358–368. doi: 10.1016/j.chom.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Chou J., Kern E.R., Whitley R.J., Roizman B.R. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- Clippinger A.J., Alwine J.C. Dynein mediates the localization and activation of mTOR in normal and human cytomegalovirus-infected cells. Genes Dev. 2012;26:2015–2026. doi: 10.1101/gad.196147.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colina R., Costa-Mattioli M., Dowling R.J., Jaramillo M., Tai L.H., Breitbach C.J., Martineau Y., Larsson O., Rong L., Svitkin Y.V. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- Cuesta R., Xi Q., Schneider R.J. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 2000;19:3465–3474. doi: 10.1093/emboj/19.13.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a012286. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Harding H.P., Raught B., Gingras A.C., Berlanga J.J., Scheuner D., Kaufman R.J., Ron D., Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- Dever T.E., Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev S.E., Terenin I.M., Andreev D.E., Ivanov P.A., Dunaevsky J.E., Merrick W.C., Shatsky I.N. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. Biol. Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J., Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews M., Sonenberg N., Hershey J.W.B., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2007. pp. 129–153. [Google Scholar]
- Dunbar T.L., Yan Z., Balla K.M., Smelkinson M.G., Troemel E.R. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe. 2012;11:375–386. doi: 10.1016/j.chom.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde N.C., Child S.J., Geballe A.P., Malik H.S. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes K.A., Dunbar T.L., Powell J.R., Ausubel F.M., Troemel E.R. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2010;107:2153–2158. doi: 10.1073/pnas.0914643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M.R., Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Feigenblum D., Schneider R.J. Modification of eukaryotic initiation factor 4F during infection by influenza virus. J. Virol. 1993;67:3027–3035. doi: 10.1128/jvi.67.6.3027-3035.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P., Everly D.N., Jr., Read G.S. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 2005;79:9651–9664. doi: 10.1128/JVI.79.15.9651-9664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetten J.V., Roy N., Gilboa E. A frameshift mutation at the NH2 terminus of the nucleoprotein gene does not affect generation of cytotoxic T lymphocyte epitopes. J. Immunol. 1991;147:2697–2705. [PubMed] [Google Scholar]
- Fontana M.F., Banga S., Barry K.C., Shen X., Tan Y., Luo Z.Q., Vance R.E. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L., Rong L., Larsson O., Koumakpayi I.H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L.A. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. USA. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia M.M., Covarrubias S., Wong W., Glaunsinger B.A. A common strategy for host RNA degradation by divergent viruses. J. Virol. 2012;86:9527–9530. doi: 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M.A., Contreras I., Hallé M., Tremblay M.L., McMaster R.W., Olivier M. Leishmania GP63 alters host signaling through cleavage-activated protein tyrosine phosphatases. Sci. Signal. 2009;2:ra58. doi: 10.1126/scisignal.2000213. [DOI] [PubMed] [Google Scholar]
- Gottwein E., Corcoran D.L., Mukherjee N., Skalsky R.L., Hafner M., Nusbaum J.D., Shamulailatpam P., Love C.L., Dave S.S., Tuschl T. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppo R., Brown B.A., Palmenberg A.C. Mutational analysis of the EMCV 2A protein identifies a nuclear localization signal and an eIF4E binding site. Virology. 2011;410:257–267. doi: 10.1016/j.virol.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi M.A., Cannella D. Apicomplexan parasites and subversion of the host cell microRNA pathway. Trends Parasitol. 2011;27:481–486. doi: 10.1016/j.pt.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Herdy B., Jaramillo M., Svitkin Y.V., Rosenfeld A.B., Kobayashi M., Walsh D., Alain T., Sean P., Robichaud N., Topisirovic I. Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat. Immunol. 2012;13:543–550. doi: 10.1038/ni.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A.G., Lorsch J.R. The Mechanism of Eukaryotic Translation Initiation: New Insights and Challenges. Cold Spring Harb. Perspect. Biol. 2012 doi: 10.1101/cshperspect.a011544. Published online July 18, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B.C., Yu S.L., Chen J.J., Chang S.Y., Yan B.S., Hong Q.S., Singh S., Kao C.L., Chen H.Y., Su K.Y. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe. 2011;9:58–69. doi: 10.1016/j.chom.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev I.D., Funari V.A., Taylor K.D., Nguyen Q., Reyes C.N., Strom S.P., Brown J., Becker C.A., Fleshner P.R., Dubinsky M. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isler J.A., Skalet A.H., Alwine J.C. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger B.W., Wise H.M., Kash J.C., Walters K.A., Wills N.M., Xiao Y.L., Dunfee R.L., Schwartzman L.M., Ozinsky A., Bell G.L. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo M., Gomez M.A., Larsson O., Shio M.T., Topisirovic I., Contreras I., Luxenburg R., Rosenfeld A., Colina R., McMaster R.W. Leishmania repression of host translation through mTOR cleavage is required for parasite survival and infection. Cell Host Microbe. 2011;9:331–341. doi: 10.1016/j.chom.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Joshi P.B., Kelly B.L., Kamhawi S., Sacks D.L., McMaster W.R. Targeted gene deletion in Leishmania major identifies leishmanolysin (GP63) as a virulence factor. Mol. Biochem. Parasitol. 2002;120:33–40. doi: 10.1016/s0166-6851(01)00432-7. [DOI] [PubMed] [Google Scholar]
- Joshi S., Kaur S., Redig A.J., Goldsborough K., David K., Ueda T., Watanabe-Fukunaga R., Baker D.P., Fish E.N., Fukunaga R., Platanias L.C. Type I interferon (IFN)-dependent activation of Mnk1 and its role in the generation of growth inhibitory responses. Proc. Natl. Acad. Sci. USA. 2009;106:12097–12102. doi: 10.1073/pnas.0900562106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurak I., Silverstein L.B., Sharma M., Coen D.M. Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress the expression of ATRX, an effector of intrinsic immunity. J. Virol. 2012;86:10093–10102. doi: 10.1128/JVI.00930-12. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22013031&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lee S., Shin J., Kim Y., Evnouchidou I., Kim D., Kim Y.K., Kim Y.E., Ahn J.H., Riddell S.R. Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat. Immunol. 2011;12:984–991. doi: 10.1038/ni.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Wilson A.C., Chao M.V., Mohr I. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes Dev. 2012;26:1527–1532. doi: 10.1101/gad.190157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M.F., Cook W.J., Roth F.P., Zhu J., Holman H., Knipe D.M., Coen D.M. Latent herpes simplex virus infection of sensory neurons alters neuronal gene expression. J. Virol. 2003;77:9533–9541. doi: 10.1128/JVI.77.17.9533-9541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczynska B., Sharma B., Eklund E.A., Fish E.N., Platanias L.C. Regulatory effects of programmed cell death 4 (PDCD4) protein in interferon (IFN)-stimulated gene expression and generation of type I IFN responses. Mol. Cell. Biol. 2012;32:2809–2822. doi: 10.1128/MCB.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad L.M., Glaunsinger B.A. Diverse virus-host interactions influence RNA-based regulation during γ-herpesvirus infection. Curr. Opin. Microbiol. 2012;15:506–511. doi: 10.1016/j.mib.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwun H.J., da Silva S.R., Qin H., Ferris R.L., Tan R., Chang Y., Moore P.S. The central repeat domain 1 of Kaposi’s sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) prevents cis MHC class I peptide presentation. Virology. 2011;412:357–365. doi: 10.1016/j.virol.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin A., Marcil A., Tettweiler G., Laforest M.J., Oberholzer U., Alarco A.M., Thomas D.Y., Lasko P., Whiteway M. Drosophila melanogaster Thor and response to Candida albicans infection. Eukaryot. Cell. 2007;6:658–663. doi: 10.1128/EC.00346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Dai J., Song M., Fitzgerald-Bocarsly P., Kiledjian M. Dcp2 decapping protein modulates mRNA stability of the critical interferon regulatory factor (IRF) IRF-7. Mol. Cell. Biol. 2012;32:1164–1172. doi: 10.1128/MCB.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri V., Helwak A., Miesen P., Santhakumar D., Borger J.G., Kudla G., Grey F., Tollervey D., Buck A.H. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. USA. 2012;109:279–284. doi: 10.1073/pnas.1114204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.J., Choi J.A., Choi H.H., Cho S.N., Kim H.J., Jo E.K., Park J.K., Song C.H. Endoplasmic reticulum stress pathway-mediated apoptosis in macrophages contributes to the survival of Mycobacterium tuberculosis. PLoS ONE. 2011;6:e28531. doi: 10.1371/journal.pone.0028531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.T., Wheelwright M., Teles R., Komisopoulou E., Edfeldt K., Ferguson B., Mehta M.D., Vazirnia A., Rea T.H., Sarno E.N. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012;18:267–273. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Han A.-P., Chen J.-J. Translation initiation control by heme-regulated eukaryotic initiation factor 2α kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 2001;21:7971–7980. doi: 10.1128/MCB.21.23.7971-7980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Xu S., Liu X., Zhang Q., Xu X., Liu M., Hua M., Li N., Yao H., Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- Malathi K., Dong B., Gale M., Jr., Silverman R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Solano L., Nombela C., Molero G., Gil C. Differential protein expression of murine macrophages upon interaction with Candida albicans. Proteomics. 2006;6(Suppl 1):S133–S144. doi: 10.1002/pmic.200500581. [DOI] [PubMed] [Google Scholar]
- McEwan D.L., Kirienko N.V., Ausubel F.M. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 2012;11:364–374. doi: 10.1016/j.chom.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C., Perez C., Mohr I. Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA. 2012;109:5627–5632. doi: 10.1073/pnas.1202829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I., Gluzman Y. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 1996;15:4759–4766. [PMC free article] [PubMed] [Google Scholar]
- Moreno J.A., Radford H., Peretti D., Steinert J.R., Verity N., Martin M.G., Halliday M., Morgan J., Dinsdale D., Ortori C.A. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M., Camarena V., Mohr I. Full resistance of herpes simplex virus type 1-infected primary human cells to alpha interferon requires both the Us11 and gamma(1)34.5 gene products. J. Virol. 2004;78:10193–10196. doi: 10.1128/JVI.78.18.10193-10196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J.A., Duerst R.J., Smith T.J., Morrison L.A. Herpes simplex virus type 2 virion host shutoff protein regulates alpha/beta interferon but not adaptive immune responses during primary infection in vivo. J. Virol. 2003;77:9337–9345. doi: 10.1128/JVI.77.17.9337-9345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmani D., Stern-Ginossar N., Sarid R., Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Navarro L., Jay F., Nomura K., He S.Y., Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.M., Rao D.S., Baltimore D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- Parrish S., Moss B. Characterization of a vaccinia virus mutant with a deletion of the D10R gene encoding a putative negative regulator of gene expression. J. Virol. 2006;80:553–561. doi: 10.1128/JVI.80.2.553-561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S., Resch W., Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc. Natl. Acad. Sci. USA. 2007;104:2139–2144. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S., Hurchalla M., Liu S.W., Moss B. The African swine fever virus g5R protein possesses mRNA decapping activity. Virology. 2009;393:177–182. doi: 10.1016/j.virol.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W.C., Sonenberg N. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- Pasieka T.J., Lu B., Crosby S.D., Wylie K.M., Morrison L.A., Alexander D.E., Menachery V.D., Leib D.A. Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J. Virol. 2008;82:5527–5535. doi: 10.1128/JVI.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C., McKinney C., Chulunbaatar U., Mohr I. Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J. Virol. 2011;85:156–164. doi: 10.1128/JVI.01778-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten J.S., Castile A.E., Kieft J.S. Mechanistic role of structurally dynamic regions in Dicistroviridae IGR IRESs. J. Mol. Biol. 2010;395:205–217. doi: 10.1016/j.jmb.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillich H., Loose M., Zimmer K.P., Chakraborty T. Activation of the unfolded protein response by Listeria monocytogenes. Cell. Microbiol. 2012;14:949–964. doi: 10.1111/j.1462-5822.2012.01769.x. [DOI] [PubMed] [Google Scholar]
- Piotrowska J., Hansen S.J., Park N., Jamka K., Sarnow P., Gustin K.E. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 2010;84:3654–3665. doi: 10.1128/JVI.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q.M., Luo J., Lin X., Pei J., Li L., Ficht T.A., de Figueiredo P. Functional analysis of host factors that mediate the intracellular lifestyle of Cryptococcus neoformans. PLoS Pathog. 2011;7:e1002078. doi: 10.1371/journal.ppat.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read G.S., Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate early) viral polypeptides. J. Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner J.M., Clyde K., Pezda A.C., Cheng B.Y., Wang T., Kumar G.R., Covarrubias S., Coscoy L., Glaunsinger B. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLoS Pathog. 2011;7:e1002150. doi: 10.1371/journal.ppat.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley K.J., Rabinowitz G.S., Yario T.A., Luna J.M., Darnell R.B., Steitz J.A. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31:2207–2221. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rothenburg S., Seo E.J., Gibbs J.S., Dever T.E., Dittmar K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat. Struct. Mol. Biol. 2009;16:63–70. doi: 10.1038/nsmb.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J.P., Mazur J., Bankhead P., Hiet M.S., Kallis S., Alvisi G. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte L.N., Eulalio A., Mollenkopf H.J., Reinhardt R., Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimon T.A., Kim M.J., Blumenthal A., Koo J., Ehrt S., Wainwright H., Bekker L.G., Kaplan G., Nathan C., Tabas I., Russell D.G. Induction of ER stress in macrophages of tuberculosis granulomas. PLoS ONE. 2010;5:e12772. doi: 10.1371/journal.pone.0012772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy F.J., Palsson-McDermott E., Hennessy E.J., Martin C., O’Leary J.J., Ruan Q., Johnson D.S., Chen Y., O’Neill L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- Shrestha N., Bahnan W., Wiley D.J., Barber G., Fields K.A., Schesser K. Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion. J. Biol. Chem. 2012;287:28738–28744. doi: 10.1074/jbc.M112.375915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin M.A., Skabkina O.V., Dhote V., Komar A.A., Hellen C.U., Pestova T.V. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky R.L., Corcoran D.L., Gottwein E., Frank C.L., Kang D., Hafner M., Nusbaum J.D., Feederle R., Delecluse H.J., Luftig M.A. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M., Jan E., Mulder A., Grassucci R.A., Sarnow P., Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Speck S.H., Ganem D. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe. 2010;8:100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck S.R., Shastri N. Non-conventional sources of peptides presented by MHC class I. Cell. Mol. Life Sci. 2011;68:1471–1479. doi: 10.1007/s00018-011-0655-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck S.R., Jiang V., Pavon-Eternod M., Prasad S., McCarthy B., Pan T., Shastri N. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science. 2012;336:1719–1723. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N., Elefant N., Zimmermann A., Wolf D.G., Saleh N., Biton M., Horwitz E., Prokocimer Z., Prichard M., Hahn G. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M.F. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J. Virol. 1977;23:751–767. doi: 10.1128/jvi.23.3.751-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallóczy Z., Jiang W., Virgin H.W., 4th, Leib D.A., Scheuner D., Kaufman R.J., Eskelinen E.L., Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl. Acad. Sci. USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.W., Mahajan-Miklos S., Ausubel F.M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I., Sorbara M.T., Vuckovic D., Ling A., Soares F., Carneiro L.A., Yang C., Emili A., Philpott D.J., Girardin S.E. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- Tellam J., Smith C., Rist M., Webb N., Cooper L., Vuocolo T., Connolly G., Tscharke D.C., Devoy M.P., Khanna R. Regulation of protein translation through mRNA structure influences MHC class I loading and T cell recognition. Proc. Natl. Acad. Sci. USA. 2008;105:9319–9324. doi: 10.1073/pnas.0801968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger V., Aranda M.A. Recessive resistance to plant viruses. Adv. Virus Res. 2009;75:119–159. doi: 10.1016/S0065-3527(09)07504-6. [DOI] [PubMed] [Google Scholar]
- Ventoso I., Sanz M.A., Molina S., Berlanga J.J., Carrasco L., Esteban M. Translational resistance of late alphavirus mRNA to eIF2α phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D., Mohr I. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 2006;20:461–472. doi: 10.1101/gad.1375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D., Arias C., Perez C., Halladin D., Escandon M., Ueda T., Watanabe-Fukunaga R., Fukunaga R., Mohr I. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol. Cell. Biol. 2008;28:2648–2658. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D., Mathews M., Mohr I. Tinkering with translation: Protein synthesis in virus-infected cells. In: Hershey J.W.B., Sonenberg N., Mathews M.B., editors. Protein Synthesis and Translational Control. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2012. pp. 299–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T., Costantino G., Poglitsch M., Rosner M., Zeyda M., Stuhlmeier K.M., Kolbe T., Stulnig T.M., Hörl W.H., Hengstschläger M. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- White J.P., Lloyd R.E. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Mohr I. A cultured affair: HSV latency and reactivation in neurons. Trends Microbiol. 2012 doi: 10.1016/j.tim.2012.08.005. Published online September 7, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.E., Pestova T.V., Hellen C.U., Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wolfson J.J., May K.L., Thorpe C.M., Jandhyala D.M., Paton J.C., Paton A.W. Subtilase cytotoxin activates PERK, IRE1 and ATF6 endoplasmic reticulum stress-signalling pathways. Cell. Microbiol. 2008;10:1775–1786. doi: 10.1111/j.1462-5822.2008.01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S., Eidenschenk C., Arnold C.N., Siggs O.M., Sun L., Brandl K., Mullen T.-M., Nemerow G.R., Moresco E.M.Y., Beutler B. Increased susceptibility to DNA virus infection in mice with a GCN2 mutation. J. Virol. 2012;86:1802–1808. doi: 10.1128/JVI.05660-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.W., Cui D., Arellano J., Dorweiler B., Harding H., Fitzgerald K.A., Ron D., Tabas I. Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat. Cell Biol. 2009;11:1473–1480. doi: 10.1038/ncb1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.W., Kutzler L., Kimball S.R., Tabas I. Toll-like receptor activation suppresses ER stress factor CHOP and translation inhibition through activation of eIF2B. Nat. Cell Biol. 2012;14:192–200. doi: 10.1038/ncb2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell J.W. DRiPs solidify: progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 2011;32:548–558. doi: 10.1016/j.it.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Manoury B., Fåhraeus R. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science. 2003;301:1371–1374. doi: 10.1126/science.1088902. [DOI] [PubMed] [Google Scholar]
- Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D.T., Back S.H., Kaufman R.J. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Zhao L., Jha B.K., Wu A., Elliott R., Ziebuhr J., Gorbalenya A.E., Silverman R.H., Weiss S.R. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]