Summary
Coronaviruses contain a positive-sense single-stranded genomic (g) RNA, which encodes nonstructural proteins. Several subgenomic mRNAs (sgmRNAs) encoding structural proteins are generated by template switching from the body transcription regulatory sequence (TRS) to the leader TRS. The process preferentially generates shorter sgmRNA. Appropriate readthrough of body TRSs is required to produce longer sgmRNAs and full-length gRNA. We find that phosphorylation of the viral nucleocapsid (N) by host glycogen synthase kinase-3 (GSK-3) is required for template switching. GSK-3 inhibition selectively reduces the generation of gRNA and longer sgmRNAs, but not shorter sgmRNAs. N phosphorylation allows recruitment of the RNA helicase DDX1 to the phosphorylated-N-containing complex, which facilitates template readthrough and enables longer sgmRNA synthesis. DDX1 knockdown or loss of helicase activity markedly reduces the levels of longer sgmRNAs. Thus, coronaviruses employ a unique strategy for the transition from discontinuous to continuous transcription to ensure balanced sgmRNAs and full-length gRNA synthesis.
Graphical Abstract
Highlights
-
•
GSK-3 inhibitor reduces coronavirus nucleocapsid phosphorylation (pN) and titer
-
•
pN recruits RNA helicase DDX1 and is crucial for long sgmRNA and gRNA synthesis
-
•
DDX1 knockdown or loss of helicase activity impairs longer sgmRNAs synthesis
-
•
pN/DDX1 bind to viral genome and assist template readthrough of viral body TRSs
To generate the full complement of genomic and subgenomic RNAs, coronaviruses transition from discontinuous to continuous transcription. Wu et al. find that phosphorylation of the viral nucleocapsid by glycogen synthase kinase-3 is required to recruit the cellular RNA helicase DDX1 and promote the switch to continuous transcription, suggesting therapeutic targets.
Introduction
Before the severe acute respiratory syndrome (SARS) pandemic, human coronaviruses (CoVs), such as 229E and OC43, were typically the causes of mild upper respiratory tract diseases or the common cold (Stadler et al., 2003). The SARS-CoV, the causative pathogen for the SARS outbreak, was the first high pathogenic human CoV to be identified, with a mortality rate of approximately 10% (Stadler et al., 2003). More recently, the Middle East respiratory syndrome CoV emerged, also a high pathogenic human CoV, with a mortality rate of nearly 50%. CoV infection has thus become an increasing threat to human society. Increased understanding of the key viral factors critical for CoV replication, as well as the underlying mechanisms, could facilitate the identification of appropriate targets for the development of antiviral strategies.
CoVs are enveloped RNA viruses containing a positive-sense single-stranded genomic (g) RNA, approximately 30 kb in size (Lai et al., 2007), which encodes nonstructural proteins involved in viral replication (Brian and Baric, 2005). Several subgenomic (sg) mRNAs are generated during viral replication, which predominantly encode structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, and some species-specific accessory proteins (Sawicki et al., 2007). All viral RNAs, including gRNAs and sgmRNAs, are coterminal, and the sgmRNAs are generated through a unique discontinuous transcription mechanism during the synthesis of negative-strand RNA (Pasternak et al., 2006, Sawicki et al., 2007). This discontinuous process is controlled by a conserved transcription regulating sequence (TRS), which is located after the leader sequence (leader TRS) and in front of each gene (body TRS). It has been suggested that through base pairing between the leader TRS and the complementary body TRS, a template-switching event, occurs to generate the discontinuous minus-strand RNAs that serve as the templates for the transcription of large amounts of discontinuous nested plus-strand sgmRNAs. The discontinuous transcription process encounters a decision problem when reaching the TRS: either transcription stops and switches to the leader TRS to produce shorter sgmRNAs, or transcription continues through the TRS to generate longer sgmRNAs and gRNAs. Greater abundance of shorter than longer sgmRNAs suggests that transcription might switch upon reaching the 3′ end of the body TRS to produce shorter sgmRNAs (Pasternak et al., 2004). However, the virus has to pass the body TRS in an appropriate proportion to produce sufficient longer sgmRNAs and gRNA essential to the life cycle. Studies have investigated several cis-regulating elements and trans-regulating factors involved in this process (Sola et al., 2011), but the molecular mechanisms controlling the switch from discontinuous to continuous transcription in CoV still remain unclear.
The most abundant viral sgmRNA encodes the viral N protein during infection. Categorized as a structural protein, the N protein forms a helical ribonucleoprotein structure through the wrapping of gRNA by the RNA chaperone domain (Spencer and Hiscox, 2006, Zúñiga et al., 2007), which is required for gRNA packaging into the virion (Hurst et al., 2005, Kuo and Masters, 2002, Narayanan et al., 2000). The N protein might also participate in the discontinuous transcription of sgmRNAs, because depletion of N from the replicon reduces the synthesis of sgmRNA, but not gRNA (Zúñiga et al., 2010).
The N protein is a highly basic protein with substantial phosphorylation modifications, predominantly at serine (Ser) residues (Calvo et al., 2005, Chen et al., 2005, Lin et al., 2007, White et al., 2007, Wu et al., 2009). Our previous study demonstrated that the major phosphorylation sites in both SARS-CoV and the mouse hepatitis virus (MHV) JHM strain (JHMV) are the Ser residues clustered within the central Ser-arginine (SR)-rich motif (Wu et al., 2009). Glycogen synthase kinase-3 (GSK-3) is the kinase responsible for the phosphorylation of this SR-rich motif and is conserved in both JHMV and SARS-CoV. Treatment with GSK-3 inhibitor reduces the phosphorylation of N protein and reduces the viral titer and cytopathic effects, suggesting that the phosphorylation of N is relevant to viral life cycle. In this study, we discovered a role of GSK-3-mediated pN in supporting the transition from discontinuous to continuous transcription to produce a full set of viral sgmRNAs and gRNAs through interaction with the RNA helicase DDX1. This mechanism ensures appropriate balance between the synthesis of longer and shorter sgmRNAs and gRNAs during CoV infection.
Results
GSK-3 Inhibitors Reduce JHMV Viral RNA Synthesis
We selected JHMV as the model virus to investigate the role of GSK-3-mediated N phosphorylation in the regulation of viral replication, first examining the kinetics for this specific N phosphorylation in the viral life cycle. Both the virions in the supernatant and the cell lysates harvested at different time points after the JHMV infection were processed for western blot analysis using antibodies (Abs) specific to total N and phospho-Ser197 N (pS197-N), which is a target residue in the SR motif phosphorylated by GSK-3 (Wu et al., 2009). The N protein in the virions was predominantly the unphosphorylated form at Ser197 (Figure 1 A, lane 1). After infection, the entry N protein degraded gradually, and we detected no phosphorylation at N-Ser197 until the newly synthesized N appeared 8 hr postinfection (p.i.) (Figure 1A). The majority of the intracellular N-Ser197 was phosphorylated, contrasting with observations for the N protein in virions.
Figure 1.
GSK-3 Inhibitor Treatment Reduces JHMV Nucleoprotein Phosphorylation at Ser197 and Viral Titer
(A) Kinetics of GSK-3-mediated N phosphorylation in cells infected with JHMV at an moi of 1 IU/cell. Proteins were harvested at the indicated time points and probed with N and pS197-N. “V” indicates virions.
(B) The total proteins were analyzed for N, pS197-N, and β-actin (loading control) by using western blotting.
(C) The viral titers of DMSO and kenpaullone-treated cells were estimated using plaque assay.
(D) Northern blotting was used to evaluate the expression levels of viral RNAs, using DIG-labeled N (upper panel) and GAPDH (bottom panel) as probes. Lanes 5 and 6 show hybridization using probes detecting minus-strand RNA. Lanes 3, 4, 7, and 8 represent the shorter exposure results, shown in lanes 1, 2, 5, and 6.
(E) The relative abundance of indicated viral RNAs in DMSO and kenpaullone-treated JHMV-infected cells was evaluated by RT-qPCR, with the DMSO control set to a value of 1. “Inf.” indicates infection; “kenp.” indicates kenpaullone.
To investigate the function of GSK-3 phosphorylated N in viral replication, we infected DBT cells pretreated with the GSK-3-specific inhibitor kenpaullone with JHMV and evaluated the effect on viral RNA and protein synthesis. The cellular RNA and protein, and the viruses in the culture supernatant at 8 hr p.i., a time point to ensure that the samples derived from one synchronous replication cycle (Frana et al., 1985), were harvested for analysis. The total N protein levels were similar with or without kenpaullone treatment. However, the anti-pS197-N signal was significantly decreased after treatment (Figure 1B, lane 2 versus lane 3), which supported the inhibitory effects of kenpaullone on N-Ser197 phosphorylation. Kenpaullone treatment reduced the viral titer in the supernatant by approximately 80% (Figure 1C), which indicated the involvement of GSK-3-mediated N phosphorylation in viral replication.
Our northern blot results revealed that kenpaullone treatment downregulated the expression of the viral RNA transcripts. The results indicated a preferential reduction of full-length gRNA and longer sgmRNAs, in comparison with the shorter sgmRNA6/7 (Figure 1D). Quantitative reverse transcription-polymerase chain reaction (RT-qPCR) analysis, by a panel of primer sets specific for each RNA transcript (as primer sequences listed in Table S1, available online), verified the inhibitory pattern (Figure 1E).
GSK-3-Mediated N Phosphorylation Upregulates the Synthesis of gRNA and Longer sgmRNAs of JHMV
To investigate the possibility that GSK-3-mediated N phosphorylation facilitates the transcription of longer viral RNAs during viral replication, we established stable cell lines overexpressing various N proteins to examine the effects on viral RNA levels. In our previous study, mutation of the priming site for GSK-3 at Ser 205 abrogated all N phosphorylation at SR motif (Wu et al., 2009). Therefore, we generated three DBT cell lines stably expressing the wild-type N (WT-N), the unphosphomimicry N (S205A-N, with Ser205 mutated to Ala), and the phosphomimicry N (3D-N, with Ser197, Ser201, and Ser205 changed to Asp). A green fluorescent protein (GFP) tag was fused at the carboxyl terminus in all of these N proteins, which enabled the microscopic detection and flow cytometry analysis of the stably expressed N in the majority of DBT cells (Figures 2 A and S1A–S1D). The equivalent expression of the N proteins and the similar growth rates of these cells were confirmed by the western blot (Figure 2B) and the MTT analysis (Figure S1E).
Figure 2.
pS197-N Upregulates The Expression of Longer RNAs, whereas S205A-N Functions as a Dominant-Negative Mutant
(A and B) (A) Microscopy and (B) western blotting confirmed the stable expression of GFP proteins or GFP-fused N proteins. Image panels show bright-field images (BF) and green fluorescent (GFP) channels.
(C) Viral RNA synthesis was determined by northern blotting, with the N gene (upper) and GAPDH (bottom) as probes.
(D) The relative abundance of indicated viral RNAs in stable cells was evaluated by RT-qPCR with GFP control was set to a value of 1.
(E) A western blot of the proteins in the infected stable cell lines. The GFP-tagged N and virus-derived N were detected using anti-N antibodies with MW close to 75 kDa and 50 kDa, respectively.
(F) The viral titers in the supernatants of stable cells infected with JHMV were determined by plaque assay.
We infected the three stable cell lines with JHMV (multiplicity of infection [moi] = 1) and harvested the RNA, proteins, and supernatants at 8 hr p.i. for analysis. In comparison with the control cells stably expressing GFP, we observed higher expression of viral gRNA and sgmRNAs in the cells expressing WT-N and 3D-N (Figure 2C, lane 1 versus lanes 2 and 4). More substantial increases in longer than shorter viral RNAs were noted and verified by RT-qPCR for specific transcripts (Figure 2D). In addition, we observed a markedly reduced synthesis of viral RNA in the cells expressing S205A-N (Figure 2C, lane 3). Consistently, we detected considerably higher expression of viral N protein and higher viral titers in the cells expressing WT-N and 3D-N than in the cells expressing S205A-N (Figure 2E, western blot; Figure 2F, viral titer analysis).
Therefore, our results suggested the involvement of GSK-3-phosphorylated N in the synthesis of viral RNA, particularly longer viral RNAs, leading to higher viral titers. It also indicated that the exogenous overexpression of unphosphorylated N (by GSK3) could function as a dominant-negative regulator during viral RNA synthesis.
Sucrose Fractions Containing the pS197-N Protein Complex Are Associated with CoV RNA Synthesis at the Early Stage of Viral Replication
Subsequently, we investigated the manner in which pS197-N upregulates the synthesis of longer viral RNAs of JHMV. It was established that the viral sgmRNA is synthesized by a discontinuous transcription mechanism during negative-strand RNA synthesis from viral gRNA. We thus examined the levels of minus-strand sgmRNA in kenpaullone-pretreated cells by hybridization with strand specific probes. The northern blot results showed parallel reductions in the amounts of longer negative-strand RNAs and positive-strand RNAs (Figure 1D), indicating that inhibition of N phosphorylation by GSK-3 suppresses transcription readthrough of the body TRSs in the viral RNA template.
To test this hypothesis, we tried to determine if any protein complex specifically interacts with pS197-N. Zonal sedimentation analysis on sucrose density gradients was used to separate protein complexes according to their molecular mass under nondenaturing conditions (evaluating at 4, 6, 8, 10, and 16 hr p.i.). We analyzed the fractions from each time point by western blotting with anti-N and anti-pS197-N Abs. The pS197-N at early time points after infection distributed into two peaks (Figure 3 A, 4 hr and 6 hr p.i.): the lighter fractions ranging from fractions 4 to 7 and the heavier fractions ranging from fractions 9 to 12. The pS197-N-containing lighter fractions decreased with time (Figure 3A, at 8, 10, and 16 hr p.i.). The precursor-product relationship between the light and heavy peaks has been confirmed by the pulse-chase experiments (Figure 3B), indicating the pS197-N containing protein complexes at heavier fractions were derived from those in the lighter fractions. We also analyzed the RNA in each fraction from cells at 6 and 10 hr p.i. by northern blot. The results suggested that the pS197-N-containing protein complexes were in the fractions containing the viral RNA transcripts with progressively increasing length (Figure 3C, 6 and 10 hr p.i.). The pS197-N-containing lighter fraction appeared to consist of the active nascent viral transcription complex that later transitioned to longer viral sgmRNAs.
Figure 3.
Lighter Fractions Containing the pS197-N-containing Protein Complex Are Involved in the Early Viral Life Cycle
(A) Whole-cell extracts prepared from JHMV-infected DBT cells at the indicated time points (cells collected at 6, 8, 10, and 16 hr p.i. were infected at moi = 1; cells collected at 4 hr p.i. were infected at moi = 10) were treated with the vehicle control or kenpaullone and then subjected to sucrose-density sedimentation analysis. The isolated fractions were analyzed using western blotting with anti-pS197-N and anti-N antibodies.
(B) JHMV-infected DBT cells (moi = 10) were metabolically labeled for 1 hr with 35S-methionine (upper panel), and then chased for 2 hr (middle panel) or 4 hr (lower panel) in cold media, and then subjected to sucrose gradients analysis and separated into 12 fractions. The immunoprecipitants brought down by anti-N Ab were analyzed by SDS-PAGE and visualized by autoradiography.
(C) Northern blots of viral RNA from (A): gradient-purified lysates at 6 hr p.i. (upper panel) and 10 hr p.i. (middle panel), and 6 hr p.i. with kenpaullone treatment (bottom panel), obtained using N as a probe.
(D) The localization of N in 8 hr p.i. cells with DMSO (middle panel) or kenpaullone treatment (lower panel) was analyzed using indirect immunofluorescence. “Inf.” indicates infection; “kenp.” indicates kenpaullone.
We then treated the cells with kenpaullone and harvested the lysates at 6 hr p.i. and 8 hr p.i. before conducting the same analyses. Without phosphorylation of N by GSK-3, the N-containing lighter fractions (before fraction 10) disappeared, with protein complex detectable only in the heavier fractions (fractions 11 and 12) (Figure 3A, bottom panels with kenpaullone treatment). Although the longer sgmRNAs were almost eliminated, the shorter sgmRNA6/7 remained unaffected, appearing only in the heavier fractions 11 and 12 (Figure 3C, bottom panel). Therefore, the transcription of sgmRNA6/7 does not require the GSK-3-mediated phosphorylation of N protein, which, however, is essential for the transcription of longer viral RNAs by forming specific protein complexes in lighter fractions.
As revealed by the immunofluorescence staining, we found a distinct distribution pattern for N in kenpaullone-treated cells compared with that in cells without drug treatment (Figure 3D). It raised a possibility that the N-containing complexes observed in the heavier fractions of kenpaullone-treated cells might not in the same compartment as those in untreated cells. We have applied the Ab against GRP78, a marker for endoplasmic reticulum (ER) membrane and ER-derived multiple membrane structures (Reddy et al., 2003), for hybridization of the sucrose fractions. The results showed GRP78 codistributed with the pS197-N containing complex in virus-infected cells, which, however, was absent in the N-containing fractions from the kenpaullone treated cells (Figure S2). The results indicated the N-containing complexes in JHMV-infected cells could be in different compartment as those treated with kenpaullone.
The RNA Helicase DDX1 Interacts with pS197-N in JHMV-Infected Cells
To identify the proteins associated with pS197-N in the complexes, we analyzed the lighter fractions in the lysate harvested at 6 hr p.i.. Immunoprecipitation (IP) of pS197-N followed by mass spectrometry analysis identified several proteins with potential functions in RNA biogenesis (Table S2). We further analyzed DDX1, a putative RNA helicase that functions in the unwinding of the duplex RNA structure, because of its possible activity in opening the extensive stem-loop structure of CoV gRNA during the discontinuous transcription process.
To validate the interaction between pS197-N and DDX1, we collected the lysates at 6 hr p.i. and subjected them to coimmunoprecipitation (coIP) with an anti-pS197-N Ab. A specific interaction between pS197-N and DDX1 was identified (Figure 4 A, lane 5), which was decreased by a prior RNase A treatment (Figure 4A, lane 6). We also analyzed the lysate from kenpaullone-treated JHMV-infected cells by IP with an anti-N Ab that recognizes both phospho- and unphospho-S197-N. As expected, kenpaullone treatment substantially reduced the amount of pS197-N (Figure 4B, lane 1 versus lane 2). When the total N was pulled down at a similar level (Figure 4B, middle panel, lanes 3 and 4), the level of DDX1 was considerably lower in the IP products from kenpaullone-treated cells than in those from untreated cells (Figure 4B, lower panel, lanes 3 and 4). These observations indicated that the interaction of N with DDX1 is predominantly contributed by pS197-N and possibly involves RNA.
Figure 4.
Confirmation of the Interaction between pS197-N and DDX1
JHMV-infected DBT lysates (moi = 1) with DMSO or kenpaullone treatment were harvested at 6 hr p.i. and subjected to the following analyses.
(A) IP-western blot analysis of infected cell lysates with or without RNase A treatment, precipitated with a pS197-N antibody.
(B) IP-western blot analysis of infected cells treated with DMSO or kenpaullone, precipitated with an anti-N antibody.
(C) The cell lysates subjected to sucrose gradients were fractionated and analyzed using western blotting to characterize the DDX1 sedimentation pattern.
(D) Subcellular fractionation was used to confirm the intracellular protein distribution. “N” denotes the nuclear fraction, and “C” indicates the cytosolic fraction. Lamin A/C and tubulin were used as control markers for the nuclear and cytosolic fractions, respectively.
In addition to coIP, we used cosedimentation analysis to verify the interaction between pS197-N and DDX1. In comparison with the mock control cells, JHMV infection changed the distribution of cellular DDX1 to a two-peak pattern in sucrose gradient fractions, which reflected the distribution pattern of pS197-N (Figure 4C, Inf. versus Mock). Kenpaullone treatment reversed this DDX1 distribution pattern (Figure 4C, Inf./kenp.). In subcellular fractionation experiments, DDX1 was predominantly localized in the nucleus in mock cells (Figure 4D, lane 1 versus lane 2). However, in JHMV-infected cells, DDX1 was mainly located in the cytosol, where pS197-N is preferentially localized (Figure 4D, lane 4 versus lane 3). We also used immunofluorescence staining to further verify the interaction between pS197-N and DDX1, showing a colocalization pattern at cytosol in JHMV-infected cells (Figure S3).
DDX1 Is Critical for the Synthesis of Longer JHMV RNAs in a Helicase Enzyme Activity-Dependent Manner
To examine the functional role of DDX1 interacting with pS197-N in viral RNA synthesis, we knocked down the expression of DDX1 in DBT cells by using two lenti-shRNAs targeting murine Ddx1, with an efficacy >80% (Figure 5 A, lanes 2 and 3 versus lane 1). Northern blot analysis revealed that DDX1 knockdown markedly reduced the synthesis of longer viral RNAs but minimally reduced the synthesis of shorter sgmRNA6/7 (Figure 5B), which was again verified by RT-qPCR analysis (Figure 5C). Meanwhile, our results indicated that shDdx1s did not affect the level of N protein encoded by sgmRNA7 (Figure 5D, lanes 2 and 3 versus lane 1), but the reduction of viral titer (Figure 5E, bars 2 and 3 versus bar 1) was consistent with that of viral gRNA (Figure 5C, sgmRNA1).
Figure 5.
ShRNA-Mediated Knockdown of Ddx1 in DBT Cells Inhibits JHMV Viral RNA Synthesis
The shLuc/DBT and shDdx1/DBT cells were (A) mock infected or (B–F) infected by JHMV at a moi of 1 (ending at 8 hr p.i.).
(A) Western blotting was used to estimate the efficiency of lentivirus-mediated DDX1 knockdown.
(B) Isolated total RNA from the infected cells was analyzed using northern blotting with N- and GAPDH-specific probes. Lanes 4–6 show hybridization using probes detecting minus-strand RNA.
(C) The relative abundance of indicated viral RNAs in cells was evaluated by RT-qPCR with shLuc/DBT was set to a value of 1.
(D) The infected cell lysates were analyzed for N using western blotting with anti-pS197-N and anti-N antibodies.
(E) The viral titers determined by plaque assay are shown in the bar graph.
(F) Expression of viral RNAs and proteins of JHMV-infected shLuc/DBT (left panel) and shDdx1/DBT (right panel). Northern hybridization for viral RNA purified from sucrose gradients by an N-specific probe and the western blot analysis for the indicated proteins.
We further examined the effects of shDdx1 on the RNA expression pattern in sucrose gradient fractions. The results showed that DDX1 knockdown attenuated the expression of viral RNAs, specifically in the fractions associated with longer viral RNA synthesis (Figure 5F, right panel, fractions 9–12), but did not reduce the levels of sgmRNA6/7 in the lighter fractions (Figure 5F, right panel, fractions 4–7). The pS197-N-containing complexes in these heavier fractions decreased, which suggested their dependence on DDX1 (Figure 5F, bottom panel). These results supported the idea that DDX1 in the complexes in these heavier fractions might play a role in regulating the synthesis of longer viral RNAs.
Subsequently, we used the gain-of-function approach to overexpress DDX1, both the wild-type and enzyme-dead constructs, and evaluated the effects on viral RNA levels. For the enzyme-dead mutant of DDX1, a conserved lysine (K) at residue 52 within the GKT motif which is necessary for ATP binding and hydrolysis (Hanson and Whiteheart, 2005, Tanner et al., 2003) was substituted by alanine (A). We constructed and delivered the shRNA-resistant myc-tagged DDX1 expression plasmids into the DBT cells, within which the endogenous DDX1 had already been knocked down by using shDdx1-1 (Figure 6 A, lanes 4–6).
Figure 6.
DDX1 Upregulates the Expression of Longer RNAs in an Enzyme Activity-Dependent Manner
(A) DBT cells were transfected with the indicated Myc-tagged DDX1 expression construct or an empty vector with (lanes 4–6) or without (lanes 1–3) shDdx1 transduction. The expression of exogenous DDX1 and endogenous DDX1 was confirmed using western blotting. “exo.” indicates exogenous; “endo.” indicates endogenous.
(B–E) shDdx1/DBT cells transfected with the indicated plasmid were infected with JHMV (moi = 1), and the samples were harvested 8 hr p.i. for the following analyses. (B) Isolated RNAs were analyzed using northern blotting with N (upper) and GAPDH (bottom) as probes. Lanes 4–6 show hybridization using probes detecting minus-strand RNA. (C) The relative abundance of indicated viral RNAs in cells was evaluated by RT-qPCR with vector transfected DBT was set to a value of 1. (D) The JHMV-infected cells were analyzed using western blotting for the indicated proteins. (E) The plaque assay was applied to determine the supernatant viral titers.
Northern blotting demonstrated that overexpression of the wild-type, but not the K52A mutant DDX1 construct, increased the synthesis of longer viral RNAs (Figure 6B, lanes 2 and 3 versus lane 1), which was verified by RT-qPCR (Figure 6C). Consistently, the levels of the N protein remained unaffected by DDX1 overexpression (Figure 6D, lanes 2 and 3 versus lane 1), but the viral titer increased in an enzyme-activity-dependent manner (Figure 6E, bar 2 versus bars 1 and 3). These results supported the idea that the helicase activity of DDX1 is critical for the stimulation of the synthesis of longer viral RNAs.
Preferential Binding of pS197-N and the DDX1 Complex to the 5′ Viral Genome to Increase the Synthesis of Longer Viral RNAs
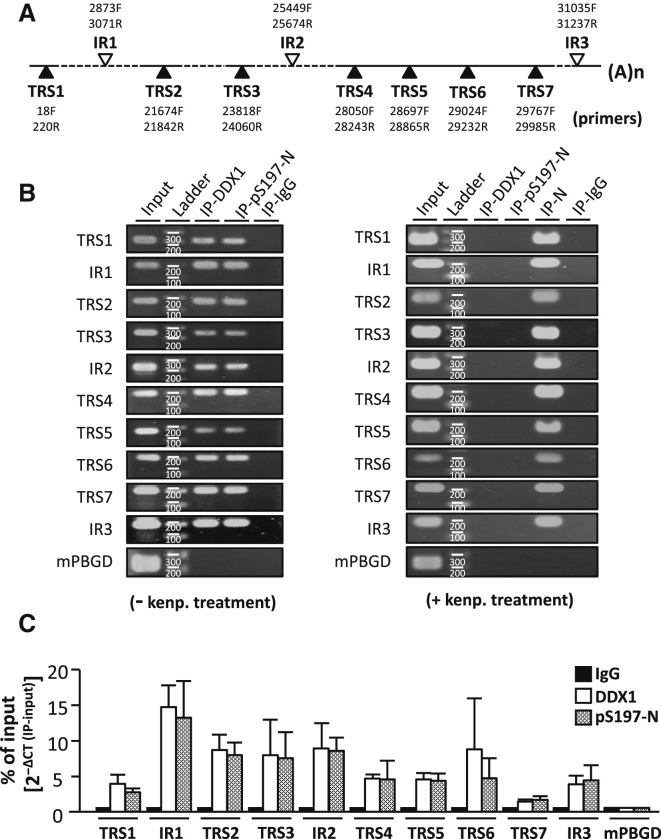
We then aimed to determine the mechanism underlying the interaction between pS197-N and DDX1 to facilitate the synthesis of longer viral RNAs. To examine if these two proteins bind to viral RNAs, particularly the TRSs, we included all of the seven TRSs and three other sites at intergenic regions (IR 1–3) as candidate targets in our analysis. We analyzed the lysates isolated from virus-infected cells by using RNA chromatin immunoprecipitation (RNA ChIP), with Abs against pS197-N and DDX1 for IP experiments. Figure 7 A shows the ten primer sets used in the RT-qPCR reaction (primer sequences in Table S3), with similar amplification efficiency for all amplicons (Table S3). The lysate from kenpaullone-treated cells was included as a negative control in RNA ChIP analysis.
Figure 7.
RNA ChIP-qPCR Analysis of the Sequences Associated with pS197-N and DDX1
(A) Schematic representation of the localization of primers. The closed triangle indicates primer sets covering the TRS motif, whereas the open triangle indicates primer sets localized between TRSs.
(B) Cells (6 hr p.i.) with DMSO (left panel) or kenpaullone treatment (right panel) were used for RNA ChIP analysis. IP was performed using specific antibodies and amplified by primers as indicated.
(C) ChIP-RT-qPCR analysis by using viral RNA from JHMV cells (6 hr p.i.). The results are expressed as percentages of input.
As the control IP with IgG and control PCR with primer set for a cellular gene PBGD (Porphobilinogen deaminase) detected no RT-PCR products, we detected specific reaction products when analyzing the virus-infected lysate by IP with anti-N or anti-pS197-N and anti-DDX1 Abs for each primer set (Figure 7B). The results showed a comparable binding pattern for pS197-N and DDX1 in all of the target sites, which was consistent with the hypothesis that they occur in the same complexes. We observed a relatively higher binding signal for both proteins in the 5′ RNA genome (except TRS1) at IR1, TRS2, TRS3, and IR2, and relatively lower signal for their binding to the 3′ RNA genome at TRS4, TRS5, TRS6, TRS7, and IR3 (with lowest binding signal for TRS7) (Figure 7C). Kenpaullone treatment significantly reduced the binding of DDX1 to all target sites (Figure 7B, right panel), which supported the idea that this binding is mediated by pS197-N. The RNA binding pattern might contribute to regulate transcription template switching and the synthesis of longer sgmRNAs and gRNA by pS197-N/DDX1 complexes.
Discussion
Studies have reported that the CoV N protein functions as a structural protein and is involved in viral transcription and replication (Almazán et al., 2004, Schelle et al., 2005, Zúñiga et al., 2010). Phosphorylation is a renowned modification of CoV N; however, little is known regarding the phosphorylated N protein’s function in viral replication. Extending on results from our previous work, in this study we demonstrate that GSK-3-mediated phosphorylation of the N protein is essential for viral RNA biogenesis. Evidence from loss-of-function (by treatment with the GSK-3 inhibitor kenpaullone) and gain-of-function (by establishing N-expressing cell lines) analyses support the role of GSK-3-phosphorylated N in facilitating the synthesis of gRNA and long sgmRNAs of JHMV (Figures 1 and 2).
Our study demonstrates the involvement of CoV N phosphorylation in regulation of the discontinuous transcription process. We identified an interacting partner for pS197-N, the DDX1 RNA helicase, which has the appropriate biochemical properties to mediate this unique function of GSK-3 phosphorylated N. DDX1 is a member of the DEAD-box protein family, which is the largest family of the superfamily 2 (SF2) helicases (Byrd and Raney, 2012). We observed that knockdown of DDX1 reduced the levels of longer viral RNAs, while overexpression of the wild-type, but not the enzyme-dead, construct of DDX1 was able to rescue. We further identified that after JHMV infection, the distribution of DDX1 changed from the nucleus to the cytosol, where viral RNA synthesis occurs. Our results provide strong evidence to support that phosphorylation modification is a unique strategy for CoV N to facilitate the synthesis of longer viral RNAs and subsequent viral progeny production, through the recruitment of cellular DDX1. As noted, DDX1 has been identified as a member of the cellular interactomes for the IBV N protein (Emmott et al., 2013), which suggests that the interaction between DDX1 and N for the regulation of viral RNA synthesis could be a general phenomenon in CoVs.
It was noteworthy that an interaction between DDX1 and the nonstructural protein 14 (nsp14) has been identified in IBV (Xu et al., 2010). We thus also analyzed whether nsp14 is present and function in the pS197-N/DDX1 complexes. The coIP results showed that expression of nsp14 has no effect on the interaction of pS197-N and DDX1 (Figures S4A and S4B). Although nsp14 could be brought down with anti-N Ab, this interaction is affected neither by S205A mutation nor by kenpaullone treatment (Figures S4A and S4B). It suggested the binding of nsp14 with N is not influenced by the GSK-3-mediated phosphorylation. Moreover, the expression of nsp14 did not affect the synthesis of all of the viral RNA transcripts (Figure S4C). The results did not support an essential role of nsp14 for the function of pS197-N/DDX1 in the discontinuous transcription process, at least for JHMV. Meanwhile, we noted that knockdown of DDX1 in the IBV-infected cells also decreased the viral titer but not the expression level of N protein as shown in this report, which supported the functional role of DDX1 identified in the current study also in IBV infected cells. Therefore, we confirmed that nsp14 could interact with DDX1, but it is independent from N phosphorylation at Ser197 and does not affect the transcription process. The interaction between nsp14 and DDX1 may function in other aspects of viral life cycle.
A further aim of this study was to elucidate the manner in which the pS197-N/DDX1 complex regulates the synthesis of long viral RNAs. We identified that the complex exhibits a unique pattern when binding to different regions of viral RNA. There shows a trend with higher binding activity for the 5′-terminal sequence (except TRS1) than 3′-terminal sequences (lowest at TRS7) of viral RNA (Figure 7C). This trend indicates the mechanism underlying the regulation of the discontinuous transcription process by the S197-N/DDX1 complex. Experimental evidence and bioinformatics analyses have indicated that the extensive stem-loop structural elements of the CoV gRNA template naturally favor the synthesis of smaller sgmRNAs (Sola et al., 2011, Van Den Born et al., 2004, van den Born et al., 2005). Studies have also identified specific long-distance RNA-RNA interactions outside the TRS as critical for the template switching process (Mateos-Gomez et al., 2013, Mateos-Gómez et al., 2011, Moreno et al., 2008). Therefore, it has been postulated that specific viral and cellular factors that assist to unwind the stem-loop RNA structure could be required for the synthesis of longer sgmRNAs (Van Den Born et al., 2004, van den Born et al., 2005). Considering that DDX1 binds to RNA to unwind the RNA duplex (Chen et al., 2002), we proposed a model for the progressive recruitment of DDX1 by pS197-N from the 3′ body TRS to the 5′ body TRS (from TRS7 to TRS2), but not the leader TRS (TRS1), which might assist the opening of the extensive stem-loop structure, increase the readthrough of the body TRSs, and induce the synthesis of long sgmRNAs (see Graphical Abstract). It is noteworthy that we observed a prominent binding signal for the viral genome at the open reading frame (ORF) 1 region (IR1 in Figure 7A). Currently, we were unable to establish the exact functional role of this increased binding; however, this region is known to localize adjacent to the internal discontinuous replication element that forms a stem-loop structure and functions during RNA synthesis in JHMV DI RNA (Kim and Makino, 1995, Lin and Lai, 1993, Repass and Makino, 1998). Thus, binding of the host helicase DDX1 to the region might promote virus RNA synthesis by unwinding the RNA secondary structure. Whether DDX1 participates in the generation of viral DI particles is a potential future area of investigation.
Our results indicate that the shorter sgmRNA6/7 is able to synthesize at unaffected levels in the absence of N phosphorylation or DDX1, which contrasts with observations for the longer sgmRNAs and gRNAs. And the decrease of viral titer caused by blocking of N phosphorylation at S197 was consistent with the reduction of sgmRNA1 level (Figures 1, 2, and 5). In sucrose gradient experiments, we observed shorter sgmRNAs in the lighter fractions, which developed into larger complexes (the heavier fractions) in pS197-N-presenting cells. However, without pS197-N (after kenpaullone treatment), the shorter sgmRNAs were well produced in the heavier fractions, although no longer RNA species could be detected. It suggested that the transcription of JHMV shorter and longer sgmRNAs might occur in two compartments and in two stages. The CoV infection is known to induce the formation of multiple membranous structures (such as double-membrane vesicles or convoluted membranes), which vary qualitatively and quantitatively during the course of an infection (Ulasli et al., 2010). Investigation of the relationship between such membranous structures and the phosphorylation status of the N protein is warranted in the future. At present, we have tried to detect the association of GRP78 with both light and heavy sucrose fractions. Most of the pS197-N-containing complexes codistributed with GRP78 among both light and heavy fractions from JHMV infected cell lysates, suggesting their localization at the ER-related membranous structures. However, in cells treated with kenpaullone, the N-containing complex is present in the heavy fractions without GRP78, indicating such complex not locating at the ER-related membranous structures (Figure S2). The findings were consistent with the idea that light and heavy fractions might be in two different compartments. However, the ultrastructural properties of the compartments are worthy to be carefully examined by future extensive high-resolution electronmicrograph analysis.
The pS197-N-containing lighter fractions are relatively prominent in the early stage of the viral life cycle (at 4 and 6 hr p.i. in Figure 3), and are predominantly associated with the synthesis of the N protein encoding the sgmRNA7 transcript. The N protein is thus the major protein synthesized in the early replication stage, and is phosphorylated by the abundant GSK-3 after synthesis. The accumulation of sufficient pS197-N might initiate the subsequent transcription of long sgmRNAs through the extensive recruitment of DDX1 to viral RNA, as a pivotal factor in switching transcription from smaller to larger sgmRNAs in subsequent viral replication cycle.
In addition to DDX1, we identified several pS197-N-interacting proteins with potential function in RNA biogenesis (Table S2). Some of them were also identified in a previous interactome analysis on the IBV (Emmott et al., 2013), including the serine/arginine-rich splicing factor 1 (SFRS1) protein. In this study, we have conducted coIP and cosedimentation analyses to confirm the interaction between pN and SFRS1 (Figures S5A and S5B). However, we observed that SFRS1 knockdown did not affect the viral N expression (Figure S5C) and viral RNA synthesis (Figures S5D and S5E), and the RNA ChIP analysis did not show notable binding of the complex to specific viral RNA sequences (Figure S5F). These results supported the specific effects of DDX1 in mediating the function of pS197-N in the synthesis of longer sgmRNAs.
The DBT cells stably expressing the unphosphomimicry S205A-N indicated that the interaction between pS197-N and DDX1 could also function as a regulator to prevent the assembly of higher-order ribonucleoprotein (RNP) complexes. Using a phospho-specific pS197-N Ab, we identified that the N protein is phosphorylated immediately after synthesis but is dephosphorylated in virions (Figure 1A). Therefore, binding with pS197-N/DDX1 might impede the packaging of gRNA into mature virions. In fact, similar dephosphorylation patterns for N proteins in assembled virions were also observed in several other viruses (Cartier et al., 1999, Ivanov et al., 2003, Law et al., 2003). One mechanism proposed for this unique pattern is that the overexpression of S205A N in cells prior to viral infection might cause the formation of higher RNP by unphosphorylated S205A-N, which prevents viral RNA transcription, including the N transcript.
It has been documented that each CoV possesses a RNA helicase, which is a product of the autoproteolysis of the ORF1ab polyprotein by 3C-like protease (Ziebuhr, 2005). Unlike DDX1, an SF2 type of RNA helicase, the viral RNA helicase is classified into the superfamily 1 (SF1). DDX1 exhibits bipolar unwinding activity (Chen et al., 2002, Li et al., 2008), and the CoV helicases demonstrate 5′ to 3′ polarity only (Adedeji et al., 2012, Seybert et al., 2000). Therefore, the two types of helicase unwind RNA substrates with distinct polarity selectivity, which indicates fundamental differences in the functions of DDX1 and CoV helicase in viral replication. The viral RNA helicase is crucial for viral replication and thus considered a candidate target for drug development (Keum and Jeong, 2012, Kumar et al., 2013). Our study indicates the critical role of the interaction between pS197-N and the cellular DDX1 in the synthesis of viral gRNA, which supports the targeting of pS197-N or DDX1 for the design of anti-CoV strategies.
Experimental Procedures
Plasmid Construction and Transfection
JHMV N (wild-type and mutants), nsp14, and mouse DDX1 (wild-type and mutant) were transfected into DBT cells by Lipofectamine 2000 (Invitrogen). Details are provided in the Supplemental Experimental Procedures.
Cell culture and establishment of stable cell lines
DBT cells and stable DBT cell lines were cultured in MEM, and 293T cells were cultured in DMEM. G418 was used to select stable cells. Details are provided in the Supplemental Experimental Procedures.
Knockdown DDX1 by Lentiviral shRNA
The TRC shRNA stocks were provided by Academia Sinica, one of the partners of The RNAi Consortium (abbreviated TRC). For Ddx1 knockdown, we used lentiviral vector pLKO_TRC005 carrying shRNAs (clone IDs TRCN0000305216 and TRCN0000305217) targeting the Ddx1 ORF. A nontargeting scrambled (SCR) shRNA was served as the negative control. Recombinant shRNA-expressing lentiviruses were produced by cotransfecting 293T cells with lentivirus-based shRNA, pCMVDR8.91 (packaging plasmid containing gag, pol and rev genes), and pMD.G (Vesicular Somatitis Virus-G envelope gene) using Lipofectamine 2000, which were harvested at 24, 48, and 72 hr after transfection, pooled, and filtered through 0.22 μm filter. Infection of DBT cells was carried out by adding the lentivirus to the medium for 16 hr, and the cells were harvested at 48 hr after infection for subsequent experiments.
RNA Extraction and Northern Blot Analysis
RNAs extracted by REzol reagent (PROtech Technologies) were resolved on 0.8% agarose/formaldehyde gel and transferred to Hybond-N nylon membranes (Amersham Biosciences). The membranes were hybridized with the DIG-labeled probes. Details are provided in the Supplemental Experimental Procedures.
Viral titer quantification
Quantification of the viral titer was determined by a plaque assay on confluent DBT cells seeded in 6-well plates. Details are provided in the Supplemental Experimental Procedures.
Sucrose Gradient Sedimentation Analysis
Cells resuspended in PBS containing 1% Triton X-100 were layered on a 10%–50% sucrose gradient. The gradients were centrifuged at 230 000 × g in a Beckman SW41 rotor for 22 hr at 16 °C, fractionated (into 12 fractions), and subjected to immunoblotting. Details are provided in the Supplemental Experimental Procedures.
Immunofluorescence Microscopy
Acetone/methanol-fixed cells were stained with appropriate primary and fluorescence-conjugated secondary antibodies. Images were visualized using a Zeiss AXIO Imager A1 microscope. Details are provided in the Supplemental Experimental Procedures.
Protein Radiolabeling and Detection
After viral infection, the cells were pulse labeled with [35S]-methionine for 1 hr and chased for 2 and 4 hr. The radioactive bands were detected by autoradiography at −80°C overnight. Details are provided in the Supplemental Experimental Procedures.
Preparation of Cell Lysates, Nuclear and Cytosol Extraction, and Immunoblotting
Cells harvested in lysis buffer were resolved using SDS-PAGE and reacted with primary antibodies. A nuclear extraction kit (Panomics) was used for subcellular fractionation. The pS197-N and pan-N antibodies were generated as described previously (Wu et al., 2009). DDX1, lamin A/C, and β-actin antibodies were purchased from Genetex. The tubulin antibody was purchased from Sigma-Aldrich. Details are provided in the Supplemental Experimental Procedures.
Coimmunoprecipitation Analysis
Two milligrams of proteins were collected from cell lysates for analysis by using IP with pS197-N or pan JHMV N antibody. Binding reactions for protein samples and specific antibodies were rotated at room temperature for 3 hr, and the mixtures were incubated for 1 hr after the addition of protein A Sepharose (GE Healthcare). The supernatants were discarded, and the resulting complexes were washed three times with a protein lysis buffer containing a protease inhibitor and a phosphatase inhibitor as described previously (Wu et al., 2009). The complexes were further analyzed using mass spectrometry or SDS-PAGE.
Protein Identification by Using Mass Spectrometry
The precipitated protein complexes from sucrose fractions were eluted by boiling and subjected to gel-assisted digestion. Trypsin-digested peptides were analyzed by LC-MS/MS using a linear ion trap Orbitrap instrument (LTQ-Orbitrap, Thermo Fisher Scientific). The mass spectral data obtained were analyzed using Proteome Discoverer Version 1.3 (Thermo Fisher Scientific). Details are provided in the Supplemental Experimental Procedures.
Quantification of the Intracellular Viral RNAs by Real-Time RT-PCR
The cDNA converted from the intracellular RNA of JHMV-infected DBT cells were subjected to qPCR to evaluate the expression of viral RNAs. Primer sets used for the quantitative analysis of gRNA and sgmRNAs from infectious JHMV are listed in Table S1. The PBGD gene was used as an internal control to normalize the signal.
RNA ChIP-qPCR Analysis
RNA ChIP was performed as described previously with some modifications (Niranjanakumari et al., 2002). Briefly, JHMV-infected cells (moi = 1 at 6 hr p.i.) were harvested, and viral RNAs associated with specific proteins were isolated by IP with corresponding antibodies for subsequent quantification by RT-qPCR. The primer sequences used are listed in Table S3. The products were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining. Quantification of binding sites was presented as percentage of input. Details are provided in the Supplemental Experimental Procedures.
Author Contributions
C.-H.W., P.-J.C., and S.-H.Y. conceived of and designed the experiments. C.-H.W. performed the experiments. C.-H.W., P.-J.C., and S.-H.Y. analyzed the data and wrote the manuscript.
Acknowledgments
Liquid chromatography-tandem mass spectrometry services were kindly provided by the Mass Spectrometry Facility at the Institute of Biological Chemistry, Academia Sinica. The RNAi reagents were obtained from the National RNAi Core Facility at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, supported by the National Core Facility Program for Biotechnology Grants of NSC (NSC 100-2319-B-001-002). This study was supported by grants from the National Science Council, Taiwan (NSC93-2751-B-002-008-Y, NSC103-2314-B-002-052-MY3), and a grant from the National Taiwan University Excellence Research Program (103R891901). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: October 8, 2014
Footnotes
Supplemental Information includes five figures, three tables, and Supplemental Experimental Procedures and can be found with this article at http://dx.doi.org/10.1016/j.chom.2014.09.009.
Supplemental Information
References
- Adedeji A.O., Marchand B., Te Velthuis A.J., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F., Galán C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd A.K., Raney K.D. Superfamily 2 helicases. Front. Biosci. 2012;17:2070–2088. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E., Escors D., López J.A., González J.M., Alvarez A., Arza E., Enjuanes L. Phosphorylation and subcellular localization of transmissible gastroenteritis virus nucleocapsid protein in infected cells. J. Gen. Virol. 2005;86:2255–2267. doi: 10.1099/vir.0.80975-0. [DOI] [PubMed] [Google Scholar]
- Cartier C., Sivard P., Tranchat C., Decimo D., Desgranges C., Boyer V. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. J. Biol. Chem. 1999;274:19434–19440. doi: 10.1074/jbc.274.27.19434. [DOI] [PubMed] [Google Scholar]
- Chen H.C., Lin W.C., Tsay Y.G., Lee S.C., Chang C.J. An RNA helicase, DDX1, interacting with poly(A) RNA and heterogeneous nuclear ribonucleoprotein K. J. Biol. Chem. 2002;277:40403–40409. doi: 10.1074/jbc.M206981200. [DOI] [PubMed] [Google Scholar]
- Chen H., Gill A., Dove B.K., Emmett S.R., Kemp C.F., Ritchie M.A., Dee M., Hiscox J.A. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J. Virol. 2005;79:1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E., Munday D., Bickerton E., Britton P., Rodgers M.A., Whitehouse A., Zhou E.M., Hiscox J.A. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J. Virol. 2013;87:9486–9500. doi: 10.1128/JVI.00321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frana M.F., Behnke J.N., Sturman L.S., Holmes K.V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P.I., Whiteheart S.W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hurst K.R., Kuo L., Koetzner C.A., Ye R., Hsue B., Masters P.S. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 2005;79:13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.I., Puustinen P., Gabrenaite R., Vihinen H., Rönnstrand L., Valmu L., Kalkkinen N., Mäkinen K. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell. 2003;15:2124–2139. doi: 10.1105/tpc.012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum Y.S., Jeong Y.J. Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem. Pharmacol. 2012;84:1351–1358. doi: 10.1016/j.bcp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.N., Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J. Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Jung Y.S., Liang P.H. Anti-SARS coronavirus agents: a patent review (2008–present) Expert Opin. Ther. Pat. 2013;23:1337–1348. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76:4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Perlman S., Anderson L.J. Coronaviridae. In: Fields Bernard N., Knipe David M., Howley Peter M., editors. Fields’ Virology. 5th ed. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1306–1335. [Google Scholar]
- Law L.M., Everitt J.C., Beatch M.D., Holmes C.F., Hobman T.C. Phosphorylation of rubella virus capsid regulates its RNA binding activity and virus replication. J. Virol. 2003;77:1764–1771. doi: 10.1128/JVI.77.3.1764-1771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Monckton E.A., Godbout R. A role for DEAD box 1 at DNA double-strand breaks. Mol. Cell. Biol. 2008;28:6413–6425. doi: 10.1128/MCB.01053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.J., Lai M.M. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J. Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Shao J., Sun M., Liu J., Xu G., Zhang X., Xu N., Wang R., Liu S. Identification of phosphorylation sites in the nucleocapsid protein (N protein) of SARS-coronavirus. Int. J. Mass Spectrom. 2007;268:296–303. doi: 10.1016/j.ijms.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gómez P.A., Zuñiga S., Palacio L., Enjuanes L., Sola I. Gene N proximal and distal RNA motifs regulate coronavirus nucleocapsid mRNA transcription. J. Virol. 2011;85:8968–8980. doi: 10.1128/JVI.00869-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez P.A., Morales L., Zuñiga S., Enjuanes L., Sola I. Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J. Virol. 2013;87:177–186. doi: 10.1128/JVI.01782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.L., Zúñiga S., Enjuanes L., Sola I. Identification of a coronavirus transcription enhancer. J. Virol. 2008;82:3882–3893. doi: 10.1128/JVI.02622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M.A. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J., Snijder E.J. Regulation of relative abundance of arterivirus subgenomic mRNAs. J. Virol. 2004;78:8102–8113. doi: 10.1128/JVI.78.15.8102-8113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J., Snijder E.J. Nidovirus transcription: how to make sense...? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Reddy R.K., Mao C., Baumeister P., Austin R.C., Kaufman R.J., Lee A.S. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J. Biol. Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- Repass J.F., Makino S. Importance of the positive-strand RNA secondary structure of a murine coronavirus defective interfering RNA internal replication signal in positive-strand RNA synthesis. J. Virol. 1998;72:7926–7933. doi: 10.1128/jvi.72.10.7926-7933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Mateos-Gomez P.A., Almazan F., Zuñiga S., Enjuanes L. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 2011;8:237–248. doi: 10.4161/rna.8.2.14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer K.A., Hiscox J.A. Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino-terminal region. FEBS Lett. 2006;580:5993–5998. doi: 10.1016/j.febslet.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N.K., Cordin O., Banroques J., Doère M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- Ulasli M., Verheije M.H., de Haan C.A., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Born E., Gultyaev A.P., Snijder E.J. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA. 2004;10:424–437. doi: 10.1261/rna.5174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E., Posthuma C.C., Gultyaev A.P., Snijder E.J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005;79:6312–6324. doi: 10.1128/JVI.79.10.6312-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.C., Yi Z., Hogue B.G. Identification of mouse hepatitis coronavirus A59 nucleocapsid protein phosphorylation sites. Virus Res. 2007;126:139–148. doi: 10.1016/j.virusres.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Yeh S.H., Tsay Y.G., Shieh Y.H., Kao C.L., Chen Y.S., Wang S.H., Kuo T.J., Chen D.S., Chen P.J. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Khadijah S., Fang S., Wang L., Tay F.P., Liu D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Sola I., Moreno J.L., Sabella P., Plana-Durán J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Cruz J.L., Sola I., Mateos-Gómez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.