Abstract
Compared with terrestrial mammals, bats have a longer lifespan and greater capacity to co-exist with a variety of viruses. In addition to cytosolic DNA generated by these viral infections, the metabolic demands of flight cause DNA damage and the release of self-DNA into the cytoplasm. However, whether bats have an altered DNA sensing/defense system to balance high cytosolic DNA levels remains an open question. We demonstrate that bats have a dampened interferon response due to the replacement of the highly conserved serine residue (S358) in STING, an essential adaptor protein in multiple DNA sensing pathways. Reversing this mutation by introducing S358 restored STING functionality, resulting in interferon activation and virus inhibition. Combined with previous reports on bat-specific changes of other DNA sensors such as TLR9, IFI16, and AIM2, our findings shed light on bat adaptation to flight, their long lifespan, and their unique capacity to serve as a virus reservoir.
Keywords: bats, STING, interferon, virus, DNA sensing, dampened
Graphical Abstract
Highlights
-
•
STING-dependent IFN activation is dampened in bats
-
•
Highly conserved serine residue (S358) is replaced in bat STING
-
•
Reversing this mutation restores STING function, IFN activation, and virus inhibition
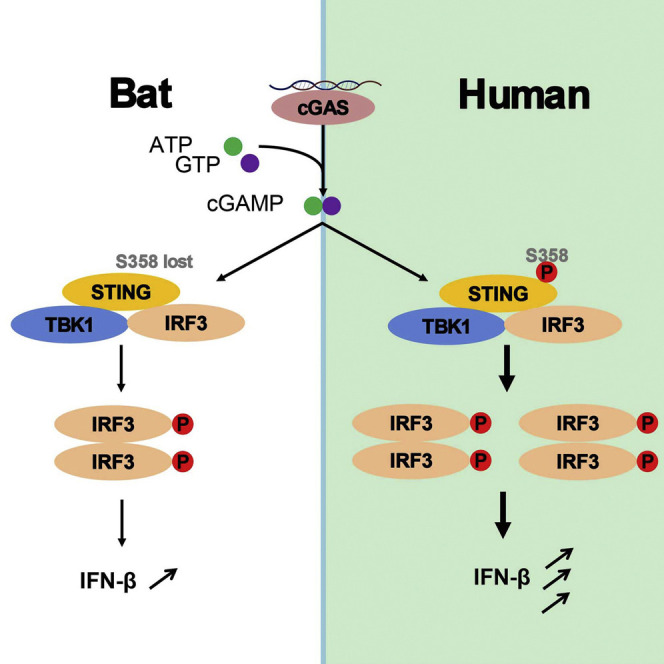
Bats co-exist with a large variety of viruses, and infection-derived cytosolic DNA could result in heightened DNA sensing and overactivation. Xie et al. show that STING-dependent IFN activation is dampened in bats due to the replacement of the highly conserved and functionally important serine residue S358.
Main Text
Bats are uniquely the only flying mammals and are found to have a positively selected oxidative phosphorylation pathway as a result of an increased metabolic capacity (Shen et al., 2010). Byproducts of oxidative metabolism and stress are known to cause DNA damage, resulting in the escape of self-DNA from the nucleus, mitochondria, or lysosomes into the cytoplasm (Barzilai et al., 2002). Bats have been increasingly linked to deadly viruses such as severe acute respiratory syndrome (SARS), Ebola virus, and henipaviruses (Wynne and Wang, 2013). Although most of these zoonotic viruses are RNA viruses, bats also harbor a variety of DNA viruses (Brook and Dobson, 2015). In addition, it is known that infection of RNA viruses can also result in cytosolic DNA due to intracellular damage (Ryan et al., 2016). Infection-derived cytosolic DNA as well as self-DNA is known to trigger robust immune responses, leading to inflammasome activation and type I interferon (IFN) induction (Schlee and Hartmann, 2016). While it is accepted that overactivation of either inflammasome or IFN can cause autoimmune diseases (Peckham et al., 2017), it is unknown how bats, while naturally maintaining a high burden of viruses and the oxidative stressors of flight, are able to regulate the response against stimulatory sensing of cytosolic DNA to avoid overactivation of innate immune pro-inflammatory pathways.
In humans, the DNA sensors of the innate immune system include AIM2 and IFI16 in inflammasome assembly (Hornung et al., 2009, Kerur et al., 2011), and TLR9, IFI16, DDX41, LSM14A, and cGAS in IFN expression (Latz et al., 2004, Li et al., 2012, Sun et al., 2013, Unterholzner et al., 2010, Zhang et al., 2011). Among these cytosolic sensors, cGAS was identified as the universal and essential DNA sensor that produces cyclic GMP-AMP (cGAMP) in response to DNA stimulation (Sun et al., 2013), which in turn binds to and activates stimulator of IFN genes (STING; also known as MITA, ERIS, and MPYS), the essential adaptor protein in multiple DNA sensing pathways (Ishikawa and Barber, 2008, Jin et al., 2008, Sun et al., 2009, Zhong et al., 2008). Following STING activation, TBK1 is recruited to STING, leading to the subsequent phosphorylation of STING and IRF3 by TBK1. This ultimately triggers the type I IFN response. Point mutation of either phosphorylation site (S358 or S366) of STING to alanine significantly impaired its ability to activate downstream IFNs (Liu et al., 2015, Tanaka and Chen, 2012, Zhong et al., 2008).
There are limited studies on bat DNA sensors despite the belief that bat cells are likely to be more at risk of cytosolic DNA exposure. A recent comparative genomics study showed that the most positively selected genes of bats seemed to be concentrated in the DNA damage checkpoint pathway and innate immunity (Zhang et al., 2013). One of these genes encodes the inflammasome sensor NLRP3. More strikingly, the entire PYHIN gene family, including AIM2 and IFI16, is lost in all bat genomes sequenced so far, implying a dampened DNA-triggered inflammasome response (Ahn et al., 2016). Bats have been shown to have a contracted type I IFN locus and different expression patterns of type III IFNs compared with those in human and mouse (Zhou et al., 2011, Zhou et al., 2016). Also, TLR9 seems to be under greater positive selection in bats compared with other mammals (Escalera-Zamudio et al., 2015). Taken together, these findings suggest that bats may have evolved to adopt a DNA sensing and IFN response mechanism in adaptation to flight, which is sufficiently different from terrestrial mammals.
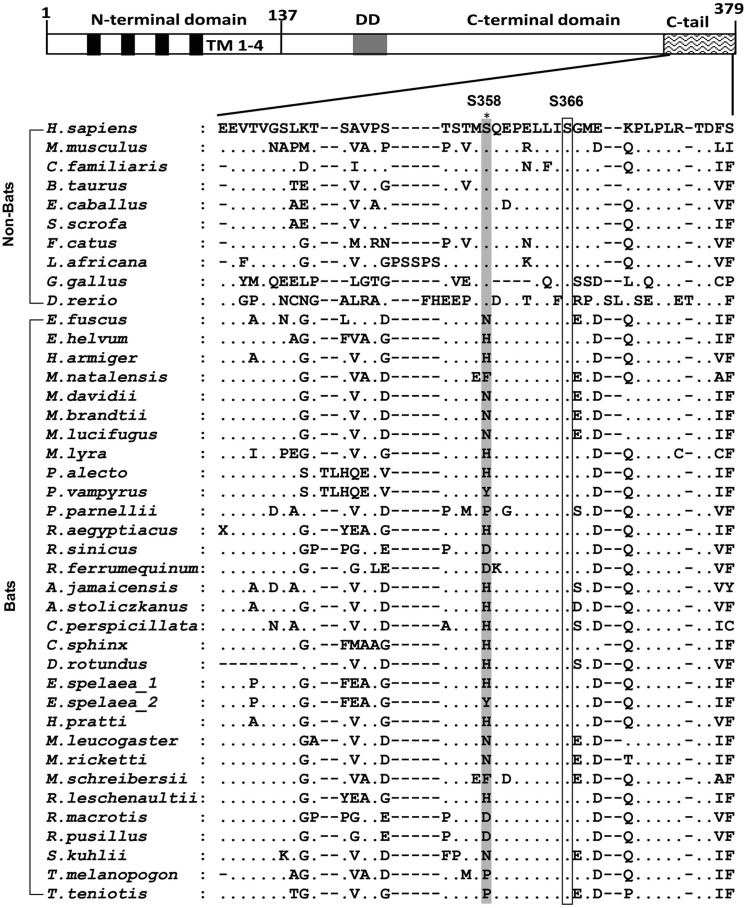
In this context, we hypothesized that cytosolic DNA, whether it is flight-induced or infection-derived, imposes strong selective pressures on the bat DNA sensors, resulting in a functionally dampened sensing mechanism and downstream IFN production to avoid overactivation on a regular basis during normal flight and/or co-existence with viruses. As STING is increasingly being recognized as the central molecule in the cytosolic DNA sensing pathway, we conducted a comprehensive sequence and functional analysis of bat STING. Sequence alignment of all available bat STING (from a total of 30 different species) with ten major non-bat mammalian STING revealed a key difference: while the S358 residue is absolutely conserved among all known non-bat mammalian STING, none of the bat STING retain the S358. Instead, this residue has been replaced by a variety of different residues at this position, including N, H, F, Y, P, D, and R (Figure 1 ). As the S358 phosphorylation site is critical for downstream IFN activation in humans and other mammals (Liu et al., 2015, Tanaka and Chen, 2012, Tsuchida et al., 2010, Zhong et al., 2008), this key residue change strongly suggests a weakened bat STING in the context of IFN activation.
Figure 1.
Mutations of S358 in Bat STING
STING domains are illustrated on top of the alignment. The highly conserved regions are boxed. Residue 358 is highlighted in gray. There are two Eonycteris spelaea STING sequences because of the polymorphism at residue 358. The full species name and accession numbers of STING or SRA data are listed in Table S1.
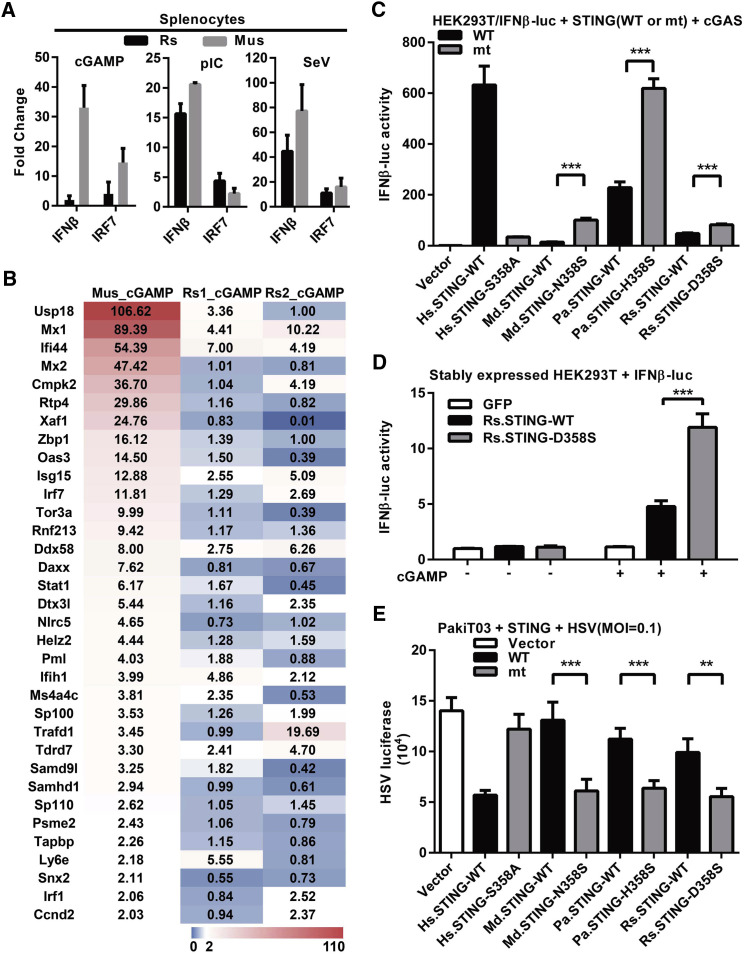
To test this, we compared the functional difference in the induction of IFNs between bat and mouse by cGAMP. Splenocytes from three individual Rhinolophus sinicus (Rs) and three laboratory mice were stimulated by cGAMP. Rs has been reported to be the reservoir host of the lethal severe acute respiratory coronavirus (SARS-CoV) (Ge et al., 2013). In contrast to a strong induction of IFNβ and IRF7 (an IFN-stimulated gene [ISG]) in mouse, transfection of cGAMP induced a much lower level of IFNβ and IRF7 mRNA in Rs bats from qPCR analysis (Figure 2 A). As controls, both poly I:C and Sendai virus treatment resulted in a comparable induction level of IFNβ and IRF7 in both cells (Figure 2A). The qPCR results were corroborated by RNA high-throughput sequencing (RNA-seq). As shown in Figure 2B, a number of mouse ISGs were strongly upregulated upon cGAMP treatment, whereas the upregulation of bat ISGs was much less, both in number and fold change. Between the two Rs bats, there were subtle differences in ISG induction, which was not unexpected considering wild-caught outbred bats were used in this study.
Figure 2.
Dampened IFN Activation and Virus Inhibition by Bat STING
(A) Splenocytes of Rhinolophus sinicus bats and mice (n = 3 cells from 3 animals each) were transfected with cGAMP (1 μg/mL) or poly I:C (1 μg/mL), or infected with SeV (100 hemagglutinin units/mL). Six hours later, the induction of IFNβ and IRF7 genes was determined by qPCR. Primers can be found in Table S2.
(B) Transcriptome next-generation sequencing of splenocyte RNAs. The differentially expressed genes (DEGs) were analyzed by RSEM at FDR (false discovery rate) < 0.05. The ISG in the DEG sets of mice and Rs bat are listed. Fold change is indicated in color from 0 to 110.
(C) Restoration of STING function by introducing S358 in bat STING. HEK293T cells were co-transfected with STING, cGAS, IFNβ promoter firefly luciferase, and renilla luciferase plasmids. Luciferase activity was determined 24 hr post-transfection. The blots showing protein levels can be found in Figure S2.
(D) cGAMP treatment of HEK293T stably expressing various STING. Cells stably expressing the indicated proteins were transfected with IFNβ promoter firefly luciferase and renilla luciferase plasmids. Six hours later, cells were permeabilized in digitonin buffer with or without 1 μg/mL cGAMP. Luciferase activity was determined 16 hr after treatments.
(E) PaKiT03 cells were transfected with indicated STING plasmids followed by infection with HSV-luciferase at MOI = 0.1 at 24 hr post-transfection. At 24 hr post-infection, HSV replication was determined by luciferase activity.
Data from (A), (C), (D), and (E) are presented as the means ± SD, n = 3, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student's t test). For (C) and (D), data represent fold change according to wells transfected with empty vector (set as 1). WT, wild-type; mt, mutant; Hs, Homo sapiens; Md, Myotis davidii; Pa, Pteropus alecto; Rs, Rhinolophus sinicus; pIC, poly I:C.
Phylogenetic analysis showed bat STING clustered with known mammalian STING (Figure S1A). qPCR analysis of mRNA levels in a range of Rs, Myotis davidii (Md), and Pteropus alecto (Pa) primary organs revealed an expression pattern not dissimilar to that found in mouse: STING was found to be expressed in a variety of tissues in bats, with highest levels in spleen and lung (Figure S1B). It can thus be concluded that phylogenetic divergence or difference in gene expression patterns between bats and mice is unlikely to be responsible for the observed reduction in STING-mediated IFN production.
We then examined whether the dampening of bat STING function by the change in residue 358 is common in other bats. In HEK293T cells, which lack endogenous cGAS and STING expression (Sun et al., 2013), STING from three representative bats, Rs, Pa, and Md, and human was overexpressed together with human cGAS and IFNβ promoter plasmids. Although polymorphism has been observed in human STING, a previous study indicated that the three variants (RGR, AQ, and HAQ) have different but comparable ability in IFN induction activities depending on the experimental conditions (Yi et al., 2013). In this study, we have confirmed their observation and used the AQ variant for further studies. Mutant human STING S358A significantly reduced STING-induced IFNβ production as reported previously (Zhong et al., 2008). Conversely, mutant bat STING X358S (X = N, H, or D) significantly restored their ability for IFNβ induction (Figure 2C). To exclude the possibility that the human cell system may affect bat STING function, we repeated the experiment in PakiT03, a Pa bat cell line that expresses reasonable amount of cGAS but a very low level of STING (Zhou et al., 2016). The pattern was essentially identical to that observed in HEK293T, with wild-type bat STING showing dampened induction of IFN and ISGs compared with the X358S mutants (Figure S2A). We also tested this dampening function by cGAMP. HEK293T cells stably expressing wild-type (D358) or mutant S358 Rs STING were stimulated with cGAMP in digitonin permeabilization solution. The S358 STING induced significantly higher IFN (Figure 2D). These results suggest that the S358 replacement is mainly responsible for dampened STING-dependent IFN activation with cGAS co-expression or cGAMP stimulation.
It is proposed that bat's exceptional ability to host viruses with few or no clinical disease is likely the result of an intricate balance between the host immune system and virus infection (Schountz, 2014, Wynne and Wang, 2013). We hypothesized that the dampened STING-IFN responses could be partially responsible for that intricate balance. In assessing the effect of different STING on herpes simplex virus (HSV) infection in PakiT03 cells, it was found that the wild-type human STING was about 2.5-fold more effective in blocking HSV replication than the S358A mutant. However, this was reversed for bat STING, in which the wild-type bat STING was less effective than the mutant STING X358S with a reduction of approximately 3-, 2.5-, and 2-fold, respectively, for Md, Pa, and Rs STING (Figure 2E).
In human STING, residues S366 and S358 are important for IRF3, but not TBK1, binding and activation (Tanaka and Chen, 2012). To understand the detailed mechanism of dampened STING-dependent IFN activation, we investigated whether this bat-specific S358 replacement universally affects IRF3 and TBK1 activation. When HEK293T cells were transfected with human or bat STING-expressing plasmids, phosphorylation of IRF3, but not TBK1, was markedly higher in cells transfected with S358 STING (Figure S2B). Similar findings were observed in bat PakiT03 cells (Figure S2C), which eventually contributed to a different downstream IFN response and in turn the observed difference in modulating HSV replication. Taken together, these results demonstrated that while bat STING maintained its antiviral defense similar to human STING, the dampening likely contributed in part to the long-term co-existence of bats and viruses.
We hypothesized that excessive exposure to cytosolic DNA in bat cells during flight and/or viral infection would pose a strong natural selection pressure to reduce activation of bat DNA sensors. In this report, we have provided genetic and functional data to support this hypothesis. We have demonstrated that bat STING is less active in IFN induction and pinpointed residue 358 as the key site of difference between bat and human STING. Experimentally, we have demonstrated the replacement of the S358 residue in different bat STING resulted in dampened downstream IFN production via IRF3 phosphorylation. To our knowledge, this is the most conclusive experimental demonstration of a key innate defense pathway that is functionally different between bats and non-bat mammals with implications that bats are more effective in peaceful co-existence with a large number of viruses.
There is abundant evidence that bats harbor more viruses per species than other mammals (Brook and Dobson, 2015, Luis et al., 2013, Olival et al., 2017). Infection-derived DNA can act as activators of DNA sensors such as cGAS and subsequently STING (Schlee and Hartmann, 2016). Constitutive activation of STING could cause severe autoimmune diseases, such as vascular and pulmonary syndrome or Aicardi-Goutieres syndrome (Barber, 2015), while some highly pathogenic viruses such as SARS-CoV are known to induce excessive inflammation eventually leading to human death (Channappanavar et al., 2016). The replacement of the serine residue at position 358 in every known bat STING is highly significant considering it is absolutely conserved in all other mammals. Our data support that the S358 replacement in bat STING dampened but did not fully diminish the functionality of STING. The nature of the weakened, but not entirely lost, functionality of STING may have profound impact for bats to maintain the balanced state of “effective response” but not “over response” against viruses. A similar finding was observed for bat IFNα, which are less in number but are constitutively expressed without stimulation (Zhou et al., 2016). Above all, this discovery helps to further our understanding of the complex mechanisms in which bats fine-tune innate defense responses against insult by viral, bacterial, or host cytosolic DNA.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-phospho-IRF3 (Ser396) | Cell Signaling Technology | Cat#4947S; RRID: AB_823547 |
| Rabbit monoclonal anti-phospho-TBK1 (Ser172) | Cell Signaling Technology | Cat#5483S; RRID: AB_10693472 |
| Rabbit monoclonal anti -TBK1 | Abcam | Cat#ab40676; RRID: AB_776632 |
| Rabbit polyclonal anti-IRF3 | proteintech | Cat#11312-1-AP; RRID: AB_2127004 |
| Mouse monoclonal anti-S tag | Abcam | Cat#ab184223 |
| Mouse monoclonal anti-beta actin | proteintech | Cat#60008-1-Ig; RRID: AB_2289225 |
| Goat anti-Mouse IgG (H+L) Secondary Antibody, HRP | Thermo Scientific | Cat#32430; RRID: AB_1185566 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP | Thermo Scientific | Cat#31460; RRID: AB_228341 |
| Bacterial and Virus Strains | ||
| Sendai virus (SeV) | Laboratory of Zhou P | Zhou et al., 2016 |
| luciferase-expressing HSV | Laboratory of Zheng C, F | Li et al., 2011 |
| Biological Samples | ||
| Splenocytes isolated from 8-10 week old BALB/c mice | this study | N/A |
| Rhinolophus sinicus splenocytes | this study | N/A |
| Rhinolophus sinicus tissues | this study | N/A |
| Tissues from 8-10 week old BALB/c mice | this study | N/A |
| Myotis davidii tissues | this study | N/A |
| Pteropus alecto tissues | this study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Complete protease cocktail inhibitor | Roche | Cat#11836170001 |
| PolyI:C (HMW) | InvivoGen | Cat#tlrl-pic |
| 2',3’-cGAMP | InvivoGen | Cat#tlrl-nacga23 |
| Lymphoprep | Axis-Shield | Cat#1114544 |
| Lipofectamine 3000 | Invitrogen | Cat#L3000015 |
| SuperSignal West Femto substrate | Thermo Scientific | Cat#34094 |
| Critical Commercial Assays | ||
| QuikChange site-directed mutagenesis kit | Stratagene | Cat#200519 |
| RNeasy mini kit | Qiagen | Cat#74134 |
| Dual-Luciferase Reporter Assay System | Promega | Cat#E1910 |
| Luciferase Assay System | Promega | Cat#E1500 |
| PrimeScript RT Master Mix | Takara | Cat#RR036A |
| SYBR Premix Ex Taq II (Tli RNaseH Plus) | Takara | Cat#RR820Q |
| Deposited Data | ||
| RNA-Seq Data | This study | SRA: PRJNA393936 |
| Eidolon helvum, Rhinolophus ferrumequinum and Pteronotus parnellii STING nucleotide sequences | This study | GenBank: MF174844–MF174846 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | Cat#CRL-11268 |
| GP2-293 | Clontech | Cat#631458 |
| PakiT03 | Laboratory of Zhou P | Zhou et al., 2016 |
| HEK293T stably transfected with GFP | This Study | N/A |
| HEK293T stably transfected with R.sinicus STING | This Study | N/A |
| HEK293T stably transfected with R.sinicus STING(D358S) | This Study | N/A |
| Experimental Models: Organisms/Strains | ||
| BALB/c mice | Beijing Vital River | Strain code 211 |
| Oligonucleotides | ||
| Primers for STING cloning and qRT-PCR, see Table S2 | This Study | N/A |
| Recombinant DNA | ||
| pQCXIH | Clontech | Cat#631516 |
| pCDNA3.1-Homo.cGAS | Laboratory of Yan-Yi Wang | Fu et al., 2017 |
| pCAGGS inserted with Homo sapiens, Myotis davidii, Pteropus alecto and Rhinolophus sinicus STING or S tag | This Study | N/A |
| pQCXIH inserted with Rhinolophus sinicus STING or GFP tag | This Study | N/A |
| Software and Algorithms | ||
| Seqman | DNASTAR | http://www.dnastar.com |
| SRA-blast | SRA-blast | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| MEGA4 | Mega | http://www.megasoftware.net/mega4/ |
| Genedoc | Genedoc | http://genedoc.software.informer.com/ |
| RSEM | RSEM | http://deweylab.github.io/RSEM/ |
| DESeq | Bioconductor | http://bioconductor.org/packages/release/bioc/html/DESeq.html |
| Prism software | GraphPad | https://www.graphpad.com |
| ImageJ | ImageJ | https://imagej.nih.gov/ij/ |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Peng Zhou (peng.zhou@wh.iov.cn).
Experimental Model and Subject Details
Ethics Statement
All animal experiments were approved by the Institutional Animal Ethical Committee of Wuhan Institute of Virology, Chinese Academy of Sciences (Serial number: WIV05201603).
Bat and Mouse Experiments
Wild-type adult BALB/c mice between ages of 8 to 10 weeks were purchased from Beijing Vital River and cared at a specific pathogen free (SPF) facility. Adult Myotis davidii and Rhinolophus sinicus captured from Taiyi cave (Xianning, China) and Pteropus alecto bats trapped in Southern Queensland, Australia were euthanized and dissected directly. Mice and bats were used without gender preference. Splenocytes of bats and mice were prepared as previously described (Zhou et al., 2011). Briefly, spleen cell were obtained by pressing spleen tissue through a cell strainer using a syringe plunger and splenocytes were collected with Lymphoprep (Axis-Shield) following manufacturer’s instructions.
Viruses and Cell Lines
HEK293T cells were maintained in DMEM + 10% FCS (Gibco). Bat PakiT03 cells were maintained in DMEM/F-12 + 10% FCS (Gibco). Splenocytes of bats and mice were maintained in RPMI-1640 + 10% FCS (Gibco). All cells were cultured at 37°C in 5% CO2. Cell lines were tested free of mycoplasma contamination and authenticated by microscopic morphologic evaluation. Sendai virus (SeV) Cantell strain was propagated in 10-day-old embryonated SPF chicken eggs at 37°C for 48 h. The HSV expressing luciferase was generously provided by Chun-Fu Zheng at Institutes of Biology and Medical Sciences, Soochow University, China.
Method Details
Plasmid Construction and Transfection
The STING sequence of Homo sapiens was amplified from HA-STING plasmid, which was generously provided by Yan-Yi Wang, Wuhan Institute of Virology, CAS, China. Myotis davidii, Pteropus alecto and Rhinolophus sinicus STING sequences were amplified from cDNA of corresponding bat spleen tissues and cloned into pCAGGS with C-terminal S-tag. STING of Rhinolophus sinicus was also cloned into pQCXIH with C-terminal GFP-tag. Various mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene). (see Table S2 for primers). Plasmids were verified by sequencing before transfection using lipofectamine 3000 (Thermo) following manufacturer’s instructions.
Virus Infection and Quantification
PakiT03 cells were infected with HSV-luciferase at MOI of 0.1 at 37°C for 1 hr. Cells were then washed with warm D-hanks and cultured in complete DMEM/F-12. At 24 hr post infection, the cells were washed by cold PBS and HSV-luciferase quantified by Luciferase Assay System (Promega) according to the manufacturer’s instructions.
HEK293T Cells Stably Expressing STING Construction
To generate retroviral vectors for transduction of Rhinolophus sinicus STING into HEK293T cells, GP2-293 Packaging Cells were plated at 6-well plate overnight at the density of 4 × 105/ml and transfected with 1.5 μg pQCXIH-R.sinicus. STING and 1.5 μg pVSV-G using lipofectamine 3000 (Thermo). At 6 hr post transfection, the media was replaced with fresh media. At 48 hr post transfection, the supernatants containing the retrovirus was collected, filtered through a 0.45 μm filter, and used to infect HEK293T cells. At 72 hr post infection, the transduced HEK293T cells were selected with 10 μg/ml hygromycin.
Dual Luciferase Assay
Plasmids with optimized amount (100 ng pCAGGS-STING; 200 ng pcDNA3.1-cGAS; 100 ng IFNβ-luc and 10 ng pRL-Tk, internal control from Promega) were transfected into HEK293T cells in 24-well plates. 24 hr later, luciferase was determined by Luciferase Assay System (Promega). The ratio of firefly to renilla luciferase signal was calculated and then normalized to the wells transfected with empty pCAGGS vector.
Quantification of Gene Expression by qRT-PCR
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN), followed by cDNA reverse transcription using PrimeScript RT Master Mix (Takara). Gene expression was determined by SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara) on StepOnePlus system. Primers were listed in Table S2.
Digitonin Permeabilization of STING Stably Expression HEK293T
cGAMP was delivered by digitonin permeabilization buffer (50 mM HEPES, pH 7, 100 mM KCl, 3 mM MgCl2, 0.1 mM DTT, 85 mM Sucrose, 0.2% BSA, 1 mM ATP, 0.1 mM GTP) containing 10 μg/ml digitonin. After incubation at 37°C for 30 min, the permeabilization buffer was replaced by complete DMEM and incubated for another 16 hr.
Western Blot
HEK293T or PakiT03 Cells were washed with cold PBS for two times, then lysed by1% NP-40 buffer supplemented with complete protease cocktail inhibitor (Roche) for 30 min on ice. Cell lysates were mixed with SDS loading buffer and denatured at 95°C for 5min. Equal amounts of denatured lysates were subjected to SDS-PAGE and transferred to PVDF membrane. The membranes were blocked with 5% BSA for 1 hr. The following primary antibodies were used at 1:1000 dilution: anti-phospho-IRF3 (CST, 4947S), anti-phospho-TBK1 (CST, 5483S), anti-TBK1 (Abcam, ab40676), anti-IRF3 (proteintech, 11312-1-AP). And the following primary antibodies were used at 1:3000 dilution: anti-S tag (Abcam, ab184223), anti-β actin (proteintech, 60008-1-Ig). After an overnight incubation with primary antibodies, the membranes were washed with TBS supplemented with 0.1% Tween-20 three times and then incubated with HRP conjugated goat anti-rabbit or goat anti-mouse secondary antibody diluted in TBST. Membranes were then washed three times and exposed using SuperSignal West Femto substrate (Thermo Scientific).
Splenocytes Stimulation and RNA Sequencing
2',3’-cGAMP and poly I:C (Invivogen) were transfected into splenocytes at 1μg/ml with lipofectamine 3000 or infected with 100 hemagglutinin units (HAU)/ml of SeV. 6 hr later, RNA was extracted and gene expression was determined by RT-qPCR. RNA-seq was conducted with 150-bp paired-end reads on an Illumina HiSeq2000 sequencer.
Sequences and NGS Data Analysis
STING sequences of Eidolon helvum, Pteronotus parnellii and Rhinolophus ferrumequinum were predicted by Genewise using Pteropus alecto STING protein as reference. Other STING sequences were either downloaded from Genbank or assembled with transcriptome data, in which case the reads of STING were picked out by SRA-blast using annotated Genbank bat STING (Eptesicus fuscus, Myotis davidii, etc) as query and assembled by Seqman (Lasergene). The Genbank or SRA accession numbers are listed in Table S1. The alignment of STING was generated by MEGA4 and edited by Genedoc. For the analysis of RNA-Seq data of splenocytes, read counts were calculated by RSEM, and differential expression analysis was conducted with DESeq at FDR (false discovery rate) < 0.05.
Quantification and Statistical Analysis
Immunoblot Band Quantitation
Quantification of band intensities was performed using ImageJ (version 1.50i).
Statistical Analysis
Data analyses were performed using GraphPad Prism 6.0 software. All data are shown as mean ± SD. Statistical analysis was performed using student’s t test with two tailed, 95% confidence. P values less than 0.05 were considered statistically significant. The “n” represents the number of animals, cells and experimental replicates carried out, and was specified in the figure legends.
Data and Software Availability
The accession number for the RNAseq data of splenocytes treatment by cGAMP reported in this paper is NCBI Short Read Archive database, SRA: PRJNA393936. The accession numbers for the Eidolon helvum, Rhinolophus ferrumequinum and Pteronotus parnellii STING nucleotide sequences are GenBank: MF174844–MF174846.
Acknowledgments
We thank Yan-Yi Wang (Wuhan Institute of Virology, CAS) for providing the human cGAS and STING plasmids, Chun-Fu Zheng (School of Basic Medical Sciences, Fujian Medical University) for the HSV-luciferase virus, and Gary Crameri and Justin Ng for leading the team that helped to collect the Pa bat tissues for this study. This work was supported in part by grants from the China Natural Science Foundation (31621061 and 81772199 to P.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB0301), and NRF2012NRF-CRP001–056 (L.-F.W.). J.C. is supported by the CAS Pioneer Hundred Talents Program.
Author Contributions
J.X., Z.-L.S., and P.Z. conceived the project, planned the experiments, and wrote the manuscript with contributions from all authors. J.X., Y.L., X.S., G.G., J.C., Y.Z., and P.Z. performed the experiments. J.X., Z.-L.S., L.-F.W., and P.Z. prepared the draft.
Published: February 22, 2018
Footnotes
Supplemental Information includes two figures and two tables and can be found with this article online at https://doi.org/10.1016/j.chom.2018.01.006.
Supplemental Information
References
- Ahn M., Cui J., Irving A.T., Wang L.F. Unique loss of the PYHIN gene family in bats amongst mammals: implications for inflammasome sensing. Sci. Rep. 2016;6:21722. doi: 10.1038/srep21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber G.N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai A., Rotman G., Shiloh Y. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair. 2002;1:3–25. doi: 10.1016/s1568-7864(01)00007-6. [DOI] [PubMed] [Google Scholar]
- Brook C.E., Dobson A.P. Bats as 'special' reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23:172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalera-Zamudio M., Zepeda-Mendoza M.L., Loza-Rubio E., Rojas-Anaya E., Mendez-Ojeda M.L., Arias C.F., Greenwood A.D. The evolution of bat nucleic acid-sensing Toll-like receptors. Mol. Ecol. 2015;24:5899–5909. doi: 10.1111/mec.13431. [DOI] [PubMed] [Google Scholar]
- Fu Y.-Z., Su S., Gao Y.-Q., Wang P.-P., Huang Z.-F., Hu M.-M., Luo W.-W., Li S., Luo M.-H., Wang Y.-Y. Human cytomegalovirus tegument protein UL82 inhibits STING-mediated signaling to evade antiviral immunity. Cell Host Microbe. 2017;21:231–243. doi: 10.1016/j.chom.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., Fitzgerald K.A. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Waterman P.M., Jonscher K.R., Short C.M., Reisdorph N.A., Cambier J.C. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N., Veettil M.V., Sharma-Walia N., Bottero V., Sadagopan S., Otageri P., Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E., Schoenemeyer A., Visintin A., Fitzgerald K.A., Monks B.G., Knetter C.F., Lien E., Nilsen N.J., Espevik T., Golenbock D.T. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang S., Zhu H., Zheng C.F. Cloning of the herpes simplex virus type 1 genome as a novel luciferase-tagged infectious bacterial artificial chromosome. Arch. Virol. 2011;156:2267–2272. doi: 10.1007/s00705-011-1094-9. [DOI] [PubMed] [Google Scholar]
- Li Y., Chen R., Zhou Q., Xu Z.S., Li C., Wang S., Mao A.P., Zhang X.D., He W.W., Shu H.B. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc. Natl. Acad. Sci. USA. 2012;109:11770–11775. doi: 10.1073/pnas.1203405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.-T., Grishin N.V. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Luis A.D., Hayman D.T.S., O'Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R.C., Mills J.N., Timonin M.E., Willis C.K.R., Cunningham A.A. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. R. Soc. B Biol. Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham D., Scambler T., Savic S., McDermott M.F. The burgeoning field of innate immune-mediated disease and autoinflammation. J. Pathol. 2017;241:123–139. doi: 10.1002/path.4812. [DOI] [PubMed] [Google Scholar]
- Ryan E.L., Hollingworth R., Grand R.J. Activation of the DNA damage response by RNA viruses. Biomolecules. 2016;6:2. doi: 10.3390/biom6010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M., Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T. Immunology of bats and their viruses: challenges and opportunities. Viruses. 2014;6:4880–4901. doi: 10.3390/v6124880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.Y., Liang L., Zhu Z.H., Zhou W.P., Irwin D.M., Zhang Y.P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. USA. 2010;107:8666–8671. doi: 10.1073/pnas.0912613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the Type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.X., Li Y., Chen L., Chen H.H., You F.P., Zhou X., Zhou Y., Zhai Z.H., Chen D.Y., Jiang Z.F. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Chen Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T., Zou J.A., Saitoh T., Kumar H., Abe T., Matsuura Y., Kawai T., Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne J.W., Wang L.F. Bats and viruses: friend or foe? PLoS Pathog. 2013;9:e1003651. doi: 10.1371/journal.ppat.1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G.H., Brendel V.P., Shu C., Li P.W., Palanathan S., Kao C.C. Single nucleotide polymorphisms of human STING can affect innate immune response to cyclic dinucleotides. PLoS One. 2013;8:e77846. doi: 10.1371/journal.pone.0077846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.J., Cowled C., Shi Z.L., Huang Z.Y., Bishop-Lilly K.A., Fang X.D., Wynne J.W., Xiong Z.Q., Baker M.L., Zhao W. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.Q., Yuan B., Bao M.S., Lu N., Kim T., Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12:959–U962. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Zhou P., Cowled C., Todd S., Crameri G., Virtue E.R., Marsh G.A., Klein R., Shi Z., Wang L.-F., Baker M.L. Type III IFNs in pteropid bats: differential expression patterns provide evidence for distinct roles in antiviral immunity. J. Immunol. 2011;186:3138–3147. doi: 10.4049/jimmunol.1003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Tachedjian M., Wynne J.W., Boyd V., Cui J., Smith I., Cowled C., Ng J.H.J., Mok L., Michalski W.P. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc. Natl. Acad. Sci. USA. 2016;113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNAseq data of splenocytes treatment by cGAMP reported in this paper is NCBI Short Read Archive database, SRA: PRJNA393936. The accession numbers for the Eidolon helvum, Rhinolophus ferrumequinum and Pteronotus parnellii STING nucleotide sequences are GenBank: MF174844–MF174846.