Abstract
Mesoporous aerogel was produced under regular atmospheric conditions using the sol–gel polymerization of tetraethyl orthosilicate with an ionic liquid as both solvent and active agent. This was then used to build a three-dimensional structure to recognize nucleotide acids. Fourier transformation infrared spectroscopy, scanning electron microscopy, 29Si solid-state nuclear magnetic resonance, and Brunauer–Emmett–Teller instruments were used to characterize this 3D aerogel, demonstrating that it had high porosity and large internal networking surface area that could capture nucleotide acids. The functionality of molecular recognition on nucleotide acids was demonstrated by immobilizing an oligonucleotide to probe its DNA target and confirming the tagged fluorescent signals by confocal laser scanning microscopy. The results indicated that the as-prepared 3D bioaerogel was capable of providing a very large surface area to capture and recognize human gene ATP5O.
Keywords: Aerogel, Molecular recognition, DNA detection, 3D biochips
1. Introduction
Several porous materials have been developed for applications in molecular recognition (e.g., polyacrylamide [1], [2], hydrogel [3], [4], nitrocellulose [5], and agarosegel [6]). The main common character of these materials as biosensor matrices is their vast 3D internal surface area, which functions as a substrate capable of immobilizing oligonucleotide probes or ligand receptors to capture more biological target molecules than traditional planar substrates. However, the problems of fragility and thermal resistance of these materials have not been thoroughly addressed, limiting the application of these materials. As we show in this contribution, silica aerogel can provide a good solution to these problems.
Silica aerogel is a material with high porosity, large surface area, low density, and low thermal conductivity. These unique characteristics give this advanced material many applications for thermal insulation, electrical batteries, nuclear waste storage, catalysis, acoustic insulation, and adsorbents [7]. In addition, due to its chemical and mechanical robustness, large internal capacity, and biocompatibility, silica aerogel also has promise as a biocompatible scaffold to immobilize or protect biological materials for virus detection [8], protein entrapment [9], protein incorporation [10], hybridization array [11], [12], and building potential matrices in the further design of biosensors. Moreover, its use for capturing comet dust has pushed this material into outer space applications [13].
This report proposes a preparation technique to synthesize mesoporous aerogel using the sol–gel process and utilize the aerogel’s high surface area and large internal porous volume for molecular recognition of nucleotide acids. Although the supercritical drying method is also a viable option to produce the aerogels [8], [11], in order to avoid the harsh conditions of high temperature and high pressure used in this technique, in this study, the aerogel was prepared by the sol–gel process with a reusable ionic liquid (IL) as the solvent and pore-forming agent. The unique properties of IL, including negligible vapor pressure and ionicity, allowed the hydrolysis and condensation of the sol–gel polymerization to produce a stable gel network without excess shrinkage and also retain the aerogel structure after the solvent extraction and freeze-drying process [9]. The as-prepared aerogel was then applied for recognition of a sequence-specific deoxynucleotide acid (DNA) target and compared the results with those from traditional planar slides.
ATP5O is known to function as a human ATP synthase and CANX as a Homo sapiens calnexin. ATP5O is also the most significantly reduced gene involved in the oxidative phosphorylation (OXPHOS) [14], which was reported in parallel with increased insulin resistance in patients with type II diabetes mellitus [15]. CANX is a molecular chaperone found primarily in the endoplasmic reticulum and is essential for proper protein folding. Correct protein assembly in the membrane is supported by CANX, which retains intermediate structures in the endoplasmic reticulum prior to ternary complex formation completion [16]. CANX is also frequently used as a house-keeper control in microarray technology [17], [18]. Therefore, ATP5O and CANX were utilized in this study as the DNA-target models for molecular recognition of nucleotide acids of the as-prepared three-dimensional aerogel biochip.
2. Experimental procedures
2.1. Materials
The raw materials and necessary solvents for preparing the ionic liquid, 1-methylimidazole, acetonitrile, 1-chlorobutane, and sodium tetrafluoroborate were all purchased from Acros (USA). The networking precursor of the aerogel, tetraethyl orthosilicate (TEOS), the solvent, methanol, and the surface-modifying reagent of aerogel, 3-glycidoxypropyltrimethoxysilane (GLYMO), were also all obtained from Acros Company. The DNA targets were PCR (polymerase chain reaction) products amplified from two human genes in total RNA: ATP5O (313 bp) and CANX (1027 bp). Table 1 lists their gene information and the primers participating in the PCR process. The 5′-end modified amino linker of the DNA-oligo probe allowed it to covalently bind either with the epoxy-modified surface of 3D aerogel or to commercial 2D glass slides (Phalanx Biotech, Hsinchu, Taiwan). Both probe and primer sequences are listed in Table 1. The conformations of DNA-probe immobilization and probe-target hybridization are shown in Fig. 1 . All RNA, PCR primers and probes were purchased from ScinoPharm (Tainan, Taiwan). The probe immobilization buffer was prepared from SSC (sodium chloride–sodium citrate buffer, Amresco), glycerol (ICN Biomedicals) and ddH2O (Milli-Q synthesis A10 system). The biochip washing buffers were made from sodium dodecyl sulfate (SDS; Amresco, USA) and SSC. The kit for the reverse transcription of total RNA to cDNA target was purchased from Invitrogen (SuperScript™ II RT, USA). The PCR purifying kit was purchased from Nobel Biotech (QIA-quick; Taiwan). SP5-Cy3 (60-mer) serves as an immobilization control probe with sequence of amine-C6-5′GCTGTAACTTATCACACCGTT TCTACAGGTTAGCTAACGAGTGT GCGCAAGTATTAAGTG-3′-Cy3, which is a complete genome of SARS coronavirus strain CV7 (NCBI Database, Accession Number DQ898174.1) labeled with Cy3 fluorescence.
Table 1.
Information on genetic targets, oligonucleotide probes, and primers adopted in this study.
| Genetic target | Length (bases) | Definition (NCBI gene bank) |
|---|---|---|
| ATP5O | 313 | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (oligomycin sensitivity conferring protein) |
| CANX | 1027 | Homo sapiens calnexin (CANX), transcript variant 1, mRNA |
| Probes | Sequences (hybridized with target on underlined bases) |
| ATP5Oc | Amine-C6-5′(GTGATTGGACGCGGTGA)-(GTCTTGACAGACATGTCAACATATTTCTCGCCAAT GCGCACAATCATTCCACCCAAGATT)3′ |
| CANXc | Amine-C6-5′(ATCGAAGAACGGCTCGC)-(TTAAAGCTC AGC TAGAAGA AAGTGAGGCATGA CATATACTG TCAACGGAGG GTGAAGGAG)3′ |
| Primers | Starting positiona | Product length |
Sequences | |
|---|---|---|---|---|
| dsDNA(bp) | ssDNA(b) | |||
| ATP5O-F1 | 229 | 524 | TTCTGCTGCATCAAAACAGAAT | |
| ATP5O-R1 | 753 | ATGGCAGAAAACCAACACTTTT | ||
| cy3-ATP5O-F2 | 440 | 313 | CGATTAAGCA CAAGGAG ATACC | |
| CANX-F1 | 3162 | 1334 | GAGGAGTGACATGAAGCATGAG | |
| CANX-R1 | 4496 | TCTGAGCTGCTGTCCACACTAT | ||
| cy3-CANX-F2 | 3469 | 1027 | GGCTTTCAAATGTACCGATGAT | |
Complementary section of nucleotide bases counted from the 5′-end of the corresponding target. Primers F1 and R1 stands for the forward and reverse primer used in the first round of PCR process to produce double-stranded cDNA, which was employed as the template of the second round of PCR process. In the second round of PCR process, only one primer, i.e., F2, was used.
Fig. 1.
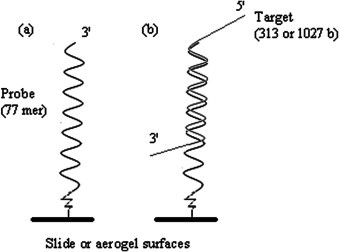
The conformations of DNA-probe immobilization and probe-target hybridization. (a) The 77-mer probes, ATP5Oc or CANXc, were immobilized on both planar slide and aerogel surfaces for comparison. (b) The upper 60-mer of the probe was designed specifically to recognize the DNA sequence of the target. The lower 17-mer provided a space to prevent the target–substrate interface from a steric hindrance.
2.2. Preparation of the aerogel template, 1-n-butyl-3-methylimidazolium tetrafluoroborate (BMIM+BF4−)
To avoid the harsh conditions in supercritical drying, the ionic liquid, 1-butyl-3-methylimidazolium tetrafluoroborate (BMIM+BF4 −, abbreviated as BMIBF), was utilized in this study as both the solvent and the template for the TEOS sol–gel polymerization. The negligible vapor pressure and ionicity of this ionic solvent made the drying process feasible in the ambient conditions to produce a stable silica gel.
To start preparing the BMIBF, a solution of 1-methylimidazole (82.0 g, 1 mol) in acetonitrile (50 g) was drop-wise added to 1-chlorobutane (370 g, 4 mol) at room temperature and stirred overnight. After removing the acetonitrile by distillation, the remaining salt, 1-n-butyl-3-methylimidazolium chloride (BMIC), was freeze-dried at −50 °C. Then 218 g (1 mol) of BMIC was added to the solution of sodium tetrafluoroborate (132 g, 1.2 mol) in acetonitrile (100 g). The solution was mechanically stirred for 2 days at room temperature. The resulting mixture was then filtered and placed under reduced pressure to remove the volatiles. The ionic liquid product, BMIBF, was finally obtained through filtration followed by freeze-drying at −50 °C. Detailed information on the above process can be found in Refs. [19], [20].
2.3. Preparation of the silica aerogel
The silica aerogel was developed using previously prepared BMIBF as the template and a solvent of the sol–gel process. In a typical run, 5.8 g (28 mmol) of TEOS, 1.9 g (58 mmol) of methanol, 3.0 g (12 mmol) of BMIBF, and 3.4 g (189 mmol) of water were initially mixed. Within 3 h, a monolithic gel was formed at room temperature. It was then cured at ambient temperature for 1 week. Then the entrapped ionic-liquid template was Soxhlet-extracted with fluxing ethanol for 1 day. The as-prepared silica aerogel was finally obtained by freeze-drying, and Fourier transformation infrared (FTIR) analysis was utilized to confirm the removal of the ionic-liquid template.
To capture the NH2-modified DNA probe, the aerogel product was ground into powder and stirred in 5% GLYMO solution for 1 h to modify the surface from —OH to epoxy functional group. The modified aerogel powder was filtered from the solution and soxhlet-extracted with ethanol for 1 day and then freeze-dried. The surface micro-structure of the modified aerogel product was then analyzed using scanning electron microscopy (SEM), and the network structure was analyzed using 29Si solid-state nuclear magnetic resonance (NMR), while the internal volume and porosity were determined using Brunauer–Emmett–Teller (BET) instruments. The results were than compared with those obtained prior to modification.
2.4. Preparation of single-stranded DNA targets
Although an asymmetric PCR process was initially tried to prepare the single-stranded (ss) human genomic targets, ATP_5O (313b) and CANX (1027b), the yield was very low. A different approach was then conducted. The double-stranded (ds) cDNA was first produced from the total RNA by the reverse-transcript (RT) PCR procedure provided by the manufacturer. The consequent dsDNA product then served as the template of the second round of asymmetrical PCR process. In the first run of regular PCR, 2 μl template was mixed with 2 μl F1 primer (10 μM), 2 μl R1 primer (10 μM), 1.25 μl dNTP (10 mM), 1 μl Taq, DNA polymerase (5 U μl−1), 5 μl PCR buffer (10×), and 36.75 μl ddH2O to make a total volume of 50 μl. As indicated in Table 1, the primers were ATP5O-F1/ATP5O-R1 or CANX-F1/CANX-R1. The PCR process was set as denaturalization step 94 °C for 5 min, annealing step 52 °C for 30 s, and extension step 72 °C for 1 min with total 40 cycles. The PCR product, dsDNA, was purified and ready for the second PCR run to asymmetrically amplify the ssDNA target.
In the asymmetrical PCR, 1 μl ds template was mixed with 2 μl Cy3-primer-F2 (10 μM), 1.25 μl dNTP (10 mM), 1 μl Taq DNA polymerase (5 U μl−1), 5 μl PCR buffer (10×), and 39.75 μl ddH2 to make a total volume of 50 μl. The cycle setting was identical with the first run. The F2 primer was cy3-ATP5O-F2 or cy3-CANX-F2 as shown in Table 1. The product of ssDNA was finally verified by 1.5% polyacrylamide gel electrophoresis (PAGE).
2.5. Pre-cleaning of slides and spotting of aerogel-dot array
The virgin slides were first cleaned by 10% NaOH in an ultrasonic shaker for 30 min and then rinsed by ddH2O. The cleaned slides were then arrayed with the functionalized aerogel dots by the procedure described below and used for optimizing the probe concentration and detecting the binding capability of the as-prepared aerogel biochip. However, from the results it was found that the aerogel particles were not quite evenly distributed in the dot on the slide: there were more in perimeter than in interior. In order to improve it for the molecular recognition tests, the slide for the experiments of Fig. 9 was further washed by acetone in the shaker for another 30 min and then baked at 70 °C for 20 min to reduce the hydrophilicity of the slide’s surface, resulting in smaller and more evenly distributed aerogel dots.
Fig. 9.
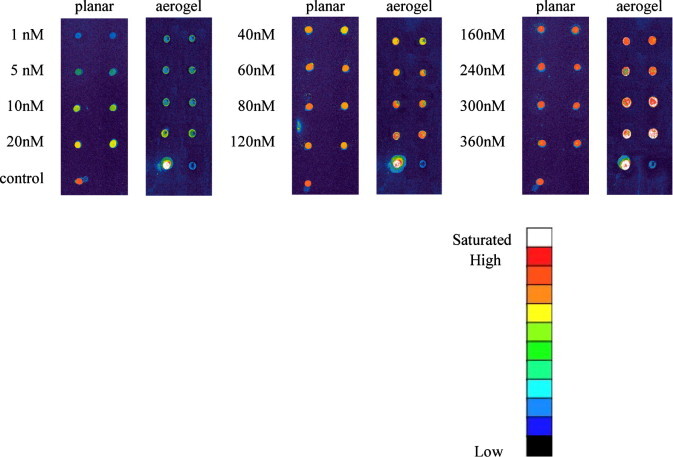
Molecular recognition test of ATP5O on aerogel. Human gene ATP5O (313b) was recognized on the aerogel surfaces over various concentrations. Two replicated columns were conducted. The results were compared side-by-side with planar slides. Probe ATP5Oc (10 μM) was immobilized on both flat slides and aerogel. Sp5-cy3 (10 μM) served as the positive and hybridization buffer as the negative controls.
For spotting the aerogel dots on the cleaned slides the epoxy-modified aerogel powder was uniformly dispersed in ddH2O to form a 10% (w/v) solution. The resulting solution was manually taken up by a 2 μl pipette and dropped onto the precleaned slide in an array aligned by a grid paper placed underneath. The slides were then baked at 100 °C for 90 min and ready for DNA-probe immobilization. Over 30 aerogel-dot arrayed slides with or without acetone-washing showed good reproducibility technique.
2.6. DNA-probe immobilization
To optimize probe concentration, Sp5-cy3 was mixed with 20× SSC and 100% glycerol to final concentrations of 0.01, 0.1, 1, 10, and 100 nM, and spotted onto the aerogel-dot array on the slide. After fluorescent scanning, 10 nM appeared to be the acceptable signal. We hence selected this concentration as the probe concentration to save material for the following experiments.
The oligonucleotide probe and positive control, Sp5-cy3, were individually mixed with 20× SSC and 100% glycerol to the final concentration of 10 nM, and dropped on both commercial and the as-prepared aerogel slides in amounts of 1 μl. The slides were then incubated in a humidified box at 30 °C for 16–17 h to immobilize the probes on the planar slides or the aerogel slides. A wash step was conducted to remove the free probes in 0.5% SDS by shaking at 80 rpm for 15 min. The slides were then immersed in the blocking buffer at 60 °C for 45 min and finally rinsed with ddH2O and spun to dry. A pre-scanning of the control spots was performed to confirm the presence of probes.
2.7. DNA probe-target hybridization and data acquisition
DNA target was mixed with 1× hybridization buffer to the final concentrations of 1, 5, 10, 20, 40, 60, 80, 120, 160, 240, 300, and 360 nM. Target solutions of 1 μl were dropped onto planar slides and aerogel dots at the previously immobilized probes. All chips were then enclosed in a humidified box and incubated at 50 °C for 16–17 h and then washed. They were initially immersed in 2× SSC/0.2× SDS at 42 °C at 80 rpm for 10 min, then changed to 2× SSC at the same conditions, and finally 0.2× SSC at room temperature. After rinsing with ddH2O and being spun dry, the slides were ready for fluorescence analysis.
The AXON 4000B scanner was employed to acquire the resulting images in an appropriate laser power and PMT setting. The excitation/emission wavelengths of Cy3 were set at 550/570 nm and the scanning resolution was 10 μm. All these parameters were fixed in this study.
2.8. Instrument
A FTIR spectrometer (Bio-Rad FTS-7) was used to verify the removal of the template from the aerogel. The BET specific surface area and the pore volume of the as-prepared aerogels were determined from the nitrogen adsorption/desorption isotherm by the Barrett–Joyner–Halenda (BJH) method using a Micrometrics ASAP 2020 analyzer. The 29Si magic angle spinning (MAS) NMR measurements for characterizing the siloxane architecture were carried out on a Bruker Msl-400 NMR spectrometer. The morphologies of the prepared aerogels were examined by a field emission (FE) SEM (LEO 1530 FEG-SEM+EDS). PCR was performed on GeneAmp_PCR, System 9700. Gel electrophoresis was analyzed on GeneFlash (Syngene, USA). The microarray scanner (GenePix 4000B, Molecular Devices) was used to excite and read the labeling fluorescence.
3. Result and discussion
Fig. 2 shows the FTIR spectrum for the as-prepared silica aerogel before and after template removal. The peak height of the template band was found to be very small after removal, revealing that the majority of the template had been removed from the wet aerogel by our extraction process. The spectrum also suggested that silica aerogel had the SiO2 structure whose O—H characteristic absorbance band was observed at 3200–3700 cm−1 and 1620–1640 cm−1 for the Si—OH vibration, 1088 cm−1 for the Si—O—Si asymmetrical stretching vibration, and 1000–1200 cm−1, 780–820 cm−1, and 430–460 cm−1 for the Si—O—Si strain stretching vibrations.
Fig. 2.
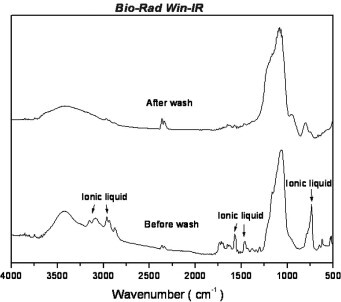
FTIR analysis of the aerogel product before and after template removal. The disappearance of the characteristic peaks in the spectrum indicates that the template was successfully removed.
Solid-state 29Si MAS NMR spectroscopy is generally used to investigate the chemical environment of 29Si atoms. According to the report of Legrand et al. [21], the resonances of the different species of Si atoms in silica are located between −80 and −110 ppm, as identified by the Qn terminology. Q0 denotes the silicon with no Si—O—Si linkage and Qn denotes the silicon with n Si—O—Si linkages, where n = 1–4. Generally, the tetra substitution (Q4) results in a characteristic peak between −100 and −110 ppm. Similarly Q3 occurs between −90 and −100 ppm and Q2 occurs between −80 and −90 ppm. Fig. 3 a shows the 29Si solid-state NMR spectrum of the as-prepared silica aerogel. Its Q4 characteristic peak appeared at −110 ppm, Q3 at −102 ppm, and Q2 at −92 ppm. Their proportion in as-prepared silica aerogel was Q4:Q3:Q2 = 55.9:39.0:5.1. This suggests that more than 87.7% of the silanol groups were condensed during the sol–gel aging process. It can be concluded that the aerogel product had a stable silica network structure. Fig. 3b shows the 29Si solid-state NMR spectrum for post-modification of hydroxyl to epoxy functional groups. Its T3 characteristic peaks appeared at −66 ppm and T2 at −57 ppm with a simultaneous Q3 proportion that had dropped to 14.6%, indicating that about 22.3% of the aerogel surface was converted to the epoxy functional group.
Fig. 3.
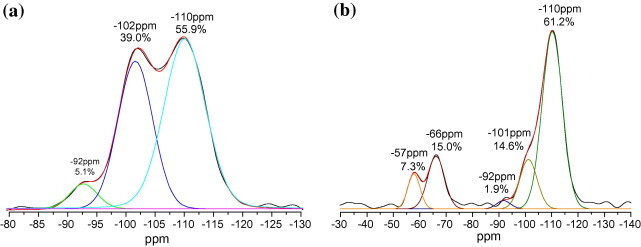
29Si solid-state NMR measurement of silica aerogel. (a) Prior to epoxy modification, Si—O substitutions were 55.9% in Q4, 39.0% in Q3, and 5.1% in Q2, and their corresponding characteristic peaks appeared at −110 ppm, −102 ppm, and −92 ppm, respectively. (b) Post-epoxy modification, Si—O substitutions were 15% in T3 and 7.3% in T2, and their characteristic peaks appeared at −66 ppm and −57 ppm, respectively. The proportion of Q3 dropped to 14.6%.
BET is an instrument utilized to probe the porosity, specific surface area, pore structure, and other physical surface prosperities of a material using an inert working gas. The extent of gas adsorption on the material is closely related to the nature of the sample and the gas. It is also dependent on the working pressure and temperature. An adsorption isotherm, which plots the amount of gas adsorption per unit weight of sample against the ratio of working pressure to the saturated vapor pressure of the gas, is usually presented as the measurement result. Fig. 4 shows the nitrogen adsorption/desorption isotherm of the silica aerogel product, which exhibited a type-IV isotherm based on the classification of Brunauer et al. [22]. In this type of material, adsorption occurs in the micropores at very low pressure, followed by adsorption in the mesopores, with capillary condensation occurring at higher pressures, leading to the hysteresis loops [23]. The measurement indicated a specific surface area of 813 m2 g−1, pore volume of 0.78 cm3 g−1, and average pore diameter of 4.2 nm. After the epoxy modification, these levels dropped to 471 m2 g−1, 0.48 cm3 g−1, and 3.7 nm, respectively. The data suggest that modified silica aerogel was still a mesoporous material. Its large surface area ensured that the detection signal of the captured biological samples on the surface can be greatly intensified.
Fig. 4.
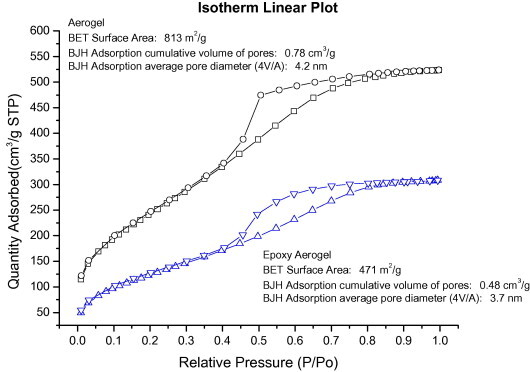
Nitrogen isotherm of the silica aerogel product. The isotherm exhibited a type-IV curve and the measurement indicated that its specific surface area was 813 m2 g−1, pore volume was 0.78 cm3 g−1, and average porosity was 4.2 nm. After the epoxy modification, these dropped to 471 m2 g−1, 0.48 cm3 g−1, and 3.7 nm.
Fig. 5 shows the magnified SEM image of silica aerogel. As shown in the image, the cluster surface is formed by a great many silica nanoparticles with an average size of around 100 nm. The interstitial space between those particles provides extensive capturing sites available for biological samples.
Fig. 5.
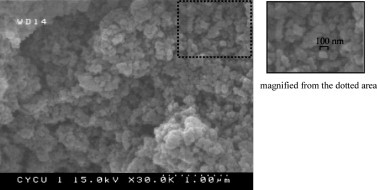
SEM image of the silica aerogel. The image shows that the surface is formed by a great number of silica nanoparticles with an average size of around 100 nm. The rough surface and interstitial space among particles provided a vast amount of capturing sites available for biological samples. The image was magnified in 30 k-folds.
Fig. 6a shows the arrayed aerogel dots on a slide ready for immobilizing the capturing probes. Fig. 6b shows the corresponding experimental design, which is with two replicated columns, especially for the experiment on the top four rows and controls on the bottom row. The positive and negative control spots validated the experimental process, normally with the positive signal on and the negative signal off.
Fig. 6.
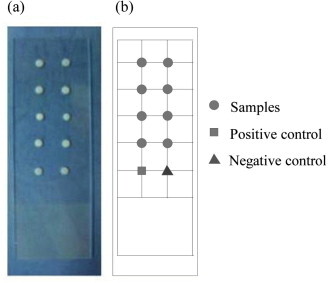
The arrayed aerogel dots on a glass slide. Left: the real object. Right: the corresponding experimental design layout. The positive and negative control spots validated the experimental process. Cy3-Sp5 served as the positive control and the immobilization buffer served as the negative control.
The binding capability of the as-prepared aerogel was initially investigated to preliminarily determine the optimal probe immobilization concentration by the test DNA, Sp5-cy3. As shown in the images of Fig. 7 , the fluorescence intensity was observed to vary with the Sp5 (60-mer) concentration of 10–0.1 μM. This can be compared with the low background generated by the aerogel at 0.01 μM of Sp5 DNA. Probe concentrations above 10 μM not only consumed excessive materials but pushed the intensity readings beyond the limit of the instrument. We thus chose 10–0.1 μM to further optimize the gene probe’s concentration for immobilization. As shown in Fig. 8 , the optimization result showed that the best performance was with 10 μM ATP5Oc, so this concentration was adopted for further tests to demonstrate the functionality of molecular recognition.
Fig. 7.
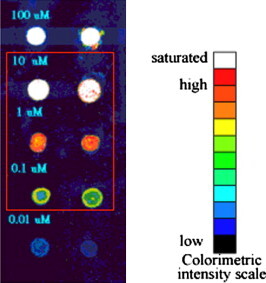
Determination of optimal probe concentration. Sp5-Cy3 (60-mer) served as the test DNA to preliminarily investigate the binding capacity of the as-prepared aerogel with DNA samples. Two replicated aerogel-dot columns were arrayed and the test DNA was loaded in a concentration titration of 100, 10, 1, 0.1, and 0.01 μM. Cy3 fluorescence intensity was presented in a 32-bit colorimetric scale, with red the most intense, black the lowest, and white beyond the analysis capability of the image scanner (saturation).
Fig. 8.
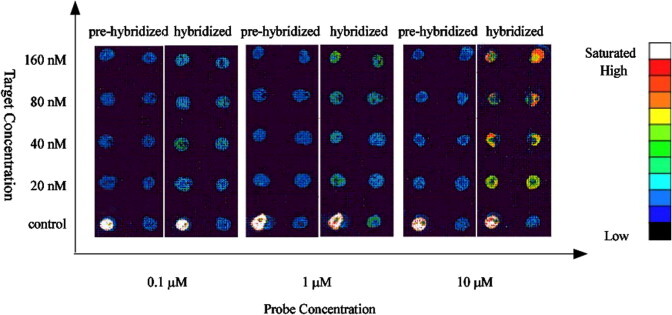
Optimization of probe immobilization concentration. ATP5Oc was immobilized on the aerogel surface in 10, 1, and 0.1 μM. The target DNA, cy3-ATP5O, was titrated in 160, 80, 40, and 20 nM. For each probe concentration, slides were scanned before and after the probe-target hybridization was performed. Positive and negative controls, which guarantee the validation of result, were set by 10 μM Sp5-cy3 and hybridization buffer, respectively.
In the beginning, the ATP5Oc-immobilized aerogel prepared with the slide without acetone-washing was utilized to recognize the target human gene ATP5O. However, a heterogeneous DNA-sample distribution was found on dots of the aerogel biochip (data not shown), indicating that the aerogel particles were unevenly distributed in the dots of the slide. This result was believed due to the slide’s surface hydrophilicity caused by the pre-cleaning procedure. In order to improve it, another recognition test was carried out with the acetone-cleaned slide and the result is shown in Fig. 9 . The dots afterward became smaller, i.e., the aerogel particles had larger contact angle with the slide, and the majority part of each dots showed more homogeneous color, indicating that a better quality of the chip was achieved. In addition, the positive control dots on the bottom left of the aerogel slides was much brighter than that of the planar slides, indicating that the capturing capacity of the aerogel slide was much larger than that of the planar slide. At low–medium concentrations, both types of slides showed similar signal intensity. At concentrations of 40–120 nM, the flat surface started to reach its accommodation saturation for target molecules, but the intensity of the aerogel spot was increased with the concentration, indicating that the amount of the captured DNA was increased accordingly. Especially, at 360 nM, the aerogel spots even more obviously appeared higher capturing ability than the planar spots.
In addition, the advantage of the larger amount of DNA captured by our aerogel biochip can be seen in Fig. 10 . Here, 1 μl of 20 nM DNA target was first added onto all top-four-row aerogel dots and then the slide was washed to remove the unbounded sample. The same procedure, including the target addition and the slide cleaning, was then applied to the top-three-row, top-two-row, and top-one-row dots for the second, third and fourth time, sequentially. Thus, the accumulated sample volume of each aerogel-spot row became 4, 3, 2 and 1 μl. After repeated washes, some aerogel dropped off from the slide to cause donut-shape spots, but the signal trend can still easily be seen. The 1 μl-sample row developed less signal intensity than that developed by the same sample-volume spot at Fig. 9 due to repeated washes. However, it is still clear that aerogel spots loaded with more sample volume significantly gained in signal intensity. The 4 μl sample row obviously had signal intensity higher than the 20 nM sample in Fig. 9. It can thus be concluded that the aerogel functions as a sample reservoir collecting more samples to intensify detection signal.
Fig. 10.
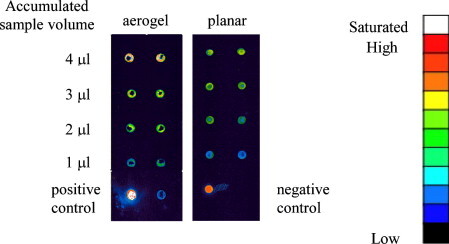
Detection sensitivity gained by addition of sample volume. ATP5O (20 nM) was loaded onto desired aerogel or planar spots each time, so that the accumulated volume was 4, 3, 2, and 1 μl on each row. A replication of two columns was conducted. Sp5-cy3 (10 M) served as positive and hybridization buffer for the negative controls.
We had also run the identical experiment on a planar slide for a side-by-side comparison. The signal was again intensified with more DNA target added on the planar slide. Therefore a certain concentration can be detected more clearly with more DNA on both types of slides. However, as 3–4 μl of sample was applied, the aerogel slide started to exhibit orange and white colors, indicating that much more target DNA was captured and confirming much higher sample-binding capacity for the aerogel slide than the planar one.
A blank test was also performed to clarify whether or not the previously observed signal should be attributed to the physical clogging of aerogel pores by DNA target, instead of to sequence specificity. The test was conducted by repeating the previous experiment on four target concentrations but on a plain aerogel surface, i.e., with no probe immobilization. The low background, as shown in Fig. 11 , indicates that the signals observed previously indeed came from the probe-target recognition.
Fig. 11.
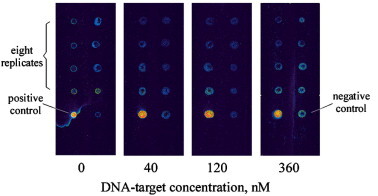
The blank test on aerogel. Repeated experiment was performed on aerogel without oligonucleotide probe immobilization. DNA target spread over the aerogel was 0, 40, 120, and 360 nM from the left to right slide with eight replicate dots on each slide. The low background suggested that the signal observed in Fig. 9 was solely from probe-target sequence recognition.
Fig. 12 shows the repeated test on a longer gene. In this test, human gene CANX (1027b) served as the DNA target and the applied conditions were identical to the previous shorter gene. As shown in the images, the longer DNA did not have a good penetration capability into the porous aerogel and thus generated a much lower intensity than flat slides since the screening effect of the aerogel had more effect on the longer gene.
Fig. 12.
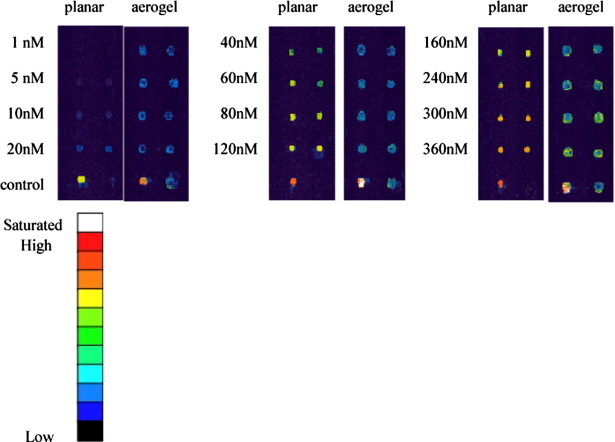
Molecular recognition test of CANX on aerogel. Human gene CANX (1027b) was recognized on the aerogel surfaces over various concentrations. Two replicated columns were conducted. The results were compared side-by-side with planar slides. The 10 μM probe CANXc was immobilized on both flat slides and aerogel, and 10 μM Sp5-cy3 served as the positive and hybridization buffer as the negative controls. The results were compared side-by-side with planar slides, which showed a better result than aerogel on a long-size DNA target.
Fig. 13 shows the recognition test for a non-specific DNA target on the as-prepared aerogel. Human gene PSMA5 (432b) was recognized on the aerogel surfaces by 10 μM ATP5Oc probe, which was immobilized on the aerogel. PSMA5 target spread over the aerogel was 0, 40, 120, and 360 nM from the left to right slides with 8 replicate dots on each slide. Sp5-cy3 (10 μM) served as the positive (bottom left) and hybridization buffer as the negative controls (bottom right). The low background level suggests that our aerogel biochips were free from non-specific sequence recognition.
Fig. 13.
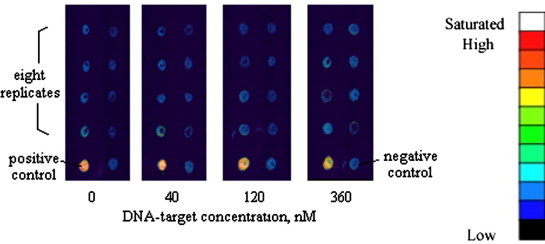
Non-specific molecular recognition test on the aerogel biochips. Human gene PSMA5 (432b) served as the target and was tested by 10 μM ATP5Oc probe immobilized on the aerogel surface. The low background suggested that our aerogel biochips were free from the non-specific sequence recognition.
4. Conclusions
In this study, mesoporous aerogels were prepared at room temperature by the sol–gel polymerization with an ionic liquid as the solvent and pore-forming agent. The as-prepared aerogel was characterized by different instruments and found that it had high porosity and large internal networking surface area. The as-prepared aerogel was further arrayed onto slides and successfully recognized a short human gene ATP5O by an immobilized oligonucleotide probe on the aerogel surface. The large capturing capacity of the porous structure was also demonstrated by comparing with a planar surface at high target concentrations. The results indicated that the as-prepared aerogel can function as a recognition substrate for nucleotide acids.
Acknowledgement
The authors gratefully acknowledge the support of the National Science Council of the Republic of China under Grant No. NSC 97-2622-E-033-008-CC1 and that of Chung Yuan Christian University, ROC under No. CYCU-97-CR-CH and CYCU-98-CR-CE.
Appendix A. Figures with essential colour discrimination
Certain figures in this article, particularly Figures 3, 4, and 6–13, are difficult to interpret in black and white. The full colour images can be found in the on-line version, at doi:10.1016/j.actbio.2009.10.001.
References
- 1.Angenendt P., Glokler J., Murphy D., et al. Toward optimized antibody microarrays: a comparison of current microarray support materials. Anal Biochem. 2002;309:253–260. doi: 10.1016/s0003-2697(02)00257-9. [DOI] [PubMed] [Google Scholar]
- 2.Kersten B., Feilner T., Kramer A., et al. Generation of Arabidopsis protein chips for antibody and serum screening. Plant Mol Biol. 2003;52:999–1010. doi: 10.1023/a:1025424814739. [DOI] [PubMed] [Google Scholar]
- 3.Rubina A., Dementieva E., Stomakhin A., et al. Hydrogel-based protein microchips: manufacturing, properties, and applications. BioTechniques. 2003;34:1008–1022. doi: 10.2144/03345rr01. [DOI] [PubMed] [Google Scholar]
- 4.Zubtsov D., Savvateeva E., Rubina A., et al. Comparison of surface and hydrogel-based protein microchips. Anal Biochem. 2007;368:205–213. doi: 10.1016/j.ab.2007.04.040. [DOI] [PubMed] [Google Scholar]
- 5.Stillman B., Tonkinson L. FAST™ slides: a novel surface for microarrays. BioTechniques. 2000;29:630–635. doi: 10.2144/00293pf01. [DOI] [PubMed] [Google Scholar]
- 6.Afanassiev V., Hanemann V., Wolfl S. Preparation of DNA and protein micro-arrays on glass slides coated with an agarose film. Nucleic Acids Res. 2000;28(12):E66. doi: 10.1093/nar/28.12.e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierre A., Pajonk G. Chemistry of aerogels and their applications. Chem Rev. 2002;102:4243–4265. doi: 10.1021/cr0101306. [DOI] [PubMed] [Google Scholar]
- 8.Power M., Hosticka B., Black E., Daitch C., Norris P. Aerogels as biosensors: viral particle detection by bacteria immobilized on large pore aerogel. J Non-Cryst Solids. 2001;285:303–308. [Google Scholar]
- 9.Li Y., Chou M., Wu T., Jinn T., Chen-Yang Y. A novel method for preparing a protein-encapsulated bioaerogel: using a red fluorescent protein as a model. Polymer. 2007;48(1):456–457. doi: 10.1016/j.actbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Wallace J., Rice J., Pietron J., Stroud R., Long J., Rolison D. Silica nanoarchitectures incorporating self-organized protein superstructures with gas-phase bioactivity. Nano Lett. 2003;3:1463–1467. [Google Scholar]
- 11.Phinney J., Conroy J., Hosticka B., Power M., Ferrance J., Landers J., et al. The design and testing of a silica sol–gel-based hybridization array. J Non-Cryst Solids. 2004;350:39–45. [Google Scholar]
- 12.Saal K., Taette T., Tulp I., Kink I., Kurg A., Maeeorg U., et al. Sol–gel films for DNA microarray applications. Mater Lett. 2006;60(15):1833–1838. [Google Scholar]
- 13.Available from: www.nasa.gov.
- 14.Mootha V., Lindgren C., Eriksson K., Subramanian A., Sihag S. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 15.Sreekumar R., Halvatsiotis P., Schimke J., Nair K. Gene expression profile in skeletal muscle of type II diabetes and the effect of insulin treatment. Diabetes. 2002;51:1913–1920. doi: 10.2337/diabetes.51.6.1913. [DOI] [PubMed] [Google Scholar]
- 16.Takizawa T., Tatematsu K., Watanabe K., Kato K., Nakanishi Y. Cleavage of calnexin caused by apoptotic stimuli: implication for the regulation of apoptosis. J Biochem. 2004;136:399–405. doi: 10.1093/jb/mvh133. [DOI] [PubMed] [Google Scholar]
- 17.York T., Plymate S., Nelson P., Eaves L., Webb H., Ware J. cDNA microarray analysis identifies genes induced in common by peptide growth factors and androgen in human prostate epithelial cells. Mol Carcinog. 2005;44(4):242–251. doi: 10.1002/mc.20139. [DOI] [PubMed] [Google Scholar]
- 18.Cammack K., Antoniou E., Hearne L., Lamberson W. Testicular gene expression in male mice divergent for fertility after heat stress. Theriogenology. 2009;71(4):651–661. doi: 10.1016/j.theriogenology.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y., Schattka J.H., Antonietti M. Room-temperature ionic liquids as template to monolithic mesoporous silica with wormlike pores via a sol–gel nanocasting technique. Nano Lett. 2004;4:477–481. [Google Scholar]
- 20.Suarez P., Dullius J., Einloft S., De Souza R., Dupont J. The use of new ionic liquids in two-phase catalytic hydrogenation reaction by rhodium complexes. Polyhedron. 1996;15:1217–1219. [Google Scholar]
- 21.Legrand A., Hommel H., Tuel A., Vidal A., Balard H., Papirer E., Levitz P., Czernichowski M., Erre R., et al. Hydroxyls of silica powders. Adv Colloid Interface Sci. 1990;33(2–4):91–330. [Google Scholar]
- 22.Brunauer S., Deming L., Deming W., Teller E. On a theory of the van der Waals adsorption of gases. J Am Chem Soc. 1940;62:1724–1732. [Google Scholar]
- 23.Hiemenz P., Rajagopalan R. 3rd ed. Marcel Dekker; New York: 1997. Principles of colloid and surface chemistry. [Google Scholar]


