Abstract
Drug-induced hypersensitivity syndrome/drug reaction with eosinophilia and systemic symptoms (DiHS/DRESS) is a potentially fatal multi-organ inflammatory disease associated with herpesvirus reactivation and subsequent onset of autoimmune diseases1–4. Pathophysiology remains elusive and therapeutic options are limited. Cases refractory to corticosteroid therapy pose a clinical challenge1,5, and approximately 30% of DiHS/DRESS patients develop complications including infections and inflammatory/autoimmune diseases1,2,5. Progress in single-cell RNA sequencing (scRNAseq) provides an opportunity to dissect human disease pathophysiology at unprecedented resolutions6, particularly in diseases lacking animal models, such as DiHS/DRESS. We performed scRNAseq on skin and blood from a refractory DiHS/DRESS case, found JAK-STAT signaling pathway as potentially targetable, and further identified that central memory CD4+ T cells were enriched with HHV6b DNA. Intervention via tofacitinib enabled disease control and tapering of other immunosuppressive agents. Furthermore, tofacitinib, as well as anti-viral agents, suppressed culprit-induced T cell proliferation in vitro, identifying the JAK-STAT pathway and herpesviruses as potential therapeutic targets in DiHS/DRESS. Thus, scRNAseq analyses guided therapeutic decisions that enabled successful therapeutic intervention in this refractory DiHS/DRESS case. Employing scRNAseq analyses in human diseases may facilitate our understanding of complicated disease pathophysiology and further provide an alternative approach in personalized medicine.
Here, we report a case of DiHS/DRESS in a 44-year-old male patient that was induced by sulfamethoxazole/trimethoprim (SMX/TMP). The patient had a severe rash with systemic symptoms which were uncontrolled despite high-dose prednisone. Attempts to taper prednisone led to severe flares and progression to toxic epidermolytic necrosis-like presentation (Fig. 1a) for which one dose of etanercept and high-dose intravenous immunoglobulin was administered with no clear improvement. Skin presentation evolved to a diffuse, psoriasis-like process, ultimately resulting in generalized exfoliative dermatitis (Fig. 1a). Cyclosporine at 400mg/day decreased skin inflammation, but tapering or inadvertent discontinuation resulted in severe flares, and the patient developed recalcitrant renal hypertension. 2 g/day of mycophenolate mofetil (MMF) was added in an effort to reduce cyclosporine dosing, but without improvement in skin inflammation, leaving the patient at high risk for developing life-threatening complications.
Figure 1. Single cell RNA sequencing analysis reveals unique skin T cell transcriptome in DiHS/DRESS.
a, Clinical presentation and course. Prednisone (PSL); intravenous immunoglobulin (IVIG); cyclosporin (CsA); mycophenolate mofetil (MMF). b, Unsupervised t-distributed stochastic neighbor (tSNE) plot displaying 4,676 cells from DiHS/DRESS skin and 13,542 skin cells from 5 healthy volunteers (HVs), coloured by shared nearest neighbor (SNN) clusters. Keratinocyte (KC); smooth muscle (SM). c, Expression levels of (x axis) cluster-defining genes in each cluster. Violin plots show the distribution of the normalized expression levels of genes and are color-coded based on cluster, as in (b). d, Numbers of differentially expressed genes (nDEGs) between DiHS/DRESS- and HV cells within each cluster projected onto the tSNE map. DEG: |log fold change| > 0.5, adjusted p-value < 0.05, Wilcoxon rank sum test. e, Pathways upregulated in DiHS/DRESS lymphoid cells (cluster 4) with representative genes in each pathway. P-value, hypergeometric test. f, A volcano plot of DEGs that are upregulated (red) or downregulated (blue) in lymphoid cells; relevant DEGs identified in the pathways are labeled (Full list in Supplementary Table 1). DiHS/DRESS cells, n=589; HV cells, n=1,148. P-value, Wilcoxon rank sum test. g, Lymphocytes (cluster 4; n=1,737 cells) were aligned across datasets using canonical correlation analysis, and projected onto a tSNE plot, color-coded based on origin (left). Expression of selected genes are projected onto the tSNE plot (top) and shown as violin plots (bottom). Violin plots show the distribution of the normalized expression levels of selected genes and dots represent individual cells.
Targeting specific immunological pathways represents a promising approach to treat inflammatory diseases. Given the recent advances in scRNAseq, its application in human diseases may enable better understanding of pathological processes6. Given the failure of conventional therapies and the lack of alternative therapeutic options in this DiHS/DRESS case, we hypothesized that scRNAseq might provide a powerful personalized medicine approach to determine transcriptomic changes that not only deepen our insight into disease mechanisms, but also enable the identification of overexpressed genes or altered pathways that might be targeted via currently available monoclonal antibodies or small-molecule inhibitors. Thus, we dissociated a skin biopsy into a single cell suspension and performed scRNAseq analysis. Five healthy volunteer (HV)-skin biopsies served as controls. After performing unsupervised clustering and tSNE plot analyses (Fig. 1b, Extended Data Fig. 1a), cluster identities were determined by the expression of established markers (Fig. 1c, Extended Data Fig. 1b), which revealed successful capture of major skin cell subsets. In comparing patient- and HV-derived cells, we identified relative abundances of keratinocytes and immune cells in DiHS/DRESS skin (Extended Data Fig. 1c) that reflected an inflammatory state. Projection of DEG numbers (nDEG) revealed broad transcriptomic changes in immune and non-immune cell types, which was most prominent in the lymphocyte cluster (Fig. 1d). Because such differences could be influenced by the relative underrepresentation of homeostatic clusters in DiHS/DRESS samples, nDEG were projected on the tSNE plots based on major cell types (e.g. keratinocytes, lymphocytes). This further demonstrated that the lymphocyte cluster was the primary cell type that exhibited the highest transcriptomic differences (Extended Data Fig. 1d). Consistent with the dense infiltration of CD4+ and CD8+ T cells as determined by immunohistochemistry (Extended Data Fig. 1e), cells that expressed CD3E and CD4 or CD8A predominated within the lymphocyte cluster (Extended Data Fig. 1f,g).
To understand the biological significances of the transcriptional changes in the lymphocyte cluster, we performed pathway enrichment analysis with DEGs obtained via unsupervised clustering analysis. We found enrichment of pathways regarding lymphocyte activation and cytokine signaling, which were in part driven by the upregulation IL2RG, JAK3 and STAT1 (Fig. 1e,f; Supplementary Table 1). IL2RG encodes the common gamma chain of cytokine receptors that are crucial for lymphocyte homeostasis and function, the signaling of which are mediated by JAK-STAT molecules, where JAK3 directly interacts with the common gamma chain7–10. Also upregulated were genes involved in cell proliferation, such as MIK67, and migration, such as CCR10 (Fig. 1e,f), whereas transcripts for potentially targetable cytokines were undetected. Subclustering the lymphocytes segregated DiHS/DRESS and HV clusters, demonstrating distinct transcriptomic differences, and further validated that the expressions of the above genes were enriched in the DiHS/DRESS cluster (Fig. 1g, Extended Data Fig. 1h). Immunofluorescence microscopy in DiHS/DRESS confirmed skin-infiltration of CCR10+ CD3+ T cells and their expression of JAK3 (Extended Data Fig. 1i,j). Furthermore, immunohistochemical staining detected phosphorylated STAT1 in mononuclear cells (Extended Data Fig. 1k), indicating that the JAK-STAT signaling pathway was active in skin-infiltrating lymphocytes. None of the genes that were upregulated in non-lymphocytes, including parenchymal cells, were directly targetable (Source Data Fig. 1d).
Given the systemic nature of DiHS/DRESS, and to explore if similar transcriptomic signatures was reflected in the blood, we performed scRNAseq of patient peripheral blood mononuclear cells (PBMCs), compared with age- and sex-matched HV PBMCs (Fig. 2a, Extended Data Fig.2a,b). Projecting nDEGs onto the tSNE plot revealed CD4+ and CD8+ lymphocyte subclusters with prominent transcriptomic changes (Fig. 2b) that were characteristic to DiHS/DRESS with proliferative gene signature as observed in skin (Fig. 2c, Extended Data Fig.2d). Single-cell T cell receptor (TCR) sequencing, which enables profiling of paired TCR chain repertoires at single T cell level, revealed a strikingly diverse TCR usage in DiHS/DRESS T cells (Extended Data Fig.2e,f), a counterintuitive finding given that DiHS/DRESS was induced by a specific drug. Nevertheless, this finding demonstrated a negative finding on clonal TCR recombination enabling the exclusion of lymphoproliferative disorders such as T cell lymphomas11,12.
Figure 2. Circulating DiHS/DRESS T cells with distinct transcriptomic profiles and enrichment of human herpesvirus 6b DNA in CD4+ central memory T cells.
a, scRNAseq analysis on PBMCs from DiHS/DRESS (6,956 cells) and an age- and sex-matched HV (7,976 cells). Individual cells are color-coded based on cluster (n=19) in a tSNE plot. tSNE projections of well-known marker genes (bottom). b, Numbers of differentially expressed genes (nDEGs) between DiHS/DRESS and HV clusters. DEG: |log fold change| > 0.5, adjusted p-value < 0.05, Wilcoxon rank sum test. c, tSNE projections of selected genes, segregated by origin. Dotted region highlights clusters with high nDEGs (CD4(3), CD8(1), and mitotic cluster). d, e, Flow cytometry analysis of PBMCs for chemokine receptor expression and memory phenotype (a representative of two independent experiments). f, Violin plots show the distributions of JAK3, STAT1, and IL2RG expression levels in clusters with high transcriptomic changes (CD4(3), CD8(1), and mitotic cluster, DiHS/DRESS, n=925 cells; HV, n=2,960 cells). Numbers indicate percentages of cells that express each gene. g, Quantitative RT-PCR of human herpesviruses (HHV) in PBMC. h, Quantitative PCR for HHV6b DNA using sorted PBMC subsets. g,h, n=1. a representative of two independent sampling point.
Unsupervised analysis revealed PBMC T cell subclusters with high nDEGs, which were characterized by high expression of CCR4 and CCR10, which function as skin-homing chemokine receptors13,14, and low expression of CCR7, which enables migration of circulating T cells15,16 (Fig. 2c, Extended Data Fig. 2d). Flow cytometry validated the increase of CCR4+CCR10+ T cells in DiHS/DRESS blood (Fig. 2d). CCR4+CCR10+ T cells were predominantly CD45RO+ CCR7– and thus of central (TCM) and effector (TEM) memory phenotypes (Fig. 2e), corresponding with the CCR4high CCR10high CCR7low, high nDEG clusters (Fig. 2b,c). CCR4+CCR10– T cells were enriched with TCM and double-negative cells with naïve T cells (Fig. 2e). Monocle trajectory analysis17,18 revealed that CCR4/CCR10-expressing CD4+ T cell clusters distributed to a later pseudotime as compared to CCR7-expressing naïve CD4+ T cell cluster (Extended Data Fig. 2g,h).
Notably, in contrast to our observations in skin, JAK-STAT genes were not detected in an unsupervised manner when the whole PBMC lymphocyte cluster was compared between DiHS/DRESS and HVs. Further comparison of clusters with high transcriptomic changes (Fig. 2b) revealed upregulated pathways regarding cytokine-mediated signaling and T cell activation (Extended Data Fig. 2i) with increased frequency of T cells that expressed JAK3 and STAT1 (Fig. 2f). These findings demonstrated that while analysis of the primary site of inflammation – skin, for this patient, is optimal for detecting targetable pathways, PBMCs can also partially reflect disease pathology, with similar features detected by using a combination of unsupervised and supervised approaches.
Contribution of herpesviruses to DiHS/DRESS pathogenesis remains controversial. However, virus reactivation occurs without immunosuppressive therapies and the emergence of virus-specific CD8+ T cells suggests that herpesvirus reactivation is an integral component of disease process4,19,20. Among herpesviruses, HHV6b reactivation is reported to occur in the majority of DiHS/DRESS cases1,4,5. We hypothesized that the refractory inflammation might reflect persistent reactivation of herpesviruses21. Quantitative PCR using patient PBMCs detected HHV6b DNA (Fig. 2g). We sorted T cells based on memory phenotypes and found that HHV6b DNA was highly enriched in CD4+ TCM (Fig. 2h). Taken together, DiHS/DRESS T cells in both skin and blood exhibited increased proliferation, distinct chemokine receptor expression, upregulated genes involved in the JAK-STAT signaling pathway, and HHV6b was primarily enriched in circulating CD4+ T cells with TCM phenotype.
Our data pointed to several potential therapeutic targets: 1) cell proliferation pathways 2) chemokine receptors 3) HHV6b and 4) the JAK-STAT pathway. MMF, which inhibits lymphocyte proliferation, had already failed to resolve skin inflammation. The occurrence of Stevens-Johnson syndrome/TEN in patients treated with mogamulizumab22, anti-CCR4 monoclonal antibody23, rendered it a less viable option. While foscarnet, cidofovir and ganciclovir are utilized in HHV6b infection24,25, none of these selectively target HHV6b, and the two former were concerning due to renal toxicity, considering the underlying kidney dysfunction caused by cyclosporine. Thus, this left JAK inhibition as the sole feasible option. After obtaining informed consent, the patient was initiated on 5 mg/day of tofacitinib, a JAK1/3 inhibitor, and increased to 10mg/day after confirming the absence of toxicity (Fig. 3a). The patient also received valganciclovir, an oral prodrug of ganciclovir, which mainly served as prophylaxis against herpesvirus infections in general, that may occur during DiHS/DRESS21 or treatment with JAK inhibitors26. Flow cytometry analysis of PBMCs two weeks after initiation of treatment revealed striking reduction of CCR4+ CCR10+ CD4+ and CD8+ T cells (Fig. 3b, Extended Data Fig. 3). Consistently, comparison of pre- and post-treatment PBMCs by scRNAseq revealed reduction of CCR4- and CCR10-expressing T cells with proliferative gene signatures (Fig. 3c–e). Importantly, treatment extinguished residual skin inflammation and ultimately allowed the reduction of prednisone to replacement doses and discontinuation of MMF and cyclosporin (Fig. 3a). Notably, the tapering and discontinuation of cyclosporine led to improved kidney function and blood pressure control. Neutrophil counts initially decreased but recovered after the discontinuation of valganciclovir. While HHV6b DNA levels transiently increased (Fig. 3f), patient skin remained clear, indicating that disease control was due to tofacitinib. Decrease in monocytes at the time of scRNAseq analysis (Fig. 3d) was transient and was unlikely to be directly related to tofacitinib.
Figure 3. Clinical improvement with JAK inhibition with reversal of DiHS/DRESS-related transcriptome.
a, Clinical course after intervention with tofacitinib and valganciclovir (VGCV) (left). Prednisone (PSL); cyclosporin (CsA); mycophenolate mofetil (MMF). Clinical presentation at 4 months after starting tofacitinib (right). b, Flow cytometry analysis for chemokine receptors and memory phenotypes in blood CD4+ T cells before and 2 weeks after intervention (representatives of 2 technical replicates). c, scRNAseq analysis on DiHS/DRESS PBMCs that were sampled before and 2 weeks after intervention. Unsupervised tSNE plot generated from merged dataset of the two time-points (n=12,516 cells). Violin plots show the distribution of the normalized expression levels of selected DiHS/DRESS-associated genes in lymphocyte and mitotic cell clusters (bottom). d, tSNE plots segregated by cellular origin (top, pre-treatment, n=7,222 cells; post-treatment, n=5,294 cells). Dotted regions highlight T cell clusters diminished after intervention. Pie charts showing relative cluster abundances pre- and post-intervention (bottom). e, Volcano plot for differentially expressed genes that were up- (red) or down-regulated (blue) by tofacitinib treatment in lymphocytes (pre-treatment, n=4,967 cells; post-treatment, n=4,320 cells; p-value, Wilcoxon rank sum test). f, Long-term follow-up of HHV6b DNA copy numbers in blood pre- and post-tofacitinib.
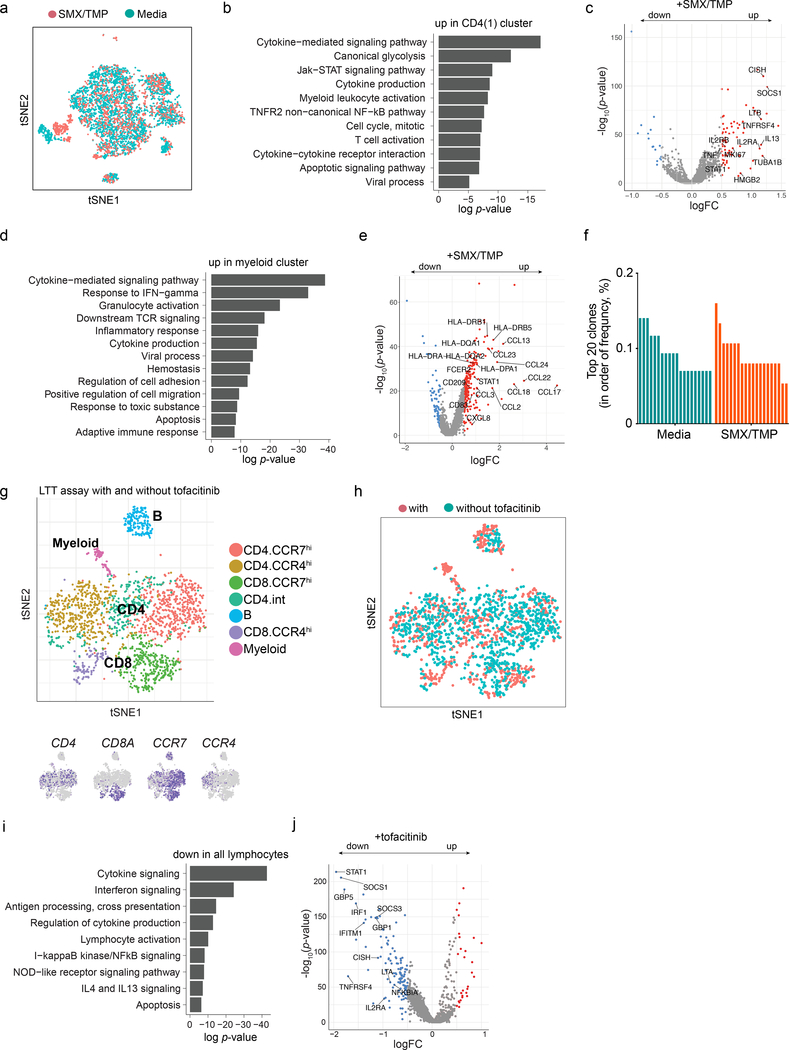
The chronic phase that we analyzed may not reflect earlier events in DiHS/DRESS. To model culprit drug-induced immune responses, we utilized the lymphocyte transformation test (LTT), which reliably measures drug-induced T cell proliferation against culprit drugs in DiHS/DRESS27,28. CFSE-labeled DiHS/DRESS CD4+ T cells that were cultured with SMX/TMP proliferated in a dose-dependent manner with formation of cell aggregates that were readily identifiable by microscopy (Fig. 4a). Comparison of cultures with and without SMX/TMP via scRNAseq analysis was performed on day 4 to capture transcriptomic changes prior to cell proliferation (Fig. 4b, Extended Data Fig. 4a), which revealed high nDEGs in the CCR7loCCR10hiCD4+ T cell cluster and monocytes (Fig. 4c). MKI67 expression identified CCR10hiCD4+ T cells as the major subset with a proliferative gene signature (Cluster 1, Fig. 4c,d). Furthermore, this CD4+ T cell cluster exhibited increased STAT1 and other genes downstream of the JAK-STAT signaling pathway including CISH and SOCS329, suggesting SMX/TMP-induced activation of the pathway in this cluster (Extended Data Fig. 4b,c). IL13, a potentially targetable cytokine30–32, was also upregulated (Extended Data Fig. 4c). SMX/TMP triggered the upregulation of CCL17 and CCL22, ligands for CCR4, as well as genes encoding MHC class II (MHC II) in monocytes, suggesting that T cell recruitment and subsequent antigen presentation is mediated by monocytes upon exposure to the culprit drug (Extended Data Fig. 4d,e).
Figure 4. SMX/TMP-induced in vitro T cell proliferation is suppressed by tofacitinib and anti-viral agents.
a, Detection of CD4+ T cell proliferation via lymphocyte transformation test. Carboxyfluorescein succinimidyl ester (CFSE)-labeled DiHS/DRESS PBMC were cultured with indicated concentrations of SMX/TMP for 6 days. Flow cytometry (left), quantification of CD4+ cells that have proliferated (center, the percentage of CFSE-diluted cells among Zombie-aqua– CD3+CD4+ cells, n=3, 3 independent experiments) and morphology of cell aggregates induced by SMX/TMP (right). b, scRNAseq tSNE analysis of PBMCs after a 4-day culture with (n=1,628 cells) or without (n=4,253 cells) SMX/TMP. c, Numbers of differentially expressed genes (nDEG) projected onto the tSNE plot (left, with SMX-TMP, n=1,628 cells; without SMX-TMP, n=4,253 cells). DEG: |log fold change| > 0.25, adjusted p-value < 0.05, Wilcoxon rank sum test. Violin plots show the distribution of the normalized expression levels of selected chemokine receptors in CD4 T cell clusters (right). d. Expression of the cell proliferation marker MIK67 across clusters. e,f, The effects of a blocking antibody against HLA-DR (e, n=5 per group) and tofacitinib and anti-viral agents (f, n=5 to 7 per group) in SMX/TMP-induced cell aggregate formation (left) and CD4+ T cell proliferation (right). Statistics are based on one-way ANOVA followed by Tukey’s (e) or Dunnett’s (f) multiple comparison test. F(4,20)=108. 4, p<0.0001 in (e); F(9,51)=37.69, p<0.0001) in (f). Centre value, mean; error bars, SD. ***p < 0.001; ****p < 0.0001; NS, not significant. All p values shown in (f) are compared to the SMX-TMP alone group (1μM ganciclovir vs. SMX-TMP, p=0.7067; 10μM ganciclovir vs. SMX-TMP, p=0.0002; 0.5μM artesunate vs. SMX-TMP, p=0.0605).
Consistent with our findings in PBMCs (Extended Data Fig. 2e,f), single-cell TCR sequencing after SMX/TMP stimulation did not reveal dominant clones (Extended Data Fig. 4f). Given this diverse TCR usage, we examined if SMX/TMP-induced CD4+ T cell proliferation required TCR-MHC II engagement. Addition of blocking antibodies against HLA-DR in LTT preparations completely inhibited CD4+ T cell proliferation (Fig. 4e). Thus, the diverse CD4+ T cell activation in response to SMX/TMP required TCR-MHC II engagement, suggesting an unconventional T cell activation process leading to activation of CD4+ T cells. This may be in line with previously suggested mechanism in drug hypersensitivities in which the culprit drug acts as a superantigen or modifies the MHC, resulting in its engagement with a wider range of TCRs20,33,34.
To determine if JAK inhibition was capable of suppressing SMX/TMP-induced T cell proliferation, LTT was performed with tofacitinib. Strikingly, all tested concentrations of tofacitinib completely suppressed CD4+ T cell proliferation (Fig. 4f) and genes associated with the JAK-STAT pathway (Extended Data Fig. 4g–j). To further test if targeting viruses could have an impact on T cell proliferation, ganciclovir, as well as artesunate, an anti-malarial with anti-HHV6 activity35, was tested in LTT. Notably, both ganciclovir and artesunate suppressed SMX/TMP-induced CD4+ T cell proliferation in dose-dependent manners (Fig. 4f). While ganciclovir could directly affect T cells by inhibiting DNA synthesis36, the similar effects we observed with artesunate supports the feasibility of targeting viruses as a strategy in the early phase of DiHS/DRESS.
A successful intervention with tofacitinib in a refractory case of DiHS/DRESS was guided by the use of scRNAseq, which demonstrated aberrant JAK-STAT activity. While early events in the disease and the contribution of viruses remain to be elucidated, LTT studies suggested that both JAK inhibition and anti-viral agents are promising targets in drug-induced phase of DiHS/DRESS. This study represents an in-depth study of a single case with findings which need to be extended to a larger cohort of patients. Nevertheless, the data suggest that single-cell omics-based approaches could be a powerful means for directing patient care in diseases with complex pathophysiology, including other inflammatory diseases and cancer.
Materials and Methods
Index patient
The index patient provided written consent to protocols that were approved by Institutional Review Board and Ethics Committee, that enabled de-identified research use of biospecimens, genetic testing results, clinical data, and standard of care (NIH protocols 07-I-0033, 93-I-0119, 15-AR-0144). Off-label use of tofacitinib to treat the index patient was submitted to, and was approved by, the Therapeutics Committee at Walter Reed National Military Medical Center on the basis that all conventional therapeutic agents had failed to control disease. Patient care and research was conducted in compliance with CARE guideline and the Declaration of Helsinki.
Sampling human materials
Sample collection and patient care was performed in accordance with protocols approved by the Institutional Review Board at National Institute of Arthritis and Musculoskeletal and Skin Diseases and National Institute of Allergy and Infectious Diseases. All individuals provided written informed consent for sample acquisition and all subsequent analyses including gene expression studies. Lesional skin biopsy was obtained from the patient and normal skin biopsy from five healthy volunteers by two overlapping 4mm punch biopsies from the flank after local anesthesia with lidocaine. Peripheral blood mononuclear cells (PBMCs) were obtained from fresh blood samples from the patient and an age- and sex-matched healthy volunteer (Supplementary Table 3). The post-treatment PBMCs were collected two weeks after initiation of the intervention.
Cell isolation and processing
PBMCs were freshly isolated using density gradient centrifugation from heparinized peripheral blood. PBMCs were either processed immediately for scRNAseq and LTT or stored at −80°C in freezing medium for later use.
All biopsied skin samples were immediately processed for scRNAseq. Samples were gently washed in PBS after removing adipose tissue with scissors and minced with scissors and disposable scalpel in a sterile tissue culture dish. Enzymatic digestion was performed in gentleMACS C tubes (Miltenyi Biotec, Auburn, CA, USA) containing 500μL of enzyme mix (Whole skin dissociation kit, Miltenyi Biotec), which were incubated in a water bath at 37 °C for 3 h under manual agitations every 15 min. Enzymes were inactivated with complete RPMI (Gibco Laboratories, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum (FBS, Gemini Biological Products, Calabasas, CA, USA) at the end of incubation. Samples were then mechanically dissociated with gentleMACS Dissociator (Miltenyi Biotec) for 1 min and spun down to collect pellets, which were suspended in 10 mL of washing buffer (PBS with 5% FBS) and gently dissociated using a 10 mL syringe. Sample was filtered through 40 μm cell strainers (BD Bioscience, San Jose, CA, USA) and spun down. Samples then underwent treatment in ACK lysis buffer (Quality Biological, Gaithersburg, MD, USA) for 1 min and neutralized with washing buffer. Samples were further filtered through 40 μm cell strainers and washed with 10 ml of washing buffer. The final samples were resuspended in 500 μL of RPMI-1640 with 20% heat-inactivated FBS before sorting.
Live cell enrichment
Live cells were enriched by fluorescence-activated cell sorting (FACS). For FACS, cells were stained with SYTOX-Green (Molecular probes, Eugene, OR, USA) and were sorted on a BD FACSAria Fusion or a BD FACSAria IIIu cell sorter (Becton, Dickinson and Company, BD Biosciences, San Jose, CA, USA). After doublet exclusion using conventional FSC-SSC gating strategy, SYTOX-Green-negative live cells were collected in a new tube with 1ml of RPMI-1640 media with 20% FBS.
Capturing, library preparation and sequencing
After centrifugation of sorted cells, they were washed and resuspended in PBS containing 0.04% bovine serum albumin (Miltenyi Biotec). Cell numbers for each sample were counted, using an automated cell counter (Cellometer Vision CBA, Nexcelom Bioscience LLC., Lawrence, MA, USA). The single-cell capturing and downstream library constructions were performed using Chromium Single Cell 5’ library or 3’ v2 library preparation kit according to the manufacturer’s protocol (10X Genomics, Pleasanton, CA, USA). Briefly, cellular suspensions were co-partitioned with barcoded gel beads to generate single-cell Gel Bead-in-Emulsion (GEMs), and poly-adenylated transcripts were reverse-transcribed. Full-length cDNA along with cell barcode identifiers were PCR-amplified, and then sequencing libraries were prepared and normalized to 3nM for loading on an Illumina HiSeq 3000 (Illumina, San Diego, CA, USA). Detailed information on sequencing results including total read count and sequencing saturation is shown in the Supplementary Table 4.
scRNAseq data analysis
scRNAseq reads were demultiplexed and aligned to the ENSEMBL GRCh38 human transcriptome to generate gene expression matrices using CellRanger (10X Genomics; sample statistics in Supplementary Table 4). We further analyzed these matrices using Seurat (v2.3.4) with default parameters, unless otherwise indicated37. We first filtered the matrices to exclude cells using a standard panel of three quality criteria: number of detected transcripts (number of Unique Molecular Identifiers; UMI), detected genes, and percent of reads mapping to mitochondrial genes (thresholds in Supplementary Table 5 chosen using analysis exemplified in Supplementary Figures 1, 2). Skin datasets from HVs were merged, log-normalized, scaled, and the variation associated with the three quality criteria regressed away. We used canonical correlation analysis (CCA) to better compare transcriptomes across different datasets37. By default, we first identified the 2,000 most variable genes within each dataset, and then generated a combined gene set after removal of unique genes to each dataset (Supplementary Table 6). The robustness of clustering was also validated by using 1000 most variable genes (Supplementary Table 7). We chose the optimal number of correlation components (CCs) for alignment using biweight midcorrelation analysis, which were verified by visualizing the distribution of cells in reduced CCA space. CCA subspaces were then reduced using the chosen number of CCs (Supplementary Table 6). We then performed tSNE analysis and graph-based clustering, using a shared nearest neighbor parameter optimized for each combined dataset. After clustering, we identified genes that marked each cluster and differentially expressed genes (DEGs) between subpopulations using the non-parametric Wilcoxon rank sum test. DiHS/DRESS, HV4 and HV5 skin samples were prepared using Chromium Single Cell 5’ library preparation kit and HV1–3 samples were processed with the 3’ library preparation kit. Independent analyses of DiHS/DRESS vs HV1–3 or HV4–5 did not result in notable differences when DEGs were generated. We visualized log-normalized expression levels in tSNE plot projections and violin plots. We used scaled log-normalized expression levels to create heatmaps. Gene ontology and gene set enrichment analysis from DEGs were performed using Metascape webtool (www.metascape.org)38.
We performed trajectory analysis on the CD4 lymphocyte subset of the merged PBMC dataset using Monocle version 2.10.117,18. We identified highly variable genes (n= 1,287) and reduced dimensionality using the DDRTree algoithm17, estimating trajectories that we visualized using pseudotime plots.
TCR V(D)J sequencing and analysis
Full length TCR V(D)J segments were enriched from amplified cDNA from 5’ libraries via PCR amplification using Chromium Single Cell V(D)J Enrichment Kit according to the manufacturer’s protocol (10x Genomics). Sequencing libraries were prepared in 3nM concentration and then sequenced on a lllumina HiSeq 3000 with 150bp paired-end reads. Demultiplexing, gene quantification, and TCR clonotype assignment were performed using CellRanger v2.1.1 (10x Genomics). Analysis was performed using Loupe V(D)J Browser v2.0.1 (10x Genomics). In brief, TCR diversity metric containing clonotype frequency and barcode information was obtained. Using barcode information, T cells with prevalent TCR clonotype were projected on tSNE plot.
Flow cytometry analysis
Freshly isolated or cultured PBMCs were stained with Zombie Aqua™ Fixable Viability Kit (BioLegend) for 20 min at room temperature and were incubated with anti-human FcR Blocking Reagent (Miltenyi Biotec, #130–059-901) and were then incubated with cell surface antibodies for 30 min on ice. The following monoclonal antibodies were used for phenotyping of freshly isolated PBMC: anti-CD3-APC-Cy7 (clone HIT3a, Biolegend, #300317), anti-CD4-APC (clone RPA-TA, Biolegend, #300514), anti-CD8- PerCP-Cy5.5 (clone SK1, Biolegend, #344709), anti-CCR4-PE-Cy7 (clone L291H4, Biolegend, #359409), anti-CCR7-FITC (clone G043H7, Biolegend, #353215), anti-CCR10-PE (clone 314305, R&D #FAB3478P-025), anti-CD45RO-BUV395 (clone UCHL1, BD Biosciences, #564292). And the following monoclonal antibodies were used for analyzing of proliferating lymphocytes within cultured PBMC: anti-CD3-APC-Cy7, anti-CD4-PE-Cy7 (RPA-TA, Biolegend, #300512), anti-CD8-BV421 (clone RPA-T8, Biolegend, #301036). Cells were acquired with an LSR ll (BD Biosciences) and data were analyzed by using FlowJo software (FlowJo, LLC, Ashland, OR, USA).
Immunohistochemistry and Immunofluorescence staining
Paraffin embedded, 5-μm formalin fixed tissue sections were dewaxed in xylene and rehydrated with distilled water. The sections were treated with heat-induced epitope retrieval technique using Cell Conditioning (CC1) buffer at pH 8.5 (Ventana Medical Systems, Tucson, AZ, USA) or Dako Target Retrieval Solution, Citrate pH 6.1 (Agilent, Santa Clara, CA, USA). Immunostaining was performed on Ventana BenchMark XT automated staining system (Ventana Medical Systems) except for phospho-STAT1, which was performed on manually. Incubation with the following primary antibodies was performed: rabbit anti-human CD3 (clone 2GV6, Ventana), rabbit anti-human CD4 (clone SP25, Ventana), rabbit anti-human CD8 (clone SP57, Ventana), and rabbit anti-human Phosphorylated-STAT1 (Ty701) (clone 58D6, Cell Signaling) monoclonal antibodies, all at 1:200 dilutions. Signal was detected using ultraView Universal DAB Detection Kit (Ventana) or SignalStain Boost IHC Detection Reagent (HRP, Rabbit) (Cell Signaling) and Dako Liquid DAB+ Substrate Chromogen System (Code K3468) (Agilent).
For immunofluorescence microscopy, 7-μm cryosections were fixed in prechilled acetone at –20 °C for 5 minutes and washed in phosphate buffered saline (PBS). After blocking for 45 minutes in PBS with 3% skim milk and 5% normal goat or donkey serum (Jackson ImmunoResearch, West Grove, PA, USA), sections were incubated overnight at 4 °C with primary antibodies: mouse anti-human CD3-Alexa Fluro488 (clone UCHT1, Biolegend, #300454, 1:100), rabbit anti-human CCR10 (polyclonal, Thermo Fisher Scientific, #PA1–21617, 1:25), rabbit IgG isotype (Thermo Fisher Scientific, #31235, 1:250), and rabbit anti-human JAK3 (clone 1A1, Bioss Inc, # bsm-54067R, 1:100). The sections were washed three times with PBS, 5 min each, then incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:400), Alexa Fluor 568-conjugated goat anti-rabbit IgG (1:400), or Alexa Fluor 568-conjugated donkey anti-rabbit IgG (1:400) antibodies (Thermo Fisher Scientific) for 45 min at room temperature. The slides were washed three times in PBS, 5 min each, and then mounted with ProLong Gold (Molecular Probes) anti-fade reagent. Fluorescence images were captured using a Zeiss Axio Image A1 fluorescent microscope with Zeiss AxioVision 4.8 software (Carl Zeiss, Minneapolis, MN, USA) and then processed with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Human herpesvirus detection
Total nucleic acid was extracted from the whole blood using the QIAsymphony SP platform using the Virus Blood 200 protocol (Qiagen, Hilden, Germany) and cell preparations were extracted using the NucliSENS easyMAG platform (bioMerieux, Boxtel, the Netherlands) with final elution volume of 50 μL. For HHV6b detection, primers for HHV6 UL38 gene (forward [5’-GGA GTG CCT GTG GGT ATT C-3’], reverse [5’-CTA AGG TGA GCC AGA TTC G-3’]) and FAM/ZEN/Iowa Black quencher-labeled TaqMan probe were used (Applied Biosystems, Foster City, CA, USA). Quantitative PCR was performed using an ABI 7500 Fast real-time PCR machine with standard cycling protocol (Applied Biosystems).
Lymphocyte transformation test (LTT)
LTT was performed using carboxyfluorescein succinimidyl ester (CFSE) dilution assay with modifications from a previously described method27,28. Isolated PBMCs were incubated with CFSE (CellTrace CFSE Cell Proliferation Kit, Invitrogen, Molecular Probes, Eugene, OR, USA) at 2μM concentration for 8 min at room temperature and the reaction was neutralized with RPMI-1640 (Gibco) containing 10% FBS. 2 × 105 PBMCs were cultivated in 96-well round bottom plate in 200ul of RPMI-1640 supplemented with 10% human AB serum (Sigma-Aldrich, St. Louis, MO, USA), 100 U/ml penicillin, 100 μg/ml streptomycin (Pen Strep, ThermoFisher Scientific, Waltham MA, USA), and 1mM nonessential amino acids in combination with or without different concentrations of utilized drugs. SMX (#S7507), TMP (#92131), artesunate (#A3731), and ganciclovir (#G2536) were all obtained from the Sigma-Aldrich and tofacitinib (CP 690550 citrate, #4556) were purchased from Tocris Bioscience (Minneapolis, MN, USA). All drugs were initially reconstituted into stock solutions (SMX, 200mg/ml; TMP 100mg/ml; artesunate, 100mM; ganciclovir, 25mM; tofacitinib, 25mg/ml) using pure dimethyl sulfoxide (Sigma-Aldrich), then diluted into target concentration in media. The concentration of SMX-TMP was 48 μg/ml unless otherwise indicated. Cultured PBMCs were harvested after incubation at 37°C for 4~6 days without changing media.
Please refer to the Life Sciences Reporting Summary for additional details.
Extended Data
Extended Data Fig. 1. Infiltration of T cells in DiHS/DRESS skin.
a, scRNAseq tSNE analysis of DiHS/DRESS and HV skin cells, colour-coded based on origin (n=18,218 cells). b, Heatmap of the top-five genes marking clusters of skin cells (x-axis; from Figure 1b). Keratinocyte (KC); smooth muscle (SM). c, Frequencies of cells in each cluster, color-coded based on origin. d, Nineteen clusters in Fig. 1b were grouped into 7 major subsets based on cell types (DiHS/DRESS cells, n=4,676; HVs cells, n=13,542). Numbers of differentially expressed genes (nDEGs) between DiHS/DRESS- and HV cells within each cell type projected onto a tSNE map. DEG: |log fold change| > 0.5, adjusted p-value < 0.05, Wilcoxon rank sum test. e, Immunohistochemical staining for CD3, CD4, and CD8 in DiHS/DRESS lesional skin. Scale bars, 100 μm. Representatives of six serial sections from one sample. f, tSNE plot for the lymphocyte subcluster, colour-coded based on origin (DiHS/DRESS cells, n=589; HV cells, n=1,148). g, tSNE projections of selected genes (n=1,737 cells). h, Frequency of cells from (f) expressing the displayed genes. i, Immunofluorescence staining with anti-CD3 (green) and anti-CCR10 (red) antibodies or rabbit IgG isotype in DiHS/DRESS lesional skin. Dotted lines denote the boundary between the epidermis (Ep) and dermis. Scale bar, 50 μm. j, Immunofluorescence staining with anti-CD3 (green) and anti-JAK3 (red) in DiHS/DRESS lesional skin. Staining in atopic dermatitis and HV skin sections are shown as comparison. Nuclear labeling with DAPI (blue). Scale bar, 50 μm. k, Hematoxylin and eosin staining (top; H&E) and immunohistochemical staining for phosphorylated STAT1 (pSTAT1) in DiHS/DRESS and HV skin. Scale bar, 20 μm. i-k, Representative of 3 independent experiments.
Extended Data Fig. 2. Characterization of DiHS/DRESS T cells in blood.
a, scRNAseq tSNE analysis of DiHS/DRESS and HV PBMCs, colour-coded based on origin (n=14,932 cells). b, Frequencies of cells in each cluster, colour-coded based on origin. c, Heatmap of the top-five genes marking PBMC clusters (from Figure 2a). d, Violin plots show the distribution of the normalized expression levels of selected genes in each lymphocyte cluster (CD4(1), n=2,322 cells; CD4(2), n=1,649 cells; CD4(3), n=1,318 cells; CD8(1), n=2,350 cells; CD8(2), n=850 cells; CD8(3), n=335 cells; mitotic, n=525 cells; Treg, n=245 cells). e, Frequencies of top-20 common T cell receptor (TCR) clonotypes in PBMCs from HV and in PBMCs and skin from DiHS/DRESS patient as determined by single-cell TCR V(D)J gene sequencing. f, Frequent clonotypes, defined as cells expressing common TCR combinations that were shared by more than 10 cells (DiHS/DRESS, n=2 clonotypes in 22 cells; HV, n=21 clonotypes in 1,466 cells), were mapped onto the tSNE plot. g. Trajectory analysis of cells in the CD4 T cell clusters (n=6,059 cells), coloured by pseudotime (top) and clusters (bottom). h, Pseudo-temporal single cell expression of indicated genes, colored by cluster. i, Pathways upregulated in DiHS/DRESS lymphoid clusters with high transcriptomic changes (CD4(3), CD8(1), and mitotic clusters, n=4,193 cells). P-value, hypergeometric test.
Extended Data Fig. 3. Reduction of circulating CD8+ effector memory T cells after tofacitinib.
Flow cytometry analysis for chemokine receptor expression and memory phenotype in DiHS/DRESS peripheral blood CD3+ CD8+ T cells pre- and 2 weeks post-treatment with tofacitinib. Right panels: Naïve, central memory (CM), and effector memory (EM) CD8+ T cells. Representatives of 2 technical replicates.
Extended Data Fig. 4. Pathway analysis of transcriptomic changes induced in CD4+ T cells and myeloid cells during LTT.
a, scRNAseq tSNE analysis of PBMCs after a 4-day culture with or without SMX/TMP, colour-coded based on origin (n=5,881 cells). b, Pathways upregulated in CD4(1) cluster (from Fig. 4b–d, n= 1,486) after SMX/TMP treatment. c, Volcano plot of up- (red) and down-regulated (blue) genes differentially expressed in SMX/TMP treated CD4(1) cells (|log fold change| > 0.5), highlighting relevant genes associated with pathways from (b). With SMX-TMP, n=418 CD4(1) cells; without SMX-TMP, n=1,068 CD4(1) cells. Full list of differentially expressed genes from the three CD4 T cell clusters (n=3,869 cells) observed in LTT (see Fig. 4b–c) is provided in Supplementary Table 2. d, Pathways upregulated in myeloid cells after SMX/TMP treatment. e. Volcano plot of differentially expressed genes in myeloid cells (n=513) by SMX/TMP treatment, labeling relevant genes in pathways from (d). f, Frequencies of top-20 common T cell receptor (TCR) clonotypes in PBMCs after a 4-day culture with or without SMX/TMP as determined by single-cell TCR V(D)J gene sequencing. g,h, scRNAseq tSNE analysis on SMX/TMP-treated PBMCs with or without tofacitinib (n=2,068 cells) colour-coded based on cluster (g, top) and origin (h). tSNE projections of selected marker genes (g, bottom). i, Pathways downregulated in CD4 lymphocytes of tofacitinib-treated PBMC, and j, Volcano plot with labeling of relevant downregulated genes. Tofacitinib-treated CD4 cells, n=825; untreated CD4 cells, n=508. P-values in b,d,I, hypergeometric test; p-values in c,e,j, Wilcoxon rank sum test.
Supplementary Material
Acknowledgements
We thank the NIAMS flow cytometry and sequencing cores for technical support, the patient and volunteers; V. Pillai, M. Taylor, and B. Higgins for their contributions. This research was supported by the Intramural Research Program of NIAMS, NCI (contract # HHSN261200800001E) and NIAID, National Institutes of Health.
Footnotes
Competing interests
All authors declare no competing interests.
Data availability
Source data for Figures 1–4 and Extended Data Figures 1, 2, and 4 are provided with the paper. The matrix and raw data for scRNAseq reported in this paper have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE132802 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132802). All other data are available from the corresponding author upon reasonable request.
References
- 1.Duong TA, Valeyrie-Allanore L, Wolkenstein P & Chosidow O Severe cutaneous adverse reactions to drugs. Lancet 390, 1996–2011 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Kano Y, et al. Sequelae in 145 patients with drug-induced hypersensitivity syndrome/drug reaction with eosinophilia and systemic symptoms: survey conducted by the Asian Research Committee on Severe Cutaneous Adverse Reactions (ASCAR). J Dermatol 42, 276–282 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Husain Z, Reddy BY & Schwartz RA DRESS syndrome: Part I. Clinical perspectives. J Am Acad Dermatol 68, 693.e691–614; quiz 706–698 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Ushigome Y, Kano Y, Ishida T, Hirahara K & Shiohara T Short- and long-term outcomes of 34 patients with drug-induced hypersensitivity syndrome in a single institution. J Am Acad Dermatol 68, 721–728 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Husain Z, Reddy BY & Schwartz RA DRESS syndrome: Part II. Management and therapeutics. J Am Acad Dermatol 68, 709.e701–709; quiz 718–720 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Shalek AK & Benson M Single-cell analyses to tailor treatments. Sci Transl Med 9(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard WJ, Lin JX & O’Shea JJ The gammac Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity 50, 832–850 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Miyazaki T, et al. Functional activation of Jak1 and Jak3 by selective association with IL-2 receptor subunits. Science 266, 1045–1047 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Russell SM, et al. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science 266, 1042–1045 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Witthuhn BA, et al. Involvement of the Jak-3 Janus kinase in signalling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature 370, 153–157 (1994). [DOI] [PubMed] [Google Scholar]
- 11.de Masson A, et al. High-throughput sequencing of the T cell receptor beta gene identifies aggressive early-stage mycosis fungoides. Science translational medicine 10(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng WK, et al. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in cutaneous T cell lymphoma. Science translational medicine 5, 214ra171 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Homey B, et al. CCL27-CCR10 interactions regulate T cell-mediated skin inflammation. Nat Med 8, 157–165 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Reiss Y, Proudfoot AE, Power CA, Campbell JJ & Butcher EC CC chemokine receptor (CCR)4 and the CCR10 ligand cutaneous T cell-attracting chemokine (CTACK) in lymphocyte trafficking to inflamed skin. The Journal of experimental medicine 194, 1541–1547 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell JJ, et al. CCR7 expression and memory T cell diversity in humans. Journal of immunology (Baltimore, Md. : 1950) 166, 877–884 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Sallusto F, Lenig D, Forster R, Lipp M & Lanzavecchia A Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nature methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picard D, et al. Drug reaction with eosinophilia and systemic symptoms (DRESS): a multiorgan antiviral T cell response. Sci Transl Med 2, 46ra62 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Musette P & Janela B New Insights into Drug Reaction with Eosinophilia and Systemic Symptoms Pathophysiology. Frontiers in medicine 4, 179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishida T, Kano Y, Mizukawa Y & Shiohara T The dynamics of herpesvirus reactivations during and after severe drug eruptions: their relation to the clinical phenotype and therapeutic outcome. Allergy 69, 798–805 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honda T, et al. Stevens-Johnson Syndrome Associated with Mogamulizumab-induced Deficiency of Regulatory T cells in an Adult T-cell Leukaemia Patient. Acta Derm Venereol 95, 606–607 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Vela M, Aris M, Llorente M, Garcia-Sanz JA & Kremer L Chemokine receptor-specific antibodies in cancer immunotherapy: achievements and challenges. Frontiers in immunology 6, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ljungman P & Singh N Human herpesvirus-6 infection in solid organ and stem cell transplant recipients. J Clin Virol 37 Suppl 1, S87–91 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Pohlmann C, et al. Cidofovir and foscarnet for treatment of human herpesvirus 6 encephalitis in a neutropenic stem cell transplant recipient. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 44, e118–120 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Curtis JR, Xie F, Yun H, Bernatsky S & Winthrop KL Real-world comparative risks of herpes virus infections in tofacitinib and biologic-treated patients with rheumatoid arthritis. Annals of the rheumatic diseases 75, 1843–1847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyfeler B & Pichler WJ The lymphocyte transformation test for the diagnosis of drug allergy: sensitivity and specificity. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology 27, 175–181 (1997). [PubMed] [Google Scholar]
- 28.Pichler WJ & Tilch J The lymphocyte transformation test in the diagnosis of drug hypersensitivity. Allergy 59, 809–820 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Villarino AV, Kanno Y & O’Shea JJ Mechanisms and consequences of Jak-STAT signaling in the immune system. Nature immunology 18, 374–384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck LA, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. The New England journal of medicine 371, 130–139 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Corren J, et al. Lebrikizumab treatment in adults with asthma. The New England journal of medicine 365, 1088–1098 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Teraki Y & Fukuda T Skin-Homing IL-13-Producing T Cells Expand in the Circulation of Patients with Drug Rash with Eosinophilia and Systemic Symptoms. Dermatology 233, 242–249 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Illing PT, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486, 554–558 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Pichler WJ, et al. Drug Hypersensitivity: How Drugs Stimulate T Cells via Pharmacological Interaction with Immune Receptors. Int Arch Allergy Immunol 168, 13–24 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Milbradt J, Auerochs S, Korn K & Marschall M Sensitivity of human herpesvirus 6 and other human herpesviruses to the broad-spectrum antiinfective drug artesunate. J Clin Virol 46, 24–28 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Battiwalla M, et al. Ganciclovir inhibits lymphocyte proliferation by impairing DNA synthesis. Biol Blood Marrow Transplant 13, 765–770 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature communications 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.