Abstract
In vitro differentiation of human embryonic stem cells (hESCs) has transformed the ability to study human development on both biological and molecular levels and provided cells for use in regenerative applications. Standard approaches for hESC culture using colony type culture to maintain undifferentiated hESCs and embryoid body (EB) and rosette formation for differentiation into different germ layers are inefficient and time-consuming. Presented here is a single-cell culture method using hESCs instead of a colony-type culture. This method allows maintenance of the characteristic features of undifferentiated hESCs, including expression of hESC markers at levels comparable to colony type hESCs. In addition, the protocol presents an efficient method for neural progenitor cell (NPC) generation from single-cell type hESCs that produces NPCs within 1 week. These cells highly express several NPC marker genes and can differentiate into various neural cell types, including dopaminergic neurons and astrocytes. This single-cell culture system for hESCs will be useful in investigating the molecular mechanisms of these processes, studies of certain diseases, and drug discovery screens.
Keywords: Developmental Biology, Issue 155, human embryonic stem cells, growth and maintenance, single cell culture, differentiation, neural progenitor cells
Introduction
Human embryonic stem cells (hESCs) have the potential to differentiate into the three primary germ layers, which then differentiate into various multipotent progenitor cell lineages. These lineages subsequently give rise to all cell types in the human body. In vitro hESC culture systems have transformed the ability to study human embryonic development and have served as a valuable tool for obtaining new insights into how these processes are regulated at the biological and molecular levels. Similarly, studies of induced pluripotent stem cells (iPSCs) generated from reprogramming somatic cells isolated from human patients provide novel insights into various diseases. In addition, progenitor and differentiated cells derived from hESCs can be useful for research involving stem cell therapy and drug screening1,2,3,4.
hESCs can be induced to differentiate into neural progenitor cells (NPCs), which are multipotential cells with an extensive self-renewal capacity. Subsequently, these cells can be differentiated into neurons, astrocytes, and oligodendrocytes5,6. NPCs also offer a cellular system for in vitro studies of neurodevelopmental biology and various neurological diseases. However, current colony type culture methods involving hESCs and their differentiation into NPCs are inefficient and often involve coculture as well as embryoid body (EB) and rosette formation5,7,8,9. These protocols exhibit lower survival rates and spontaneous differentiation and are more time-consuming.
Presented here is an improved and robust culture system that is easily scalable and uses high density single-cell type culture of hESCs10. The inclusion of Roh-kinase (ROCK) inhibitor contributed to significantly enhanced survival efficiency during single cell type culture of hESC10,11,12,13,14. In this culture system, hESCs can be easily maintained and expanded. In addition, the protocol presents an efficient method to generate NPCs from single-cell type culture of hESCs, which allows the production of highly pure NPCs. Inhibition of BMP/TGFβ/activin signaling pathways with ALK inhibitors efficiently induce differentiation of single-cell type hESCs into NPCs15,16, which then can be induced to differentiate into functional neural lineages, such as dopaminergic neurons and astrocytes.
In summary, the single-cell type culture protocol using hESCs offers an attractive model to study the differentiation of these cells into various lineages, including NPCs. This protocol is easily scalable and therefore suitable for generating cells for research involving regenerative therapy and drug screening.
Protocol
1. Preparation of hESC-qualified Basement Membrane Matrix-coated Plates
Slowly thaw the hESC-qualified basement membrane matrix (see Table of Materials) solution at 4 °C for at least 2–3 h or overnight to avoid formation of a gel.
To prepare basement membrane matrix-coated plates, dilute matrix in cold DMEM/F12 to a 2% final concentration. Mix well and coat each well of a 6 well plate with 1 mL of the diluted matrix solution.
-
Incubate the basement membrane matrix-coated plates at room temperature (RT) for at least 3 h or at 4 °C overnight.
NOTE: Plates with basement membrane matrix can be stored at 4 °C for 1 week before the matrix solution is removed and plates are used.
Materials
| Name of Material/Equipment | Company | Catalog Number | Comments/Description |
|---|---|---|---|
| CO2 incubator | Thermo Fisher | 4110 | |
| Centrifuge | DAMON/ICE | 428–6759 | |
| H9 (WA09) human embryonic stem cell line | WiCell | WA09 | |
| 6-well plates | Corning | 3516 | |
| 35mm μ-dishes | ibidi | 81156 | Cell culture dish |
| mTeSR1 | Stemcell Technologies | 85850 | hESC cultue medium |
| Stemdiff Neural Progenitor medium | Stemcell Technologies | 5833 | NPC culture medium |
| Dispase | Stemcell Technologies | 7923 | Cell detachement solution |
| Cryostor CS 10 | Stemcell Technologies | 7930 | Cell freezing solution |
| DMEM/F12 | Thermo Fisher | 10565–018 | |
| DMEM | Thermo Fisher | 10569–010 | |
| Knockout DMEM | Thermo Fisher | 10829018 | |
| Knockout Serum Replacement | Thermo Fisher | 10828028 | |
| N2 supplement | Thermo Fisher | 17502001 | |
| B27 supplement | Thermo Fisher | 17504044 | |
| B27 supplement (-Vit A) | Thermo Fisher | 12587010 | |
| Accutase | Innovative Cell Technologies | AT104–500 | Cell detachement solution |
| Fetal Bovine Serum | Fisher Scientific | SH3007003HI | |
| NEAA | Thermo Fisher | 11140050 | |
| ROCK inhibitor | Tocris | 1254 | |
| Dorsomorphin | Tocris | 3093 | |
| SB431542 | Tocris | 1614 | |
| EGF | Peprotech | AF-100–16A | |
| bFGF | Peprotech | 100–18C | |
| Poly-L-ornithine | Sigma Aldrich | P3655 | |
| Laminin | Sigma Aldrich | L2020 | |
| Neurobasal | Thermo Fisher | 21103049 | |
| GlutaMax | Thermo Fisher | 35050061 | Glutamine 100X |
| SHH | Applied Biological Materials | Z200617 | |
| FGF8 | Applied Biological Materials | Z101705 | |
| Ascorbic Acid | Sigma Aldrich | A4403 | |
| BDNF | Applied Biological Materials | Z100065 | |
| GDNF | Applied Biological Materials | Z101057 | |
| IGF | Peprotech | 100–11 | |
| Activin A | R&D system | 338-AC-050 | |
| Heregulin β-1 | Peprotech | 100–3 | |
| Corning hESC-qulified Matrix (Magrigel) | Corning | 354277 | Basement membrane matrix |
| Geltrex matrix | Thermo Fisher | A1569601 | Basement membrane matrix |
2. Adaptation of Colony type hESCs to Single-cell hESC Culture
To passage the feeder-free cultures of colony type H9 (WA09) hESCs grown on basement membrane matrix, aspirate the medium from the wells (Figure 1A)9.
Wash 1x with 1 mL of DPBS. Add 1 mL of dispase solution (1 U/mL) per well and incubate at 37 °C for 20 min.
Remove the dispase, wash the cells 1x gently with 2 mL of DMEM/F12, remove the medium, and add 2 mL of DMEM/F12 to each plate.
Gently detach colonies by gently pipetting up and down and transfer to a 15 mL tube.
Centrifuge the pellets for 2 min at 370 × g and aspirate the medium.
To dissociate the cell pellets into single cells, add 2 mL of cell detachment solution (1x concentration, see Table of Materials) and incubate at 37 °C for 10 min.
Centrifuge cells for 2 min at 370 × g and remove the detachment solution.
Add fresh mTeSR1 human ESC medium and dissociate the cells into single cells by gentle pipetting up and down.
To adapt colony type hESCs to a single-cell type culture, plate approximately 1.5–2.0 × 106 hESCs into each well of the basement membrane matrix-coated 6 well plate in 2 mL of mTeSR1 containing 10 μM ROCK inhibitor for 24 h (Figure 1A).
After 24 h, replace the hESC medium with fresh mTeSR1 without ROCK inhibitor and allow the hESCs to grow as a single-cell type for 3 days. Change the medium daily.
-
On day 4, when cultures reach nearly 100% confluency, dissociate cells in detachment solution, then replate as described in step 2.6.
NOTE: The ROCK inhibitor improves cell survival during the initial 24 h of single-cell type hESC culture.
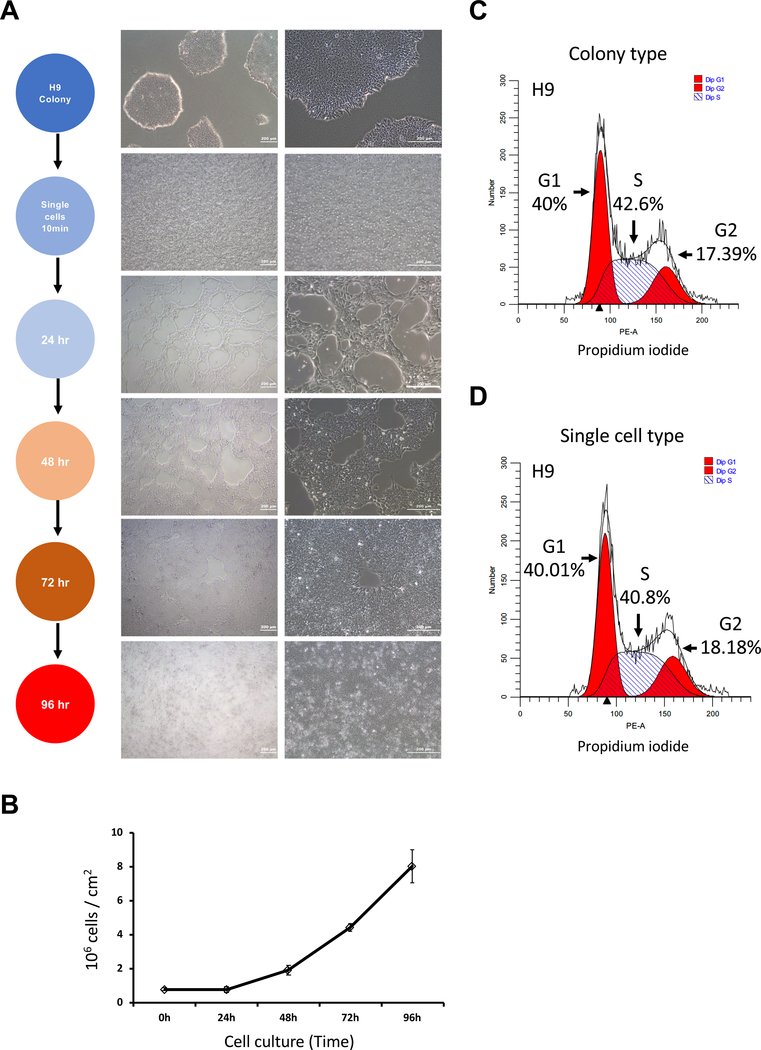
Figure 1: Adaptation of colony type hESCs to single-cell type culture.
(A) Representative phase contrast images of single cell cultures of H9 hESCs at different times after plating on 2% basement membrane matrix-coated dishes. Low (left) and high (right) magnification. Top panel: representative image of colony type hESCs. Other panels show representative images of cultures at different times during the adaptation to single-cell type hESCs. Scale bar = 200 μm. (B) Growth curves of H9 hESCs were monitored in 2% basement membrane matrix-coated plates with 10 μM ROCK inhibitor for the first 24 h during the single-cell culture condition. (C,D) Cell cycle analysis of colony type (C) and single-cell type (D) H9 hESCs by flow cytometry.
3. Embryoid Body Formation and Differentiation into Three Germ Layers (Figure 2)
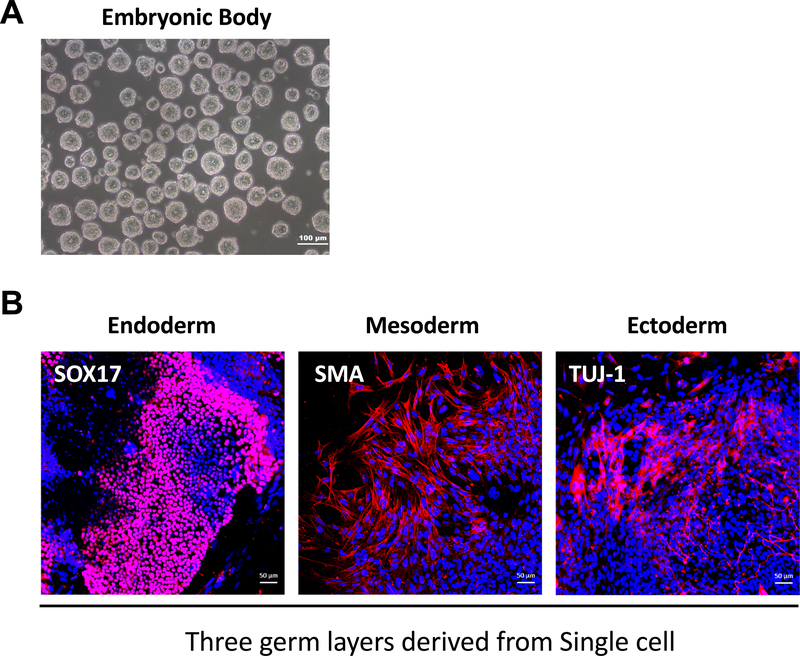
Figure 2: In vitro differentiation of adapted single-cell type hESCs into three germ layers.
(A) Representative phase images of embryonic bodies (EB) derived from single-cell type of hESCs. Scale bar = 100 μm. (B) Immunofluorescent images of differentiated hESCs analyzed for the expression of the three different germ layer markers: SOX17 (endoderm), SMA (mesoderm), and Tuj-1 (ectoderm). Nuclei were stained with DAPI. Scale bar = 50 μm.
To form EBs, first resuspend cells in 3 mL of mTeSR1 medium with 10 μM ROCK inhibitor during the first 24 h, then incubate overnight into 60 mm low attachment dishes in a 37 °C incubator to allow aggregation.
After 24 h, the small EBs are transferred to a 15 mL tube. Let the EBs settle to the bottom of the tube and gently remove the medium with a pipette. Transfer EBs into EB medium (knockout-DMEM supplemented with 20% knockout serum replacement, 1x glutamine supplement [see Table of Materials], 1% NEAA [non-essential amino acids; see Table of Materials], and 0.2% β-mercaptoethanol) and allow them to expand in low attachment dishes for 7 days. The medium can be changed every other day as described above (Figure 2A).
On day 7, collect the EBs from the dishes and transfer them into a 15 mL tube. Gently remove the medium with a pipette and transfer the EBs to a basement membrane matrix-coated 6 well plate.
-
Allow the EBs to attach to the plate and incubate for 12 days in EB medium, during which they will differentiate into the three germ layers (Figure 2B). Change the medium every other day.
NOTE: During aggregation of cells, the low attachment dish helps avoid EB attachment.
4. Differentiation of Single-cell Type hESCs Into NPCs (Figure 3)
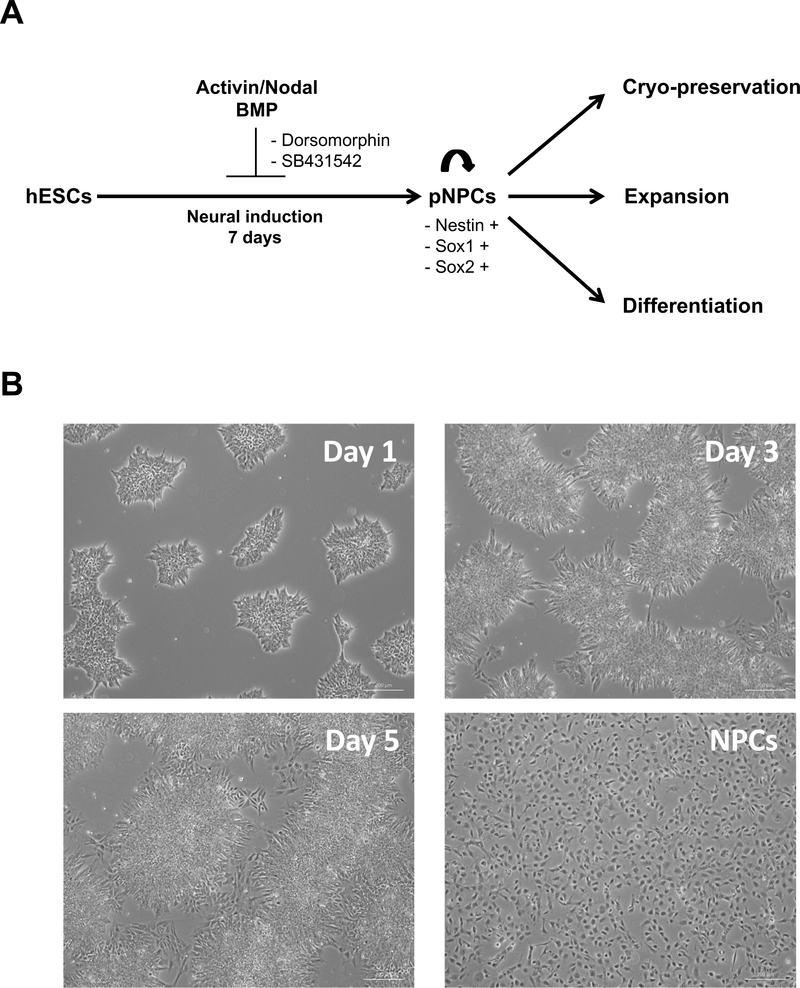
Figure 3: Differentiation of single-cell type hESCs into neural progenitor cells by direct differentiation.
(A) Schematic of the differentiation protocol of hESCs into neural progenitor cells (NPCs). hESCs were treated with dorsomorphin (DMH) and SB431542 (SB) 1 day after plating. (B) Representative phase contrast images of cell morphology during neural differentiation. Scale bar = 200 μm.
To induce NPC differentiation, dissociate single-cell type hESCs with 1 mL of detachment solution (1x) and incubate for 10 min at 37 °C.
Centrifuge the cells for 2 min at 370 × g and remove the detachment solution supernatant. Add 1 mL of DMEM/F12 and resuspend cells by gentle pipetting.
Plate cells on a basement membrane matrix-coated 6 well plate at a density of 2 × 105 cells/well in 2 mL of mTeSR1 containing 10 μM ROCK inhibitor.
After 24 h, replace the culture medium with neural induction medium (DMEM with 1% B27 minus vitamin A) supplemented with 1 μM dorsomorphin and 5 μM SB431542.
-
Change the medium every other day during the first 4 days of neural induction, then every day until reaching confluence at day 7 (Figure 3B).
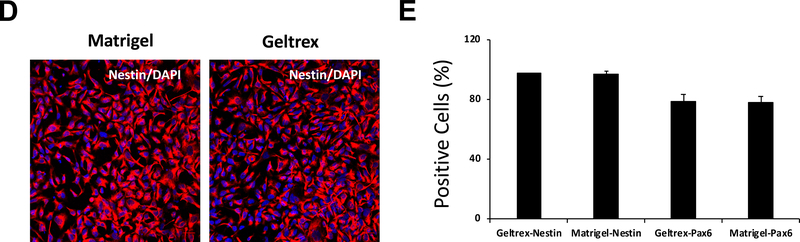
NOTE: (1) Dorsomorphin inhibits the BMP pathway by targeting ALK2,3,6 receptors and also inhibits AMPK; SB431542 is an inhibitor of the TGFβ/Activin pathway by targeting ALK5,7. (2) The same protocol was tested with another ESC line (WA01), which yielded similar results as WA0916. (3) We have generally used Matrigel for the basement membrane matrix; however, cells can be cultured on Geltrex and then differentiated into NPCs with very similar results as indicated by the expression of several NPC markers (Figure 4D,E). Handle Geltrex in the same manner as Matrigel (see section 1).
Figure 4: hESC culture on different basement membrane matrix products.
(A) hESCs cultured on Matrigel or Geltrex exhibited an ability to grow and differentiate into NPCs that was similar to the single cell-culture. Cells were stained with a NESTIN antibody. Nuclei were stained with DAPI. Scale bar = 50 μm. (B) hESCs cultured on Matrigel or Geltrex showed similar potential to differentiate into NPCs as indicated by the percentage of NESTIN- and PAX6-positive cells. Cells were analyzed by flow cytometry at day 7 of NPC differentiation.
5. NPC Expansion and Cryopreservation
After 7 days of neural induction, dissociate cells with 1 mL of detachment solution (1x) and incubate for 10 min at 37 °C.
Centrifuge the cells for 2 min at 370 × g and remove the detachment solution supernatant. Add 1 mL of NPC expansion medium (see Table of Materials) and resuspend cells by gentle pipetting.
Plate 1 × 105 cells in 2 mL of NPC expansion medium on basement membrane matrix-coated 6 well plates.
Passage NPCs multiple times, as necessary, when cultures reach nearly 90% confluency. During the first 3–4 passages, add 10 μM ROCK inhibitor during the initial 24 h to prevent cell death. After these passages, cells can be passaged and cultured without ROCK inhibitor. Change the medium every other day during the expansion.
- Cryopreserve NPCs when cultures reach confluency.
- For cryopreservation, dissociate cells in 1 mL of detachment solution (1x) for 5 min at 37 °C, then centrifuge suspension for 2 min at 370 × g to remove the detachment solution.
- Add cell freezing solution (see Table of Materials) to dissociated NPCs at 1 × 106 cells/mL and distribute 1 mL aliquots to cryovials.
- Place the cryovials into freezing container and store them overnight in a −80 °C freezer.
Store the cryovials in the liquid nitrogen.
6. Preparation of poly-L-ornithine (PLO) and Laminin Coated Plates
To prepare PLO/laminin coated plates, dilute the PLO stock solution into PBS or water to a final concentration of 10 μg/mL. Mix well and coat each well of a 6 well plate with 1 mL of diluted PLO solution.
Incubate for at least 2 h at 37 °C or overnight at 4 °C.
Wash each well with PBS.
-
Dilute laminin stock solution in cold PBS or water at 10 μg/mL. Mix the laminin solution well. Remove the PBS and immediately coat each well with 1 mL of the laminin solution. Keep plates at 4 °C and use within 1 week. Wash with PBS and immediately add culture medium.
NOTE: Do not let the PLO/laminin coated plates dry out.
7. Differentiation of hESC-derived NPCs Into Dopaminergic Neurons (Figure 6A)
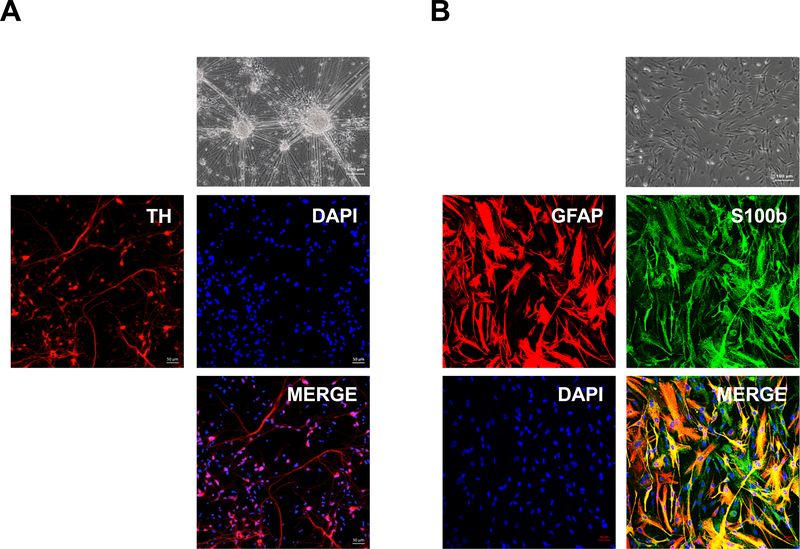
Figure 6: Dopaminergic neuron and astrocyte differentiation of NPCs derived from single-cell type hESCs.
(A) Representative phase contrast image of dopaminergic neurons (top panel). Scale bar = 100 μm. Differentiated cells were stained with antibodies against the dopaminergic neuron marker TH (tyrosine hydroxylase) as indicated. Nuclei were stained with DAPI. Scale bar = 50 μm. (B) Representative phase contrast image of astrocytes (top panel). Scale bar = 100 μm. Differentiated cells were stained with antibodies against the astrocyte marker GFAP (glial fibrillary acidic protein) and S100-B (S100 calcium binding protein B), as indicated. Nuclei were stained with DAPI. Scale bar = 50 μm.
Plate NPCs in PLO/laminin-coated dishes in NPC expansion medium at a density of approximately 50%.
After 24 h, change the medium to DA1 medium (neurobasal medium containing 2 mM glutamine supplement, 1x NEAA, 1x B27, 200 ng/mL SHH, and 100 ng/mL FGF8) and change the medium every other day.
Passage cultures when they reach confluency. Dissociate cells with 1 mL of detachment solution (1x) and incubate for 10 min at 37 °C.
Centrifuge the cells for 2 min at 370 × g and remove the detachment solution supernatant. Add 1 mL of DA1 medium and resuspend the cells by gentle pipetting.
Plate 1 × 105 cells in 2 mL of DA1 medium on PLO/laminin-coated 6 well plates.
-
After 10 days, change to DA2 medium (neurobasal medium containing 2 mM glutamine supplement, 1x NEAA, 1x B27, 200 μM ascorbic acid, 20 ng/mL BDNF, and 20 ng/mL GNDF) and change medium every other day for an additional 20 days (Figure 6A).
NOTE: Add fresh ascorbic acid daily to the DA2 medium. Within the first 14 days, the differentiated cells should be passed when they reach 90% confluency, but no longer after that.
8. Differentiation of ESC-derived NPCs into Astrocytes
Plate neural progenitor cells on PLO/laminin-coated plates in NPC expansion medium at a density of approximately 50%.
After 24 h, change the medium to astrocyte medium (DMEM/F12 medium including 1:100 N2, 1:100 B27, 200 ng/mL IGF, 10 ng/mL activin A, 10 ng/mL heregulin β-1, and 8 ng/mL bFGF) and change the medium every other day.
Passage cultures when they reach confluency. Dissociate cells with 1 mL of detachment solution (1x) and incubate for 10 min at 37 °C.
Centrifuge the cells for 2 min at 370 × g and remove the detachment solution supernatant. Add 1 mL of astrocyte medium and resuspend the cells by gentle pipetting.
Plate 1 × 105 cells in 2 mL of astrocyte medium on PLO/laminin-coated 6 well dishes.
Allow differentiation to continue for a total of 5–6 weeks (Figure 6B).
9. Immunofluorescence Staining
Culture cells in 35 mm μ-dishes as described above for each cell type. Aspirate the medium and add 1 mL of 4% PFA solution per dish and incubate for 20 min at RT.
-
Wash 2x for 5 min with PBS.
NOTE: Once the cells are fixed, the assay plates can be sealed with parafilm and stored at 4 °C. Stain with an antibody within 1 week after fixation.
Add 300 μL of blocking solution (goat or donkey serum in PBS) per dish and incubate at RT for 30–60 min.
Wash the dish 2x with PBS for 5 min.
Add 300 μL of primary antibody (Table 1) solution (diluted in blocking solution) and incubate at RT for 1.5 h or overnight at 4 °C.
Wash 2x with PBS for 10 min.
Add 300 μL of fluorescent-conjugated secondary antibody (Table 1) in blocking solution and incubate in the dark for 1 h at RT.
Wash the cells 2x for 10 min with PBS.
Add Hoechst staining solution (1 μM final concentration in PBS) to stain nuclei. Incubate the cells in the dark for 5 min at RT.
Wash cells 1x with PBS for 5 min and then add 500 μL of PBS. Keep dishes in the dark, then observe fluorescence with a confocal microscope (Figure 2B, Figure 5C, Figure 6A, and Figure 7B).
Table 1.
Antibodies used in immunocytochemistry and FACS analysis.
| Primary Antibodies | Species | Dilution | Catalog Number | Company |
|---|---|---|---|---|
| OCT4 | Mouse | 1:1000 | sc-5279 | Santa Cruz |
| TRA 1-81 | Mouse | 1:500 | sc-21706 | Santa Cruz |
| SSEA-1 | Mouse | 1:500 | sc-21702 | Santa Cruz |
| SOX1 | Goat | 1:500 | AF-3369 | R&D |
| SOX2 | Rabbit | 1:1000 | sc-20088 | Santa Cruz |
| SMA | Rabbit | 1:500 | ab-5694 | Abcam |
| Tuj1 | Mouse | 1:1000 | T8578 | Sigma |
| NESTIN | Mouse | 1:1000 | ab-22035 | Abcam |
| OTX2 | Mouse | 1:250 | ab-21990 | Abcam |
| hNCAM | Mouse | 1:200 | sc-106 | Santa Cruz |
| hPAX6 | Mouse | 1:250 | 561664 | BD |
| TH | Mouse | 1:500 | T1299 | Sigma |
| S100b | Mouse | 1:250 | ab4066 | Abcam |
| GFAP | Rabbit | 1:1000 | ab7260 | Abcam |
| Secondary Antibodies | ||||
| Anti-mouse | Goat | 1:1000 | AF 488 or 647 | Life Technologies |
| Anti-rabbit | Goat | 1:1000 | AF 488 or 647 | Life Technologies |
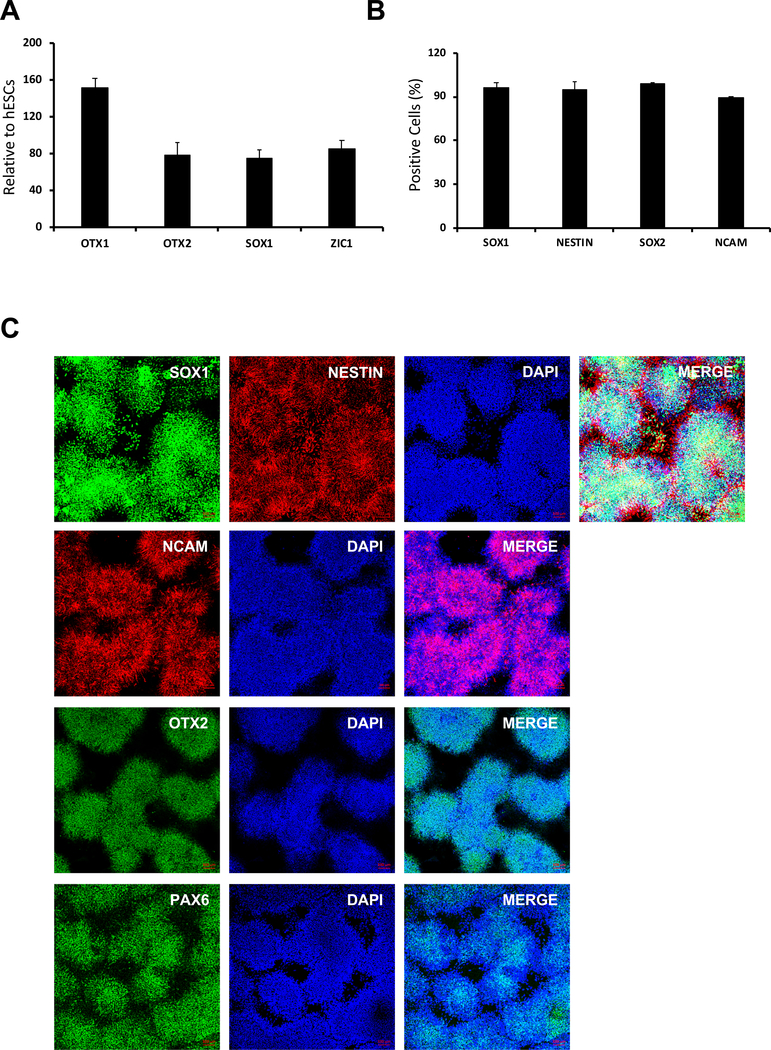
Figure 5: Expression of NPC markers.
(A) After 7 days of neural differentiation, expression of NPC marker genes (i.e., OTX1, OTX2, SOX1, and ZIC1) was analyzed by QRT-PCR. Values were normalized to GAPDH and calculated relative to the values of hESCs (p < 0.05). (B) The percentage of SOX1-, NESTIN-, SOX2-, and NCAM-positive cells was determined by flow cytometry at day 7 of NPC differentiation. (C) At day 7 NPC differentiation, cells were stained with antibodies against the neural markers SOX1, NESTIN, NCAM, OTX2, and PAX6. Nuclei were stained with DAPI. Scale bar = 100 μm.
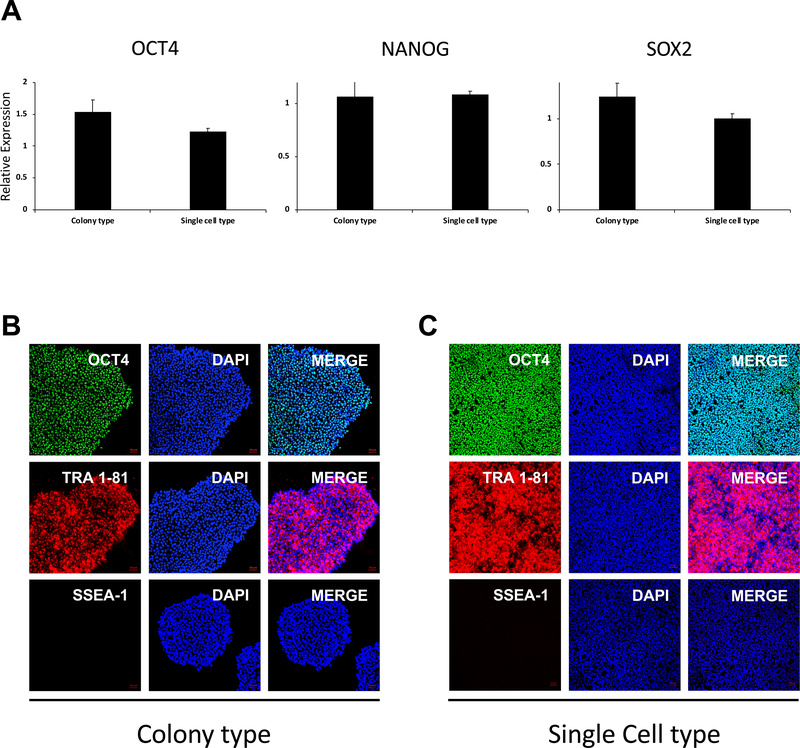
Figure 7: Characterization of single cell type hESCs.
(A) hESCs adapted to single-cell type culture were analyzed for the expression of ESC markers (i.e., OCT4, NANOG, and SOX2) by QRT-PCR. Values were normalized to GAPDH (p < 0.05). (B) Immunofluorescent images of hESCs stained for the expression of the pluripotency markers OCT4, TRA-1-81, and SSEA-1. Nuclei were stained with DAPI. Scale bar = 50 μm.
Representative Results
Presented here is an improved protocol for the maintenance and expansion of single-cell type culture of hESCs and their efficient differentiation into neural progenitor cells, which subsequently differentiates into various downstream neural lineages, including dopaminergic neurons and astrocytes.
Representative phase contrast images show cell morphology at different steps during the adaptation of colony type hESCs to the single-cell type culture (Figure 1A). Through the single-cell culture condition, it was found that the adapted hESCs were able to be maintained at high density, then easily and efficiently subcultured when reaching confluency (Figure 1A,B). These cells retained the cell cycle characteristics (i.e., a short G1 phase and high proportion of cells in S phase) typical of colony type hESCs (Figure 1C,D). They also expressed the ESC markers (i.e., OCT4, TRA 1-81, SOX2, and NANOG) at levels comparable to those of colony type hESCs as indicated by QRT-PCR and immunostaining analysis (Figure 7, Table 2). Moreover, it was shown that single-cell hESCs were able to form embryoid bodies containing cells from all three germ layers: endoderm (SOX17 expression), mesoderm (SMA expression) and ectoderm (Tuj-1 expression) (Figure 2).
Table 2.
List of primers used in QRT-PCR analysis.
| Primer name | Primer sequence |
|---|---|
| OCT4 | F: GGAAGGTATTCAGCCAAACG |
| R: CTCCAGGTTGCCTCTCACTC | |
| NANOG | F: GGTTCCAGAACCAGAGAATGA |
| R: ATTGGAAGGTTCCCAGTCG | |
| SOX1 | F: CCTTAGGTTTCCCCTCGCTTT |
| R: CAGGCTGAATTCGGTTCTCATT | |
| OTX1 | F: AAGATCAACCTGCCGGAGTCT |
| R: CGTGAATTGGCCACTGCTTT | |
| OTX2 | F: TGGAAGCACTGTTTGCCAAG |
| R: TAAACCATACCTGCACCCTCG | |
| ZIC1 | F: AACCCCAAAAAGTCGTGCAAC |
| R: TCCTCCCAGAAGCAGATGTGA | |
| GAPDH | F: CCCATCACCATCTTCCAGGAG |
| R: CTTCTCCATGGTGGTGAAGACG |
Next, it was demonstrated that single-cell type hESCs efficiently differentiated into neural progenitor cells using an NPC protocol (Figure 3A), as indicated by the loss of typical hESC morphology and appearance of NPC morphology (Figure 3B)16. NPC differentiation was supported by the increased expression of the signature NPC markers (i.e., SOX1, OTX2, ZIC1, and OTX1; Figure 5A, Table 2) and confirmed by immunostaining and FACS analysis. The same analysis also showed that more than 90% of the cells stained positive for SOX1, PAX6, and NCAM protein (Figure 5B,C). To examine the ability of single-cell hESC-derived NPCs to differentiate into various downstream neural lineages, the differentiation of these cells into dopaminergic neurons and astrocytes was examined. As shown in Figure 6A,B, single-cell hESC-derived NPCs were able to differentiate into dopaminergic neurons and astrocytes, as indicated by the appearance of characteristic morphologies and expression of lineage-specific dopaminergic markers (i.e., TH; Figure 6A) and astrocyte markers (i.e., GFAP and S100B; Figure 6B).
Discussion
Scalable and efficient methods for the differentiation of hESCs into various lineages and the generation of sufficient numbers of differentiated cells are important criteria for drug screening and stem cell therapy. Various single-cell passing methods have been published, in which cells are cultured in the presence of ROCK inhibitor or other small molecules to improve survival, but the final products of these culture methods are colony type hESCs17,18,19,20,21. The single-cell ESC protocol, which is partially based on previously published methods19,20,21,22, successfully generates and maintains single cell-type hESC cultures and prevents colony type hESC culture. It includes high density single cell plating, multicellular association, monolayer growth, and efficient subculture (Figure 1). The latter was achieved by the addition of ROCK inhibitor during the initial 24 h of single-cell type culture of hESCs, which improves cell survival17,18,19,20,21. This protocol is more easily scalable and allows expansion of these cells for therapeutic applications in drug screening and stem cell.
It is further demonstrated that single-cell type hESCs can efficiently differentiate to the NPC lineage (Figure 3) without use of an intermediate stage, such as EB and rosette formation23,24,25. High neural conversion from single-cell type hESCs was achieved through the inhibition of BMP/TGFβ/activin signaling pathways by treatment with the ALK inhibitors, dorsomorphin, and SB43154215,16,26. With this protocol, the adapted single-cell type hESCs can efficiently differentiate into NPCs without the need for EB and rosette formation (Figure 5) and or be induced to differentiate into dopaminergic neurons and astrocytes (Figure 6).
In summary, single-cell type culture of hESCs provides a rapid and efficient system to study the molecular mechanisms that regulate multistep differentiation to various lineages. Specifically, this protocol utilized NPCs and described their subsequent differentiation into additional neural lineages, such as astrocytes and dopaminergic neurons. The protocol provides a platform for simple, robust, and scalable production of progenitor and differentiated cells that can be suitable for basic studies, drug screening, and applications in regenerative medicine.
Acknowledgments
We thank Dr. Carl D. Bortner (NIEHS) for his assistance with the FACS analysis. This research was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, the National Institutes of Health, Z01-ES-101585 to AMJ.
Footnotes
Disclosures
The authors declare no conflicts of interest.
Video Link
The video component of this article can be found at https://www.jove.com/video/60571/
References
- 1.Thomson JA et al. Embryonic stem cell lines derived from human blastocysts. Science. 282 (5391), 1145–1147 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Rosler ES et al. Long-term culture of human embryonic stem cells in feeder-free conditions. Developmental Dynamics. 229 (2), 259–274 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Mallon BS, Park KY, Chen KG, Hamilton RS, McKay RD Toward xeno-free culture of human embryonic stem cells. The International Journal of Biochemistry & Cell Biology. 38 (7), 1063–1075 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman LM, Carpenter MK Characterization and culture of human embryonic stem cells. Nature Biotechnology. 23 (6), 699–708 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Yan Y et al. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Translational Medicine. 2 (11), 862–870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goncalves JT, Schafer ST, Gage FH Adult Neurogenesis in the Hippocampus: From Stem Cells to Behavior. Cell. 167 (4), 897–914 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Watanabe K et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 25 (6), 681–686 (2007). [DOI] [PubMed] [Google Scholar]
- 8.International Stem Cell I et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nature Biotechnology. 25 (7), 803–816 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Hartung O, Huo H, Daley GQ, Schlaeger TM Clump passaging and expansion of human embryonic and induced pluripotent stem cells on mouse embryonic fibroblast feeder cells. Current Protocols in Stem Cell Biology. 14 (1), Chapter 1 Unit 1C 10 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Chen KG et al. Non-colony type monolayer culture of human embryonic stem cells. Stem Cell Research. 9 (3), 237–248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Hou Z, Gulbranson DR, Thomson JA Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 7 (2), 240–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Krawetz R, Liu S, Meng G, Rancourt DE ROCK inhibitor improves survival of cryopreserved serum/feeder-free single human embryonic stem cells. Human Reproduction. 24 (3), 580–589 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Ohgushi M et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 7 (2), 225–239 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Pakzad M et al. Presence of a ROCK inhibitor in extracellular matrix supports more undifferentiated growth of feeder-free human embryonic and induced pluripotent stem cells upon passaging. Stem Cell Reviews and Reports. 6 (1), 96–107 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Chambers SM et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature Biotechnology. 27 (3), 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon K et al. GLIS3 Transcriptionally Activates WNT Genes to Promote Differentiation of Human Embryonic Stem Cells into Posterior Neural Progenitors. Stem Cells. 37 (2), 202–215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emre N et al. The ROCK inhibitor Y-27632 improves recovery of human embryonic stem cells after fluorescence-activated cell sorting with multiple cell surface markers. PLoS ONE. 5 (8), e12148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanna J et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proceedings of the National Academy of Sciences of the United States of America. 107 (20), 9222–9227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha K et al. Surface-engineered substrates for improved human pluripotent stem cell culture under fully defined conditions. Proceedings of the National Academy of Sciences of the United States of America. 108 (46), 18714–18719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsutsui H et al. An optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cells. Nature Communications. 2, 167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y et al. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proceedings of the National Academy of Sciences of the United States of America. 107 (18), 8129–8134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS ONE. 3 (2), e1565 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DS et al. Highly pure and expandable PSA-NCAM-positive neural precursors from human ESC and iPSC-derived neural rosettes.PLoS ONE. 7 (7), e39715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Hu J, Zhou L, Pollard SM, Smith A Interplay between FGF2 and BMP controls the self-renewal, dormancy and differentiation of rat neural stem cells. Journal of Cell Science. 124 (Pt 11), 1867–1877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou JM, Chu JX, Chen XJ An improved protocol that induces human embryonic stem cells to differentiate into neural cells in vitro. Cell Biology International. 32 (1), 80–85 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Smith JR et al. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Developmental Biology. 313 (1), 107–117 (2008). [DOI] [PubMed] [Google Scholar]