Abstract
Tip growth of pollen tubes and root hairs occurs via rapid polar growth. These rapidly elongating cells require tip-focused endomembrane trafficking for the deposition and recycling of proteins, membranes, and cell wall materials. Most of the image-based data published to date are subjective and non-quantified. Quantitative and comparative descriptors of these highly dynamic processes have been a major challenge, but are highly desirable for genetic and chemical genomics approaches to dissect this biological network. To address this problem, we screened for small molecules that perturbed the localization of a marker for the Golgi Ras-like monomeric G-protein RAB2:GFP expressed in transgenic tobacco pollen. Semi-automated high-throughput imaging and image analysis resulted in the identification of novel compounds that altered pollen tube development and endomembrane trafficking. Six compounds that caused mislocalization and varying degrees of altered movement of RAB2:GFP-labeled endomembrane bodies were used to generate a training set of image data from which to quantify vesicle dynamics. The area, velocity, straightness, and intensity of each body were quantified using semi-automated image analysis tools revealing quantitative differences in the phenotype caused by each compound. A score was then given to each compound enabling quantitative comparisons between compounds. Our results demonstrate that image analysis can be used to quantitatively evaluate dynamic subcellular endomembrane phenotypes induced by bioactive chemicals, mutations, or other perturbing agents as part of a strategy to quantitatively dissect the endomembrane network.
Key words: endomembranes, vesicle trafficking, quantification, pollen, chemical biology
INTRODUCTION
Endomembrane trafficking is a dynamic network of processes that is essential for abiotic stress response, pathogen response, cytokinesis, cell expansion, tip growth, and development (Samuels et al., 1995; Wick et al., 2003). The endomembrane system consists of discrete membrane-bound compartments that are interconnected within complex trafficking pathways. These pathways are highly coordinated and regulated to ensure proper localization and allocation of proteins, cell wall components, and other vital materials for proper growth and development (Roberts, 1994; Miller et al., 1997; Fürthauer and González-Gaitán, 2009).
Pollen tubes are single, elongated cells that undergo a specialized form of polar growth known as tip growth—a mode of growth shared by root hairs, trichomes, and neurons (Hepler et al., 2001). Tip growth requires high levels of tip-focused endomembrane trafficking to deposit lipids, proteins, and cell wall materials to facilitate rapid growth (Hepler et al., 2001). Pollen tubes must form an elongating tube that travels from the surface of the stigma to deliver the sperm to the ovule (Lord, 2000). This rapid tip growth is facilitated by the coordination of the actin cytoskeleton and a highly active secretory system (Hepler et al., 2001).
Within the Ras superfamily of proteins, the Rab family of small GTPases is evolutionarily conserved and regulates the budding, transport, and fusion of vesicles from intracellular donor compartments to target compartments, thus mediating intracellular trafficking (Stenmark and Olkkonen, 2001; Nielsen et al., 2008; Ebine et al., 2011). When bound to GDP, RABs are inactive; however, when bound to GTP, they are activated and interact with downstream effector proteins (Stenmark and Olkkonen, 2001). Although many RAB proteins are found in yeast, plants, and mammals, of the three, RAB2 is found only in mammals and plants (Rutherford and Moore, 2002). The Arabidopsis thaliana ortholog RAB B1 and the mammalian RAB2 are involved with vesicle maturation of the anterograde trafficking pathway from the endoplasmic reticulum (ER) to the Golgi stacks and localizes primarily in the Golgi (Rutherford and Moore, 2002). In Nicotiana tobaccum (tobacco), RAB2 is found predominantly in pollen grains, pollen tubes, and root hairs, probably due to the high secretory demands of the two cell types. Transcripts are detected in floral buds and lateral roots, though this might be due to their presence in pollen and root hairs (Cheung et al., 2002). When fused to GFP, RAB2 is observed in Golgi stacks that are highly active within the germinating pollen grain as well as the elongating tube (Cheung et al., 2002). This localization can be exploited to study endomembrane trafficking in a developing pollen tube. Because the ER and Golgi are major components along the secretory pathway, it can be inferred that, if agents such as bioactive small molecules perturb this pathway, global secretion will be significantly impaired, allowing the effect of the small molecules to be identified by microscopy.
Qualitative descriptions of trafficking are useful but very limited in the number of comparisons that can be made and conclusions that can be drawn. Since endomembrane trafficking is a physical phenomenon that can be observed by microscopy, it can be quantified and subjected to statistical analysis. Indeed, our ability to quantify distribution, sizes, and rates of endomembrane compartments in many organisms and tissue types has been growing over the past two decades (Pyke and Leech, 1991; Derksen et al., 1995; Logan et al., 2003; Rink et al., 2005; Pochynyuk et al., 2007; Ketelaar et al., 2008; Sparkes et al., 2008; Kato et al., 2010; Collinet et al., 2010; Salomon et al., 2010). Earlier studies used various methods to quantify trafficking that were limited by the available technology. Initial investigations utilized transmission electron microscopy and manual measurement of organelles. This was used to infer growth rates based on mathematical models (Ketelaar et al., 2008) or by calculating organelle distribution (Derksen et al., 1995). Although informative, these studies observed frozen or fixed tissue rather than live cells, which provide a more accurate depiction of cellular processes.
The advent of GREEN FLOURESCENT PROTEIN (GFP) made quantitative analyses of living cells possible. Live imaging in combination with image analysis gave a more comprehensive description of organelle size and distribution (Pochynyuk et al., 2007). Nevertheless, these approaches used still frame images—snapshots in time that omit any temporal dynamics. To account for this, video data can capture movements and morphological changes of cellular compartments in real time while simultaneously measuring several features in animal cells (Collinet et al., 2010) and endomembrane trafficking in plant pathogen-infected leaf epidermal cells (Salomon et al., 2010). To visualize and quantify video images from cells displaying rapid growth and highly dynamic trafficking, we used germinating pollen as a model system. Our present study combines quantitative video analysis with bioactive chemicals to rapidly and controllably perturb trafficking, permitting the quantitative and comparative description of compounds affecting intracellular trafficking in pollen grains.
There are several challenges to be overcome in understanding endomembrane trafficking in the sporophyte and gametophyte. Classical genetic approaches are limited by functional gene redundancy because mutation of a single gene can result in a lack of phenotype due to compensation. Alternatively, when an essential gene is mutated, embryo lethality can result. These obstacles can be circumvented by a chemical genomics approach that uses small molecules to temporarily modulate protein function. Small molecules often bind to proteins, transiently inhibiting proper function. This marriage of synthetic chemistry and biology coupled with the grand scale of genomics is a powerful approach that produces large amounts of useful phenotypic data prior to genetics (Hicks and Raikhel, 2012). However, the powers of chemical biology and genetics are both limited by an inability to accurately quantify trafficking processes to obtain objective descriptors to compare the dynamics in the presence of different inhibitors or mutations. For example, due to the high-throughput nature of chemical screening, data analysis should be quantitative and automatable to accommodate large image data sets.
To address the pressing need to develop quantitative image analysis and comparative descriptors, we report a chemical genomics approach coupled to semi-automated image acquisition and image analysis to identify chemical compounds with differential effects on endomembrane trafficking. Utilizing these compounds as a training set to develop our approach, we quantified their effects on vesicle traits such as size, velocity, straightness, and intensity as quantitative descriptors. Furthermore, we developed a scoring system that permitted a comparison of differential intracellular dynamics induced by each of the molecules—a critical step towards fully automated image characterization of endomembrane dynamics.
RESULTS
A Pollen-Specific Chemical Screen Reveals Eight Classes of Morphological and Developmental Phenotypes
To discover novel compounds that allow the dissection of endomembrane trafficking pathways and to identify a set of compounds to generate a training data set for quantification, a chemical genomic screen was used to identify small molecules that affect pollen tube morphology and development. Because of their rapid growth and ease at which vesicle movement is visualized, Nicotiana tobaccum (tobacco) pollen was an ideal model system in which to perform this screen. Compounds were screened for those that inhibited germination or altered tube morphology, both of which are dependent upon proper tip-focused vesicle trafficking. In a developing pollen tube, vesicle traffic follows a distinct reverse fountain flow in which cargo moves towards the growing tip on the sides of the tube and cargo moving from the tip moves down the center of the tube (Pierson and Cresti, 1992). Several small molecules were found that disrupted this distinct trafficking pattern.
Previously, more than 46 000 compounds were screened in pollen at a concentration of 50–100 µM (Robert et al., 2011). Of these, 360 compounds were found to cause morphological or developmental defects in pollen, demonstrating their bioactivity (Robert et al., 2008). Small molecules were categorized as bioactive if they had an observable effect on morphology, development, or localization of a fluorescent marker. A small molecule was considered to be bioactive if pollen grains did not germinate or there were defects in tube morphology such as isodiametric tip growth. In this current study, the 360 compounds were serially diluted to 25–50 and 10–20 µM, depending on the molecular weight of the compound, to determine the minimal effective concentrations and to uncover possible latent phenotypes (Figure 1 ). This permitted us to focus our screens on the resulting 225 compounds that inhibited germination and that remained bioactive at lower concentrations, thereby determining effective concentrations for potential compounds of interest (Supplemental Table 1).
Figure 1.
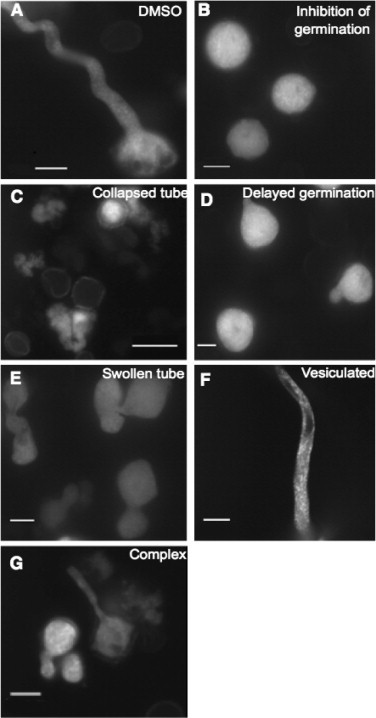
Morphological Phenotypes of Pollen Expressing Rab2:GFP.
(A) Control treatment treated with DMSO.
(B) Inhibition of germination.
(C) Collapsed pollen tubes.
(D) Short tubes due to delayed growth.
(E) Swollen pollen tubes.
(F) Vesiculated tubes.
(G) A complex phenotype in which multiple phenotypes are demonstrated. All images were taken using a 40× objective. Concentration of compounds ranged from 10 to 50 µM, depending on the compound used. Scale bar = 20 µm.
Identification of Compounds that Affect Endomembrane Trafficking
To test the effect of these 225 bioactive compounds on endomembrane trafficking in tobacco pollen, a secondary screen was performed using a transgenic tobacco line expressing RAB2:GFP in pollen (Cheung et al., 2002) to visualize the potential effects on RAB2 trafficking at two concentrations: 25–50 and 10–20 μM. Images were collected for each compound, analyzed using the Imaris image analysis software package, and organized into groups that exhibited the same or similar phenotypes (Figure 1). Table 1 shows the number of compounds resulting in each phenotype observed. To focus our efforts on compounds that showed the most dramatic effects, a subgroup of six compounds were selected which caused disruptions in the localization of RAB2:GFP associated with endomembranes (Figure 2 ). The compounds that comprised this subgroup were termed RAB2 effectors (RAEs). Compared to untreated controls, disruptions in the localization of RAB2:GFP were manifested as agglomerations (Figure 3 ) which we termed ‘RAB2 bodies’. The bodies were presumed to be derived from Golgi stacks and ER. RAEs 6 and 7 elicited similar phenotypes, suggesting a similar mode of action and structure. Structure–activity relationship (SAR) analysis indicated that both RAEs 6 and 7 have a nitro group along with a nitrile group next to a double bond. These compounds disrupted RAB2:GFP movement or localization and therefore globally altered trafficking in the cell, allowing measurement of these changes.
Table 1.
Phenotypes Resulting from Different Concentrations of Library Compounds.
| Phenotype | 50-100 μM | 25-50 μM | 10-20 μM |
|---|---|---|---|
| Germination Inhibition | 112 | 76 | 37 |
| Collapsed Tube | 51 | 27 | 12 |
| Delayed Growth | 40 | 30 | 23 |
| Swollen Tube | 4 | 9 | 5 |
| Vesiculated Tube | 8 | 0 | 8 |
| Complex Phenotype | 27 | 14 | 13 |
| TOTAL | 290 | 156 | 98 |
Number of compounds resulting in different phenotypes are indicated.
Figure 2.
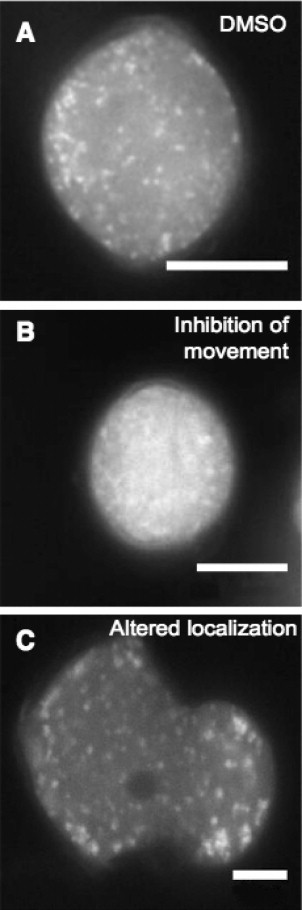
RAEs Alter the Localization and Movement of RAB2:GFP.
(A) Control DMSO-treated tobacco pollen expressing RAB2:GFP.
(B) RAB2:GFP localization in a pollen grain and tube following inhibition of trafficking by RAE2.
(C) Tobacco pollen showing defects in normal localization of RAB2:GFP caused by treatment with RAE7. Images are taken using a 63× objective. Scale bar = 10 µm.
Figure 3.
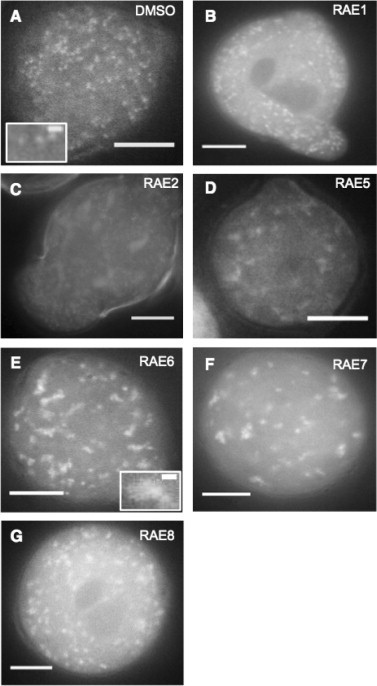
RAE Phenotypes. RAB2:GFP in pollen treated with (A) DMSO, (B) 36 µM RAE1, (C) 38 µM RAE2, (D) 35 µM RAE5, (E) 78 µM RAE6, (F) 32 µM RAE7, (G) 28 µM RAE7. All images were taken using a 63× objective. Scale bar = 10 µm. Insets indicate zoomed image of the respective labeled body. Scale bar within inset = 1 µm.
Thirty-second time-lapse videos were taken of untreated pollen tubes and pollen tubes treated with the candidate compounds that affected RAB2:GFP localization or movement (Figure 4 and Supplemental Movies 1–6). Although the images of the automated confocal microscope were of lower quality compared to available manual confocal instruments, our eventual goal was to fully automate image capture and analysis; thus, we developed our approach utilizing the automated instrument. Quantitative comparison of the phenotypes induced by the RAEs demonstrated that the semi-automated method can be used to study endomembrane trafficking pathways by disrupting RAB2:GFP movement and localization. RAEs resulted in either moderately or drastically decreased movement.
Figure 4.
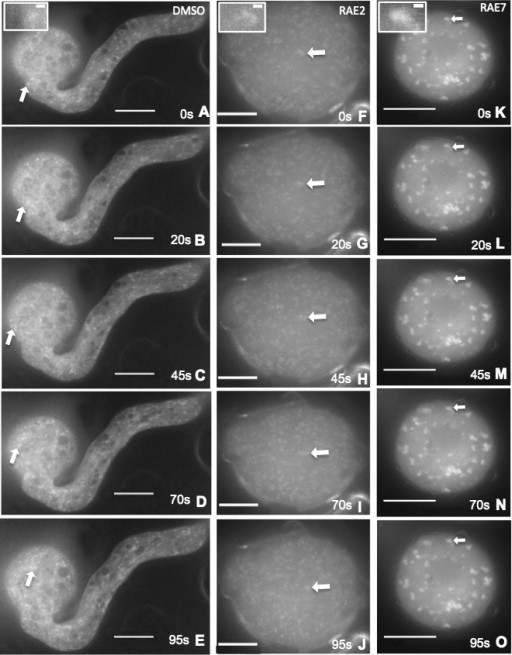
Movement of RAB2:GFP Is Inhibited by RAEs.(A–O) Selected frames from time-lapse videos of pollen tubes. (A–E) DMSO-treated RAB2:GFP-labeled pollen. Scale bar = 15 µm. (F–J) RAE2-treated RAB2:GFP-labeled pollen. Scale bar = 10 µm. (K–O) RAE7-treated RAB2:GFP-labeled pollen. Scale bar = 20 µm. Phenotypes are shown at 0, 20, 45, 70, and 95 s after acquisition. Arrows denote the position of the same RAB2 body in each frame. Insets indicate zoomed image of the respective labeled body. Scale bar within inset = 1 µm.
Quantification of RAB2 Bodies Reveals Distinct Groupings and Latent Effects
The observed phenotypes had varying degrees of severity, which could not be sufficiently described using qualitative descriptors. We therefore quantified the area, velocity, straightness of movement, and intensity of the RAB2:GFP agglomerations using the segmentation function in the Imaris image analysis software package. Values were quantified as means (Table 2 ). However, determining the mean values of RAB2 body measurement in pollen tubes was insufficient to fully understand this system. Since both the Golgi bodies and the RAB2 bodies were highly dynamic and displayed a broad range of sizes, movements, movement patterns, and intensities, we presented the area, velocity, straightness, and intensity measurements as cumulative distribution plots (Sparkes et al., 2008) to better describe organelle movement. By permitting a perspective on the variation, these plots provided a more informative comparison of organelle movement (Figure 5 ).
Table 2.
Quantitative Values Describing the Effect of Each RAE.
| Compound | Structure | Average area of RAB2 bodies (um2) | % Area of UT RAB2 Bodies | Average velocity of RAB2 bodies (um/s) | % Velocity of UT of RAB2 bodies | Straightness | % Straightness of UT | % Intensity of UT | Difference Index |
|---|---|---|---|---|---|---|---|---|---|
| Untreated (UT) | No compound | 1.926 | 100 | 0.911 | 100 | 0.525 | 100 | 100 | 1 |
| RAE1 (5271226) Chembridge |  |
4.4 | 228 | 0.289 | 31.7 | 0.313 | 60 | 137 | 1.88 |
| RAE2 (5532951) Chembridge | 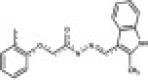 |
4.247 | 221 | 0.308 | 33.8 | 0.532 | 101 | 160 | 2.14 |
| RAE5 (6396311) Chembridge | 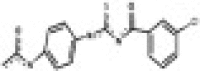 |
5.155 | 268 | 0.109 | 11.9 | 0.122 | 23 | 111 | 2.12 |
| RAE 6 (LAT033E05) LATCA | 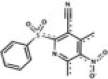 |
7.372 | 383 | 0.046 | 5 | 0.107 | 20 | 191 | 4.59 |
| RAE7 (LAT035F05) LATCA | 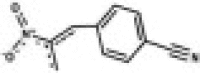 |
7.73 | 401 | 0.048 | 5.2 | 0.105 | 20 | 218 | 5.22 |
| RAE8 (LAT045A04) LATCA |  |
6.991 | 363 | 0.068 | 7.4 | 0.077 | 15 | 207 | 4.37 |
Quantification of RAB2 bodies induced by RAEs.
Figure 5.
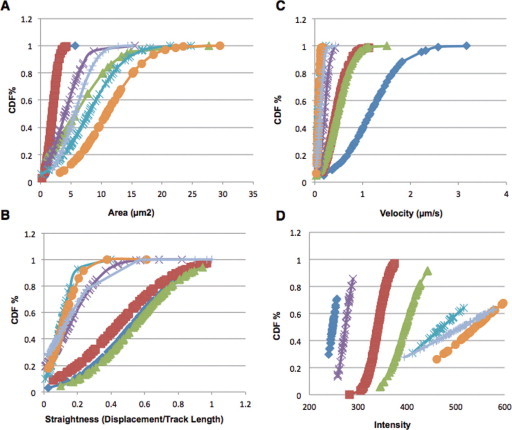
Cumulative Distribution Function (CDF) of the Four Features Measured—Area, Velocity, Straightness, and Intensity—of All Six RAEs Plus an Untreated Control.
(A) The CDF for the average area.
(B) The CDF for the average velocity.
(C) The CDF for average straightness.
(D) The CDF for the average intensity. In all plots, blue diamonds represent the untreated control. Red squares represent RAE1. Green triangles represent RAE2. Purple Xs represent RAE5. Teal asterisks represent RAE6. Orange circles represent RAE7. Gray crosses represent RAE8. All bodies measured were within the pollen grain.
The cumulative distribution plot of the areas indicates a trend that is also inferred by the mean values. In the case of RAEs 1 and 2, the localization of RAB2:GFP was altered, causing the formation of RAB2 bodies that were 228% and 221% larger than control Golgi stacks, respectively. RAEs 1 and 2 also had a range of velocity values between approximately 0.03 and 1.5 µm s–1, which was about half the range of the untreated Golgi bodies (Figure 5A). RAEs 5, 6, 7, and 8 resulted in RAB2 bodies that ranged from 268% to 401% larger than the mean of untreated Golgi bodies. They also exhibited lower variance (Figure 5B), indicating that the individual agglomerations were of similar size.
Changes in velocity were also apparent. Initially, RAEs 1 and 2 resulted in slightly reduced speed of the RAB2 bodies, whereas RAEs 5, 6, 7, and 8 dramatically reduced speed. Similarly to area measurements, velocity was measured and reported as a percentage of the velocity of the untreated Golgi bodies (Figure 5B). RAEs 1 and 2 moved at only 31% and 37% of the velocity of the untreated Golgi bodies, respectively. The quantification of RAEs 5, 6, 7, and 8 supported our initial observation of a dramatic reduction in speed. The RAB2 bodies produced by treatment with the RAEs traveled at between 11.9% and 5% of the velocity of untreated Golgi bodies. Upon quantification of the data, we noted that the compounds formed natural groups of weak effectors (RAEs 1 and 2) and strong effectors (RAEs 5, 6, 7, and 8).
Beyond the speed of movement, the pattern of movement was a key component of dynamic organelle behavior. A robust descriptor of movement pattern was the straightness of trajectory of each body. The straightness was determined by the ratio of displacement over the track length. Values closer to one indicated a tendency towards linear movement, whereas those farther from one indicated movements resembling short-distance random or Brownian motion (Figure 5C). Similarly to our previous results, there was a distinct grouping and similarity between RAEs 1 and 2, which had greater values for straightness. This indicated more directed movement, whereas RAEs 5, 6, 7, and 8 stimulated a tendency towards random motion.
Lastly, we quantified the intensity of RAB2:GFP fluorescence (Figure 5D). The untreated pollen showed the lowest RAB2:GFP intensity followed closely by RAE 5. RAEs 1 and 2 resulted in an intermediate level of intensity, at 137% and 160% of the untreated, respectively. RAEs 6, 7, and 8 resulted in the highest levels of intensities, ranging from 191% to 218% of untreated RAB2:GFP intensity levels.
It stands to reason that some of these parameters are most likely correlated. To confirm our hypothesis, the Pearson correlation coefficients for every parameter combination were calculated (Figure 6 ). Area was positively correlated with intensity (R = 0.715), suggesting that a the amount of RAB2:GFP increased with the size of RAB2 bodies. As to be expected, straightness was negatively correlated with velocity (R = 0.569). Bodies traveling in a straight line moved faster than those exhibiting Brownian motion. Less obviously, intensity and straightness were negatively correlated (R = 0.571). In other words, vesicles that moved in a more directed manner tended to have lower intensities. All other parameters showed little to no correlation.
Figure 6.
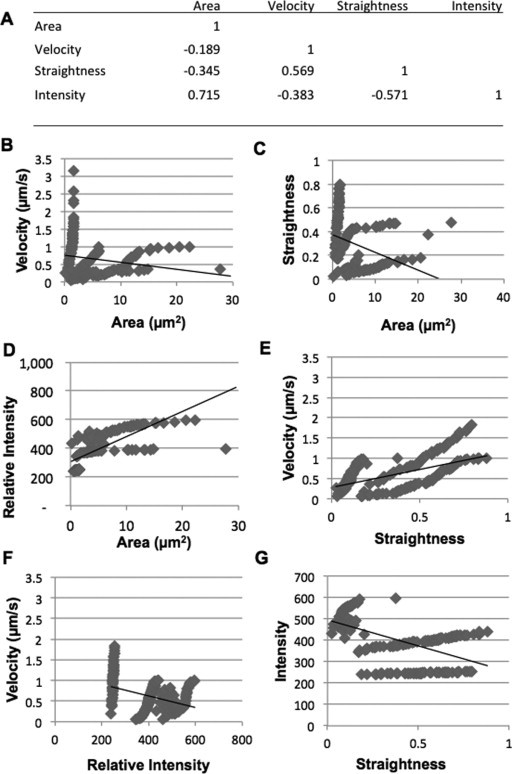
Correlation of Trafficking Features.
(A) A correlation matrix showing the Pearson correlation coefficients showing the relationships of all features.
(B) The correlation between velocity and area.
(C) The correlation between straightness and area.
(D) The correlation between intensity and area.
(E) The correlation between velocity and straightness.
(F) The correlation between velocity and intensity.
(G) The correlation between intensity and straightness.
The Difference Score Provides a Quantitative Measure of Bioactivity
To understand the relative global effects of the RAEs on trafficking, we developed a quantitative score that incorporated all the descriptors of RAB2:GFP dynamics. Our ‘difference index’ was developed to give an integrated quantitative value to describe the degree to which each compound affected the area, velocity, straightness, and intensity of each agglomeration (Table 2). This straightforward index was the sum of the squares of the normalized value for each measure multiplied by an assigned weighting factor. Because it was difficult to determine whether one feature might play a larger role in the identity of a compound, each feature was given equal weight. However, our approach can permit alterations in the weighting of particular phenotypes as necessary.
DISCUSSION
We utilized tobacco pollen as an efficient system to screen for chemicals that inhibit endomembrane trafficking. In a previous study (Drakakaki et al., 2011), more than 46 000 chemicals were screened in tobacco pollen. Of these compounds, 360 were found to be bioactive, affecting morphology or development of the pollen tube. Our goal was to utilize this screening system to identify compounds among the 360 that would perturb the secretory pathway in pollen. We reasoned that the tracking of Golgi bodies that were perturbed in movement and other descriptors would offer an ideal model to develop a video-imaging approach to quantify and compare the dynamics of trafficking. Golgi compartments are relatively easy to visualize and are mobile within the pollen grain and tube in this rapidly growing organism. In the past, imaging fixed tissue and single still frame images have limited quantitative studies of endomembrane dynamics. The development of powerful computational tools such as commercially available Imaris image analysis software packages have allowed for the quantification of trafficking parameters in living pollen tubes over time, providing a more accurate measure of in planta conditions.
Six compounds were identified that altered the localization and/or movement of RAB2:GFP. Upon quantification of size, velocity, straightness, and intensity, the compounds RAEs 1, 2, 5, 6, 7, and 8 formed two phenotypic classes. The first group caused modest changes in size, velocity, straightness, and intensity, whereas the second caused dramatic changes in these descriptors. The first group, which comprised RAEs 1, 2, and 5, resulted in smaller agglomerations whose movement was modestly reduced compared to the untreated controls. This was reflected in their difference indices, which were 1.88, 2.14, and 1.12, respectively. Because this is a relative index, the score of the untreated pollen is 1.00. Greater deviations from 1.00 indicate greater relative differences in dynamics compared to the untreated condition. Interestingly, RAE 5 initially grouped with RAEs 6, 7, and 8 but received a score of 2.12, which grouped it with the weaker compounds RAEs 1 and 2. More specifically, whereas RAE 5 was similar to RAEs 6 and 7 in terms of percent velocity and percent straightness of untreated RAB2 Golgi bodies, it was much similar to RAEs 1 and 3 in terms of percent area of normal RAB2 Golgi bodies and essentially unchanged in percent intensity of normal (Table 2). RAE 5 RAB2 bodies were of size and intensity similar to the untreated bodies, but were less mobile and directional.
The second group consisted of RAEs 6, 7, and 8, which gave rise to relatively large agglomerations, drastically reduced movement and directionality, and greater intensity. Their difference index scores of 4.59, 5.22, and 4.37, respectively, indicated that these compounds resulted in overall endomembrane phenotypes with major differences compared to untreated pollen. This type of analysis indicates the value of quantification of vesicle dynamics in a non-biased manner, permitting more precise phenotype classification.
These differences in the degree of effect could be caused by a variety of mechanisms on which we could only speculate here. RAEs 1, 2, and 5 could be targeting proteins that have a minor role in regulating the localization of RAB2:GFP and therefore only cause a minor defect, whereas RAEs 6, 7, and 8 may be targeting proteins that play a major role in regulation of RAB2:GFP localization and endomembrane trafficking, thereby causing a dramatic defect. Alternatively, RAEs 1, 2, and 5 may effect trafficking via reversible target binding, whereas RAEs 6, 7, and 8 could bind irreversibly to their target(s), resulting in stronger phenotypes.
Interestingly, RAEs 6 and 7, two of the more potent compounds, have a nitrile and nitro group in common. The nitrile group can undergo a nucleophilic reaction that is facilitated by the nitro group to irreversibly bind with proteins (Beck, 1978; Habibi et al., 2012). Likewise, RAE 8 contains a functional group containing N-ethylmaleimide (NEM). NEM is a Michael acceptor, which adds nucleophiles such as the thiols in cysteine residues and is a commonly used cysteine modifier (Crankshaw and Grant, 2001) which could lead to irreversible target binding. This could very well be the case with RAE8. In fact, soluble N-ethylmaleimide-sensitive factor adaptor protein receptors (SNAREs) are, by definition, sensitive to NEM, suggesting that the target of RAE 8 might conceivably be a specific SNARE (Söllner et al., 1993). Because RAE 8 is predicted to be an irreversible inhibitor, Multidimensional Protein Identification Technology (MudPIT) would be a promising approach for target identification. However, it is beyond our scope to develop an image analysis approach to quantify endomembrane dynamics. Although it is intriguing to consider the reversibility and targets of RAEs, this is beyond our goal of developing a technically robust video-imaging tool to quantify trafficking.
This approach can be used for the rapid characterization and management of small molecules. This method has the ability to streamline chemical genetic screens and aid in experimental design. It is important to note that other automatic or semi-automatic image acquisition modalities can be used as well as freely available image analysis software. When analyzing data, the limitations of scale are determined by the resolution of the image and the frame rate at which the video data were collected, not necessarily the image analysis software. One advantage of our approach to classifying small molecules is the difference score. This will help facilitate the classification of compounds into meaningful clusters based on bioactivity as a quantitative measure of vesicle dynamics (Drakakaki et al., 2011). Though each feature value was given equal weight, one could increase the weight of any given feature, tailoring this score to any application. However, the difference index can only take into account phenotypes that can be quantified and the number of features measured determines the usefulness and flexibility. In addition, the score is highly dependent on the control phenotype and can only describe changes in preexisting conditions. It is not helpful in describing new phenotypes that were not represented in the control condition.
Among the six RAEs, 6 and 7 are the only previously characterized compounds. RAE 6, also known as 3-Pyridinecarbonitrile, 4,6-dimethyl-5-nitro-2-(phenylsulfonyl), is an inhibitor of severe acute respiratory syndrome coronovirus (SARS-CoV), a main protease (Mpr°) that is essential to the lifecycle of the SARS-CoV (Lu et al., 2006). This suggests that the target in tobacco might be a protease. RAE 7, a substituted pyrrole derivative, is an androgen receptor inhibitor, suggesting that its target in tobacco might be a hormone or other type of receptor (Matsunaga et al., 2006). Analyzing the known effects of RAEs 6 and 7 in mammalian systems could shed some light on their possible targets in pollen.
Overall, our study has contributed several important advances in the evaluation of vesicle trafficking dynamics: (1) we have focused on the quantification of video images to capture true real-time dynamics in vivo; (2) we examined the correlations of individual parameters such as velocity and straightness of movement; (3) the summary value score permitted a quantitative and comparative measure of overall vesicle dynamics; (4) the quantification of dynamic behaviors will make it possible to move towards the increased automation of chemical and genetic screens that can be focused directly on changes in vesicle dynamics, permitting the more efficient linkage of cellular and developmental phenotypes.
Quantitative analyses such as these provide a deeper understanding of highly dynamic intracellular movement by providing data that are objective. The difference index gives a single quantitative value that can immediately give insight into the degree of compound effects. A subset of compounds acts transiently, causing a range of phenotypes that vary in degree and duration. Excluding temporal changes excludes potentially important data. Our approach allows quantification and observation of temporal dynamics that occur as a result of transient binding of small molecules.
Full automation of image analysis and quantification of vesicle dynamics will become essential with increased throughput and the dramatically larger resultant image data sets. Quantitative descriptions such as these will facilitate the development of realistic predictive mathematical models of endomembrane trafficking in response to small molecules. Quantitative analysis allows for the identification of the dominant biological forces that contribute to a given biological phenomenon in addition to focusing the biological questions that are subsequently asked (Phillips and Milo, 2009).
Chemical genomics, though still a young field, is starting to gain popularity for dissecting dynamic biological processes. Quantitative analysis will extract comparative information for the elucidation of endomembrane trafficking pathways and their effects on organism development. This connection will allow understanding of the developmental consequences of changes in endomembrane dynamics at the cellular level.
METHODS
Semi-Automated Image-Based Screen
Chemical libraries were screened as indicated previously to derive a set of 360 compounds inhibiting pollen germination (Drakakaki et al., 2011). For the present study, chemical stocks were in 100% DMSO and distributed in 96-well clear-bottom plates at concentrations of 5 mg ml–1 (50–100 μM), 0.5 mg ml–1 (25–50 μM), and 0.05 mg ml–1 (10–20 μM). For the primary screen, wild-type Nicotiana tobaccum pollen was harvested at dehiscence. Thirty anthers from six flowers were harvested and allowed to germinate in 14 ml of GM2 medium (18% sucrose, 0.01% boric acid, 5 μM CaCl2, 5 μM Ca(NO3)2, and 1 mM MgSO4 (pH 6.5–7.0). Pollen was then vortexed to release the pollen grains from the anthers. The pollen suspension was added to the 96-well clear-bottom plates and incubated at room temperature in the presence of the appropriate compound on a covered orbital shaker for 3 h. Images of pollen tubes were acquired using the BD Pathway automated microscope (BD Biosciences) with the 40× and 63× objective. The sample was imaged using 470 nm/30 excitation filters and 520 nm/30 for GFP.
A custom semi-automated program was created for a 96-well plate using autofocus for the primary screen. Bright field images were then viewed and screened for defects in morphology as previously described (Robert et al., 2008). The compounds that were found to cause defects in pollen development were screened again using transgenic tobacco pollen expressing the RAB2:GFP fusion protein under the LAT52 pollen-specific promoter (Cheung et al., 2002). Still images as well as 4–5-s time-lapse movies containing 20 frames were captured over a period of approximately 2 min. Compounds were screened based on their effect on RAB2:GFP localization and movement. Of the 290 compounds that initially caused defects in pollen tube development, six were found to consistently affect the localization and movement of RAB2:GFP and were termed RAEs. RAE1 (Benzoic acid, 2-methyl-, 2-[(3-bromo-4-hydroxyphenyl)methylene] hydrazide, Chembridge 5271226) and RAE 2 (Acetic acid, 2-(2-methylphenoxy)-, 2-[(2-methyl-1H-indol-3-yl)methylene]hydrazide, Chembridge 5532951) both caused small agglomerations and decreased movement of the RAB2 bodies. RAE 5(Benzamide, N-[[[4-(acetylamino)phenyl]amino]thioxomethyl]-3-chloro-, Chembridge no. 6396311), RAE 6 (3-Pyridinecarbonitrile, 4,6-dimethyl-5-nitro-2-(phenylsulfonyl), LATCA no. LAT033E05), RAE 7 (Benzonitrile, 4-[(1E)-2-nitro-1-propen-1-yl]-, LATCA no. LAT035F05), and RAE 8 (1H-Pyrrole-2,5-dione, 1-(3,4,5-trimethoxyphenyl), LATCA no. LAT045A04) caused larger bodies and reduced movement to a greater degree. All confocal images were taken at the same gain and magnification so as not to artificially increase or decrease the intensity.
Image Analysis
Images were analyzed using Imaris (version 7.3.1, Bitplane, South Windsor, CT) with the ‘spots’ software package. RAB2:GFP bodies were quantified using the automatic threshold function provided by the ‘spots’ algorithm builder. The number of bodies measured per pollen grain ranged from 66 to 220. The mask value used was between 13 and 14, which determined the regions that the algorithm would measure. Segmentation along with region growing was used to quantify the fluorescent RAB2:GFP bodies. The average area of three replicates was recorded in μm2. Three biological replicates were used in quantifying the average speed of RAB2:GFP bodies using the ‘spots’ tracking function. The automatic settings were used for creating the segmentation algorithm. Because of the small size of the bodies measured, the maximum distance traveled between frames was 2 μm. Once the bodies were identified and tracked, an additional filter included in the spots package was used to exclude those bodies that were unable to be tracked more than 15 frames. Area, velocity, straightness, and intensity were represented as cumulative distribution plots. The plots were developed by determining the average of each parameter for each spot over three replicates. The mean and standard deviation for all spots in a movie were calculated and used in determining the z score. This z score, also called the CDF%, was plotted against the average of the selected parameter.
From these average values, a difference index was created. The following equation was used to calculate the difference score:
where a is the area of the organelles, or in this case, RAB2 bodies, b is the average velocity of all RAB2 bodies, c is the average intensity of all RAB2 bodies, and d is the average straightness of all rab2 bodies. Each normalized value was given an equal weight of 0.25. The sum of the squares of the normalized values was then found for each feature. The resulting number is the difference index.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by the National Science Foundation’s Integrative Graduate Education and Research Traineeship (DGE-0903667) to N.U.; (DGE-0504249) to M.Q.B.; and the National Science Foundation Grant (MCB-0817916) to N.V.R. and G.R.H.
Acknowledgments
We would like to thank Drs Imogen Sparkes, Zenbiao Yang, and Sean Cutler, as well as members of the Raikhel lab for helpful discussions concerning this project and manuscript. No conflict of interest declared.
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Oxford University Press on behalf of CSPB and IPPE, SIBS, CAS.
Supplementary Material
List of Bioactive Compounds, Their Compound Number, Supplier, Structure, Name, Availability, and Phenotypes at Dilutions of 1:02 and 1:05.
REFERENCES
- Cheung A.Y., Chen C.Y., Glaven R.H., de Graaf B.H.J., Vidali L., Hepler P.K., Wu H. Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell Online. 2002;14:945–962. doi: 10.1105/tpc.000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinet C., Stöter M., Bradshaw C.R., Samusik N., Rink J.C., Kenski D., Habermann B., Buchholz F., Henschel R., Mueller M.S. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- Crankshaw M.W., Grant G.A. Modification of cysteine. Curr. Protoc. Protein Sci. 2001 doi: 10.1002/0471140864.ps1501s03. Chapter 15, Unit 15.1. [DOI] [PubMed] [Google Scholar]
- Derksen J., Rutten T., Lichtscheidl I.K., De Win A.H.N., Pierson E.S., Rongen G. Quantitative analysis of the distribution of organelles in tobacco pollen tubes: implications for exocytosis and endocytosis. Protoplasma. 1995;188:267–276. [Google Scholar]
- Drakakaki G., Robert S., Szatmari A.M., Brown M.Q., Nagawa S., Van Damme D., Leonard M., Yang Z., Girke T., Schmid S.L. Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl Acad. Sci. U S A. 2011;108:17850–17855. doi: 10.1073/pnas.1108581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., Fujimoto M., Okatani Y., Nishiyama T., Goh T., Ito E., Dainobu T., Nishitani A., Uemura T., Sato MH. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 2011;13:853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- Fürthauer M., González-Gaitán M. Tales of 1001 functions: the multiple roles of membrane trafficking in development. Traffic. 2009;10:781–782. doi: 10.1111/j.1600-0854.2009.00931.x. [DOI] [PubMed] [Google Scholar]
- Habibi D., Ogloff N., Jalili R.B., Yost A., Weng A.P., Ghahary A., Ong C.J. Borrelidin, a small molecule nitrile-containing macrolide inhibitor of threonyl–tRNA synthetase, is a potent inducer of apoptosis in acute lymphoblastic leukemia. Investigational New Drugs. 2012;30:1361–1370. doi: 10.1007/s10637-011-9700-y. [DOI] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 2001;17:159–187. doi: 10.1146/annurev.cellbio.17.1.159. [DOI] [PubMed] [Google Scholar]
- Hicks G., Raikhel N. Small molecules present large opportunities in plant biology. Annu. Rev. Plant Biol. 2012;63:261–282. doi: 10.1146/annurev-arplant-042811-105456. [DOI] [PubMed] [Google Scholar]
- Kato N., He H., Steger A.P. A systems model of vesicle trafficking in Arabidopsis pollen tubes. Plant Physiol. 2010;152:590–601. doi: 10.1104/pp.109.148700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Galway M.E., Mulder B.M., Emons A.M.C. Rates of exocytosis and endocytosis in Arabidopsis root hairs and pollen tubes. J. Microscopy. 2008;231:265–273. doi: 10.1111/j.1365-2818.2008.02031.x. [DOI] [PubMed] [Google Scholar]
- Logan D.C., Scott I., Tobin A.K. The genetic control of plant mitochondrial morphology and dynamics. Plant J. 2003;36:500–509. doi: 10.1046/j.1365-313x.2003.01894.x. [DOI] [PubMed] [Google Scholar]
- Lord E. Adhesion and cell movement during pollination: cherchez la femme. Trends Plant Sci. 2000;5:368–373. doi: 10.1016/s1360-1385(00)01744-1. [DOI] [PubMed] [Google Scholar]
- Lu I.L., Mahindroo N., Liang P.H., Peng Y.H., Kuo C.J., Tsai K.C., Hsieh H.P., Chao Y.S., Wu S.Y. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J. Med. Chem. 2006;49:5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- Matsunaga, N., Ito, M., and Hitaka, T. (2006). Substituted pyrrole derivatives as androgen receptor inhibitors and their preparation, pharmaceutical compositions, and use for treatment of hormone-sesative cancers such as prostate cancer. 2006. Patent no. WO 2006064944.
- Miller D.D., de Ruijter N.C.A., Emons A.M.C. From signal to form: aspects of the cytoskeleton-plasma membrane-cell wall continuum in root hair tips. J. Exp. Bot. 1997;48:1881–1896. [Google Scholar]
- Nielsen E., Cheung A.Y., Ueda T. The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 2008;147:1516–1526. doi: 10.1104/pp.108.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R., Milo R. A feeling for the numbers in biology. Proc. Natl Acad. Sci. U S A. 2009;106:21465–21471. doi: 10.1073/pnas.0907732106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochynyuk O., Staruschenko A., Bugaj V., Lagrange L., Stockand J.D. Quantifying RhoA facilitated trafficking of the epithelial Na+ channel toward the plasma membrane with total internal reflection fluorescence-fluorescence recovery after photobleaching. J. Biol. Chem. 2007;282:14576–14585. doi: 10.1074/jbc.M701348200. [DOI] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1991;96:1193–1195. doi: 10.1104/pp.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl Acad. Sci. U S A. 2008;105:8464–8469. doi: 10.1073/pnas.0711650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K. The plant extracellular matrix: in a new expansive mood. Curr. Opin. Cell Biol. 1994;6:688–694. doi: 10.1016/0955-0674(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Rutherford S., Moore I. The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 2002;5:518–528. doi: 10.1016/s1369-5266(02)00307-2. [DOI] [PubMed] [Google Scholar]
- Salomon S., Grunewald D., Stüber K., Schaaf S., MacLean D., Schulze-Lefert P., Robatzek S. High-throughput confocal imaging of intact live tissue enables quantification of membrane trafficking in Arabidopsis. Plant Physiol. 2010;154:1096–1104. doi: 10.1104/pp.110.160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels A.L., Giddings T.H., Jr, Staehelin L.A. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J. Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T., Whiteheart S.W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P., Rothman J.E. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Teanby N.A., Hawes C. Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J. Exp. Bot. 2008;59:2499–2512. doi: 10.1093/jxb/ern114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Olkkonen V.M. The Rab GTPase family. Genome Biol. 2001;2:1–7. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick P., Gansel X., Oulevey C., Page V., Studer I., Dürst M., Sticher L. The expression of the t-SNARE AtSNAP33 is induced by pathogens and mechanical stimulation. Plant Physiol. 2003;132:343–351. doi: 10.1104/pp.102.012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
UNCITED REFERENCES
- Nebenführ A., Ritzenthaler C., Robinson D.G. Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol. 2002;130:1102–1108. doi: 10.1104/pp.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chow T.F., Puckrin R.S., Alfred S.E., Korir A.K., Larive C.K., Cutler S.R. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat Chem Biol. 2007;11:716–721. doi: 10.1038/nchembio.2007.32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Bioactive Compounds, Their Compound Number, Supplier, Structure, Name, Availability, and Phenotypes at Dilutions of 1:02 and 1:05.


