Abstract
It is more than 50 years since the lysosome was discovered. Since then its hydrolytic machinery, including proteases and other hydrolases, has been fairly well identified and characterized. Among these are the cysteine cathepsins, members of the family of papain-like cysteine proteases. They have unique reactive-site properties and an uneven tissue-specific expression pattern. In living organisms their activity is a delicate balance of expression, targeting, zymogen activation, inhibition by protein inhibitors and degradation. The specificity of their substrate binding sites, small-molecule inhibitor repertoire and crystal structures are providing new tools for research and development. Their unique reactive-site properties have made it possible to confine the targets simply by the use of appropriate reactive groups. The epoxysuccinyls still dominate the field, but now nitriles seem to be the most appropriate “warhead”. The view of cysteine cathepsins as lysosomal proteases is changing as there is now clear evidence of their localization in other cellular compartments. Besides being involved in protein turnover, they build an important part of the endosomal antigen presentation. Together with the growing number of non-endosomal roles of cysteine cathepsins is growing also the knowledge of their involvement in diseases such as cancer and rheumatoid arthritis, among others. Finally, cysteine cathepsins are important regulators and signaling molecules of an unimaginable number of biological processes. The current challenge is to identify their endogenous substrates, in order to gain an insight into the mechanisms of substrate degradation and processing. In this review, some of the remarkable advances that have taken place in the past decade are presented. This article is part of a Special Issue entitled: Proteolysis 50 years after the discovery of lysosome.
Keywords: Cysteine cathepsin, Protein inhibitor, Cystatin, Small-molecule inhibitor, Mechanism of interaction, Biological function
Highlights
► Current advances in the field of cysteine cathepsins and their regulation. ► Cysteine cathepsin activity as a delicate balance of various factors. ► Structure of cysteine cathepsins and their mechanism of interaction with inhibitors. ► Inhibition of cysteine cathepsins by protein and small-molecule inhibitors. ► The increased expression of cysteine cathepsins implicated in various diseases.
1. Introduction
In 1955, Christian de Duve discovered a distinct class of a single population of homogeneous granules containing five acidic hydrolases. It was proposed to refer to these granules as lysosomes, because of their richness in terms of hydrolytic enzymes [1]. The finding that these hydrolases act on different substrates suggested that the lysosomes, also known as “suicide bags”, might play a role in intracellular digestion at acidic pH. Lysosomes were then discovered in several cell types and found to be involved in the degradation of extracellular material taken up by endocytosis and intracellular material taken up by autophagy. Thus, in a short time, the lysosome concept was quickly accepted [2], [3], [4], [5]. In parallel, based on these findings, numerous lysosomal hydrolases were isolated, characterized, and their mechanisms of action were established. With the discovery of the ubiquitin-proteasome system [6] it was concluded that there are two major systems for intracellular protein degradation: the lysosomal system and the ubiquitin-proteasome system. It is well established that the endosomal/lysosomal system has numerous functions in normal and pathological processes, including the survival function [7].
In the lysosomal system, protein degradation is a result of the combined random and limited action of various proteases (also termed peptidases or proteolytic enzymes). In order to achieve the efficient degradation of biological macromolecules within the lysosomes, lysosomes contain a number of diverse hydrolases, including proteases, amylases, lipases and nucleases. Among the approximately 50 known lysosomal hydrolases, of particular importance are the aspartic, serine and cysteine proteases [8]. In addition to the aspartic cathepsin D, cysteine cathepsins have a key role among the lysosomal proteases. They belong to the clan CA of cysteine peptidases, which are widely distributed among living organisms, and represent one of the most investigated groups of enzymes. More specifically, they are members of the C1 family of papain-like enzymes, the largest and the best characterized family of cysteine peptidases, which include, among others, papain and related plant enzymes, proteases in parasites and helminths, insect-homologs of papain, trematode cysteine proteases, viral proteases, and lysosomal cysteine cathepsins [9]. This chapter will focus on human cysteine cathepsins, which have been the subject of numerous reviews in the past [10], [11], [12], [13], [14], [15], [16], [17], [18].
2. Cysteine cathepsins
The name cathepsin, which is derived from the Greek kathepsein (to digest), was proposed for the protease that was active in a slightly acidic environment [19]. Later, the name cathepsin was introduced for the serine proteases cathepsins A and G, the aspartic proteases cathepsins D and E, and the lysosomal cysteine cathepsins. There are 11 human cysteine cathepsins, i.e., the cathepsins B, C, F, H, K, L, O, S, V, X and W, existing at the sequence level; this was confirmed by a bioinformatic analysis of the draft sequence of the human genome [20]. Lysosomal cathepsins require a reducing, slightly acidic environment, such as found in the lysosomes, in order to be optimally active. Therefore, cysteine cathepsins were initially considered as intracellular enzymes, responsible for the non-specific, bulk proteolysis in the acidic environment of the endosomal/lysosomal compartment, where they degrade intracellular and extracellular proteins [17], [21]. However, this view is rapidly changing.
2.1. Localization and main properties
The majority of cathepsins are ubiquitously expressed in human tissues such as cathepsins B, H, L, C, X, F, O and V. Their expression profile indicates that these enzymes are involved in a normal cellular protein degradation and turnover. In contrast, cathepsins K, W and S show a restricted cell or tissue-specific distribution, indicating their more specific roles. For example, cathepsin K is highly expressed in osteoclasts, in most epithelial cells and in the synovial fibroblasts in rheumatoid arthritis joints [22]. Among the matrix-degrading enzymes, cathepsin K is the only enzyme for which an essential role in bone resorption has been unambiguously documented in mice and humans [23]. Cathepsin W (also lymphopain) is predominantly expressed in CD8+ lymphocytes and natural killer (NK) cells [24], [25]. However, when cathepsin W expression has been downregulated using the shRNA approach, its essential role in the process of cytotoxity has been excluded [26]. Cathepsin S is predominantly expressed in the professional antigen-presenting cells (APCs), such as the dendritic cells (DCs) and B-cells [27]. In addition, cathepsin V (also named L2) is highly homologous to cathepsin L, but in contrast to the ubiquitously expressed cathepsin L, its expression is restricted to thymus and testis [28], [29]. However, recent studies have shown that active cathepsins are also localized in other cellular compartments, such as the nucleus, cytoplasm and plasma membrane. It was thus shown that the catalytically active cathepsin L variants localized to the nucleus play a role in the regulation of cell-cycle progression [30] and the proteolytic processing of the N-terminus of the histone H3 tail [31], [32]. In addition, it was shown to interact with the histones H2A.Z, H2B, H3 and the protease inhibitor stefin B [33]. These studies and the nuclear localization of cathepsin F [34] and possibly other cathepsins, provide the first evidence that histones may be very important substrates of cathepsins, thus opening up a new role for these enzymes.
Moreover, there is sufficient evidence that the specific physiological functions of the cathepsins might be at least partially attributed to their differences in localization inside and outside the cells [9], [35], [36]. This is consistent with the recent advances in the field of proteases that led to a new concept, based on proteases as important signaling molecules involved in the controlling mechanisms in many normal and pathological processes [37], [38], [39].
Cysteine cathepsins are optimally active in a slightly acidic pH and are mostly unstable at neutral pH. When cathepsins are outside the lysosomes or extracellularly they can be relatively rapidly irreversibly inactivated at neutral pH [40]. An exception is cathepsin S, which is stable at a neutral or slightly alkaline pH, thus retaining most of its activity [41]. The inactivation of cathepsin L, the most unstable of the cathepsins at neutral pH, was found to be a first-order process and the rate of the process decreased with the substrate concentration that conferred some protection [42]. In cathepsin B, an irreversible loss of activity accompanied by structural changes was observed. The enzyme was destabilized by increasing the ionic strength and the content of organic solvent [43]. However, after the binding of heparin to cathepsin B through an interaction with its occluding loop, the enzyme was partially protected from alkaline-pH-induced inactivation as a consequence of the restriction of the enzyme flexibility, which makes stronger contacts within the molecule [44], [45]. In addition, it was recently suggested that catalase may participate in the protection of extracellular cysteine cathepsins against peroxidation [46].
The extracellular localization of cysteine cathepsins often coincides with their increased expression and/or activity, indicating that pH is not the sole factor responsible for their activity. Twenty years ago, it was demonstrated that cathepsin B, from normal or tumor tissues degraded purified extracellular-matrix components, type IV collagen, laminin and fibronectin, under both acid and neutral pH [47]. Cathepsins with strong elastolytic and collagenolytic activities are now known to be chiefly responsible for the remodeling of the extracellular matrix (ECM), thus contributing to various pathologies. The ECM consists of elastins, collagens and proteoglycans, which have all been identified as the cathepsin's substrates [48]. Elastin was found to be cleaved by the cathepsins K, L, S, F, V and B, although they differ in their elastolytic activitiy. Cathepsin V was thus found to be the most potent enzyme, followed by the cathepsins K, S, F, L and B [49], [50]. It was further demonstrated that the elastolytic activities of the cathepsins V and K could be inhibited by ubiquitously expressed glycosaminoglycans (GAGs), such as chondroitin sulfate, through the formation of cathepsin-GAG complexes. In contrast, cathepsin S, which does not form complexes with chondroitin sulfate, is not inhibited, thus suggesting that there might exist some specific regulation of the elastolytic activities of cathepsins by GAGs.
The most potent mammalian collagenase is cathepsin K (also known as cathepsin O2) which, in contrast to the cathepsins B, L and S, is highly expressed in osteoclasts [51], [52], suggesting that it plays a special role in bone resorption under normal and pathological conditions [52]. Cathepsin K forms collagenolytically highly active complexes with GAGs, with different GAGs competing for the binding to the enzyme [53]. Some of them, such as chondroitin and keratan sulfate, enhance the collagenolytic activity of cathepsin K, whereas heparan sulfate or heparin selectively inhibits this activity. Moreover, GAGs potentially inhibit the collagenase activity of cathepsins L and S [53]. In addition, the specific inhibition of the collagenase activity of cathepsin K by negatively charged polymers, such as polyglutamates and oligonucleotides, without affecting the overall proteolytic activity of the protease was observed [54]. It was suggested that during the interaction between cathepsin K and polymers, collagen degradation can be specifically inhibited, while its non-collagenolytic activities remain unchanged.
Although there is major evidence that various proteins can be degraded in vitro by cathepsins in acidic pH, there are scarce data about their intracellular physiological substrates. One of the main reasons for this is the cathepsin's instability and the loss of activity due to irreversible unfolding [40], [55]. The first, well-established, intracellular substrate of cathepsins was the proapoptotic Bcl-2 homolog protein Bid, which was initially found to be processed by the lysosomal extract, thereby triggering cytochrome c release from mitochondria and apoptosis [56]. There is significant redundancy among the cathepsins in cleaving Bid, as in in vitro experiments, and in a cell-free system Bid was found to be efficiently processed by the cathepsins B, K, L and H to a proapoptotic form [57]. This finding was confirmed in various cellular models [57], [58]. Moreover, it was found that, in addition to Bid, cathepsins simultaneously degrade the antiapoptotic Bcl-2 family proteins, such as Bcl-2, Bcl-xL, Mcl-1 and XIAP, thereby triggering apoptosis in a synergistic manner [58]. However, there are numerous biological processes in which cathepsins play an important role, but their substrates have not been identified so far. This part of the review will not focus on the well-known basic properties of cysteine cathepsins, which are presented elsewhere [9].
3. Structure and specificity
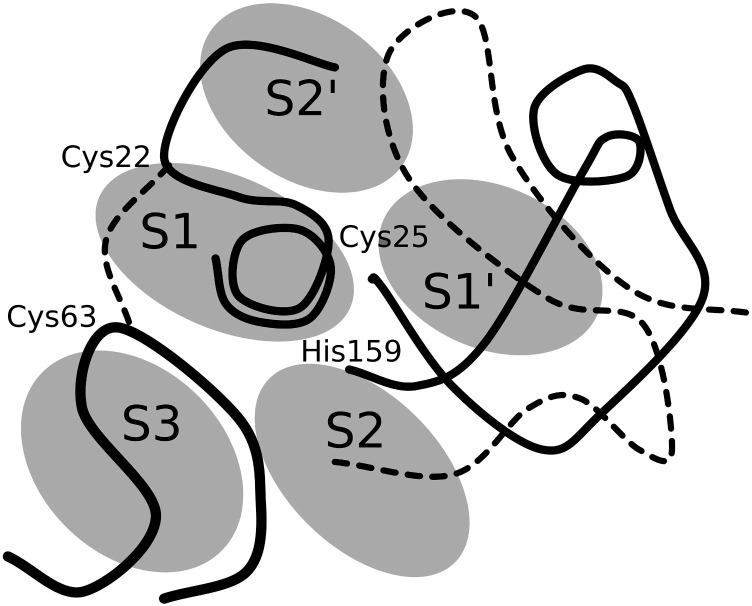
Understanding the interactions between cysteine cathepsins and their substrates has been and remains the main challenge in the research on cysteine cathepsins. Papain served as the model in the pioneering work of Schechter and Berger [59], in which they proposed the nomenclature for the positions of the substrate residues (P) and the subsites (S) where they bind to the surface of a protease. The positions and subsites were numbered in both directions from the scisille bond between the residues P1 and P1′ onwards, where the non-primed side refers to the N-terminal part and the primed side to the C-terminal part of the substrate. In their pioneering work, they found that adding residues to peptides longer than 7 alanine residues does not affect the kinetics of their degradation and concluded that there are seven substrate residues binding into seven subsites from S4 to S3′. Three decades later, we revised the definition of the substrate binding sites using an insight provided by the crystal structures of the complexes with small-molecule, substrate-mimicking inhibitors [11]. These structures show that a substrate binds along the active-site cleft in an extended conformation (Fig. 1 ).
Fig. 1.
3D schematic representation of the substrate-binding sites of papain-like proteases along the active-site cleft. The representation is based on the proposed revised definition of substrate-binding sites based on the crystal structures of substrate-mimicking inhibitors bound to the protease's active sites [11]. The substrate-binding sites of the papain-like proteases are located on the left (S1, S3, S2′) and right (S2, S1′) side of the active-site cleft in accordance with the standard view orientation. According to papain numbering, L-domain loops include residues Gln19-Cys25 and Arg59-Tyr67 whereas R-domain loops contain residues Leu134-His159 and Asn175-Ser205, respectively. The active-site residues Cys25 and His159 and the disulphide Cys22-Cys63 are indicated.
Cysteine cathepsins exhibit broad specificity, thus cleaving their substrates preferentially after basic or hydrophobic residues. This is true not only for synthetic but also for protein substrates and consistent with their roles in intracellular protein degradation [17]. The information about their specificity is mostly gathered on a case-by-case basis, which limits their applicability. Nevertheless, within the past decade some substantial progress has been made in the way we understand the specificity of cysteine cathepsins, in the variety of approaches applied and in the study cases where specificity has been utilized to extract new knowledge.
3.1. Structures of cathepsins
The crystal structure of papain, a cysteine protease from Carica papaya [60], [61], was among the first dozen protein crystal structures to be determined. Together with actinidin [62], these two structures provided the first insight into their three-dimensional (3D) structure. Later developments enabled the isolation of cysteine cathepsins, such as cathepsin B, H, L, S, X, C, from various tissues, while the rest of the cathepsins were expressed in various expression systems. In the 1990s the crystal structure of human cathepsin B was determined [63]; this was followed by cathepsins L [64], K [65], [66], H [67], X [68], V [69], C [70], [71], S [72] and F [73].
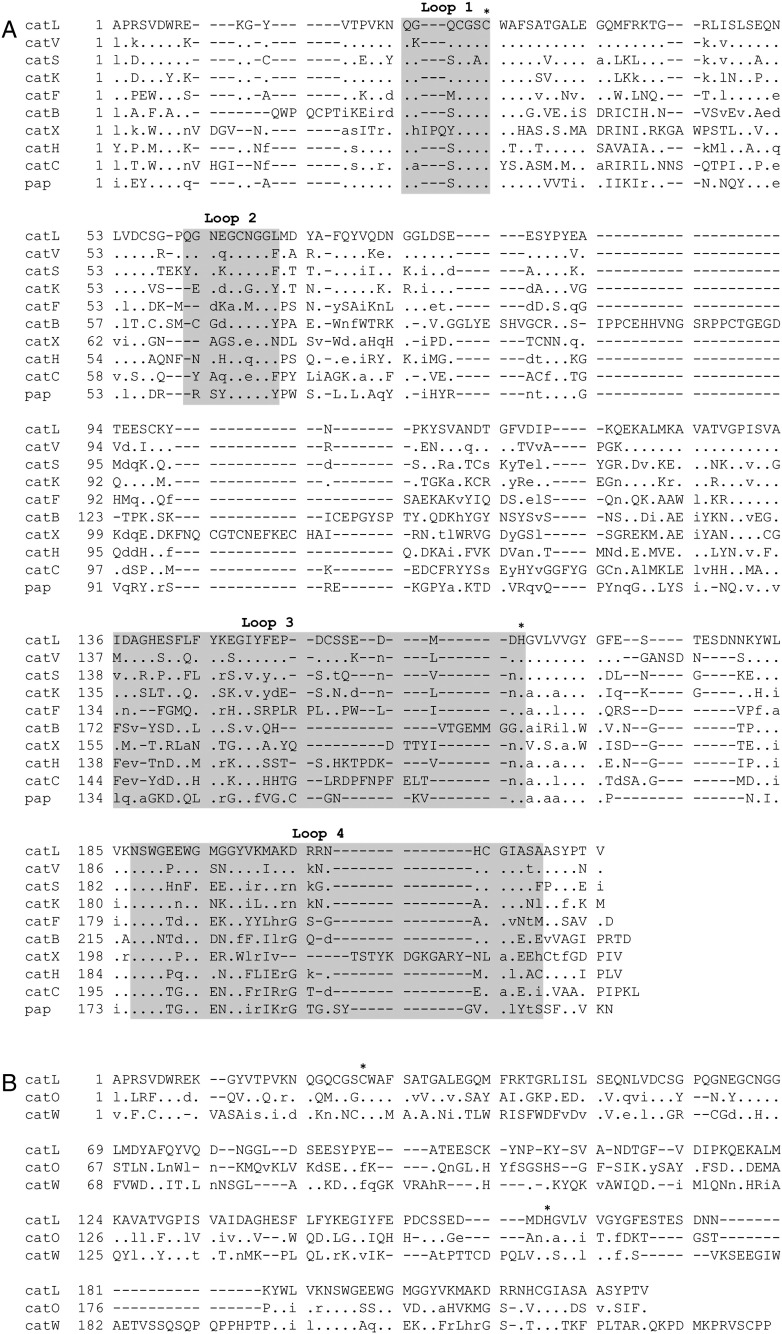
A 3D-based sequence alignment of the mature form of the nine cysteine cathepsins with a known 3D structure exhibits conservation of the active-site residues (Cys25 and His163, cathepsin L numbering), those residues interacting with the main chain of the bound substrate, (Gln19, Gly68, Trp183), the N-terminus Pro2 and certain Cys residues as well (Fig. 2A). Even though the 3D structures of the remaining two human cysteine cathepsins, O and W, are still unknown, a 3D sequence alignment carried out independently using the crystal structure of human cathepsin L as a template [74] shows the same conservation pattern (Fig. 2B).
Fig. 2.
3D-based sequence alignment of the mature form of cysteine cathepsins. (A) The structural alignment of eight human cysteine cathepsins, porcine cathepsin H and papain, as a representative member of the papain-family, was done with the program MAIN [363]. The PDB codes of the structures are as follows: cathepsin B (1huc) [63], cathepsin C (1k3b) [70], cathepsin F (1m6d) [73], cathepsin H (8pch) [67], cathepsin K (1mem) [65], cathepsin L (1icf) [74], cathepsin S (1glo) [72], cathepsin V (1fh0) [69], cathepsin X (1ef7) [68], and papain (9pap) [61]. According to cathepsin L numbering, the residues that form the substrate-binding site, within the L-domain loops (Q19-C25, Q60-L69) and R-domain loops (I136-H163, N187-A214), are boxed and highlighted in light gray. (B) The structure-based sequence alignment of human cathepsins O and W was guided using the known 3D structure of human cathepsin L (1icf) [74] using the Expresso (3D-Coffee) program [364]. In both cases the leading sequence was human cathepsin L (1icf) [74] and it is shown in uppercase letters whereas the non-conserved residues are indicated in lowercase letters. Identical residues are shown as dots and the gaps are marked by the dash symbol. Active site residues Cys25 and His163 are indicated by an asterisk.
The papain fold is shown in Fig. 3 using cathepsin L [74], as a typical representative of the cysteine cathepsin endopeptidase. It is composed of two domains, referred to as the left (L-) and right (R-), in accordance with the standard view. The L-domain contains three α-helices. The longest, i.e., the vertical helix, known also as the central helix, is over 30 residues long. The R-domain is a kind of β-barrel with the front strand(s) forming a coiled structure. At the bottom, the barrel is enclosed by an α-helix. The reactive site histidine is located at the top of the sheet forming the barrel. The two-domain interface opens at the top, forming the active-site cleft. In its center are the reactive site residues, Cys25 and His163, each coming from a different domain. The reactive site cysteine is located at the N-terminus of the central helix of the L-domain, whereas the histidine is located within the β-barrel residues of the R-domain. These two catalytic residues form the thiolate-imidazolium ion pair, which is essential for the proteolytic activity of the enzymes.
Fig. 3.
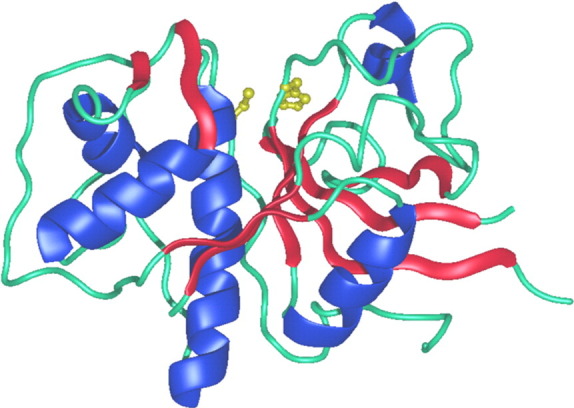
Fold of the mature form of cysteine cathepsins–endopeptidases. The fold of the two-chain form of native cathepsin L (1icf) [74] is shown in the green ribbon representation. The parts corresponding to the secondary structure elements, α-helices and β-sheets, are shown in blue and red color, respectively. The side chains of the reactive-site cysteine (Cys25) and histidine (His163) are indicated in a ball-and-stick representation.
The active-site surface is composed of the residues coming from four loops located on both domains (Fig. 1, Fig. 2A). The two L-domain loops are short and cross-connected with a disulfide bond. The two R-domain loops are much larger and form the lid of the β-barrel hydrophobic core. The substrate binds along the active-site cleft in an extended conformation [11], [75]. Its side chains make alternating contacts with the L- and R-domains. The two loops on the L-domain and their bridged interface provide the surface for the side chains of the P3, P1 and P2′ residues, whereas the loops and residues of the R-domain provide the binding surface for the P2 and P1′ residues. An exception to this is the P2 residue. According to our model [75], the P2 residue interacts with both domains. Its main-chain atoms form an antiparallel hydrogen-bonding ladder with Gly68, while the side-chain atoms bind into the S2 pocket formed at the two-domain interface. The alternate positioning of the side chains suggests that there may be a cooperativity effect between the residues oriented toward the surface, in particular the P1 and P2′ appear to be close enough to be able to interact. However, the experimental evidence for the latter is still lacking.
Most cysteine cathepsins exhibit a predominantly endopeptidase activity, whereas cathepsins X and C are exopeptidases only. Cathepsin C is an aminodipeptidase [70] and cathepsin H is, in addition to an endopeptidase, an aminopeptidase [67]. Cathepsin B is an endopeptidase and a carboxydipeptidase [63], whereas cathepsin X is a carboxymonopeptidase [68]. The crystal structures revealed that exopeptidases contain additional structural features that modify the active-site cleft [63], [67], [70] (Fig. 4 ). Whereas in endopeptidases (cathepsins F, L, K, S and V) the active-site cleft extends along the whole length of the two-domain interface, in exopeptidases (cathepsins B, C, H and X) additional features reduce the number of binding sites [75]. In the case of carboxypeptidases, substrate binding is governed by the loops that fill the active-site cleft on the primed sides. These loops in both carboxypeptidases, the occluding loop of cathepsin B and the mini-loop of cathepsin X, utilize histidine residue(s) to dock the C-terminal carboxylic group of the peptidyl substrate. In contrast, cathepsins H and C utilize regions from their propeptide parts in order to provide features that fill the active-site cleft on the non-primed sides. They use carboxylic groups of the main- and side-chain residues to dock the positively charged N-terminus of the peptidyl substrates. In cathepsin H the carboxylic group is located at the C-terminus of an eight-residues-long peptide [67], part of the propeptide, termed the mini-chain, which remains attached to the body of the enzyme after its activation. However, in cathepsin C, the carboxylic group comes from the D1 side chain positioned at the N-terminus of a whole additional domain, called the exclusion domain, which fills the active-site cleft [70]. Interestingly, in both aminopeptidases, glycosylation appears crucial for stabilizing the structure of these additional features and simultaneously fills the active-site cleft to tighten the substrate binding.
Fig. 4.
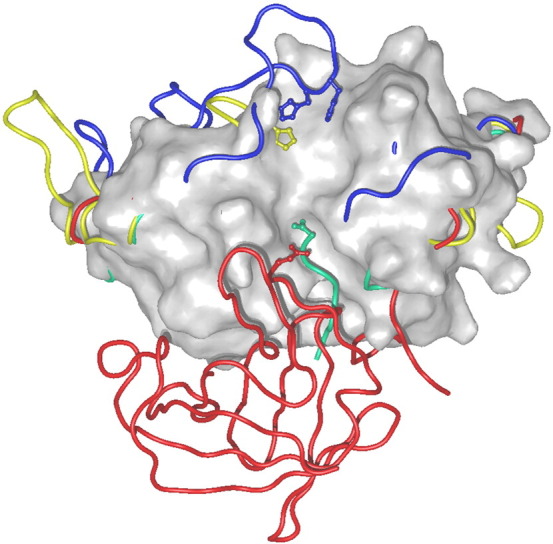
Additional features of cysteine cathepsins–exopeptidases. Chain traces of exopeptidases cathepsins B, X, H and C are indicated in blue, yellow, green and red color, respectively, over the surface of the endopeptidase cathepsin L (1icf) [74].
3.2. Understanding substrate binding
While in the exopeptidases additional features inserted into the endopeptidase scaffold define how many residues will bind on the primed or non-primed side of the active-site cleft, understanding the preferences for the peptidyl substrate side chains is a significantly more subtle matter.
The 3D structures have shown that the substrates bind in the active-site cleft in an extended conformation. The superposition of the structures provided support for only three well-defined substrate binding sites, S2, S1 and S1′, where the substrate residues interact with the enzyme by the main- and side-chain interactions (Fig. 1). Among these only the S2 site forms a pocket. In the positions outside this region, there are only side-chain interactions. The spread of residue positions across the active-site cleft on the non-primed side demonstrates that the S3 and S4 binding sites are not really sites. More appropriately, they are described as areas in which the substrate residues individually find their most favorable binding position [11], [75]. On the primed binding side, there is still a limited structural insight into the substrate binding. Data are provided by the synthetic epoxysuccinyl-based inhibitors CA030, CA074 and NS134 complexes with cathepsin B [76], [77], [78]. Their binding revealed the position of the P1′ and P2′ residues. However, the information therein is also limited. On one hand it is redundant as the P1′-like and P2′-like residues are essentially the same in all structures. On the other hand, their information is biased. The P2′-like residue docks its C-terminal carboxylic group against the side chains of the His110 and His111 positioned within the occluding loop of the cathepsin B. Since the occluding loop is specific for cathepsin B, in other cathepsins the positioning of the P2′ residue may be different.
Hence, there are only three defined substrate binding sites (S2, S1, S1′), where the amino acid residues of the substrate interact with the main- and side-chain atoms at the surface of the enzyme (Fig. 1). In the case of cathepsin B the main-chain interactions are offered by the occluding loop histidines, while in the others there are no recognizable exposed residues provided to serve as a dock for the main-chain atoms of the substrate residues beyond S2 and S1′. Therefore, these sites were recommended to be termed as areas [11]. Clearly, in the absence of docking partners for the main-chain atoms, these binding areas provide more opportunities for different binders. They may, however, reduce the selectivity of the substrate.
3.3. Substrate binding sites
The substrate specificity of a protease is a delicate interplay of the interactions between the surface of the protease and its substrates. Studies trying to reveal this interplay are based on two complementary approaches: (i) studies of the structure–function relationship and (ii) studies of the substrate specificity, and a combination of these two approaches has been used for the past 20 years to tackle the specificity of cysteine cathepsins for target proteins. In the first approach, the residues of interest located in the sequence and 3D structure of a protease are mutated to reveal their relevance and role in certain interactions. In the second approach, the enzymes are left intact, but the substrates are manipulated in size and composition. The synthetic substrates include naturally occurring amino acid residues as well as non-natural amino acid residues. Within the frame of these studies are also the degradation studies of true protein substrates by proteomic approaches. The latter are increasing in importance because in in vitro experiments they get as close to the naturally occurring events as is currently possible.
3.3.1. Structure–function relationship
Although considerable information about protease function can be gained through a structure–function relationship, studies of cysteine cathepsins were mainly performed using the substrate-specificity approach [10], [11], [79]. The majority of studies performed in the 1990s were mostly focused on cathepsin B, which was the first cathepsin for which the crystal structure was determined [63]. The structure of the enzyme [63] revealed that part of the S2 pocket is formed by Glu245, a unique residue found only in cathepsin B but not in other cathepsins. Its mutation to glutamine showed the importance of processing the substrate with an arginine in the P2 position, but had no effect on the phenylalanine substrates [80]. Another example is a mutation at the S1 site of cathepsin B, which made the site more favorable for glycine, corresponding to the glycine-favorable papaya protease IV (glycyl endopeptidase) [81]. Although not entirely linked with the subsite specificity, the mutations of the histidine residues in the occluding loop, as well as the deletion of the loop, revealed the importance of the loop and the two histidine residues for the endopeptidase activity of cathepsin B [82], [83]. A similar attempt was made to understand the differences between the S2 sites of the cathepsins L and K [84]. Double mutations of the S2 pocket of cathepsins K (Y67L/L205A) and L (L67Y/A205L) induce a switch of their enzyme specificity towards small selective inhibitors and peptidyl substrates, confirming the crucial role of residues at positions 67 and 205. However, both mutants, the cathepsin-K-like cathepsin L as well as the cathepsin-L-like cathepsin K, were not capable of degrading collagen. This indicated that introducing the proline tolerance alone did not convert the cathepsin L to a collagenase, while abolishing the proline selectivity in cathepsin K prevented it from degrading the collagen. A similar follow-up study addressed the cathepsin K chondroitin binding site [85]. These example studies demonstrated that we have a basic understanding of how specificity is encoded in the structure. However, this understanding is still not sufficient to enable us to predict the selectivity for substrates based on the sequence and atomic structure of the cysteine cathepsins.
3.3.2. Discovery of selective substrates
With the switch from in vitro biochemical studies to physiological studies in a complex biological millieu, the access to highly selective substrates was found to be crucial for monitoring the activities of individual cysteine cathepsins in biological samples. The focus on substrate studies therefore shifted to the discovery of substrates specific for individual cathepsins. These studies use peptidyl libraries combined with the positional scanning of individual positions, mostly one at a time [86]. The survey below is a summary of a few examples, which are indicative of the extensive research in the past decade. The summary is organized on the basis of individual cathepsins.
Cathepsin B. Although numerous studies on cathepsin B specificity have been performed in the past [79], [80], [87], a recent high-throughput study demonstrated that arginine is indeed the most suitable residue in the P1 position [88]. Moreover, based on the screening of the P3 to P1′ sites, the Abz-GIVRAK(Dnp)-OH peptide was found to contain the most favorable residues for cathepsin B.
Cathepsin X. The development of substrates and their analysis [89] were crucial for clarifying that cathepsin X is a carboxymonopeptidase only. As a result, the question about its additional dicarboxypeptidase [90] and endopeptidase activities has been resolved. The ambiguity very likely resulted from the presence of impurities in the samples, either obtained by isolation from a natural source (cathepsin B) or by expression in the proform with the leftovers of the cathepsin L as the activating enzyme.
Cathepsin K. The substrate specificity of cathepsin K has also been extensively studied as the enzyme is an important drug target for the treatment of osteoporosis. Among the mammalian cysteine cathepsins, cathepsin K was found to have a unique preference for a proline residue in the P2 position, the primary determinant of its substrate specificity [91]. As a result of such screening, the highly selective Abz-HPGGPQ-EDDnp substrate was identified. The selectivity of the substrate primarily depends on the S2 and S2′ site specificities and the ionization state of the histidine in the P3 position. Its hydrolysis was completely abolished in the cathepsin-K-deficient cell lysates, indicating its usefulness for monitoring cathepsin K activity in physiological fluids and cell lysates. Another screen of the S3–S3′ site specificity using Abz-KLRFSKQ-EDDnp as the template showed that the requirements of the S3–S1 sites are more restricted than those of the S1′–S3′ sites [92]. In addition, it was found that positively charged residues are favored in the P1 position, with glycine being almost equally well accepted. At the S3 site, positively charged residues were readily accepted, whereas the other sites had less pronounced preferences. However, when a proline was used in the P2 position it conferred specificity for cathepsin K [92].
Cathepsin S. When screening cathepsin S using a template based on the Ii sequence, clear preferences for groups of amino acid residues were observed in the positions P3, P1 and P1′ [93]. Based on these results, highly cathepsin-S-selective peptides were developed. On this basis, the authors suggested that the observed cleavage sites in Ii might be of relevance for its in vivo processing [93]. In a follow-up study, it was shown that the specificity of cathepsin S was mainly determined by the S2, S1′, and S3′ sites. At the P2 position the hydrophobic valine, methionine and norleucine residues were clearly preferred. Similarly, hydrophobic branched side chains were preferred at the P1′ position. The resulting GRWHTVGLRWE-Lys(Dnp)-DArg-NH2 substrate with the cleavage site between Gly7 and Leu8 was claimed to be selective for cathepsin S more so than for cathepsins B, H, L and X [94]. The same authors have also confirmed that the substrate processing in endosomal fractions of antigen-presenting cells was abolished in the presence of LHVS (morpholinurea-leucine-homophenylalanin-vinyl sulfone-phenyl), a selective inhibitor of cathepsin S. In a similar attempt to distinguish cathepsin S from other cathepsins, a substrate was derived on the basis of its putative physiological substrates, i.e., the insulin β-chain and the class-II-associated invariant chain (CLIP) [95]. The resulting Abz-LEQ-EDDnp (Abz = ortho-aminobenzoic acid; EDDnp = N-[[2,4-dinitrophenyl]ethylenediamine]) peptide was capable of differentiating cathepsin S from the cathepsins L, B, V and K.
Cathepsin V. When a substrate screen was made using cathepsins L and V, it was found that the cathepsin V S1 and S3 sites exhibit a broad specificity, while cathepsin L has a preference for positively charged residues in the same positions. The S2 sites of both enzymes were found to require hydrophobic residues with a preference for Phe and Leu. On the other hand, the S1′ and S2′ sites in both cathepsins were found to be less specific [96].
The listed examples lack a systematic approach that would not be restrained by the limited variability of the substrate composition, nor by the confined list of targets used in the selection process. However, novel approaches are on the way. An attempt that was considerably less restrained by substrate selection was the high-throughput proteomic study of cathepsin K specificity using an oligopeptidyl library derived from natural proteins [97]. Unrestrained by the length of the peptide, the screen revealed cathepsin K's preference for the aspartate at the P5′ position. This specificity is believed to be relevant for the recognition of collagen substrates. Yet another way to determine the substrate specificity of cysteine cathepsins is to use a peptide phage library. Very recently, it has been reported that a cathepsin-L-like enzyme from the tick Rhipicephalus (Boophilus) microplus has a preference for leucine or arginine at the P1 position [98]. Similar attempts will certainly shed light on the novel aspects of the substrate specificity of cysteine cathepsins.
3.4. Small-molecule inhibitors of cysteine cathepsins
The discovery of an epoxysuccinyl-based inhibitor E-64 (L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane), was very important for the research on cysteine cathepsins [99]. It selectively alkylates the reactive site cysteine and remains covalently bound to the enzyme. Because it reacts almost exclusively with the reactive site cysteine of papain-like proteases, it has immediately become a widely used indicator of the proteolytic activity of cysteine cathepsins. Together with its derivatives that have improved cell-permeability properties, it was of immense importance for progress in understanding the function and properties of cysteine cathepsins, ranging from a titration of their activity to an assessment of their activity in cell lysates, cultures and living organisms. E-64 is a non-selective inhibitor of all the cysteine cathepsins, with the exception of cathepsin C, which is only weakly inhibited. In addition, E-64 also efficiently inhibits calpain [100]. The exploitation of its scaffold led to the seminal contribution of Katunuma's group, who developed the first specific inhibitors of cathepsin B, CA030, CA074 and their analogs [101], [102]. The crystal structure of cathepsin B with CA030 (ethyl-ester of epoxysuccinyl-L-Ile-L-Pro-OH), a CA074 analog [76] and later with CA074 in a complex with bovine cathepsin B [77], have shown that these inhibitors bind into the primed side (sites S1′ and S2′) of the active-site cleft in the direction of the substrate. These structures have confirmed that the carboxylic group of proline is indeed responsible for binding to the occluding loop histidines, His110 and His111, which were held responsible for the binding of the C-terminal carboxylic group of the peptidyl substrate. The alignment of the E-64 and CA030 binding geometries indicated that the epoxysuccinyl group possesses an internal symmetry with two C-terminal carboxylic groups to which both amino acid residues can be attached. The idea of using epoxysuccinyl as a building block that allows simultaneous access to the non-primed as well as the primed side of the active-site cleft was successfully adopted in the development of inhibitors of cathepsin B [103], [104] as well as the inhibitors of cathepsins K, L and S [105], [106]. The predicted binding geometry of the double-headed inhibitors has been confirmed by the crystal structures of papain-CLIK complexes as the model for cathepsin L [107] and the cathepsin B-NS-134 complex [78]. There are several, more detailed, papers on this subject worthy of further reading [75], [108], [109].
Additional uses of the epoxysuccinyl-based inhibitors include the activity-based probes developed by Bogyo's group, mentioned below. Leupeptin, which is found in bacteria, was another inhibitor known from the 1970s [110]. It is an aldehyde that covalently binds to nucleophilic serine and cysteine residues at the active site of the cysteine and serine proteases. Due to its protease-class non-specific interactions it was less widely used than E-64. However, its reaction with the active site of cysteine proteases is reversible and allows a reactivation of the enzyme, while the attached residues can provide selectivity [111], [112]. There are a number of reactive groups, also called “warheads”, which interact with more than one class of protease. For example, one of the pioneers of the synthesis of protease inhibitors, Elliott Shaw, exploited diazomethane as the reactive group [113], [114], [115].
The covalent interactions with the active site either point into the non-primed region or, as in the case of the epoxysuccinyl group, can extend to both sides. The epoxysuccinyl group is a useful concept for the inhibition of endopeptidases (see above) as well as for the carboxypeptidases cathepsins B [101], [102], [103], [104] and X [116]. However, for the aminopeptidases cathepsins C and H this concept would be difficult to apply due to their two C-termini. To achieve selectivity for cathepsin-C dipeptidyl constructs with acyloxymethyl ketones, fluoromethyl ketones and vinyl sulfones were therefore developed [117]. On the other hand, the selectivity for cathepsin H would be confined to a single binding subsite, S1. Inhibitors with limited selectivity were also synthesized using chloro- and fluoro-methyl ketones as the reactive groups attached to a single amino acid residue [114].
For endopetidases and carboxypeptidases no such restraints on the non-primed side of the active-site cleft apply. Therefore, there were numerous developments. The list of reactive groups used is rather extensive: acyloxymethyl ketones [118], vinyl sulfones [119], [120], nitriles [121], [122], azepanone-based [123] and allyl-sulfones [124]. More information can be found in an excellent overview [125]. There are also constructs that extend across the active-site cleft without a covalent interaction and are not degraded, e.g., the cyclic ketones [126]. Lately, nitriles have been shown to give the best results and they have also been used in all clinical candidates [127]. The versatility of the small-molecule compounds is not confined like that of the peptidyl substrates and it does not directly address the subsite specificity in terms of the biological function and the role of cysteine cathepsins; therefore, this review does not attempt to provide any in-depth look at their selectivity. It should be mentioned that a number of inhibitors have been crystallized in complexes with cysteine cathepsins and their structures have been determined. At present there are 160 deposited structures corresponding to cathepsins, mainly they are cysteine proteases in complexes with small-molecule inhibitors. The drug-discovery processes, predominantly targeting cathepsins K, L and S, are underway. A few of these developments have already entered clinical trials [37].
3.5. Activity-based probes
Activity-based probes (ABPs) are substances that interact with the active site of an enzyme to report on its activity. Their sophisticated chemistry makes it possible to characterize the enzyme function directly in native biological systems [128]. ABPs usually carry a reporter, such as biotin, or a quencher-fluorophore pair, which gets activated upon a reaction with the target enzyme.
Fluorescent probes based on the epoxysuccinyl- and acyloxymethyl-ketone reactive groups enabled imaging in cells as well as non-invasive optical imaging of cathepsins B and L in mice [129], [130]. In addition, ABPs for cathepsin X have also become available [116].
Until recently, ABPs targeting cysteine proteases were suicidal inhibitors, which remained attached to the enzyme after forming a covalent bond with the reactive site cysteine. However, a novel approach termed “reverse design”, built on the existing selective inhibitors of cysteine cathepsins and their subsequent conversion into substrates as the activity probes of proteases in vivo, was established [131]. The conversion of the inhibitors of cathepsins K and S by the replacement of the active-site-directed functional electrophylic group with a peptidyl unit and with the addition of a fluorophore and a quencher resulted in cell-permeable ABPs, which revealed the endolysosomal localization of cathepsins L and S. In these studies cathepsins B, L, K and S were used to evaluate the selectivity of the probes. Due to the non-suicidal nature of the activity probe, the physiology of the cells under investigation is potentially less disturbed. Moreover, such activity-based probes can be used for the validation of the effectiveness of activity-modulating compounds such as drug candidates in cells and in in vivo systems in a time-dependent manner [132].
3.6. Protein substrate hydrolysis
It has been some time since the test for proteolytic activity was based on the degradation of insulin. Nowadays, we try to understand how proteases hydrolyze their physiological substrates. The recent knowledge about the biological targets of cysteine cathepsins has largely come from identifying the origin of genetic disorders and, consequently, gene-knockout studies. There is a significant redundancy of cysteine cathepsins due to their co-localization and broad specificity. Although most of the cysteine cathepsins do not have a specific role, they possess a proteolytic potential to cleave a number of substrates. However, it is likely that specific roles are linked to specific interactions between proteases and substrates. For example, cathepsins B, H, K, L, and S cleave kininogen, thereby releasing the kinin peptide; however, only cathepsin K was shown to be able to cleave kinin at the naturally occurring cleavage site, supporting the above idea [133], [134].
It should be kept in mind that cysteine cathepsins co-localize and therefore work in concert. Due to these limitations, only those specific, non-redundant roles could be revealed and confirmed by the absence of a particular cathepsin. Here we have made an attempt to divide the protein substrate hydrolysis into classical, i.e., confined to endosomes/lysosomes or secretory lysosomes, and non-classical, i.e., taking place outside these organelles, and correspondingly named the substrates.
As seen below, apart from the cathepsin roles in antigen presentation and in granule protease activation, most of the specific physiological roles under investigation take place in non-classical locations and environments. Among these are the collagen degradation by cathepsin K, thyroglobulin processing and thyroid hormone release by various cathepsins, histone H3 processing by mouse cathepsin L and the role of cathepsin X in integrin processing and T-cell signaling.
A rather simple mechanism of the classical pathway appears to be that of the cathepsin C interaction with substrates. Its role is to gradually remove two consecutive residues from protein N-termini. As shown by the gene-knockout studies, a number of serine proteases from immune and inflammatory cells are activated by cathepsin C in the granules in this way [135].
Cysteine cathepsins also play a crucial role in adaptive immunity. Cathepsin S is crucial for the processing of the invariant chain in bone-marrow-derived antigen-presenting cells such as dendritic cells and macrophages [136], while the human V and mouse L are required for Ii processing in thymus. In addition, cathepsins generate peptides, which are presented to MHC class-II molecules. Cathepsin L was found indispensable for the generation of a specific peptides repertoire in cortical thymic epithelial cells, which is required for CD4+ T cell positive selection in thymus [137]. Considering the role of cathepsin S, an attempt was made to reveal the substrate specificity of cathepsin S by screening a peptide library [93]. For the purposes of validation the pattern of the cleavage sites on the Ii polypeptide was compared with the results of the peptide library screen, and, not surprisingly, some similarities were found. However, the cleavage sites in the invariant chain were numerous, so the question remains as to whether cathepsin S simply chops the Ii into smaller peptides or does cathepsin S perform a specific cleavage. Since cathepsins V/L were found to be crucial for the processing of the thymus MHC class-II Ii complex, it is evident that several proteases can do the same job. More recently, a new pathway for prohormone processing mediated by the secretory vesicle cathepsin L was described [138], [139].
Glycosaminoglycans play a crucial role in the formation of complexes between cysteine cathepsins and their protein substrates. GAGs have thus been shown to regulate the collagenolytic activity of human cysteine cathepsins [53], [140], [141]. Furthermore, for the collagenase activity of cathepsin K, its complex formation with chondroitin sulfate is mandatory [140]. In the cartilage, cathepsin K alone is capable of providing the source for GAGs by removing them from aggrecan [142]. The insight into the mechanism of this rather unique relationship was revealed by the crystal structure of the chondroitin sulfate-cathepsin K complex and the accompanying biochemical study [143]. In the crystal, several cathepsin K molecules bind to a single chondroitin sulfate at a site distant from the active site. This work suggested that in the cathepsin K-hydrolysis of collagen the role of chondroitin sulfate is to bring together and align several cathepsin K molecules in a way that makes possible the cleavage of the triple collagen helix. The chondroitin thus provides a scaffold for the association of cathepsin K molecules. This effect is not shared by cathepsin L [85]. Mutations in cathepsin K, which modified the chondroitin binding site in cathepsin K into cathepsin-L-like, decreased the collagenolytic activity of the protein. In the crystal structure of the complex between cathepsin-L-like cathepsin K and chondroitin sulfate, the latter still binds to cathepsin K; however, not in the same way as in the structure with native cathepsin K. To summarize, these studies have revealed the relevance of chondroitin sulfate binding to cathepsin K for collagene proteolysis. Nevertheless, modifying the binding site for chondroitin into cathepsin-L-like has not removed the collagenase activity. This indicates that cathepsin K possesses additional features that enable it to cleave collagen efficiently.
Gelatin, partially degraded and unfolded collagen, can, however, be cleaved by several cathepsins [140]. Similarly, cathepsins L and S can release GAGs from cartilage proteins such as aggrecan [142] and there is evidence that cathepsin L participates in bone resorption [144]. These findings suggest that a number of cathepsins participate in bone resorption and remodeling. However, it has only been shown for cathepsin K that its presence in the organism is vital for the osteoclast resorption and that its inhibition has a strong potential to treat osteoporosis. Quite possibly it is the only cathepsin capable of unwinding the collagen triple helix.
A further insight into the activity of cathepsin K was provided by kinetic and spectroscopic studies, which provided evidence for there being two states of cathepsin K, depending on the pH of the media [145]. This is reminiscent of the cathepsin B exo- and endopeptidyl activity [146], which was believed to be a result of the dissociation of ionizing groups in the binding cleft. When the structure of cathepsin B became available [63], it was evident that the occluding loop must be displaced in order to enable the endopeptidase activity of the enzyme. This was finally confirmed by mutations in the occluding loop, including the removal of its parts [82], [83]. In contrast to the exopeptidase cathepsin B, the cathepsin K fold does not provide an obvious structural feature on the surface of cathepsin K, which would trigger a change of state and thereby affect the substrate kinetics in a pH-dependent manner. As a possible mechanism, the oligomerization of cathepsin K has not been excluded by the experimental data. However, it remains open whether this behavior is consistent with the model of cathepsin K to form a “beads on a string” structure, as earlier suggested [143].
Another example that sheds light on substrate hydrolysis by cathepsins is hormone release in the degradation of thyroglobulin [147]. Despite a general belief to the contrary, it was shown that cathepsins are capable of processing thyroglobulin under neutral and oxidizing conditions. Cathepsins B, K, L and S were thus found to generate distinct fragments from thyroglobulin. Moreover, the thyroid hormone thyroxine was liberated by the action of cathepsin S under extracellular conditions, while cathepsins B, K and L were most efficient under endo-lysosomal conditions, suggesting that the biological role of a protease may depend on the local environment.
Yet another non-classical role involves cathepsin L in the cleavage of histone H3 in the nucleus [31]. Based on the assumption that this was a rather specific interaction, it was anticipated that the binding of a model substrate peptide into the active site might provide an insight into the specificity of this interaction. A rather long peptide (derived from the histone H3 sequence 19–33) was therefore co-crystallized with an inactive mutant of the human cathepsin L [148]. However, only the binding of three residues into the non-primed binding sites at the cathepsin L surface could have been satisfactorily resolved. Nevertheless, this work should encourage further attempts to gain a more detailed structural insight into the interactions between cysteine cathepsins and their physiological substrates.
Cathepsins were also found to participate in processes such as the modulation of T-cell migration. This process combines cathepsin targeting outside endosomes and subsequent proteolysis at the target location. The extracellular function of cathepsin X, the enzyme suggested to be involved in this process, may include binding to integrins via an RGD motif in its proregion, which further suggests an activation mechanism of the enzyme at a location outside the endosome. It has been further suggested that the attachment of migrating cells to extracellular matrix components is modulated [149]. Moreover, cathepsin X was found to cleave β2 integrin, thereby demonstrating another way of regulating T-cell migration [150]. There are indications that cathepsin X removes the C-terminal residues of LFA-1, a β2 integrin, thereby increasing its affinity for the adaptor protein talin [151], [152]. The subsequent cleavages result in the intermediate as well as the high-affinity LFA-1. The intermediate affinity form can associate with α-actinin 1, while the high-affinity form is crucial for the T-cell migration. Although not described in more details, cysteine cathepsins were shown to participate in the regulation of antimicrobial molecules by inactivation of the secretory leucoprotease inhibitor [153] and, even though in part disputed [154], in the processing of viruses, such as cathepsin L in the processing of the corona virus [155].
4. Regulation
Under normal conditions in healthy organisms, cells use various mechanisms to prevent potentially harmful and uncontrolled proteolytic activity, including the compartmentalization of cathepsins within the lysosome or other organelles, zymogen activation, pH, the regulation of their activities by small-molecule inhibitors and various endogenous protein inhibitors, or a combination of all these factors for an optimal enzyme function [14], [21], [156]. Lysosomal cathepsins contribute to these processes by the irreversible cleavage of the peptide bonds with limited or random proteolysis accompanying the protein degradation. In the remainder of the review we will focus on zymogen activation and the regulation by protein inhibitors, although other aspects are also discussed.
4.1. Zymogen activation
Lysosomal cathepsins are synthesized as preproenzymes. The cleavage of the N-terminal signal peptide occurs during the passage to the endoplasmic reticulum in parallel with the N-linked glycosylation. After the removal of the signal peptide, the propeptide assists in the proper folding of the enzyme and targeting to the endosomes/lysosomes using a specific mannose-6-phosphate receptor (M6PR) pathway, simultaneously acting as an inhibitor to prevent any inappropriate proteolytic activity of the zymogen [157], [158], [159]. Dissociation of the bound, immature M6PR occurs in the mildly acidic environment of the endosomes. After removal of the N-terminal propeptide, the mature proteolytically active cathepsins are released. These mature enzymes occur as single-chain or double-chain forms, without affecting their catalytic activity. The two-chain enzymes remain connected by disulphide bridges [160].
Further progress in understanding the nature of zymogen activation was obtained from the procathepsin structures. The crystal structures of human and rat procathepsin B [161], [162], [163], human procathepsin L [164], human procathepsin K [165], [166] and human procathepsin X [167] showed that the propeptide chain folds on the surface of the enzyme in an extended conformation and runs through the active-site cleft, in the opposite direction to the substrate, thereby blocking the access of the latter to the active site, which is already formed in the zymogen (Fig. 5 ). In the structure of most proenzymes, the hydrophobic interactions, salt bridges and hydrogen-bonding interaction in the propeptide and between the propeptide and the mature enzyme exists. The exception is the structure of cathepsin X, in which the propeptide binds covalently to the mature enzyme with a disulphide bridge between the cysteine residue in the propeptide and the active-site cysteine, thus preventing any autocatalytic processing [167]. However, it can be processed in vitro under reducing conditions by cathepsin L [168].
Fig. 5.
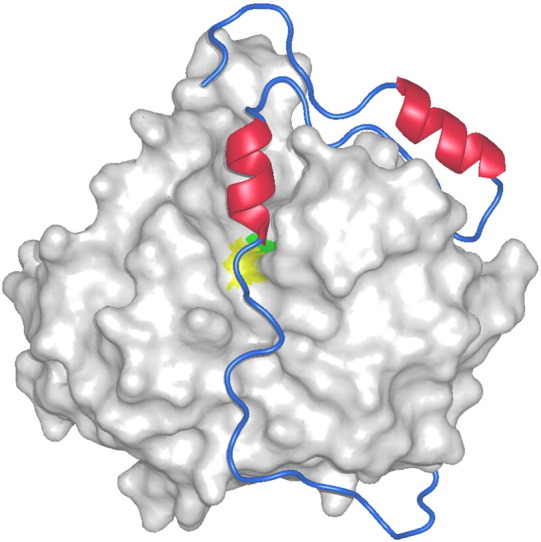
Procathepsin B fold. The propeptide part of human procathepsin B (2pbh) [163] is shown as a chain trace over the surface of the mature part of the enzyme. The α-helices are shown in red color. The surfaces of the reactive-site residues, cysteine and histidine, are marked with yellow and green color, respectively.
In other cathepsins the proteolytic removal of the N-terminal propeptide, resulting in the activation of the enzyme, is catalyzed by different proteases such as cathepsin D and various cysteine proteases [169], [170], or autocatalytically under acidic conditions [171], [172], [173], [174]. Some early studies suggested that autocatalytic processing is a unimolecular process [171], [172], while others proposed inter- and intra-molecular mechanisms [173], [175]. This enigma has been recently clarified. It is now clear that the auto-activation of cathepsins is a combination of a unimolecular and a bimolecular process [176]. Procathepsin B possesses a low catalytic activity that is, nevertheless, sufficient to trigger the autocatalytic activation of the zymogen. This activity is the result of the propeptide dissociation from the active-site cleft as the first step in this process, which is in fact the only unimolecular step in the process [177]. In the next step, which is already bimolecular, this catalytically active zymogen molecule processes and activates another procathepsin molecule in one or more steps. The mature cathepsin molecules generated in this way then initiate a chain reaction leading to a rapid activation of the remaining procathepsin molecules. Taken together, only endopeptidases such as the cathepsins B, H, L, S and K can be activated by the removal of the propeptide, whereas the true exopeptidases, such as the cathepsins C and X, required endopeptidases, such as the cathepsins L and S, for their activation [178].
The autocatalytic activation of cathepsins is substantially accelerated in the presence of negatively charged molecules, such as GAGs and dextran sulfate, suggesting that GAGs are involved in the in vivo processing of cathepsins [179]. Similarly, GAGs accelerate the autocatalytic removal of the propeptide and the subsequent activation of cathepsin B. These findings suggest that GAGs may play a physiological role in the activation of procathepsin B [179]. The results were confirmed by the autocatalytic activation of procathepsin S in the presence of GAGs [180].
Propeptides, which are removed during the processing of cysteine cathepsins, have been shown to be potent inhibitors of their cognate enzymes in vitro. The first such example was the synthetic propeptide of cathepsin B, which was found to inhibit cathepsin B at pH 6.0 with Ki = 0.4 nM, whereas at pH 4.0 the inhibition was much weaker [181]. This further suggested that during the activation at acidic pH the propeptide dissociates from the surface of the enzyme, thereby facilitating the activation, and this was then later experimentally confirmed [176]. Subsequently, the recombinant cathepsin L propeptide was found to inhibit cathepsin L with Ki = 0.088 nM and cathepsin S with Ki = 44.6 nM, whereas no inhibition of cathepsin B or papain was observed, providing the first evidence for propeptides acting as selective cathepsin inhibitors [182]. Further studies confirmed these findings and demonstrated that propeptides exhibit a limited selectivity of inhibition against their cognate enzymes [183]. Moreover, cathepsin S propeptide was found to be rapidly degraded by cathepsin L [184]. The latter, linked with the acidic, pH-induced unfolding and dissociation of the procathepsin B propeptide [176], suggests that the propeptides released during the zymogen activation are degraded, and as a result lose their inhibitory role. However, it should be noted that in the procathepsin structure the propeptide is covalently bound to the enzyme (Fig. 5), whereas all the in vitro studies mentioned above were performed with isolated peptides. Therefore, these studies do not necessarily reflect the in vivo situation. For more in-depth information about the structural aspects and function of the propeptides see [160], [185], [186].
In spite of their similar biological function, the propeptides of cysteine cathepsins show very little similarity in their amino acid sequence. This is in striking contrast to the strong overall homology between the majority of their mature forms. However, this can probably be explained by the need for a selective inhibition of their cognate enzymes during trafficking to the endolysosomal compartments and subsequent degradation after the processing to prevent the sustained inhibition of the cathepsins in the lysosomes. A sequence analyses of the family of papain-like cysteine proteases led to the recognition of two distinct subfamilies, designated as cathepsin-L-like and cathepsin-B-like proteases, which can be distinguished by both the propeptide and mature enzyme structure [186], [187]. The main difference between the subfamilies exists in the sequence of the propeptides and their length [17]. Propeptides of the cathepsin L subfamily (cathepsins L, V, K, S, W, F and H) contain a propeptide of about 100 residues, with two conserved motifs: a highly conserved ERFNIN motif and the GNFD motif. The former is lacking in the cathepsin B subfamily, while the propeptides of cathepsins C, O and X lack the ERFNIN motif. The propeptide of cathepsin X contains only 38 residues and is the shortest of the cathepsins [188], [189]. In contrast, the cathepsin F propeptide with 251 residues is the longest of the human cathepsins and contains, in addition, an N-terminal cystatin-like domain [190], [191]. The propeptide of cathepsin C contains 206 residues [192]. Interestingly, after the removal of the 87 propeptide residues during proteolytic activation, the remaining 119 residues become part of the structure of the mature cathepsin C responsible for its dipeptidyl peptidase activity, known as the exclusion domain [70].
4.2. Protein inhibitors of cysteine proteases
The major regulators of the mature cysteine cathepsins are their endogenous protein inhibitors, cystatins, thyropins and serpins, among others. Basically, they are competitive, reversible, tight-binding inhibitors, preventing substrate binding to the same active site. Some basic principles of the kinetics of the interaction between inhibitors and proteases, as well as their physiological role, are discussed. In short, based on their physiological role, they are divided into emergency and regulatory inhibitors [14]. Typical emergency inhibitors are cystatins, which are separated from their target enzymes and primarily act on escaped proteases [14] or proteases of invading pathogens [193]. This was also demonstrated for thyropins [194]. Regulatory inhibitors not only block but also modulate the protease activity. Moreover, they are often co-localized with their target enzymes and are classified into threshold-type, buffer-type and delay-type inhibitors. Buffer-type inhibitors keep proteases complexed in the absence of substrates and release them in the presence of substrates, thus preventing inappropriate proteolysis. Propeptides of lysosomal cathepsins most likely belong to this type of inhibitors (reviewed in [17], [181]). All these four types of inhibitors (emergency, threshold, buffer and delay) are important to keep the necessary balance under normal physiological conditions [14].
4.3. Classification and main properties
The first classification of the protein inhibitors of papain-like cysteine proteases, the cystatin superfamily, into three families was based on proteins homologous to chicken cystatin, the first inhibitory protein with a known amino acid sequence [195]. In addition, to close the sequence relationship with at least a 50% sequence identity [196], the new criteria included the absence or presence of two or nine disulphide bonds [197]. However, the rapidly increasing number of cystatins from various origins resulted in a new division into four types on the basis of the presence of one, two or three copies of the cystatin-like domains and the presence or absence of disulphide bonds [198]. While the first three types, namely, stefins, cystatin and kininogens, are inhibitory proteins [198], [199], [200], [201], [202], the fourth type consists of non-inhibitory homologs of cystatins such as fetuins [203] and histidine-rich glycoprotein [204], containing two cystatin-like domains [205]. However, in evolution they lost their inhibitory activity due to mutations in the structurally important regions [206]. A very important step was the introduction of the MEROPS database as an integrative source of information, firstly the peptidases [207], [208]. Then, for protein peptidase inhibitors, a single system of nomenclature on the basis of similarities at the amino acid sequence level and three-dimensional structures was introduced [209]. They were grouped into clans, families and individual inhibitors, correspondingly. The cystatin family is assigned to three subfamilies, I25A, I25B and I25C of clan IH, while thyropins are assigned to family I31 of clan IX [209], [210]. The classification of protein peptidase inhibitors is continually under revision as can be seen from the MEROPS website at http://merops.sanger.ac.uk. Therefore, the cystatins will be discussed, grouped into types [197], [198], which is more convenient with respect to the present status in the field.
4.3.1. Stefins (type 1 cystatins)
Stefins are single-chain proteins of ~ 100 amino acid residues, synthesized without signal peptide and they lack carbohydrates and disulphide bonds. They are primarily intracellular proteins [199], but can also be detected in body fluids [211]. Stefins are present in most major eukaryotic subgroups [206]. Originally, two representatives of this group, stefins A and B, were found in various mammals, including humans [199]. In addition, bovine stefin C has been identified as the first Trp-containing stefin with a prolonged N-terminus [212]. Stefins belong to the subfamily I25A of the cystatin protein family [210].
4.3.2. Cystatins (type 2 cystatins)
Cystatins are more widely distributed proteins than stefins [202], [206]. They are single-chain proteins of ~ 115 amino acid residues. In contrast to stefins, cystatins contain a signal peptide responsible for secretion through the cell membrane to the extracellular milieu, recognized as extracellular proteins [199], [200], [211]. Currently, seven members of this type of inhibitor have been identified: cystatin C, salivary cystatins (cystatins S, SA and SN), cystatin D, cystatin E/M and cystatin F (leukocystatin). All type 2 cystatins contain two highly conserved intra-molecular disulphide bridges, with the exception of human cystatin F, which possesses an additional disulphide bridge, thus stabilizing the N-terminal part of the protein [213]. The human type 2 cystatins are grouped into subfamily I25B of the cystatin family I25 [210].
4.3.3. Kininogens (type 3 cystatins)
Kininogens have been known for several decades as the precursor molecules of the kinins found predominantly in the blood plasma of mammals and in some other species. Although kinins could be released through two major pathways involving either the plasma kallikrein-kinin system or tissue kallikrein, an alternative route may involve cysteine cathepsins [214]. Kininogens are multifunctional and multidomain glycoproteins comprising three distinct types: high-molecular-weight kininogen (HK), low-molecular-weight kininogen (LK) and T-kininogen (TK), an acute phase protein only found in rats [215]. Both human HK and LK are products of the same gene, resulting from the alternative mRNA splicing [216], while TK is encoded by the TK-gene [217]. The mature kininogens are single-chain proteins with multiple domains [218], [219]. After cleavage by kallikreins, they release the kinin segment and are converted into two-chain proteins, a heavy- and a light-chain [218]. The heavy chains of HK and LK have an identical amino acid sequence, whereas the light-chain of HK is much longer than that of LK [199], [200]. The molecular mass of LK is 51 kDa [220] and 84 kDa for HK [221]. The heavy chains of HK and LK are composed of three tandemly repeated type 2 cystatin-like domains (domains 1, 2, 3) containing eight disulphide bridges. Only the second and third domains inhibit the papain-like proteases [222]. The inhibitory domains 2 and 3 are more closely related than domain 1. Both HK and LK bind two molecules of various cysteine proteases including cathepsins and cruzipain with high affinity [220], [221]. In contrast, in vitro studies showed that the aspartic protease cathepsin D inactivates the third domain of the human kininogen and cystatin C, suggesting a role for cathepsin D in regulating the activity of cysteine cathepsins at acidic pH [223]. It was recently demonstrated that HK regulates the endothelial function, thus serving as a cardioprotective peptide [224]. Some other studies showed some novel function of kininogens, such as the regulation of angiogenesis (reviewed in [225]). Like type 2 cystatins, the kininogens are grouped into subfamily I25B of the cystatin family I25 [210].
4.3.4. Thyropins
The discovery that a fragment of the p41 invariant chain (p41Ii) associated with MHC class-II molecules inhibits cathepsin L [194], [226], [227] suggested the appearance of a new family of protein inhibitors of cysteine cathepsins. This finding was confirmed by the discovery of a new protein, equistatin, isolated from the sea anemone Actinia equina, strongly inhibiting cathepsin L and papain [228] and human cathepsin D [229]. The determined amino acid sequences of the equistatin [228] and p41 fragment [227] showed no homology to cystatins, but a significant homology to thyroglobulin type-1 domains, which are present in prohormone thyroglobulin [230]. Therefore, the name thyropins has been suggested for this new group of proteins [231]. In equistatin, as a three-domain protein, the N-terminal domain 1 is responsible for the inhibition of papain-like proteases [228], whereas the second domain inhibits cathepsin D [229], [232]. Thyroglobulin type-1 domains are also present in several functionally unrelated proteins [14]. Thyropins are grouped into family I31 of clan IX [210]. More details can be found in a recent review [233].
4.3.5. Other protein inhibitors
Some serpins, as typical inhibitors of serine proteins, can also inhibit cysteine proteases in cross-class inhibition [14]. The human squamous cell carcinoma antigen-1 (SCCA1) is a potent inhibitor of the cathepsins K, L and S [234], whereas hurpin specifically inhibits only cathepsin L [235]. Similarly, the serpin endopin 2C selectively inhibits cathepsin L compared to elastase [236]. There are numerous other cystatins or cystatin-related proteins that are expressed in other tissues and types in humans and other mammals [237].
Several genes, including CRES (cystatin-related epididymal spermatogenic), testatin, cystatin-T, have been identified, but they lack the highly conserved QXVXG region crucial for cysteine cathepsin's inhibition. These genes are primarily expressed in the reproductive tract, suggesting a cell-specific and regulatory function different from cystatins. CRES proteins are classified as a new subgroup within the type 2 cystatins (reviewed in [238]). Certainly, this group of proteins deserves more detailed investigations in order to clarify their function.
Gene-encoding cystatins have also been found in various parasitic organisms. Their function is presumably to inhibit their own and host proteases to enable parasite development. Cystatins have been found in several ticks, which constitute the main vector of Lyme disease in the USA and Europe. The two-cystatin transcripts are encoded by two different genes in the tick Ixodes scapularis. Both expressed homologous salivary proteins, sialostatin L [239] and sialostatin L2 [240], strongly inhibited, almost equally, cathepsin L with Ki = 4.7 nM and cathepsin V with Ki = 57 nM, while no inhibition of cathepsins B, X and C was observed. Cathepsin L as collagenolytic enzyme hydrolyzes ECM proteins, such as collagen and elastin, and it was shown that its collagenolytic activity can be inhibited by chicken cystatin [241]. Consequently, it was demonstrated that sialostatin L, by inhibiting the collagenolytic activity of cathepsin L, displays an anti-inflammatory role and reduces the proliferation of cytotoxic T-lymphocytes [239].
There are many other parasitic organisms containing specific parasite-derived inhibitors, such as Plasmodium [242], [243], Trypanosoma cruzi [244], human filarial nematodes [245], and other parasitic organisms [193], among others. Although there are numerous other cystatins from plants and other organisms, it is beyond the scope of this review.
5. Mechanism of inhibition of cysteine cathepsins
Cystatins and stefins are tight-reversible binding protein inhibitors of papain-like cysteine proteases (reviewed in [10], [14], [199], [200], [202], [237]). The interaction with their target enzymes can be very tight, a Ki value of ~10 fM has been determined for the interaction between cystatin C and papain [246]. An important step in the elucidation of the molecular mechanism by which these protein inhibitors inhibit their target enzymes was the determination of the crystal structure of chicken cystatin [247] and the successfully prepared recombinant human stefin B [248] in a complex with S-carboxymethylated papain [249]. Both inhibitors, cystatin and stefin B, consist of a long, five-turn α-helix and a five-stranded antiparallel β-pleated sheet, with an additional helix or strand, respectively [247], [249]. These structural data provided strong evidence that there are three regions crucial for the interaction with their cognate enzymes: the amino terminus and two hairpin loops. An exposed first β-hairpin loop (comprising a highly conserved QVVAG region or similar sequence QXVXG) is flanked on either side by the projecting N-terminal segment and the highly conserved second β-hairpin loop, which contains the highly conserved Pro-Trp dipeptide. Both hairpin loops and the amino terminus form a tripartite, mainly hydrophobic, wedge-shaped edge, containing most of the conserved residues and segments involved in binding, highly complementary to the active-site cleft of papain. The N-terminal truncated forms of chicken cystatin confirmed the crucial importance for binding of the residues preceding the conserved Gly9 residue [250]. The hydrophobic side-chain interactions stabilized the complex. In cystatin and stefin complexes the interactions in the S2 subsite strengthen the complexes with papain considerably [250], [251], [252]. However, it was reported that in spite of considerable similarity between the structural feature of free stefin A in solution [253] and the crystal of the homologous protein stefin B in complex with papain [249], there are some important differences in the regions that are crucial for protein binding, such as the five N-terminal residues and the second binding loop [253]. Similarly, the comparison of the chicken cystatin structure determined by X-ray crystallography [247] and NMR [254] showed significant differences in the structurally variable segments of the polypeptide chain. These findings suggested that the binding of cystatin/stefin type inhibitors cannot be satisfactorily explained solely on the basis of the stefin B-papain complex [249]. The crystal structure of stefin A, in complex with the papain-like exopeptidase cathepsin H showed some distinct differences [255]. The N-terminal residue of stefin A adopted the form of a hook, which slightly displays a cathepsin H mini-chain and distorted a small part of the structure. The recently determined crystal structure of human stefin A–cathepsin B complex indicated that the papain-like part of the cathepsin B structure remains unmodified upon the binding of stefin A, whereas the occluding loop residues are displaced [256]. Consequently, cathepsin B can bind certain ligands along the whole interdomain interphase, such as inhibitors or substrates. On the other hand, cathepsin B is poorly inhibited by kininogens due to the residues His110-His111 positioned within its occluding loop. The His110Ala cathepsin B mutant, unlike its His111Ala mutant, does not hydrolyze kininogens but forms a tight-binding complex [257].
Although the interaction between cystatins and their target proteases is primarily non-specific, cystatins are able to discriminate between endo- and exopeptidases, as a consequence of the differences in the structures of the interacting regions of the enzymes, as already explained. However, it was reported that mouse stefin A variants discriminate between papain-like endopeptidases, such as cathepsins L and S, and the exopeptidases cathepsins B, C and H. The interaction with exopeptidases is several orders of magnitude weaker compared to the human, porcine and bovine stefins [258]. Namely, bovine stefins A, B and C inhibit rapidly and tightly endopeptidases such as papain, cathepsin L and S in the pM range, whereas cathepsin B does so very poorly [212], [259], [260]. Furthermore, it was shown that human cystatin C, stefins B and chicken cystatin strongly inhibit the endopeptidase cruzipain from T. cruzi with Ki = 1.4–72 pM [261].
Among the thyropins, it is the only very potent inhibitor of human origin, the p41 fragment of which inhibits human cathepsin L with Ki = 1.7 pM [227]. This fragment was isolated in a complex with cathepsin L [194] and its crystal structure was determined [74]. The structure of the p41 fragment demonstrates a new fold consisting of two sub-domains connected by disulphide bonds. The wedge-shaped and three-loop arrangement of this fragment bound to the active-site cleft of cathepsin L is reminiscent of the binding of cystatins [74], thus demonstrating the first example of convergent evolution observed in cysteine protease inhibitors, similar to chagasin [262]. Recently, it has been shown that the p41 fragment inhibits human cathepsins V, K, S and F and mouse cathepsin L with Ki values in the nM range [263] in addition to human cathepsin L [227] and cruzipain [226]. These findings suggest that the regulation of the proteolytic activity of most of the cysteine cathepsins by the p41 fragment has an important and wideranging control mechanism of antigen presentation [263]. Similar to the p41 fragment, equistatin binds rapidly and tightly to papain and cathepsin L, but with a lower affinity to cathepsin B [228].
Some equilibrium constants for the dissociation of the complexes between human cystatins and the p41 fragment and their target papain-like enzymes are summarized in Table 1 . The affinity in Ki values is due to the differences in the active-site regions of the endo- and exopeptidases, as discussed above.
Table 1.
Interaction between some representative lysosomal cysteine proteases (human cathepsins, cruzipain and papain) and selected members of the cystatin family (human cystatins, chicken cystatin) and thyropins (p41 fragment).
| Ki (nM) |
|||||
|---|---|---|---|---|---|
| Inhibitor | Cathepsin B | Cathepsin H | Cathepsin L | Cruzipain | Papain |
| Stefin A | 8.2 | 0.31 | 1.3 | 0.0072 | 0.019 |
| Stefin B | 73 | 0.58 | 0.23 | 0.060 | 0.12 |
| Cystatin C | 0.27 | 0.28 | < 0.005 | 0.014 | 0.00001 |
| Cystatin D | > 1000 | 7.5 | 18 | n.d. | 1.2 |
| Cystatin E/M | 32 | n.d. | n.d. | n.d. | 0.39 |
| Cystatin F | > 1000 | n.d. | 0.31 | n.d. | 1.1 |
| Cystatin S | n.d. | n.d. | n.d. | n.d. | 108 |
| Cystatin SA | n.d. | n.d. | n.d. | n.d. | 0.32 |
| Cystatin SN | 19 | n.d. | n.d. | n.d. | 0.016 |
| Chicken cystatin | 1.7 | 0.06 | 0.019 | 0.001 | 0.005 |
| L-kininogen | 600 | 0.72 | 0.017 | 0.041 | 0.015 |
| p41 fragment | > 1000 | 5.3 | 0.002 | 0.058 | 1.40 |
6. Evolution of cystatins
Early studies of the evolution of the cystatin superfamily were based on a small sample of taxonomic diversity and diversity within the cystatin superfamily [200], [264]. Since then, the number of representatives across the mammals, plants, viruses and parasites increased mainly due to the accumulation of mammalian and plant genomic sequences. In addition, several genes, including CRES (cystatin-related epididymal spermatogenic), testatin and cystatin-T related to the cystatins, were identified but lack consensus sites that are important for inhibition [238]. Gradually, the numerous proteins containing cystatin domain(s) have been discovered, resulting in a challenging problem of their comprehensive evolutionary classification. A detailed analysis of more than 2100 prokaryotic and eukaryotic genomes provided strong evidence for the emergence of the cystatin superfamily in the ancestors of eukaryotes [206]. A primordial gene duplication resulted in only two ancestral eukaryotic lineages, stefins and cystatins. While stefins remained the intracellular proteins responsible for regulation of an endogenous protein turnover, cystatins gained a signal peptide and emerged as extracellular inhibitors responsible for regulation of an exogenous protein turnover involved in host-pathogen/parasite predator interactions. Therefore, stefins may function as inhibitors of endogenous cysteine proteases, whereas cystatins function as inhibitors of exogenous cysteine proteases. It is noteworthy that intracellular stefins are more conserved than very divergent extracellular cystatins. Moreover, the first diversification of cystatins occurred in the ancestors of vertebrates when the first orthologous families emerged. This indicates that functional diversification has occurred in multicellular eukaryotes, often resulting in a loss of the ancestral inhibitory activity, gaining novel functions in innate immunity. Furthermore, the observed distribution of stefins and cystatins in bacterial genomes is very limited. This suggests that the bacterial stefins and cystatins may serve as emergency inhibitors, thus enabling the survival of bacteria in the host, defending them from the host's proteolytic activity by inhibiting cysteine cathepsins.
7. Physiological role
Originally, cysteine cathepsins were believed to participate exclusively in terminal protein degradation during necrotic and autophagic cell death. However, nowadays it is well established that they execute numerous specific functions participating in important physiological processes, such as MHC-II-mediated antigen presentation, bone remodeling, keratinocyte differentiation, prohormones activation, and others [13], [17], [18]. One of the most intriguing fields is the processes of adaptive immunity in which antigen processing and presentation play a major role. Cysteine cathepsins such as cathepsins L, S [27], [265], [266], cathepsin F [267] and human cathepsin V [268], play a crucial role by degrading the invariant chain and the responsible generated antigenic peptides during MHC class-II restricted antigen presentation, which are recognized by CD4+ T cells [269]. It has been found that cathepsin V is highly homologous to cathepsin L and exclusively expressed in human thymus and testis. This suggests that cathepsin V is a protease that controls the generation of αβ-CLIP complexes in human thymus in analogy to cathepsin L in mice [268]. The observed inhibition of several cysteine cathepsins by a p41 fragment might be an important and general control mechanism for antigen presentation [263]. For further reading see [270], [271], [272]. There are several other physiological states, such as bone remodeling, angiogenesis and prohormone activation, among others, in which cathepsins and their endogenous inhibitors play an important role [9], [17], [18], [139], [273]. However, an increase in the level of cathepsin expression and activity appears to be implicated in the development of various pathological conditions, such as neurological disorders, cardiovascular diseases, obesity, inflammatory diseases, such as rheumatoid arthritis, and cancer, among others. It was recently shown that cathepsin K deficiency induces structural and metabolic changes linked to learning and memory deficits [274].
Obesity is an important risk factor for various metabolic and cardiovascular disorders, the so-called “metabolic syndrome” where several studies indicate the role of cathepsins L, S and K [275], [276]. It was documented that the pharmacological inhibition of cathepsins L resulted in a reduced body weight in mice [277], [278].
In cardiovascular diseases a crucial role is played by cysteine cathepsins, with their strong elastolytic and collagenolytic activity in ECM remodeling. Consequently, they are implicated in the development of arterial enlargements and aneurysm formation. Cysteine cathepsins are differently expressed in various stages of atherosclerosis. Cathepsins K and S were found as the first cathepsins expressed in human atherosclerotic lesions [279]. On the other hand, deficiencies of both enzymes in a mouse model reduce atherosclerosis [280], [281]. Furthermore, it was demonstrated that cathepsins B, K and S are expressed in cerebral aneurysms, thus promoting their progression by the degradation of ECM in arterial walls. In contrast to the upregulated expression of these cathepsins, cystatin C expression was reduced with aneurysm progression, suggesting that an imbalance between cathepsins and their inhibitors may be responsible for the breakdown of ECM in arterial walls and the rupture of cerebral aneurysms [282]. Recently, it was found that leukocytic cathepsin S deficiency results in a markedly altered plaque morphology, reduced apoptosis and decreased collagen deposition, which might be critical for plaque stability [283]. The pathological mechanisms that are responsible for the degradation of arterial ECM are still not well understood. For further reading, these reviews are suggested [48], [283].
There are several genetic disorders due to mutations in genes of cysteine cathepsins and their protein inhibitors cystatins. Genetic studies revealed that loss-of-function mutations in the cathepsin C (DPPI) gene results in early-onset periodontitis and palmoplantar keratosis, characteristics of Haim-Munk and Papillon-Lefevre syndromes [284]. The location of missense mutations suggests, based on structural studies, how they disrupt the fold and function of cathepsin C [70]. In the cathepsin K gene there are at least fifteen known mutations in humans, thus resulting in cathepsin-K deficiency and leading to pycnodysostosis [285]. Interestingly, mutation Y212C causes the appearance of enzyme activity only against type-I collagen, preserving its overall activity [286]. However, this mutation prevents the formation of complexes between cathepsin K and chondroitin sulfate, which is crucial for collagenolytic activity.
Similarly, genetic disorders were also observed in protein inhibitors cystatins. The abnormal formation of the human cystatin-C variant (L68Q), formerly the γ-trace protein, resulted in cerebral haemorrage with amyloidosis [287], now known as hereditary cystatin-C amyloid angiopathy — HCCAA (reviewed in [288]). Our discovery that human γ-trace is a protein inhibitor of cysteine cathepsins, human cystatin [195], [289], then termed cystatin C [290], was crucial for the understanding of the formation of oligomeric proteins. Further studies revealed that the dimerization of native human cystatin C occurs through a three-dimensional domain-swapping mechanism, and suggest the mechanism of its aggregation in the brain arteries of HCCAA patients [291], [292]. However, the fibril formation of tetramers of human stefin B under in vitro conditions involves a previously unknown process of intramolecular contacts termed “hand shaking”, which occurs concurrently with the trans-to-cys isomerization of Pro74 [293]. A recent review about the mechanisms of amyloid fibril formation is focused on domain-swapping [294].
The mutations on the cystatin B (stefin B) gene are involved in the progressive myoclonus epilepsy of Unverricht-Lundborg Disease (EPM1), an autosomal recessive neurodegenerative disorder [295]. Several mutations have been detected in the stefin-B gene and one of them affects the conserved QVVAG region, which is crucial for the binding to cysteine cathepsins [295]. Nevertheless, for functional studies it is necessary to take into consideration the species differences between humans and mice [296], [297]. Cathepsin F is a widely expressed lysosomal enzyme. However, it is the only cysteine cathepsin whose inactivation based on cathepsin F−/− mice results into a neurodegenerative disease termed neuronal ceroid lipofuscinosis (NCL) and suggested its possible relation to late-onset NCL [268]. However, adult-onset patients did not reveal any mutations to date [298], [299].
7.1. Cysteine cathepsins in cancer
Cathepsins, in particular cathepsin B, were first linked to cancer as long as 30 years ago [300]. It is now clear that the cathepsins have an important role in both tumor progression and invasion, which is supported by the numerous clinical reports and results from experimental mouse-cancer models [301], [302], [303]. Elevated cathepsin expression and/or activity has been shown to be associated with cancer progression in a number of different types of tumors [304], [305], [306], [307], [308], [309]. Moreover, the level of cathepsin expression positively correlated with a poor prognosis for cancer patients and was suggested to be a potential prognostic marker [310], [311], [312]. In addition, serum cathepsin levels have been positively correlated with metastasis [313]. The roles of cathepsins in the individual tumor-biological processes were also confirmed ex vivo using cell-based systems. As such, the cathepsins B, L and some others have been shown to promote the migration and invasion of tumor cells [314], [315], [316], [317], [318], [319]. In addition to their well-known function associated with the ECM degradation and remodeling in the tumor microenvironment, cathepsins were suggested to participate in the proteolytic cascade activation (Fig. 6 ). The activation of pro-uPA and other proteases, which was demonstrated in vitro, would thus potentiate their effect on the dissolution of the tumor matrix and the basement membrane [320], [321], [322], [323], which is a prerequisite for invasion and metastasis. To fulfill these functions cathepsins have to be localized at the extracellular space, in order to have access to the bulk of potential targets and processes affecting tumor progression. As a confirmation of cathepsin secretion, increased serum levels of these proteases in cancer patients have been detected [304], [324], [325], [326], [327], [328]. Despite the extracellular functions of cysteine cathepsins, intracellular cathepsins could participate in tumor progression, affecting processes acting both as pro-tumorigenic and anti-tumorigenic [329]. As an example of intracellular pro-tumorigenic activity, cathepsins could participate in intracellular collagen degradation [330], [331], [332], [333]. Moreover, this role of cysteine cathepsins in normal and malignant extracellular matrix degradation is in agreement with the finding that the endocytic transmembrane glycoprotein uPA receptor-associated protein directs the collagen IV for lysosomal delivery and degradation [333]. On the other hand, cathepsins have recently attracted considerable attention as potential mediators in programmed cell death (apoptosis) [38], a key process in cancer development and progresion [334]. However, evidence has been accumulated for multiple scenarios of how cysteine cathepsins promote cell death. For example, after their release from the lysosomes they could engage the mitochondrial pathway of apoptosis by the activation of caspases indirectly through the degradation of the antiapoptotic Bcl-2 family members and the proteolytic activation of Bid (Fig. 6). Moreover, their ability to cleave XIAP suggests their involvement in the control of apoptosis, both upstream and downstream of mitochondria [58].
Fig. 6.
Mechanisms of the cysteine cathepsins involvement in cancer progression. Extracellular cysteine cathepsins promote tumor progression by initiating a proteolytic cascade that results in the activation of a urokinase-type plasminogen activator (uPA), matrix metalloproteinases (MMPs) and plasminogen. Collectively, active proteases, including cysteine cathepsins themselves, can degrade all of the components of the extracellular matrix, promoting tumor progression and metastasis. The degradation of cell-adhesion proteins (e.g., E-cadherin) would additionally increase the disseminative ability of tumor cells. Alternatively, translocated into the cytosol cysteine cathepsins are shown to act as initiator proteases in lysosome-mediated cell death through the degradation of antiapoptotic Bcl-2 family members and the proteolytic activation of Bid, engaging the mitochondrial pathway of apoptosis.
Another level of complexity is introduced by the fact that the proteolytic activity of cysteine cathepsins is regulated by their endogenous protein inhibitors, stefins, cystatins and serpins [14]. It has been found that a higher level of cathepsin inhibitors in different types of cancer correlates with a favorable prognosis for cancer patients [335], [336]. On the other hand, an increased concentration of stefins in the body fluids of carcinoma patients, correlated significantly with shorter survival times [337], [338]. Although a higher level of cathepsin inhibitors may counterbalance the up-regulation of cysteine cathepsins, their role in cancer remains poorly defined, and this calls for further investigations to be performed [339].
Additional evidence confirming the important role of cathepsins in cancer progression was provided by the use of cathepsins-knockout technology and transgenic cancer models in small animals. In the same year, two research groups have independently shown the effect of major cathepsin/cathepsins depletion on tumor development, progression, and metastasis in two murine cancer models: (i) pancreatic islet cell carcinoma (RIP1-Tag2) [340] and (ii) MMTV-PyMT mouse mammary cancer [341], as summarized in Table 2 . The depletion of cathepsin B in both cancer models resulted in a significantly reduced tumor burden and a delay in the development of high-grade carcinomas. Furthermore, while in the RIP1-Tag2 model a significant 44% decrease in the proliferation of the tumor cells was detected, the proliferation index was reduced in 10-week-old mice by 12%, but not in mice that were 14 weeks old. In contrast to the RIP1-Tag2 model, no differences in tumor vascularization and apoptosis have been detected in the MMTV-PyMT model of breast cancer [341], which was confirmed by a follow-up report [342]. Moreover, it has been shown that the genetic ablation of cathepsin B in primary MMTV-PyMT tumor cells induces the membrane translocation of another carboxypeptidase, cathepsin X, that restores the invasive capability of tumor cells and annihilates the effect of the principal protease deletion at the level of lung metastasis [341]. This first report on such a cathepsin-compensation effect has been further confirmed by the use of double cathepsin B and X knockout mice crossed with the MMTV-PyMT mouse model, providing evidence for the synergistic anticancer effects of both carboxypeptidases [342].
Table 2.
Summary of the effects of cathepsin B deletion on MMTV-PyMT and RIP-Tag2 tumorigenesis.
| MMTV-PyMT breast cancer | RIP1-Tag2 pancreatic cancer | |
|---|---|---|
| Tumor volume | 40% decrease | 72% decrease |
| Histopathological progression | Significant delay of high-grade carcinomas development | Significant delay of invasive carcinomas development |
| Cell proliferation | Early stage cancer: 12% decrease late stage cancer; no change |
44% decrease |
| Apoptosis | No change | 229% increase |
| Vascularizalion | No change | 56% decrease |
Despite the important role of cathepsins from tumor cells, there is an increasing body of evidence confirming the up-regulation of cysteine cathepsins by cells of the tumor microenvironment, such as macrophages [302], [340], [341]. The cancer state is currently viewed as a product of its microenvironment [343]. Therefore, new technologies that enable the targeting of the tumor microenvironment would represent an efficient approach to cancer prevention and intervention [344]. Indeed, such a technology was recently provided by the development of a novel, targeted, drug-delivery system enabling the targeting of both the tumors and their microenvironment [345]. Moreover, the targeting of JPM-565, a small-molecule inhibitor of cysteine cathepsins, which was very potent in the treatment of pancreatic islet tumors in a mouse model [346], lacked any efficacy in a mouse breast-cancer model due to the very poor bioavailability of the compound [347]. The respective anti-tumor therapy using JPM-565 proved to be very successful, resulting in a significant reduction in tumor growth [345], thereby proving the concept of stroma targeting and the cathepsins as potent targets in cancer treatment. Therefore, the application of selective inhibitors of cathepsins offers new possibilities for the diagnosis and treatment of human diseases.
7.2. Cysteine cathepsins in rheumatoid arthritis
Rheumatoid arthritis (RA) is characterized by chronic synovial joint inflammation and the infiltration of an activated CD4+ T cell and APC, such as dendritic cells and macrophages [348]. It involves a progressive destruction of the articular cartilage, eventually leading to a loss of joint function. In addition to metalloproteases, such as collagenases and aggrecanases, lysosomal cysteine cathepsins B and L have been identified in synovial fluids [349], [350], [351], [352]. Furthermore, the involvement of cathepsin B and L in bone degradation was demonstrated by the selective inhibition of these enzymes [353]. In addition, the expression of the potent ECM degrading enzyme, cathepsin K, was found in synovial fibroblasts from RA patients to correlate with the severity of the disease [354]. There have been reports of an interesting finding that the chondroitin sulfate found in synovial fluids stabilizes the cathepsin K activity and subsequently increases its collagenolytic activity [355]. Very recently, it was reported that neither cathepsin B nor cathepsin H serum levels were associated with this disease, while the progression of destruction was associated with high serum levels of the inhibitors stefin A and B and not cystatin C [356]. In addition, it was found that cathepsin B and significantly elevated cathepsin S were detected in the synovial fluid of RA patients [357]. Significantly, an elevated level of cathepsin S is consistent with its critical role in the immune response. Recent developments in medical treatments may contribute to more effective and safe treatments of rheumatoid arthritis and ostheoarthritis using specific inhibitors [358], [359].
8. Perspectives
Over the past two decades we have witnessed tremendous advances in the understanding of the properties and structures of cysteine cathepsins and their endogenous protein inhibitors cystatins; although most of our knowledge has emerged only recently. The understanding of the basic biology of these proteins under normal and pathological conditions is constantly improving, although additional rules have yet to be more precisely established. The application of genomic and proteomic approaches to identify new substrates of individual cathepsins uncovers new roles for these enzymes in vivo [360], [361]. New strategies for the in vivo imaging of cysteine cathepsin activity for designing selective enzyme activity-based probes are in progress. Computational approaches based on the three-dimensional structure of domain–domain interactions involving cysteine cathepsins and their protein inhibitors should provide a more comprehensive view of this biological system [362]. By exploring the insights into the cysteine cathepsins biology of humans, new emerging therapies in various diseases, such as cancer, inflammatory diseases, neurological disorders, cardiovascular diseases, and parasitic diseases, are developing with a high probability of being incorporated into clinical trials.
Acknowledgements
The authors are grateful to Dr. Urška Repnik for valuable discussions and to Dr. Iztok Dolenc and Dejan Suban for their assistance in bibliography formatting. This work was supported by grants from the Slovene Research Agency (J1-2307 to V.T., P1-0048 to D.T., J1-9520 and J3-2258 to V.S., and P1-0140 and J1-3602 to B.T.) and by the FP7 projects LIVIMODE and MICROENVIMET (to B.T.).
Footnotes
This article is part of a Special Issue entitled: Proteolysis 50 years after the discovery of lysosome.
Contributor Information
Vito Turk, Email: vito.turk@ijs.si.
Dušan Turk, Email: dusan.turk@ijs.si.
References
- 1.De Duve C., Pressman B.C., Gianetto R., Wattiaux R., Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 1955;60:604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Duve C. The lysosome turns fifty. Nat. Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- 3.Bainton D.F. The discovery of lysosomes. J. Cell Biol. 1981;91:66–76. doi: 10.1083/jcb.91.3.66s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Duve C. Lysosomes revisited. Eur. J. Biochem. 1983;137:391–397. doi: 10.1111/j.1432-1033.1983.tb07841.x. [DOI] [PubMed] [Google Scholar]
- 5.De Duve C. Lysosomes, a new group of cytoplasmic particles. In: Hayashi T., editor. Subcellular Particles. The Ronald Press Co.; New York: 1959. pp. 128–159. [Google Scholar]
- 6.A. Ciechanover, Intracellular protein degradation: From a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting, Biochim. Biophys. Acta 1824 (2012) 2–12. [DOI] [PubMed]
- 7.Turk B., Turk V. Lysosomes as “suicide bags” in cell death: myth or reality? J. Biol. Chem. 2009;284:21783–21787. doi: 10.1074/jbc.R109.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brix K. Lysosomal proteases: revival of the sleeping beauty. In: Saftig P., editor. Lysosomes. Springer; Georgetown, TX: 2005. pp. 50–59. [Google Scholar]
- 9.Barrett A.J., Rawlings N.D., Woessner J.F., editors. Handbook of Proteolytic Enzymes. 2nd Ed. Elsevier; Amsterdam: 2004. [Google Scholar]
- 10.Turk B., Turk V., Turk D. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biol. Chem. 1997;378:141–150. [PubMed] [Google Scholar]
- 11.Turk D., Guncar G., Podobnik M., Turk B. Revised definition of substrate binding sites of papain-like cysteine proteases. Biol. Chem. 1998;379:137–147. doi: 10.1515/bchm.1998.379.2.137. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson D.P. Cysteine peptidases of mammals: their biological roles and potential effects in the oral cavity and other tissues in health and disease. Crit. Rev. Oral Biol. Med. 2002;13:238–275. doi: 10.1177/154411130201300304. [DOI] [PubMed] [Google Scholar]
- 13.Turk V., Turk B., Gunčar G., Turk D., Kos J. Lysosomal cathepsins: structure, role in antigen processing and presentation, and cancer. Adv. Enzyme Regul. 2002;42:285–303. doi: 10.1016/s0065-2571(01)00034-6. [DOI] [PubMed] [Google Scholar]
- 14.Turk B., Turk D., Salvesen G.S. Regulating cysteine protease activity: essential role of protease inhibitors as guardians and regulators. Curr. Pharm. Des. 2002;8:1623–1637. doi: 10.2174/1381612023394124. [DOI] [PubMed] [Google Scholar]
- 15.Stoka V., Turk B., Turk V. Lysosomal cysteine proteases: structural features and their role in apoptosis. IUBMB Life. 2005;57:347–353. doi: 10.1080/15216540500154920. [DOI] [PubMed] [Google Scholar]
- 16.Conus S., Simon H.U. Cathepsins: key modulators of cell death and inflammatory responses. Biochem. Pharmacol. 2008;76:1374–1382. doi: 10.1016/j.bcp.2008.07.041. [DOI] [PubMed] [Google Scholar]
- 17.Turk B., Turk D., Turk V. Lysosomal cysteine proteases: more than scavengers. Biochim. Biophys. Acta. 2000;1477:98–111. doi: 10.1016/s0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 18.Vasiljeva O., Reinheckel T., Peters C., Turk D., Turk V., Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr. Pharm. Des. 2007;13:387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- 19.Willstätter R., Bamann E. Über die proteasen der magenschleimhaut. Erste abhandlung über die enzyme der leukocyten. Hoppe-Seyler's Z. Physiol. Chem. 1929;180:127–143. [Google Scholar]
- 20.Rossi A., Deveraux Q., Turk B., Sali A. Comprehensive search for cysteine cathepsins in the human genome. Biol. Chem. 2004;385:363–372. doi: 10.1515/BC.2004.040. [DOI] [PubMed] [Google Scholar]
- 21.Turk V., Turk B., Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salminen-Mankonen H.J., Morko J., Vuorio E. Role of cathepsin K in normal joints and in the development of arthritis. Curr. Drug Targets. 2007;8:315–323. doi: 10.2174/138945007779940188. [DOI] [PubMed] [Google Scholar]
- 23.Asagiri M., Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40:251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 24.Linnevers C., Smeekens S.P., Brömme D. Human cathepsin W, a putative cysteine protease predominantly expressed in CD8+ T-lymphocytes. FEBS Lett. 1997;405:253–259. doi: 10.1016/s0014-5793(97)00118-x. [DOI] [PubMed] [Google Scholar]
- 25.Wex T., Wex H., Hartig R., Wilhelmsen S., Malfertheiner P. Functional involvement of cathepsin W in the cytotoxic activity of NK-92 cells. FEBS Lett. 2003;552:115–119. doi: 10.1016/s0014-5793(03)00895-0. [DOI] [PubMed] [Google Scholar]
- 26.Stoeckle C., Gouttefangeas C., Hammer M., Weber E., Melms A., Tolosa E. Cathepsin W expressed exclusively in CD8+ T cells and NK cells, is secreted during target cell killing but is not essential for cytotoxicity in human CTLs. Exp. Hematol. 2009;37:266–275. doi: 10.1016/j.exphem.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Hsing L.C., Rudensky A.Y. The lysosomal cysteine proteases in MHC class II antigen presentation. Immunol. Rev. 2005;207:229–241. doi: 10.1111/j.0105-2896.2005.00310.x. [DOI] [PubMed] [Google Scholar]
- 28.Santamaria I., Velasco G., Cazorla M., Fueyo A., Campo E., Lopez-Otin C. Cathepsin L2, a novel human cysteine proteinase produced by breast and colorectal carcinomas. Cancer Res. 1998;58:1624–1630. [PubMed] [Google Scholar]
- 29.Bromme D., Li Z., Barnes M., Mehler E. Human cathepsin V functional expression, tissue distribution, electrostatic surface potential, enzymatic characterization, and chromosomal localization. Biochemistry. 1999;38:2377–2385. doi: 10.1021/bi982175f. [DOI] [PubMed] [Google Scholar]
- 30.Goulet B., Baruch A., Moon N.S., Poirier M., Sansregret L.L., Erickson A., Bogyo M., Nepveu A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell. 2004;14:207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- 31.Duncan E.M., Muratore-Schroeder T.L., Cook R.G., Garcia B.A., Shabanowitz J., Hunt D.F., Allis C.D. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos-Rosa H., Kirmizis A., Nelson C., Bartke T., Saksouk N., Cote J., Kouzarides T. Histone H3 tail clipping regulates gene expression. Nat. Struct. Mol. Biol. 2009;16:17–22. doi: 10.1038/nsmb.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ceru S., Konjar S., Maher K., Repnik U., Krizaj I., Bencina M., Renko M., Nepveu A., Zerovnik E., Turk B., Kopitar-Jerala N. Stefin B interacts with histones and cathepsin L in the nucleus. J. Biol. Chem. 2010;285:10078–10086. doi: 10.1074/jbc.M109.034793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maubach G., Lim M.C., Zhuo L. Nuclear cathepsin F regulates activation markers in rat hepatic stellate cells. Mol. Biol. Cell. 2008;19:4238–4248. doi: 10.1091/mbc.E08-03-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zavašnik-Bergant T., Turk B. Cysteine cathepsins in the immune response. Tissue Antigens. 2006;67:349–355. doi: 10.1111/j.1399-0039.2006.00585.x. [DOI] [PubMed] [Google Scholar]
- 36.Brix K., Dunkhorst A., Mayer K., Jordans S. Cysteine cathepsins: cellular roadmap to different functions. Biochimie. 2008;90:194–207. doi: 10.1016/j.biochi.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Turk B. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 38.Stoka V., Turk V., Turk B. Lysosomal cysteine cathepsins: signaling pathways in apoptosis. Biol. Chem. 2007;388:555–560. doi: 10.1515/BC.2007.064. [DOI] [PubMed] [Google Scholar]
- 39.Turk B., Stoka V. Protease signalling in cell death: caspases versus cysteine cathepsins. FEBS Lett. 2007;581:2761–2767. doi: 10.1016/j.febslet.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 40.Turk B., Bieth J.G., Bjork I., Dolenc I., Turk D., Cimerman N., Kos J., Colic A., Stoka V., Turk V. Regulation of the activity of lysosomal cysteine proteinases by pH-induced inactivation and/or endogenous protein inhibitors, cystatins. Biol. Chem. Hoppe Seyler. 1995;376:225–230. doi: 10.1515/bchm3.1995.376.4.225. [DOI] [PubMed] [Google Scholar]
- 41.Kirschke H., Wiederanders B., Bromme D., Rinne A. Cathepsin S from bovine spleen. Purification, distribution, intracellular localization and action on proteins. Biochem. J. 1989;264:467–473. doi: 10.1042/bj2640467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turk B., Dolenc I., Turk V., Bieth J.G. Kinetics of the pH-induced inactivation of human cathepsin L. Biochemistry. 1993;32:375–380. doi: 10.1021/bi00052a046. [DOI] [PubMed] [Google Scholar]
- 43.Turk B., Dolenc I., Zerovnik E., Turk D., Gubensek F., Turk V. Human cathepsin B is a metastable enzyme stabilized by specific ionic interactions associated with the active site. Biochemistry. 1994;33:14800–14806. doi: 10.1021/bi00253a019. [DOI] [PubMed] [Google Scholar]
- 44.Almeida P.C., Nantes I.L., Chagas J.R., Rizzi C.C., Faljoni-Alario A., Carmona E., Juliano L., Nader H.B., Tersariol I.L. Cathepsin B activity regulation. Heparin-like glycosaminogylcans protect human cathepsin B from alkaline pH-induced inactivation. J. Biol. Chem. 2001;276:944–951. doi: 10.1074/jbc.M003820200. [DOI] [PubMed] [Google Scholar]
- 45.Costa M.G., Batista P.R., Shida C.S., Robert C.H., Bisch P.M., Pascutti P.G. How does heparin prevent the pH inactivation of cathepsin B? Allosteric mechanism elucidated by docking and molecular dynamics. BMC Genomics. 2010;11(Suppl 5):S5. doi: 10.1186/1471-2164-11-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herve-Grepinet V., Veillard F., Godat E., Heuze-Vourc'h N., Lecaille F., Lalmanach G. Extracellular catalase activity protects cysteine cathepsins from inactivation by hydrogen peroxide. FEBS Lett. 2008;582:1307–1312. doi: 10.1016/j.febslet.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Buck M.R., Karustis D.G., Day N.A., Honn K.V., Sloane B.F. Degradation of extracellular-matrix proteins by human cathepsin B from normal and tumour tissues. Biochem. J. 1992;282:273–278. doi: 10.1042/bj2820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lutgens S.P., Cleutjens K.B., Daemen M.J., Heeneman S. Cathepsin cysteine proteases in cardiovascular disease. FASEB J. 2007;21:3029–3041. doi: 10.1096/fj.06-7924com. [DOI] [PubMed] [Google Scholar]
- 49.Yasuda Y., Li Z., Greenbaum D., Bogyo M., Weber E., Bromme D. Cathepsin V, a novel and potent elastolytic activity expressed in activated macrophages. J. Biol. Chem. 2004;279:36761–36770. doi: 10.1074/jbc.M403986200. [DOI] [PubMed] [Google Scholar]
- 50.Bromme D., Okamoto K., Wang B.B., Biroc S. Human cathepsin O2, a matrix protein-degrading cysteine protease expressed in osteoclasts. Functional expression of human cathepsin O2 in Spodoptera frugiperda and characterization of the enzyme. J. Biol. Chem. 1996;271:2126–2132. doi: 10.1074/jbc.271.4.2126. [DOI] [PubMed] [Google Scholar]
- 51.Drake F.H., Dodds R.A., James I.E., Connor J.R., Debouck C., Richardson S., Lee-Rykaczewski E., Coleman L., Rieman D., Barthlow R., Hastings G., Gowen M. Cathepsin K, but not cathepsins B, L, or S, is abundantly expressed in human osteoclasts. J. Biol. Chem. 1996;271:12511–12516. doi: 10.1074/jbc.271.21.12511. [DOI] [PubMed] [Google Scholar]
- 52.Lecaille F., Bromme D., Lalmanach G. Biochemical properties and regulation of cathepsin K activity. Biochimie. 2008;90:208–226. doi: 10.1016/j.biochi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 53.Li Z., Yasuda Y., Li W., Bogyo M., Katz N., Gordon R.E., Fields G.B., Bromme D. Regulation of collagenase activities of human cathepsins by glycosaminoglycans. J. Biol. Chem. 2004;279:5470–5479. doi: 10.1074/jbc.M310349200. [DOI] [PubMed] [Google Scholar]
- 54.Selent J., Kaleta J., Li Z., Lalmanach G., Bromme D. Selective inhibition of the collagenase activity of cathepsin K. J. Biol. Chem. 2007;282:16492–16501. doi: 10.1074/jbc.M700242200. [DOI] [PubMed] [Google Scholar]
- 55.Turk B., Dolenc I., Lenarcic B., Krizaj I., Turk V., Bieth J.G., Bjork I. Acidic pH as a physiological regulator of human cathepsin L activity. Eur. J. Biochem. 1999;259:926–932. doi: 10.1046/j.1432-1327.1999.00145.x. [DOI] [PubMed] [Google Scholar]
- 56.Stoka V., Turk B., Schendel S.L., Kim T.H., Cirman T., Snipas S.J., Ellerby L.M., Bredesen D., Freeze H., Abrahamson M., Bromme D., Krajewski S., Reed J.C., Yin X.M., Turk V., Salvesen G.S. Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J. Biol. Chem. 2001;276:3149–3157. doi: 10.1074/jbc.M008944200. [DOI] [PubMed] [Google Scholar]
- 57.Cirman T., Oresic K., Mazovec G.D., Turk V., Reed J.C., Myers R.M., Salvesen G.S., Turk B. Selective disruption of lysosomes in HeLa cells triggers apoptosis mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J. Biol. Chem. 2004;279:3578–3587. doi: 10.1074/jbc.M308347200. [DOI] [PubMed] [Google Scholar]
- 58.Droga-Mazovec G., Bojic L., Petelin A., Ivanova S., Romih R., Repnik U., Salvesen G.S., Stoka V., Turk V., Turk B. Cysteine cathepsins trigger caspase-dependent cell death through cleavage of Bid and antiapoptotic Bcl-2 homologues. J. Biol. Chem. 2008;283:19140–19150. doi: 10.1074/jbc.M802513200. [DOI] [PubMed] [Google Scholar]
- 59.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 60.Drenth J., Jansonius J.N., Koekoek R., Swen H.M., Wolthers B.G. Structure of papain. Nature. 1968;218:929–932. doi: 10.1038/218929a0. [DOI] [PubMed] [Google Scholar]
- 61.Kamphuis I.G., Kalk K.H., Swarte M.B., Drenth J. Structure of papain refined at 1.65 A resolution. J. Mol. Biol. 1984;179:233–256. doi: 10.1016/0022-2836(84)90467-4. [DOI] [PubMed] [Google Scholar]
- 62.Baker E.N., Dodson E.J. Crystallographic refinement of the structure of actinidin at 1.7 Å resolution by fast fourier least-squares methods. Acta Crystallogr., Sect. A. 1980;36:559–572. [Google Scholar]
- 63.Musil D., Zucic D., Turk D., Engh R.A., Mayr I., Huber R., Popovic T., Turk V., Towatari T., Katunuma N., Bode W. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J. 1991;10:2321–2330. doi: 10.1002/j.1460-2075.1991.tb07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujishima A., Imai Y., Nomura T., Fujisawa Y., Yamamoto Y., Sugawara T. The crystal structure of human cathepsin L complexed with E-64. FEBS Lett. 1997;407:47–50. doi: 10.1016/s0014-5793(97)00216-0. [DOI] [PubMed] [Google Scholar]
- 65.McGrath M.E., Klaus J.L., Barnes M.G., Bromme D. Crystal structure of human cathepsin K complexed with a potent inhibitor. Nat. Struct. Biol. 1997;4:105–109. doi: 10.1038/nsb0297-105. [DOI] [PubMed] [Google Scholar]
- 66.Zhao B.G., Janson C.A., Amegadzie B.Y., Dalessio K., Griffin C., Hanning C.R., Jones C., Kurdyla J., McQueney M., Qiu X.Y., Smith W.W., AbdelMeguid S.S. Crystal structure of human osteoclast cathepsin K complex with E-64. Nat. Struct. Biol. 1997;4:109–111. doi: 10.1038/nsb0297-109. [DOI] [PubMed] [Google Scholar]
- 67.Guncar G., Podobnik M., Pungercar J., Strukelj B., Turk V., Turk D. Crystal structure of porcine cathepsin H determined at 2.1 angstrom resolution: location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function. Structure. 1998;6:51–61. doi: 10.1016/s0969-2126(98)00007-0. [DOI] [PubMed] [Google Scholar]
- 68.Guncar G., Klemencic I., Turk B., Turk V., Karaoglanovic-Carmona A., Juliano L., Turk D. Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease. Structure. 2000;8:305–313. doi: 10.1016/s0969-2126(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 69.Somoza J.R., Zhan H.J., Bowman K.K., Yu L., Mortara K.D., Palmer J.T., Clark J.M., McGrath M.E. Crystal structure of human cathepsin V. Biochemistry. 2000;39:12543–12551. doi: 10.1021/bi000951p. [DOI] [PubMed] [Google Scholar]
- 70.Turk D., Janjic V., Stern I., Podobnik M., Lamba D., Dahl S.W., Lauritzen C., Pedersen J., Turk V., Turk B. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001;20:6570–6582. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olsen J.G., Kadziola A., Lauritzen C., Pedersen J., Larsen S., Dahl S.W. Tetrameric dipeptidyl peptidase I directs substrate specificity by use of the residual pro-part domain. FEBS Lett. 2001;506:201–206. doi: 10.1016/s0014-5793(01)02911-8. [DOI] [PubMed] [Google Scholar]
- 72.Turkenburg J.P., Lamers M., Brzozowski A.M., Wright L.M., Hubbard R.E., Sturt S.L., Williams D.H. Structure of a Cys25 → Ser mutant of human cathepsin S. Acta Crystallogr. Sect. D. 2002;58:451–455. doi: 10.1107/s0907444901021825. [DOI] [PubMed] [Google Scholar]
- 73.Somoza J.R., Palmer J.T., Ho J.D. The crystal structure of human cathepsin F and its implications for the development of novel immunomodulators. J. Mol. Biol. 2002;322:559–568. doi: 10.1016/s0022-2836(02)00780-5. [DOI] [PubMed] [Google Scholar]
- 74.Guncar G., Pungercic G., Klemencic I., Turk V., Turk D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 1999;18:793–803. doi: 10.1093/emboj/18.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turk D., Turk B., Turk V. Papain-like lysosomal cysteine proteases and their inhibitors: drug discovery targets? Biochem. Soc. Symp. 2003;70:15–30. doi: 10.1042/bss0700015. [DOI] [PubMed] [Google Scholar]
- 76.Turk D., Podobnik M., Popovic T., Katunuma N., Bode W., Huber R., Turk V. Crystal-structure of cathepsin-B inhibited with CA030 at 2.0-angstrom resolution - a basis for the design of specific epoxysuccinyl inhibitors. Biochemistry. 1995;34:4791–4797. doi: 10.1021/bi00014a037. [DOI] [PubMed] [Google Scholar]
- 77.Yamamoto A., Hara T., Tomoo K., Ishida T., Fujii T., Hata Y., Murata M., Kitamura K. Binding mode of CA074, a specific irreversible inhibitor, to bovine cathepsin B as determined by X-ray crystal analysis of the complex. J. Biochem. 1997;121:974–977. doi: 10.1093/oxfordjournals.jbchem.a021682. [DOI] [PubMed] [Google Scholar]
- 78.Stern I., Schaschke N., Moroder L., Turk D. Crystal structure of NS-134 in complex with bovine cathepsin B: a two-headed epoxysuccinyl inhibitor extends along the entire active-site cleft. Biochem. J. 2004;381:511–517. doi: 10.1042/BJ20040237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lalmanach G., Serveau C., Brillard-Bourdet M., Chagas J.R., Mayer R., Juliano L., Gauthier F. Conserved cystatin segments as models for designing specific substrates and inhibitors of cysteine proteinases. J. Protein Chem. 1995;14:645–653. doi: 10.1007/BF01886903. [DOI] [PubMed] [Google Scholar]
- 80.Hasnain S., Hirama T., Huber C.P., Mason P., Mort J.S. Characterization of cathepsin B specificity by site-directed mutagenesis - importance of Glu(245) in the S2-P2 specificity for arginine and its role in transition-state stabilization. J. Biol. Chem. 1993;268:235–240. [PubMed] [Google Scholar]
- 81.Fox T., Mason P., Storer A.C., Mort J.S. Modification of S1 subsite specificity in the cysteine protease cathepsin-B. Protein Eng. 1995;8:53–57. doi: 10.1093/protein/8.1.53. [DOI] [PubMed] [Google Scholar]
- 82.Nägler D.K., Storer A.C., Portaro F.C.V., Carmona E., Juliano L., Menard R. Major increase in endopeptidase activity of human cathepsin B upon removal of occluding loop contacts. Biochemistry. 1997;36:12608–12615. doi: 10.1021/bi971264+. [DOI] [PubMed] [Google Scholar]
- 83.Illy C., Quraishi O., Wang J., Purisima E., Vernet T., Mort J.S. Role of the occluding loop in cathepsin B activity. J. Biol. Chem. 1997;272:1197–1202. doi: 10.1074/jbc.272.2.1197. [DOI] [PubMed] [Google Scholar]
- 84.Lecaille F., Chowdhury S., Purisima E., Bromme D., Lalmanach G. The S2 subsites of cathepsins K and L and their contribution to collagen degradation. Protein Sci. 2007;16:662–670. doi: 10.1110/ps.062666607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cherney M.M., Lecaille F., Kienitz M., Nallaseth F.S., Li Z.Q., James M.N.G., Bromme D. Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions. J. Biol. Chem. 2011;286:8988–8998. doi: 10.1074/jbc.M110.126706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Choe Y., Leonetti F., Greenbaum D.C., Lecaille F., Bogyo M., Bromme D., Ellman J.A., Craik C.S. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 2006;281:12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- 87.Bromme D., Bonneau P.R., Lachance P., Storer A.C. Engineering the S2 subsite specificity of human cathepsin S to a cathepsin L- and cathepsin B-like specificity. J. Biol. Chem. 1994;269:30238–30242. [PubMed] [Google Scholar]
- 88.Cotrin S.S., Puzer L., Judice W.A.D., Juliano L., Carmona A.K., Juliano M.A. Positional-scanning combinatorial libraries of fluorescence resonance energy transfer peptides to define substrate specificity of carboxydipeptidases: assays with human cathepsin B. Anal. Biochem. 2004;335:244–252. doi: 10.1016/j.ab.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 89.Puzer L., Cotrin S.S., Cezari M.H.S., Hirata I.Y., Juliano M.A., Stefe L., Turk D., Turk B., Juliano L., Carmona A.K. Recombinant human cathepsin X is a carboxymonopeptidase only: a comparison with cathepsins B and L. Biol. Chem. 2005;386:1191–1195. doi: 10.1515/BC.2005.136. [DOI] [PubMed] [Google Scholar]
- 90.Klemencic I., Carmona A.K., Cezari M.H.S., Juliano M.A., Juliano L., Guncar G., Turk D., Krizaj I., Turk V., Turk B. Biochemical characterization of human cathepsin X revealed that the enzyme is an exopeptidase, acting as carboxymonopeptidase or carboxydipeptidase. Eur. J. Biochem. 2000;267:5404–5412. doi: 10.1046/j.1432-1327.2000.01592.x. [DOI] [PubMed] [Google Scholar]
- 91.Lecaille F., Weidauer E., Juliano M.A., Bromme D., Lalmanach G. Probing cathepsin K activity with a selective substrate spanning its active site. Biochem. J. 2003;375:307–312. doi: 10.1042/BJ20030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alves M.F.M., Puzer L., Cotrin S.S., Juliano M.A., Juliano L., Bromme D., Carmona A.K. S3 to S3′ subsite specificity of recombinant human cathepsin K and development of selective internally quenched fluorescent substrates. Biochem. J. 2003;373:981–986. doi: 10.1042/BJ20030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ruckrich T., Brandenburg J., Cansier A., Muller M., Stevanovic S., Schilling K., Wiederanders B., Beck A., Melms A., Reich M., Driessen C., Kalbacher H. Specificity of human cathepsin S determined by processing of peptide substrates and MHC class II-associated invariant chain. Biol. Chem. 2006;387:1503–1511. doi: 10.1515/BC.2006.188. [DOI] [PubMed] [Google Scholar]
- 94.Lutzner N., Kalbacher H. Quantifying cathepsin S activity in antigen presenting cells using a novel specific substrate. J. Biol. Chem. 2008;283:36185–36194. doi: 10.1074/jbc.M806500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oliveira M., Torquato R.J.S., Alves M.F.M., Juliano M.A., Bromme D., Barros N.M.T., Carmona A.K. Improvement of cathepsin S detection using a designed FRET peptide based on putative natural substrates. Peptides. 2010;31:562–567. doi: 10.1016/j.peptides.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 96.Puzer L., Cotrin S.S., Alves M.F.M., Egborge T., Araujo M.S., Juliano M.A., Juliano L., Bromme D., Carmona A.K. Comparative substrate specificity analysis of recombinant human cathepsin V and cathepsin L. Arch. Biochem. Biophys. 2004;430:274–283. doi: 10.1016/j.abb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 97.Schilling O., Overall C.M. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 2008;26:685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
- 98.Clara R.O., Soares T.S., Torquato R.J.S., Lima C.A., Watanabe R.O.M., Barros N.M.T., Carmona A.K., Masuda A., Vaz Junior I.S., Tanaka A.S. Boophilus microplus cathepsin L-like (BmCL1) cysteine protease: specificity study using a peptide phage display library. Vet. Parasitol. 2011;181:291–300. doi: 10.1016/j.vetpar.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 99.Hanada K., Tamai M., Yamagishi M., Ohmura S., Sawada J., Tanaka I. Isolation and characterization of E-64, a new thiol protease inhibitor. Agric. Biol. Chem. 1978;42:523–528. [Google Scholar]
- 100.Parkes C., Kembhavi A.A., Barrett A.J. Calpain inhibition by peptide epoxides. Biochem. J. 1985;230:509–516. doi: 10.1042/bj2300509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Towatari T., Nikawa T., Murata M., Yokoo C., Tamai M., Hanada K., Katunuma N. Novel epoxysuccinyl peptides — a selective inhibitor of cathepsin B, in vivo. FEBS Lett. 1991;280:311–315. doi: 10.1016/0014-5793(91)80319-x. [DOI] [PubMed] [Google Scholar]
- 102.Murata M., Miyashita S., Yokoo C., Tamai M., Hanada K., Hatayama K., Towatari T., Nikawa T., Katunuma N. Novel epoxysuccinyl peptides — selective inhibitors of cathepsin B, in vitro. FEBS Lett. 1991;280:307–310. doi: 10.1016/0014-5793(91)80318-w. [DOI] [PubMed] [Google Scholar]
- 103.Schaschke N., Assfalg-Machleidt I., Machleidt W., Turk D., Moroder L. E-64 analogues as inhibitors of cathepsin B. On the role of the absolute configuration of the epoxysuccinyl group. Bioorg. Med. Chem. 1997;5:1789–1797. doi: 10.1016/s0968-0896(97)00105-3. [DOI] [PubMed] [Google Scholar]
- 104.Schaschke N., Assfalg-Machleidt I., Machleidt W., Moroder L. Substrate/propeptide-derived endo-epoxysuccinyl peptides as highly potent and selective cathepsin B inhibitors. FEBS Lett. 1998;421:80–82. doi: 10.1016/s0014-5793(97)01538-x. [DOI] [PubMed] [Google Scholar]
- 105.Katunuma N., Matsui A., Inubushi T., Murata E., Kakegawa H., Ohba Y., Turk D., Turk V., Tada Y., Asao T. Structure-based development of pyridoxal propionate derivatives as specific inhibitors of cathepsin K in vitro and in vivo. Biochem. Biophys. Res. Commun. 2000;267:850–854. doi: 10.1006/bbrc.1999.1953. [DOI] [PubMed] [Google Scholar]
- 106.Katunuma N., Murata E., Kakegawa H., Matsui A., Tsuzuki H., Tsuge H., Turk D., Turk V., Fukushima M., Tada Y., Asao T. Structure based development of novel specific inhibitors for cathepsin L and cathepsin S in vitro and in vivo. FEBS Lett. 1999;458:6–10. doi: 10.1016/s0014-5793(99)01107-2. [DOI] [PubMed] [Google Scholar]
- 107.Tsuge H., Nishimura T., Tada Y., Asao T., Turk D., Turk V., Katunuma N. Inhibition mechanism of cathepsin L-specific inhibitors based on the crystal structure of papain-CLIK148 complex. Biochem. Biophys. Res. Commun. 1999;266:411–416. doi: 10.1006/bbrc.1999.1830. [DOI] [PubMed] [Google Scholar]
- 108.Gour-Salin B.J., Lachance P., Magny M.C., Plouffe C., Menard R., Storer A.C. E64 trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane analogs as inhibitors of cysteine proteinases — investigation of S-2 subsite interactions. Biochem. J. 1994;299:389–392. doi: 10.1042/bj2990389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Watanabe D., Yamamoto A., Tomoo K., Matsumoto K., Murata M., Kitamura K., Ishida T. Quantitative evaluation of each catalytic subsite of cathepsin B for inhibitory activity based on inhibitory activity-binding mode relationship of epoxysuccinyl inhibitors by X-ray crystal structure analyses of complexes. J. Mol. Biol. 2006;362:979–993. doi: 10.1016/j.jmb.2006.07.070. [DOI] [PubMed] [Google Scholar]
- 110.Hori M., Hemmi H., Suzukake K., Hayashi H., Uehara Y., Takeuchi T., Umezawa H. Biosynthesis of leupeptin. J. Antibiot. 1978;31:95–98. doi: 10.7164/antibiotics.31.95. [DOI] [PubMed] [Google Scholar]
- 111.Yasuma T., Oi S., Choh N., Nomura T., Furuyama N., Nishimura A., Fujisawa Y., Sohda T. Synthesis of peptide aldehyde derivatives as selective inhibitors of human cathepsin L and their inhibitory effect on bone resorption. J. Med. Chem. 1998;41:4301–4308. doi: 10.1021/jm9803065. [DOI] [PubMed] [Google Scholar]
- 112.Wood W.J.L., Patterson A.W., Tsuruoka H., Jain R.K., Ellman J.A. Substrate activity screening: a fragment-based method for the rapid identification of nonpeptidic protease inhibitors. J. Am. Chem. Soc. 2005;127:15521–15527. doi: 10.1021/ja0547230. [DOI] [PubMed] [Google Scholar]
- 113.Crawford C., Mason R.W., Wikstrom P., Shaw E. The design of peptidyldiazomethane inhibitors to distinguish between the cysteine proteinases calpain II, cathepsin L and cathepsin B. Biochem. J. 1988;253:751–758. doi: 10.1042/bj2530751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Angliker H., Wikstrom P., Kirschke H., Shaw E. The inactivation of the cysteinyl exopeptidases cathepsin H and cathepsin C by affinity-labeling reagents. Biochem. J. 1989;262:63–68. doi: 10.1042/bj2620063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shaw E., Mohanty S., Colic A., Stoka V., Turk V. The affinity-labeling of cathepsin-S with peptidyl diazomethyl ketones — comparison with the inhibition of cathepsin L and calpain. FEBS Lett. 1993;334:340–342. doi: 10.1016/0014-5793(93)80707-2. [DOI] [PubMed] [Google Scholar]
- 116.Paulick M.G., Bogyo M. Development of activity-based probes for cathepsin X. ACS Chem. Biol. 2011;6:563–572. doi: 10.1021/cb100392r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kam C.M., Gotz M.G., Koot G., McGuire M., Thiele D., Hudig D., Powers J.C. Design and evaluation of inhibitors for dipeptidyl peptidase I (Cathepsin C) Arch. Biochem. Biophys. 2004;427:123–134. doi: 10.1016/j.abb.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 118.Bromme D., Smith R.A., Coles P.J., Kirschke H., Storer A.C., Krantz A. Potent inactivation of cathepsin S and cathepsin L by peptidyl (acyloxy)methyl ketones. Biol. Chem. Hoppe Seyler. 1994;375:343–347. doi: 10.1515/bchm3.1994.375.5.343. [DOI] [PubMed] [Google Scholar]
- 119.Bromme D., Klaus J.L., Okamoto K., Rasnick D., Palmer J.T. Peptidyl vinyl sulphones: a new class of potent and selective cysteine protease inhibitors - S2P2 specificity of human cathepsin O2 in comparison with cathepsins S and L. Biochem. J. 1996;315:85–89. doi: 10.1042/bj3150085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mendieta L., Pico A., Tarrago T., Teixido M., Castillo M., Rafecas L., Moyano A., Giralt E. Novel peptidyl aryl vinyl sulfones as highly potent and selective inhibitors of cathepsins L and B. ChemMedChem. 2010;5:1556–1567. doi: 10.1002/cmdc.201000109. [DOI] [PubMed] [Google Scholar]
- 121.Asaad N., Bethel P.A., Coulson M.D., Dawson J.E., Ford S.J., Gerhardt S., Grist M., Hamlin G.A., James M.J., Jones E.V., Karoutchi G.I., Kenny P.W., Morley A.D., Oldham K., Rankine N., Ryan D., Wells S.L., Wood L., Augustin M., Krapp S., Simader H., Steinbacher S. Dipeptidyl nitrile inhibitors of cathepsin L. Bioorg. Med. Chem. Lett. 2009;19:4280–4283. doi: 10.1016/j.bmcl.2009.05.071. [DOI] [PubMed] [Google Scholar]
- 122.Frizler M., Stirnberg M., Sisay M.T., Gütschow M. Development of nitrile-based peptidic inhibitors of cysteine cathepsins. Curr. Top. Med. Chem. 2010;10:294–322. doi: 10.2174/156802610790725452. [DOI] [PubMed] [Google Scholar]
- 123.Kerns J.K., Nie H., Bondinell W., Widdowson K.L., Yamashita D.S., Rahman A., Podolin P.L., Carpenter D.C., Jin Q., Riflade B., Dong X.Y., Nevins N., Keller P.M., Mitchell L., Tomaszek T. Azepanone-based inhibitors of human cathepsin S: optimization of selectivity via the P2 substituent. Bioorg. Med. Chem. Lett. 2011;21:4409–4415. doi: 10.1016/j.bmcl.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 124.Gotz M.G., Caffrey C.R., Hansell E., McKerrow J.H., Powers J.C. Peptidyl allyl sulfones: a new class of inhibitors for clan CA cysteine proteases. Bioorg. Med. Chem. 2004;12:5203–5211. doi: 10.1016/j.bmc.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 125.Powers J.C., Asgian J.L., Ekici O.D., James K.E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 126.Marquis R.W., Ru Y., Zeng J., Trout R.E.L., LoCastro S.M., Gribble A.D., Witherington J., Fenwick A.E., Garnier B., Tomaszek T., Tew D., Hemling M.E., Quinn C.J., Smith W.W., Zhao B.G., McQueney M.S., Janson C.A., D'Alessio K., Veber D.F. Cyclic ketone inhibitors of the cysteine protease cathepsin K. J. Med. Chem. 2001;44:725–736. doi: 10.1021/jm000320t. [DOI] [PubMed] [Google Scholar]
- 127.Patterson A.W., Wood W.J.L., Hornsby M., Lesley S., Spraggon G., Ellman J.A. Identification of selective, nonpeptidic nitrile inhibitors of cathepsin S using the substrate activity screening method. J. Med. Chem. 2006;49:6298–6307. doi: 10.1021/jm060701s. [DOI] [PubMed] [Google Scholar]
- 128.Cravatt B.F., Wright A.T., Kozarich J.W. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 129.Blum G., Mullins S.R., Keren K., Fonovic M., Jedeszko C., Rice M.J., Sloane B.F., Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat. Chem. Biol. 2005;1:203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- 130.Blum G., von Degenfeld G., Merchant M.J., Blau H.M., Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat. Chem. Biol. 2007;3:668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 131.Watzke A., Kosec G., Kindermann M., Jeske V., Nestler H.P., Turk V., Turk B., Wendt K.U. Selective activity-based probes for cysteine cathepsins. Angew. Chem. Int. Ed. 2008;47:406–409. doi: 10.1002/anie.200702811. [DOI] [PubMed] [Google Scholar]
- 132.Caglic D., Globisch A., Kindermann M., Lim N.H., Jeske V., Juretschke H.P., Bartnik E., Weithmann K.U., Nagase H., Turk B., Wendt K.U. Functional in vivo imaging of cysteine cathepsin activity in murine model of inflammation. Bioorg. Med. Chem. 2011;19:1055–1061. doi: 10.1016/j.bmc.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 133.Godat E., Lecaille F., Desmazes C., Duchene S., Weidauer E., Saftig P., Bromme D., Vandier C., Lalmanach G. Cathepsin K: a cysteine protease with unique kinin-degrading properties. Biochem. J. 2004;383:501–506. doi: 10.1042/BJ20040864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lecaille F., Vandier C., Godat E., Herve-Grepinet V., Bromme D., Lalmanach G. Modulation of hypotensive effects of kinins by cathepsin K. Arch. Biochem. Biophys. 2007;459:129–136. doi: 10.1016/j.abb.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 135.Pham C.T.N., Ley T.J. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakagawa T., Roth W., Wong P., Nelson A., Farr A., Deussing J., Villadangos J.A., Ploegh H., Peters C., Rudensky A.Y. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280:450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- 137.Honey K., Nakagawa T., Peters C., Rudensky A. Cathepsin L regulates CD4(+) T cell selection independently of its effect on invariant chain: a role in the generation of positively selecting peptide ligands. J. Exp. Med. 2002;195:1349–1358. doi: 10.1084/jem.20011904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Funkelstein L., Hook V. The novel role of cathepsin L for neuropeptide production illustrated by research strategies in chemical biology with protease gene knockout and expression. Methods Mol. Biol. 2011;768:107–125. doi: 10.1007/978-1-61779-204-5_5. [DOI] [PubMed] [Google Scholar]
- 139.Hook V., Funkelstein L., Lu D., Bark S., Wegrzyn J., Hwang S.R. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li Z.Q., Hou W.S., Escalante-Torres C.R., Gelb B.D., Bromme D. Collagenase activity of cathepsin K depends on complex formation with chondroitin sulfate. J. Biol. Chem. 2002;277:28669–28676. doi: 10.1074/jbc.M204004200. [DOI] [PubMed] [Google Scholar]
- 141.Nascimento F.D., Rizzi C.C.A., Nantes I.L., Stefe L., Turk B., Carmona A.K., Nader H.B., Juliano L., Tersariol I.L.S. Cathepsin X binds to cell surface heparan sulfate proteoglycans. Arch. Biochem. Biophys. 2005;436:323–332. doi: 10.1016/j.abb.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 142.Hou W.S., Li Z.Q., Buttner F.H., Bartnik E., Bromme D. Cleavage site specificity of cathepsin K toward cartilage proteoglycans and protease complex formation. Biol. Chem. 2003;384:891–897. doi: 10.1515/BC.2003.100. [DOI] [PubMed] [Google Scholar]
- 143.Li Z.Q., Kienetz M., Cherney M.M., James M.N.G., Bromme D. The crystal and molecular structures of a cathepsin K: chondroitin sulfate complex. J. Mol. Biol. 2008;383:78–91. doi: 10.1016/j.jmb.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 144.Kakegawa H., Nikawa T., Tagami K., Kamioka H., Sumitani K., Kawata T., Drobnic-Kosorok M., Lenarcic B., Turk V., Katunuma N. Participation of cathepsin L on bone-resorption. FEBS Lett. 1993;321:247–250. doi: 10.1016/0014-5793(93)80118-e. [DOI] [PubMed] [Google Scholar]
- 145.Novinec M., Kovacic L., Lenarcic B., Baici A. Conformational flexibility and allosteric regulation of cathepsin K. Biochem. J. 2010;429:379–389. doi: 10.1042/BJ20100337. [DOI] [PubMed] [Google Scholar]
- 146.Polgar L., Csoma C. Dissociation of ionizing groups in the binding cleft inversely controls the endo- and exopeptidase activities of cathepsin B. J. Biol. Chem. 1987;262:14448–14453. [PubMed] [Google Scholar]
- 147.Jordans S., Jenko-Kokalj S., Kuhl N.M., Tedelind S., Sendt W., Bromme D., Turk D., Brix K. Monitoring compartment-specific substrate cleavage by cathepsins B, K, L, and S at physiological pH and redox conditions. BMC Biochem. 2009;10:23. doi: 10.1186/1471-2091-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Adams-Cioaba M.A., Krupa J.C., Xu C., Mort J.S., Min J.R. Structural basis for the recognition and cleavage of histone H3 by cathepsin L. Nat. Commun. 2011;2:197. doi: 10.1038/ncomms1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lechner A.M., Assfalg-Machleidt I., Zahler S., Stoeckelhuber M., Machleidt W., Jochum M., Nagler D.K. RGD-dependent binding of procathepsin X to integrin alpha(v)beta(3) mediates cell-adhesive properties. J. Biol. Chem. 2006;281:39588–39597. doi: 10.1074/jbc.M513439200. [DOI] [PubMed] [Google Scholar]
- 150.Obermajer N., Repnik U., Jevnikar Z., Turk B., Kreft M., Kos J. Cysteine protease cathepsin X modulates immune response via activation of beta(2) integrins. Immunology. 2008;124:76–88. doi: 10.1111/j.1365-2567.2007.02740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jevnikar Z., Obermajer N., Doljak B., Turk S., Gobec S., Svajger U., Hailfinger S., Thome M., Kos J. Cathepsin X cleavage of the beta(2) integrin regulates talin-binding and LFA-1 affinity in T cells. J. Leukocyte Biol. 2011;90:99–109. doi: 10.1189/jlb.1110622. [DOI] [PubMed] [Google Scholar]
- 152.Jevnikar Z., Obermajer N., Pecar-Fonovic U., Karaoglanovic-Carmona A., Kos J. Cathepsin X cleaves the beta 2 cytoplasmic tail of LFA-1 inducing the intermediate affinity form of LFA-1 and alpha-actinin-1 binding. Eur. J. Immunol. 2009;39:3217–3227. doi: 10.1002/eji.200939562. [DOI] [PubMed] [Google Scholar]
- 153.Taggart C.C., Lowe G.J., Greene C.M., Mulgrew A.T., O'Neill S.J., Levine R.L., McElvaney N.G. Cathepsin B, L, and S cleave and inactivate secretory leucoprotease inhibitor. J. Biol. Chem. 2001;276:33345–33352. doi: 10.1074/jbc.M103220200. [DOI] [PubMed] [Google Scholar]
- 154.Simmons G., Bertram S., Glowacka I., Steffen I., Chaipan C., Agudelo J., Lu K., Rennekamp A.J., Hofmann H., Bates P., Pohlmann S. Different host cell proteases activate the SARS-coronavirus spike-protein for cell–cell and virus–cell fusion. Virology. 2011;413:265–274. doi: 10.1016/j.virol.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang I.C., Bosch B.J., Li F., Li W.H., Lee K.H., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J.M., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Twining S.S. Regulation of proteolytic activity in tissues. Crit. Rev. Biochem. Mol. Biol. 1994;29:315–383. doi: 10.3109/10409239409083484. [DOI] [PubMed] [Google Scholar]
- 157.Saftig P., Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 158.Schroder B.A., Wrocklage C., Hasilik A., Saftig P. The proteome of lysosomes. Proteomics. 2010;10:4053–4076. doi: 10.1002/pmic.201000196. [DOI] [PubMed] [Google Scholar]
- 159.Hasilik A., Wrocklage C., Schroder B. Intracellular trafficking of lysosomal proteins and lysosomes. Int. J. Clin. Pharmacol. Ther. 2009;47:S18–S33. doi: 10.5414/cpp47018. [DOI] [PubMed] [Google Scholar]
- 160.Wiederanders B., Kaulmann G., Schilling K. Functions of propeptide parts in cysteine proteases. Curr. Protein Pept. Sci. 2003;4:309–326. doi: 10.2174/1389203033487081. [DOI] [PubMed] [Google Scholar]
- 161.Cygler M., Sivaraman J., Grochulski P., Coulombe R., Storer A.C., Mort J.S. Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion. Structure. 1996;4:405–416. doi: 10.1016/s0969-2126(96)00046-9. [DOI] [PubMed] [Google Scholar]
- 162.Turk D., Podobnik M., Kuhelj R., Dolinar M., Turk V. Crystal structures of human procathepsin B at 3.2 and 3.3 Angstroms resolution reveal an interaction motif between a papain-like cysteine protease and its propeptide. FEBS Lett. 1996;384:211–214. doi: 10.1016/0014-5793(96)00309-2. [DOI] [PubMed] [Google Scholar]
- 163.Podobnik M., Kuhelj R., Turk V., Turk D. Crystal structure of the wild-type human procathepsin B at 2.5 A resolution reveals the native active site of a papain-like cysteine protease zymogen. J. Mol. Biol. 1997;271:774–788. doi: 10.1006/jmbi.1997.1218. [DOI] [PubMed] [Google Scholar]
- 164.Coulombe R., Grochulski P., Sivaraman J., Menard R., Mort J.S., Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15:5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 165.Sivaraman J., Lalumiere M., Menard R., Cygler M. Crystal structure of wild-type human procathepsin K. Protein Sci. 1999;8:283–290. doi: 10.1110/ps.8.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.LaLonde J.M., Zhao B.G., Janson C.A., D'Alessio K.J., McQueney M.S., Orsini M.J., Debouck C.M., Smith W.W. The crystal structure of human procathepsin K. Biochemistry. 1999;38:862–869. doi: 10.1021/bi9822271. [DOI] [PubMed] [Google Scholar]
- 167.Sivaraman J., Nagler D.K., Zhang R.L., Menard R., Cygler M. Crystal structure of human procathepsin X: a cysteine protease with the proregion covalently linked to the active site cysteine. J. Mol. Biol. 2000;295:939–951. doi: 10.1006/jmbi.1999.3410. [DOI] [PubMed] [Google Scholar]
- 168.Nagler D.K., Zhang R.L., Tam W., Sulea T., Purisima E.O., Menard R. Human cathepsin X: a cysteine protease with unique carboxypeptidase activity. Biochemistry. 1999;38:12648–12654. doi: 10.1021/bi991371z. [DOI] [PubMed] [Google Scholar]
- 169.Nissler K., Kreusch S., Rommerskirch W., Strubel W., Weber E., Wiederanders B. Sorting of non-glycosylated human procathepsin S in mammalian cells. Biol. Chem. 1998;379:219–224. doi: 10.1515/bchm.1998.379.2.219. [DOI] [PubMed] [Google Scholar]
- 170.Nishimura Y., Kawabata T., Furuno K., Kato K. Evidence that aspartic proteinase is involved in the proteolytic processing event of procathepsin L in lysosomes. Arch. Biochem. Biophys. 1989;271:400–406. doi: 10.1016/0003-9861(89)90289-0. [DOI] [PubMed] [Google Scholar]
- 171.Rowan A.D., Mason P., Mach L., Mort J.S. Rat procathepsin B. Proteolytic processing to the mature form in vitro. J. Biol. Chem. 1992;267:15993–15999. [PubMed] [Google Scholar]
- 172.Mach L., Mort J.S., Glossl J. Maturation of human procathepsin B. Proenzyme activation and proteolytic processing of the precursor to the mature proteinase, in vitro, are primarily unimolecular processes. J. Biol. Chem. 1994;269:13030–13035. [PubMed] [Google Scholar]
- 173.Menard R., Carmona E., Takebe S., Dufour E., Plouffe C., Mason P., Mort J.S. Autocatalytic processing or recombinant human procathepsin L. Contribution of both intermolecular and unimolecular events in the processing of procathepsin L in vitro. J. Biol. Chem. 1998;273:4478–4484. doi: 10.1074/jbc.273.8.4478. [DOI] [PubMed] [Google Scholar]
- 174.Jerala R., Zerovnik E., Kidric J., Turk V. pH-induced conformational transitions of the propeptide of human cathepsin L. A role for a molten globule state in zymogen activation. J. Biol. Chem. 1998;273:11498–11504. doi: 10.1074/jbc.273.19.11498. [DOI] [PubMed] [Google Scholar]
- 175.McQueney M.S., Amegadzie B.Y., Dalessio K., Hanning C.R., McLaughlin M.M., McNulty D., Carr S.A., Ijames C., Kurdyla J., Jones C.S. Autocatalytic activation of human cathepsin K. J. Biol. Chem. 1997;272:13955–13960. doi: 10.1074/jbc.272.21.13955. [DOI] [PubMed] [Google Scholar]
- 176.Rozman J., Stojan J., Kuhelj R., Turk V., Turk B. Autocatalytic processing of recombinant human procathepsin B is a bimolecular process. FEBS Lett. 1999;459:358–362. doi: 10.1016/s0014-5793(99)01302-2. [DOI] [PubMed] [Google Scholar]
- 177.Pungercar J.R., Caglic D., Sajid M., Dolinar M., Vasiljeva O., Pozgan U., Turk D., Bogyo M., Turk V., Turk B. Autocatalytic processing of procathepsin B is triggered by proenzyme activity. FEBS J. 2009;276:660–668. doi: 10.1111/j.1742-4658.2008.06815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dahl S.W., Halkier T., Lauritzen C., Dolenc I., Pedersen J., Turk V., Turk B. Human recombinant pro-dipeptidyl peptidase I (cathepsin C) can be activated by cathepsins L and S but not by autocatalytic processing. Biochemistry. 2001;40:1671–1678. doi: 10.1021/bi001693z. [DOI] [PubMed] [Google Scholar]
- 179.Caglic D., Pungercar J.R., Pejler G., Turk V., Turk B. Glycosaminoglycans facilitate procathepsin B activation through disruption of propeptide-mature enzyme interactions. J. Biol. Chem. 2007;282:33076–33085. doi: 10.1074/jbc.M705761200. [DOI] [PubMed] [Google Scholar]
- 180.Vasiljeva O., Dolinar M., Pungercar J.R., Turk V., Turk B. Recombinant human procathepsin S is capable of autocatalytic processing at neutral pH in the presence of glycosaminoglycans. FEBS Lett. 2005;579:1285–1290. doi: 10.1016/j.febslet.2004.12.093. [DOI] [PubMed] [Google Scholar]
- 181.Fox T., de Miguel E., Mort J.S., Storer A.C. Potent slow-binding inhibition of cathepsin B by its propeptide. Biochemistry. 1992;31:12571–12576. doi: 10.1021/bi00165a005. [DOI] [PubMed] [Google Scholar]
- 182.Carmona E., Dufour E., Plouffe C., Takebe S., Mason P., Mort J.S., Menard R. Potency and selectivity of the cathepsin L propeptide as an inhibitor of cysteine proteases. Biochemistry. 1996;35:8149–8157. doi: 10.1021/bi952736s. [DOI] [PubMed] [Google Scholar]
- 183.Guay J., Falgueyret J.P., Ducret A., Percival M.D., Mancini J.A. Potency and selectivity of inhibition of cathepsin K, L and S by their respective propeptides. Eur. J. Biochem. 2000;267:6311–6318. doi: 10.1046/j.1432-1327.2000.01730.x. [DOI] [PubMed] [Google Scholar]
- 184.Maubach G., Schilling K., Rommerskirch W., Wenz I., Schultz J.E., Weber E., Wiederanders B. The inhibition of cathepsin S by its propeptide — specificity and mechanism of action. Eur. J. Biochem. 1997;250:745–750. doi: 10.1111/j.1432-1033.1997.00745.x. [DOI] [PubMed] [Google Scholar]
- 185.Groves M.R., Coulombe R., Jenkins J., Cygler M. Structural basis for specificity of papain-like cysteine protease proregions toward their cognate enzymes. Proteins. 1998;32:504–514. [PubMed] [Google Scholar]
- 186.Cygler M., Mort J.S. Proregion structure of members of the papain superfamily. Mode of inhibition of enzymatic activity. Biochimie. 1997;79:645–652. doi: 10.1016/s0300-9084(97)83497-9. [DOI] [PubMed] [Google Scholar]
- 187.Karrer K.M., Peiffer S.L., DiTomas M.E. Two distinct gene subfamilies within the family of cysteine protease genes. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nagler D.K., Menard R. Human cathepsin X: a novel cysteine protease of the papain family with a very short proregion and unique insertions. FEBS Lett. 1998;434:135–139. doi: 10.1016/s0014-5793(98)00964-8. [DOI] [PubMed] [Google Scholar]
- 189.Santamaria I., Velasco G., Pendas A.M., Fueyo A., Lopez-Otin C. Cathepsin Z, a novel human cysteine proteinase with a short propeptide domain and a unique chromosomal location. J. Biol. Chem. 1998;273:16816–16823. doi: 10.1074/jbc.273.27.16816. [DOI] [PubMed] [Google Scholar]
- 190.Wang B., Shi G.P., Yao P.M., Li Z.Q., Chapman H.A., Bromme D. Human cathepsin F. Molecular cloning, functional expression, tissue localization, and enzymatic characterization. J. Biol. Chem. 1998;273:32000–32008. doi: 10.1074/jbc.273.48.32000. [DOI] [PubMed] [Google Scholar]
- 191.Nagler D.K., Sulea T., Menard R. Full-length cDNA of human cathepsin F predicts the presence of a cystatin domain at the N-terminus of the cysteine protease zymogen. Biochem. Biophys. Res. Commun. 1999;257:313–318. doi: 10.1006/bbrc.1999.0461. [DOI] [PubMed] [Google Scholar]
- 192.Paris A., Strukelj B., Pungercar J., Renko M., Dolenc I., Turk V. Molecular cloning and sequence analysis of human preprocathepsin C. FEBS Lett. 1995;369:326–330. doi: 10.1016/0014-5793(95)00777-7. [DOI] [PubMed] [Google Scholar]
- 193.Klotz C., Ziegler T., Danilowicz-Luebert E., Hartmann S. Cystatins of parasitic organisms. Adv. Exp. Med. Biol. 2011;712:208–221. doi: 10.1007/978-1-4419-8414-2_13. [DOI] [PubMed] [Google Scholar]
- 194.Ogrinc T., Dolenc I., Ritonja A., Turk V. Purification of the complex of cathepsin L and the MHC class II-associated invariant chain fragment from human kidney. FEBS Lett. 1993;336:555–559. doi: 10.1016/0014-5793(93)80875-u. [DOI] [PubMed] [Google Scholar]
- 195.Turk V., Brzin J., Longer M., Ritonja A., Eropkin M., Borchart U., Machleidt W. Protein inhibitors of cysteine proteinases. III. Amino-acid sequence of cystatin from chicken egg white. Hoppe-Seyler's Z. Physiol. Chem. 1983;364:1487–1496. doi: 10.1515/bchm2.1983.364.2.1487. [DOI] [PubMed] [Google Scholar]
- 196.Dayhoff M.O., Barker W.C., Hunt L.T. National Biomedical Research Foundation; Washington, D.C.: 1978. Protein Superfamilies. [Google Scholar]
- 197.Barrett A.J., Fritz H., Grubb A., Isemura S., Jarvinen M., Katunuma N., Machleidt W., Mulleresterl W., Sasaki M., Turk V. Nomenclature and classification of the proteins homologous with the cysteine-proteinase inhibitor chicken cystatin. Biochem. J. 1986;236:312-312. doi: 10.1042/bj2360312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rawlings N.D., Barrett A.J. Evolution of proteins of the cystatin superfamily. J. Mol. Evol. 1990;30:60–71. doi: 10.1007/BF02102453. [DOI] [PubMed] [Google Scholar]
- 199.Turk V., Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285:213–219. doi: 10.1016/0014-5793(91)80804-c. [DOI] [PubMed] [Google Scholar]
- 200.Barrett A.J., Rawlings N.D., Davies M.E., Machleidt W., Salvensen G., Turk V. Cysteine proteinase inhibitors of cystatin superfamily. In: Barrett A.J., Salvesen G., editors. Proteinase Inhibitors. Elsevier; Amsterdam: 1986. pp. 519–569. [Google Scholar]
- 201.Abrahamson M., Alvarez-Fernandez M., Nathanson C.M. Cystatins. Biochem. Soc. Symp. 2003;70:179–199. doi: 10.1042/bss0700179. [DOI] [PubMed] [Google Scholar]
- 202.Turk V., Stoka V., Turk D. Cystatins: biochemical and structural properties, and medical relevance. Front. Biosci. 2008;13:5406–5420. doi: 10.2741/3089. [DOI] [PubMed] [Google Scholar]
- 203.Reynolds J.L., Skepper J.N., McNair R., Kasama T., Gupta K., Weissberg P.L., Jahnen-Dechent W., Shanahan C.M. Multifunctional roles for serum protein fetuin-A in inhibition of human vascular smooth muscle cell calcification. J. Am. Soc. Nephrol. 2005;16:2920–2930. doi: 10.1681/ASN.2004100895. [DOI] [PubMed] [Google Scholar]
- 204.Jones A.L., Hulett M.D., Parish C.R. Histidine-rich glycoprotein: a novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol. Cell Biol. 2005;83:106–118. doi: 10.1111/j.1440-1711.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 205.Brown W.M., Dziegielewska K.M. Friends and relations of the cystatin superfamily — new members and their evolution. Protein Sci. 1997;6:5–12. doi: 10.1002/pro.5560060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kordis D., Turk V. Phylogenomic analysis of the cystatin superfamily in eukaryotes and prokaryotes. BMC Evol. Biol. 2009;9:266. doi: 10.1186/1471-2148-9-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rawlings N.D., Barrett A.J. MEROPS: the peptidase database. Nucleic Acids Res. 1999;27:325–331. doi: 10.1093/nar/27.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rawlings N.D., Barrett A.J., Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rawlings N.D., Tolle D.P., Barrett A.J. Evolutionary families of peptidase inhibitors. Biochem. J. 2004;378:705–716. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rawlings N.D. Peptidase inhibitors in the MEROPS database. Biochimie. 2010;92:1463–1483. doi: 10.1016/j.biochi.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 211.Abrahamson M., Barrett A.J., Salvesen G., Grubb A. Isolation of six cysteine proteinase inhibitors from human urine. Their physicochemical and enzyme kinetic properties and concentrations in biological fluids. J. Biol. Chem. 1986;261:1282–1289. [PubMed] [Google Scholar]
- 212.Turk B., Krizaj I., Kralj B., Dolenc I., Popovic T., Bieth J.G., Turk V. Bovine stefin C, a new member of the stefin family. J. Biol. Chem. 1993;268:7323–7329. [PubMed] [Google Scholar]
- 213.Ni J., Fernandez M.A., Danielsson L., Chillakuru R.A., Zhang J.L., Grubb A., Su J., Gentz R., Abrahamson M. Cystatin F is a glycosylated human low molecular weight cysteine proteinase inhibitor. J. Biol. Chem. 1998;273:24797–24804. doi: 10.1074/jbc.273.38.24797. [DOI] [PubMed] [Google Scholar]
- 214.Veillard F., Lecaille F., Lalmanach G. Lung cysteine cathepsins: intruders or unorthodox contributors to the kallikrein-kinin system? Int. J. Biochem. Cell Biol. 2008;40:1079–1094. doi: 10.1016/j.biocel.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 215.DeLa Cadena R.A.D., Colman R.W. Structure and functions of human kininogens. Trends Pharmacol. Sci. 1991;12:272–275. doi: 10.1016/0165-6147(91)90569-e. [DOI] [PubMed] [Google Scholar]
- 216.Kitamura N., Kitagawa H., Fukushima D., Takagaki Y., Miyata T., Nakanishi S. Structural organization of the human kininogen gene and a model for its evolution. J. Biol. Chem. 1985;260:8610–8617. [PubMed] [Google Scholar]
- 217.Kakizuka A., Kitamura N., Nakanishi S. Localization of DNA sequences governing alternative mRNA production of rat kininogen genes. J. Biol. Chem. 1988;263:3884–3892. [PubMed] [Google Scholar]
- 218.Muller-Esterl W., Iwanaga S., Nakanishi S. Kininogens revisited. Trends Biochem. Sci. 1986;11:336–339. [Google Scholar]
- 219.Kaufmann J., Haasemann M., Modrow S., Muller-Esterl W. Structural dissection of the multidomain kininogens. Fine mapping of the target epitopes of antibodies interfering with their functional-properties. J. Biol. Chem. 1993;268:9079–9091. [PubMed] [Google Scholar]
- 220.Turk B., Stoka V., Bjork I., Boudier C., Johansson G., Dolenc I., Colic A., Bieth J.G., Turk V. High-affinity binding of two molecules of cysteine proteinases to low-molecular-weight kininogen. Protein Sci. 1995;4:1874–1880. doi: 10.1002/pro.5560040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Turk B., Stoka V., Turk V., Johansson G., Cazzulo J.J., Bjork I. High-molecular-weight kininogen binds two molecules of cysteine proteinases with different rate constants. FEBS Lett. 1996;391:109–112. doi: 10.1016/0014-5793(96)00611-4. [DOI] [PubMed] [Google Scholar]
- 222.Salvesen G., Parkes C., Abrahamson M., Grubb A., Barrett A.J. Human low-Mr kininogen contains three copies of a cystatin sequence that are divergent in structure and in inhibitory activity for cysteine proteinases. Biochem. J. 1986;234:429–434. doi: 10.1042/bj2340429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lenarcic B., Krasovec M., Ritonja A., Olafsson I., Turk V. Inactivation of human cystatin C and kininogen by human cathepsin D. FEBS Lett. 1991;280:211–215. doi: 10.1016/0014-5793(91)80295-e. [DOI] [PubMed] [Google Scholar]
- 224.Kolte D., Osman N., Yang J., Shariat-Madar Z. High molecular weight kininogen activates B(2) receptor signaling pathway in human vascular endothelial cells. J. Biol. Chem. 2011;286:24561–24571. doi: 10.1074/jbc.M110.211557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lalmanach G., Naudin C., Lecaille F., Fritz H. Kininogens: More than cysteine protease inhibitors and kinin precursors. Biochimie. 2010;92:1568–1579. doi: 10.1016/j.biochi.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 226.Bevec T., Stoka V., Pungercic G., Cazzulo J.J., Turk V. A fragment of the major histocompatibility complex class II-associated p41 invariant chain inhibits cruzipain, the major cysteine proteinase from Trypanosoma cruzi. FEBS Lett. 1997;401:259–261. doi: 10.1016/s0014-5793(96)01443-3. [DOI] [PubMed] [Google Scholar]
- 227.Bevec T., Stoka V., Pungercic G., Dolenc I., Turk V. Major histocompatibility complex class II-associated p41 invariant chain fragment is a strong inhibitor of lysosomal cathepsin L. J. Exp. Med. 1996;183:1331–1338. doi: 10.1084/jem.183.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lenarcic B., Ritonja A., Strukelj B., Turk B., Turk V. Equistatin, a new inhibitor of cysteine proteinases from Actinia equina, is structurally related to thyroglobulin type-1 domain. J. Biol. Chem. 1997;272:13899–13903. doi: 10.1074/jbc.272.21.13899. [DOI] [PubMed] [Google Scholar]
- 229.Lenarcic B., Turk V. Thyroglobulin type-1 domains in equistatin inhibit both papain-like cysteine proteinases and cathepsin D. J. Biol. Chem. 1999;274:563–566. doi: 10.1074/jbc.274.2.563. [DOI] [PubMed] [Google Scholar]
- 230.Molina F., Bouanani M., Pau B., Granier C. Characterization of the type-1 repeat from thyroglobulin, a cysteine-rich module found in proteins from different families. Eur. J. Biochem. 1996;240:125–133. doi: 10.1111/j.1432-1033.1996.0125h.x. [DOI] [PubMed] [Google Scholar]
- 231.Lenarcic B., Bevec T. Thyropins — new structurally related proteinase inhibitors. Biol. Chem. 1998;379:105–111. [PubMed] [Google Scholar]
- 232.Strukelj B., Lenarcic B., Gruden K., Pungercar J., Rogelj B., Turk V., Bosch D., Jongsma M.A. Equistatin, a protease inhibitor from the sea anemone Actinia equina, is composed of three structural and functional domains. Biochem. Biophys. Res. Commun. 2000;269:732–736. doi: 10.1006/bbrc.2000.2356. [DOI] [PubMed] [Google Scholar]
- 233.Mihelic M., Turk D. Two decades of thyroglobulin type-1 domain research. Biol. Chem. 2007;388:1123–1130. doi: 10.1515/BC.2007.155. [DOI] [PubMed] [Google Scholar]
- 234.Schick C., Pemberton P.A., Shi G.P., Kamachi Y., Cataltepe S., Bartuski A.J., Gornstein E.R., Bromme D., Chapman H.A., Silverman G.A. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry. 1998;37:5258–5266. doi: 10.1021/bi972521d. [DOI] [PubMed] [Google Scholar]
- 235.Welss T., Sun J.R., Irving J.A., Blum R., Smith A.I., Whisstock J.C., Pike R.N., von Mikecz A., Ruzicka T., Bird P.I., Abts H.F. Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis. Biochemistry. 2003;42:7381–7389. doi: 10.1021/bi027307q. [DOI] [PubMed] [Google Scholar]
- 236.Hwang S.R., Stoka V., Turk V., Hook V. Resistance of cathepsin L compared to elastase to proteolysis when complexed with the serpin endopin 2C, and recovery of cathepsin L activity. Biochem. Biophys. Res. Commun. 2006;340:1238–1243. doi: 10.1016/j.bbrc.2005.12.130. [DOI] [PubMed] [Google Scholar]
- 237.Turk V., Turk B. Lysosomal cysteine proteases and their protein inhibitors: recent developments. Acta Chim. Slov. 2008;55:727–738. [Google Scholar]
- 238.Cornwall G.A., Hsia N. A new subgroup of the family 2 cystatins. Mol. Cell. Endocrinol. 2003;200:1–8. doi: 10.1016/s0303-7207(02)00408-2. [DOI] [PubMed] [Google Scholar]
- 239.Kotsyfakis M., Sa-Nunes A., Francischetti I.M.B., Mather T.N., Andersen J.F., Ribeiro J.M.C. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. J. Biol. Chem. 2006;281:26298–26307. doi: 10.1074/jbc.M513010200. [DOI] [PubMed] [Google Scholar]
- 240.Kotsyfakis M., Karim S., Andersen J.F., Mather T.N., Ribeiro J.M.C. Selective cysteine protease inhibition contributes to blood-feeding success of the tick Ixodes scapularis. J. Biol. Chem. 2007;282:29256–29263. doi: 10.1074/jbc.M703143200. [DOI] [PubMed] [Google Scholar]
- 241.Maciewicz R.A., Etherington D.J., Kos J., Turk V. Collagenolytic cathepsins of rabbit spleen: a kinetic analysis of collagen degradation and inhibition by chicken cystatin. Collagen Relat. Res. 1987;7:295–304. doi: 10.1016/s0174-173x(87)80035-3. [DOI] [PubMed] [Google Scholar]
- 242.Rennenberg A., Lehmann C., Heitmann A., Witt T., Hansen G., Nagarajan K., Deschermeier C., Turk V., Hilgenfeld R., Heussler V.T. Exoerythrocytic Plasmodium parasites secrete a cysteine protease inhibitor involved in sporozoite invasion and capable of blocking cell death of host hepatocytes. PLoS Pathog. 2010;6:e1000826. doi: 10.1371/journal.ppat.1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rosenthal P.J. Falcipains and other cysteine proteases of malaria parasites. Adv. Exp. Med. Biol. 2011;712:30–48. doi: 10.1007/978-1-4419-8414-2_3. [DOI] [PubMed] [Google Scholar]
- 244.Monteiro A.C.S., Abrahamson M., Lima A., Vannier-Santos M.A., Scharfstein J. Identification, characterization and localization of chagasin, a tight-binding cysteine protease inhibitor in Trypanosoma cruzi. J. Cell Sci. 2001;114:3933–3942. doi: 10.1242/jcs.114.21.3933. [DOI] [PubMed] [Google Scholar]
- 245.Gregory W.F., Maizels R.M. Cystatins from filarial parasites: evolution, adaptation and function in the host-parasite relationship. Int. J. Biochem. Cell Biol. 2008;40:1389–1398. doi: 10.1016/j.biocel.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 246.Lindahl P., Abrahamson M., Bjork I. Interaction of recombinant human cystatin C with the cysteine proteinases papain and actinidin. Biochem. J. 1992;281:49–55. doi: 10.1042/bj2810049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bode W., Engh R., Musil D., Thiele U., Huber R., Karshikov A., Brzin J., Kos J., Turk V. The 2.0 A X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 1988;7:2593–2599. doi: 10.1002/j.1460-2075.1988.tb03109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jerala R., Trstenjak M., Lenarcic B., Turk V. Cloning a synthetic gene for human stefin B and its expression in Escherichia coli. FEBS Lett. 1988;239:41–44. doi: 10.1016/0014-5793(88)80541-6. [DOI] [PubMed] [Google Scholar]
- 249.Stubbs M.T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., Turk V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990;9:1939–1947. doi: 10.1002/j.1460-2075.1990.tb08321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Machleidt W., Thiele U., Laber B., Assfalgmachleidt I., Esterl A., Wiegand G., Kos J., Turk V., Bode W. Mechanism of inhibition of papain by chicken egg white cystatin. Inhibition constants of N-terminally truncated forms and cyanogen bromide fragments of the inhibitor. FEBS Lett. 1989;243:234–238. doi: 10.1016/0014-5793(89)80135-8. [DOI] [PubMed] [Google Scholar]
- 251.Bjork I., Ylinenjarvi K. Interaction of chicken cystatin with inactivated papains. Biochem. J. 1989;260:61–68. doi: 10.1042/bj2600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Abrahamson M., Ritonja A., Brown M.A., Grubb A., Machleidt W., Barrett A.J. Identification of the probable inhibitory reactive sites of the cysteine proteinase-inhibitors human cystatin C and chicken cystatin. J. Biol. Chem. 1987;262:9688–9694. [PubMed] [Google Scholar]
- 253.Martin J.R., Craven C.J., Jerala R., Kroonzitko L., Zerovnik E., Turk V., Waltho J.P. The three-dimensional solution structure of human stefin A. J. Mol. Biol. 1995;246:331–343. doi: 10.1006/jmbi.1994.0088. [DOI] [PubMed] [Google Scholar]
- 254.Engh R.A., Dieckmann T., Bode W., Auerswald E.A., Turk V., Huber R., Oschkinat H. Conformational variability of chicken cystatin. Comparison of structures determined by X-ray diffraction and NMR-spectroscopy. J. Mol. Biol. 1993;234:1060–1069. doi: 10.1006/jmbi.1993.1659. [DOI] [PubMed] [Google Scholar]
- 255.Jenko S., Dolenc I., Guncar G., Dobersek A., Podobnik M., Turk D. Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases. J. Mol. Biol. 2003;326:875–885. doi: 10.1016/s0022-2836(02)01432-8. [DOI] [PubMed] [Google Scholar]
- 256.Renko M., Pozgan U., Majera D., Turk D. Stefin A displaces the occluding loop of cathepsin B only by as much as required to bind to the active site cleft. FEBS J. 2010;277:4338–4345. doi: 10.1111/j.1742-4658.2010.07824.x. [DOI] [PubMed] [Google Scholar]
- 257.Naudin C., Lecaille F., Chowdhury S., Krupa J.C., Purisima E., Mort J.S., Lalmanach G. The occluding loop of cathepsin B prevents its effective inhibition by human kininogens. J. Mol. Biol. 2010;400:1022–1035. doi: 10.1016/j.jmb.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 258.Mihelic M., Teuscher C., Turk V., Turk D. Mouse stefins A1 and A2 (Stfa1 and Stfa2) differentiate between papain-like endo- and exopeptidases. FEBS Lett. 2006;580:4195–4199. doi: 10.1016/j.febslet.2006.06.076. [DOI] [PubMed] [Google Scholar]
- 259.Turk B., Ritonja A., Bjork I., Stoka V., Dolenc I., Turk V. Identification of bovine stefin A, a novel protein inhibitor of cysteine proteinases. FEBS Lett. 1995;360:101–105. doi: 10.1016/0014-5793(95)00060-m. [DOI] [PubMed] [Google Scholar]
- 260.Turk B., Colic A., Stoka V., Turk V. Kinetics of inhibition of bovine cathepsin S by bovine stefin-B. FEBS Lett. 1994;339:155–159. doi: 10.1016/0014-5793(94)80405-2. [DOI] [PubMed] [Google Scholar]
- 261.Stoka V., Nycander M., Lenarcic B., Labriola C., Cazzulo J.J., Bjork I., Turk V. Inhibition of cruzipain, the major cysteine proteinase of the protozoan parasite, Trypanosoma cruzi, by proteinase-inhibitors of the cystatin superfamily. FEBS Lett. 1995;370:101–104. doi: 10.1016/0014-5793(95)00798-e. [DOI] [PubMed] [Google Scholar]
- 262.Wang S.X., Pandey K.C., Scharfstein J., Whisstock J., Huang R.K., Jacobelli J., Fletterick R.J., Rosenthal P.J., Abrahamson M., Brinen L.S., Rossi A., Sali A., McKerrow J.H. The structure of chagasin in complex with a cysteine protease clarifies the binding mode and evolution of an inhibitor family. Structure. 2007;15:535–543. doi: 10.1016/j.str.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 263.Mihelic M., Dobersek A., Guncar G., Turk D. Inhibitory fragment from the p41 form of invariant chain can regulate activity of cysteine ceathepsins in antigen presentation. J. Biol. Chem. 2008;283:14453–14460. doi: 10.1074/jbc.M801283200. [DOI] [PubMed] [Google Scholar]
- 264.Muller-Esterl W., Fritz H., Kellermann J., Lottspeich F., Machleidt W., Turk V. Genealogy of mammalian cysteine proteinase-inhibitors — common evolutionary origin of stefins, cystatins and kininogens. FEBS Lett. 1985;191:221–226. doi: 10.1016/0014-5793(85)80012-0. [DOI] [PubMed] [Google Scholar]
- 265.Villadangos J.A., Ploegh H.L. Proteolysis in MHC class II antigen presentation: who's in charge? Immunity. 2000;12:233–239. doi: 10.1016/s1074-7613(00)80176-4. [DOI] [PubMed] [Google Scholar]
- 266.Beers C., Burich A., Kleijmeer M.J., Griffith J.M., Wong P., Rudensky A.Y. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J. Immunol. 2005;174:1205–1212. doi: 10.4049/jimmunol.174.3.1205. [DOI] [PubMed] [Google Scholar]
- 267.Shi G.P., Bryant R.A.R., Riese R., Verhelst S., Driessen C., Li Z.Q., Bromme D., Ploegh H.L., Chapman H.A. Role for cathepsin F in invariant chain processing and major histocompatibility complex class II peptide loading by macrophages. J. Exp. Med. 2000;191:1177–1185. doi: 10.1084/jem.191.7.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Tang C.H., Lee J.W., Galvez M.G., Robillard L., Mole S.E., Chapman H.A. Murine cathepsin F deficiency causes neuronal lipofuscinosis and late-onset neurological disease. Mol. Cell. Biol. 2006;26:2309–2316. doi: 10.1128/MCB.26.6.2309-2316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Cresswell P. Invariant chain structure and MHC class II function. Cell. 1996;84:505–507. doi: 10.1016/s0092-8674(00)81025-9. [DOI] [PubMed] [Google Scholar]
- 270.Colbert J.D., Matthews S.P., Miller G., Watts C. Diverse regulatory roles for lysosomal proteases in the immune response. Eur. J. Immunol. 2009;39:2955–2965. doi: 10.1002/eji.200939650. [DOI] [PubMed] [Google Scholar]
- 271.Conus S., Simon H.U. Cathepsins and their involvement in immune responses. Swiss Med. Wkly. 2010;140:4–11. doi: 10.4414/smw.2010.13042. [DOI] [PubMed] [Google Scholar]
- 272.C. Watts, The endosome-lysosome pathway and information generation in the immune system, Biochim. Biophys. Acta, 1824 (2012), 13–20. [DOI] [PMC free article] [PubMed]
- 273.Lecaille F., Kaleta J., Bromme D. Human and parasitic papain-like cysteine proteases: their role in physiology and pathology and recent developments in inhibitor design. Chem. Rev. 2002;102:4459–4488. doi: 10.1021/cr0101656. [DOI] [PubMed] [Google Scholar]
- 274.Dauth S., Sirbulescu R.F., Jordans S., Rehders M., Avena L., Oswald J., Lerchl A., Saftig P., Brix K. Cathepsin K deficiency in mice induces structural and metabolic changes in the central nervous system that are associated with learning and memory deficits. BMC Neurosci. 2011;12:74. doi: 10.1186/1471-2202-12-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Taleb S., Clement K. Emerging role of cathepsin S in obesity and its associated diseases. Clin. Chem. Lab. Med. 2007;45:328–332. doi: 10.1515/CCLM.2007.083. [DOI] [PubMed] [Google Scholar]
- 276.Naour N., Rouault C., Fellahi S., Lavoie M.E., Poitou C., Keophiphath M., Eberle D., Shoelson S., Rizkalla S., Bastard J.P., Rabasa-Lhoret R., Clement K., Guerre-Millo M. Cathepsins in human obesity: changes in energy balance predominantly affect cathepsin S in adipose tissue and in circulation. J. Clin. Endocrinol. Metab. 2010;95:1861–1868. doi: 10.1210/jc.2009-1894. [DOI] [PubMed] [Google Scholar]
- 277.Yang M., Zhang Y., Pan J., Sun J., Liu J., Libby P., Sukhova G.K., Doria A., Katunuma N., Peroni O.D., Guerre-Millo M., Kahn B.B., Clement K., Shi G.P. Cathepsin L activity controls adipogenesis and glucose tolerance. Nat. Cell Biol. 2007;9:U130–U970. doi: 10.1038/ncb1623. [DOI] [PubMed] [Google Scholar]
- 278.Lafarge J.-C., Clément K., Guerre-Millo M. Cathepsins S, L, and K and their pathophysiological relevance in obesity. Clin. Rev. Bone Miner. Metab. 2011;9:133–137. [Google Scholar]
- 279.Sukhova G.K., Shi G.P., Simon D.I., Chapman H.A., Libby P. Expression of the elastolytic cathepsins S and K in human atheroma and regulation of their production in smooth muscle cells. J. Clin. Invest. 1998;102:576–583. doi: 10.1172/JCI181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Rodgers K.J., Watkins D.J., Miller A.L., Chan P.Y., Karanam S., Brissette W.H., Long C.J., Jackson C.L. Destabilizing role of cathepsin S in murine atherosclerotic plaques. Arterioscler. Thromb. Vasc. Biol. 2006;26:851–856. doi: 10.1161/01.ATV.0000203526.75772.4b. [DOI] [PubMed] [Google Scholar]
- 281.Lutgens E., Lutgens S.P.M., Faber B.C.G., Heeneman S., Gijbels M.M.J., de Winther M.P.J., Frederik P., van der Made I., Daugherty A., Sijbers A.M., Fisher A., Long C.J., Saftig P., Black D., Daemen M., Cleutjens K. Disruption of the cathepsin K gene reduces atherosclerosis progression and induces plaque fibrosis but accelerates macrophage foam cell formation. Circulation. 2006;113:98–107. doi: 10.1161/CIRCULATIONAHA.105.561449. [DOI] [PubMed] [Google Scholar]
- 282.Aoki T., Kataoka H., Ishibashi R., Nozaki K., Hashimoto N. Cathepsin B, K, and S are expressed in cerebral aneurysms and promote the progression of cerebral aneurysms. Stroke. 2008;39:2603–2610. doi: 10.1161/STROKEAHA.107.513648. [DOI] [PubMed] [Google Scholar]
- 283.Lafarge J.C., Naour N., Clement K., Guerre-Millo M. Cathepsins and cystatin C in atherosclerosis and obesity. Biochimie. 2010;92:1580–1586. doi: 10.1016/j.biochi.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 284.Toomes C., James J., Wood A.J., Wu C.L., McCormick D., Lench N., Hewitt C., Moynihan L., Roberts E., Woods C.G., Markham A., Wong M., Widmer R., Ghaffar K.A., Pemberton M., Hussein I.R., Temtamy S.A., Davies R., Read A.P., Sloan P., Dixon M.J., Thakker N.S. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat. Genet. 1999;23:421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- 285.Motyckova G., Fisher D.E. Pycnodysostosis: role and regulation of cathepsin K in osteoclast function and human disease. Curr. Mol. Med. 2002;2:407–421. doi: 10.2174/1566524023362401. [DOI] [PubMed] [Google Scholar]
- 286.Hou W.S., Bromme D., Zhao Y.M., Mehler E., Dushey C., Weinstein H., Miranda C.S., Fraga C., Greig F., Carey J., Rimoin D.L., Desnick R.J., Gelb B.D. Characterization of novel cathepsin K mutations in the pro and mature polypeptide regions causing pycnodysostosis. J. Clin. Invest. 1999;103:731–738. doi: 10.1172/JCI653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Grubb A., Lofberg H. Human gamma-trace, a basic microprotein: amino acid sequence and presence in the adenohypophysis. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3024–3027. doi: 10.1073/pnas.79.9.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Olafsson I., Grubb A. Hereditary cystatin C amyloid angiopathy. Amyloid. 2000;7:70–79. doi: 10.3109/13506120009146827. [DOI] [PubMed] [Google Scholar]
- 289.Brzin J., Popovic T., Turk V., Borchart U., Machleidt W. Human cystatin, a new protein inhibitor of cysteine proteinases. Biochem. Biophys. Res. Commun. 1984;118:103–109. doi: 10.1016/0006-291x(84)91073-8. [DOI] [PubMed] [Google Scholar]
- 290.Barrett A.J., Davies M.E., Grubb A. The place of human gamma-trace (cystatin-C) amongst the cysteine proteinase-inhibitors. Biochem. Biophys. Res. Commun. 1984;120:631–636. doi: 10.1016/0006-291x(84)91302-0. [DOI] [PubMed] [Google Scholar]
- 291.Janowski R., Kozak M., Jankowska E., Grzonka Z., Grubb A., Abrahamson M., Jaskolski M. Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Biol. 2001;8:316–320. doi: 10.1038/86188. [DOI] [PubMed] [Google Scholar]
- 292.Kaeser S.A., Herzig M.C., Coomaraswamy J., Kilger E., Selenica M.L., Winkler D.T., Staufenbiel M., Levy E., Grubb A., Jucker M. Cystatin C modulates cerebral beta-amyloidosis. Nat. Genet. 2007;39:1437–1439. doi: 10.1038/ng.2007.23. [DOI] [PubMed] [Google Scholar]
- 293.Jenko Kokalj S., Guncar G., Stern I., Morgan G., Rabzelj S., Kenig M., Staniforth R.A., Waltho J.P., Zerovnik E., Turk D. Essential role of proline isomerization in stefin B tetramer formation. J. Mol. Biol. 2007;366:1569–1579. doi: 10.1016/j.jmb.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 294.Zerovnik E., Stoka V., Mirtic A., Guncar G., Grdadolnik J., Staniforth R.A., Turk D., Turk V. Mechanisms of amyloid fibril formation — focus on domain-swapping. FEBS J. 2011;278:2263–2282. doi: 10.1111/j.1742-4658.2011.08149.x. [DOI] [PubMed] [Google Scholar]
- 295.Joensuu T., Kuronen M., Alakurtti K., Tegelberg S., Hakala P., Aalto A., Huopaniemi L., Aula N., Michellucci R., Eriksson K., Lehesjoki A.E. Cystatin B: mutation detection, alternative splicing and expression in progressive myclonus epilepsy of Unverricht-Lundborg type (EPM1) patients. Eur. J. Hum. Genet. 2007;15:185–193. doi: 10.1038/sj.ejhg.5201723. [DOI] [PubMed] [Google Scholar]
- 296.Reiser J., Adair B., Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J. Clin. Invest. 2010;120:3421–3431. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Puente X.S., Sanchez L.M., Overall C.M., Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat. Rev. Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 298.Siintola E., Lehesjoki A.E., Mole S.E. Molecular genetics of the NCLs — status and perspectives. Biochim. Biophys. Acta, Mol. Basis Dis. 2006;1762:857–864. doi: 10.1016/j.bbadis.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 299.Jalanko A., Braulke T. Neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta, Mol. Cell Res. 2009;1793:697–709. doi: 10.1016/j.bbamcr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 300.Sloane B.F., Dunn J., Honn K.V. Lysosomal cathepsin B: correlation with metastatic potential. Science. 1981;212:1151–1153. doi: 10.1126/science.7233209. [DOI] [PubMed] [Google Scholar]
- 301.Turk V., Kos J., Turk B. Cysteine cathepsins (proteases) — on the main stage of cancer? Cancer Cell. 2004;5:409–410. doi: 10.1016/s1535-6108(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 302.Mohamed M.M., Sloane B.F. Cysteine cathepsins: multifunctional enzymes in cancer. Nat. Rev. Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 303.Gocheva V., Joyce J.A. Cysteine Cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6:60–64. doi: 10.4161/cc.6.1.3669. [DOI] [PubMed] [Google Scholar]
- 304.Gabrijelcic D., Svetic B., Spaic D., Skrk J., Budihna M., Dolenc I., Popovic T., Cotic V., Turk V. Cathepsin B, H and L in human breast carcinoma. Eur. J. Clin. Chem. Clin. Biochem. 1992;30:69–74. [PubMed] [Google Scholar]
- 305.Hirano T., Takeuchi S. Serum cathepsin-B levels and urinary-excretion of cathepsin-B in the patients with colorectal-cancer — possible indicators for tumor malignancy. Int. J. Oncol. 1994;4:151–153. doi: 10.3892/ijo.4.1.151. [DOI] [PubMed] [Google Scholar]
- 306.Kos J., Smid A., Krasovec M., Svetic B., Lenarcic B., Vrhovec I., Skrk J., Turk V. Lysosomal proteases cathepsins D, B, H, L and their inhibitors stefins A and B in head and neck cancer. Biol. Chem. Hoppe Seyler. 1995;376:401–405. doi: 10.1515/bchm3.1995.376.7.401. [DOI] [PubMed] [Google Scholar]
- 307.Khan A., Krishna M., Baker S.P., Banner B.F. Cathepsin B and tumor-associated laminin expression in the progression of colorectal adenoma to carcinoma. Mod. Pathol. 1998;11:704–708. [PubMed] [Google Scholar]
- 308.Fernandez P.L., Farre X., Nadal A., Fernandez E., Peiro N., Sloane B.F., Shi G.P., Chapman H.A., Campo E., Cardesa A. Expression of cathepsins B and S in the progression of prostate carcinoma. Int. J. Cancer. 2001;95:51–55. doi: 10.1002/1097-0215(20010120)95:1<51::aid-ijc1009>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 309.Talieri M., Papadopoulou S., Scorilas A., Xynopoulos D., Arnogianaki N., Plataniotis G., Yotis J., Agnanti N. Cathepsin B and cathepsin D expression in the progression of colorectal adenoma to carcinoma. Cancer Lett. 2004;205:97–106. doi: 10.1016/j.canlet.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 310.Campo E., Munoz J., Miquel R., Palacin A., Cardesa A., Sloane B.F., Emmertbuck R. Cathepsin B expression in colorectal carcinomas correlates with tumor progression and shortened patient survival. Am. J. Pathol. 1994;145:301–309. [PMC free article] [PubMed] [Google Scholar]
- 311.Lah T.T., Cercek M., Blejec A., Kos J., Gorodetsky E., Somers R., Daskal I. Cathepsin B, a prognostic indicator in lymph node-negative breast carcinoma patients: comparison with cathepsin D, cathepsin L, and other clinical indicators. Clin. Cancer Res. 2000;6:578–584. [PubMed] [Google Scholar]
- 312.Scorilas A., Fotiou S., Tsiambas E., Yotis J., Kotsiandri F., Sameni M., Sloane B.F., Talieri M. Determination of cathepsin B expression may offer additional prognostic information for ovarian cancer patients. Biol. Chem. 2002;383:1297–1303. doi: 10.1515/BC.2002.146. [DOI] [PubMed] [Google Scholar]
- 313.Hirano T., Manabe T., Takeuchi S. Serum cathepsin B levels and urinary excretion of cathepsin B in the cancer patients with remote metastasis. Cancer Lett. 1993;70:41–44. doi: 10.1016/0304-3835(93)90072-h. [DOI] [PubMed] [Google Scholar]
- 314.Krueger S., Haeckel C., Buehling F., Roessner A. Inhibitory effects of antisense cathepsin B cDNA transfection on invasion and motility in a human osteosarcoma cell line. Cancer Res. 1999;59:6010–6014. [PubMed] [Google Scholar]
- 315.Krueger S., Kellner U., Buehling F., Roessner A. Cathepsin L antisense oligonucleotides in a human osteosarcoma cell line: effects on the invasive phenotype. Cancer Gene Ther. 2001;8:522–528. doi: 10.1038/sj.cgt.7700341. [DOI] [PubMed] [Google Scholar]
- 316.Bervar A., Zajc I., Sever N., Katunuma N., Sloane B.F., Lah T.T. Invasiveness of transformed human breast epithelial cell lines is related to cathepsin B and inhibited by cysteine proteinase inhibitors. Biol. Chem. 2003;384:447–455. doi: 10.1515/BC.2003.050. [DOI] [PubMed] [Google Scholar]
- 317.Premzl A., Zavasnik-Bergant V., Turk V., Kos J. Intracellular and extracellular cathepsin B facilitate invasion of MCF-10A neoT cells through reconstituted extracellular matrix in vitro. Exp. Cell Res. 2003;283:206–214. doi: 10.1016/s0014-4827(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 318.Yang Z., Cox J.L. Cathepsin L increases invasion and migration of B16 melanoma. Cancer Cell Int. 2007;7:8. doi: 10.1186/1475-2867-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tu C., Ortega-Cava C.F., Chen G.S., Fernandes N.D., Cavallo-Medved D., Sloane B.F., Band V., Band H. Lysosomal cathepsin B participates in the podosome-mediated extracellular matrix degradation and invasion via secreted lysosomes in v-Src fibroblasts. Cancer Res. 2008;68:9147–9156. doi: 10.1158/0008-5472.CAN-07-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Kobayashi H., Moniwa N., Sugimura M., Shinohara H., Ohi H., Terao T. Effects of membrane-associated cathepsin B on the activation of receptor-bound prourokinase and subsequent invasion of reconstituted basement membranes. Biochim. Biophys. Acta. 1993;1178:55–62. doi: 10.1016/0167-4889(93)90109-3. [DOI] [PubMed] [Google Scholar]
- 321.Kobayashi H., Schmitt M., Goretzki L., Chucholowski N., Calvete J., Kramer M., Gunzler W.A., Janicke F., Graeff H. Cathepsin B efficiently activates the soluble and the tumor cell receptor-bound form of the proenzyme urokinase-type plasminogen activator (Pro-uPA) J. Biol. Chem. 1991;266:5147–5152. [PubMed] [Google Scholar]
- 322.Ikeda Y., Ikata T., Mishiro T., Nakano S., Ikebe M., Yasuoka S. Cathepsins B and L in synovial fluids from patients with rheumatoid arthritis and the effect of cathepsin B on the activation of pro-urokinase. J. Med. Invest. 2000;47:61–75. [PubMed] [Google Scholar]
- 323.Guo M., Mathieu P.A., Linebaugh B., Sloane B.F., Reiners J.J., Jr. Phorbol ester activation of a proteolytic cascade capable of activating latent transforming growth factor-β. A process initiated by the exocytosis of cathepsin B. J. Biol. Chem. 2002;277:14829–14837. doi: 10.1074/jbc.M108180200. [DOI] [PubMed] [Google Scholar]
- 324.Mort J.S., Leduc M.S. The combined action of two enzymes in human serum can mimic the activity of cathepsin B. Clin. Chim. Acta. 1984;140:173–182. doi: 10.1016/0009-8981(84)90342-5. [DOI] [PubMed] [Google Scholar]
- 325.Leto G., Tumminello F.M., Pizzolanti G., Montalto G., Soresi M., Gebbia N. Lysosomal cathepsins B and L and stefin A blood levels in patients with hepatocellular carcinoma and/or liver cirrhosis: potential clinical implications. Oncology. 1997;54:79–83. doi: 10.1159/000227666. [DOI] [PubMed] [Google Scholar]
- 326.Warwas M., Haczynska H., Gerber J., Nowak M. Cathepsin B like activity as a serum tumour marker in ovarian carcinoma. Eur. J. Clin. Chem. Clin. Biochem. 1997;35:301–304. doi: 10.1515/cclm.1997.35.4.301. [DOI] [PubMed] [Google Scholar]
- 327.Sebzda T., Saleh Y., Gburek J., Warwas M., Andrzejak R., Siewinski M., Rudnicki J. Total and lipid-bound plasma sialic acid as diagnostic markers in colorectal cancer patients: correlation with cathepsin B expression in progression to Dukes stage. J. Exp. Ther. Oncol. 2006;5:223–229. [PubMed] [Google Scholar]
- 328.Herszényi L., Farinati F., Cardin R., István G., Molnár L.D., Hritz I., De Paoli M., Plebani M., Tulassay Z. Tumor marker utility and prognostic relevance of cathepsin B, cathepsin L, urokinase-type plasminogen activator, plasminogen activator inhibitor type-1, CEA and CA 19–9 in colorectal cancer. BMC Cancer. 2008;8:194. doi: 10.1186/1471-2407-8-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Vasiljeva O., Turk B. Dual contrasting roles of cysteine cathepsins in cancer progression: apoptosis versus tumour invasion. Biochimie. 2008;90:380–386. doi: 10.1016/j.biochi.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 330.Sameni M., Moin K., Sloane B.F. Imaging proteolysis by living human breast cancer cells. Neoplasia. 2000;2:496–504. doi: 10.1038/sj.neo.7900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Sameni M., Dosescu J., Sloane B.F. Imaging proteolysis by living human glioma cells. Biol. Chem. 2001;382:785–788. doi: 10.1515/BC.2001.094. [DOI] [PubMed] [Google Scholar]
- 332.Szpaderska A.M., Frankfater A. An intracellular form of cathepsin B contributes to invasiveness in cancer. Cancer Res. 2001;61:3493–3500. [PubMed] [Google Scholar]
- 333.Kjøller L., Engelholm L.H., Høyer-Hansen M., Danø K., Bugge T.H., Behrendt N. uPARAP/endo180 directs lysosomal delivery and degradation of collagen IV. Exp. Cell Res. 2004;293:106–116. doi: 10.1016/j.yexcr.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 334.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 335.Kos J., Lah T.T. Cysteine proteinases and their endogenous inhibitors: target proteins for prognosis, diagnosis and therapy in cancer (Review) Oncol. Rep. 1998;5:1349–1361. doi: 10.3892/or.5.6.1349. [DOI] [PubMed] [Google Scholar]
- 336.Jedeszko C., Sloane B.F. Cysteine cathepsins in human cancer. Biol. Chem. 2004;385:1017–1027. doi: 10.1515/BC.2004.132. [DOI] [PubMed] [Google Scholar]
- 337.Kos J., Krasovec M., Cimerman N., Nielsen H.J., Christensen I.J., Brunner N. Cysteine proteinase inhibitors stefin A, stefin B, and cystatin C in sera from patients with colorectal cancer: relation to prognosis. Clin. Cancer Res. 2000;6:505–511. [PubMed] [Google Scholar]
- 338.Kos J., Werle B., Lah T., Brunner N. Cysteine proteinases and their inhibitors in extracellular fluids: markers for diagnosis and prognosis in cancer. Int. J. Biol. Markers. 2000;15:84–89. doi: 10.1177/172460080001500116. [DOI] [PubMed] [Google Scholar]
- 339.Yu W.F., Liu J.A., Shi M.A., Wang J.A., Xiang M.X., Kitamoto S., Wang B., Sukhova G.K., Murphy G.F., Orasanu G., Grubb A., Shi G.P. Cystatin C deficiency promotes epidermal dysplasia in K14-HPV16 transgenic mice. PLoS One. 2010;5:e13973. doi: 10.1371/journal.pone.0013973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Gocheva V., Zeng W., Ke D.X., Klimstra D., Reinheckel T., Peters C., Hanahan D., Joyce J.A. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Vasiljeva O., Papazoglou A., Krueger A., Brodoefel H., Korovin M., Deussing J., Augustin N., Nielsen B.S., Almholt K., Bogyo M., Peters C., Reinheckel T. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66:5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- 342.Sevenich L., Schurigt U., Sachse K., Gajda M., Werner F., Muller S., Vasiljeva O., Schwinde A., Klemm N., Deussing J., Peters C., Reinheckel T. Synergistic antitumor effects of combined cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2497–2502. doi: 10.1073/pnas.0907240107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Mueller M.M., Fusenig N.E. Friends or foes — bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 344.Jain R.K., Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 2010;7:653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Mikhaylov G., Mikac U., Magaeva A.A., Itin V.I., Naiden E.P., Psakhye I., Babes L., Reinheckel T., Peters C., Zeiser R., Bogyo M., Turk V., Psakhye S.G., Turk B., Vasiljeva O. Ferri-liposomes as a novel MRI-visible drug delivery system for targeting tumours and their microenvironment. Nat. Nanotechnol. 2011;6:594–602. doi: 10.1038/nnano.2011.112. [DOI] [PubMed] [Google Scholar]
- 346.Joyce J.A., Baruch A., Chehade K., Meyer-Morse N., Giraudo E., Tsai F.Y., Greenbaum D.C., Hager J.H., Bogyo M., Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 347.Schurigt U., Sevenich L., Vannier C., Gajda M., Schwinde A., Werner F., Stahl A., von Elverfeldt D., Becker A.K., Bogyo M., Peters C., Reinheckel T. Trial of the cysteine cathepsin inhibitor JPM-OEt on early and advanced mammary cancer stages in the MMTV-PyMT-transgenic mouse model. Biol. Chem. 2008;389:1067–1074. doi: 10.1515/BC.2008.115. [DOI] [PubMed] [Google Scholar]
- 348.Feldmann M., Brennan F.M., Maini R.N. Rheumatoid arthritis. Cell. 1996;85:307–310. doi: 10.1016/s0092-8674(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 349.Lenarcic B., Gabrijelcic D., Rozman B., Drobnic-Kosorok M., Turk V. Human cathepsin B and cysteine proteinase-inhibitors (CPIs) in inflammatory and metabolic joint diseases. Biol. Chem. Hoppe Seyler. 1988;369:257–261. [PubMed] [Google Scholar]
- 350.Gabrijelcic D., Annanprah A., Rodic B., Rozman B., Cotic V., Turk V. Determination of cathepsin B and cathepsin H in sera and synovial fluids of patients with different joint diseases. J. Clin. Chem. Clin. Biochem. 1990;28:149–153. doi: 10.1515/cclm.1990.28.3.149. [DOI] [PubMed] [Google Scholar]
- 351.Lemaire R., Huet G., Zerimech F., Grard G., Fontaine C., Duquesnoy B. Selective induction of the secretion of cathepsins B and L by cytokines in synovial fibroblast-like cells. Br. J. Rheumatol. 1997;36:735–743. doi: 10.1093/rheumatology/36.7.735. [DOI] [PubMed] [Google Scholar]
- 352.Hashimoto Y., Kakegawa H., Narita Y., Hachiya Y., Hayakawa T., Kos J., Turk V., Katunuma N. Significance of cathepsin B accumulation in synovial fluid of rheumatoid arthritis. Biochem. Biophys. Res. Commun. 2001;283:334–339. doi: 10.1006/bbrc.2001.4787. [DOI] [PubMed] [Google Scholar]
- 353.Hill P.A., Buttle D.J., Jones S.J., Boyde A., Murata M., Reynolds J.J., Meikle M.C. Inhibition of bone resorption by selective inactivators of cysteine proteinases. J. Cell. Biochem. 1994;56:118–130. doi: 10.1002/jcb.240560116. [DOI] [PubMed] [Google Scholar]
- 354.Hou W.S., Li Z.Q., Gordon R.E., Chan K., Klein M.J., Levy R., Keysser M., Keyszer G., Bromme D. Cathepsin K is a critical protease in synovial fibroblast-mediated collagen degradation. Am. J. Pathol. 2001;159:2167–2177. doi: 10.1016/S0002-9440(10)63068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Aznavoorian S., Moore B.A., Alexander-Lister L.D., Hallit S.L., Windsor L.J., Engler J.A. Membrane type I-matrix metalloproteinase-mediated degradation of type I collagen by oral squamous cell carcinoma cells. Cancer Res. 2001;61:6264–6275. [PubMed] [Google Scholar]
- 356.Jorgensen I., Kos J., Krasovec M., Troelsen L., Klarlund M., Jensen T.W., Hansen M.S., Jacobsen S., Grp D.R.D.S., Grp T.S. Serum cysteine proteases and their inhibitors in rheumatoid arthritis: relation to disease activity and radiographic progression. Clin. Rheumatol. 2011;30:633–638. doi: 10.1007/s10067-010-1585-1. [DOI] [PubMed] [Google Scholar]
- 357.Pozgan U., Caglic D., Rozman B., Nagase H., Turk V., Turk B. Expression and activity profiling of selected cysteine cathepsins and matrix metalloproteinases in synovial fluids from patients with rheumatoid arthritis and osteoarthritis. Biol. Chem. 2010;391:571–579. doi: 10.1515/BC.2010.035. [DOI] [PubMed] [Google Scholar]
- 358.Wang D., Bromme D. Drug delivery strategies for cathepsin inhibitors in joint diseases. Expert Opin. Drug Delivery. 2005;2:1015–1028. doi: 10.1517/17425247.2.6.1015. [DOI] [PubMed] [Google Scholar]
- 359.Yasuda Y., Kaleta J., Bromme D. The role of cathepsins in osteoporosis and arthritis: rationale for the design of new therapeutics. Adv. Drug Delivery Rev. 2005;57:973–993. doi: 10.1016/j.addr.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 360.Lopez-Otin C., Overall C.M. Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 361.Lopez-Otin C., Bond J.S. Proteases: multifunctional enzymes in life and disease. J. Biol. Chem. 2008;283:30433–30437. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Stoka V., Turk V. A structural network associated with the kallikrein-kinin and renin-angiotensin systems. Biol. Chem. 2010;391:443–454. doi: 10.1515/BC.2010.046. [DOI] [PubMed] [Google Scholar]
- 363.D. Turk, Weiterentwicklung eines programms fuer molekuelgraphik und elektrondichte-manipulation und seine anwendung auf verschiedene protein-strukturaufklaerungen. PhD Thesis, Technische Universität, München (1992).
- 364.Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]