Abstract
Chemical biology is an emerging field that enables the study and manipulation of biological systems with probes whose reactivities provide structural insights. The opportunistic fungal pathogen Cryptococcus neoformans possesses a polysaccharide capsule that is a major virulence factor, but is challenging to study. We report here the synthesis of a hydroxylamine-armed fluorescent probe that reacts with reducing glycans and its application to study the architecture of the C. neoformans capsule under a variety of conditions. The probe signal localized intracellularly and at the cell wall–membrane interface, implying the presence of reducing-end glycans at this location where the capsule is attached to the cell body. In contrast, no fluorescence signal was detected in the capsule body. We observed vesicle-like structures containing the reducing-end probe, both intra- and extracellularly, consistent with the importance of vesicles in capsular assembly. Disrupting the capsule with DMSO, ultrasound, or mechanical shear stress resulted in capsule alterations that affected the binding of the probe, as reducing ends were exposed and cell membrane integrity was compromised. Unlike the polysaccharides in the assembled capsule, isolated exopolysaccharides contained reducing ends. The reactivity of the hydroxylamine-armed fluorescent probe suggests a model for capsule assembly whereby reducing ends localize to the cell wall surface, supporting previous findings suggesting that this is an initiation point for capsular assembly. We propose that chemical biology is a promising approach for studying the C. neoformans capsule and its associated polysaccharides to unravel their roles in fungal virulence.
Keywords: chemical biology, polysaccharide, vesicles, biosynthesis, glycobiology, extracellular vesicles, virulence factor, fungi, aminooxy fluorescent probes, capsule, fungal pathogen, glucuronoxylomannan, reducing glycans
Introduction
Cryptococcus neoformans is a major opportunistic human pathogen causing 1 million infections annually and ∼600,000 deaths (1). The fungal infection often occurs in early childhood and is followed by either clearance or a state of latency (2, 3). However, if the host immune system becomes compromised through senescence, HIV/AIDS, or is chemically induced for solid organ transplantation, the infection can re-emerge and cause lethal cryptococcosis or fungal meningitis (1, 4). C. neoformans has also been shown to create biofilms that can adhere to medical devices (5). A further problem is that current anti-fungal drugs often fail to clear an infection in hosts with impaired immunity. A better understanding of the biology of C. neoformans may allow the generation of new therapies to combat this infectious disease.
A major virulence factor of C. neoformans is the polysaccharide capsule that surrounds the cell body. Polysaccharides are also secreted into the environment and host tissues during infection, known as exopolysaccharide. The cryptococcal capsule is composed of a variety of constituents, including galactoxylomannan (GalXM),5 mannoproteins, β-glucans, and glucuronoxylomannan (GXM) (6). GXM predominates and contributes to virulence by interfering with host immunity and protecting the fungal cells. The role of these polysaccharides during infection has been implicated in interference and prevention of phagocytosis, inhibition of leukocyte migration, and cytokine production (7–9). The cell wall of C. neoformans is also made up of several polysaccharides, including β-(1→3) and β-(1→6) glucan, α-(1→3) glucan, chitin, and chitosan (10, 11).
The study of the capsule and its shed exopolysaccharides is challenging because, apart from antibodies, we lack tools to probe the fundamental nature of the organization, architecture, and structure of the capsule (12). The capsule is composed predominantly of water, which makes it vulnerable to desiccation. Also, little is known regarding how the capsule is synthesized, transported, and assembled extracellularly. NMR analysis completed by Cherniak et al. (13) defined the chemical structure of the GXM as a linear backbone of α-(1→3)-mannose bearing β-(1→2) and β-(1→4) xylose branches and β-(1→2) glucuronic acid branches, and a 6-O-acetyl substitution along the mannan backbone. It has been proposed that the glucuronic acid residues mediate divalent cation bridge interactions intramolecularly that help give rise to a polysaccharide architecture through a matrix of self-aggregation (14). Evidence that the capsular polysaccharide consists of a higher-order branched matrix comes from static and dynamic light scattering, viscosity analysis, and high-resolution microscopy (15).
A fundamental question in C. neoformans biology is how the capsule is assembled and organized on the cell surface (16). However, several glycosyltransferase enzymes required for the biosynthesis of the GXM capsule have been identified by Doering and co-workers (17–19). There is uncertainty over the mechanism of capsule enlargement with two proposed models that are nonmutually exclusive. The first is termed “proximal growth” (20), in which the polysaccharide chain is increased in size by incorporation of polysaccharide at the cell body, displacing pre-existing molecules to the outer edge (17), i.e. a mechanism consistent with prokaryotic bacteria capsule assembly (21). The second is termed “distal growth,” where addition of new polysaccharide is incorporated at the capsule edge, with older material remaining close to the cell body (12).
Despite the mechanism of capsule assembly not being elucidated, it is evident that the cell wall and capsule are capable of dynamic reorganization, as shown during budding events from immunofluorescence studies, such that the mother cell reorganizes its capsule to facilitate the emerging bud, although the exact mechanisms that govern this process remain unclear (12). Several studies suggest intracellular biosynthesis, followed by vesicular transport across the cell wall (23, 24). The importance of vesicles in C. neoformans is further demonstrated by mutations in the C. neoformans secretory system known as cap mutants (23, 25). These mutants lack capsular GXM but secrete other polysaccharides thus suggesting the presence of other transport mechanisms independent of that for the capsule, possibly a process with similarities to that of bacterial capsule biosynthesis (21, 26, 27). It is also unclear how the capsule attachment to the cell wall occurs or whether the cell wall provides the skeleton for proteins or other chemical moieties that then mediate its attachment (10).
Chemical glycobiology is an emerging and exciting sub-discipline of chemical biology that promises to allow further study of the cryptococcal capsule biosynthesis, assembly, and function. Unlike the study of proteins, carbohydrates are not template-encoded, which complicates research in understanding change in their structure and function (28). The introduction of chemical reporter groups on glycans provides new strategies to study an organism's glycome. Chemical glycobiology has proven the ability to image live cells and organisms using metabolic oligosaccharide engineering followed by bio-orthogonal ligation chemistry (29–32). To gain insight into the distribution of reducing sugars in C. neoformans, we synthesized a hydroxylamine-armed fluorescent probe (reducing end probe, R.E probe). Hydroxylamines, also commonly known as aminooxys, have been used extensively in chemical biology to label glycans, whole-cell glycan cells, and to create site-selective conjugates of antibodies and other proteins using aldehyde tags (33–36). Under chemical or biological settings, hydroxylamine nucleophiles are known to react with a high selectively with the reducing end of glycans (which contain a transient aldehyde), to a give the corresponding oxime conjugate (Fig. 1B) (33, 35–38). This stable adduct, when attached to a fluorophore, would allow imaging of cryptococcal cells and give insight into the distribution of reducing ends in the cell, cell wall, and capsule.
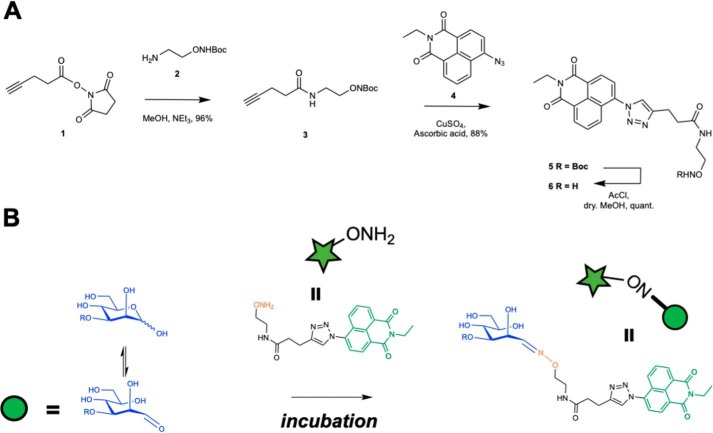
Figure 1.
Hydroxylamine-armed fluorescent probe to study the distribution of reducing sugars in C. neoformans. A, synthesis of the R.E probe. B, proposed mechanism of action of the R.E armed fluorescent probe. The R.E probe reacts preferentially with aldehydes to form a stable oxime adduct that can be used to image the localization of reducing ends in the capsule and inside the cell. The hydroxylamine functionality (ONH2) is converted into an oxime (ON) after reacting with the reducing end glycan.
This structure–activity-based probe allowed insight into capsule biosynthesis, structure, and assembly. We used this probe to determine the localization of reducing end glycans at the capsule–cell wall interface and to visualize the presence of both intra- and extracellular polysaccharide-containing vesicles. We also use the probe to examine the effect of different cellular processing techniques on capsular architecture and that the shed EPS contains reducing ends.
Results
Synthesis of hydroxylamine-armed fluorescent probe
To synthesize the hydroxylamine arm of the fluorescent probe, we started from commercially available ethanolamine. After a five-step synthesis, an N-Boc protected hydroxylamine derivative 2 was prepared (Fig. 1A) (39, 40). This was then coupled with compound 1, an N-hydroxysuccinimide–activated ester of propargylacetic acid (41); after purification by flash column chromatography, compound 3 was isolated in a 96% yield. The fluorescent arm of the hydroxylamine-armed probe was prepared from 4-bromo-1,8-naphthalic anhydride, following known literature procedures to give compound 4 (42). The two arms of the probe were then combined using copper click chemistry to produce compound 5 in 88% yield after flash chromatography. Subsequently, the N-Boc protecting group was removed using acetyl chloride in anhydrous methanol to produce the hydroxylamine-armed fluorescent probe 6.
To confirm the reactivity of the R.E probe for reducing glycans, we performed control experiments by incubating the oxime probe with a saccharide that contained a reducing end (d-mannose) and one that did not (methyl α-d-mannopyranoside) (Figs. S2 and S3). Using mass spectrometry (MS) and NMR, we confirmed the oxime conjugate only formed in the presence of reducing glycans.
Incubation of the R.E probe with C. neoformans does not alter fluorescent properties
To confirm that C. neoformans was not metabolizing the R.E probe and altering its optical properties, we incubated the probe with four different C. neoformans cell samples for 24 h, washed cells, and performed analysis using a fluorescence spectrometer. We chose H99 (ATCC 208821) as a reference strain and two mutant strains CAP59Δ and CAP67Δ. In H99 cells, we were also intrigued to explore whether capsule architecture is altered by melanization of the cell wall. To do so, we melanized cells and probed them with the R.E probe to learn whether the distribution of reducing ends is altered by this change in cell wall architecture.
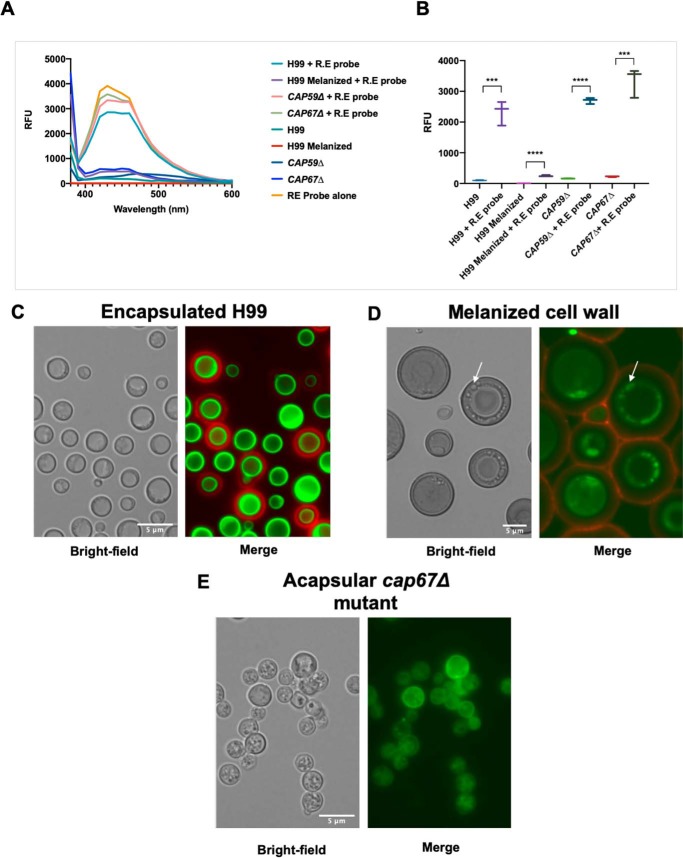
Spectrometric analysis confirmed the incorporation of the probe into C. neoformans cells (Fig. 2A) with characteristic emission maxima of 440–470 nm (excitation 360 nm). The encapsulated H99, CAP59Δ, and CAP67Δ cells displayed higher levels of fluorescence compared with that of control cells that were not incubated with the R.E probe (Fig. 2A). In contrast, melanized cells displayed no fluorescence emission when excited at this wavelength, likely due to signal quenching of signal by the melanin (43). Encapsulated H99, CAP59Δ, and CAP67Δ cells also displayed higher relative fluorescence units (RFU) at the emission maxima of the probe (Fig. 2B).
Figure 2.
Incubation of hydroxylamine-armed fluorescent probe with C. neoformans. A, fluorescence spectrum of C. neoformans cells incubated with and without hydroxylamine-armed probe (R.E probe). Those that have been incubated with the R.E probe have increased emission maxima at ∼453 nm (excitation 360 nm) compared with encapsulated H99, CAP59Δ, or CAP67Δ with the exception of C. neoformans with encapsulated H99 with a melanized cell wall. Melanization of the cell wall is likely quenching the florescence of the probe. B, RFU of C. neoformans cells incubated with the R.E probe are significantly higher at emission maxima (440 nm) of the probe (excitation 360 nm). Error bars represent 95% confidence interval. Statistical significance was determined using unpaired t test (***, p ≤ 0.001; ****, p ≤ 0.0001). Experiments were performed in triplicate. C, encapsulated H99: the capsule shown by anti-GXM Mab 18B7-AF594 (red) displays no reactivity to the R.E probe 6 (green) and is localized at the cell wall–membrane interface. D, melanized C. neoformans cells display the same localization of the R.E probe. The R.E probe appears intracellularly possible in vesicular bodies and displays no reactivity toward the capsule, despite the melanization of the cell wall. E, acapsular mutant C. neoformans cap67Δ incubated with the R.E probe shows bright fluorescence intensity coming from the cytoplasm and the cell wall–membrane interface. Labeling of cytoplasm occurs in spherical vesicle-like structures in acapsular cap67Δ and melanized H99 cells. Scale, 5 μm.
Visualizing the reducing-end probe distributions in C. neoformans
Incubation of the reducing-end probe 6 with C. neoformans strain H99 in rich or minimal media revealed that the probe localized near the cell membrane–cell wall interface. Incubations with R.E probe 6 (green) and directly conjugated anti-glucuronoxylomannan (GXM) antibody 18B7 (18B7-AF594, red) allowed visualization of the capsule, revealing no probe labeling throughout the capsular GXM (Fig. 2C). In melanized fungal cells, we observed stronger fluorescence from the R.E probe in organelles or vesicular bodies around the periphery of a large vacuole inside the cells (Fig. 2D). Acapsular mutant CAP67Δ showed no staining by 18B7-AF594, as expected from the absence of capsular GXM, but it did manifest fluorescence in the cytoplasm and at the cell wall–membrane interface after labeling with the R.E probe (Fig. 2E). These structures could reflect the cytoplasmic accumulation of polysaccharide-containing vesicles associated with the acapsular phenotype (44). The presence of R.E probe labeling inside the cytoplasm of the melanized and acapsular mutants suggests the probe can penetrate the cell wall and membrane and react with reducing sugars in the cytoplasmic space. This phenomenon was less evident in encapsulated H99 cells (Fig. 2C). It is possible that in melanized cells, the strong cell wall fluorescence observed in nonmelanized cells is quenched by the pigment, thus allowing better visualization of cytoplasmic fluorescence and revealing internal details. In addition, we noted that the cell wall of melanized cells contained localized fluorescent signals (Fig. 2D) that could reflect vesicles crossing the cell wall, which have been shown to carry polysaccharide (24).
Incubating Cryptococcus sp. with the reducing-end probe
To examine the capsule architecture across the Cryptococcus genus, we incubated two Cryptococcus gattii strains representing serotypes B (ATCC 24065) (Fig. 3D) and C (ATCC 32608) (Fig. 3C), an environmental strain Cryptococcus albidus (Fig. 3B), which has been characterized as having a capsule akin to a serotype A strain (46), and a serotype D strain C. neoformans (ATCC 24067) (Fig. 3A) with the R.E probe. To visualize the capsule perimeter, we used India ink staining, which is excluded by the capsule, and the capsule can be exhibited by a halo around the cells. Although well-documented phenotypic differences were observed (47), including differences in capsule diameter, globose, oblong and elliptical cell shapes, all cryptococcal species analyzed reacted to the R.E probe in a conserved manner, with the R.E probe localizing at the capsule–wall interface (Fig. 3) and the capsule displaying no reactivity. Furthermore, the incubation of the R.E probe with the various strains tested resulted in a similar level of fluorescence at the emission maxima of the probe (Fig. 3E).
Figure 3.
Localization of reducing end polysaccharides is maintained across the Cryptococcus genus as labeled by the R.E probe. Incubations with the R.E probe (green), followed by India ink staining allows visualization of the capsule perimeter and where the reducing-ends reside. Any of the Cryptococcal sp. tested resulted in a similar staining pattern, suggesting that capsule architecture and biosynthesis are maintained across species. A, C. neoformans (ATCC 24067). B, C. albidus. C, C. gattii (ATCC 32608). D, C. gattii (ATCC 24065). E, incubation with the R.E probe increases RFU at emission of probe (excitation 360 nm). Error bars represent 95% confidence interval, Statistical significance was determined using unpaired t test (***, p ≤ 0.001; ****, p ≤ 0.0001). Experiments were performed in triplicate. Scale, 5 μm.
Capsule perturbation alters probe reactivity
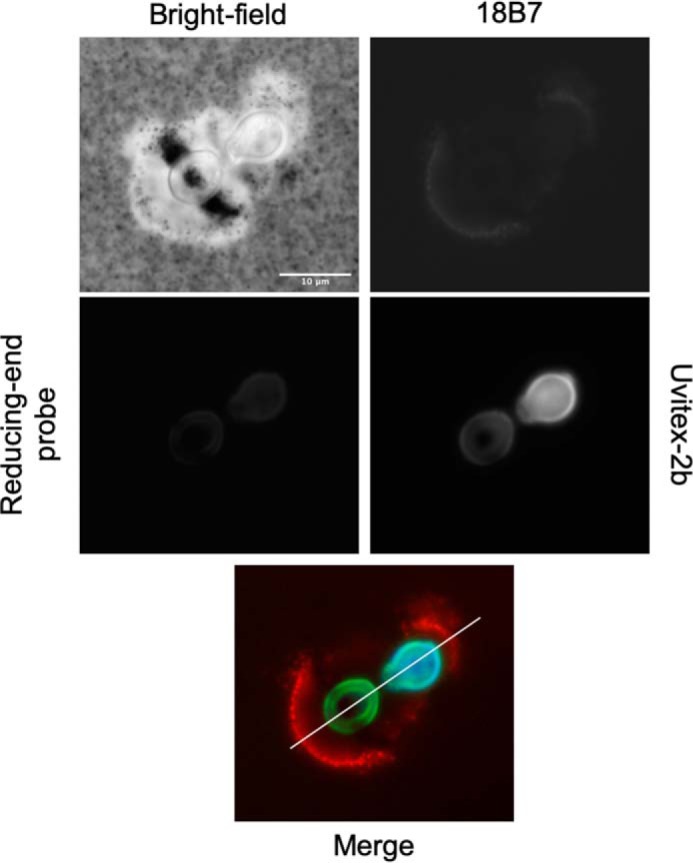
To better understand the localization of R.E probe 6, 3-day-old H99 encapsulated cells grown in minimal media were processed in three ways: DMSO, sonication, and French press. DMSO is used to remove the capsule from the cell (48). French press passage has been used to isolate cryptococcal cell walls from capsular polysaccharide (49). Sonication was previously shown to strip the capsule (50). We hypothesized that these treatments may perturb the capsule structure, which could affect the R.E probe labeling and provide information on selectivity. We also hypothesized that some of the cellular processing methods might expose GXM reducing ends that could then be labeled with the R.E probe 6 (Fig. 4). DMSO-treated H99 cells lost the large majority of their GXM capsular polysaccharide, as evident by a diminished signal from the anti-GXM 18B7-AF568 conjugate (Fig. 4). Treating C. neoformans cells with DMSO altered the localization of the R.E probe compared with that of the control. The R.E probe signal became stronger inside the cell, suggesting the cell membrane had been permeabilized by the DMSO treatment, and this affected the permeability and reactivity. Using cell wall–specific Uvitex-2b allowed the visualization of the cell wall by its selectivity for chitin staining (51). In the DMSO-treated cells the Uvitex-2b (Blue) fluorescence signal was less sharp compared with that of the control, suggesting the cell wall was also affected in DMSO-treated cells. Importantly, both dyes were altered in a manner independent from each other (Fig. 4), indicating labeling of chitin or chitosan was not the predominant localization of the R.E probe.
Figure 4.

Capsule perturbation alters probe reactivity. H99 C. neoformans cells were grown in capsule-inducing minimal media for 3 days. Subsequently, the cells were independently processed with three different methods: DMSO, French press, and sonication. Thereafter, the cells were incubated with the fluorescent R.E probe (labels reducing carbohydrates, green) overnight, washed three times, stained for 30 min with Uvitex-2b (stains cell wall chitin, blue), and an 18B7-Alexa Fluor 594 conjugate (stains capsule, red) for 1 h. Scale, 10 μm.
Incubation of the R.E probe with French press-treated cells caused the greatest signal disruption of the R.E probe, suggesting the greatest quantities of reducing polysaccharide were exposed by this method. This can be visualized by clusters of sheared capsule polysaccharides that reacted with the R.E probe and were not associated with any cell. We interpret this result as implying that the mechanical distribution caused by French press treatment exposed the reducing ends of polysaccharides that were once localized only at the cell wall, or that French press treatment can cause the breakage of glycosidic bonds due to high temperature and pressures (49). The colocalization signal from 18B7-AF568 and the R.E probe showed that the R.E linker labeled the GXM reducing ends (Fig. 4). In contrast, the Uvitex-2b signal was not affected significantly by French press treatment (Fig. 4). Treating cells with a horn sonicator for 30 s (20 watts) caused the shearing of the capsule, as visualized by the smaller diameter of the capsule compared with the control cells. The localization of the R.E probe was unaffected by this cell-treatment strategy as was the Uvitex-2b binding, suggesting that the cell wall is labeled by the R.E probe. However, sonication also reduced a zone of clearance typically observed between the cell wall and the 18B7-AF568 conjugate (Fig. 4, Merge, Sonication, inset) (50). Furthermore, in sonicated cells, the R.E probe signal colocalized with 18B7-AF568 in a yellow band around the cell body (Fig. 4, Merge, Sonication, inset).
Extracellular vesicular bodies in CAP59Δ and CAP67Δ mutants
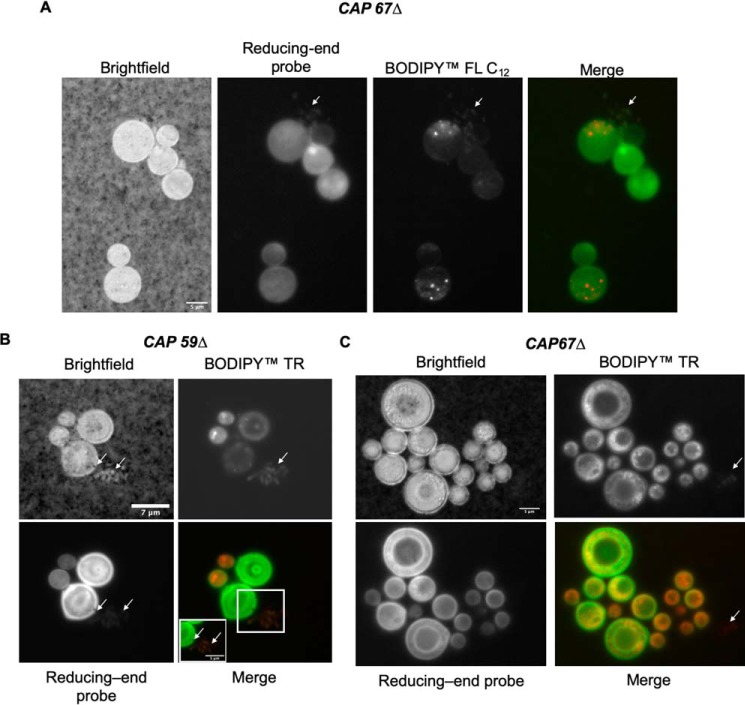
The ability of the reducing-end probe to permeate into the cell and label what appeared to be glycan containing vesicular bodies prompted further investigation. Using two lipophilic dyes BODIPY TR methyl ester (BODIPY TR) and BODIPY FL C12 (BODIPY C12), we visualized the presence of structures consistent with extracellular vesicular structures or organelles in both CAP59Δ and CAP67Δ mutants (Fig. 5). Furthermore, we visualized extracellular glycan-containing vesicles in both CAP59Δ and CAP67Δ mutants as the R.E probe and lipid dye colocalized extracellularly (Fig. 5, A and B). Although these acapsular strains do not secrete GXM, they do release GalXM (52). We also visualized extracellular structures that stained with BODIPY TR but not with the R.E probe, which presumably represents nonglycan-containing vesicles (Fig. 5C). Intracellular staining was difficult to resolve as either lipophilic dye caused substantial staining, possibly labeling mitochondria, Golgi apparatus, and vesicular bodies. Despite this, the R.E probe did appear to colocalize with the lipophilic dyes, possibly in vesicles or Golgi apparatus. It was also apparent that the R.E probe did not enter the vacuole of the cells or that it was quenched in that environment.
Figure 5.
Secretion of vesicular bodies in CAP59Δ and CAP67Δ mutants. A, incubation of CAP67Δ with BODIPY FL C12 (lipophilic dye, red) and the R.E probe shows vesicle secretion (arrows). B, CAP59Δ cells incubated with BODIPY TR (lipophilic dye, red) also show signs of secretion of glycan-containing vesicles (inset and arrows). C, CAP67Δ cells incubated with the BODIPY TR and R.E probe show signs of colocalization; however, BODIPY TR also stains nonglycan-containing vesicles (arrows) and shows signs of secretion of nonglycan-containing vesicular bodies (arrow). Scale denoted in bright field: A, 5 μm; B, 7 μm (inset, 5 μm); and C, 5 μm.
Dynamic reorganization of C. neoformans polysaccharide capsule and cell–cell wall to facilitate budding
Previously, observations of gentle pressure to India ink–stained C. neoformans cells caused perpendicular lines to the budding axis suggesting a complex organization of the capsule (53). Hence, we wondered whether we would gain further information about capsular organization using the R.E probe. We subjected encapsulated H99 cells grown in minimal media for 2 days to gentle pressure after staining with the R.E probe, 18B7, and Uvitex-2b (Fig. 6) to simultaneously stain for reducing end glycans, capsule, and cell wall, respectively. We observed the same perpendicular India ink lines to the budding axis as reported previously (53), but we also observed that the capsule, cell wall, and R.E probe had become polarized along the budding axis. This suggests that the innermost layer of GXM is attached to the cell wall in a very strong manner, possibly through covalent bonds to either the chitin or glucans. The density of capsule is altered as a result of budding, which allows India ink to penetrate deeper in perpendicular lines compared with to that of the budding axis. The Uvitex-2b staining correlates to this change in capsule density as the chitin polarizes along the budding axis.
Figure 6.
Dynamic reorganization of C. neoformans polysaccharide capsule and cell–cell wall to facilitate budding. India ink staining perpendicular to emerging bud correlates with reorganization of chitin cell wall and capsule. 18B7 and R.E probe both have been polarized along the budding axis (line merge). This causes a change in capsule density perpendicular to the budding axis allowing India ink staining to penetrate deeper when gentle pressure is applied to the coverslip. Scale, 10 μm.
Applying shear pressure to understand probe localization
To examine the localization of the R.E probe further, we applied shear pressure to the glass coverslip of encapsulated H99 cells previously labeled with 18B7, Uvitex-2b and the R.E probe. The shear pressure caused some cells to burst or fragment (Fig. 7, arrows), but also to appear to remove capsular material from cells. This experiment further confirmed that the reducing end probe localized at the base of the GXM polymer at the capsule–cell wall interface (Fig. 7). This implies that the capsule in encapsulated cells is organized in a more stable manner than a simple collection of self-aggregated GXM polymers and with strong attachment to cell wall chitin or glucans either through direct linkage to the polysaccharide or proteins that embedded in the cell wall (14, 54).
Figure 7.

Applying shear pressure to understand probe localization. Application of shear pressure to glass slides caused a population of cells to rupture. Several fragments of cells can be seen (arrows), where the capsule can be seen attached to the cell wall, and also where the R.E probe localization occurs. Scale, 10 μm.
Incubation of EPS and CPS fractions with the reducing-end probe
To further understand the location and abundance of reducing ends in the capsule of C. neoformans and its EPS and CPS (Fig. 8A), we prepared three fractions for investigation and a chitin sample as a positive control. The EPS fraction contained two fractions corresponding to a >10 fraction and a >100 kDa fraction. The EPS was isolated through ultrafiltration membrane discs. While the CPS fraction was prepared by DMSO treatment of the C. neoformans cell pellet. Each fraction was then incubated with the R.E probe overnight at 30 °C in the dark and subsequently dialyzed (molecular mass cutoff, 3.5 kDa) to remove the unreacted probe. Aliquots from each sample were then compared with controls to see whether there was an increase in fluorescence postincubation, corresponding to the linker reacting with the reducing end of the sample. All samples that were incubated with the reducing-end linker showed at least a >2-fold increase in RFU compared with controls, consistent with the notion that the linker reacts with reducing sugars (Fig. 8B) and forms an oxime conjugate. This experiment also revealed that the EPS fractions shed from C. neoformans into the extracellular space contain reducing ends, something not previously known.
Figure 8.
Exopolysaccharides of C. neoformans contain reducing ends. A, R.E probe was incubated with CPS, EPS (>10 kDa and >100 kDa), chitin (positive control), or alone for 24 h. The incubation was repeated in three independent experiments. Following dialysis, fluorescence emission spectra were recorded (excitation 360 nm) and compared with their nonincubated controls, showing an increase in fluorescence at the probe's emission maxima, indicating the R.E probe is forming a conjugate with the reducing end of various C. neoformans EPS and CPS isolates. Polysaccharides that were incubated with the R.E probe show >2-fold increase in RFU compared with controls, revealing that the shed exopolysaccharide contains reducing ends. Error bars represent 95% confidence intervals. Statistical significance was determined using unpaired t test (****, p ≤ 0.0001). B, hypothesis: if reducing ends are present in the exopolysaccharide of C. neoformans, they would form stable conjugates with the R.E probe, which could be determined using a fluorescence spectrometer. Glycan notification followed the Symbol Nomenclature for Glycans (SNFG). C, 1H NMR analysis of >10- and >100-kDa exopolysaccharide isolates revealed the presence of aromatic peaks in the region of δ 7.7–8.6 ppm, which are not present in the original EPS fraction. Exploring C. neoformans with hydroxylamine-armed probe 28 aromatic signals are characteristic of the R.E probe. Characteristic structural reporter groups of anomeric mannose protons can be visualized from δ 5.5 to 4.9 ppm.
To further establish that the incubated EPS samples did indeed react with the linker, we used 1H NMR analysis to confirm the presence of the probe's aromatic peaks at δ 7.7–8.6 ppm in the spectra of the >10- and >100-kDa EPS fractions (Fig. 8C). However, we were unable to obtain an 1H NMR of the CPS and chitin carbohydrate signal when samples were run in deuterium oxide, and this is likely due to low solubility of chitin in an aqueous solvent and the heterogeneity of the CPS sample (Fig. S1).
Discussion
We describe the synthesis of a hydroxylamine-armed fluorescent probe that selectively labels reducing sugars, and we describe the use of this probe to study the capsule architecture and biosynthesis in C. neoformans. The series of reactions performed yielded significant insights into capsular architecture and biosynthesis and confirmed that this probe has great potential to help answer outstanding questions in the field. One striking result was the lack of reactivity of the R.E probe with the body of the polysaccharide capsule. Instead, the probe revealed that reducing end glycans are mostly localized at the cell wall–capsule interface. This suggests that these groups are involved in capsule attachment to the cellular surface and/or to serve as sites for the origin of capsular assembly. This result would be consistent with a model of capsular assembly occurring at the cell wall and extending away radially. A precedent for this type of capsular biosynthesis and assembly can be found in both eukaryotic and prokaryotic cells. For example, in Escherichia coli there are over 80 capsular serotypes, but despite this diversity, there is little variation in capsular biosynthesis or assembly (21). In Gram-negative bacteria, it is proposed that multiprotein complexes are used to span the cell envelope for the transportation of the capsule to the cell surface (55, 56). In this work, we revealed that despite species differences, the R.E probe did not react with the capsule body, suggesting conservation of capsule assembly and biosynthesis processes conserved across the Cryptococcus sp.
The most striking observation was the absence of R.E. probe reactivity in the body of the capsule. The most straightforward interpretation of this observation is that there are no reducing ends in the body of capsule. However, certain caveats suggest caution in making inferences from this negative result until confirmed by independent approaches. First, GXM macromolecules have mass in the millions of Da (57), making reducing end sugars a very small constituent of the molecule, and the addition of a single fluorophore may not be detectable relative to the strong signal from the cell wall and cytoplasm. Second, the capsule does contain lipids (51, 58), which can quench fluorescence (59), and it is conceivable that R.E. probe fluorescence is quenched, as may occur in the melanized cell wall. Third, and least probable, would be an accessibility constraint the precluded reactivity. Although GXM macromolecules appear to be oligodendrimers with a very dense core (15), the lack of accessibility explanation is less likely given that the R.E. probe reacted with exopolysaccharide. Mindful of these caveats, we proceed with the most parsimonious interpretation of the results and consider the implications of no R.E. probe reactivity in the capsule for its assembly and structure.
An internal location near the cell wall for the synthesis of the capsule was proposed based on the analysis of antibody-labeled capsule polysaccharide movements (60). A subsequent study proposed that capsular enlargement occurred through distal extension, although these mechanisms were not mutually exclusive (12). Our observations are consistent with the hypothesis of Frases et al. (61) that the distance from the cell body to the capsule is spanned by the length of a single GXM polymer (Fig. 9), which is also supported by the finding that capsule size is regulated at the polymer level (62). The orientation of this polysaccharide can be deduced by the reactivity of the R.E probe such that the capsule's reducing end faces into the extracellular space (Fig. 9). This observation suggests a model where some capsule biosynthesis may occur at the cell wall, possibly through enzymes that spool the growing polysaccharide chain out into the extracellular milieu. The vesicular polysaccharide-containing vesicles may contain the seed motif required for cell wall–associated enzymes for the material to elongate the structure. An analogous motif to the core pentasaccharide is required in N-glycan biosynthesis in humans (63). Taken together with our results that reducing ends occur at the cell wall–capsule interface, this adds evidence supporting the “proximal growth” capsular assembly hypothesis.
Figure 9.
Diagram illustrating three hypotheses for capsule assembly in C. neoformans. Polysaccharide molecules are shown in a linear manner for simplicity, with the localization of the reducing ends a key point for this depiction. The most parsimonious interpretation of the R.E. probe data supports hypothesis 1.
The synthesis of capsular polysaccharides was shown to occur within vesicular bodies, and the export of these vesicles across the cell wall is necessary for the export of polysaccharides and capsule assembly (24, 64, 65). Furthermore, extracellular vesicles are implicated in diverse functions, including cell communication and transport of virulence factors (66, 67). We gained additional evidence of the importance of vesicles in C. neoformans capsule biosynthesis in both acapsular mutants CAP67Δ, CAP59Δ, and melanized H99 cells. CAP59Δ and CAP67Δ are believed to have defective transport mechanisms for capsule assembly and manifested R.E probe in spherical vesicle structures in the cytoplasm that were not observed in H99-encapsulated cells. We suspect that these structures contained high quantities of reducing glycans, possibly capsular polysaccharides that accumulated in the cytoplasm as a result of the export defect associated with the acapsular phenotype (44). Vesicular structures were also observed inside melanized C. neoformans cells around the periphery of a large vacuole. Furthermore, the punctate pattern of fluorescence observed in the melanized cell wall could reflect polysaccharide-carrying vesicles across this structure. It is noteworthy that the R.E probe was able to pass through the melanized cell wall and that it was not sequestered in melanin due to its aromatic and hydrophobic nature. Despite the defective mechanisms for vesicle transport in the CAP59Δ and CAP67Δ mutants, we observed vesicle secretion in these mutants of both glycan-containing vesicles and other vesicles not stained by the R.E probe. Although glycan-containing vesicles could represent partial capsular assembly, other vesicles could contain proteins and RNA, which have recently been shown to be important in virulence of C. gattii (67). Intracellular staining with lipophilic dyes was more complex and difficult to resolve; although colocalization did appear, staining of other cellular organelles was present, and due to the presence of large vacuoles in cells, it made total resolution difficult.
Our observations with the R.E probe add to the wealth of data indicating the importance of vesicles in polysaccharide transport and capsule assembly. Furthermore, we believe that the R.E probe could be of great use to delve further into the mechanism of polysaccharide-containing vesicle transport and polysaccharide integration into the capsule itself utilizing microscopic techniques.
Further evidence of the capsule's highly-organized and dynamic structure was revealed by using a technique previously described by Zaragoza et al. (53). In this work, we observed the same ring-like channels observed in the work by Zaragoza et al. (53). Furthermore, the cell wall chitin, reducing ends, and distal capsule (as stained by 18B7) polarize along the budding axis. Together, this suggests a coordinated alteration to both the cell wall, and capsular architecture is necessary during the budding process.
Several questions about C. neoformans EPS revolve around its function and origin. It has been posited that EPS is the loss of capsular polysaccharide due to rearrangement or that it is unincorporated capsular polysaccharides (11, 68, 69). Although Frases et al. (69) showed a number of physiochemical differences between EPS and capsular polysaccharides, no report has observed reducing ends in EPS. Incubation of a >100- and >10-kDa exopolysaccharide fraction confirmed the presence of reducing ends in the polysaccharides that are shed into the extracellular space. This has particularly important implications for vaccine development, as it provides a functional group that may be exploited in creating glycoconjugates for vaccine development. Furthermore, the presence of a reducing group in soluble exopolysaccharide could greatly facilitate their attachment to surfaces such as beads for immunological assays and purification of antibodies using affinity chromatography.
We also gained insight into how different cellular processing methods affect the polysaccharide capsule as visualized through microscopy. Overall, processing the C. neoformans cells with DMSO, French press, and sonication resulted in different effects on capsule size and membrane-wall integrity as measured by R.E probe reactivity. DMSO-treated cells had absent or reduced capsules, and there were indications that the cell membrane was permeabilized. Similarly, sonicated cells displayed reduced capsule size, but in contrast, membrane structure had not been affected. In the sonicated cells, it was also possible to visualize the colocalization of the R.E probe and the 18B7AF568 conjugate in the capsular polysaccharide. French press exposed reducing ends of the capsule that were previously associated with the cell wall or inaccessible and were then labeled by the R.E probe and 18B7AF568, providing further confirmation that the probe can react with GXM.
Given that reducing ends (aldehydes) are among the most reactive groups in polysaccharides, the lack of R.E probe labeling in the capsule suggests that this could be a strategy to reduce capsular reactivity to compounds in soil and/or phagosomes after ingestion by phagocytic predators such as amoeba. For example, polysaccharide-reducing groups can react with protein amino groups in the Maillard reaction, which could potentially generate toxic compounds or produce stresses that damage the capsule. Microbial polysaccharides are vulnerable to depolymerization by reactive oxygen and nitrogen species, and the presence of reactive reducing groups enhances their susceptibility to a myriad of microbicidal compounds produced by the oxidative burst of neutrophils, macrophage, and amoeba (22).
In summary, our results establish a proof of concept that chemical biology provides new tools to study the capsule of C. neoformans. For example, the R.E probe could be used to label extracellular vesicles containing polysaccharide and thus facilitate their purification. In further studies, we intend to identify and characterize the glycan structures containing reducing ends and hope to possibly use insights for the development of novel therapeutics against C. neoformans.
Experimental procedures
General synthetic methods
Unless otherwise noted, all reactions containing air- and moisture-sensitive reagents were carried out under an inert atmosphere of nitrogen in oven-dried glassware with magnetic stirring. N2-flushed stainless cannulas or plastic syringes were used to transfer air- and moisture-sensitive reagents. All reactions were monitored by thin-layer chromatography (TLC) on Merck DC-Alufolien plates precoated with silica gel 60 F254. Visualization was performed with UV-light (254 nm) fluorescence quenching. Evaporation in vacuo refers to the removal at 40 °C, unless otherwise stated, of volatiles on a Buchi rotary evaporator with an integrated vacuum pump.
Chromatography
Silica gel flash chromatography was carried out using Davisil LC60A (40–63 μm) silica gel or with automated flash chromatography systems, Buchi Reveleris® X2 (UV 200–500 nm and ELSD detection, Reveleris® silica cartridges 40 μm, BÜCHI Labortechnik AG) and Biotage® SP4 HPFC (UV 200–500 nm, Biotage® SNAP KP-Sil 50-μm irregular silica, Biotage AB).
Synthetic materials
All chemicals for the synthesis were purchased from commercial suppliers (Acros, Carbosynth Ltd., Thermo Fisher Scientific, Glycom A/S, Merck, Sigma, and VWR) and used without purification. Anhydrous dichloromethane and tetrahydrofuran were obtained from a PureSolv-ENTM solvent purification system (Innovative Technology Inc.). All other anhydrous solvents were used as purchased from Sigma in AcroSeal® bottles.
Instrumentation
1H NMR (400, 500, or 600 MHz) and 13C NMR (101 MHZ or 125 MHz) spectra were recorded on Varian-Inova or Bruker spectrometers at 25 °C in chloroform-d1 (CDCl3), methanol-d4 (CD3OD), and water-d2 (D2O), and 1H NMR spectra were standardized against the residual solvent peak (CDCl3, δ = 7.26 ppm; CD3OD, δ = 3.31 ppm; D2O, δ = 4.79 ppm; d6-DSS δ = 0.0 ppm, or internal tetramethylsilane, δ = 0.00 ppm). Bruker instrumentation spectrometers were equipped with Avance II console and triple resonance, TCI cryogenic probe with the z axis pulsed field. 13C NMR spectra were standardized against the residual solvent peak (CDCl3, δ = 77.16 ppm; CD3OD, δ = 49.00 ppm. All 13C NMR are 1H decoupled. All NMR data are represented as follows: chemical shift (δ ppm) and multiplicity (s = singlet; d = doublet; t = triplet; q = quartet; dd = doublet of doublets; ddd = doublet of doublets of doublets; dt = doublet of triplets; m = multiplet; br = broad signal; ad = apparent doublet; and at = apparent triplet), coupling constant in Hz, integration. Assignments were aided by homonuclear 1H-1H (COSY, TOCSY, and 1H-13C heteronuclear (HSQC, HMBC) two-dimensional correlation spectroscopies. 13C chemical shifts were reported with one digit after the decimal point, unless an additional digit was reported to distinguish overlapping peaks. Software used for data processing was MestReNova, version 11.0.0–17609 (MestReLab Research S.L.). NMR analysis of EPS fractions were conducted with 64, 128, or 256 scans and an FID size of 16,384 points. Standard Bruker pulse sequences were used to collect the 1D data (p3919gp and zggpw5). Data were processed in Topspin (Bruker version 3.5) by truncating the FID to 8192 points, using a squared cosine bell window function, and zero filling to 65,536 points. Relaxation times for EPS fractions were 1 or 2 s. High-resolution MS data were recorded on a Waters micromass LCT LC-Tof instrument using electrospray ionization (ESI) in either positive or negative mode. Low-resolution MS experiments were recorded on a Waters micromass Quattro Micro LC-MS/MS instrument using ESI in either positive or negative mode. Synthetic procedures for the synthesis of the RE probe can be found in supporting information.
Growth conditions
C. neoformans serotype A strain H99 (American Type Culture Collection (ATCC) 208821), C. neoformans serotype D (ATCC 24067), C. gattii strains serotype B (ATCC 24065) and C (ATCC 32608), CAP59Δ, CAP67Δ, and C. albidus were grown for 48 h at 30 °C in capsule-inducing media composed of 10 mm MgSO4, 29.3 mm KH2PO4, 13 mm glycine, 3 μm thiamine-HCl, adjusted to pH 5.5, and supplemented with 15 mm (regular minimal media) dextrose, with or without 1 mm l-DOPA (melanized cells).
Cellular processing techniques
DMSO extraction was carried out as described previously (45). Ultrasonication was carried out as described previously (50). French press was carried out as reported previously (49).
Hydroxylamine-armed probes physicochemical properties incubation with C. neoformans
C. neoformans cells were grown inoculated in Sabouraud dextrose medium for 2 days and then transferred to capsule-inducing minimal media for 3 days. Cells were pelleted (2000 rpm, 5 min) and washed with PBS three times. The cells were then incubated with the reducing end probe overnight in the dark at 30 °C. Cells were pelleted and washed to the remove excess probe. The spectrometric analysis was completed using a SpectraMax M5 microplate reader. Statistical analysis was performed in GraphPad Prism (Version 8, GraphPad Software, La Jolla, CA).
Isolation of EPS samples (10 and 100 kDa)
EPS isolation followed published procedure (14). Briefly, fungal cells were separated from culture supernatants by centrifugation at 4000 × g (15 min, 5 °C). The supernatant fluids were collected and centrifuged again at 15,000 × g (15 min, 4 °C) to remove smaller debris. The resulting supernatant was concentrated ∼20-fold using an Amicon (Millipore, Danvers, MA) ultrafiltration cell (with a cutoff of 100 kDa and a total capacity of 200 ml) with stirring and Biomax polyethersulfone ultrafiltration discs (63.5 mm). A nitrogen (N2) stream was used as the pressure gas. After the supernatant was concentrated, a thick, translucent film was observed in close association with the ultrafiltration disc and was covered by a concentrated fluid phase. The fluid phase was discarded, and the viscous layer was collected with a cell scraper for storage at room temperature. Fractions that were passed through the 100-kDa filtration discs were filtered through 10-kDa membranes, resulting again in film formation. It is important to note that the filtration cutoff is a hydrodynamic exclusion criterion, and the molecules in these fractions are smaller but are retained by the filter. Subsequently, aliquots of both the 10- and 100-kDa fractions were subjected to lyophilization in order to approximate the amount of EPS material.
Fluorescent microscopy
H99, CAP59Δ, or CAP67Δ C. neoformans cells were grown and inoculated in Sabouraud dextrose medium for 2 days and then transferred to capsule-inducing minimal media for 3 days. Samples were blocked in blocking buffer (1% bovine serum albumin (BSA)). Thereafter, the cells were incubated with or without the 2.5 μm of the fluorescent reducing-end probe overnight at 37 °C in the dark. If desired, a 1:1000 (1 mm) dilution of BODIPYTM FL C12 was incubated for 5 h after the addition of the reducing end probe and left overnight. Cells were pelleted and washed three times to remove excess probe or dye with PBS plus 1% BSA and stained for 20 min with or without Uvitex-2b and washed three times with PBS. If desired, a 1 mm (1:1000) CellTraceTM BODIPYTM TR methyl ester was added 1 h prior to imaging and then incubated with 1 μg/ml IgG1 18B7-Alexa Fluor 594 conjugate for 1 h at 37 °C. The Channel exposures used for microscopy were as follows: FITC (800 ms) (excitation/emission 498:516 nm), TRITC (600 ms) (excitation/emission 540:580 nm), and 4,6-diamidino-2-phenylindole (50 ms) (excitation/emission 350:450 nm). Images were collected with an Olympus AX70 microscope, photographed with a QImaging Retiga 1300 digital camera using the QCapture Suite Version 2.46 software (QImaging, Burnaby, British Columbia, Canada), and processed with ImageJ or Fiji (National Institutes of Health).
Polysaccharide labeling and fluorescence reading
CPS, 100 kDa EPS, 10 kDa EPS, and chitin fractions (∼1.18 μmol, 1 eq) were incubated with hydroxylamine-armed probe 1 (1 mg, 2.36 for μmol, 2 eq) in 1 ml of PBS (100 mm, pH 4) for 24 h and shaken at 30 °C in the dark (set up in triplicate). The reaction mixtures were then transferred to dialysis tubing (1- or 3.5-kDa molecular mass cutoff, Spectrum Laboratories, Inc.) and dialyzed against distilled water for 48 h at room temperature, in the dark, with water replacement every 6 h. Three aliquots of each fraction were then transferred to an opaque plate reader to analyze fluorescence. Fluorescence in different fractions was measured with the SpectraMax M5 microplate reader (Molecular Devices). All measurements were performed at 37 °C with an excitation wavelength of 360 nm, an emission wavelength of 440 nm, and a cutoff emission filter of 435 nm. Wells were set up in triplicate, and 30 readings were taken per well with a 5-s mix time prior to reading. Photomultiplier tube sensitivity was set to high. Graph generation and statistical analysis were complete using GraphPad Prism (Version 8, GraphPad Software, La Jolla, CA).
Generating India ink equatorial rings in C. neoformans polysaccharide
Cells were prepared as described above. 2 μl of India ink was added to 8 μl of a solution of cells on a glass slide. The coverslip was placed on top, and a firm pressure was applied.
Rupture of Cryptococcus neoformans cells
Cells were prepared as described above, with India ink staining. Shear pressure was applied by moving the coverslip back and forth four times.
Author contributions
C. J. C., R. J. C., L. G., M. P. W., A. B., S. O., and A. C. conceptualization; C. J. C., R. J. C., L. G., M. P. W., and A. B. data curation; C. J. C., R. J. C., L. G., M. P. W., A. B., S. O., and A. C. formal analysis; C. J. C., R. J. C., S. O., and A. C. funding acquisition; C. J. C., R. J. C., L. G., M. P. W., and A. B. investigation; C. J. C., R. J. C., L. G., M. P. W., and A. B. methodology; C. J. C., S. O., and A. C. writing-original draft; C. J. C., R. J. C., L. G., M. P. W., S. O., and A. C. writing-review and editing; S. O. and A. C. supervision; S. O. and A. C. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Yannick Ortin for NMR support, Daniel Quinn Smith for the contribution of editing the figures, and the Johns Hopkins Biomolecular NMR Center.
This work was supported by Irish Research Council Postgraduate Award GOIPG/2016/998 (to C. J. C.), by JHU CFAR National Institutes of Health/NIAID Grants P30AI094189 (to R. J. B. C.) and AI007417 (to M. P. W.), by National Institutes of Health Grant GM00728847 (to A. B.), by Science Foundation Ireland Award 13/IA/1959 (to S. O.), and by National Institutes of Health Grants AI052733 16 and HL059842 19 (to A. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and supporting Refs. 1–7.
- GalXM
- galactoxylomannan
- GXM
- glucuronoxylomannan
- R.E probe
- reducing-end probe, hydroxylamine-armed fluorescent probe
- CPS
- capsular polysaccharide
- EPS
- exopolysaccharide
- RFU
- relative fluorescence unit
- TRITC
- tetramethylrhodamine isothiocyanate
- ESI
- electrospray ionization.
References
- 1. Park B. J., Wannemuehler K. A., Marston B. J., Govender N., Pappas P. G., and Chiller T. M. (2009) Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23, 525–530 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 2. Garcia-Hermoso D., Janbon G., and Dromer F. (1999) Epidemiological evidence for dormant Cryptococcus neoformans infection. J. Clin. Microbiol. 37, 3204–3209 10.1128/JCM.37.10.3204-3209.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldman D. L., Khine H., Abadi J., Lindenberg D. J., Pirofski La., Niang R., and Casadevall A. (2001) Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107, E66 10.1542/peds.107.5.e66 [DOI] [PubMed] [Google Scholar]
- 4. Singh N., Dromer F., Perfect J. R., and Lortholary O. (2008) Cryptococcosis in solid organ transplant recipients: current state of the science. Clin. Infect. Dis. 47, 1321–1327 10.1086/592690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martinez L. R., and Casadevall A. (2015) Biofilm formation by Cryptococcus neoformans. Microbiol. Spectr. 3, 10.1128/microbiolspec.MB-0006-2014 [DOI] [PubMed] [Google Scholar]
- 6. Caballero, Van Dyke M. C., and Wormley F. L. Jr. (2018) A call to arms: quest for a cryptococcal vaccine. Trends Microbiol. 26, 436–446 10.1016/j.tim.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vecchiarelli A., Retini C., Pietrella D., Monari C., Tascini C., Beccari T., and Kozel T. R. (1995) Downregulation by cryptococcal polysaccharide of tumor necrosis factor α and interleukin-1β secretion from human monocytes. Infect. Immun. 63, 2919–2923 10.1128/IAI.63.8.2919-2923.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong Z. M., and Murphy J. W. (1995) Effects of the two varieties of Cryptococcus neoformans cells and culture filtrate antigens on neutrophil locomotion. Infect. Immun. 63, 2632–2644 10.1128/IAI.63.7.2632-2644.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Retini C., Vecchiarelli A., Monari C., Tascini C., Bistoni F., and Kozel T. R. (1996) Capsular polysaccharide of Cryptococcus neoformans induces proinflammatory cytokine release by human neutrophils. Infect. Immun. 64, 2897–2903 10.1128/IAI.64.8.2897-2903.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O'Meara T. R., and Alspaugh J. A. (2012) The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25, 387–408 10.1128/CMR.00001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bose I., Reese A. J., Ory J. J., Janbon G., and Doering T. L. (2003) A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2, 655–663 10.1128/EC.2.4.655-663.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zaragoza O., Telzak A., Bryan R. A., Dadachova E., and Casadevall A. (2006) The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol. Microbiol. 59, 67–83 10.1111/j.1365-2958.2005.04928.x [DOI] [PubMed] [Google Scholar]
- 13. Cherniak R., Valafar H., Morris L. C., and Valafar F. (1998) Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5, 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nimrichter L., Frases S., Cinelli L. P., Viana N. B., Nakouzi A., Travassos L. R., Casadevall A., and Rodrigues M. L. (2007) Self-aggregation of Cryptococcus neoformans capsular glucuronoxylomannan is dependent on divalent cations. Eukaryot. Cell 6, 1400–1410 10.1128/EC.00122-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cordero R. J., Frases S., Guimaräes A. J., Rivera J., and Casadevall A. (2011) Evidence for branching in cryptococcal capsular polysaccharides and consequences on its biological activity. Mol. Microbiol. 79, 1101–1117 10.1111/j.1365-2958.2010.07511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Casadevall A., Coelho C., Cordero R. J. B., Dragotakes Q., Jung E., Vij R., and Wear M. P. (2019) The capsule of Cryptococcus neoformans. Virulence 10, 822–831 10.1080/21505594.2018.1431087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sommer U., Liu H., and Doering T. L. (2003) An α-1,3-mannosyltransferase of Cryptococcus neoformans. J. Biol. Chem. 278, 47724–47730 10.1074/jbc.M307223200 [DOI] [PubMed] [Google Scholar]
- 18. Klutts J. S., and Doering T. L. (2008) Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J. Biol. Chem. 283, 14327–14334 10.1074/jbc.M708927200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klutts J. S., Levery S. B., and Doering T. L. (2007) A β-1,2-xylosyltransferase from Cryptococcus neoformans defines a new family of glycosyltransferases. J. Biol. Chem. 282, 17890–17899 10.1074/jbc.M701941200 [DOI] [PubMed] [Google Scholar]
- 20. Cordero R. J., Bergman A., and Casadevall A. (2013) Temporal behavior of capsule enlargement by Cryptococcus neoformans. Eukaryot. Cell 12, 1383–1388 10.1128/EC.00163-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Whitfield C. (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 10.1146/annurev.biochem.75.103004.142545 [DOI] [PubMed] [Google Scholar]
- 22. Duan J., and Kasper D. L. (2011) Oxidative depolymerization of polysaccharides by reactive oxygen/nitrogen species. Glycobiology 21, 401–409 10.1093/glycob/cwq171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoneda A., and Doering T. L. (2006) A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 17, 5131–5140 10.1091/mbc.e06-08-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodrigues M. L., Nimrichter L., Oliveira D. L., Frases S., Miranda K., Zaragoza O., Alvarez M., Nakouzi A., Feldmesser M., and Casadevall A. (2007) Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 6, 48–59 10.1128/EC.00318-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zaragoza O., Rodrigues M. L., De Jesus M., Frases S., Dadachova E., and Casadevall A. (2009) The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 68, 133–216 10.1016/S0065-2164(09)01204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Godinho R. M., Crestani J., Kmetzsch L., Araujo Gde., S., Frases S., Staats C. C., Schrank A., Vainstein M. H., and Rodrigues M. L. (2014) The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci. Rep. 4, 6198 10.1038/srep06198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kmetzsch L., Joffe L. S., Staats C. C., de Oliveira D. L., Fonseca F. L., Cordero R. J., Casadevall A., Nimrichter L., Schrank A., Vainstein M. H., and Rodrigues M. L. (2011) Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol. Microbiol. 81, 206–218 10.1111/j.1365-2958.2011.07686.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seeberger P. H., and Werz D. B. (2007) Synthesis and medical applications of oligosaccharides. Nature 446, 1046–1051 10.1038/nature05819 [DOI] [PubMed] [Google Scholar]
- 29. Bertozzi C. R., and Kiessling L. L. (2001) Chemical glycobiology. Science 291, 2357–2364 10.1126/science.1059820 [DOI] [PubMed] [Google Scholar]
- 30. Saxon E., and Bertozzi C. R. (2000) Cell surface engineering by a modified Staudinger reaction. Science 287, 2007–2010 10.1126/science.287.5460.2007 [DOI] [PubMed] [Google Scholar]
- 31. Sletten E. M., and Bertozzi C. R. (2011) From mechanism to mouse: a tale of two bioorthogonal reactions. Acc. Chem. Res. 44, 666–676 10.1021/ar200148z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang P. V., Chen X., Smyrniotis C., Xenakis A., Hu T., Bertozzi C. R., and Wu P. (2009) Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew. Chem. Int. Ed. Engl. 48, 4030–4033 10.1002/anie.200806319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu P., Shui W., Carlson B. L., Hu N., Rabuka D., Lee J., and Bertozzi C. R. (2009) Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proc. Natl. Acad. Sci. U.S.A. 106, 3000–3005 10.1073/pnas.0807820106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zeng Y., Ramya T. N., Dirksen A., Dawson P. E., and Paulson J. C. (2009) High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods 6, 207–209 10.1038/nmeth.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kool E. T., Crisalli P., and Chan K. M. (2014) Fast α nucleophiles: structures that undergo rapid hydrazone/oxime formation at neutral pH. Org. Lett. 16, 1454–1457 10.1021/ol500262y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spears R. J., and Fascione M. A. (2016) Site-selective incorporation and ligation of protein aldehydes. Org. Biomol. Chem. 14, 7622–7638 10.1039/C6OB00778C [DOI] [PubMed] [Google Scholar]
- 37. Hermanson G. T. (2013) Bioconjugate Techniques, 3rd. Ed., pp. 155–157, Elsevier Inc., New York [Google Scholar]
- 38. Morgan R. K., and Cohen M. S. (2015) A clickable aminooxy probe for monitoring cellular ADP-ribosylation. ACS Chem. Biol. 10, 1778–1784 10.1021/acschembio.5b00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dal Molin M., Verolet Q., Colom A., Letrun R., Derivery E., Gonzalez-Gaitan M., Vauthey E., Roux A., Sakai N., and Matile S. (2015) Fluorescent flippers for mechanosensitive membrane probes. J. Am. Chem. Soc. 137, 568–571 10.1021/ja5107018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jobe K., Brennan C. H., Motevalli M., Goldup S. M., and Watkinson M. (2011) Modular “click” sensors for zinc and their application in vivo. Chem. Commun. 47, 6036–6038 10.1039/c1cc11213a [DOI] [PubMed] [Google Scholar]
- 41. Horatscheck A., Wagner S., Ortwein J., Kim B. G., Lisurek M., Beligny S., Schütz A., and Rademann J. (2012) Benzoylphosphonate-based photoactive phosphopeptide mimetics for modulation of protein-tyrosine phosphatases and highly specific labeling of SH2 domains. Angew. Chem. Int. Ed. Engl. 51, 9441–9447 10.1002/anie.201201475 [DOI] [PubMed] [Google Scholar]
- 42. Zhang C., Liu Z., Li Y., He W., Gao X., and Guo Z. (2013) In vitro and in vivo imaging application of a 1,8-naphthalimide-derived Zn2+ fluorescent sensor with nuclear envelope penetrability. Chem. Commun. 49, 11430–11432 10.1039/c3cc46862c [DOI] [PubMed] [Google Scholar]
- 43. Perez-Dulzaides R., Camacho E., Cordero R. J. B., and Casadevall A. (2018) Cell wall dyes interfere with Cryptococcus neoformans melanin deposition. Microbiology 164, 1012–1022 10.1099/mic.0.000682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. García-Rivera J., Chang Y. C., Kwon-Chung K. J., and Casadevall A. (2004) Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3, 385–392 10.1128/ec.3.2.385-392.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bryan R. A., Zaragoza O., Zhang T., Ortiz G., Casadevall A., and Dadachova E. (2005) Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot. Cell 4, 465–475 10.1128/EC.4.2.465-475.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ikeda R., Matsuyama H., Nishikawa A., Shinoda T., and Fukazawa Y. (1991) Comparison of serological and chemical characteristics of capsular polysaccharides of Cryptococcus neoformans var. neoformans serotype A and Cryptococcus albidus var. albidus. Microbiol. Immunol. 35, 125–138 10.1111/j.1348-0421.1991.tb01540.x [DOI] [PubMed] [Google Scholar]
- 47. Kwon-Chung K. J., Fraser J. A., Doering T. L., Wang Z., Janbon G., Idnurm A., and Bahn Y. S. (2014) Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect. Med. 4, a019760 10.1101/cshperspect.a019760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gates M. A., Thorkildson P., and Kozel T. R. (2004) Molecular architecture of the Cryptococcus neoformans capsule. Mol. Microbiol. 52, 13–24 10.1111/j.1365-2958.2003.03957.x [DOI] [PubMed] [Google Scholar]
- 49. Cook W. L., Felton F. G., Muchmore H. G., and Rhoades E. R. (1969) Cell wall differences of patient and soil isolates of Cryptococcus neoformans. Med. Mycol. 7, 257–260 10.1080/00362177085190471 [DOI] [PubMed] [Google Scholar]
- 50. Strother C. (2019) Cryptococcus neoformans: New Techniques in an Old Field. Ph.D. thesis, Johns Hopkins University [Google Scholar]
- 51. Nicola A. M., Frases S., and Casadevall A. (2009) Lipophilic dye staining of Cryptococcus neoformans extracellular vesicles and capsule. Eukaryot. Cell 8, 1373–1380 10.1128/EC.00044-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. De Jesus M., Nicola A. M., Rodrigues M. L., Janbon G., and Casadevall A. (2009) Capsular localization of the Cryptococcus neoformans polysaccharide component galactoxylomannan. Eukaryot. Cell 8, 96–103 10.1128/EC.00331-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zaragoza O., McClelland E. E., Telzak A., and Casadevall A. (2006) Equatorial ring-like channels in the Cryptococcus neoformans polysaccharide capsule. FEMS Yeast Res. 6, 662–666 10.1111/j.1567-1364.2006.00070.x [DOI] [PubMed] [Google Scholar]
- 54. Reese A. J., Yoneda A., Breger J. A., Beauvais A., Liu H., Griffith C. L., Bose I., Kim M. J., Skau C., Yang S., Sefko J. A., Osumi M., Latge J. P., Mylonakis E., and Doering T. L. (2007) Loss of cell wall α(1–3)-glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 63, 1385–1398 10.1111/j.1365-2958.2006.05551.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cuthbertson L., Kos V., and Whitfield C. (2010) ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 74, 341–362 10.1128/MMBR.00009-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bushell S. R., Mainprize I. L., Wear M. A., Lou H., Whitfield C., and Naismith J. H. (2013) Wzi is an outer membrane lectin that underpins group 1 capsule assembly in Escherichia coli. Structure 21, 844–853 10.1016/j.str.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McFadden D. C., De Jesus M., and Casadevall A. (2006) The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J. Biol. Chem. 281, 1868–1875 10.1074/jbc.M509465200 [DOI] [PubMed] [Google Scholar]
- 58. Sebolai O. M., Pohl C. H., Botes P. J., Strauss C. J., van Wyk P. W., Botha A., and Kock J. L. (2007) 3-Hydroxy fatty acids found in capsules of Cryptococcus neoformans. Can. J. Microbiol. 53, 809–812 10.1139/W07-045 [DOI] [PubMed] [Google Scholar]
- 59. MacDonald R. I. (1990) Characteristics of self-quenching of the fluorescence of lipid-conjugated rhodamine in membranes. J. Biol. Chem. 265, 13533–13539 [PubMed] [Google Scholar]
- 60. Pierini L. M., and Doering T. L. (2001) Spatial and temporal sequence of capsule construction in Cryptococcus neoformans. Mol. Microbiol. 41, 105–115 10.1046/j.1365-2958.2001.02504.x [DOI] [PubMed] [Google Scholar]
- 61. Frases S., Pontes B., Nimrichter L., Viana N. B., Rodrigues M. L., and Casadevall A. (2009) Capsule of Cryptococcus neoformans grows by enlargement of polysaccharide molecules. Proc. Natl. Acad. Sci. U.S.A. 106, 1228–1233 10.1073/pnas.0808995106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoneda A., and Doering T. L. (2008) Regulation of Cryptococcus neoformans capsule size is mediated at the polymer level. Eukaryot. Cell 7, 546–549 10.1128/EC.00437-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dwek R. A. (1996) Glycobiology: toward understanding the function of sugars. Chem. Rev. 96, 683–720 10.1021/cr940283b [DOI] [PubMed] [Google Scholar]
- 64. Rodrigues M. L., Nimrichter L., Oliveira D. L., Nosanchuk J. D., and Casadevall A. (2008) Vesicular trans-cell wall transport in fungi: a mechanism for the delivery of virulence-associated macromolecules? Lipid Insights 2, 27–40 10.4137/lpi.s1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Oliveira D. L., Nimrichter L., Miranda K., Frases S., Faull K. F., Casadevall A., and Rodrigues M. L. (2009) Cryptococcus neoformans cryo-ultramicrotomy and vesicle fractionation reveals an intimate association between membrane lipids and glucuronoxylomannan. Fungal Genet. Biol. 46, 956–963 10.1016/j.fgb.2009.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Silva V. K. A., Rodrigues M. L., and May R. C. (2019) Deciphering fungal extracellular vesicles: from cell biology to pathogenesis. Curr. Clin. Microbiol. Rep. 6, 89–97 10.1007/s40588-019-00128-1 [DOI] [Google Scholar]
- 67. Bielska E., Sisquella M. A., Aldeieg M., Birch C., O'Donoghue E. J., and May R. C. (2018) Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 9, 1556 10.1038/s41467-018-03991-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. García-Rodas R., Cordero R. J., Trevijano-Contador N., Janbon G., Moyrand F., Casadevall A., and Zaragoza O. (2014) Capsule growth in Cryptococcus neoformans is coordinated with cell cycle progression. mBio. 5, e00945–14 10.1128/mBio.00945-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Frases S., Nimrichter L., Viana N. B., Nakouzi A., and Casadevall A. (2008) Cryptococcus neoformans capsular polysaccharide and exopolysaccharide fractions manifest physical, chemical, and antigenic differences. Eukaryot. Cell 7, 319–327 10.1128/EC.00378-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.