Abstract
Strigolactones (SLs) are terpenoid-derived plant hormones that regulate various developmental processes, particularly shoot branching, root development, and leaf senescence. The SL receptor has an unusual mode of action. Upon binding SL, it hydrolyzes the hormone, and then covalently binds one of the hydrolytic products. These initial events enable the SL receptor DAD2 (in petunia) to interact with the F-box protein PhMAX2A of the Skp-Cullin-F-box (SCF) complex and/or a repressor of SL signaling, PhD53A. However, it remains unclear how binding and hydrolysis structurally alters the SL receptor to enable its engagement with signaling partners. Here, we used mutagenesis to alter DAD2 and affect SL hydrolysis or DAD2's ability to interact with its signaling partners. We identified three DAD2 variants whose hydrolytic activity had been separated from the receptor's interactions with PhMAX2A or PhD53A. Two variants, DAD2N242I and DAD2F135A, having substitutions in the core α/β hydrolase-fold domain and the hairpin, exhibited hormone-independent interactions with PhMAX2A and PhD53A, respectively. Conversely, the DAD2D166A variant could not interact with PhMAX2A in the presence of SL, but its interaction with PhD53A remained unaffected. Structural analyses of DAD2N242I and DAD2D166A revealed only small differences compared with the structure of the WT receptor. Results of molecular dynamics simulations of the DAD2N242I structure suggested that increased flexibility is a likely cause for its SL-independent interaction with PhMAX2A. Our results suggest that PhMAX2A and PhD53A have distinct binding sites on the SL receptor and that its flexibility is a major determinant of its interactions with these two downstream regulators.
Keywords: plant hormone, signal transduction, receptor, mutagenesis, crystallography, molecular dynamics, DAD2, Petunia hybrida, phytohormone signaling, strigolactone
Introduction
Strigolactones (SLs)5 are a group of terpenoid-derived plant hormones, characterized by a butenolide ring linked to a tricyclic lactone by an enol–ether bridge. They were initially identified as exogenous signals that promote the germination of parasitic weeds, as well as symbiosis between host plants and arbuscular mycorrhizal fungi (1–3). In plants, SLs regulate various developmental processes, particularly shoot branching, root development, and leaf senescence (4–9).
The SL signal is perceived by an α/β hydrolase-fold protein that possesses a canonical Ser-His-Asp catalytic triad in its active site cavity (10). Orthologs of the SL receptor have been identified from a number of species, including petunia (DECREASED APICAL DOMINANCE 2; DAD2 (10)), rice (DWARF14; OsD14 (11, 12)), Arabidopsis (Arabidopsis thaliana D14; AtD14 (13)), and pea (RAMOSUS3; RMS3 (14)). The catalytic triad allows the SL receptor to slowly hydrolyze the SL molecule at the enol–ether linkage, resulting in the generation of a tricyclic lactone (ABC-ring) and a butenolide product (D-ring (10, 12, 14, 15)). SL hydrolysis by the SL receptor leads to the covalent modification of the catalytic histidine residue as shown for AtD14, OsD14, and RMS3 (14, 16, 17).
Numerous studies have shown that the receptor has a lower melting temperature (Tm) in the presence of SL (10, 14, 18–20). In response to SL, the receptor interacts with an F-box protein of the Skp1-Cullin1-F-box (SCF) ubiquitin ligase complex: petunia (MORE AXILLARY GROWTH 2A, PhMAX2A); Arabidopsis (MAX2); rice (DWARF3, D3) (10, 16, 19)) and the repressors of SL signaling: petunia (PhD53A); rice (DWARF53, D53); Arabidopsis (Suppressor of MAX2-Like, SMXL 6/7/8) (21–25). The interaction between the receptor, SCF ubiquitin ligase complex, and the SL repressor protein, leads to degradation of the repressor protein in an ubiquitin-dependent manner via the 26S proteasomal pathway, resulting in the activation of the SL signaling cascade (26).
Several research groups have crystallized the SL receptor in the presence of SL and SL-intermediates, with none of these ligand-associated receptor structures showing a substantial change in conformation compared with the apo-receptor structures (12, 15, 19, 27). In 2015, Zhao et al. (19) reported that the OsD14 receptor, in the presence of the synthetic SL analogue, GR24, showed a similar deuterium exchange rate as that observed for the receptor alone, which is consistent with the structural evidence of minimal conformational change. The OsD14 receptor does, however, show increased deuterium exchange in the presence of both GR24 and the rice F-box protein D3. These results suggested that the SL receptor undergoes conformational change in the presence of both SL and the F-box protein and that binding of the SL ligand alone may not lead to a conformational change.
Consistent with the findings for the rice receptor of Zhao et al. (19), structural characterization of the Arabidopsis receptor AtD14 in complex with the F-box protein D3 and ASK1 (ARABIDOPSIS SKP1-LIKE1, another component of the SCF complex and required for efficient expression of the F-box protein) showed that AtD14 underwent a conformational change in its four-helix-lid domain (Fig. S1) upon interaction with D3. A SL hydrolysis intermediate called CLIM (Covalently-Linked Intermediate Molecule, composed of the D-ring from the SL ligand) was proposed to be trapped within the cavity of the bound AtD14 receptor and covalently linked to the catalytic His and Ser residues (16). Based on the structure of the AtD14-D3-ASK1 protein complex, it was proposed that SL hydrolysis and/or binding of the D-ring to the catalytic histidine residue of AtD14 play a role in generation of the active conformation of AtD14, allowing interaction with D3 (16). Using MS, Yao et al. (16) showed that the D-ring covalently binds to AtD14 both in vitro and in vivo, although the attachment was detected only on the catalytic His residue of the AtD14 receptor (16). A similar modification was also detected on the catalytic His residue of the pea SL receptor, RMS3, after it was incubated with GR24 (14). These findings suggest a possible role of D-ring binding in the formation of the active conformation of the SL receptor for interaction with the SCF complex.
In contrast with the hydrolysis-dependent mechanism of SL signaling discussed above, recent work by Seto et al. (28) suggests a hydrolysis-independent mechanism of SL signaling. Based on their observations of altered Tm of the AtD14 receptor in the presence of a range of different ligands over time, the authors suggested that an intact SL molecule probably leads to or induces the active signaling state of the AtD14 receptor (28). The authors further showed that a catalytic amino acid mutant, AtD14D218A, defective in SL hydrolysis, could bind an intact SL molecule with high affinity and cause a small Tm change. The AtD14D218A mutant, in the presence of SL, could also interact with the MAX2-ASK1 complex and SMXL7 in yeast two-hybrid assays (28). When expressed in planta, this mutant could complement the Atd14-1 knockout mutant but not the Atd14 max4 (receptor and SL biosynthesis) double mutant (28). These findings led to the hypothesis that SL hydrolysis by D14 is not essential for the initiation of SL signal transduction. In a study conducted by Shabek et al. (29), the rice F-box protein D3 was shown to regulate the hydrolytic activity of OsD14 via its C-terminal α-helix, and the authors also proposed that the D3 protein can bind different conformations of the OsD14 receptor (open or closed) via its C-terminal α-helix, suggesting that the F-box protein might be involved in the selection of hydrolysis-dependent or hydrolysis-independent signaling mechanisms by binding the receptor in different conformations.
Despite these advances we still do not completely understand how hydrolysis alters the WT receptor to allow interaction with signaling partners. We know that some mutations can dispense with the requirement for hydrolysis to enable interaction with signaling partners. Here we aimed to use mutagenesis to generate mutants with the ability to interact with the F-box protein PhMAX2A or the target protein PhD53A, even in the absence of hormone. Biochemical and structural analysis of these mutants was undertaken to provide insight into how the receptor becomes capable of interacting with partners, without the need for including a ligand that, in WT, is hydrolyzed during perception.
Results
Mutations on the lid domain and core α/β hydrolase-fold altered interaction with PhMAX2A and PhD53A
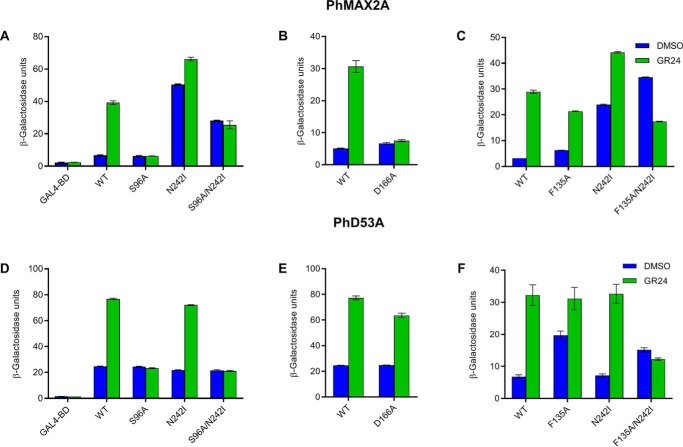
To understand the relative contributions of the different functions of the DAD2 SL receptor, we used mutagenesis to identify variants of DAD2 showing changes in interactions with binding partners, and/or altered enzyme activity. Two approaches were used. The first was to use random mutagenesis together with a yeast two-hybrid screen for mutants that could interact with binding partners in the absence of hormone. The other approach was to specifically mutate residues based on their location within the receptor or how well they were conserved in orthologous proteins. Of the mutants generated (Fig. S2 and Fig. 1), DAD2N242I, DAD2D166A, DAD2F135A were selected for further analysis as these mutants had altered interactions with binding partners. We also included the active site mutant DAD2S96A, as well as the DAD2S96A/N242I and DAD2F135A/N242I double mutants (Fig. 1, Figs. S1–S3). The DAD2S96A catalytic site mutant had previously been shown to have lost its hydrolytic activity and also the ability to interact with the F-box protein, PhMAX2A, or to complement the dad2 mutant background (Fig. 1A; also see Ref. 10).
Figure 1.
Interaction of DAD2 mutants with PhMAX2A and PhD53A. A–C, interaction of DAD2 mutants with PhMAX2A. D–F, interaction of DAD2 mutants with PhD53A. Interactions in each figure were quantified in the presence and absence of GR24 (10 μm for A–C and 1 μm for D–F) using yeast two-hybrid β-gal liquid culture assays. All data shown are mean ± S.E.; n = 3 technical replicates. Western blotting analyses confirming the presence of the fusion proteins in yeast are shown in Fig. S3.
The DAD2N242I mutant was able to interact with PhMAX2A in the absence of GR24 (Fig. 1A). This interaction with PhMAX2A in the absence of hormone could still occur even in combination with the active-site mutation (DAD2S96A/N242I). In the presence of GR24, the overall interaction level for this double mutant with PhMAX2A was reduced compared with that of DAD2WT. The DAD2F135A mutant, similar to DAD2WT, could not interact with PhMAX2A in the absence of GR24, although the double mutant DAD2F135A/N242I was able to interact with PhMAX2A in the absence of GR24 (Fig. 1C). The DAD2F135A/N242I double mutant also showed a lower level of interaction with PhMAX2A in the presence of GR24, compared with the levels shown in the absence of GR24. The DAD2D166A mutant showed the same baseline interaction as the negative control with PhMAX2A in the absence or presence of GR24 (Fig. 1B).
We also investigated the ability of these mutants to interact with the target repressor protein PhD53A. DAD2WT interaction with PhD53A is induced by GR24 and this interaction is retained by both DAD2N242I and DAD2D166A mutants (Fig. 1, D and E). The DAD2F135A and DAD2F135A/N242I mutants both showed a strong interaction with PhD53A in the absence of GR24 (Fig. 1F). However, DAD2F135A/N242I showed an overall reduced ability to interact with PhD53A in the presence of GR24 compared with DAD2WT. Neither the DAD2S96A nor the DAD2S96A/N242I mutants were able to interact with PhD53A (Fig. 1D).
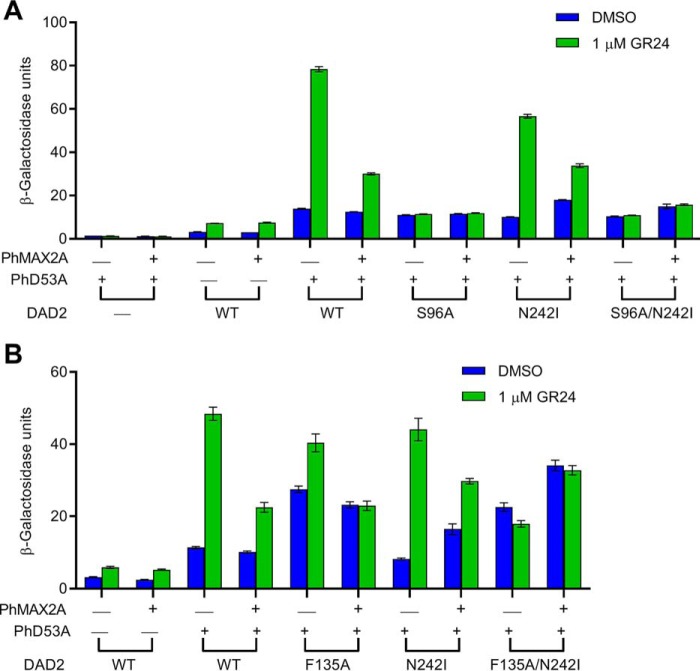
To examine the interaction between DAD2 and PhD53A in the presence of PhMAX2A, we generated yeast expressing all three proteins (Fig. 2). The presence of PhMAX2A reduced the strength of interaction between DAD2WT and PhD53A in the presence of GR24 (Fig. 2). This decrease was also observed for the DAD2 mutants, DAD2N242I and DAD2F135A, but not DAD2S96A, DAD2S96A/N242I, and DAD2F135A/N242I (Fig. 2). In the absence of GR24, PhMAX2A enhanced the interaction between DAD2F135A/N242I and PhD53A, and also between DAD2N242I and PhD53A (Fig. 2B).
Figure 2.
Interactions of DAD2 mutants with PhD53A in the presence and absence of PhMAX2A. A and B denote experiments conducted on separate occasions. Protein-protein interactions were quantified by assaying β-gal activity in the absence and presence of 1 μm GR24. WT DAD2 is indicated by WT. All data shown are mean ± S.E.; n = 3 technical replicates. Western blotting analyses confirming the presence of the fusion proteins in yeast are shown in Fig. S3.
DAD2 mutants have altered melting temperature compared with DAD2WT
We investigated the thermal stability of the DAD2 mutants (DAD2F135A, DAD2D166A, DAD2N242I, and DAD2S96A/N242I) in the presence and absence of GR24, using differential scanning fluorimetry (DSF) assays. DAD2F135A/N242I was not included as it was not possible to express stable soluble protein. Consistent with our previous results (10, 30), DAD2WT was observed to show a shift (ΔTm) of −8.9 °C in its melting temperature, from 57.2 °C in the absence of GR24 to 48.3 °C in the presence of GR24 (Table 1 and Fig. S4). Compared with DAD2WT, the DAD2 mutants, DAD2F135A, DAD2D166A, and DAD2N242I, had lower melting temperatures in both the presence and absence of GR24 (Fig. S4 and Table 1). The overall ΔTm for these mutants was observed to be greater than for DAD2WT (−16.7 °C for DAD2F135A, −13.0 °C for DAD2D166A, and −13.6 °C for DAD2N242I; Table 1). The DAD2S96A/N242I mutant also had a lower Tm in the absence of GR24; however, no changes were observed in the melting temperature of DAD2S96A/N242I in the presence of GR24 (Fig. S4 and Table 1), implying that this double mutant does not interact with GR24. Taken together, our results indicate that the DAD2F135A, DAD2D166A, and DAD2N242I mutants are thermally less stable than DAD2WT.
Table 1.
Melting temperatures of DAD2WT and DAD2 mutants in the presence and absence of GR24
| Protein | Melting temperature (Tm, °C) |
Difference (ΔTm, °C) | |
|---|---|---|---|
| DMSO | GR24 | ||
| DAD2WT | 57.2 | 48.3 | −8.9 |
| DAD2F135A | 53.0 | 36.3 | −16.7 |
| DAD2D166A | 56.0 | 43.0 | −13.0 |
| DAD2N242I | 51.2 | 37.6 | −13.6 |
| DAD2S96A/N242I | 48.2 | 48.5 | +0.3 |
DAD2N242I and DAD2D166A are catalytically similar to DAD2WT
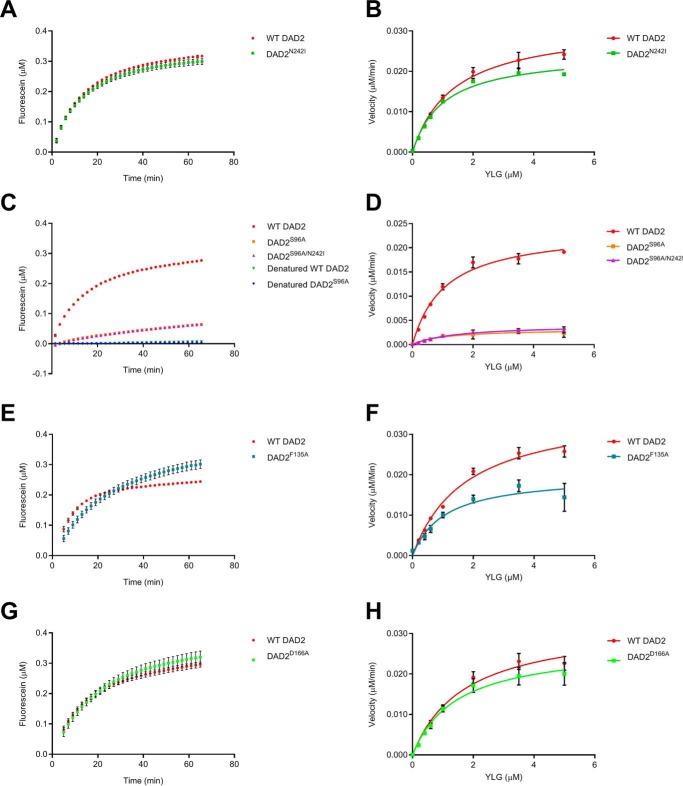
We examined whether the DAD2 mutants are catalytically distinct from DAD2WT, using a SL analogue, yoshimulactone green (YLG) as the substrate (31). We have previously shown that DAD2WT displays a two-step hydrolysis mechanism, similar to AtD14, OsD14, and RMS3, using YLG; therefore all kinetic analyses in this study were conducted using a 15-min time point of the pre-steady-state phase (14, 29–31). Here, we refer to kcat as the rate constant of the pre-steady-state phase and the K½ as the YLG concentration that gives half the maximal velocity (Vmax (14, 30)). DAD2N242I and DAD2D166A have similar kinetics to DAD2WT (Fig. 3, A and G, and Table 2) and DAD2F135A has a reduced kcat (Table 2 and Fig. 3, E and F).
Figure 3.
Hydrolysis of YLG by DAD2 mutants. A, C, E, and G, time course hydrolysis of 1 μm YLG by DAD2 mutants (0.34 μm protein) over 60 min. Denatured DAD2WT (WT DAD2) and DAD2S96A (S96A) were used as negative controls. B, D, F, and H, hydrolysis of YLG by DAD2 mutants (0.34 μm protein) at various YLG concentrations. Each data point is the mean ± S.E. of three technical replicates.
Table 2.
Kinetic parameters of DAD2WT and DAD2 mutants
| Protein | K½ | Vmax | Kcat |
|---|---|---|---|
| μm | μm/min | min−1 | |
| DAD2WT | 1.36 ± 0.17 | 0.032 | 0.093 ± 0.004 |
| DAD2S96A | 1.06 ± 0.57 | 0.003 | 0.010 ± 0.002 |
| DAD2F135A | 1.05 ± 0.31 | 0.020 | 0.059 ± 0.006 |
| DAD2D166A | 1.46 ± 0.33 | 0.027 | 0.080 ± 0.007 |
| DAD2N242I | 1.03 ± 0.11 | 0.025 | 0.073 ± 0.003 |
| DAD2S96A/N242I | 1.44 ± 0.35 | 0.004 | 0.012 ± 0.001 |
We have previously shown that the catalytic triad mutant of DAD2 (DAD2S96A) is hydrolytically inactive when examined using TLC with GR24 as the substrate (10). Unexpectedly, weak hydrolytic activity toward the YLG substrate was detected in DAD2S96A and DAD2S96A/N242I (Table 2 and Fig. 3, C and D). The observed weak YLG hydrolysis by DAD2S96A was abolished when this mutant was subjected to heat denaturation (Fig. 3C). However, it should be noted that YLG is a nonnatural substrate (31). The observed weak hydrolysis of YLG by DAD2S96A may therefore not be entirely relevant to SL hydrolysis by DAD2.
DAD2N242I exhibits structural flexibility
To determine whether the N242I mutation has altered the conformation of DAD2, allowing it to interact with PhMAX2A in the absence of GR24, we solved the crystal structure of DAD2N242I at 1.58 Å resolution, with two molecules of the mutant protein per asymmetric unit. Both molecules displayed the classical apo-SL receptor structure, with no major conformational change observed in either of the two protein molecules (Fig. 4 and Fig. S5). A MES molecule was bound within the catalytic cavity of the two DAD2N242I molecules, where MES interacts with the catalytic Ser-96 and His-246 residues through hydrogen bonds (Fig. S6). DSF results showed that there was no significant shift in the melting temperature of DAD2WT and DAD2N242I in the presence of MES (Fig. S7), suggesting that MES binds to DAD2 with low affinity. It is likely that MES was present within the internal cavity of the protein in the crystal structure, because of its high concentration (0.1 m) in the crystallization solution.
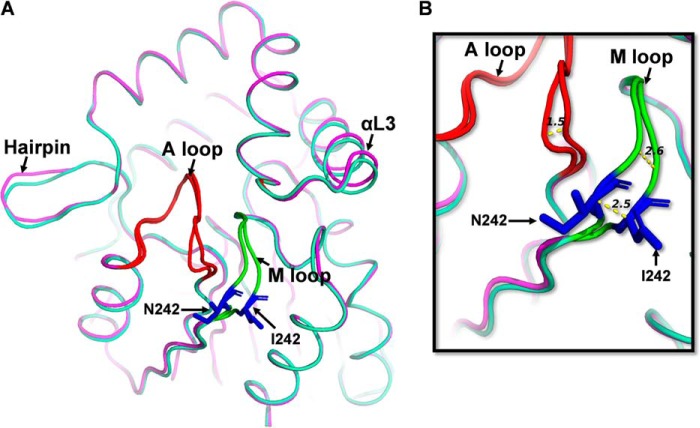
Figure 4.
Structural comparison of DAD2WT and molecule A of DAD2N242I. A, structure of molecule A of DAD2N242I protein (light green) superimposed onto the structure of DAD2WT (magenta, PDB ID 4DNP) (10). The Asn-242 and Ile-242 residues are shown in stick representation in blue, the A loop in red, and the M loop in fluorescent green. B, the displacement of A and M loops (in Å, shown as yellow dotted lines) of molecule A of DAD2N242I compared with that of DAD2WT. Comparison of molecule B of DAD2N242I and DAD2WT is shown in Fig. S5. The DAD2N242I protein also included a C89Q mutation to facilitate crystallization.
Detailed comparisons of the DAD2N242I structure with the structure of DAD2WT highlighted small differences particularly in two core loops of both molecules of DAD2N242I. The first loop, referred to here as the activation loop (A loop), consists of residues Thr-214–Pro-221 and shapes the entrance to the catalytic cavity. The second loop (Leu-241–Pro-248), referred to here as the mutation loop (M loop) containing the mutated N242I residue, connects the β7 sheet to the α11 helix (Fig. 4 and Fig. S5). In molecule A of the DAD2N242I crystal structure, the A and M loops were displaced compared with those of DAD2WT. In particular, a displacement of up to 2.5 Å was observed for the mutated residue at position 242 between DAD2WT and DAD2N242I (Fig. 4). Similar displacements, although of smaller amplitude, were also observed in molecule B of the asymmetric unit (Fig. S5). Apart from these two loops, other small displacements were also observed in the N-terminal end of the protein, the hairpin and the lid domain (αL1 and αL3 helices) of DAD2N242I compared with DAD2WT (Fig. 4 and Fig. S5).
The DAD2N242I mutant is more dynamic than DAD2WT
Based on the observed differences between DAD2N242I and DAD2WT, it was hypothesized that DAD2N242I might possess increased structural flexibility compared with DAD2WT. To test this hypothesis, molecular dynamics (MD) simulations were performed using both DAD2WT and DAD2N242I. Examination of MD trajectories revealed similar overall movements, except in a few regions, particularly the αL2 helix-loop-αL3 helix region in the lid domain and the N termini of the proteins (Fig. 5 and Movie S1). The residues of these regions showed larger deviations from their initial positions in the DAD2N242I structures than in DAD2WT structures (Fig. 5). The root-mean-square fluctuation (RMSF), which quantifies the average deviation of the atomic positions of the residues from their average position during a simulation, also indicated that the residues comprising the αL2 helix-loop-αL3 helix region of the lid domain and the N terminus of DAD2N242I have higher fluctuations than those of DAD2WT (Fig. S8). By contrast, the residues comprising the A and M loops showed no significant differences in RMSF between the two models (Fig. 5 and Fig. S8).
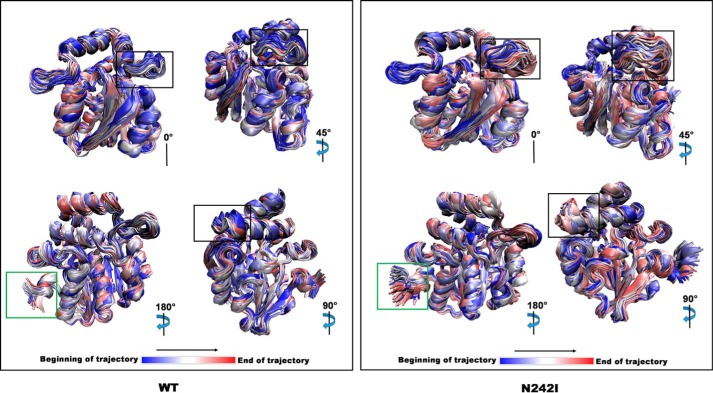
Figure 5.
Visualization of DAD2WT and DAD2N242I MD simulation trajectories. Subset (first, every 100th and last) of structures present within the DAD2WT (WT, left) and DAD2N242I (N242I, right) MD simulation trajectories. The colors represent the start (blue), middle (white), and the end (red) of the simulation. The degree of rotation specified for each figure of WT or N242I is with respect to the structure shown in the top left corner. The αL2 helix-loop-αL3 helix region of the lid domain (highlighted in the black box) and the N terminus of the protein (highlighted in the green box) show the most differences in movement between the WT and N242I simulations.
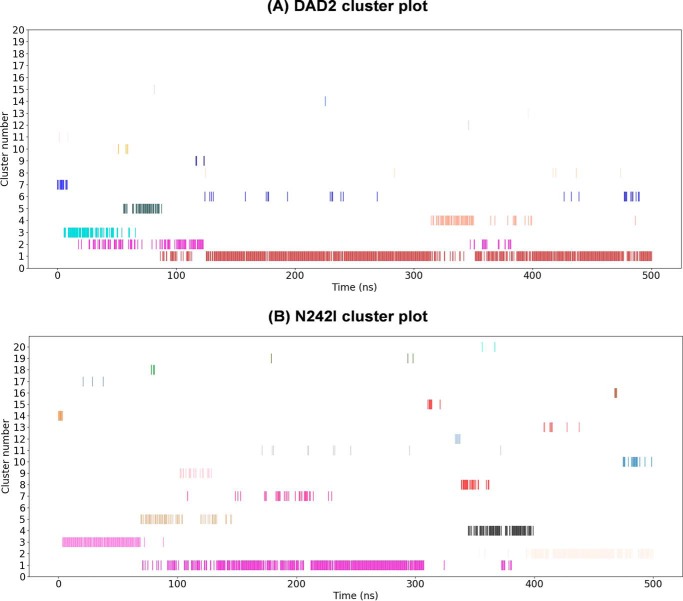
A cluster analysis (in which the different structures that occur during the simulation were grouped based on their similarity as quantified by their RMSD (32, 33)) indicated that DAD2N242I displays more flexibility than DAD2WT, based on the observed number of clusters, the size of clusters, and the duration for which the clusters were present in the simulation (Fig. 6). For DAD2WT, the most populated cluster, cluster 1, was populated for 84% of the simulation (∼420 ns), and was continuously occupied for extended periods of time (Fig. 6A). The other clusters were only present transiently. By contrast, DAD2N242I adopted several metastable conformations during the simulation, which was revealed by the appearance of several relatively short-lived clusters throughout the simulation (Fig. 6B). Furthermore, the most stable conformation adopted by DAD2N242I, cluster 1, was present only for ∼230 ns (i.e. 46%) of the total simulation. Taken together, the results from the MD simulations suggest that DAD2N242I is more flexible than DAD2WT and exhibits more dynamic states than DAD2WT.
Figure 6.
Cluster analysis of DAD2WT and DAD2N242I MD simulations. Cluster time series plots of DAD2WT (A) and DAD2N242I (B) showing cluster occupation versus the simulation time (in ns). Each cluster is represented as a horizontal series of lines in a specific color where each line represents occupancy of that cluster at that point in time. Clusters are ordered from the most (1) to the least (20) populated. The cluster analysis highlights that DAD2N242I is more dynamic as it occupies several clusters throughout the simulation, whereas DAD2WT mostly exists in a single stable conformation represented by cluster 1. The cluster program identified 15 clusters for DAD2WT and 26 clusters for DAD2N242I at the specified cut-off of 0.15 nm. The DAD2WT plot depicts all of the identified 15 clusters, whereas the DAD2N242I plot depicts the most populated 20 clusters from the identified 26 clusters.
The D166A mutation does not affect the overall conformation of the protein
As described previously, the DAD2D166A mutant retains its hydrolytic activity and interaction with PhD53A, but loses the ability to interact with PhMAX2A. To investigate the structural changes that could be responsible for this observation, we solved the crystal structure of DAD2D166A at 1.52 Å resolution, with two molecules of the mutant protein in the asymmetric unit. Both DAD2D166A molecules displayed a WT-like structure (10), with no structural changes (Fig. S9). Detailed comparison of the DAD2D166A structure with that of DAD2WT (PDB ID 4DNP (10)) showed only minor differences between the two proteins, specifically in the αL1 helix, αL3 helix, and the loop containing the mutated Ala-166 residue (referred to here as the D loop) of DAD2D166A (Fig. S9).
To gain an understanding of the possible role of the D166A mutation, we examined the corresponding residue (Asp-167) in the orthologous protein AtD14 both in the unbound (open) state (12) and in the F-box-bound (closed) state (16). In the open state, the Asp-167 residue is located within the loop that connects the αL2 helix and αL3 helix of the lid domain, whereas in the closed state, this residue becomes part of the newly formed extended coil (Fig. S10). Moreover, the Asp-167 residue changes from being completely exposed in the open state, to being completely buried in the closed state. In the closed state, the side chain of Asp-167 forms two hydrogen bonds with the main chain atoms of the following valine residue (Val-168). Although Asp-167 is located within the interaction surface where the F-box protein has been reported to bind to the receptor (16), the buried side chain of this residue does not form a direct interaction with the F-box protein. This information suggests that disruption of electrostatic interactions with adjacent residues may be responsible for the effects of the DAD2D166A mutation.
Discussion
In this work we aimed to address how the SL receptor gains the ability to interact with signaling partners during SL signal reception and transduction. We identified three mutants (DAD2N242I, DAD2D166A, and DAD2F135A) in which we altered the ability of the receptor to interact with PhMAX2A and/or PhD53A in the presence or absence of SL. These mutants generally altered the interaction with one signaling partner, whereas not altering interaction with the other (Fig. S11), indicating that the SCF complex and PhD53A protein bind the receptor using different interaction surfaces and may affect the conformation of the receptor in different ways.
The N242I mutation enabled the protein to bypass the requirement for a ligand in the interaction with PhMAX2A but not with PhD53A, which was still dependent on SL (Fig. 1). The crystal structure of DAD2N242I showed displacements of two loops compared with DAD2WT (Fig. 4 and Fig. S5). The displacements near the site of the mutation (M and A loops) were thought to be due to direct effects of the mutation. In MD simulations, we observed that DAD2N242I was more dynamic than DAD2WT, with more short-lived transient conformations than DAD2WT, which displayed a single conformation for most of the simulation (Fig. 6). The most dynamic region of the mutant protein was found to be the αL2 helix-loop-αL3 helix region of the lid domain (Movie S1 and Fig. S1). This region of the lid domain is in close proximity to the M loop, where the shift of the M loop may allow this region to become more flexible, resulting in the observed movement in the MD simulation.
In the AtD14-D3-ASK1 complex reported by Yao et al. (16) the αL2 helix-loop-αL3 helix region of the AtD14 receptor is part of the interaction surface and residues from this region such as Ala-160–Val-164, Glu-174, and Arg-177 are involved in the interaction with the F-box D3 protein. Based on these observations and the movements of this region of the DAD2N242I model, we propose that it is the increased flexibility of the αL2 helix-loop-αL3 helix region that allows DAD2N242I to interact with PhMAX2A. We predict that the movement of this region exposes key residues, perhaps transiently, including residues that are buried under the αL3 lid helix, and makes them available for interaction with PhMAX2A. The flexibility of this region could also facilitate the conformational changes required for the interaction between the DAD2N242I mutant and PhMAX2A protein.
It is possible that the N242I mutation does not affect the flexibility of regions of the mutant protein involved in, or required for, binding of the PhD53A protein, which is fulfilled by the presence of SL. Indeed the DAD2S96A/N242I mutant, which has severely reduced hydrolytic activity, is unable to interact with PhD53A in the presence of SL. However, it can still interact with PhMAX2A with or without SL, suggesting that SL alters the flexibility of different regions of the receptor protein to enable interaction with signaling partners. This is consistent with the altered Tm observed for the mutant receptor in the absence of SL and is consistent with the observations of Zhao et al. (19). The SL-independent interaction detected between DAD2N242I and PhD53A in the presence of PhMAX2A in yeast two-hybrid experiments (Fig. 2B) also suggests that the binding of the F-box protein enables the mutant receptor to undergo further conformational change(s) that favors the recruitment of the target protein, PhD53A, to the SCF complex.
Similar to DAD2N242I, the DAD2F135A mutant has hormone-independent interactions, in this case with PhD53A, whereas its interaction with PhMAX2A is still dependent on SL (Fig. 1). In the absence of SL, the interaction with PhD53A remained unaffected by the presence or absence of PhMAX2A (Fig. 2). However, in combination with the N242I mutation, we observed an increase in hormone-independent interaction between the double mutant (DAD2F135A/N242I) and PhD53A in the presence of PhMAX2A (Fig. 2), suggesting that the double mutant is forming a ternary complex with both proteins. In the DSF assays, DAD2F135A showed a lower Tm than DAD2WT (Table 1 and Fig. S4). It is possible that, like the DAD2N242I mutant, both DAD2F135A and DAD2F135A/N242I have increased flexibility in specific regions of the receptor that enable these mutants to interact, respectively, with one or two of the downstream partners (Fig. S11). However, we have not been able to solve the structures of these mutants and larger conformational changes remain a possibility for being responsible for their altered interactions with signaling partners, and altered kinetic properties in the case of DAD2F135A.
The D166A mutation led to a loss of interaction with PhMAX2A in the presence of SL without affecting interaction with PhD53A (Fig. 1). Like DAD2N242I, this mutant has WT-like hydrolytic activity, was found to be thermally less stable than the WT receptor, and there were no obvious conformational changes in the DAD2D166A crystal structure (Fig. 3, Tables 1 and 2, and Figs. S4 and S9). The Asp-166 residue is situated in the loop that connects the αL2 lid helix to the αL3 lid helix and is conserved in most identified SL receptor orthologs, but is not directly involved in the interaction with the F-box protein. Instead it is likely that replacement of Asp-166 with an Ala residue has its effect by preventing key electrostatic interactions required for the receptor to adopt the closed state in which it interacts with the F-box protein.
Our work using MD simulations suggests the hypothesis that flexibility of certain regions of the receptor can affect the ability to interact with signaling partners, although we consider it unlikely that we have fully identified all regions involved (Fig. S11). With respect to the SCF complex, Zhao et al. (19) and Yao et al. (16) have identified regions of the SL receptor that are involved in the interaction with the F-box protein. These regions include the helices of the lid and the loop that shapes the entrance of the cavity (referred to here as the A loop). In the crystal structure of the AtD14-D3-ASK1 complex by Yao et al. (16), the A loop becomes disordered, whereas the lid helices undergo changes and move toward each other to close the entrance of the cavity to create the interaction surface. In our previous work, we also showed that a residue of the A loop interacts with a SL receptor antagonist, tolfenamic acid, effectively locking the receptor in the open form (30). The flexibility of this loop may therefore also be involved in the ability of the receptor to interact with signaling partners.
In this and other work (19, 28), it is evident that SL signaling can be activated in the absence of the ligand or hydrolytic activity of the receptor. However, phylogenetic comparisons show the catalytic triad is conserved in all SL receptors examined (34). It is possible that hydrolysis may still be involved in removing SL from the cell after it is perceived, although it is currently unknown if other proteins can also fulfil this role or if hydrolysis and/or the catalytic triad is required for another function. Furthermore, the hydrolytic products of the receptor do not have biological activity, at least with respect to altering branching in plants (10, 15, 35, 36), however, it remains possible that these products may still have a role that is currently unknown.
Perception of SL appears to be a surprisingly complex process (26). The data presented here demonstrate that it is possible to bypass the need for ligand hydrolysis by introducing mutations that have an effect on the flexibility of specific regions of the SL receptor that are involved in interactions with downstream signaling partners. This is consistent with the findings of Seto et al. (28) and Zhao et al. (19). Such a hydrolysis-independent mechanism may also be regulated by the SCF complex as suggested by Shabek et al. (29). A major goal to shed further light on the changes occurring in the receptor upon binding of its partners is to solve the structure of a complex containing the receptor as well as the F-box protein and a target (e.g. the repressor protein D53) of the receptor-SCF complex.
Experimental procedures
Mutagenesis of DAD2
Amino acid mutations were introduced into the bacterial codon-optimized ORF of DAD2. Random mutagenesis was performed using an error-prone PCR polymerase using the GeneMorph® II Random EZClone Domain Mutagenesis kit according to the manufacturer's instructions (Agilent Technologies). Site-directed mutagenesis was performed using the QuikChange® Lightning Site-directed Mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions.
A C89Q mutation was introduced into the DAD2D166A, DAD2F135A, and DAD2N242I constructs to obtain protein crystals with good diffraction (30). DAD2D166A used in the DSF and YLG hydrolysis assays also contained the C89Q mutation. The C89Q mutation in DAD2 does not alter the function of the proteins in terms of SL hydrolysis (see Fig. S12) and SL binding (30).
Expression and purification of recombinant proteins
DAD2 and its mutants were expressed as fusion proteins with a cleavable, N-terminal His6-MBP tag in Escherichia coli Rosetta gami-2 (DE3) cells (Novagen, USA) overnight at 20 °C. The cells were harvested and homogenized twice using an EmusiFlex C3 (Avestin) at 10,000–15,000 p.s.i. The soluble fraction was purified by metal affinity chromatography using a 5-ml HisTrapTM HP column (GE Healthcare) and eluted with 20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 300 mm imidazole. The eluted protein was dialyzed against 20 mm Tris-HCl (pH 8.0), 50 mm NaCl, and 1 mm DTT (for proteins used for crystallization, dialysis was performed in the same buffer without DTT) at 4 °C for 16 h and further purified by anion-exchange using a 5-ml HiTrapTM Q HP column (GE healthcare). Proteins were eluted using a continuous salt gradient of 0–500 mm in 20 mm Tris-HCl (pH 8.0) and 1 mm DTT (proteins used for crystallization were eluted in the same condition without DTT). The His6-MBP tag was then cleaved using tobacco etch virus protease at a protease:protein ratio of 1:50 (w/w) at 4 °C by dialyzing against 20 mm Tris-HCl (pH 8), 150 mm NaCl, 50 mm l-arginine, 50 mm l-glutamic acid, 1 mm DTT, and 1 mm EDTA. The cleaved protein was then dialyzed against the same buffer lacking 1 mm DTT and 1 mm EDTA for 2 h at 4 °C before being subjected to another round of metal affinity chromatography. The flow-through containing purified protein was collected, concentrated to ∼6 mg/ml, and stored at −80 °C. For the DAD2D166A mutant, anion-exchange chromatography was not performed. Prior to crystallization, YLG hydrolysis, and DSF assays, the proteins were buffer-exchanged into 20 mm Tris-HCl (pH 8.0) and 150 mm NaCl (proteins used for YLG hydrolysis and DSF assays were exchanged into buffer containing 1 mm DTT) using SuperdexTM 75 100/300 GL column (GE Healthcare) and concentrated, if required for an experiment.
Crystallization
The crystallization experiments were performed by a hanging drop vapor diffusion method at room temperature. For DAD2N242I, the initial crystal hits were obtained in two conditions, the first comprising 0.1 m MES (pH 6.5) and 1.6 m magnesium sulfate, and the second comprising 0.1 m sodium cacodylate (pH 6.5) and 1.4 m sodium acetate. Crystals used for data collection were obtained in a condition containing 0.9 μl of 7 mg/ml of DAD2N242I in the presence of 0.1 m MES buffer (pH 6.5) with 1.58 m MgSO4 or 1.72 m MgSO4. For DAD2D166A, initial crystal hits were obtained in several conditions of the Morpheus screen (Molecular Dimension Ltd.). The condition containing 0.1 m Trizma-Bicine (pH 8.5), 30 mm MgCl2, 30 mm CaCl2, and 20% (v/v) PEG 500 monomethyl ether, 10% (v/v) PEG 20000 was further refined and crystals used for data collection were obtained in conditions containing 0.9 μl of 5 mg/ml of DAD2D166A in the presence of 0.1 m Trizma-acetate-Bicine (pH 8.5), 30 mm MgCl2, 30 mm CaCl2, and 18–22% PEG 500 monomethyl ether, 9–11% PEG 20000. The crystals used for data collection were obtained in a condition comprised of 0.1 m Trizma-acetate-Bicine (pH 8.5), 30 mm MgCl2, 30 mm CaCl2, and 20% (v/v) PEG 500 monomethyl ether, 10% PEG 20000. A small amount of DAD2F135A was obtained and crystallized in 0.1 m Trizma-acetate-Bicine (pH 8.5), 30 mm MgCl2, 30 mm CaCl2, and 12% (v/v) PEG 500 monomethyl ether, 6% PEG 20000.
Data collection, processing, and structure determination
Complete datasets were collected at the Australian synchrotron (Victoria, Australia). The dataset for the DAD2N242I protein crystal was collected on the MX2 beamline to a resolution of 1.58 Å. The dataset for the DAD2D166A crystals was collected on the MX1 beamline to a resolution of 1.52 Å. Data indexing and integration were performed using iMOSFLM or XDS (37, 38). The intensities were merged using aimless/pointless from the CCP4 suite (39–41). The number of molecules in the asymmetric unit were determined using Matthew's coefficient from the CCP4 suite. Structures were solved by molecular replacement using the crystal structure of WT DAD2 (DAD2WT; PDB ID 4DNP) as the model in PHASER MR from CCP4 suite (42, 43). Structure refinement and subsequent cycles of model building were performed in REFMAC5 and COOT, respectively (44, 45). The final models were refined using the optimized parameters obtained from the PDB redo server. The statistics for data collection and refinement are listed in Table S1. Crystals obtained with DAD2F135A showed only poor diffraction and were not analyzed further.
MD simulations
The initial coordinates for the DAD2WT and DAD2N242I models were obtained from their respective crystal structures (DAD2, PDB ID 4DNP, and DAD2N242I, PDB ID 6UH8, molecule A of the asymmetric unit). The crystallographic water and any ligands were removed from the PDB file. The simulations were performed using the Gromacs software, version 2016.1 (SCR_014545 (46)) and the Amber force field, FF99SB (47). Periodic boundary conditions were used with a cubic simulation box with sides of 2 nm from the solute in all directions. The systems were solvated using the TIP3P (48) water model. All amino acids were assumed to be in their standard protonation state at physiological pH (∼7), resulting in a net charge of −4 for DAD2WT and −5 for DAD2N242I. Sodium ions were added to ensure an overall net neutral system. All simulations were performed using a 2-fs integration time step with the covalent bond lengths involving hydrogen atoms constrained using LINCS (49). Nonbonded interactions were calculated explicitly up to a cut-off of 1.2 nm. Outside this, long-range electrostatic interactions were calculated using Particle Mesh Ewald summation (50). The temperature was maintained using the Velocity Rescale Thermostat (51) and for the NPT simulations, constant pressure was maintained using the Berendsen barostat (52). The systems underwent a two-stage energy minimization comprising 5000 steps using the steepest descent algorithm followed by 5000 steps with the conjugate gradient algorithm. The systems were then heated from 50 to 298 K over 250 ps at a heating rate of 1 K/ps in the NVT ensemble, followed by a short (250 ps) equilibration simulation under NPT conditions (298 K, 1 atm). Production simulations were then performed in the NPT ensemble for 500 ns at 298 K and 1 atm pressure.
Analysis of MD simulation data
The MD simulation data were analyzed using Gromacs (version 2016.1; SCR_014545 (46)) tools and visualized using Visual Molecular Dynamics (VMD) (version 1.9.3; SCR_001820 (53)). For cluster analysis, only every 100th frame was considered to make the all-by-all RMSD calculations required for cluster analysis tractable. Clustering was performed using the Gromaos algorithm and a root-mean-square deviation (RMSD) cut-off of 0.15 nm. This cut-off value was selected as it gave a manageable number of clusters (i.e. less than 50). Occupation of the most populated 20 clusters was then plotted against the time series of the simulation using an in-house Python script. For RMSF analysis, the main trajectory files of the two models were used and the analysis was performed using the RMSF tool of Gromacs.
Differential scanning fluorimetry
DSF experiments were performed as described in Hamiaux et al. (10). Reaction mixtures were set up in 384 multiwell plates at a molar ratio of 25:1 (GR24:protein). Each reaction, in a final volume of 20 μl, consisted of 250 μm (rac)-GR24, 10 μm protein, and ×10 SyproTM Tangerine protein dye in 20 μm Tris-HCl (pH 8.0), 150 mm NaCl, and 2.5% DMSO. The final reaction mixture for the DMSO control consisted of the same assay components without (rac)-GR24.
DSF assays were also performed using MES as a test compound. Various concentrations of MES were tested (0.25–128 mm). The experiment was setup in a similar method as described above with MES instead of (rac)-GR24.
YLG hydrolysis assays
The hydrolytic activity of DAD2 mutants was determined using YLG (TCI Chemicals) according to the methods of Tsuchiya et al. (31) with some modifications. To determine the time course hydrolysis of YLG, each reaction mixture (in a final volume of 100 μl) consisted of 0.34 μm protein and 1 μm YLG in 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 1% DMSO. The YLG control consisted of 1 μm YLG in the same buffer. For kinetics analyses, assay reactions were performed at seven YLG concentrations (0, 0.2, 0.4, 0.6, 1.2, 3.5, and 5 μm) in triplicate. Protein concentration and reaction buffer were the same as the time course hydrolysis assays. All experiments were conducted in three technical replicates. Fluorescence was monitored using a FLUOStar® Omega microplate reader (BMG LabTech) with 485 ± 12 nm excitation filter and 520 ± 10 nm emission filter every 2 min over a 60-min interval. The gain was set to 920, and the number of flashes to 20. Fluorescence units were converted to fluorescein concentrations using fluorescein standard curves. Velocity (μm/min) at each YLG concentration was calculated as the amount of fluorescein produced (μm) per minute. Michaelis-Menten analyses were determined by nonlinear regression in GraphPad Prism using the velocity at the 15-min time point.
Yeast two-hybrid assays
Yeast two-hybrid experiments were performed by β-gal liquid culture assay according to methods from the Clontech Yeast Protocols Handbook (2009). DAD2WT and DAD2 mutants were cloned into a pBD vector, whereas PhMAX2A and PhD53A were cloned into a pAD vector for expression in yeast PJ69-4 (54, 55). The pURA3-GW vector was used to express PhMAX2A as a third untagged protein when required in the yeast assays. Yeast transformants were subjected to Western blot analysis to confirm the presence of the binding domain and activation domain fusion proteins (30). The strength of DAD2-PhMAX2A and DAD2-PhD53A interactions compared with DMSO controls were quantified using orthonitro-phenyl-β-galactopyranoside (Sigma-Aldrich) as the substrate in the presence of 10 and 1 μm (rac)-GR24, respectively.
Accession numbers
The crystal structure of DAD2N242I has been deposited in the Protein Data Bank under PDB ID 6UH8, and the crystal structure of DAD2D166A under PDB ID 6UH9.
Author contributions
H. W. L., P. S., C. H., and K. C. S. data curation; H. W. L., P. S., B. J. J., R. S. D., Z. L., C. H., T. C., J. R. A., and K. C. S. formal analysis; H. W. L., P. S., B. J. J., R. S. D., Z. L., and T. C. investigation; H. W. L., P. S., and K. C. S. visualization; H. W. L., P. S., B. J. J., R. S. D., C. H., and J. R. A. methodology; H. W. L., P. S., and K. C. S. writing-original draft; B. J. J. and K. C. S. conceptualization; B. J. J. and K. C. S. funding acquisition; B. J. J., C. H., J. R. A., and K. C. S. writing-review and editing; R. S. D. validation; C. H., T. C., J. R. A., R. D. N., and K. C. S. supervision; K. C. S. project administration.
Supplementary Material
Acknowledgments
We thank members of the Laboratory of Structural Biology (University of Auckland) for help with X-ray data collection, and Colm Carraher and Donald Hunter for comments on the manuscript. Crystallography data were collected on the MX1 and MX2 beamlines at the Australian Synchrotron, part of ANSTO (Victoria, Australia).
This work was supported by the Marsden Fund from Royal Society Te Apārangi Contract PAF1301, by Plant and Food Research, and Rutherford Discovery Fellowship Grant 15-MAU-001 (to J. R. A.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S12, Movie S1, and Table S1.
The atomic coordinates and structure factors (codes 6UH8 and 6UH9) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SL
- strigolactone
- DAD2
- Decreased Apical Dominance 2
- MAX2A
- More Axillary Growth 2A
- DSF
- differential scanning fluorimetry
- YLG
- yoshimulactone green
- MD
- molecular dynamics
- RMSF
- root-mean-square fluctuation
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- PDB
- Protein Data Bank
- MBP
- maltose-binding protein
- RMSD
- root-mean-square deviation
- β-gal
- β-galactosidase.
References
- 1. Cook C. E., Whichard L. P., Turner B., Wall M. E., and Egley G. H. (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190 10.1126/science.154.3753.1189 [DOI] [PubMed] [Google Scholar]
- 2. Yokota T., Sakai H., Okuno K., Yoneyama K., and Takeuchi Y. (1998) Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry 49, 1967–1973 10.1016/S0031-9422(98)00419-1 [DOI] [Google Scholar]
- 3. Akiyama K., Matsuzaki K., and Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- 4. Gomez-Roldan V., Fermas S., Brewer P. B., Puech-Pagès V., Dun E. A., Pillot J. P., Letisse F., Matusova R., Danoun S., Portais J. C., Bouwmeester H., Becard G., Beveridge C. A., Rameau C., and Rochange S. F. (2008) Strigolactone inhibition of shoot branching. Nature 455, 189–194 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- 5. Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., and Yamaguchi S. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- 6. Kapulnik Y., Delaux P.-M., Resnick N., Mayzlish-Gati E., Wininger S., Bhattacharya C., Séjalon-Delmas N., Combier J.-P., Bécard G., Belausov E., Beeckman T., Dor E., Hershenhorn J., and Koltai H. (2011) Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- 7. Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., Cardoso C., Lopez-Raez J. A., Matusova R., Bours R., Verstappen F., and Bouwmeester H. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155, 721–734 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Snowden K. C., Simkin A. J., Janssen B. J., Templeton K. R., Loucas H. M., Simons J. L., Karunairetnam S., Gleave A. P., Clark D. G., and Klee H. J. (2005) The decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17, 746–759 10.1105/tpc.104.027714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamada Y., Furusawa S., Nagasaka S., Shimomura K., Yamaguchi S., and Umehara M. (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240, 399–408 10.1007/s00425-014-2096-0 [DOI] [PubMed] [Google Scholar]
- 10. Hamiaux C., Drummond R. S., Janssen B. J., Ledger S. E., Cooney J. M., Newcomb R. D., and Snowden K. C. (2012) DAD2 Is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 10.1016/j.cub.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 11. Arite T., Umehara M., Ishikawa S., Hanada A., Maekawa M., Yamaguchi S., and Kyozuka J. (2009) D14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424 10.1093/pcp/pcp091 [DOI] [PubMed] [Google Scholar]
- 12. Zhao L. H., Zhou X. E., Wu Z. S., Yi W., Xu Y., Li S., Xu T. H., Liu Y., Chen R. Z., Kovach A., Kang Y., Hou L., He Y., Xie C., Song W., et al. (2013) Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439 10.1038/cr.2013.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chevalier F., Nieminen K., Sanchez-Ferrero J. C., Rodríguez M. L., Chagoyen M., Hardtke C. S., and Cubas P. (2014) Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26, 1134–1150 10.1105/tpc.114.122903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Saint Germain A., Clavé G., Badet-Denisot M.-A., Pillot J.-P., Cornu D., Le Caer J.-P., Burger M., Pelissier F., Retailleau P., Turnbull C., Bonhomme S., Chory J., Rameau C., and Boyer F.-D. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 12, 787–794 10.1038/nchembio.2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura H., Xue Y. L., Miyakawa T., Hou F., Qin H. M., Fukui K., Shi X., Ito E., Ito S., Park S. H., Miyauchi Y., Asano A., Totsuka N., Ueda T., Tanokura M., and Asami T. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4, 2613 10.1038/ncomms3613 [DOI] [PubMed] [Google Scholar]
- 16. Yao R., Ming Z., Yan L., Li S., Wang F., Ma S., Yu C., Yang M., Chen L., Chen L., Li Y., Yan C., Miao D., Sun Z., Yan J., et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473 10.1038/nature19073 [DOI] [PubMed] [Google Scholar]
- 17. Yao R., Wang L., Li Y., Chen L., Li S., Du X., Wang B., Yan J., Li J., and Xie D. (2018) Rice DWARF14 acts as an unconventional hormone receptor for strigolactone. J. Exp. Botany 69, 2355–2365 10.1093/jxb/ery014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waters M. T., Scaffidi A., Flematti G., and Smith S. M. (2015) Substrate-induced degradation of the α/β-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 8, 814–817 10.1016/j.molp.2014.12.020 [DOI] [PubMed] [Google Scholar]
- 19. Zhao L.-H., Zhou X. E., Yi W., Wu Z., Liu Y., Kang Y., Hou L., de Waal P. W., Li S., Jiang Y., Scaffidi A., Flematti G. R., Smith S. M., Lam V. Q., Griffin P. R., et al. (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25, 1219–1236 10.1038/cr.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abe S., Sado A., Tanaka K., Kisugi T., Asami K., Ota S., Kim H. I., Yoneyama K., Xie X., Ohnishi T., Seto Y., Yamaguchi S., Akiyama K., Yoneyama K., and Nomura T. (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. U.S.A. 111, 18084–18089 10.1073/pnas.1410801111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang Y., Ward S., Li P., Bennett T., and Leyser O. (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28, 1581–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., Meng X., Liu G., Yu H., Yuan Y., Yi W., Zhao L., Ma H., He Y., Wu Z., et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 10.1038/nature12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., Wu F., Mao H., Dong W., Gan L., Ma W., Gao H., Chen J., Yang C., et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 10.1038/nature12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., Meng X., Wang Y., Smith S. M., and Li J. (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142 10.1105/tpc.15.00605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J. P., Abbas A., Leyser O., and Nelson D. C. (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Machin D. C., Hamon-Josse M., and Bennett T. (2019) Fellowship of the rings: a saga of strigolactones and other small signals. New Phytol. 225, 621–636 [DOI] [PubMed] [Google Scholar]
- 27. Kagiyama M., Hirano Y., Mori T., Kim S. Y., Kyozuka J., Seto Y., Yamaguchi S., and Hakoshima T. (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160 10.1111/gtc.12025 [DOI] [PubMed] [Google Scholar]
- 28. Seto Y., Yasui R., Kameoka H., Tamiru M., Cao M., Terauchi R., Sakurada A., Hirano R., Kisugi T., Hanada A., Umehara M., Seo E., Akiyama K., Burke J., Takeda-Kamiya N., et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun. 10, 191 10.1038/s41467-018-08124-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shabek N., Ticchiarelli F., Mao H., Hinds T. R., Leyser O., and Zheng N. (2018) Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature 563, 652–656 10.1038/s41586-018-0743-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hamiaux C., Drummond R. S. M., Luo Z., Lee H. W., Sharma P., Janssen B. J., Perry N. B., Denny W. A., and Snowden K. C. (2018) Inhibition of strigolactone receptors by N-phenylanthranilic acid derivatives: structural and functional insights. J. Biol. Chem. 293, 6530–6543 10.1074/jbc.RA117.001154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsuchiya Y., Yoshimura M., Sato Y., Kuwata K., Toh S., Holbrook-Smith D., Zhang H., McCourt P., Itami K., Kinoshita T., and Hagihara S. (2015) Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868 10.1126/science.aab3831 [DOI] [PubMed] [Google Scholar]
- 32. Abramyan T. M., Snyder J. A., Thyparambil A. A., Stuart S. J., and Latour R. A. (2016) Cluster analysis of molecular simulation trajectories for systems where both conformation and orientation of the sampled states are important. J. Comp. Chem. 37, 1973–1982 10.1002/jcc.24416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Phillips J. L., Colvin M. E., and Newsam S. (2011) Validating clustering of molecular dynamics simulations using polymer models. BMC Bioinformatics 12, 445 10.1186/1471-2105-12-445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Delaux P.-M., Xie X., Timme R. E., Puech-Pages V., Dunand C., Lecompte E., Delwiche C. F., Yoneyama K., Bécard G., and Séjalon-Delmas N. (2012) Origin of strigolactones in the green lineage. New Phytol. 195, 857–871 10.1111/j.1469-8137.2012.04209.x [DOI] [PubMed] [Google Scholar]
- 35. Boyer F.-D., de Saint Germain A., Pillot J.-P., Pouvreau J.-B., Chen V. X., Ramos S., Stévenin A., Simier P., Delavault P., Beau J.-M., and Rameau C. (2012) Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol. 159, 1524–1544 10.1104/pp.112.195826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Umehara M., Cao M., Akiyama K., Akatsu T., Seto Y., Hanada A., Li W., Takeda-Kamiya N., Morimoto Y., and Yamaguchi S. (2015) Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant Cell Physiol. 56, 1059–1072 10.1093/pcp/pcv028 [DOI] [PubMed] [Google Scholar]
- 37. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Battye T. G. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. W. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Evans P. R. (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 10.1107/S090744491003982X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr 62, 72–82 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- 42. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 45. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of Macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 46. Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., and Lindahl E. (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- 47. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Physics 79, 926–935 10.1063/1.445869 [DOI] [Google Scholar]
- 49. Hess B., Bekker H., Berendsen H. J., and Fraaije J. G. (1997) LINCS: a linear constraint solver for molecular simulations. J. Comp. Chem. 18, 1463–1472 [DOI] [Google Scholar]
- 50. Di Pierro M., Elber R., and Leimkuhler B. (2015) A stochastic algorithm for the isobaric–isothermal ensemble with Ewald summations for all long range forces. J. Chem. Theory Comput. 11, 5624–5637 10.1021/acs.jctc.5b00648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bussi G., Donadio D., and Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Physics 126, 014101 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- 52. Berendsen H. J. C., Postma J. P. M., Gunsteren W. F. v., DiNola A., and Haak J. R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Physics 81, 3684–3690 10.1063/1.448118 [DOI] [Google Scholar]
- 53. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 54. Maier R., Brandner C., Hintner H., Bauer J., and Onder K. (2008) Construction of a reading frame-independent yeast two-hybrid vector system for site-specific recombinational cloning and protein interaction screening. BioTechniques 45, 235–244 10.2144/000112897 [DOI] [PubMed] [Google Scholar]
- 55. James P., Halladay J., and Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.